Abstract
Monoclonal antibodies (MAbs) directed against epitopes in the V2 domain of human immunodeficiency virus type 1 gp120 often possess neutralizing activity, but these generally are highly type specific, neutralize only laboratory isolates, or have low potency. The most potent of these is C108g, directed against a type-specific epitope in HXB2 and BaL gp120s, which is glycan dependent and, in contrast to previous reports, dependent on intact disulfide bonds. This epitope was introduced into two primary Envs, derived from a neutralization-sensitive (SF162) and a neutralization-resistant (JR-FL) isolate, by substitution of two residues and, for SF162, addition of an N-linked glycosylation site. C108g effectively neutralized both variant Envs with considerably higher potency than standard MAbs against the V3 and CD4-binding domains and the broadly neutralizing MAbs 2G12 and 2F5. These amino acid substitutions also introduced the epitope recognized by a second V2-specific MAb, 10/76b, but this MAb possessed potent neutralizing activity only in the absence of the glycan required for C108g reactivity. In contrast to other gp120-specific neutralizing MAbs, C108g did not block binding of soluble Env proteins to either the CD4 or the CCR5 receptor, but studies with a fusion-arrested Env indicated that C108g neutralized at a step preceding the one blocked by the gp41-specific MAb, 2F5. These results indicate that the V1/V2 domain possesses targets that mediate potent neutralization of primary viral isolates via a novel mechanism and suggest that inclusion of carbohydrate determinants into these epitopes may help overcome the indirect masking effects that limit the neutralizing potency of antibodies commonly produced after infection.
A major mechanism of resistance to antibody-mediated neutralization of primary human immunodeficiency virus (HIV) isolates is the blocking of antibody binding to common neutralization targets in native Env complexes by glycans present in several regions of gp120 (6, 22, 44). Evidence for a major role for the V1/V2 domain in this effect is provided by studies showing that deletion of V1 and V2 sequences increases the overall sensitivity of various HIV and simian immunodeficiency virus isolates to neutralization (5, 19) and that the V1/V2 domain contains the primary determinant of the very large difference in neutralization sensitivity of two related primary isolates, SF162 and JR-FL (27). Mutations in the V1/V2 domain have also been shown to influence multiple aspects of viral phenotype and tropism (11, 20, 21, 25, 30, 34, 36, 39, 42, 46), suggesting that in addition to its role in protecting against antibody-mediated neutralization, this region has a specific function necessary for infection. These observations raise the question of whether the V1/V2 domain contains epitopes that can function as effective targets for viral neutralization, particularly in viral envelopes in which the more common neutralization targets are masked.
Earlier studies provided some evidence that V2 epitopes can function as neutralization determinants; however, those studies did not suggest that antibodies against this region are important components of the protective neutralizing response or that the V2 domain is a useful vaccine target. The initial monoclonal antibodies (MAbs) isolated against this region were generated by immunizing mice with HXB2-derived gp120. Many of these were directed against discontinuous epitopes and had limited cross-reactivity and relatively weak neutralizing activities (17, 24, 36). Rats immunized with HXB2 gp120 produced MAbs that recognized both linear and conformationally dependent discontinuous epitopes in the V2 domain (23, 35, 45). While some of the MAbs against the linear epitopes possessed stronger neutralizing activity for lab-adapted viruses, these MAbs were highly type specific for viruses with the IIIB and related V2 sequences. A separate study of MAbs isolated from transgenic mice producing human immunoglobulins that were immunized with recombinant SF162 gp120 (rgp120) described a series of relatively potent MAbs directed against highly type-specific linear epitopes in V1 and one MAb that recognized a fairly conserved linear epitope in V2 that possessed only low neutralizing activity (15).
Other studies have examined V1/V2-dependent MAbs isolated from HIV-infected humans. One report described a human MAb (697D) against a relatively conserved conformational V2 epitope that possessed neutralizing activity for some primary isolates but not for laboratory-adapted viruses (14). However, subsequent studies indicated that the neutralizing activity of this MAb was quite weak. Four Fabs derived from a phage library of human heavy- and light-chain sequences from an asymptomatic HIV type 1 (HIV-1)-seropositive human recognized a distinct class of epitopes that appeared to involve both the V2 loop and the CD4-binding site (8), one of which possessed neutralizing activity for several laboratory-adapted viruses.
A V2-specific MAb (C108g) isolated from a chimpanzee infected with the HXB2 isolate (40, 43) provided the strongest indication that the V2 domain was a potentially useful vaccine target. This antibody was directed against a type-specific, glycan-dependent epitope restricted to the HXB2 and BaL viral isolates and neutralized these isolates at considerably lower antibody concentrations than any other MAb tested (41, 43, 45). The neutralizing activity of C108g for those isolates was synergistic with that of MAbs directed against the V3 loop or the CD4-binding site (41). The high type specificity of C108g precluded its evaluation against other viral isolates. However, studies using affinity-purified human antibodies isolated from infected individuals indicated that some humans produce more broadly reactive V2-directed antibodies with relatively potent neutralizing activities for multiple primary isolates (18, 28), suggesting the presence of appropriate conserved targets in this domain. Considering that epitope masking by the V1/V2 domain may strongly limit the sensitivity of common neutralization epitopes in typical primary isolates (27), directing the immune response toward potent neutralization targets in the blocking domain itself may provide a possible approach to overcoming this problem.
The focus of the present study was to more fully define the structure of the C108g epitope and to examine its ability to mediate neutralization when presented in the context of prototypical primary Envs. A fuller definition of this sensitive V2 neutralization target and identification of factors that contribute to its exceptional neutralization potency and novel mechanism might facilitate the design of synthetic or recombinant antigens expressing more common variants of this epitope that are capable of inducing antibodies with broadly neutralizing activity for primary viral isolates.
MATERIALS AND METHODS
Viruses and viral neutralization assays.
Neutralization activity was determined with a single-cycle infectivity assay using virions generated from the Env-defective luciferase-expressing HIVNL4-3 genome pseudotyped with molecularly cloned HIV Env as previously described (27). Wild-type JR-FL and SF162 Envs were expressed from pcDNA3.1(zeo)-based expression plasmids (Invitrogen) as previously described (27). Mutations to introduce amino acid substitutions required for C108g reactivity were generated by QuikChange site-directed mutagenesis (Stratagene) on subclones of the relevant env genes, sequenced, and placed into these expression plasmids on suitable restriction fragments. The pCAGGS expression plasmid for JR-FL Env bearing the “SOS” Cys mutations (2) was provided by James Binley. The C108g epitope was introduced into this plasmid by exchange of an appropriate restriction fragment from the JRFL(GKV) variant.
Antibodies and recombinant Env proteins.
C108g was obtained from an Epstein-Barr virus-transformed B-cell line derived from a chimpanzee infected with IIIB virus (43) and purified by adsorption to protein A beads. 10/76b was from a hybridoma cell line generated by Jane McKeating (23) and provided by the National Institute for Biological Standards and Control, Hertfordshire, United Kingdom. This antibody was purified by adsorption to an affinity column containing rabbit anti-rat immunoglobulin (Ig). All other MAbs and polyclonal antisera from pooled HIV-positive human sera (HIVIG) were produced in our laboratories or obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health. Recombinant gp120BaL and a full-length single-chain gp120-CD4 complex (FLSC), consisting of the D1D2 domains of CD4 fused via a 20-amino-acid linker to the C terminus of BaL gp120, were produced by stable transfection of 293 cells with plasmids expressing the codon-optimized BaL gp120 and FLSC sequences (10), provided by Timothy Fouts at the Institute of Human Virology. These Env proteins were purified from cell culture medium by lectin chromatography with Galanthus nivalis snowdrop agglutinin (Sigma-Aldrich, St. Louis, MO) as previously described (13).
Antibody binding assays to soluble gp120.
Sheep anti-HIV-1 gp120 C5 polyclonal antibody (Cliniqa Corp., Fallbrook, CA) at 10 μg/ml in sodium bicarbonate buffer, pH 9.8, was adsorbed onto enzyme-linked immunosorbent assay (ELISA) plates (Falcon) overnight at 37°C. After blocking with 2% dry milk in phosphate-buffered saline (PBS; pH 7.4), the plates were incubated with a solution of BaL rgp120 at 2 μg/ml in PBS overnight at 37°C. Prior to capture, the rgp120 was either reduced by treatment with 1% dithiothreitol (DTT) in PBS for 5 h at 37°C, followed by acetylation with 2% iodoacetamide for 1 h at 37°C, or deglycosylated by digestion with a 1:25 dilution of peptide N-glycosidase F (New England Biolabs) for 6 h at 37°C. Binding of MAbs was for 1 h at 37°C in the presence of 2% dry milk-PBS; wells were washed with PBS-0.1% Tween 20, and bound MAbs were detected using alkaline phosphatase-conjugated anti-IgG (Zymed Laboratories, South San Francisco, CA), followed by addition of substrate (1 mg/ml p-nitrophenol in diethanolamine buffer [pH 9.8]). A405 measurements were taken at various time points using a Spectra SLT ELISA plate reader (TECAN Instruments, Research Triangle Park, NC). Relative affinities were determined by comparing the concentration of MAb required to achieve 50% maximal binding to rgp120.
Inhibition of binding of Env proteins to cells expressing HIV receptors.
The inhibition by various MAbs of binding of soluble Env proteins to U87 cells expressing either CD4 or CCR5 was measured by flow cytometry. A total of 5 × 105 cells were incubated with BaL rgp120 or BaL FLSC at 1 μg/ml in PBS containing 10% fetal bovine serum (FACS buffer), both in the presence and in the absence of blocking reagent at 37°C for 1 h with gentle agitation. Cells were washed in 1 ml of ice-cold FACS buffer, treated with biotin-labeled HIVIG at 50 μg/ml in FACS buffer at 4°C for 1 h, washed, and treated with R-phycoerythrin-conjugated streptavidin in FACS buffer at 4°C for 30 min, washed again with FACS buffer, and fixed with 3% paraformaldehyde in PBS. Bound Env protein was then quantitated by flow cytometry, using a FACScalibur flow cytometer (BD Biosciences, San Jose, CA) and CellQuest software for acquisition and analysis.
Neutralization assays of disulfide-shackled (SOS) Env pseudovirions.
Single-round infections of U87.CD4.CCR5 cells were performed with luciferase-expressing virions pseudotyped with JR-FL(GKV).SOS Env as previously described (2). Briefly, virus was preincubated at 37°C for 1 h either in the presence or in the absence of test MAbs at concentrations above their 90% neutralizing dose (ND90) for JR-FL Env in standard assays and then added to cells in the presence of Polybrene (20 μg/ml) for 2 h. The medium was then replaced with fresh medium with and without MAbs and incubated at 37°C for an additional 2 h to allow binding. The virus was then activated by reducing the intrasubunit disulfide bond by exposure to 12.5 mM DTT for 10 min, and luciferase activity was measured 2 days later as described above. All assays were performed in duplicate and were repeated for a total of at least four replicates.
RESULTS
Biochemical properties of the C108g epitope.
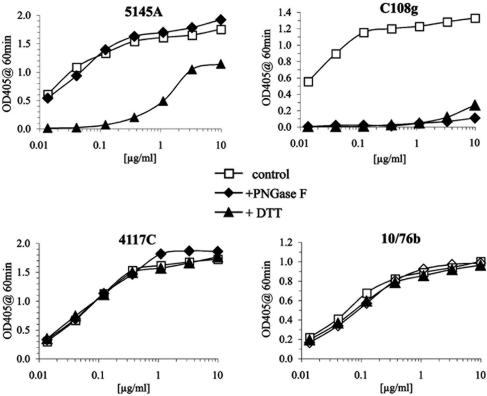
C108g was previously reported to be directed against a glycan-dependent epitope that survived reduction of disulfide bonds (43). The particular glycan required was subsequently shown to be located at position 160 (numbering based on that of the HXB2 sequence) (45), a highly conserved, but not invariant, glycosylation site. The initial characterization of the biochemical properties of the C108g epitope was performed using only a single, very high concentration of antibody (43), which obscured possible changes in the affinity of the MAb to the modified antigen. The sensitivity of C108g to reduction and deglycosylation was reexamined in a more quantitative manner by performing a titration of the reactivity of C108g with BaL rgp120 captured on an ELISA plate (Fig. 1). Control antibodies tested in parallel included 5145A, directed against a disulfide-dependent epitope in the CD4-binding domain; 4117C, directed against a linear V3 epitope; and 10/76b, directed against a linear V2 epitope also present in the HXB2 and BaL isolates.
FIG. 1.
Effects of reduction and deglycosylation on stability of epitopes in the V2 and other domains. Recombinant BaL gp120 was captured on wells of a 96-well ELISA plate either untreated (open squares) or after deglycosylation with peptide N-glycosidase F (PNGase F; closed diamonds) or reduction by DTT (closed triangles). The reactivity of the indicated MAbs to control or treated rgp120 was titrated for 1 h at 37°C and bound antibody detected with alkaline phosphatase-conjugated anti-IgG, followed by incubation with alkaline phosphatase substrate. OD405, optical density at 405 nm.
As expected, the reactivities of 4117C and 10/76b with BaL gp120 were unaffected by either deglycosylation or reduction of rgp120. Reactivity of 5145A was also unaffected by deglycosylation but was reduced over 100-fold by reduction. The residual signal may be due to a small amount of partially nonreduced material, since this reactivity was not observed for samples treated with DTT for 24 h (data not shown). Either reduction or deglycosylation abolished C108g reactivity, with the reduced sample producing only a trace of a signal at the highest MAb concentration tested (20 μg/ml). Similar results were obtained for C108g and 10/76b with HXB2-derived rgp120. These results confirmed that the C108g epitope was highly dependent on the presence of N-linked glycans and showed that, in contrast to the original report, this epitope was also dependent on the presence of intact disulfide bonds.
Insertion of the C108g epitope into SF162 and JR-FL Envs.
Prior studies showed that C108g binds weakly to synthetic peptides that include the V2-derived sequence STSIRGKV, located at residues 162 to 169 of HXB2 gp120 (43), adjacent to the N-linked glycan at position 160 that was essential for C108g binding to gp120 (45). Although the apparent affinity of C108g for these peptides was very low (2 × 107 liters/mole versus 5.5 × 1010 liters/mole for rgp120), this reactivity indicates that residues within this sequence contribute to the epitope. A comparison of the sequence of this region for the two C108g-reactive Envs (HXB2 and BaL) and two prototypical C108g-nonreactive primary Envs (JR-FL and SF162) suggested residues likely to be involved in the C108g epitope (Fig. 2). JR-FL differed from the C108g-reactive sequences only at positions 167 and 168 (DE instead of GK), while SF162 differed both in this region (NKM in place of GKV at positions 167 to 169) and at the position 160 glycosylation site. In place of the N-linked glycosylation signal NIT at position 160 to 162, SF162 Env contained KVT and thus was lacking the glycan required for C108g reactivity.
FIG. 2.
Sequences of the V2 regions of the Env proteins used in this study. The region in HXB2 V2 containing glycosylation site 160 and the adjoining C108g-reactive peptide sequence is boxed, and N-linked glycosylation motifs are underlined. The mutated residues in this region for JR-FL and SF162 are in italics. The SF16Z(NI+GKV) variant contained both the NI and the G-V mutations.
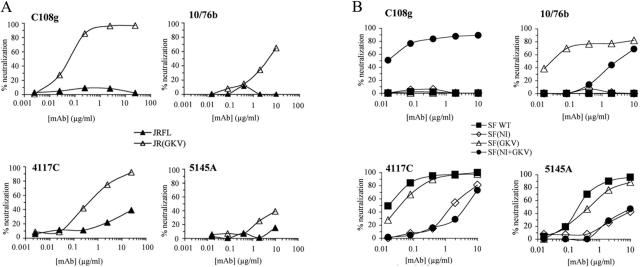
To examine the effects of these polymorphisms on the C108g epitope, JR-FL and SF162 env genes were modified by site-specific mutagenesis. For JR-FL, the DE at positions 167/168 was converted to GK to produce the JR(GKV) variant. For SF162, the N-linked glycosylation site at position 160 was introduced by converting KV at positions 160 and 161 to NI and NKM at positions 167 to 169 to GKV. These modifications were made separately [to generate the SF(NI) and SF(GKV) variants] and together [to generate the SF(NI+GKV) variant]. The effects of these changes on the expression of both the C108g and 10/76b epitopes were then examined by titration of the neutralizing activity of these MAbs for luc-expressing virions pseudotyped with the various Env proteins.
For JR-FL Env, the DE-to-GK substitution resulted in expression of both epitopes (Fig. 3A). Neutralization of this variant by C108g was quite potent, with an ND50 of 0.049 μg/ml, while 10/76b neutralized only weakly, with an ∼100-fold higher ND50 (5.1 μg/ml). For SF162 Env, the NKM-to-GKV substitution resulted in expression of the 10/76b epitope in high-affinity form (ND50 = 0.032 μg/ml), but the C108g epitope was not detectably expressed (Fig. 3B). Restoration of the 160 glycan alone did not result in expression of either epitope. Introducing both alterations into SF162 Env resulted in expression of the C108g epitope in high-affinity form (ND50 = 0.014 μg/ml), but the inclusion of the glycan significantly interfered with 10/76b reactivity, resulting in an ∼200-fold increase in ND50 (ND50 = 6.5 μg/ml) compared to that for the SF(GKV) variant.
FIG. 3.
(A) Effects of mutations in the JR-FL V2 domain on neutralization by V2-specific MAbs C108g and 10/76b and MAbs to the V3 region (4117C) and CD4-binding domain (5145A). Neutralization assays were performed with env deleted, luc-expressing NL4-3 provirus pseudotypes with either the parental JR-FL Env (closed triangles) or the JR(GKV) variant expressing the C108g epitope in the JR-FL background (open triangles). (B) Effects of mutations in the SF162 V2 domain on neutralization by V2-specific MAbs C108g and 10/76b and MAbs to the V3 region and CD4-binding domains. Neutralization was determined as in panel A for luc-expressing viral pseudotypes with parental SF162 Env (closed squares) or with the SF162 variants containing the glycosylation site at position 160 [SF(NI), open diamonds], the changes at positions 167 to 169 [SF(GKV), open triangles], or the combination of these changes [SF(NI+GKV), closed circles] that expressed the C108g epitope. WT, wild type.
V2 modifications required for expression of the C108g epitope affect the overall neutralization sensitivity of SF162 and JR-FL Envs.
The JR-FL V1/V2 domain exerts a strong inhibitory effect on neutralization by antibodies to sites commonly recognized during infection by HIV-1, including the V3 loop and CD4-binding domain (27). To determine whether the changes required for expression of the C108g and 10/76b epitopes affected this epitope masking, the sensitivity of the modified Envs to MAbs against a number of known neutralization epitopes outside the V2 domain was compared to that of the parental Envs.
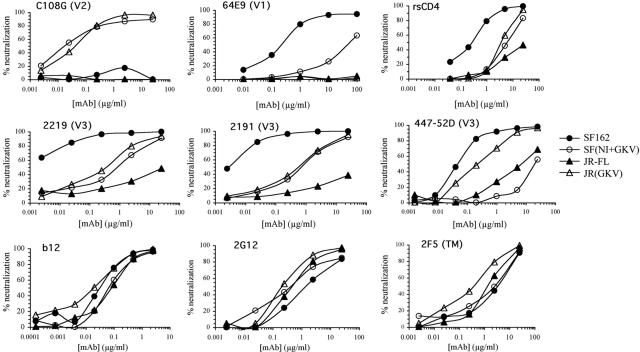
The DE-to-GK substitution in the JR-FL Env resulted in a substantial increase in sensitivity to neutralization by anti-V3 MAbs (Fig. 3A and 4 and Table 1). In most cases, ND50s were not obtained for the parental JR-FL at the highest concentration tested (25 μg/ml), and thus the extent of the increased sensitivity of the variant could only be calculated as a lower limit. These ranged from greater than 25-fold for 4117C to greater than 55-fold for 2219. The increase was 54-fold for 447-52D, the only anti-V3 MAb for which an ND50 was obtained for the parental JR-FL Env. While this increase in neutralization sensitivity to anti-V3 MAbs was significant, the ND50s against JR(GKV) were still hundreds-fold higher than those against the highly sensitive chimeric JR-FL Env with the V1/V2 domain for many of these MAbs (27), indicating that other sequence differences between JR-FL and SF162 V1/V2 domains accounted for the majority of this blocking effect. These V2 substitutions had only a small effect on the neutralization sensitivity to a typical anti-CD4bs MAb (5145A) or to a series of broadly neutralizing MAbs against conserved sites (IgGb12, 2G12, and 2F5), for which the ND50s against the variant were reduced by only two- to fourfold (Table 1).
FIG. 4.
Analysis of neutralizing activities of MAbs against various domains for the viruses pseudotyped with the two parental Envs SF162 (closed circles) and JR-FL (closed triangles) and with the two C108g-expressing variants SF(NI+GKV) (open circles) and JR(GKV) (open triangles). The MAb used and the domain it recognizes are indicated at the top of each panel.
TABLE 1.
Summary of neutralization endpoints (ND50s)a for SF162 and JR-FL variants
| Domain and MAb | ND50 (μg/ml)
|
|||||
|---|---|---|---|---|---|---|
| SF162 | SF(NI) | SF(GKV) | SF(NI+GKV) | JR-FL | JR(GKV) | |
| V1 | ||||||
| 58E1 | 0.60 | 30 | 2.2 | >100 | >100 | >100 |
| 64E4 | 0.28 | 10 | 0.90 | 68 | >100 | >100 |
| 45D1 | 0.60 | >30 | 3.0 | >100 | >100 | >100 |
| V2 | ||||||
| C108g | >25 | >25 | >10 | 0.014 | >25 | 0.049 |
| 10/76b | >25 | >25 | 0.032 | 6.5 | >10 | 5.1 |
| V3 | ||||||
| 4117C | 0.011 | 1.3 | 0.035 | 5.4 | >25 | 0.99 |
| 447-52D | 0.015 | 5.5 | 0.090 | 21 | 6.5 | 0.12 |
| 2191 | 0.0045 | 0.14 | 0.020 | 0.9 | >25 | 0.80 |
| 2219 | 0.0080 | 0.35 | 0.017 | 1 | >25 | 0.45 |
| Other | ||||||
| 5145A | 0.20 | >25 | 0.42 | 20 | >25 | 25 |
| sCD4 | 0.82 | 4.5 | 1.6 | 3.3 | 8.4 | 5.6 |
| IgGb12 | 0.035 | 0.065 | 0.055 | 0.065 | 0.082 | 0.022 |
| 2G12 | 1.1 | 0.76 | 1.2 | 1.20 | 0.65 | 0.43 |
| 2F5 | 2.6 | 5.3 | 0.60 | 2.4 | 1.4 | 0.78 |
Concentration of MAb that reduced infectivity by 50%. Values for samples for which ND50s were not obtained are listed as >X, where X is the highest antibody concentration tested.
The addition of an N-linked glycosylation site at position 160 of SF162 Env resulted in a more significant decrease in neutralization sensitivity to some anti-V3 MAbs (Fig. 3B and 4 and Table 1). The effect was greatest for MAbs 447-52D (336-fold increase in ND50) and 4117C (130-fold increase), while more modest increases (31- to 44-fold) were obtained for MAbs 2191 and 2219. The addition of this glycosylation site also caused a significant decrease in sensitivity to several MAbs directed against sites in the C terminus of the V1 domain of SF162, with ND50s 35- to 50-fold higher for the SF(NI) variant than for the parental SF162 Env. The GKV variant was also more resistant to the anti-V1 MAbs (3- to 5-fold), and these effects appeared to be additive for the combined SF(NI+GKV) variant, for which the ND50 were >160-fold higher than for the parental Env. These effects contrasted with those seen for MAbs to highly conserved domains (IgGb12, 2G12, and 2F5), for which addition of the glycosylation site at position 160 had only a slight effect (an about two- to threefold increase) on neutralization sensitivity.
The SF(GKV) mutation also resulted in small increases in neutralization sensitivity to V3 MAbs (2- to 6-fold increases in ND50s), and the combined NI+GKV substitution resulted in an additional ∼3- to 6-fold increase in ND50 for the anti-V3 MAbs over the insertion of the glycosylation site alone (Fig. 3B and 4 and Table 1). Although the separate NI and GKV substitutions had only a small effect on neutralization by CD4-binding domain-specific MAb 5145A, the combined NI+GKV substitutions had a more substantial effect, reducing sensitivity to this MAb by 50-fold. These results showed that both positions involved in the expression of the C108g epitope contributed to the ability of the V1/V2 domain to inhibit neutralization by antibodies to the V3 and CD4-binding domains.
In contrast to other gp120-specific MAbs, C108g does not block receptor binding of soluble Env proteins.
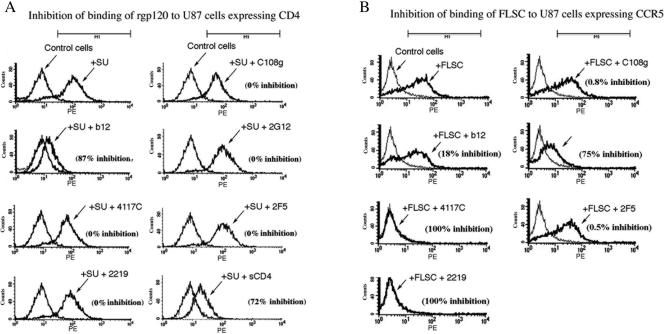
The potent neutralization by C108g suggested that mechanistic studies of the effect of this MAb on HIV-1 Env function might provide information on the role of the V2 domain during infection. An essential role of gp120 is binding to the specific cell surface receptors that determine cellular tropism and mediate fusion. One possible mechanism of C108g neutralization was inhibition of binding of virions to either CD4 or CCR5 on the cell surface. This was tested by assaying the ability of C108g to block the binding of soluble Env proteins to U87 cells expressing either of the receptors. Binding assays to cells expressing CD4 were performed with recombinant BaL gp120. Since the ability of gp120 to bind to CCR5 requires activation by prior binding to CD4, CCR5-binding assays were performed with an activated CD4-gp120 fusion protein, FLSC (10, 16).
Binding of gp120 or FLSC was assayed by fluorescence-activated cell sorter analysis in the presence or absence of MAbs at relatively high concentrations. As shown in Fig. 5A, binding of gp120 to cells expressing CD4 was inhibited significantly by b12 (87% inhibition) and sCD4 (72% inhibition) but not by any of the other MAbs tested. On the other hand, binding of FLSC to cells expressing CCR5 was completely blocked by two V3-specific MAbs and significantly blocked by 2G12 (75% inhibition), but not by b12, 2F5, or C108g (Fig. 5B). Thus, while the neutralizing activity of the other gp120-specific MAbs could be ascribed to inhibition of binding to either the CD4 or CCR5 receptor, C108g did not block binding of soluble pg120 ligands to either of the receptors.
FIG. 5.
Fluorescence-activated cell sorter assays of the inhibition of binding of either gp120 to U87 cells expressing CD4 (panel A) or of FLSC to cells expressing CCR5 (panel B) by MAbs or sCD4. Env proteins were used at 1.0 μg/ml, MAbs at 20 μg/ml, and sCD4 at 100 μg/ml. Binding of Env proteins to cells was detected by subsequent staining with biotinylated HIVIG, followed by R-phycoerythrin-conjugated streptavidin and quantitated on a Becton-Dickinson FACScalibur flow cytometer. The MAb used and the extent of inhibition compared to the control in the absence of antibody, measured as the decrease in mean fluorescence intensity compared to the control, are indicated in each panel.
The C108g-sensitive step occurs prior to the step blocked by the introduction of an intersubunit disulfide bond in Env.
Env constructs were recently described in which the association of HIV-1 SU and TM was stabilized by the introduction of an intersubunit disulfide bond (SOS-Env) (3, 31, 32). Pseudovirions containing SOS-Env attach to cells expressing CD4 and coreceptor but require triggering with reducing agent for fusion to occur (2). Further studies suggested that prior to such triggering virus entry was arrested after CD4 and coreceptor engagement and that the arrested complex was insensitive to gp120-targeting inhibitors but still neutralized by gp41-specific inhibitors, including peptides that prevent six-helix bundle formation and the gp41-specific MAb 2F5 (1).
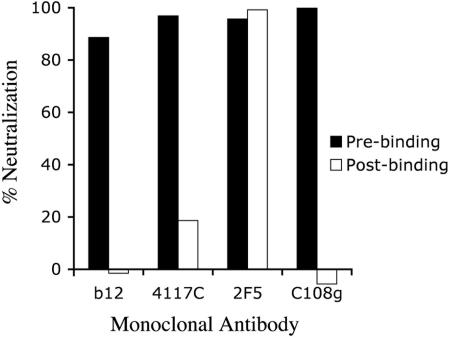
Pseudovirions containing a JR-FL(GKV)-derived SOS Env were constructed and tested for sensitivity to neutralization by MAbs against the CD4-binding domain (b12), the V3 loop (4117C), gp41 (2F5), and V2 (C108g). The pseudovirions were exposed to neutralizing concentrations of MAbs either prior to binding to cells or postbinding, and the bound virions were then activated by exposure to 5 mM DTT for 10 min. Whereas all of the MAbs neutralized efficiently when present prior to binding, only 2F5 was capable of neutralizing the SOS-arrested virions (Fig. 6). These results indicated that the C108g-sensitive step in Env function occurred prior to the step that was blocked by the introduction of the SOS intersubunit disulfide bond.
FIG. 6.
Pre- and postbinding neutralization assays of SOS-shackled pseudovirions. Virus pseudotyped with the JR(GKV).SOS Env was either preincubated with MAbs and then added to cells (prebinding neutralization conditions) or allowed to bind to U87/CD4/CCR5 target cells for 2 h and then treated with the indicated antibodies for 2 h at 37°C (postbinding conditions). The virus and antibodies were then removed and the bound virus activated for fusion by brief exposure to DTT. After washing, the cells were incubated for an additional 2 days and assayed for luciferase levels. Antibodies were used at the following concentrations: 4117C, 19 μg/ml; C108g, 5 μg/ml; 2F5, 12 μg/ml; b12, 3.5 μg/ml.
DISCUSSION
The studies described in this paper demonstrate the ability of an epitope in the V2 domain to mediate potent neutralization of R5 Envs that are relatively resistant to MAbs against the dominant neutralization regions due to V1/V2-mediated epitope masking. The neutralization potency of C108g for JR-FL and SF162 variants containing this epitope was considerably higher than that of V3-specific MAbs tested; in the case of the SF162 variant, ND50s for C108g ranged from ∼70-fold lower than those of 2191 and 2219 to as much as 1,500-fold lower than that of 447-52D. This ratio was lower but still significant for the JR-FL variant, with ND50 ratios ranging from 2.4-fold for 447-52D to 22-fold for 4117C. The neutralizing potency of C108g was also considerably greater than that of several of the rare broadly neutralizing MAbs, with ND50s 86-fold lower than that of 2G12 and 171-fold lower than that of 2F5 for the SF162 variant; for the JR-FL variant, the corresponding ratios were 9-fold and 6-fold. C108g also neutralized more potently than standard ligands for the CD4-binding domain, such as sCD4 and the 5145A MAb, and its potency was higher than that of IgGb12 for the SF162 variant (ND50 lower by 4.6-fold) and only slightly lower than that of IgGb12 for the JR-FL variant (ND50 higher by 2.2-fold).
The potent neutralization by C108g was particularly intriguing in view of the fact that the sequences targeted by this MAb were major contributing factors toward the V1/V2 domain masking of epitopes in the V3 and CD4-binding domains. The addition of the 160 glycan to SF162 Env significantly reduced the neutralization potency of several MAbs directed against the V3 domain (ND50s increased by 30- to 366-fold). Substitution of the acidic residues DE at positions 167 and 168 in the JR-FL sequence by GK increased the neutralization sensitivity of the JR-FL variant to the anti-V3 MAbs by over 50-fold in several cases, whereas replacement of NKM at position 167 to 169 in the SF162 sequence by GKV reduced the neutralization sensitivity of the V3-specific MAbs by 2- to 6-fold. While the effect of the insertion of the glycosylation site into the SF162 sequence could be ascribed to steric hindrance of binding by the N-linked glycan at position 160, the substitutions at positions 167 and 168 were not significantly bulkier than the originally residues, and thus they affected neutralization by a different mechanism than that of the proposed “glycan shield” (44). This suggests that the V1/V2 masking effect may involve specific interactions between charged residues in addition to general steric effects due to carbohydrates.
In addition to its effects on more distal sites such as the V3 domain, the glycan at position 160 also masked epitopes in the adjacent V1 and V2 domains. The substitution of G and V at positions 167 and 169 of SF162 gp120 resulted in 10/76b reactivity, and the neutralization potency of 10/76b for this Env was almost as potent as that of C108g for the SF(NI+GKV) Env (Table 1). However, whereas C108g neutralization required the glycan at position 160, the neutralization activity of 10/76b was attenuated more than 200-fold by the presence of this glycan. Since 10/76b bound equally well to native and deglycosylated monomeric rgp120 (Fig. 1), this effect appeared to be due to conformational masking in the native Env oligomer rather than to direct modification of the epitope. The addition of the glycan at position 160 to SF162 Env also reduced the potency of several type-specific MAbs directed against the V1 domain by 35- to 50-fold. Thus, the glycan at position 160 has a strong masking effect on potentially sensitive neutralization targets in the V1, V2, and V3 domains. The unusual ability of C108g to neutralize in the presence of this glycan suggests that recognition of components of the glycan in addition to peptide sequences is an effective strategy for bypassing these masking effects.
The identification of potential vaccine targets in the V2 domain would be facilitated by a better understanding of the function that is being inhibited by neutralizing antibodies such as C108g and 10/76b. Interpretation of prior mechanistic studies of HIV-1 neutralization by antibodies is complicated by the fact that cells often contain ancillary adhesion factors for HIV-1, such as heparan sulfate proteoglycans (4, 26, 33) and sulfated lactosylceramide (12). These factors can mediate binding of certain strains of virus through interactions with V3 domains (26) that are sensitive to inhibition by gp120-specific antibodies (7, 38). Such nonspecific binding was not observed for the soluble Env proteins and U87 cells used in the present study, and the experiments shown in Fig. 5 clearly indicated that reagents directed against the CD4-binding domain block binding of gp120 to cellular CD4, while 2G12 and antibodies directed against the V3 loop block binding of activated gp120 to CCR5. These binding-inhibitory activities of MAbs against the V3 and CD4-binding domains were consistent with previous reports (29, 37, 38), while the ability of 2G12 to inhibit binding to CCR5 has not been previously described.
The inability of C108g to inhibit binding of the soluble Env proteins to either the CD4 or CCR5 receptor indicates that the site targeted by C108g is not directly involved in the recognition of either receptor by these proteins. One possibility suggested by this result is that C108g neutralizes at a postbinding step. However, unlike the TM-specific MAb 2F5, which also inhibits viral entry postbinding (2, 9) but is believed to block six-helix bundle formation (1), C108g failed to neutralize infection mediated by a receptor-bound, fusion-arrested SOS-Env construct (Fig. 6), indicating that C108g blocks a step prior to that blocked by 2F5. Mutations in the V2 domain of HXB2 Env, including the conversion of residue K168 to L, have been shown to reduce the stability of the association between gp120 and gp41 (36). It is therefore possible that the V2 domain in general, and the region targeted by C108g and 10/76b in particular, may be involved in conformational changes in Env following receptor binding that facilitate the disengagement of gp120 from gp41, and that binding of antibodies to these sites may interfere with these rearrangements. However, it is also possible that the lack of inhibition of receptor binding by C108g is a reflection of the soluble Env proteins used in these assays, but that in the native oligomeric complexes present in intact virions additional interactions occur between the C108g epitope and other Env regions that are involved in receptor binding that may interfere with such interactions.
Defining additional neutralization targets that are insensitive to epitope masking effects is important for HIV vaccine development. The regulation of HIV-1 neutralization sensitivity by epitope masking poses a serious dilemma for the design of an effective vaccine, since the common classes of antibodies generated in response to infection or immunization with standard immunogens (i.e., antibodies directed against the V3 and CD4-binding domains) are directed against neutralization targets that are strongly affected by epitope masking. Consequently, even if the problem of epitope variability is avoided or overcome, binding of these antibodies to intact virus will be inhibited by such masking, thus reducing the effectiveness of vaccines targeting these epitopes. The ability of the V2 domain to mask multiple regions of gp120 suggests that portions of the V2 domain itself must be exposed on the surface of intact virions, consistent with the potent neutralizing activity of C108g. Such exposed sites could present attractive vaccine targets that are not affected by the general masking effects that limit other domains of gp120.
A major limitation in the use of the C108g epitope itself as a vaccine target is its restricted distribution in natural viral sequences. This is due primarily to the polymorphic residues at positions 167 to 169, since the glycosylation site at position 160 is present in a large majority of known HIV-1 sequences. The Gly at position 167 is critical, since C108g does not recognize gp120 derived from the NL4-3 clone, which is similar to the HXB2 sequence in this region except for a Gly-to-Asp substitution at position 167, and substitution of Asp for Gly at this position in the minimal peptide results in loss of recognition by C108g (43). The presence of Gly at this position is rare, present in only two known isolates, IIIB and BaL. The great majority of other sequences contain Asp at this site, while a smaller group of isolates contains an Asn at this site. The other residues in this region are less variable and may be less important for immunoreactivity. Thus, it may be possible to generate a broadly reactive V2-specific response with potent neutralizing activity with an immunogen that expresses the critical structural region of V2 defined by C108g, including the glycan at position 160, but that contains the consensus residue at position 167. The requirement of C108g binding on the maintenance of intact disulfide bonds also indicates that there is a strong conformational context to this structure that is currently undefined. A more complete elucidation of the structure of this epitope and that of its underlying functional domain may provide additional insight needed for the rational design of immunogens capable of inducing potently neutralizing antibodies that recognize more conserved forms of this epitope.
Acknowledgments
These studies were supported by U.S. Public Health Service grants AI46283 and AI50452 to A.P., AI151987 to S.C.K., and AI36085 and HL59725 to S.Z.-P. Hybridoma cells producing 10/76b were provided by the EU Programme EVA/MRC Centralised Facility for AIDS Reagents, NIBSC, United Kingdom (grants QLK2-CT-1999-00609 and GP828102).
We acknowledge the invaluable contributions made by our colleague Sam Kayman to these studies and to the overall work of our lab during the past 16 years. Sam passed away from complications of lung cancer on 9 January 2005.
REFERENCES
- 1.Abrahamyan, L. G., R. M. Markosyan, J. P. Moore, F. S. Cohen, and G. B. Melikyan. 2003. Human immunodeficiency virus type 1 Env with an intersubunit disulfide bond engages coreceptors but requires bond reduction after engagement to induce fusion. J. Virol. 77:5829-5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Binley, J. M., C. S. Cayanan, C. Wiley, N. Schulke, W. C. Olson, and D. R. Burton. 2003. Redox-triggered infection by disulfide-shackled human immunodeficiency virus type 1 pseudovirions. J. Virol. 77:5678-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binley, J. M., R. W. B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bobardt, M. D., A. C. Saphire, H. C. Hung, X. Yu, B. Van der Schueren, Z. Zhang, G. David, and P. A. Gallay. 2003. Syndecan captures, protects, and transmits HIV to T lymphocytes. Immunity 18:27-39. [DOI] [PubMed] [Google Scholar]
- 5.Cao, J., N. Sullivan, E. Desjardin, C. Parolin, J. Robinson, R. Wyatt, and J. Sodroski. 1997. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J. Virol. 71:9808-9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng-Mayer, C., A. Brown, J. Harouse, P. A. Luciw, and A. J. Mayer. 1999. Selection for neutralization resistance of the simian/human immunodeficiency virus SHIVSF33A variant in vivo by virtue of sequence changes in the extracellular envelope glycoprotein that modify N-linked glycosylation. J. Virol. 73:5294-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demaria, S., and Y. Bushkin. 1996. Soluble CD4 induces the binding of human immunodeficiency virus type 1 to cells via the V3 loop of glycoprotein 120 and specific sites in glycoprotein 41. AIDS Res. Hum. Retrovir. 12:281-290. [DOI] [PubMed] [Google Scholar]
- 8.Ditzel, H. J., J. M. Binley, J. P. Moore, J. Sodroski, N. Sullivan, L. S. Sawyer, R. M. Hendry, W. P. Yang, C. F. Barbas III, and D. R. Burton. 1995. Neutralizing recombinant human antibodies to a conformational V2- and CD4-binding site-sensitive epitope of HIV-1 gp120 isolated by using an epitope-masking procedure. J. Immunol. 154:893-906. [PubMed] [Google Scholar]
- 9.Finnegan, C. M., W. Berg, G. K. Lewis, and A. L. DeVico. 2002. Antigenic properties of the human immunodeficiency virus transmembrane glycoprotein during cell-cell fusion. J. Virol. 76:12123-12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fouts, T. R., R. Tuskan, K. Godfrey, M. Reitz, D. Hone, G. K. Lewis, and A. L. DeVico. 2000. Expression and characterization of a single-chain polypeptide analogue of the human immunodeficiency virus type 1 gp120-CD4 receptor complex. J. Virol. 74:11427-11436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freed, E. O., and M. A. Martin. 1994. Evidence for a functional interaction between the V1/V2 and C4 domains of human immunodeficiency virus type 1 envelope glycoprotein gp120. J. Virol. 68:2503-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furuta, Y., K. Eriksson, B. Svennerholm, P. Fredman, P. Horal, S. Jeansson, A. Vahlne, J. Holmgren, and C. Czerkinsky. 1994. Infection of vaginal and colonic epithelial cells by the human immunodeficiency virus type 1 is neutralized by antibodies raised against conserved epitopes in the envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA 91:12559-12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilljam, G. 1993. Envelope glycoproteins of HIV-1, HIV-2, and SIV purified with Galanthus nivalis agglutinin induce strong immune responses. AIDS Res. Hum. Retrovir. 9:431-438. [DOI] [PubMed] [Google Scholar]
- 14.Gorny, M. K., J. P. Moore, A. J. Conley, S. Karwowska, J. Sodroski, C. Williams, S. Burda, L. J. Boots, and S. Zolla-Pazner. 1994. Human anti-V2 monoclonal antibody that neutralizes primary but not laboratory isolates of human immunodeficiency virus type 1. J. Virol. 68:8312-8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He, Y., W. J. Honnen, C. P. Krachmarov, M. Burkhart, S. C. Kayman, J. Corvalan, and A. Pinter. 2002. Efficient isolation of novel human monoclonal antibodies with neutralizing activity against HIV-1 from transgenic mice expressing human Ig loci. J. Immunol. 169:595-605. [DOI] [PubMed] [Google Scholar]
- 16.He, Y., P. D'Agostino, and A. Pinter. 2003. Analysis of the immunogenic properties of a single-chain polypeptide analogue of the HIV-1 gp120-CD4 complex in transgenic mice that produce human immunoglobulins. Vaccine 21:4421-4429. [DOI] [PubMed] [Google Scholar]
- 17.Ho, D. D., M. S. C. Fung, Y. Cao, X. L. Li, C. Sun, T. W. Chang, and N. C. Sun. 1991. Another discontinuous epitope on glycoprotein gp120 that is important in human immunodeficiency virus type 1 neutralization is identified by a monoclonal antibody. Proc. Natl. Acad. Sci. USA 88:8949-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honnen, W. J., Z. Wu, S. C. Kayman, and A. Pinter. 1996. Potent neutralization of a macrophage-tropic HIV-1 isolate by antibodies against the V1/V2 domain of gp120, p. 289-297. In E. N. F. Brown, D. Burton, and J. Mekalanos (ed.), Vaccines 1996: molecular approaches to the control of infectious diseases. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Johnson, W. E., H. Sanford, L. Schwall, D. R. Burton, P. W. Parren, J. E. Robinson, and R. C. Desrosiers. 2003. Assorted mutations in the envelope gene of simian immunodeficiency virus lead to loss of neutralization resistance against antibodies representing a broad spectrum of specificities. J. Virol. 77:9993-10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koito, A., G. Harrowe, J. A. Levy, and C. Cheng-Mayer. 1994. Functional role of the V1/V2 region of human immunodeficiency virus type 1 envelope glycoprotein gp120 in infection of primary macrophages and soluble CD4 neutralization. J. Virol. 68:2253-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolchinsky, P., E. Kiprilov, P. Bartley, R. Rubinstein, and J. Sodroski. 2001. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J. Virol. 75:3435-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ly, A., and L. Stamatatos. 2000. V2 loop glycosylation of the human immunodeficiency virus type1 SF162 envelope Facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J. Virol. 74:6769-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKeating, J. A., C. Shotton, J. Cordell, S. Graham, P. Balfe, N. Sullivan, M. Charles, M. Page, A. Blomstedt, S. Olofsson, S. C. Kayman, Z. Wu, A. Pinter, C. Dean, J. Sodroski, and R. A. Weiss. 1993. Characterization of neutralizing monoclonal antibodies to linear and conformation-dependent epitopes within the first and second variable domains of human immunodeficiency virus type 1 gp120. J. Virol. 67:4932-4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore, J. P., Q. J. Sattentau, H. Yoshiyama, M. Thali, M. Charles, N. Sullivan, S.-W. Poon, M. S. Fung, F. Traincard, M. Pincus, G. Robey, J. E. Robinson, D. D. Ho, and J. Sodroski. 1993. Probing the structure of the V2 domain of human immunodeficiency virus type 1 surface glycoprotein gp120 with a panel of eight monoclonal antibodies: human immune response to the V1 and V2 domains. J. Virol. 67:6136-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morikita, T., Y. Maeda, S.-I. Fijii, S. Matsushita, K. Obaru, and K. Takatsuki. 1997. The V1/V2 region of human immunodeficiency virus type 1 modulates the sensitivity to neutralization by soluble CD4 and cellular tropism. AIDS Res. Hum. Retrovir. 13:1291-1299. [DOI] [PubMed] [Google Scholar]
- 26.Ohshiro, Y., T. Murakami, K. Matsuda, K. Nishioka, K. Yoshida, and N. Yamamoto. 1996. Role of cell surface glycosaminoglycans of human T cells in human immunodeficiency virus type-1 (HIV-1) infection. Microbiol. Immunol. 40:827-835. [DOI] [PubMed] [Google Scholar]
- 27.Pinter, A., W. J. Honnen, Y. He, M. K. Gorny, S. Zolla-Pazner, and S. C. Kayman. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 78:5205-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinter, A., W. J. Honnen, S. C. Kayman, O. Troshev, and Z. Wu. 1998. Potent neutralization of primary HIV-1 isolates by antibodies directed against epitopes present in the V1/V2 domain of HIV-1 gp120. Vaccine 16:1803-1808. [DOI] [PubMed] [Google Scholar]
- 29.Posner, M. R., T. Hideshima, T. Cannon, M. Mukherjee, K. H. Mayer, and R. A. Byrn. 1991. An IgG human monoclonal antibody that reacts with HIV-1/GP120, inhibits virus binding to cells, and neutralizes infection. J. Immunol. 146:4325-4332. [PubMed] [Google Scholar]
- 30.Ross, T. M., and B. R. Cullen. 1998. The ability of HIV type 1 to use CCR-3 as a coreceptor is controlled by envelope V1/V2 sequences acting in conjunction with a CCR-5 tropic V3 loop. Proc. Natl. Acad. Sci. USA 95:7682-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanders, R. W., L. Schiffner, A. Master, F. Kajumo, Y. Guo, T. Dragic, J. P. Moore, and J. M. Binley. 2000. Variable-loop-deleted variants of the human immunodeficiency virus type 1 envelope glycoprotein can be stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits. J. Virol. 74:5091-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanders, R. W., M. Vesanen, N. Schuelke, A. Master, L. Schiffner, R. Kalyanaraman, M. Paluch, B. Berkhout, P. J. Maddon, W. C. Olson, M. Lu, and J. P. Moore. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 76:8875-8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saphire, A. C., M. D. Bobardt, Z. Zhang, G. David, and P. A. Gallay. 2001. Syndecans serve as attachment receptors for human immunodeficiency virus type 1 on macrophages. J. Virol. 75:9187-9200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shieh, J. T., J. Martin, G. Baltuch, M. H. Malim, and F. Gonzalez-Scarano. 2000. Determinants of syncytium formation in microglia by human immunodeficiency virus type 1: role of the V1/V2 domains. J. Virol. 74:693-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shotton, C., C. Arnold, Q. Sattentau, J. Sodroski, and J. A. Mckeating. 1995. Identification and characterization of monoclonal antibodies specific for polymorphic antigenic determinants within the V2 region of the human immunodeficiency virus type 1 envelope glycoprotein. J. Virol. 69:222-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullivan, N., M. Thali, C. Furman, D. D. Ho, and J. Sodroski. 1993. Effect of amino acid changes in the V1/V2 region of the human immunodeficiency virus type 1 gp120 glycoprotein on subunit association, syncytium formation, and recognition by a neutralizing antibody. J. Virol. 67:3674-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 38.Ugolini, S., I. Mondor, P. W. Parren, D. R. Burton, S. A. Tilley, P. J. Klasse, and Q. J. Sattentau. 1997. Inhibition of virus attachment to CD4+ target cells is a major mechanism of T cell line-adapted HIV-1 neutralization. J. Exp. Med. 186:1287-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vella, C., D. King, N. N. Zheng, H. Fickenscher, J. Breuer, and R. S. Daniels. 1999. Alterations in the V1/V2 domain of HIV-2CBL24 glycoprotein 105 correlate with an extended cell tropism. AIDS Res. Hum. Retrovir. 15:1399-1402. [DOI] [PubMed] [Google Scholar]
- 40.Vijh-Warrier, S., E. Murphy, I. Yokoyama, and S. A. Tilley. 1995. Characterization of the variable regions of a chimpanzee monoclonal antibody with potent neutralizing activity against HIV-1. Mol. Immunol. 32:1081-1092. [DOI] [PubMed] [Google Scholar]
- 41.Vijh-Warrier, S., A. Pinter, W. J. Honnen, and S. A. Tilley. 1996. Synergistic neutralization of human immunodeficiency virus type 1 by a chimpanzee monoclonal antibody against the V2 domain of gp120 in combination with monoclonal antibodies against the V3 loop and the CD4-binding site. J. Virol. 70:4466-4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, N. 1997. The role of gp120 V1/V2 sequence diversity in HIV-1 phenotype and antibody neutralization. Ph.D. dissertation. Sackler Institute of Graduate Biomedical Sciences, New York University School of Medicine, New York, N.Y.
- 43.Warrier, S. V., A. Pinter, W. J. Honnen, M. Girard, E. Muchmore, and S. A. Tilley. 1994. A novel, glycan-dependent epitope in the V2 domain of human immunodeficiency virus type 1 gp120 is recognized by a highly potent, neutralizing chimpanzee monoclonal antibody. J. Virol. 68:4636-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 45.Wu, Z., S. C. Kayman, W. Honnen, K. Revesz, H. Chen, S. Vijh-Warrier, S. A. Tilley, J. McKeating, C. Shotton, and A. Pinter. 1995. Characterization of neutralization epitopes in the V2 region of human immunodeficiency virus type 1 gp120: role of glycosylation in the correct folding of the V1/V2 domain. J. Virol. 69:2271-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the structure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]