To be successful and have the opportunity to replicate properly, a virus has to thwart or, just the reverse, boost many systems of the host cell. Given that host cells have evolved to eliminate these hostile parasites, a fierce battle ensues. There is now compelling evidence that enveloped virions released from infected cells will carry the vestiges of this battle both internally and externally. The focus of the present minireview will be the incorporation of the host cellular proteins into or onto the newly formed viruses. The roles of a few of these host cellular proteins have been studied, albeit very recently, because of their implication in the biology of some viruses. But for the vast majority, only the observation of their incorporation has been reported in the scientific literature.
The various studies in this research area have been conducted with eukaryotic cells infected primarily with RNA enveloped viruses. Human immunodeficiency virus type 1 (HIV-1) has been the most extensively studied in this respect, but other viruses have also contributed to a better understanding of this intriguing phenomenon. A list of the molecules that have been reported to be embedded in some enveloped viruses is shown in Table 1. The processes through which cellular proteins are acquired by viruses are still ill defined. Here we will describe and discuss the roles, or potential roles, that the major cellular proteins found associated with enveloped viruses may play in their life cycles. First, the internally associated host cell molecules will be described, and in a second section, the cell surface constituents found included within the envelopes of different viruses will be introduced.
TABLE 1.
Host cellular molecules acquired by some enveloped viruses
| Virusa | Host moleculeb | Function | Incorporated | Reference |
|---|---|---|---|---|
| EBV | HLA-DR | Antigen presentation | Yes | 54 |
| HCMV | CD55 | Complement control protein | Yes | 101 |
| CD59 | Complement control protein | Yes | 101 | |
| Annexin II | Calcium-binding protein | Yes | 119 | |
| PPA2, PP1 | Serine/threonine protein phosphatase | Yes | 2, 68 | |
| β2-microglobulin | β-globulin, MHC-1 | Yes | 45 | |
| VV | α-Tubulin | Cytoskeletal protein | No | 26 |
| HSP70 | HSPs, chaperone | No | 26 | |
| HSP90 | HSPs, chaperone | No | 26 | |
| CypA | Immunophilin | Yes | 26 | |
| CD46 | Complement control protein | Yes | 55, 110 | |
| CD55 | Complement control protein | Yes | 55, 110 | |
| CD59 | Complement control protein | Yes | 55, 110 | |
| CD29 | Integrin β chain | Yes | 55, 110 | |
| CD71 | Transferrin receptor | Yes | 55, 110 | |
| CD81 | Tetraspanin | Yes | 55, 110 | |
| MHC-1 | Antigen presentation | Yes | 55, 110 | |
| DHBV | p23 | Chaperone of HSP90 | Yes | 52 |
| HSP90 | HSPs, chaperone | Yes | 52 | |
| VSV | CypA | Immunophilin | Yes | 16 |
| MPMV | Tsg101 | Vesicular transport protein | Yes | 41 |
| Nedd4 | Vesicular transport protein | Yes | 41 | |
| HTLV-1 | Nedd4 | Vesicular transport protein | Yes | 12 |
| CD55 | Complement control protein | Yes | 101 | |
| CD59 | Complement control protein | Yes | 101 | |
| MLV | Actin | Cytoskeletal protein | Yes | 73, 114 |
| α-Tubulin | Cytoskeletal protein | Yes | 114 | |
| Clathrin | Vesicular transport protein | Yes | 114 | |
| Nucleolin | Nucleolar phosphoprotein | Yes | 5 | |
| Ubiquitin | Protein degradation and sorting | Yes | 39 | |
| APOBEC3G | Cytosine deaminase | Yes | 49 | |
| MMLV | CypA | Immunophilin | No | 107 |
| Ubiquitin | Protein degradation and sorting | Yes | 78 | |
| Staufen | RNA binding protein | Yes | 72 | |
| HSP70 | HSPs, chaperone | No | 46 | |
| HIV-1 | CD45 | Membrane phosphatase | No | 75 |
| CD46 | Complement control protein | Yes | 91, 92 | |
| CD55 | Complement control protein | Yes | 92, 101 | |
| CD59 | Complement control protein | Yes | 92, 101 | |
| Actin | Cytoskeletal protein | Yes | 80, 81 | |
| Pin1 | Peptidyl-prolyl isomerase | Yes | 80 | |
| tRNA synthetase | Ligase | Yes | 47 | |
| GAPDH | Aldehyde oxidoreductases | Yes | 78 | |
| MAPK ERK2 | Serine/threonine kinase | Yes | 25 | |
| HSP27 | HSPs, chaperone | No | 46 | |
| HSP40 | HSPs, chaperone | No | 46 | |
| HSP60 | HSPs, chaperone | Yes | 46 | |
| HSP70 | HSPs, chaperone | Yes | 46 | |
| HSC70 | HSPs, chaperone | Yes | 46 | |
| HSP90 | HSPs, chaperone | No | 46 | |
| CypA | Immunophilin | Yes | 37, 107 | |
| CypB | Immunophilin | No | 18, 37 | |
| FKBP12 | Protein degradation and sorting | Yes | 20 | |
| Ubiquitin | Vesicular transport protein | Yes | 78 | |
| Tsg101 | Vesicular transport protein | Yes | 112 | |
| Tal | Vesicular transport protein | Yes | 3 | |
| VPS28 | Vesicular transport protein | Yes | 11, 104, 113 | |
| AIP1/ALIX | Vesicular transport protein | Yes | 113 | |
| VPS4B | Vesicular transport protein | Yes | 113 | |
| VPS37B | Vesicular transport protein | No | 104 | |
| APOBEC3G | Cytosine deaminase | Yes | 53, 66, 99 | |
| APOBEC3F | Cytosine deaminase | Yes | 58, 115, 123 | |
| UNG | Uracyl-DNA glycosylase | Yes | 64, 86, 118 | |
| Staufen | RNA binding protein | Yes | 72 | |
| HLA-DR | Antigen presentation | Yes | 21-24, 90 | |
| ICAM-1 | Adhesion molecule | Yes | 35, 36, 67, 82, 89 | |
| Ezrin/myoesin/cofilin | Cytoskeletal proteins | Yes | 80, 81 | |
| Thy-1 | GP1-anchored protein | Yes | 75 | |
| GM1 | Ganglioside | Yes | 83 | |
| Various other cell surface constituents | 76, 109 | |||
| HIV-2 | HSP70 | HSPs, chaperone | Yes | 46 |
| CypA | Immunophilin | No | 37 | |
| UNG | Uracyl-DNA glycosylase | Yes | 86 | |
| Staufen | RNA binding protein | Yes | 72 | |
| HLA-DR | Antigen presentation | Yes | 4, 76 | |
| MHC-I | Antigen presentation | Yes | 4 | |
| SIV | CypA | Immunophilin | Yesc | 19 |
| CypB | Immunophilin | Yes | 37 | |
| Ubiquitin | Protein degradation and sorting | Yes | 79 | |
| AIP1/ALIX | Vesicular transport protein | Yes | 113 | |
| APOBEC3G | Cytosine deaminase | Yes | 13, 61, 62, 98, 120 | |
| UNG | Uracyl-DNA glycosylase | No | 86 | |
| Influenza virus | GM1 | Ganglioside | Yes | 83 |
| Parainfluenza virus | GM1 | Ganglioside | Yes | 83 |
| HSV | GM1 | Ganglioside | Yes | 83 |
EBV, Epstein-Barr virus; DHBV, duck hepatitis virus; MPMV, Mason-Pfizer monkey virus; HSV, herpes simplex virus.
GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MAPK ERK2, mitogen-activated protein kinase extracellular signal-regulated kinase 2.
Not associated with all SIV strains.
HOST CELL PROTEINS INSERTED INTO VIRIONS
(i) Tsg101 and other components of the MVB pathway.
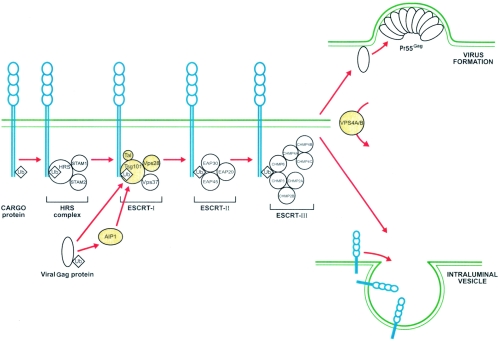
One of the major breakthroughs in the understanding of the complex interplay between enveloped viruses and the host cell machinery is certainly comprehension of the viral budding process (71). Recent studies have revealed how enveloped RNA viruses highjack the vesicular cellular machinery for their own purposes (85). Figure 1 depicts how such viruses can usurp this cellular pathway normally used to create vesicles that bud into late-endosomal compartments, which are better known as multivesicular bodies (MVB).
FIG. 1.
Schematicrepresentation of the intimate link between viral components and the human MVB pathway. Cargo proteins to be sorted are first monoubiquitinated and then bound to the Hrs complex in the early endosomes. After fusion of the early endosomes and the sorting endosomes, the cargo proteins are initially transferred to the ESCRT-I complex and then relayed successively to the ESCRT-II and ESCRT-III complexes. Finally, under the action of the VPS4A/B enzyme, cargo proteins are released into an intraluminal vesicle in formation. Monoubiquitinated structural proteins of enveloped viruses mimic cellular cargo proteins and enter the MVB pathway to be released at the site of virus budding. Proteins highlighted in yellow are incorporated within different retroviruses, as described in the text. Ub, ubiquitin; STAM, signal-transducing adapter molecule; EAP, ELL-associated proteins; CHMP, charged multivesicular body protein.
To take advantage of the MVB pathway, enveloped RNA viruses such as retroviruses, rhabdoviruses, and filoviruses possess conserved sequences in their structural proteins called late domains (L domains) (38). Each L domain is able to bind to specific cellular factors that redirect the structural proteins of the nascent viruses into the MVB pathway of the infected cell, thereby orchestrating the budding and egress of virions, using the same cellular vesicular machinery as cellular endosomes (85).
Enveloped RNA viruses like Ebola viruses, HIV-1, and human T-cell leukemia virus type 1 (HTLV-I) get access to this sorting machinery by binding to the Tsg101 subunit of the human endosomal complex required for transport I (ESCRT-I) via the L domains of their structural proteins. It has been proposed that viral proteins are targeted to the MVB machinery possibly by virtue of a single ubiquitin linked to their L domains, which would explain their binding to Hrs and Tsg101 (38). Indeed, fairly large amounts of free ubiquitin were detected in purified preparations of simian immunodeficiency virus (SIV), HIV-1, murine leukemia virus (MLV), and Moloney MLV (MMLV) and in avian leucosis virus (41, 78, 79, 88). While usurping the MVB pathway, these viruses apparently incorporate structures of the cellular sorting machinery within the newly formed entities. Tsg101 incorporation within HIV-1 particles remains the best-known example and has been reported in numerous studies (32, 48, 57, 104, 113). Tsg101 has been found in purified Mason-Pfizer monkey virus preparations as well, along with the Nedd4 enzyme, an ubiquitin ligase containing a WW sequence with binding capacity to the PPXY L domain. The above proteins have also been found embedded in HTLV-I viral particles (12, 41). Tal, a novel E3 ubiquitin ligase recently described in yeasts and mammalian cells, has been shown to be incorporated within HIV-1-like particles (3). This ligase binds to Tsg101 in a bivalent mode and, when bound, mediates multiple monoubiquitination of Tsg101, which disables the sorting activity of Tsg101 and is thought to free the cargo protein.
Other cellular protein subunits of all three ESCRT complexes have been reported to be incorporated intobudding virions. VPS28, a subunit of ESCRT-I that binds to Tsg101, was detected within HIV-1 particles (11, 104, 113). Tsg101 also interacts with the AIP1/ALIX subunit of the ESCRT-II complex, and the latter is recruited into both HIV-1 and SIV (113). Of note, equine infectious anemia virus (EIAV) p9 protein is capable of binding AIP1/ALIX, which may presumably be packaged into EIAV virions (102). The subunit of the ESCRT-III complex,VPS4B, seems to be acquired by HIV-1 (113). This protein is involved in the very last steps of vesicle formation and has a role in releasing ESCRT complexes from the newly formed vesicles. Nevertheless, not all parts of these large multiprotein complexes are retained in viruses, as exemplified by the reported failure to detect VPS37B in purified HIV-1 preparations (104).
(ii) APOBEC3G, a protein of the RNA-editing machinery.
Among the many threats that the RNA viruses must overcome inside cells is the cellular RNA-editing machinery. Products of the mRNA-editing gene family are specialized for deamination of cytidine on RNA, causing a switch from C to U in mRNA. A mechanism of protection exploited by viruses has been unraveled, principally with the discovery of the relationship between APOBEC3G and the HIV-1 Vif protein (49, 61, 66, 99). The APOBEC3G protein (also termed CEM15) has the ability to get packaged within HIV-1. Being in close proximity to the viral genomic RNA, this editing enzyme therefore has the chance to catalyze the deamination from C to U on the minus strand of the viral nucleic acid. Therefore, upon plus-strand synthesis of the viral genome during the reverse transcription phase, an A nucleotide is inserted into the plus strand at the many spots where the C to U deamination has occurred on the minus strand. As a result, many nonviable mutations are created and the newly synthesized virions are thus inactivated. This G-to-A hypermutation phenomenon occurring throughout the HIV-1 genome has been noticed for quite a while (111).
The precise role of the auxiliary Vif protein of HIV-1 in the virus life cycle and/or pathogenesis has long been an enigma. But recently Vif was brought into the spotlight by the discovery that it can associate with APOBEC3G in lymphoid cells. Vif binds to APOBEC3G inside infected cells in order to prevent this protein from being packaged within nascent virions (53, 61, 99). Convincing evidence seems to indicate that Vif strongly diminishes the APOBEC3G intracellular pool. Here it is interesting to highlight that, in sharp contrast to the usual observation that viruses associate with cellular proteins, in this case the virus evolved to minimize interaction with and incorporation of a specific cellular component. In this regard, we can assume that this phenomenon supports the hypothesis that the incorporation process of host-encoded proteins is specific, at least to a certain extent. It also underlines the fact that association with particular cellular proteins can be highly detrimental to the virus and, therefore, that the process of host cell protein incorporation can be a very critical issue for viruses.
The mechanism of APOBEC3G incorporation within HIV-1 cells is still ill defined, though a few hints are starting to emerge. The first studies reported a potential interaction of the APOBEC3G N terminus with the nucleocapsid protein of HIV-1 (27). Whether the two zinc coordination motifs contained within the APOBEC3G are directly implicated in this intimate association remains controversial (1, 27). The same applies to the possible bridging of viral or cellular RNA between APOBEC3G and the Gag protein (27, 105). Since APOBEC3G can be incorporated into various types of viruses having different Gag sequences (HIV-1, MLV, SIV, and EIAV), it has been proposed that the link between Gag and APOBEC3G may not rely exclusively on sequence but also on some specific structural motifs (27). Human APOBEC3G activity is not restricted to HIV-1, as it can inactivate HIV-2, SIV, EIAV, and MLV (27, 61). Furthermore, the effect of Vif on the incorporation of APOBEC3G is species specific. For example, the Vif protein does not prevent the packaging of mouse and African green monkey APOBEC proteins within HIV-1 particles produced by transient transfection in 293T cells, as they do not bind to the Vif protein like their human homolog does (66). Finally, APOBEC3F, another member of the mRNA-editing enzyme family incorporated into HIV-1, was very recently demonstrated to inhibit viral replication in the same way as APOBEC3G (58, 115, 123). It will be interesting to evaluate whether other members of this family carry out a similar activity on RNA viruses (10).
(iii) UNGs and Staufen.
A variety of viruses encode uracil-DNA glycosylases (UNGs) or dUTPases to block uracil incorporation within the viral DNA (30). This is the case for herpesviruses, poxviruses, and some nonprimate retroviruses. As for primate lentiviruses that do not encode such enzymes, one of them, HIV-1, does incorporate host cellular UNG within the viral particles (64, 118). In contrast, HIV-2 and SIVMAC fail to package this enzyme and may have evolved different, yet-unknown strategies to achieve a similar goal (86). The mechanism of UNG incorporation is still under investigation, but it appears that it might occur through association with the viral Vpr protein, integrase, and reverse transcriptase enzymes, individually or in cooperation (64, 86).
Interactions between host proteins and the genetic material of retroviruses also seem to be necessary for the encapsidation of genomic RNA. Staufen is a double-stranded RNA-binding protein that is enclosed within HIV-1, HIV-2, and MMLV (72). The amount of genomic RNA included within the released viral particles is correlated with the amount of Staufen protein incorporated, suggesting a role in viral RNA packaging. The Staufen protein was recently shown to cosediment with the viral Pr55Gag precursor protein and to associate directly with Pr55Gag. This link involves the Staufen dsRBD3 domain in collaboration with the C-terminal domain (28).
(iv) Cyclophilins and other prolyl isomerases.
Expressed in all organisms, from bacteria to primates, cyclophilins catalyze the isomerization of peptidyl-prolyl bonds, a rate-limiting step in protein folding. They also function as chaperones, having a broad subcellular distribution. Cyclophilin A (CypA), an abundant cytosolic protein found in all tissues examined, is best known for its ability to bind cyclosporine A (CsA). CypA is involved in T-cell activation and is thought both to provide a chaperone activity and to maintain proper protein conformation. It has been known for over a decade that CypA is efficiently inserted within HIV-1 at a ratio of 1 CypA molecule to 10 Gag molecules (37, 107), which represents approximately 250 molecules per virion. The immunosuppressive drug CsA inhibits incorporation of CypA into HIV-1. However, the closely related retroviruses HIV-2 and SIV do not incorporate CypA, with the exception of the chimpanzee-specific SIVcpz (19). It has also been detected inside vaccinia virus (VV) and vesicular stomatitis virus (VSV). Interestingly, the ability of HIV-1 to package this host-derived molecule has been investigated for various viral subtypes. Viruses of the outlier O group incorporate CypA in amounts similar to that of viruses belonging to the M group, but their infectivity does not rely on it (18, 19, 37). Further analyses indicated that all five O-group isolates tested incorporated CypA in a CsA-sensitive way, while their infectivity strongly depends on it in only three cases (116). CypA is initially packaged inside the virus by a direct interaction between its hydrophobic binding pocket and the proline-rich flexible exposed loop located within the amino-terminal domain of the viral capsid (CA) protein (14). Nevertheless, its affinity for the CA molecule is weak, and during maturation it has been observed to relocate to the viral surface (95).
Several studies have focused on the role of CypA in the HIV-1 life cycle (97). Most importantly, it is the only incorporated cellular protein shown to be critical for viral infectivity. Moreover, infectivity is finely tuned by host CypA expression levels (121). It efficiently catalyzes the cis-trans-isomerization of a peptide bond on CA (15). As for its exact role in infectivity, some authors have argued for a role in early events of the replication cycle, such as uncoating, and others for a role in late events, such as maturation. Since it has been observed that Gag assembles in the absence of CypA, it has been proposed that CypA is required at a step between Gag assembly and virion morphogenesis, possibly for conformational changes (103). Moreover, in vitro studies provided evidence that CypA does not efficiently destabilize assembled CA at the molar ratio observed in the virion, and the authors concluded it was unlikely to serve as an uncoating factor (44, 117). Their data suggest that CypA more likely exerts its effect by facilitating the coordinated rearrangement of CA subunits during the maturation process.
On the other hand, arguing against a role in late events are the facts that assembly occurs in the presence of CsA and that disruption of the Gag-CypA interaction still allows for assembly and budding to give particles with the proper number of Gag proteins. Moreover, it has been shown that the core stability is due to protein-protein contacts between the CA subunits without involvement of CypA, which can bind only to an aggregated form of immature CA and not a dissociated one. Thus, binding of CypA could very well serve only as a means of entry into the virion (14). Moreover, incorporation of a catalytically inactive form of CypA is sufficient for efficient infection, and thus the isomerase activity is not involved in virus infectivity (96). Spinoculated CypA-deficient viruses enter target cells efficiently but fail to infect them, which also points to a postentry event (93).
Although group O viruses do not require CypA for replication, the fact that they package all CypA into mature HIV-1 particles suggests that they evolved from a virus which was at one time CypA dependent. Thus, interestingly, the study of incorporation of a host protein supports the hypothesis that the group M and O viruses were transmitted to humans on two separate occasions from nonhuman primates, as has previously been suggested (19).
In addition to enhancing infectivity, a number of other roles have been suggested for the incorporated CypA. V3 loop peptides derived from HIV-1 macrophage- and T-cell-tropic external envelope gp120 bind with high affinity to the active site of CypA (33), pointing to a possible role in virus attachment. In other regards, it has been proposed that CypA could be a mediator in the initial attachment of HIV-1 to the host cell plasma membrane (100) through its interaction with heparans expressed at the cell surface (94). In fact, at least one CypA isoform has been detected outside the viral membrane (69). It is yet unclear how the cytosolic protein might penetrate the viral membrane, but the existence of several isoforms differentially located within the virion points to possible posttranslational modifications. Moreover, binding affinity to the CA protein strongly decreases as the CA matures, which could allow dissociation and relocation of the cyclophilin. However, the possible contribution of CypA to virus attachment is difficult to reconcile with the differential dependence of the infectivity of certain O-group viruses on CypA (116). In other regards, CD147, a transmembrane glycoprotein of the immunoglobulin superfamily, has also been identified as a receptor for CypA, and it would interact with it downstream of the CypA-heparin interaction (87). A previous study indicated that CypA is also important for the de novo synthesis of the viral protein Vpr, and in the absence of its activity, Vpr-mediated cell cycle arrest is completely lost in HIV-1-infected T cells (122). Moreover, in human cells it reduces HIV-1 sensitivity to restriction factors present within host cells (108).
When packaged in viruses of the Rhabdoviridae family, such as VSV, CypA seems to act differently. For example, although CypA is important for VSV infection, it acts at the level of primary transcription, helping in the proper folding of the N protein (16). Interestingly, the prevailing virulent NJ strain of VSV has a critical dependence on CypA, whereas the less virulent and less widespread IND serotype does not. As for VV, a member of the Poxviridae family, Castro and coworkers have found CypA packaged in viral cores, with approximately 156 molecules per single VV particle (26). They have speculated that CypA could either mediate the transport of virus proteins to virosomes, catalyze conformational changes in virus proteins important to the assembly process, or participate in the uncoating of viral cores.
Other prolyl isomerases have been studied for their incorporation into viruses, including FK-506-binding proteins (FKBPs), parvulins (e.g., pin1), and other cyclophilins. Among the latter, cyclophilin B (CypB), which is targeted to the endoplasmic reticulum, is not incorporated into HIV-1 in vivo, unlike cytosolic CypA (18, 37). FKBP12 has been detected inside HIV-1 at an average of 25 molecules per virion (20). Even though the specificity of FKBPs is much higher than that of CypA and one of their best substrate sequences contains Phe-Pro, which is known as an HIV-1 protease-specific cleavage site (20, 50), the relevance of FKBP12 to the virus biology remains to be established.
(v) HSPs.
Some chaperone heat shock proteins (HSPs) have been found incorporated into enveloped viruses. HSP70 has been detected within certain retroviruses but not in either VV or MMLV (26, 46). This stress-inducible chaperone protein has been estimated to be present in HIV-1 particles at a ratio of up to one molecule per polymerase protein and to be incorporated into the related HIV-2ROD strain and three SIV strains (46). HSP60 has been observed in HIV-1 by certain authors but not by others (6, 46, 78, 80). As for HSP90, another chaperone that acts in cooperation with certain partner proteins, such as p23 and HSP70, but is not incorporated into HIV-1 and VV, its presence has been inferred inside duck hepatitis B virus at a ratio of two to four copies per virion (52). It is deemed to be incorporated through association with the viral polymerase, along with p23. It is noteworthy that certain studies have reported cyclophilins functioning as part of a cellular chaperoning complex along with HSPs (16).
HOST CELL PROTEINS LOCATED ON VIRUSES
(i) VV.
VV has been extensively studied for its usefulness in vaccination strategies. Successful incorporation of foreign antigens has been achieved within recombinant VV, and in some cases it resulted in superior vaccine efficacy (40, 56). Naturally occurring association of host cell components with VV has also been described recently and constitutes a mechanism of protection for this virus against the host's complement immune system. Four types of virus particles are produced in VV-infected cells: intracellular mature virus, intracellular enveloped virus, cell-associated enveloped virus, and extracellular enveloped virus (EEV). The EEVs acquire host complement control proteins in their lipid bilayers, which can potently reduce the activity of the immune complement system and thus virolysis (55, 110). These proteins include CD46, CD55, and CD59. Four other membrane proteins unrelated to complement control were found as well, i.e., CD29, CD71, CD81, and major histocompatibility complex class I (MHC-I), although no specific role in the virus biology has been attributed to the latter proteins. The incorporation of the above complement control proteins appears to be virus strain and cell type specific. Additionally, the association of such host molecules with the virus envelope depends on which intracellular cell membrane the envelope comes from. Envelopes derived from the trans-Golgi network or membranes downstream carry complement control proteins, but these are not found associated with viruses originating from the endoplasmic reticulum, intermediate compartments, or cis- or medial-Golgi bodies(55).
(ii) HCMV.
Human cytomegalovirus (HCMV) has been occasionally reported to associate with cellular proteins. A first study reported the association of β2-microglobulin with the virus (45). This association was discovered while investigating factors inhibiting the detection of this virus in urine. It was shown that, following inoculation of HCMV into urine, β2-microglobulin binds to the virus. This association was not therefore occurring while the virus was budding but rather with cell-free viruses.
(iii) HIV-1.
Numerous host membrane proteins have been shown to be inserted within mature HIV-1 particles. The functionality of some specific virus-bound host cell surface proteins has been characterized, but the role and function of most remain to be elucidated. A complete description of host cell surface constituents embedded in HIV-1 has already been published (76, 77, 109). This section will focus only on molecules found within HIV-1 and, in some instances, other primate lentiviruses which have been shown to play a potential role in the viral life cycle.
(iv) MHC-I.
Incorporation of MHC-I and particularly the Cw4 allele within X4-tropic primary and laboratory isolates of HIV-1 has been reported to exert a profound influence on both virus infectivity and susceptibility to neutralizing antibodies (31). The effect was found to be associated with changes in viral envelope conformation.
(v) MHC-II.
A very common cellular antigen found on HIV-1 is the HLA-DR isotype of MHC-II. Beyond its natural association with the T-cell receptor, HLA-DR interacts with the CD4 glycoprotein. Interestingly, it has been reported that HIV-1-associated HLA-DR has the capacity to interact with the CD4 molecule on target cells. Even though this interaction is relatively weak, this additional virus-cell interaction in cooperation with the normal gp120-CD4 association increases the binding efficacy and kinetics of HIV-1 infection roughly twofold (21, 22). This increment in virus infectivity was illustrated with viruses produced in naturally HLA-DR-expressing cells as well as in transiently transfected cells. In other regards, the MHC molecules are intimately implicated in adaptive immunity, presenting antigenic peptides to the peptide-specific T cell. This function of antigen presentation by virus-associated HLA-DR is currently under investigation, for either cell activation, anergy, and/or apoptosis. Actually, this aspect has been addressed in only one study that reported the ability of HLA-DR-containing viruses to present superantigens and activate primary T cells (90).
(vi) ICAM-1.
The cellular adhesion molecule ICAM-1 is a major constituent of the HIV-1, HIV-2, and SIV envelope. This molecule has a very strong affinity for its major cellular ligand, LFA-1, and as with the HLA-DR protein, ICAM-1 influences the level of HIV-1 infectivity. If HLA-DR modestly influences the rate of HIV-1 infection, virus-associated ICAM-1 possesses a much more substantial impact on virus infectivity. Its incorporation within HIV-1 allows for 5- to 10-fold-increased infectivity in target T cells (35). Furthermore, its ligand, LFA-1, can switch to a high-affinity state following activation signals through a conformational change of the protein. Levels of up to nearly 100-fold can be reached when ICAM-1-bearing virions are used to infect target cells expressing LFA-1 under an activated form (36). Note that these studies were performed with progeny virus produced by cells expressing ICAM-1 upon transfection.
Virus entry studies, including subcellular fractionation experiments with primary human T lymphocytes, clearly illustrated that the acquisition of ICAM-1 by nascent HIV-1 may modify the entry route of the virus within the target T cell. The ICAM-1-carrying particles are more likely to release their material within the cell cytosol, instead of being endocytosed, as are virions lacking host ICAM-1 (106). The direct fusion of HIV-1 particles with the cell cytoplasmic membrane gives the virus direct access to the cytosol. This entry route is known to establish a more productive infection. In contrast, the intracellular endocytosis of this virus is proposed to lead to the degradation of the incoming particle and to a nonproductive infection (65).
One study extended these observations to a more relevant and physiological model of human tonsil tissue explants. In this experimental model, infection with ICAM-1-bearing HIV-1 particles led to a more robust infection than that with virions lacking host ICAM-1 (a 5- to 10-fold increase) (17). Antibody-mediated activation of LFA-1 resulted in a more profound depletion of CD4+ T lymphocytes within the tonsil tissue explants. Progeny viruses present in the culture supernatant following the explant infection contained high levels of host cell ICAM-1 within their envelopes, illustrating the capacity of the virus to acquire this adhesion molecule when produced in human secondary lymphoid organs.
Few studies have put in evidence the fact that cell-derived ICAM-1 within HIV-1 provides protection against neutralizing antibodies (34, 51, 59, 89). For example, sera from HIV-1-seropositive patients were less potent in inhibiting ICAM-1-bearing viruses than viruses devoid of it, though neutralization resistance was also seen with some monoclonal antibodies to the envelope proteins (34, 51, 89). In one study, this protection conferred by host cellular ICAM-1 within HIV-1 particles was most noticeable when the target cell LFA-1 was in an activated state (34). It is thus clear that this knowledge about the influence of host molecules incorporated within HIV-1 and their impact on protection against circulating antibodies is of great value for the design of vaccine strategies based on induction of neutralizing antibodies.
The cellular ICAM-1 present on HIV-1 might also interfere with the action of the new generation of fusion peptide inhibitors, as was indicated when peripheral blood mononuclear cells (PBMCs) from healthy donors were infected with primary isolates of HIV-1 either lacking or bearing host ICAM-1 in the presence of T-20 (8). Data showed that ICAM-1-bearing HIV-1 particles were about twofold less sensitive to the inhibiting action of the T-20 peptide than were isogenic viruses devoid of host ICAM-1 (8). In an experimental setting where the LFA-1 ligand on PBMCs was brought to its activated state, the decrease in sensitivity reached four- to fivefold. This increased resistance to the antiviral efficacy of T-20 for primary isolates of HIV-1 that are known to carry host ICAM-1 is believed to be the result of higher fusion kinetics for these viruses.
How HIV-1 and other lentiviruses manage to acquire cellular membrane proteins within their envelopes is not well established yet. But like many processes in biology, the incorporation of cellular proteins into the external lipid bilayer of a lentivirus likely occurs through several different mechanisms, as it seems to be cell type and virus strain dependent, at least under in vitro conditions (23, 24, 67, 82). One study reported a direct interaction between virion-incorporated HLA-DR and a 43-amino-acid domain located in the cytoplasmic tail of HIV-1 transmembrane envelope glycoprotein gp41 (84). In contrast, incorporation of ICAM-1 was found to be independent of HIV-1 envelope glycoproteins (7).
(vii) Proteins interacting with the cellular cytoskeleton.
Lentiviruses intimately interact with the actin microfilament meshwork of the cellular cytoskeleton (60). Indeed, actin microfilaments have been detected in purified preparations of HIV-1 along with ezrin, moesin, and cofilin (80, 81). The three latter consist of linker proteins between the plasma membrane and the actin filaments. Many membrane proteins incorporated within HIV-1 can be bound to the cytoskeleton actin network. The fact that some cytoskeleton structures are packaged within HIV-1 led to the hypothesis that selective inclusion of cytoskeleton-bound cell surface proteins may occur during the budding process. This hypothesis is supported by the recent demonstration that in transfection systems the selective uptake of host ICAM-1 is due to an interaction between the cytoplasmic tail of ICAM-1 and the virus-encoded Pr55Gag polyprotein (9). Given that the connection between ICAM-1 and the cortical actin was demonstrated on one hand and that the interaction between HIV-1 Gag and filamentous actin was confirmed on the other hand, it was proposed that the insertion of Gag, actin, and ICAM-1 within nascent virions would be in the form of a multimolecular complex (9).
(viii) Lipid raft molecules.
Another possible explanation for the differential incorporation of the cell membrane proteins onto the surfaces of these enveloped viruses is that the newly formed viral entities depart from the host cell by passing through a specific region of the cytoplasmic bilayer. The association of a cell surface molecule with the virus would depend on whether it is clustered at the virus budding site or not. Evidence that such a phenomenon is plausible has accumulated for few viruses over the years. The cellular membranes contain detergent-resistant lipids that are organized in patches and are enriched in sphingolipids, cholesterol, and glycerophospholipids. These structures, called lipid rafts, are known to include specific membrane proteins, while excluding others, and to play key roles in intracellular signaling processes. It is becoming clear that some enveloped viruses assemble and bud through these unique lipidic regions. Influenza virus, Ebola virus, measles virus, and HIV-1 have all been demonstrated to egress from infected cells through such specialized microdomains (29). Obviously, a virus budding through a lipid raft would incorporate cellular membrane molecules that are part of the raft, while excluding those outside the raft. Measles virus, for example, was demonstrated to incorporate at least some parts of the components of the lipid rafts (63). Moreover, for HIV-1 and SIV, treatment of virus stocks with β-cyclodextrin, a drug known to deplete cholesterol from membrane rafts, results in the creation of pores in the viral envelope and consequently a loss of infectivity (43). The above two studies support the concept of virus budding through raft structures. A study performed by Nguyen and colleague brought more direct proofs that support this working model (75). This group utilized HIV-1 harvested from infected Jurkat-transformed T cells to evaluate the possible presence of some raft-associated cellular membrane proteins within the viral envelope. They discovered that the glycosylphosphatidylinositol (GPI)-linked proteins Thy-1 and CD59 are incorporated efficiently within HIV-1. It should be pointed out that GPI-linked proteins are especially concentrated within lipid rafts on the cytoplasmic membrane of eukaryotic cells. Furthermore, a specific marker of lipid rafts, the ganglioside GM1, was found embedded in the external lipid bilayer of the virus preparation as well. This raft marker was also identified in envelopes of parainfluenza virus, influenza virus, Epstein-Barr virus, and herpes simplex virus (83). In contrast, the transmembrane protein CD45 that localizes outside the lipid rafts failed to be acquired by the excreted HIV-1 viral particles.
Interestingly, a quite different scenario seems to operate for HIV-1 emerging from macrophages. Indeed, HIV-1 formation in this particular cell type seems to occur by budding through intracellular vesicles. A recent hypothesis, called the Trojan exosome hypothesis, describes the similarity between newly formed HIV-1 and exosomes produced by macrophages and proposes that viruses in formation pass through the exosome-excreting pathway (42). One recent study supported this hypothesis by showing that the array of host molecules carried by HIV-1 particles produced in macrophages is a reflection of molecules expressed on the surfaces of exosomes rather than molecules normally present within lipid rafts on the cell surface (74).
(ix) Complement control proteins.
Complement control proteins are in most cases GPI-linked membrane proteins (70). And, as mentioned above, they are regularly found in lipid rafts and are more susceptible to incorporation into enveloped viruses. The complement control family members CD46, CD55, and CD59 were readily detected in envelopes of HCMV, HTLV-I, and HIV-1 (91, 92, 101). Just as for VV, the acquisition of complement control proteins such as CD46, CD55, and CD59 was shown to give protection from lysis and attack by complement for all these viruses.
CONCLUSION
To the best of our knowledge, there is no mention of incorporation of host-derived proteins in the following enveloped-virus families: Orthomyxoviridae, Bunyaviridae, Coronaviridae, Arenaviridae, Flaviviridae, and Togaviridae. It seems unlikely that this phenomenon does not occur in these groups, and the fact that it has not been mentioned might simply reflect the fact that they have not yet been studied.
It is somewhat difficult to distinguish features shared by the various host molecules found embedded within budding viruses. It is noteworthy, however, that all proteins indicated in this paper to be found inside virions are cytosolic molecules and that, likewise, host constituents found on the surfaces of virions are derived from cellular membranes, in accordance with what is known of the budding process. Moreover, most virus-anchored intracellular constituents appear to be part of very basic cell biology mechanisms: members of the MVB pathway, cyclophilins, chaperones, heat-shock proteins, or cytoskeleton subunits, for example. Otherwise, no other obvious trend has been observed, especially with the cell membrane components inserted within the viral envelope, except for the observation that some are clearly concentrated in lipid rafts.
Specific key roles or auxiliary roles in the virus life cycle have been demonstrated for few cellular proteins associated with viruses (or, one could say, plundered by viruses in the war against their target cells), and the vast majority of them remain orphans in this regard. This observation leads us to propose that some host cell-derived molecules may simply be stowaways and are carried by viruses inadvertently.
Describing the host cellular proteins associated with viruses might have looked fruitless initially. In fact, up to now many cellular proteins found within viruses do not have a known role in the life cycles of viruses. But just as described above for APOBEC3G or the cyclophilins found within the retroviruses, once comprehension of a cellular protein function is established in relation to virus biology it opens the door for promising new therapies. Indeed, a better understanding of this research topic may lead to the development of novel vaccine strategies or the creation of totally new classes of antiviral drugs.
Acknowledgments
We acknowledge numerous contributions from various laboratories that were not cited in the present review due to space limitations.
The work presented in this review from our laboratory is supported by grants to M.J.T. from the Canadian Institutes of Health Research (CIHR) HIV/AIDS Research Program (HOP-14438) and the CIHR New Emerging Team Program (HSD-63191). M.J.T. is the recipient of the Canada Research Chair in Human Immuno-Retrovirology (senior level).
REFERENCES
- 1.Alce, T. M., and W. Popik. 2004. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 279:34083-34086. [DOI] [PubMed] [Google Scholar]
- 2.Allal, C., C. Buisson-Brenac, V. Marion, C. Claudel-Renard, T. Faraut, P. Dal Monte, D. Streblow, M. Record, and J. L. Davignon. 2004. Human cytomegalovirus carries a cell-derived phospholipase A2 required for infectivity. J. Virol. 78:7717-7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amit, I., L. Yakir, M. Katz, Y. Zwang, M. D. Marmor, A. Citri, K. Shtiegman, I. Alroy, S. Tuvia, Y. Reiss, E. Roubini, M. Cohen, R. Wides, E. Bacharach, U. Schubert, and Y. Yarden. 2004. Tal, a Tsg101-specific E3 ubiquitin ligase, regulates receptor endocytosis and retrovirus budding. Genes Dev. 18:1737-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arthur, L. O., J. W. Bess, Jr., R. C. Sowder II, R. E. Benveniste, D. L. Mann, J. C. Chermann, and L. E. Henderson. 1992. Cellular proteins bound to immunodeficiency viruses: implications for pathogenesis and vaccines. Science 258:1935-1938. [DOI] [PubMed] [Google Scholar]
- 5.Bacharach, E., J. Gonsky, K. Alin, M. Orlova, and S. P. Goff. 2000. The carboxy-terminal fragment of nucleolin interacts with the nucleocapsid domain of retroviral Gag proteins and inhibits virion assembly. J. Virol. 74:11027-11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartz, S. R., C. D. Pauza, J. Ivanyi, S. Jindal, W. J. Welch, and M. Malkovsky. 1994. An Hsp60 related protein is associated with purified HIV and SIV. J. Med. Primatol. 23:151-154. [DOI] [PubMed] [Google Scholar]
- 7.Beauséjour, Y., and M. J. Tremblay. 2004. Envelope glycoproteins are not required for insertion of host ICAM-1 into human immunodeficiency virus type 1 and ICAM-1-bearing viruses are still infectious despite a suboptimal level of trimeric envelope proteins. Virology 324:165-172. [DOI] [PubMed] [Google Scholar]
- 8.Beauséjour, Y., and M. J. Tremblay. 2004. Susceptibility of HIV type 1 to the fusion inhibitor T-20 is reduced on insertion of host intercellular adhesion molecule 1 in the virus membrane. J. Infect. Dis. 190:894-902. [DOI] [PubMed] [Google Scholar]
- 9.Beauséjour, Y., and M. J. Tremblay. 2004. Interaction between the cytoplasmic domain of ICAM-1 and Pr55Gag leads to acquisition of host ICAM-1 by human immunodeficiency virus type 1. J. Virol. 78:11916-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bishop, K. N., R. K. Holmes, A. M. Sheehy, and M. H. Malim. 2004. APOBEC-mediated editing of viral RNA. Science 305:645. [DOI] [PubMed] [Google Scholar]
- 11.Bishop, N., and P. Woodman. 2001. TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J. Biol. Chem. 276:11735-11742. [DOI] [PubMed] [Google Scholar]
- 12.Blot, V., F. Perugi, B. Gay, M.-C. Prevost, L. Briant, F. Tangy, H. Abriel, O. Staub, M.-C. Dokhelar, and C. Pique. 2004. Nedd4.1-mediated ubiquitination and subsequent recruitment of Tsg101 ensure HTLV-1 Gag trafficking towards the multivesicular body pathway prior to virus budding. J. Cell Sci. 117:2357-2367. [DOI] [PubMed] [Google Scholar]
- 13.Bogerd, H. P., B. P. Doehle, H. L. Wiegand, and B. R. Cullen. 2004. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl. Acad. Sci. USA 101:3770-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.BonHomme, M., S. Wong, C. Carter, and S. Scarlata. 2003. The pH dependence of HIV-1 capsid assembly and its interaction with cyclophilin A. Biophys. Chem. 105:67-77. [DOI] [PubMed] [Google Scholar]
- 15.Bosco, D. A., E. Z. Eisenmesser, S. Pochapsky, W. I. Sundquist, and D. Kern. 2002. Catalysis of cis/trans isomerization in native HIV-1 capsid by human cyclophilin A. Proc. Natl. Acad. Sci. USA 99:5247-5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bose, S., M. Mathur, P. Bates, N. Joshi, and A. K. Banerjee. 2003. Requirement for cyclophilin A for the replication of vesicular stomatitis virus New Jersey serotype. J. Gen. Virol. 84:1687-1699. [DOI] [PubMed] [Google Scholar]
- 17.Bounou, S., J. E. Leclerc, and M. J. Tremblay. 2002. Presence of host ICAM-1 in laboratory and clinical strains of human immunodeficiency virus type 1 increases virus infectivity and CD4+-T-cell depletion in human lymphoid tissue, a major site of replication in vivo. J. Virol. 76:1004-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braaten, D., H. Ansari, and J. Luban. 1997. The hydrophobic pocket of cyclophilin is the binding site for the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 71:2107-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braaten, D., E. K. Franke, and J. Luban. 1996. Cyclophilin A is required for the replication of group M human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus SIVCPZGAB but not group O HIV-1 or other primate immunodeficiency viruses. J. Virol. 70:4220-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Briggs, C. J., D. E. Ott, L. V. Coren, S. Oroszlan, and J. Tozser. 1999. Comparison of the effect of FK506 and cyclosporin A on virus production in H9 cells chronically and newly infected by HIV-1. Arch. Virol. 144:2151-2160. [DOI] [PubMed] [Google Scholar]
- 21.Cantin, R., J. F. Fortin, G. Lamontagne, and M. Tremblay. 1997. The acquisition of host-derived major histocompatibility complex class II glycoproteins by human immunodeficiency virus type 1 accelerates the process of virus entry and infection in human T-lymphoid cells. Blood 90:1091-1100. [PubMed] [Google Scholar]
- 22.Cantin, R., J. F. Fortin, G. Lamontagne, and M. Tremblay. 1997. The presence of host-derived HLA-DR1 on human immunodeficiency virus type 1 increases viral infectivity. J. Virol. 71:1922-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cantin, R., J. F. Fortin, and M. Tremblay. 1996. The amount of host HLA-DR proteins acquired by HIV-1 is virus strain- and cell type-specific. Virology 218:372-381. [DOI] [PubMed] [Google Scholar]
- 24.Cantin, R., G. Martin, and M. J. Tremblay. 2001. A novel virus capture assay reveals a differential acquisition of host HLA-DR by clinical isolates of human immunodeficiency virus type 1 expanded in primary human cells depending on the nature of producing cells and the donor source. J. Gen. Virol. 82:2979-2987. [DOI] [PubMed] [Google Scholar]
- 25.Cartier, C., M. Deckert, C. Grangeasse, R. Trauger, F. Jensen, A. Bernard, A. Cozzone, C. Desgranges, and V. Boyer. 1997. Association of ERK2 mitogen-activated protein kinase with human immunodeficiency virus particles. J. Virol. 71:4832-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castro, A. P., T. M. Carvalho, N. Moussatche, and C. R. Damaso. 2003. Redistribution of cyclophilin A to viral factories during vaccinia virus infection and its incorporation into mature particles. J. Virol. 77:9052-9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cen, S., F. Guo, M. Niu, J. Saadatmand, J. Deflassieux, and L. Kleiman. 2004. The interaction between HIV-1 Gag and APOBEC3G. J. Biol. Chem. 279:33177-33184. [DOI] [PubMed] [Google Scholar]
- 28.Chatel-Chaix, L., J. F. Clement, C. Martel, V. Beriault, A. Gatignol, L. DesGroseillers, and A. J. Mouland. 2004. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol. Cell. Bio. 24:2637-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chazal, N., and D. Gerlier. 2003. Virus entry, assembly, budding, and membrane rafts. Microbiol. Mol. Biol. Rev. 67:226-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen, R., H. Wang, and L. M. Mansky. 2002. Roles of uracil-DNA glycosylase and dUTPase in virus replication. J. Gen. Virol. 83:2339-2345. [DOI] [PubMed] [Google Scholar]
- 31.Cosma, A., D. Blanc, J. Braun, C. Quillent, C. Barassi, C. Moog, S. Klasen, B. Spire, G. Scarlatti, E. Pesenti, A. G. Siccardi, and A. Beretta. 1999. Enhanced HIV infectivity and changes in GP120 conformation associated with viral incorporation of human leucocyte antigen class I molecules. AIDS 13:2033-2042. [DOI] [PubMed] [Google Scholar]
- 32.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Endrich, M. M., and H. Gehring. 1998. The V3 loop of human immunodeficiency virus type-1 envelope protein is a high-affinity ligand for immunophilins present in human blood. Eur. J. Biochem. 252:441-446. [DOI] [PubMed] [Google Scholar]
- 34.Fortin, J. F., R. Cantin, M. G. Bergeron, and M. J. Tremblay. 2000. Interaction between virion-bound host intercellular adhesion molecule-1 and the high-affinity state of lymphocyte function-associated antigen-1 on target cells renders R5 and X4 isolates of human immunodeficiency virus type 1 more refractory to neutralization. Virology 268:493-503. [DOI] [PubMed] [Google Scholar]
- 35.Fortin, J. F., R. Cantin, G. Lamontagne, and M. Tremblay. 1997. Host-derived ICAM-1 glycoproteins incorporated on human immunodeficiency virus type 1 are biologically active and enhance viral infectivity. J. Virol. 71:3588-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fortin, J. F., R. Cantin, and M. J. Tremblay. 1998. T cells expressing activated LFA-1 are more susceptible to infection with human immunodeficiency virus type 1 particles bearing host-encoded ICAM-1. J. Virol. 72:2105-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franke, E. K., H. E. Yuan, and J. Luban. 1994. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372:359-362. [DOI] [PubMed] [Google Scholar]
- 38.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 40.Gomez, C. E., and M. Esteban. 2001. Recombinant proteins produced by vaccinia virus vectors can be incorporated within the virion (IMV form) into different compartments. Arch. Virol. 146:875-892. [DOI] [PubMed] [Google Scholar]
- 41.Gottwein, E., J. Bodem, B. Muller, A. Schmechel, H. Zentgraf, and H. G. Krausslich. 2003. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J. Virol. 77:9474-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gould, S. J., A. M. Booth, and J. E. Hildreth. 2003. The Trojan exosome hypothesis. Proc. Natl. Acad. Sci. USA 100:10592-10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graham, D. R., E. Chertova, J. M. Hilburn, L. O. Arthur, and J. E. Hildreth. 2003. Cholesterol depletion of human immunodeficiency virus type 1 and simian immunodeficiency virus with beta-cyclodextrin inactivates and permeabilizes the virions: evidence for virion-associated lipid rafts. J. Virol. 77:8237-8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grattinger, M., H. Hohenberg, D. Thomas, T. Wilk, B. Muller, and H. G. Krausslich. 1999. In vitro assembly properties of wild-type and cyclophilin-binding defective human immunodeficiency virus capsid proteins in the presence and absence of cyclophilin A. Virology 257:247-260. [DOI] [PubMed] [Google Scholar]
- 45.Grundy, J. E., J. A. McKeating, and P. D. Griffiths. 1987. Cytomegalovirus strain AD169 binds beta 2 microglobulin in vitro after release from cells. J. Gen. Virol. 68:777-784. [DOI] [PubMed] [Google Scholar]
- 46.Gurer, C., A. Cimarelli, and J. Luban. 2002. Specific incorporation of heat shock protein 70 family members into primate lentiviral virions. J. Virol. 76:4666-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Halwani, R., S. Cen, H. Javanbakht, J. Saadatmand, S. Kim, K. Shiba, and L. Kleiman. 2004. Cellular distribution of Lysyl-tRNA synthetase and its interaction with Gag during human immunodeficiency virus type 1 assembly. J. Virol. 78:7553-7564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hammarstedt, M., and H. Garoff. 2004. Passive and active inclusion of host proteins in human immunodeficiency virus type 1 Gag particles during budding at the plasma membrane. J. Virol. 78:5686-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 50.Harrison, R. K., and R. L. Stein. 1990. Substrate specificities of the peptidyl prolyl cis-trans isomerase activities of cyclophilin and FK-506 binding protein: evidence for the existence of a family of distinct enzymes. Biochemistry 29:3813-3816. [DOI] [PubMed] [Google Scholar]
- 51.Hioe, C. E., L. Bastiani, J. E. Hildreth, and S. Zolla-Pazner. 1998. Role of cellular adhesion molecules in HIV type 1 infection and their impact on virus neutralization. AIDS Res. Hum. Retrovir. 39(Suppl. 3):S247-S254. [PubMed] [Google Scholar]
- 52.Hu, J., D. O. Toft, and C. Seeger. 1997. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. EMBO J. 16:59-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kao, S., M. A. Khan, E. Miyagi, R. Plishka, A. Buckler-White, and K. Strebel. 2003. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 77:11398-11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knox, P. G., and L. S. Young. 1995. Epstein-Barr virus infection of CR2-transfected epithelial cells reveals the presence of MHC class II on the virion. Virology 213:147-157. [DOI] [PubMed] [Google Scholar]
- 55.Krauss, O., R. Hollinshead, M. Hollinshead, and G. L. Smith. 2002. An investigation of incorporation of cellular antigens into vaccinia virus particles. J. Gen. Virol. 83:2347-2359. [DOI] [PubMed] [Google Scholar]
- 56.Kwak, H., W. Mustafa, K. Speirs, A. J. Abdool, Y. Paterson, and S. N. Isaacs. 2004. Improved protection conferred by vaccination with a recombinant vaccinia virus that incorporates a foreign antigen into the extracellular enveloped virion. Virology 322:337-348. [DOI] [PubMed] [Google Scholar]
- 57.Licata, J. M., M. Simpson-Holley, N. T. Wright, Z. Han, J. Paragas, and R. N. Harty. 2003. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J. Virol. 77:1812-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liddament, M. T., W. L. Brown, A. J. Schumacher, and R. S. Harris. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14:1385-1391. [DOI] [PubMed] [Google Scholar]
- 59.Losier, M., J. F. Fortin, R. Cantin, M. G. Bergeron, and M. J. Tremblay. 2003. Virion-bound ICAM-1 and activated LFA-1: a combination of factors conferring resistance to neutralization by sera from human immunodeficiency virus type 1-infected individuals independently of the disease status and phase. Clin. Immunol. 108:111-118. [DOI] [PubMed] [Google Scholar]
- 60.Luftig, R. B., and L. D. Lupo. 1994. Viral interactions with the host-cell cytoskeleton: the role of retroviral proteases. Trends Microbiol. 2:178-182. [DOI] [PubMed] [Google Scholar]
- 61.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 62.Mangeat, B., P. Turelli, S. Liao, and D. Trono. 2004. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 279:14481-14483. [DOI] [PubMed] [Google Scholar]
- 63.Manie, S. N., S. Debreyne, S. Vincent, and D. Gerlier. 2000. Measles virus structural components are enriched into lipid raft microdomains: a potential cellular location for virus assembly. J. Virol. 74:305-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mansky, L. M., S. Preveral, L. Selig, R. Benarous, and S. Benichou. 2000. The interaction of Vpr with uracil DNA glycosylase modulates the human immunodeficiency virus type 1 in vivo mutation rate. J. Virol. 74:7039-7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marechal, V., F. Clavel, J. M. Heard, and O. Schwartz. 1998. Cytosolic Gag p24 as an index of productive entry of human immunodeficiency virus type 1. J. Virol. 72:2208-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 67.Martin, G., and M. J. Tremblay. 2004. HLA-DR, ICAM-1, CD40, CD40L, and CD86 are incorporated to a similar degree into clinical human immunodeficiency virus type 1 variants expanded in natural reservoirs such as peripheral blood mononuclear cells and human lymphoid tissue cultured ex vivo. Clin. Immunol. 111:275-285. [DOI] [PubMed] [Google Scholar]
- 68.Michelson, S., P. Turowski, L. Picard, J. Goris, M. P. Landini, A. Topilko, B. Hemmings, C. Bessia, A. Garcia, and J. L. Virelizier. 1996. Human cytomegalovirus carries serine/threonine protein phosphatases PP1 and a host-cell derived PP2A. J. Virol. 70:1415-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Misumi, S., T. Fuchigami, N. Takamune, I. Takahashi, M. Takama, and S. Shoji. 2002. Three isoforms of cyclophilin A associated with human immunodeficiency virus type 1 were found by proteomics by using two-dimensional gel electrophoresis and matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Virol. 76:10000-10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miwa, T., and W. C. Song. 2001. Membrane complement regulatory proteins: insight from animal studies and relevance to human diseases. Int. Immunopharmacol. 1:445-459. [DOI] [PubMed] [Google Scholar]
- 71.Morita, E., and W. I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395-425. [DOI] [PubMed] [Google Scholar]
- 72.Mouland, A. J., J. Mercier, M. Luo, L. Bernier, L. DesGroseillers, and E. A. Cohen. 2000. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J. Virol. 74:5441-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nermut, M. V., K. Wallengren, and J. Pager. 1999. Localization of actin in Moloney murine leukemia virus by immunoelectron microscopy. Virology 260:23-34. [DOI] [PubMed] [Google Scholar]
- 74.Nguyen, D. G., A. Booth, S. J. Gould, and J. E. Hildreth. 2003. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J. Biol. Chem. 278:52347-52354. [DOI] [PubMed] [Google Scholar]
- 75.Nguyen, D. H., and J. E. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ott, D. E. 1997. Cellular proteins in HIV virions. Rev. Med. Virol. 7:167-180. [DOI] [PubMed] [Google Scholar]
- 77.Ott, D. E. 2002. Potential roles of cellular proteins in HIV-1. Rev. Med. Virol. 12:359-374. [DOI] [PubMed] [Google Scholar]
- 78.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278:111-121. [DOI] [PubMed] [Google Scholar]
- 79.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder III, Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72:2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ott, D. E., L. V. Coren, D. G. Johnson, B. P. Kane, R. C. Sowder III, Y. D. Kim, R. J. Fisher, X. Z. Zhou, K. P. Lu, and L. E. Henderson. 2000. Actin-binding cellular proteins inside human immunodeficiency virus type 1. Virology 266:42-51. [DOI] [PubMed] [Google Scholar]
- 81.Ott, D. E., L. V. Coren, B. P. Kane, L. K. Busch, D. G. Johnson, R. C. Sowder III, E. N. Chertova, L. O. Arthur, and L. E. Henderson. 1996. Cytoskeletal proteins inside human immunodeficiency virus type 1 virions. J. Virol. 70:7734-7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Paquette, J. S., J. F. Fortin, L. Blanchard, and M. J. Tremblay. 1998. Level of ICAM-1 surface expression on virus producer cells influences both the amount of virion-bound host ICAM-1 and human immunodeficiency virus type 1 infectivity. J. Virol. 72:9329-9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pickl, W. F., F. X. Pimentel-Muinos, and B. Seed. 2001. Lipid rafts and pseudotyping. J. Virol. 75:7175-7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Poon, D. T. K., L. V. Coren, and D. E. Ott. 2000. Efficient incorporation of HLA class II onto human immunodeficiency virus type 1 requires envelope glycoprotein packaging. J. Virol. 74:3918-3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pornillos, O., J. E. Garrus, and W. I. Sundquist. 2002. Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 12:569-579. [DOI] [PubMed] [Google Scholar]
- 86.Priet, S., J. M. Navarro, N. Gros, G. Querat, and J. Sire. 2003. Differential incorporation of uracil DNA glycosylase UNG2 into HIV-1, HIV-2, and SIVMAC viral particles. Virology 307:283-289. [DOI] [PubMed] [Google Scholar]
- 87.Pushkarsky, T., G. Zybarth, L. Dubrovsky, V. Yurchenko, H. Tang, H. Guo, B. Toole, B. Sherry, and M. Bukrinsky. 2001. CD147 facilitates HIV-1 infection by interacting with virus-associated cyclophilin A. Proc. Natl. Acad. Sci. USA 98:6360-6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Putterman, D., R. B. Pepinsky, and V. M. Vogt. 1990. Ubiquitin in avian leukosis virus particles. Virology 176:633-637. [DOI] [PubMed] [Google Scholar]
- 89.Rizzuto, C. D., and J. G. Sodroski. 1997. Contribution of virion ICAM-1 to human immunodeficiency virus infectivity and sensitivity to neutralization. J. Virol. 71:4847-4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rossio, J. L., J. Bess, Jr., L. E. Henderson, P. Cresswell, and L. O. Arthur. 1995. HLA class II on HIV particles is functional in superantigen presentation to human T cells: implications for HIV pathogenesis. AIDS Res. Hum. Retrovir. 11:1433-1439. [DOI] [PubMed] [Google Scholar]
- 91.Saifuddin, M., T. Hedayati, J. P. Atkinson, M. H. Holguin, C. J. Parker, and G. T. Spear. 1997. Human immunodeficiency virus type 1 incorporates both glycosyl phosphatidylinositol-anchored CD55 and CD59 and integral membrane CD46 at levels that protect from complement-mediated destruction. J. Gen. Virol. 78:1907-1911. [DOI] [PubMed] [Google Scholar]
- 92.Saifuddin, M., C. J. Parker, M. E. Peeples, M. K. Gorny, S. Zolla-Pazner, M. Ghassemi, I. A. Rooney, J. P. Atkinson, and G. T. Spear. 1995. Role of virion-associated glycosylphosphatidylinositol-linked proteins CD55 and CD59 in complement resistance of cell line-derived and primary isolates of HIV-1. J. Exp. Med. 182:501-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Saphire, A. C., M. D. Bobardt, and P. A. Gallay. 2002. Cyclophilin A plays distinct roles in human immunodeficiency virus type 1 entry and postentry events, as revealed by spinoculation. J. Virol. 76:4671-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Saphire, A. C., M. D. Bobardt, and P. A. Gallay. 1999. Host cyclophilin A mediates HIV-1 attachment to target cells via heparans. EMBO J. 18:6771-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Saphire, A. C., M. D. Bobardt, and P. A. Gallay. 2000. Human immunodeficiency virus type 1 hijacks host cyclophilin A for its attachment to target cells. Immunol. Res. 21:211-217. [DOI] [PubMed] [Google Scholar]
- 96.Saphire, A. C., M. D. Bobardt, and P. A. Gallay. 2002. trans-Complementation rescue of cyclophilin A-deficient viruses reveals that the requirement for cyclophilin A in human immunodeficiency virus type 1 replication is independent of its isomerase activity. J. Virol. 76:2255-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Scarlata, S., and C. Carter. 2003. Role of HIV-1 Gag domains in viral assembly. Biochim. Biophys. Acta 1614:62-72. [DOI] [PubMed] [Google Scholar]
- 98.Schrofelbauer, B., D. Chen, and N. R. Landau. 2004. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. USA 101:3927-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 100.Sherry, B., G. Zybarth, M. Alfano, L. Dubrovsky, R. Mitchell, D. Rich, P. Ulrich, R. Bucala, A. Cerami, and M. Bukrinsky. 1998. Role of cyclophilin A in the uptake of HIV-1 by macrophages and T lymphocytes. Proc. Natl. Acad. Sci. USA 95:1758-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Spear, G. T., N. S. Lurain, C. J. Parker, M. Ghassemi, G. H. Payne, and M. Saifuddin. 1995. Host cell-derived complement control proteins CD55 and CD59 are incorporated into the virions of two unrelated enveloped viruses. Human T cell leukemia/lymphoma virus type I (HTLV-I) and human cytomegalovirus (HCMV). J. Immunol. 155:4376-4381.7594597 [Google Scholar]
- 102.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 103.Streblow, D. N., M. Kitabwalla, and C. D. Pauza. 1998. Gag protein from human immunodeficiency virus type 1 assembles in the absence of cyclophilin A. Virology 252:228-234. [DOI] [PubMed] [Google Scholar]
- 104.Stuchell, M. D., J. E. Garrus, B. Muller, K. M. Stray, S. Ghaffarian, R. McKinnon, H.-G. Krausslich, S. G. Morham, and W. I. Sundquist. 2004. The human endosomal complex required for transport I (ESCRT-I) and its role in HIV-1 budding. J. Biol. Chem. 279:36059-36071. [DOI] [PubMed] [Google Scholar]
- 105.Svarovskaia, E. S., H. Xu, J. L. Mbisa, R. Barr, R. J. Gorelick, A. Ono, E. O. Freed, W. S. Hu, and V. K. Pathak. 2004. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 279:35822-35828. [DOI] [PubMed] [Google Scholar]
- 106.Tardif, M. R., and M. J. Tremblay. 2003. Presence of host ICAM-1 in human immunodeficiency virus type 1 virions increases productive infection of CD4+ T lymphocytes by favoring cytosolic delivery of viral material. J. Virol. 77:12299-12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Thali, M., A. Bukovsky, E. Kondo, B. Rosenwirth, C. T. Walsh, J. Sodroski, and H. G. Gottlinger. 1994. Functional association of cyclophilin A with HIV-1 virions. Nature 372:363-365. [DOI] [PubMed] [Google Scholar]
- 108.Towers, G. J., T. Hatziioannou, S. Cowan, S. P. Goff, J. Luban, and P. D. Bieniasz. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9:1138-1143. [DOI] [PubMed] [Google Scholar]
- 109.Tremblay, M. J., J. F. Fortin, and R. Cantin. 1998. The acquisition of host-encoded proteins by nascent HIV-1. Immunol. Today 19:346-351. [DOI] [PubMed] [Google Scholar]
- 110.Vanderplasschen, A., E. Mathew, M. Hollinshead, R. B. Sim, and G. L. Smith. 1998. Extracellular enveloped vaccinia virus is resistant to complement because of incorporation of host complement control proteins into its envelope. Proc. Natl. Acad. Sci. USA 95:7544-7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vartanian, J. P., A. Meyerhans, B. Asjo, and S. Wain-Hobson. 1991. Selection, recombination, and G→A hypermutation of human immunodeficiency virus type 1 genomes. J. Virol. 65:1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Pro. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 114.Wang, M. Q., W. Kim, G. Gao, T. A. Torrey, H. C. Morse III, P. De Camilli, and S. P. Goff. 2003. Endophilins interact with Moloney murine leukemia virus Gag and modulate virion production. J. Biol. 3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wiegers, K., and H. G. Krausslich. 2002. Differential dependence of the infectivity of HIV-1 group O isolates on the cellular protein cyclophilin A. Virology 294:289-295. [DOI] [PubMed] [Google Scholar]
- 117.Wiegers, K., G. Rutter, U. Schubert, M. Grattinger, and H. G. Krausslich. 1999. Cyclophilin A incorporation is not required for human immunodeficiency virus type 1 particle maturation and does not destabilize the mature capsid. Virology 257:261-274. [DOI] [PubMed] [Google Scholar]
- 118.Willetts, K. E., F. Rey, I. Agostini, J. M. Navarro, Y. Baudat, R. Vigne, and J. Sire. 1999. DNA repair enzyme uracil DNA glycosylase is specifically incorporated into human immunodeficiency virus type 1 viral particles through a Vpr-independent mechanism. J. Virol. 73:1682-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wright, J. F., A. Kurosky, E. L. Pryzdial, and S. Wasi. 1995. Host cellular annexin II is associated with cytomegalovirus particles isolated from cultured human fibroblasts. J. Virol. 69:4784-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xu, H., E. S. Svarovskaia, R. Barr, Y. Zhang, M. A. Khan, K. Strebel, and V. K. Pathak. 2004. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl. Acad. Sci. USA 101:5652-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yin, L., D. Braaten, and J. Luban. 1998. Human immunodeficiency virus type 1 replication is modulated by host cyclophilin A expression levels. J. Virol. 72:6430-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zander, K., M. P. Sherman, U. Tessmer, K. Bruns, V. Wray, A. T. Prechtel, E. Schubert, P. Henklein, J. Luban, J. Neidleman, W. C. Greene, and U. Schubert. 2003. Cyclophilin A interacts with HIV-1 Vpr and is required for its functional expression. J. Biol. Chem. 278:43202-43213. [DOI] [PubMed] [Google Scholar]
- 123.Zheng, Y. H., D. Irwin, T. Kurosu, K. Tokunaga, T. Sata, and B. M. Peterlin. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78:6073-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]