Abstract
Adult human enteroviral heart disease is often associated with the detection of enteroviral RNA in cardiac muscle tissue in the absence of infectious virus. Passage of coxsackievirus B3 (CVB3) in adult murine cardiomyocytes produced CVB3 that was noncytolytic in HeLa cells. Detectable but noncytopathic CVB3 was also isolated from hearts of mice inoculated with CVB3. Sequence analysis revealed five classes of CVB3 genomes with 5′ termini containing 7, 12, 17, 30, and 49 nucleotide deletions. Structural changes (assayed by chemical modification) in cloned, terminally deleted 5′-nontranslated regions were confined to the cloverleaf domain and localized within the region of the deletion, leaving key functional elements of the RNA intact. Transfection of CVB3 cDNA clones with the 5′-terminal deletions into HeLa cells generated noncytolytic virus (CVB3/TD) which was neutralized by anti-CVB3 serum. Encapsidated negative-strand viral RNA was detected using CsCl-purified CVB3/TD virions, although no negative-strand virion RNA was detected in similarly treated parental CVB3 virions. The viral protein VPg was detected on CVB3/TD virion RNA molecules which terminate in 5′ CG or 5′ AG. Detection of viral RNA in mouse hearts from 1 week to over 5 months postinoculation with CVB3/TD demonstrated that CVB3/TD virus strains replicate and persist in vivo. These studies describe a naturally occurring genomic alteration to an enteroviral genome associated with long-term viral persistence.
The six serotypes of the group B coxsackieviruses (CVB1-6) are enteroviruses (Picornaviridae, species HEV-B) (53). The CVB genome is a single-stranded RNA molecule, 7,400 nucleotides (nt) in length, that is encapsidated within an icosahedral shell (74). The 11 viral proteins (83) are encoded by a single open reading frame which is flanked on the 5′ and 3′ termini by nontranslated regions (NTRs) (20). The CVB induce numerous human illnesses, including inflammatory heart disease, pancreatitis, and aseptic meningitis and may also trigger the onset of type 1 diabetes (5, 31, 72, 81, 82). The CVB were recognized as causes of human heart disease shortly after their description early in the 1950s (26, 27) and remain the enteroviruses most commonly associated with human cardiomyopathies on the grounds of isolation and serology (5, 6, 34, 62, 88). Meta-analysis shows an association between enteroviruses and myocarditis in 23% of cases (7), although this association is more variable in cases of dilated cardiomyopathy, a serious disease that often leads to a failing heart (61). Mouse models of CVB-induced pancreatitis (94, 110), myocarditis (45, 52, 110), myositis (104, 105), and rapid-onset type 1 diabetes (31) which facilitate the study of these diseases have been developed.
Enterovirus infections are generally considered to be acute events, with symptoms and virus titers peaking within a few days postinoculation (p.i.) and with virus being cleared by the adaptive immune response (17). However, enterovirus infections can persist under conditions of immunodeficiency (48, 51, 65, 71). Infectious enteroviruses are frequently isolated from pediatric cases of human myocarditis, but isolation from adult cases of the disease has been exceedingly rare (34, 62, 88), perhaps because pediatric cases may be more likely to be diagnosed during the acute infection. Despite an apparent absence of infectious enterovirus that can be isolated from adult cardiomyopathies, the detection of positive- and negative-strand enteroviral RNA in approximately 20% of adults with myocarditis or dilated cardiomyopathy by reverse transcription-PCR (RT-PCR) (87, 116) as well as the detection of enteroviral protein in cardiomyopathic heart muscle (4, 60) provides evidence that enteroviruses can persist and replicate in the heart in the apparent absence of infectious virus. CVB can persist for long periods of time in different strains of mice (22, 55, 69, 95, 104). Interestingly, cardiovirulent CVB3 strains replicate for a longer period in mouse heart tissue than do less-virulent or avirulent strains (21, 109, 110), and cardiovirulent viral RNA can persist for weeks or months in murine heart tissue in the absence of isolable infectious virus (95, 104). Prolonged, low-level replication of CVB in cardiomyocytes has been proposed as a pathogenic mechanism by which host gene expression may be altered in the heart muscle, presumably by inducing changes in host cell RNA transcription and translation (114, 115).
The picornavirus 5′ NTR performs critical functions in both viral translation and RNA replication (reviewed in reference 13). Within a specific picornavirus group, the 5′ NTR is of a defined length: for example, the 5′ NTR of human enterovirus B varies little in length at approximately 740 nucleotides long (20). Naturally occurring deletions or insertions of more than one or two bases have not been observed in enteroviral 5′ NTRs. The cloning of an infectious CVB3 cDNA that lacked the initial two 5′ uridine residues was reported (56), but sequence analysis of the progeny viral RNA revealed that this terminal deletion (TD) was repaired during initial expression of the virus.
The 5′ NTR contains extensive secondary structures, consisting of six defined domains of stem-loop secondary structures (59, 101, 120). The first of these is domain I, which folds in two-dimensional depictions into a trifoliate or cloverleaf-type structure (see Fig. 5A) (3). In CVB3, domain I is defined by a stem formed when nt 2 to 9 pair with nucleotides 80 to 87 (stem a). Within the domain, three stem-loop structures (b to d) radiate from a single junction-loop (96). A wealth of information shows that the cloverleaf (domain I) is essential for enterovirus RNA replication (2, 3, 11, 84, 106, 111, 118). Many mutations engineered into domain I are considered lethal, as assessed by their failure to produce viral plaques or to cause a cytopathic effect (CPE) upon indicator monolayer cells in culture (3). Mutations induced in the cloverleaf, or deletions of the entire domain, result in ablation of negative-strand RNA replication in cell extracts (11) or Xenopus laevis oocytes (36). Mutational analysis of poliovirus (PV) domain I indicates that deletions are likely to reduce replication, perhaps reducing it to the level at which CPE is not observed (3). Mutations in stem-loop b of domain I can prevent cytopathic virus replication (2, 3) and RNA replication (3, 84), while mutations induced in stem-loop d have been shown to prevent binding of the PV 3CD protein and to decrease initiation of negative-strand RNA replication (11, 36). Stem-loop b of the PV1 cloverleaf binds poly(rC) binding protein (PCBP) (84) which completes a ternary complex including the cloverleaf and the viral 3CD protein (which binds stem-loop d) (35, 42). This ternary complex competes with a complex essential for initiation of viral protein synthesis that is formed between PCBP and domain IV in the internal ribosome entry site (IRES) (76, 89); this competition may reduce translational initiation of positive-strand RNA such that replication of negative-strand viral RNA is not impeded by the presence of translating ribosomal complexes (10, 36). An interaction of domain I with the 3′ NTR has also been shown to be necessary for negative-strand viral synthesis (11, 42), an interaction which may be mediated through an interaction of PCBP with poly(A) binding protein (42). Encapsidation of newly synthesized positive-strand viral RNA molecules has been suggested to be linked to viral RNA replication via interaction with components of the replication complex (79) which may involve domain I in this process.
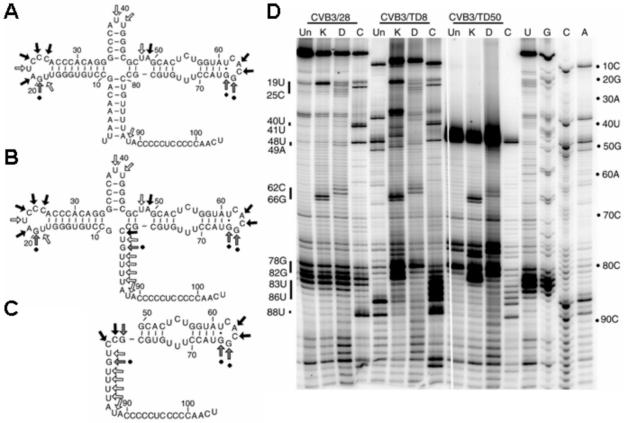
FIG. 5.
5′-terminal deletions of domain I maintain remaining cloverleaf structure as assessed by chemical modification. (A-C) Chemically modified sites for nt 1 to 104 of CVB3/28 (A), nt 1 to 93 of TD8 (B), and nt 1 to 51 of TD50 (C). Arrows indicate sites of kethoxal (gray arrows with dots at ends), DMS (black arrows), and CMCT (open arrows) modification. (D) Representative 12% sequencing gel of primer extension for domain 1 on RNA samples using primer PE108. RNA was prepared in vitro from subclones containing the 5′ NTRs of pRibozCVB3/28 (CVB3/28), pRibozCVB3/TD8 (CVB3/TD8), and pRibozCVB3/TD50 (CVB3/TD50). Un, unmodified RNA; K, kethoxal-modified RNA; D, DMS-modified RNA; C, CMCT-modified RNA. Representative modified positions are indicated. Primer extension with dideoxynucleotides was also performed to generate a sequence ladder (U, G, C, A).
The conundrum of enteroviral RNA persistence in heart tissue in the absence of detectable infectious virus remains without adequate explanation. Although never clearly demonstrable in animals or humans, numerous reports have documented that under certain circumstances, defective interfering (DI) picornavirus populations can be generated in cell culture (for examples, see references 23 and 24). The molecular lesion in the DI picornaviral quasispecies was demonstrated to be deletions of various sizes in the capsid protein (P1) coding region by electron microscopy (63) and, later, by use of infectious PV cDNAs which had been engineered to contain typical capsid protein coding region deletions (40, 49, 50). However, in this report, we describe a new class of naturally occurring, replication-competent enterovirus genomes with deletions of various lengths of the 5′ terminus that impinge upon the sequences comprising domain I. The presence of these TDs in CVB genomes correlates with efficient encapsidation of negative-strand viral RNA, a finding not previously reported with picornaviruses.
MATERIALS AND METHODS
Cells and virus stocks.
The cardiovirulent strain, CVB3/28 (109), was used throughout this work. CVB3 stocks were prepared in HeLa cell cultures from transfections with bacterial plasmids containing cDNA genomes as described previously (18). CVB3/28 was titer on HeLa cell monolayers by use of a 50% tissue culture infective dose (TCID50) as described previously (112). Cell debris was cleared from standard virus stocks by low-speed centrifugation. Concentration of CVB3/TD strains (described below) was required since transfections of cloned CVB3/TD genomes generated low viral titers. Therefore, 50 to 100 ml of cleared cell lysates of the CVB3/TD virus transfections (pass 1) were routinely concentrated by ultracentrifugation, resuspending the pelleted material and sedimenting virus again through a 30% sucrose column (made in 10 mM Tris-HCl [pH 7.5]-1 M NaCl) into a glycerol button at 120,000 × g at 8°C for 15 to 19 h. Titers of the CVB3/TD strains were established using enzymatic amplification as described below. Adult murine cardiomyocytes (ACM) were isolated from C3H/HeJ mice and placed in culture as described previously (112, 113). For passage of CVB3 in ACM cultures, 1 × 106 ACM were inoculated with 2 × 106 TCID50 units of CVB3, then cultured for 48 to 72 h (ACM1) (112). Sequential passages (ACM2-5) were prepared using 50% of centrifugally cleared supernatant (112) from the previous passage to inoculate fresh ACM cultures. For detection of cytopathic effect of ACM1-5, 1 × 105 HeLa cells were inoculated with 20% of the cleared supernatant for 30 min at 37°C, washed three times with 0.1 M NaCl, and refed with fresh medium. E. coli Stable2, DH5alpha (Invitrogen, Carlsbad, CA), or SURE2 (Stratagene, La Jolla, CA) was used for plasmid transformation and propagation as described previously (18).
Inoculation of mice.
A/J male mice (6 weeks old; Jackson Laboratories, Bar Harbor, ME) were inoculated intraperitoneally (i.p.) with 5 × 104 TCID50 units of CVB3 in 0.1 ml of unsupplemented media. C3H/HeJ male mice (4 weeks old; Jackson Laboratories) were inoculated i.p. with 1 × 105 TCID50 units in 0.1 ml of unsupplemented media. Infectious titers in heart tissues were determined as described previously (21, 108). B6.CB17-Prkdcscid/SzJ (4-month-old) and C3H/HeJ (4-week-old) male mice (Jackson Laboratories) were inoculated i.p. with 5 × 104 relative TCID50 (rTCID50) units (described below) of the specific CVB3/TD strain. One-quarter of the heart tissue was frozen for isolation of nucleic acids by homogenization in Trizol as described below (Invitrogen), and one-quarter was used for culture. Half of the heart tissue was fixed in formalin for paraffin embedding, sectioning, and staining with hematoxylin and eosin (112).
Cell culture of heart homogenates.
Twenty-five to fifty milligrams of heart tissue was homogenized in 0.4 ml cell culture medium, frozen and thawed, and cleared by centrifugation. HeLa cells (1 × 105) were inoculated with 50% of the heart homogenate for 30 min at 37°C, washed three times with 0.1 M NaCl, and refed with fresh medium. Cultures were harvested after 72 h by freeze-thaw lysis and cleared by centrifugation. Virus was passed by infection of 2 × 105 HeLa cells with 50% of the supernatant and harvested as described above.
RT-PCR and viral sequence analysis.
Viral RNA from cell culture stocks or homogenates of murine heart was isolated using Trizol LS reagent (Invitrogen) per the manufacturer's instructions. cDNA was transcribed with Superscript II (Invitrogen) in 20-μl reaction mixtures. Briefly, cDNA was synthesized in a 20-μl reaction volume containing RNA from 0.25 ml of virus stock or 5 × 104 HeLa cells inoculated 48 h previously with a CVB3/TD strain. Reaction mixtures contained RNA, an antisense primer (3END or E1; Table 1) at 0.125 optical density at 260 nm (OD260) units/ml, 0.5 mM deoxynucleoside triphosphates (dNTPs), and 100 U/μl Superscript II in a buffer containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, and 2 mM dithiothreitol (DTT). RT reaction mixtures were incubated at 42°C for 60 min; RT was then inactivated by incubation at 75°C for 10 min. After chilling on ice, RT reaction mixtures were diluted fivefold with nuclease-free water.
TABLE 1.
Names, annealing sites, and sequences of primers used in this study
| Primer type and namea | Genomic locationb strand | Nucleotide sequence (5′ to 3′)c |
|---|---|---|
| CVB3-specific primers | ||
| 3END | 7399-7377, − | CGCACCGAATGCGGAGAATTTAC |
| 5PUFF | 1235-1254, + | ATGCAGTACCACTACTTAGG |
| 998 | 1021-998, − | CGTTATGGTGGAGTTACCTAATGT |
| E1 | 644-627, − | CACCGGATGGCCAATCCA |
| E2 | 450-464, + | TCCGGCCCCTGAATG |
| E3 | 563-537, − | ACACGGACACCCAAAGTAGTCGGTTCC |
| E3REV | 537-562, + | GGAACCGACTACTTTGGGTGTCCGTG |
| E7 | 139-118, − | TGACTGTTGATCGGTGTGTGTT |
| E8 | 63-82, + | ACGGTACCTTTGTGCGCCTG |
| ID9 | 3201-3220, + | CTAGACTCTGCCAATACGAG |
| KS1 | 692-668, − | GCTAAGTGGTATAAACCCAACAAAG |
| KS2 | 303-325, + | CCCCAGTGTAGATCAGGTCGATG |
| PE62 | 82-63, − | CAGGCGCACAAAGTACCGT |
| PE108 | 128-109, − | CGGTGTGTGTTACTTCTAAG |
| PE127 | 403-383, − | GAGCGTCCCATGGGTTTCCCC |
| PE128 | 484-464, − | TCCGCAGTTAGGATTAGCCGC |
| PE129 | 261-241, − | AGGTTTTTCGAAGTAGTTGGC |
| PE130 | 578-558, − | TAGGAATAAAATGAAACACGG |
| PE162 | 182-163, − | GGTAACAGAAGTGCTTGATC |
| PE227 | 244-228, − | TGGGCCGGATAACGAACG |
| PE351 | 371-352, − | GCCAACGCAGCCACCGCCAC |
| PE627 | 647-628, − | AGTCACCGGATGGCCAATCC |
| RN | 5528-5509, − | CACCAACCTCTTGATCATTC |
| S | 1-20, + | TTAAAACAGCCTGTGGGTTG |
| S1 | 1-28, + | TTAAAACAGCCTGTGGGTTGATCCCACC |
| S2 | 21-49, + | ATCCCACCCACAGGGCCCATTGGGCGCTA |
| S3 | 33-61, + | GGGCCCATTGGGCGCTAGCACTCTGGTAT |
| S4 | 45-74, + | CGCTAGCACTCTGGTATCACGGTACCTTTG |
| Primers for cloning | ||
| DC-TAIL | AATCGATCTGTCGACCCCCCCCCC | |
| 5CMV | GGGAATATTCTGCTTCGCGATGTACGGGCCAGAT | |
| 3CMV | AAAGCGGCCGCCGCGGAATTTCGATAAGCCAGTAAGCAGTG | |
| 28RIBOZ1 | 1-20, + | ATGAGGCCGAAAGGCCGAAAACCCGGTATCCCGGGTTCTTAAAACAGCCTGTGGGTTG |
| 28RIBOZ2 | GACACTGATCCGCGGGTGTTTTAACTGATGAGGCCGAAAGGCCGAAAACCCGGTATC | |
| 28RIBOZPCR | CCACTAGTTCTAGACACTGATCCGCGGG | |
| S2TD8 | 8-36, + | ATAACCGCGGAGCCTGTGGGTTGATCCCACCCATAGGGC |
| S2TD13 | 13-41, + | ATAACCGCGGTGGGTTGATCCCACCCATAGGCGGCCATT |
| S2TD18 | 18-45, + | ATAACCGCGGTTGATCCCACCCATAGGGCCCATTGGGC |
| S2TD31 | 31-57, + | ATAACCGCGGTAGGGCCCATTGGGCGCTAGCACTCTG |
| S2TD50 | 50-76, + | ATAACCGCGGCACTCTGGTATCACGGTACCTTTGTG |
| TD8RIBOZ1 | 8-23, + | ATGAGGCCGAAAGGCCGAAAACCCGGTATCCCGGGTTCAGCCTGTGGGTTGATCCACCC |
| TD8RIBOZ2 | GACACTGATCCGCGGGCACAGGCTCTGATGAGGCCGAAAGGCCGAAAACCCGGTATC | |
| TD50RIBOZ1 | 50-69, + | ATGAGGCCGAAAGGCCGAAAACCCGGTATCCCGGGTTCGCACTCTGGTATCACGGTAC |
| TD50RIBOZ2 | GACACTGATCCGCGGGCAGAGTGCCTGATGAGGCCGAAAGGCCGAAAACCCGGTATC | |
| Primers for amplification of GADPH | ||
| 5-GAPDH | AGGTGAAGGTCGGAGTCAACG | |
| 3-GAPDH | GGTGAAGACGCCAGTGGACTC |
With the exception of DC-Tail, 5CMV, 3CMV, 28RIBOZPCR, and 5- and 3-GADPH, sequences of primers are based on the CVB3/28 genome (GenBank accession no. AY752944).
Nucleotide position numbering is as for CVB3/28 (Genbank accession no. AY752944). Positive is same sense as genomic RNA, negative is antisense to genomic RNA.
Underlined sequences are not genomic cDNA.
For shorter amplimers (<1 kb), cDNA was amplified in a PCR mixture containing 20% of the total cDNA, 0.2 U/μl Taq DNA polymerase (Promega, Madison, WI) (18), sense and antisense primers each at 0.125 OD260 units/ml, 0.16 mM dNTPs, 1 mM MgCl2, 50 mM KCl, 10 mM Tris (pH 9), and 0.1% Triton X-100. Reaction mixtures were subjected to one cycle in a Minicycler (MJ Research, Waltham, MA) for 1 min at 94°C, 1 min at 50 to 57°C, and 1 min at 72°C and then to 35 cycles of 30 s at 94°C, 30 s at 50 to 57°C, and 35 s at 72°C, finishing with 10 min at 72°C. Annealing temperatures varied with the primers employed. Amplimers were analyzed on 1.5% agarose gels in 40 mM Tris-acetate (pH 8)-1 mM EDTA; DNA was visualized using SYBR green (Amresco, Solon, OH) with UV illumination using the NucleoVision gel documentation system (Nucleo Tech, San Mateo, CA) and software (GelExpert version 3.5).
For long-distance (LD) PCR (to amplify amplimers 1 to 8 kb in length), Elongase enzyme mix (Invitrogen) was used in 50-μl reaction volumes, containing 10 μl of diluted cDNA, 1 μl of Elongase polymerase mix (Invitrogen), 0.4 mM dNTPs, sense and antisense primers at 0.125 OD260 units/ml, 60 mM Tris-SO4 (pH 9.1), 18 mM (NH4)2SO4, and 2 mM MgSO4. LD PCR was carried out using the following cycling conditions: 1 min at 94°C; 1 cycle of 30 s at 94°C, 30 s at 50 to 57°C, and 30 s at 68°C; 35 cycles of 30 s at 94°C, 30 s at 50 to 57°C, and 7 min and 30 s at 68°C; and a finish of 10 min at 68°C. Amplimers were analyzed on 1% agarose gels. Southern blots of gels were probed with oligomeric sequences located within the expected amplimer. Oligomeric probes were 5′ end labeled with [γ-32P]ATP (3,000 Ci/mmol; Amersham, Piscataway, NJ) as described previously (25). Using primer E1 for reverse transcription and E1 and E2 for amplification (Table 1), the sensitivity of detection of RNA from virus was 600 to 700 molecules of in vitro-transcribed CVB3 RNA (data not shown). DNA sequencing was carried out from cloned DNA or from amplified fragments at the core facility at the University of Nebraska at Lincoln or manually using the Thermonuclease Sequenase Radiolabeled Terminator cycle sequencing kit (USB Corp., Cleveland, OH) per instructions in the kit with analysis on denaturing gels as described previously (44).
Determination of CVB3/TD strain titers by real-time RT-qPCR.
CVB3/TD strains did not produce CPE; we therefore quantitated the amount of CVB3/TD in stocks using reverse transcriptase mediated real-time quantitative PCR (qPCR) (43). T7 RNA polymerase transcripts (positive-sense) transcripts were synthesized from a BamHI-digested plasmid containing nt 302 to 691 of the pCVB3/28 genome in pCR2.1-TOPO (Invitrogen). Transcripts were treated with RNase-free DNase I (Ambion, Austin, Tex.) and purified by ethanol precipitation, and concentration was determined spectrophotometrically. Complementary DNAs were synthesized from the dilutions of RNA transcripts or from CVB3/TD viral RNA (from 10 μl of virus stock) in reverse transcriptase reactions as described above containing the primer E1 (Table 1). cDNA reactions were diluted fivefold with water, and 10 percent of cDNA was used with E1 and E2 (Table 1) at 0.125 OD260 units/ml in DyNAmo SYBR green qPCR mix (Finnzyme, Finland) according to the manufacturer's instructions. Cycling times were as follows: 1 cycle at 95°C for 15 min; 45 cycles at 95°C for 20 s, 55°C for 20 s, and 72°C for 20 s; and a final extension at 72°C for 10 min. qPCRs to generate a standard curve were carried out using an Opticon 2 DNA engine (MJ Research). As the titer of the CVB3/TD stocks cannot be measured by cytopathic effect, the concentration of positive-strand viral RNA in each stock was determined and compared to the concentration in a CVB3/28 stock of known titer. Comparison of results from CVB3/TD RNAs with the parental CVB3/28 of known TCID50 titer determined the relative titer of the concentrated CVB3/TD stocks. Because qPCR measures genome copies, not infectious units, we devised the term rTCID50 to describe CVB3/TD titers. Real-time PCR quantitation of the parental CVB3/28 at concentrations of 2 × 104 to 2 × 107 generated numbers of molecules from 230 to 95 per TCID50, generating an average number of positive-strand viral genomes of 173 (standard deviation, 57.6), corresponding to the rTCID50 unit. This ratio of positive-strand genomes per infectious unit falls within the range reported for PVs (30 to 1,000 particles/PFU [93]), assuming that each viral particle contains a positive-strand genome.
Determination of 5′-terminal sequences of viral genomes.
The 5′-terminal sequences of genomic RNAs of a stock of CVB3/28 and a lysate of 1.5 × 105 cells of the fifth passage of heart homogenate of AJ mice at day 18 p.i. (AJ18 P5) were reverse transcribed using primer 998 (Table 1) with SuperScript II reverse transcriptase as described above. RNA was hydrolyzed using 0.2 N NaOH at 37°C for 20 min, then neutralized by addition of Tris-HCl (pH 7.5) and 0.2 N HCl. Following purification of single-strand cDNA using Geneclean (Qbiogene, Carlsbad, CA), G tailing of the cDNA was carried out using 1.5 U/μl terminal deoxynucleotide transferase (TdT; Promega) in cacodylate buffer (1 M potassium cacodylate, 0.25 M Tris-HCl (pH 7.6), 2 mM dithiothreitol, 1 mM dGTP, 2 mM MnCl2) at 30°C for 1.5 h (107). Tailed cDNA was diluted 100-fold with sterile deionized H2O and purified using Geneclean (Qbiogene). PCRs were carried out as described above for shorter fragments using Taq DNA polymerase and the primers DC-Tail and E1 (Table 1). PCR-amplified fragments were purified (StrataPrep PCR purification kit; Stratagene) and ligated into pPCR-Script-Amp using the pPCR-Script-Amp kit (Stratagene) according to the manufacturer's instructions. Plasmids containing the 5′-NTR sequences were then sequenced using the internal primer E7 (Table 1).
Cloning and transfection of 5′-TD CVB3 genomes.
An infectious clone of CVB3/28 (109) with an upstream cytomegalovirus (CMV) promoter in a plasmid vector was generated by amplifying the CMV promoter from pcDNA3.1 (Invitrogen) with primers 5CMV and 3CMV (Table 1) to add a NotI site downstream. This DNA was ligated into the 1.9-kb PvuII/SspI restriction fragment of pCR-Script-Amp (Stratagene) to generate pCRCMV. The 8.5-kb PvuI-StuI restriction fragment of pCVB3-28 (109) containing the full-length infectious cDNA copy of the CVB3 genome with an upstream T7 RNA polymerase promoter was cloned into the 1.7-kb PvuII-PvuI fragment of pCR-Script-Amp (Stratagene) to generate pCRCVB3-28. The 1.1-kb NotI-PvuI fragment of pCRCMV (containing the CMV promoter) was then ligated into the 9.3-kb NotI-PvuI fragment of pCRCVB3/28 to generate an infectious CVB3 genome with an upstream CMV promoter and T7 RNA polymerase promoter (pCMVCVB3-28). To clone the 5′-terminally deleted 5′ NTRs into the pCMVCVB3-28 cDNA genome, cDNAs from pass 5 HeLa cultures of AJ day 18 heart homogenate (as described above) were amplified with primer 998 and with primer S2TD8, S2TD13, S2TD18, S2TD31, or S2TD50 (Table 1). Amplimers were ligated into pCMVCVB3-28 using a SacII site inserted into each of the S2 primers and the SmaI site at nucleotide 891 of the CVB3/28 sequence (GenBank accession no. AY752944) to generate the recombinant deleted genomes (CVB3/TD). The CVB3/TD8 (7 nt deleted), CVB3/TD13 (12 nt deleted), CVB3/TD18 (17 nt deleted), CVB3/TD31 (30 nt deleted), and CVB3/TD50 (49 nt deleted) genomes begin at CVB3 nt 8, 13, 18, 31, and 50 respective to the parental CVB3 sequence (see Fig. 4B).
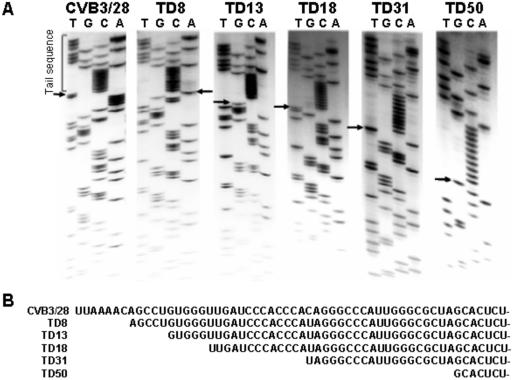
FIG. 4.
Multiple deletions from the 5′ end exist in cardiac viral populations late in infection. (A) cDNA was dG tailed with TdT, then enzymatically amplified with primers DC-TAIL and E1 (Table 1). Amplimers were cloned and then sequenced using primer E7. Parental CVB3/28 RNA was assayed similarly for comparison. Arrows indicate the 5′ termini of the CVB3/TD genomes. (B) Sequences of 5′ ends of CVB3/28 and CVB3/TD8, -13, -18, -31, and -50.
To generate parental CVB3/28, and CVB3/TD8 and TD50 genomes without extraneous nucleotides at the 5′ end from T7 or CMV promoter-driven transcripts, a ribozyme (41) that cleaves at the correct base was designed for each 5′ end. Three successive PCRs were used to add 77 nt containing a ribozyme sequence to the 5′ end of the genome. Specifically, nt 1 to 644 of the infectious CVB3/28 (109) cDNA genome were amplified with 28RIBOZ1 and E1 (Table 1). This PCR product was then amplified with 28RIBOZ2 and E3 (Table 1). This was subsequently amplified with 28RIBOZPCR and E3 (Table 1). A SacII-BstBI digest of the final amplified cDNA was ligated into the SacII-BstBI fragment of pCMVCVB3-28 to generate pRibozCVB3/28. In a similar fashion, a ribozyme was added upstream of the CVB3/TD8 cDNA genome by amplifying nt 1 to 637 of the CVB3/TD8 cDNA genome with TD8RIBOZ1 and E1 (Table 1), then amplifying the product with TD8RIBOZ2 and E3 (Table 1), and finally amplifying the last product with 28RIBOZPCR and E3 (Table 1). The SacII-BstBI digested product was ligated into the pCMVCVB3/TD8 plasmid as described directly above to generate pRibozCVB3/TD8. For the TD50 ribozyme, the first two amplifications were performed using TD50RIBOZ1 and TD50RIBOZ2 followed by the final PCR and cloning as described to generate pRibozCVB3/TD50. The 5′ NTR and upstream 100 bp of the full-length genome in each final clone were sequenced to verify the ribozyme and 5′-end sequences. Virus stocks were generated in HeLa cell culture by DNA transfection as described above.
Structure determination of RNA domain I in the CVB3 5′ NTR.
Experimental determination of RNA structure was performed using chemical modification analysis (33a, 72a). T7 RNA transcripts of ribozyme pCVB3-28, TD8, and TD50 were generated from subclones of the genome linearized by digestion with Ecl136II (cleaves at nt 749 in the CVB3/28 genome). For chemical modification reactions, 15 μg RNA was denatured at 80°C in 100 μl of dimethylsulfate (DMS)/kethoxal buffer (40 mM potassium-cacodylate [pH 7.2], 10 mM MgCl2, 50 mM NH4Cl, 0.75 mM DTT) for β-ethoxy-α-ketobutyraldehyde (kethoxal) or DMS modifications or in 50 μl of carboiimide metho-p-toluene sulfonate (CMCT) buffer (40 mM potassium borate [pH 8.0], 10 mM MgCl2, 50 mM NH4Cl, 0.75 mM DTT) for 1-cyclohexyl-3-(morpholinoethyl)CMCT modification. Reaction mixtures were slowly cooled to 42°C and then chilled on ice. For the kethoxal modification, 5 μl of a 1.5 M solution of kethoxal (USB Corp.) was added to the RNA and incubated at 37°C for 30 min. For the DMS modification, 2 μl of a 20% DMS solution in 95% ethanol was added to the RNA and incubated at 37°C for 10 min. For the CMCT modification, 50 μl of a solution containing 42 mg/ml of CMCT dissolved in CMCT buffer was added to the RNA and incubated for 10 min at 37°C. Kethoxal reactions were stopped by adding 50 μl of 150 mM sodium acetate-250 mM potassium borate, pH 7.0. DMS reactions were stopped by adding 25 μl of 1 M Tris-HCl (pH 7.5)-1 M β-mercaptoethanol-0.1 M EDTA, and CMCT reactions were stopped by adding 300 μl of 95% ethanol. Modified RNA was recovered by ethanol precipitation. Sites of modification were identified by primer extension with reverse transcriptase (120). Primers were 5′ labeled using [γ-32P]ATP and T4 polynucleotide kinase (Promega) (25) with 100 pmol of oligonucleotide, 100 μCi of [γ-32P]ATP (4,000 Ci/mmol), and 10 U of T4 polynucleotide kinase for 40 min at 37°C and inactivated for 20 min at 60°C. Oligonucleotides used for the primer extension reactions were PE62, PE108, PE127, PE128, PE129, PE130, PE162, PE227, PE351, PE627, and E1 (Table 1). For the primer extension reactions, a labeled oligonucleotide was annealed to 1 μg of modified or unmodified RNA in annealing buffer (50 mM Tris HCl [pH 8.3], 40 mM KCl) by heating at 80°C for 2 min followed by cooling to 42°C. Primer extension mixtures were composed of 2 μl of annealing mixture, 2 μl of extension mixture (100 mM Tris [pH 8.3], 80 mM KCl, 12 mM MgCl2, 4 mM of each dNTP) and 1 U of avian myeloblastosis virus (AMV) reverse transcriptase (Life Sciences, St. Petersburg, FL). To generate a sequence ladder, 2 μl of an annealing mixture containing unmodified RNA was added to each of four reaction volumes containing one of the four different termination mixtures (50 mM Tris HCl [pH 8.3], 40 mM KCl, 6 mM MgCl2, 1 mM of each dNTP, 0.1 mM of one dideoxynucleotide triphosphate) and 1 U of AMV reverse transcriptase. All reaction mixtures were incubated at 42°C for 20 min and then stopped with 2 μl 95% formamide containing 20 mM EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol. Reaction mixtures were frozen at −70°C for no longer than 1 week prior to electrophoresis. For each primer extension, samples were electrophoresed on 12% polyacrylamide/urea sequencing gel in Tris-borate EDTA buffer at 60 W for both 4 and 6 h. Modified bases were located by comparison to the sequencing tracts, and then the intensities of bands of unmodified and modified RNAs were compared to locate positions in which chemical modification resulted in termination of the elongating cDNA.
Strand-specific hybridization to virus RNA.
CVB3 strains collected by ultracentrifugation were resuspended in 2% of the original volume of 100 mM NaCl-10 mM Tris (pH 7.5)-1 mM EDTA. Virus was treated with 0.5 U/μl RNase A and 200 U/μl RNase T1 (RNase Cocktail; Ambion) at 37°C for 30 min. Viral RNA was prepared with Trizol LS (Invitrogen). T7 RNA (positive-sense) and T3 RNA (negative-sense) polymerase runoff transcripts were synthesized from an XhoI (T7)- or NotI (T3)-digested plasmid containing nt 1 to 2011 of the pCVB3/28 genome cloned in the NotI and XhoI sites of pBlueScriptII SK+ (Stratagene) and treated with RNase-free DNase I (Ambion). Viral RNA and transcripts were blotted in duplicate on Nytran-N (Schleicher and Schuell Bioscience, Keene, NH) (117). Primer E1 was used for detection of positive-strand viral RNA; primer 5Puff was used to detect negative-strand RNA (Table 1). Primers were labeled with [γ-32P]ATP using T4 kinase as described previously (25), to obtain a specific radioactivity of 1 to 2 × 108 cpm/μg for each probe. Each blot was washed twice for 10 min in 3 M tetramethyl ammonium chloride at 60°C (29). Hybridization signals on autoradiograms were quantified using Kodak 1D image analysis software (Eastman Kodak, Rochester, NY).
To determine whether negative-strand RNA is encapsidated, virus stocks (CVB3/28, CVB3/TD8, and CVB3/TD50) were purified by CsCl density centrifugation. Virions in cell culture lysates were concentrated by ultracentrifugation through 30% sucrose as described, chloroform extracted, and then collected again through sucrose. Virus preparations were resuspended in 100 mM NaCl, brought to 1.34 g/ml in CsCl, and then centrifuged at 45,000 rpm in a Beckman SW55Ti rotor for 45 h at 25°C. Density of gradient fractions was determined by measuring refractive indices. Fractions were diluted fivefold in sterile deionized water. The titers of fractions of the CVB3/28 gradient were determined on HeLa cell monolayers by TCID50 analysis (112) to determine the peak of infectivity. Parental CVB3/28 infectious titer peaked at 1.348 g/ml, while the CVB3/TD8 titer peaked at 1.331 g/ml and the CVB3/TD50 titer peaked at 1.344 g/ml as determined by measuring the optical density at 260 nm (infectious titer could not be determined by CPE). Three micrograms of RNA prepared from peak fractions with Trizol (Invitrogen), T7 and T3 RNA polymerase transcripts, or mixtures of transcripts (99:1, 50:50, and 1:99, respectively) were annealed with biotinylated oligonucleotide KS1 (for positive-strand RNA isolation) or KS2 (for negative-strand RNA isolation) in 0.1 M NaCl at 75°C for 5 min and slow cooled to anneal. The RNA:biotinylated primer complex was isolated using 10 to 15 μg of Dynabeads M-280 streptavidin (Dynal Biotech, New York, NY) according to the manufacturer's protocol. After rinsing the beads with diethyl pyrocarbonate-treated water, complexes were disassociated at 95°C for 5 min, and beads were removed magnetically to free RNA of the biotinylated oligonucleotide. An RT-PCR assay of the Dynabead rinses demonstrated no detectable RNA. Purified RNA was used for the synthesis of cDNA with KS1 (for positive-strand RNA reverse transcription) or with KS2 (for negative-strand RNA reverse transcription) as described above. Ten percent of the purified cDNA was amplified as described with KS1 and KS2, electrophoresed on 1.5% agarose gels, and visualized with UV as described.
Western blot analysis of viral proteins in infected cells.
HeLa cells were inoculated with parental CVB3/28 at a multiplicity of infection of 50 or with equivalent rTCID50 of the CVB3/TD virus as quantified by real-time qPCR, washed three times with 0.1 M NaCl, and refed with fresh medium. Cell lysates were harvested at various times after inoculation in reducing Laemmli buffer (58) and then were electrophoresed in 14% polyacrylamide gels with sodium dodecyl sulfate (Novex, San Diego, CA) and blotted onto Immobilon-P membranes (Millipore, Bedford, MA) (19). Blots were probed with a 1:1,500 dilution of the primary polyclonal horse anti-CVB3 neutralizing antibody (American Type Culture Collection, Manassas, VA), which detects CVB3 capsid protein VP1 (18). The primary antibody was detected using peroxidase-conjugated rabbit anti-horse immunoglobulin G (Jackson ImmunoResearch Laboratories, West Grove, PA) at a dilution of 1:125,000. Results were visualized using ECL-Plus and Hyperfilm (Amersham, Arlington Heights, IL). NucleoVision Gel Documentation System and software (GelExpert version 3.5; Nucleo Tech) were used for analysis.
For detection of VPg, RNase-treated virus stocks were prepared by CsCl density centrifugation. Briefly, freeze-thaw lysates of HeLa cells infected with virus were treated with RNase A/T1 (as described above) to remove residual cellular RNA. Virions in lysates were concentrated by ultracentrifugation through a 30% sucrose cushion as described, chloroform extracted, and then collected again by centrifugation though sucrose. Virus was resuspended in 100 mM NaCl, brought to 1.34 g/ml in CsCl, and centrifuged to equilibrium at 30,000 rpm in a Beckman SW41 rotor for 72 h at 8°C. CsCl-purified virus was diluted fivefold in water, and RNA was extracted with Trizol (Invitrogen). A 1.4-μg portion of each RNA sample was treated with 0.5 U/μl RNase A and 200 U/μl RNase T1 (RNase Cocktail; Ambion) and/or proteinase K (5 μg/ml), or 1.4 μg was left untreated. Samples were diluted with an equal volume of 2× reducing Laemmli buffer (58), electrophoresed in 14% polyacrylamide gels with sodium dodecyl sulfate (Novex) and blotted onto Immobilon-P (Millipore) (19). A rabbit polyclonal antibody against PV1 VPg was the kind gift of Bert Semler (University of California—Irvine): antibody N10 was raised against the 10 amino terminal amino acids of VPg (73). Western blots were probed with a 1:500 dilution of either anti-VPg antibody, followed by peroxidase-conjugated goat anti-rabbit immunoglobulin G (Jackson ImmunoResearch Laboratories) at a dilution of 1:150,000. The secondary antibody was detected using ECL-Plus as described above.
Protein covalently linked to RNA from pRibozCVB3/TD8 and TD50 virions was prepared for amino-terminal sequence analysis by electrophoresis of viral RNA in sodium dodecyl sulfate-containing 14% polyacrylamide gels (Novex) at 125 V for 30 to 40 min followed by electrophoretic blotting to Immobilon-P membranes (Millipore) at 15 V for 30 min. Blots were washed three times with phosphate-buffered saline. Blots were stained in Coomassie blue solution until bands clearly appeared. The blots were washed three times with water for 5 min each and dried. Protein was sequenced by Edman degradation at the University of Nebraska Medical Center Protein Structure Core Facility (Omaha, NE).
RESULTS
Generation of noncytopathic virus following inoculation of primary ACM cultures.
Following inoculation of ACM cultures with CVB3, cultures were examined for evidence of CPE; none was observed (data not shown). Cardiomyocytes appeared as shown previously (112). Aliquots of frozen and thawed supernatants of ACM passages of CVB3 (ACM1-5) were inoculated onto HeLa cell monolayers to assay for the presence of infectious virus. Lysis of the HeLa cell monolayer inoculated with ACM1 and ACM2 was observed within 48 h of inoculation (Fig. 1A). However, no CPE was observed in HeLa cultures inoculated with ACM3-5; these cultures appeared by examination using light microscopy to be identical to uninfected, control HeLa cell monolayers (Fig. 1A). The infectious titer of CVB3 in ACM1, measured in HeLa cell monolayers, was low (1 × 103 to 3 × 103 TCID50/ml), while the titer in the ACM2 supernatant was not measurable (<74 TCID50/ml) (data not shown). This was consistent with a rapidly diminishing ability to replicate in these cells and/or with incomplete removal of residual inoculum virus in ACM1 by washing. To determine whether viral RNA was detectable in the HeLa cultures inoculated with lysates from ACM1-5, total nucleic acids were isolated from HeLa cell monolayers inoculated 24 h previously. Using primers within a conserved region of the 5′ NTR to amplify the viral RNA, RT-PCR analysis showed that CVB3 RNA was readily detectable in ACM1-5-infected HeLa cell cultures despite the lack of CPE in ACM3-5 (Fig. 1B). This was confirmed by probing a Southern blot of the amplified DNA using an internal oligomeric probe (Fig. 1C). These results showed that CVB3 RNA had been transferred from ACM to HeLa cells despite a lack of CPE in HeLa cells inoculated with ACM3-5.
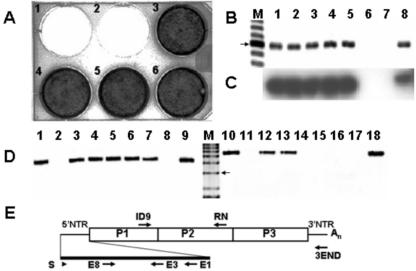
FIG. 1.
Passage of CVB3 in ACM on HeLa cells shows that viral RNA persists without apparent CPE. (A) Lanes 1 to 5, HeLa cells infected with supernatants of ACM passage 1 to 5; lane 6, uninfected HeLa cells. Cells were fixed and stained with crystal violet. (B) Agarose gel of amplimers generated by RT-PCR from HeLa cell cultures as shown in panel A. Total RNA was isolated from each ACM supernatant-infected cell culture for use as template in an RT-PCR with primers E1 and E8. Expected size for an E1/E8 amplimer is 582 bp. Lanes 1 to 5, HeLa cells infected with ACM passages 1 to 5; lane 6, uninfected HeLa cells; lane 7, HeLa cells without RNA; lane 8, HeLa cells with CVB3/28 RNA. The arrow indicates a 600-bp band. (C) Gel shown in panel B was analyzed by Southern blotting and probed with T4 kinase 32P-labeled primer E3. Film was exposed for 16 h at −75°C. (D) Agarose gel of amplimers generated by RT-PCR from HeLa cell cultures with primers ID9 and 3END (4.2 kb) (lanes 1 to 9) or with primers S and RN (5.5 kb) (lanes 10 to 18). The arrow indicates a 3-kb band. Lanes 1 and 10, HeLa cells infected with CVB3/28; lanes 2 and 11, HeLa cells only; lanes 3 to 7 and 12 to 16, ACM passages 1 to 5; lanes 8 and 17, HeLa cells without viral RNA; lanes 9 and 18, HeLa cells with CVB3/28 RNA. (E) Relative positions in the CVB3 genome of primers used.
Previous work had documented that deletions of the capsid protein coding region in different picornaviral genomes can give rise to DI virus strains in cell culture (23, 63); this could be recapitulated by genetic engineering of infectious cDNA clones of the PV genome (40, 50). We therefore screened CVB3 RNA isolated from ACM1-5-inoculated HeLa cells for deletions by nonquantitative LD RT-PCR analysis with primers annealed internally or terminally to both ends of the CVB3 RNA. Using a primer (ID9) located near the middle of the CVB3 genome at nt 3298 and the 3END primer, a single band with the expected size of 4.2 kb was repeatedly generated for ACM1-5-infected HeLa cells (Fig. 1D, lanes 3 to 7) as well as for the positive virus control (Fig. 1D, lane 1). These and other amplifications using different primers (such as E8 and 3END) located within the CVB3 genome (data not shown) failed to reveal evidence of deletions in these viral RNA populations. However, when the ACM1-5-infected HeLa cell RNAs were amplified using an internal primer (RN) and a 5′-terminal specific primer (S) (Table 1), the expected amplimer of 5.5 kb was generated only from ACM1- and ACM2-infected HeLa cells and the parental CVB3 control (Fig. 1D, lanes 12, 13, and 10, respectively); no product was obtained from ACM3-5-infected HeLa cell RNA despite numerous repeated attempts (Fig. 1D, lanes 14 to 16). We repeated the experiment using S4 and 3END with similar results (data not shown). The parental (control) CVB3 virus and ACM1- and ACM2-infected HeLa cell RNAs produced the expected size of amplimer, but no amplimer was detected using RNAs from ACM3-5-infected HeLa cultures. These data indicated that the failure to induce CPE by the ACM3-5 virus populations was linked to an apparent alteration of the 5′ termini of these genomes, which did not permit amplification using primers targeted to the 5′-terminal sequences.
Generation of noncytopathic CVB3 strains during replication in the mouse heart.
Having observed that a noncytolytic CVB3 quasispecies had evolved following infection of, and passage in, ACM, we tested whether this were reproducible in vivo. For this experiment, we used A/J mice, which are known to permit long-term viral RNA persistence (92), and C3H/HeJ mice, which supply a model of acute CVB-induced myocarditis (112). Mice were inoculated i.p. with CVB3/28 and then were killed at different times p.i. to ascertain how long virus persisted in the heart tissue of each mouse strain. Hearts were excised and homogenized in tissue culture medium, after which the presence of virus in the supernatants was assayed on HeLa cells or CVB3 RNA was detected by RT-PCR. Infectious virus was waning in hearts of A/J mice by 14 days p.i. (Table 2), and by 28 days p.i., cytopathic infectious CVB3 was no longer detectable in A/J mouse hearts. However, RT-PCR analysis of heart RNA in these same mice detected viral RNA in all mice through 28 days p.i. and in one mouse of a group of nine at 53 days p.i. (Table 2). Complete absence of cytopathic CVB3 in hearts of C3H/HeJ mice was observed at 21 days p.i., 1 week earlier than in A/J mice (Table 2), although CVB3 RNA was detected in four of four hearts at 21 days p.i. by RT-PCR. These results were consistent with, and confirmed past reports of, CVB RNA persisting in murine hearts after the time that infectious virus can no longer be detected (92, 95).
TABLE 2.
HeLa cell passage of CVB3-infected A/J mouse heart homogenates replicates CVB3 in the absence of cytopathic effecta
| Characteristic | No. of mice with indicated characteristic/total no. of mice in group on day p.i.:
|
|||||
|---|---|---|---|---|---|---|
| 4 | 8 | 14 | 21 | 28 | 53 | |
| A/J mice | ||||||
| CPE present | 3/3 | 2/2 | 2/5 | 1/3 | 0/5 | 0/9 |
| Viral RNA detected by RT-PCR | 3/3 | 2/2 | 5/5 | 3/3 | 5/5 | 1/9 |
| C3H/HeJ mice | ||||||
| CPE present | ND | ND | 4/4 | 0/4 | ND | ND |
| Viral RNA detected by RT-PCR | ND | ND | 4/4 | 4/4 | ND | ND |
Homogenated hearts taken from mice at day 4, day 8, day 14, day 21, day 28, and day 53 p.i. were assayed on HeLa cell monolayers and examined for CPE. Total RNA purified from HeLa cell monolayers was used for RT-PCR with primers E1 and E2, and amplimers were detected by agarose gel electrophoresis and staining as described in Materials and Methods. ND, not done.
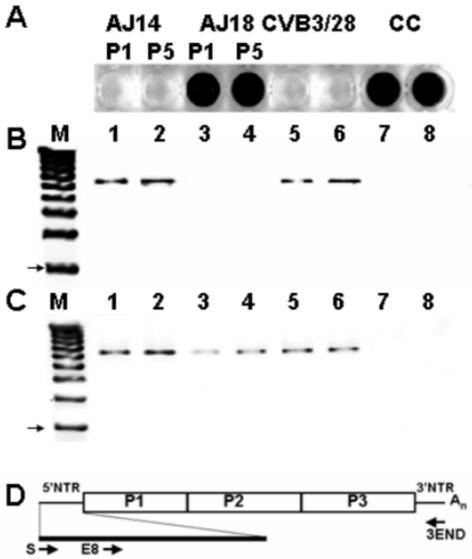
To test the hypothesis that 5′-terminal deletions could occur in CVB3 genomes when replicating in vivo, we inoculated A/J mice with CVB3, then assayed hearts at 14 and 18 days p.i. for the presence of virus by inoculating heart homogenate supernatants onto HeLa cell monolayers (AJ14 and AJ18). In this experiment, cytolytic CVB3 was present in hearts at day 14 p.i. (Fig. 2A, AJ14) but was inapparent on day 18 (Fig. 2A, AJ18). By comparison, infection of HeLa cell monolayers with parental CVB3 lysed all cells (Fig. 2A, CVB3/28). Uninfected HeLa cell cultures (Fig. 2A, CC) appeared by light microscopy to be identical to cells inoculated with heart homogenate from a day 18 mouse (AJ18) (data not shown). HeLa cells inoculated with AJ14 and AJ18 heart homogenates were frozen and thawed after 48 h in culture, and then 50% of the cleared supernatant was passed onto fresh cultures. After five such passages, the virus from AJ14 hearts retained its lytic phenotype, while virus from AJ18 remained noncytopathic on HeLa cells (Fig. 2A, compare AJ14 and AJ18).
FIG. 2.
Infection of HeLa cells with homogenate of CVB3-infected AJ heart at day 18 does not induce cytopathic effect but does replicate CVB3 RNA. (A) HeLa cultures inoculated with homogenates of hearts at days 14 p.i. (AJ14) and 18 p.i. (AJ18) passed once (P1) and five times (P5) in HeLa as described in Materials and Methods, inoculated with parental virus (CVB3/28) or uninfected cell culture (CC). Cells were stained with crystal violet. (B and C) Total RNA purified from HeLa-passaged AJ14, AJ18, CVB3/28-inoculated HeLa cells, and uninfected cells were used for RT-PCR with the primer 3END and with primers S (B) or E8 (C) (Table 1). The arrow indicates a 3-kb band. Lane M, 1-kb DNA ladder; lanes 1 and 2, AJ14 P1 and 5; lanes 3 and 4, AJ18 P1 and 5; lanes 5 and 6, CVB3/28; lanes 7 and 8, uninfected cell culture. (D) Relative positions in the CVB3 genome of primers used.
To determine whether CVB3 RNA in AJ14- and AJ18-infected HeLa cell cultures had altered 5′ genomic termini, RNA was isolated from these cultures, and cDNA was amplified using the 5′-terminal primer S and the 3′-terminal primer 3END (Fig. 2B). Genome-length amplimers of approximately 7.4 kb were observed in HeLa cells infected with AJ14 (Fig. 2B, lanes 1 and 2) or with parental CVB3 (Fig. 2B, lanes 5 and 6) for both passes 1 and 5. However, no amplification was obtained from RNA from cells inoculated with AJ18 (Fig. 2B, lanes 3 and 4), identical to the expected negative signal from uninfected control HeLa cells (Fig. 2B, lanes 7 and 8). To determine whether viral RNA was present in AJ18-infected HeLa cells, we performed amplifications using a primer specific for nt 63 to 82 (E8 [Table 1]) and the 3END primer (Fig. 2D). We detected a band of between 7 and 8 kb in both AJ14- and AJ18-infected HeLa cell cultures for passages 1 and 5 as well as for the positive control, CVB3-infected HeLa cells. Similar results were obtained when heart homogenates of C3H/HeJ mice (Table 2) were directly assayed by RT-PCR or were first passed onto HeLa cell cultures: in the absence of CPE (day 21), the 5′-terminal primer S failed to prime, although primers located at nt 64 or further within the 5′ NTR generated amplimers (data not shown). These results indicated that CVB3 can generate a population of virus with significantly altered 5′ genomic termini during replication in the murine heart. Further, the results indicated that the altered 5′ termini appeared as an apparent function of the viral quasispecies becoming noncytopathic when assayed in a virus-sensitive cell culture system.
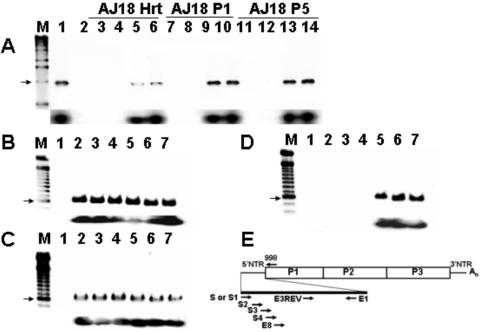
To map the 5′-sequence alteration or deletion, five overlapping 5′-end primers and a constant internal antisense primer (998 [Table 1]) were used to amplify viral RNA from AJ day 18 heart homogenates and AJ18-infected HeLa cell cultures (Fig. 3A). The lack of amplification with primers S (Fig. 3A, lanes 3, 7, and 11) and S2 (Fig. 3A, lanes 4, 8, and 12) and the presence of amplification with primers S3 (Fig. 3A, lanes 5, 9, and 13) and S4 (Fig. 3A, lanes 6, 10, and 14) demonstrated that the defect was localized to approximately the first 50 nucleotides of the genome. That the amplified cDNA was from CVB3 was confirmed by detection of amplimers generated by each primer set on a Southern blot with an internal oligomeric probe (Fig. 3A); no amplification in AJ18 heart or passages with the S and S2 primers was detected by Southern blotting (Fig. 3A, lanes 3, 4, 7, 8, 11, and 12). For a control of amplification, RNA from HeLa cells inoculated with CVB3/28 were used as templates for RT-PCR with each primer set and analyzed by gel electrophoresis and Southern blotting (Fig. 3B). RNA from the first passage of AJ14-infected HeLa cells (Fig. 2) was similarly amplified and analyzed (Fig. 3C). As expected from earlier results which showed the CVB3 RNA in these cells maintained an intact 5′ terminus, all primer pairs amplified cDNA and were indistinguishable from amplifications of RNA from CVB3/28-infected cultures. Amplification of cDNA from RNA of the fifth passage of AJ18 virus in HeLa cells was successful only with the 5′ primer S3, located at nt 33 to 61, and S4, located at nt 45 to 74 (Fig. 3D); primers located between nt 1 and 49 (S, S1, or S2 [Table 1]) failed to amplify. This failure was confirmed by Southern blotting (Fig. 3D). These results suggested that the extent of the alteration to the 5′ terminus might range from approximately 30 to 50 bases inward from the 5′ end.
FIG. 3.
Primers at the 5′ end of the viral RNA sequence fail to amplify cDNA from day 18 heart homogenates or heart homogenate passages. (A) RT-PCR of RNA from AJ d18 p.i. heart homogenate (AJ18 Hrt) or from cultures of homogenate passed once (AJ18 P1) and five times (AJ18 P5)t in HeLa cells. cDNA was amplified using different 5′ primers and a constant downstream primer, 998. Lane M, 1-kb marker (the arrow shows a 1-kb band); lane 1, RT-PCR of CVB3/28 RNA with S-998 amplimer (1,021 bp); lane 3, negative control RT-PCR; lanes 3, 7, and 11, primer S; lanes 4, 8, and 12, primer S2; lanes 5, 9, and 13, primer S3; lanes 6, 10, and 14, primer S4. Southern blot analysis (lower panel) with primer E3REV confirms that amplified cDNA is CVB3. (B-D) RT-PCR analysis shows that 5′ termini can be amplified from 105 HeLa cells infected with parental CVB3/28 (B) and AJ14 P5 (RNA heart homogenate from a mouse killed 14 days p.i. and passed five times in HeLa cells) (C) but not from AJ18 P5 (D). Bottom portions of panels B to D show Southern blots of gel probed with E3REV. Lanes M, 100-bp markers (each arrow shows a 600-bp band). Lane 1, negative control; lanes 2 to 7, infected cell RNA. Different 5′ end primers were assayed with the downstream primer E1. Lanes 1 and 2, 5′ primer is S; lane 3, 5′ primer is S1; lane 4, 5′ primer is S2; lane 5, 5′ primer is S3; lane 6, 5′ primer is S4; lane 7, 5′ primer is E8 (Table 1). (E) Relative positions in the CVB3 genome of primers used.
RNA from noncytopathic CVB3 derived following isolation from infected mouse heart tissue has 5′-terminal deletions.
Deletion and/or significant changes in the 5′-terminal sequence could explain the preceding observations for AJ18-infected HeLa cell cultures. To determine the nature of the 5′ ends, we cloned and sequenced 5′-terminal sequences of the noncytopathic viruses of the AJ18-infected HeLa cultures. Using RNA obtained from an AJ18 heart homogenate which had been passaged five times in HeLa cells (Fig. 2 and 3, AJ18 P5), cDNA was transcribed using the primer E1 (Table 1), then tailed with G residues using TdT. This cDNA population was enzymatically amplified using the primers DC-Tail (Table 1) and E1. The resultant double-strand DNA was cloned and sequenced. A total of 23 clones containing 5′-NTR sequences were randomly selected for sequencing, revealing five different-sized deletions from the 5′ terminus beginning at nt 8 (three clones), 13 and 18 (four clones each), and 31 and 50 (six clones each) (Fig. 4). Deletions of other sizes were not isolated. Sequencing gel data in Fig. 4 show typical results. The C tail is clearly evident in each sequence as a C ladder, followed by the first 5′ nucleotide. The cumulative results are presented in Fig. 4B in which the specific derived 5′-terminal sequences are shown; the CVB3 strains with terminally deleted 5′ ends are denoted as CVB3/TD, followed by the nucleotide at which the genome begins (thus, a 7-nt deletion is termed CVB3/TD8). Of five different deletion classes, just two (CVB3/TD18 and -31) terminated with a uridine residue (Fig. 4B); the other genomes terminated in A or G. Due to the tailing, genomes beginning with C would appear to begin with the second nucleotide of the genome. The only deletion cloned which might have had a C as the first residue is TD8.
Complete sequence analysis of the cloned CVB3/TD 5′-NTR sequences revealed two nucleotide substitutions by comparison to the parental CVB3/28: a C→U transition at nt 31 in the CVB3/TD8, CVB3/TD13, and CVB3/18 clones and another at nt 124 in all CVB3/TD clones. These changes were found in each of the clones sequenced for each NTR. Computer-aided analysis (MFOLD 3.1) (67, 121) predicted that the mutation at nt 31 would not disrupt secondary structure, as this change in stem-loop b of the cloverleaf is base paired to a G at nt 13. This change also occurs in other CVB3 genomes (56, 108), consistent with a commonly occurring sequence variation. The change at nt 124 was not predicted to be in a base-paired region of domain II (33).
Defining the RNA secondary structure of the altered CVB3/TD genomes.
Using a structure predicted for the parental CVB3 domain I (Fig. 5A, nt 1 to 100) (120), we modeled the structure in the shortest (CVB3/TD8, nt 1 to 93) (Fig. 5B) and longest (CVB3/TD50, nt 1 to 50) (Fig. 5C) deletions. Chemical modification of CVB3 RNA of the 5′ NTR was utilized to test the validity of these structures (typical data are shown in Fig. 5D). Under the conditions used for analysis, the chemicals modify single-stranded bases that are accessible to solvent. Kethoxal modifies G, DMS modifies C and A, and CMCT modifies U and, to some extent, G (102). Thus, modified positions are shown not to be base paired. The modification data (Fig. 5A to C) are compatible with the predicted structures in all but the most 3′ 12 nucleotides which may be involved in a tertiary interaction (W. Tapprich and J. Bailey, personal communication). For both parental CVB3/28 (Fig. 5A) and CVB3/TD8 (Fig. 5B), the loop region of stem-loop b was chemically accessible, beginning with a CMCT modification at position 19U. Nucleotides 19U, 20G, 21A, 22U, 23C, 24C, and 25C form a hairpin loop in the cloverleaf, and all are exposed to solvent. The stem region of stem-loop c was protected, as is position 39A in the loop region. A very light modification at 40U was followed by a strong hit at base 41U. Modifications at nt 48U and 49A confirm the bulge-loop in stem d. This entire sequence containing stem a, stem-loop b, and stem c is missing in TD50 (Fig. 5C), but from here to the end of stem-loop d, all three molecules display the same structure. After the 48U-49A bulge-loop, the RNA then closes for the remainder of the stem region in stem-loop d. Nucleotides 54 to 56 in this stem are predicted to be available for modification as part of an internal loop opposite nt 71 to 73. However, nuclear magnetic resonance analysis indicates that these bases participate in noncanonical pyrimidine-pyrimidine pairs (32, 80). Our probing results are consistent with this arrangement for all three molecules. The proposed tetraloop at the end of stem-loop d was exposed, with modifications between 63A and 65G. In TD8 (Fig. 5B), nt 80 to 86, which cannot be base-paired to nt 2 to 7 to create stem a, were accessible as might be expected. Similarly, in TD50 (Fig. 5C), nt 78 to 86 cannot be base paired to nt 2 to 7 and 46 to 47, and again these positions showed modification. Interestingly, in both TD8 and TD50, as in CVB3/28, the long stretch of presumably single-stranded bases between nt 89 and 104 was protected from modification. Thus, despite the dramatic deletions, the higher-order folding interactions in this region were maintained. We also probed the structure of the rest of the 5′ NTR from CVB3/28, TD8, and TD50. Outside of domain I, we were unable to detect any structural differences (data not shown).
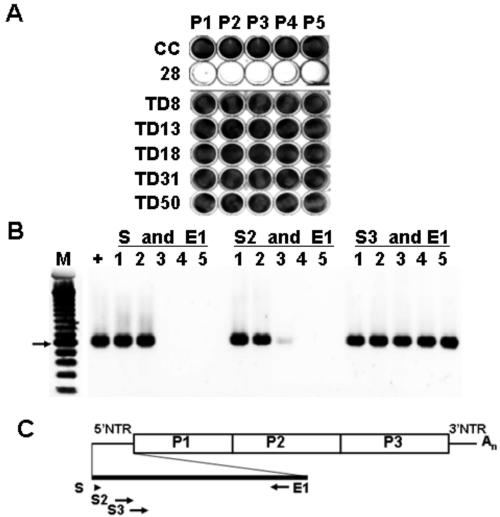
Characterization of CVB3/TD strains in cell culture.
To determine the role of the 5′ deletions in the noncytopathic phenotype, recombinant infectious CVB3/TD cDNA clones containing the 5′-end deletions were constructed using the infectious cDNA copy of the CVB3/28 genome (109). Each CVB3/TD virus strain produced viral RNA in HeLa cells transfected with the CVB3/TD virus cDNA clones as measured by RT-PCR assay (using primers S3 and E1 [Table 1]) (Fig. 6B) but did not produce CPE in HeLa cells (Fig. 6A) even after five passages in HeLa cells. The RT-PCR results displayed in Fig. 6B used RNA from the fifth passage in HeLa cells as the template for RT and demonstrate that the deletions are not repaired by passage in successive replications in cell culture (compare S3 and E1 with S and E1 or S2 and E1). RT-PCR amplification of cDNA with a range of 5′-terminal primers indicates that stocks of progeny CVB3/TD8 and TD13 from the infectious cDNA clones could be amplified with the primers S and S2 (Table 1) despite the fact that these primers did not amplify cDNA from the mixed population of genomes present in the HeLa cell culture passage of the AJ18 heart homogenates (compare results shown in Fig. 6B, lanes 1 and 2, and Fig. 3D, lanes 2 and 4). It is not surprising that these primers can amplify cDNA from the clonal, concentrated populations of CVB3/TD8 and TD13; primer S (nt 1 to 20) and primer S2 (nt 21 to 49) substantially overlap the remaining sequences of the 5′ ends of these TD genomes. Clearly the 7- and 13-nucleotide deletions were present in the AJ18 population, as these deletion mutants were originally cloned from amplified, tailed cDNA generated from an AJ18 HeLa culture (Fig. 4). The lack of amplification with the S and S2 primers (Fig. 3D) seen in pass 5 cultures of AJ18 is likely to be due to a smaller template population. In order to obtain sufficient cDNA for tailing and cloning, a threefold-greater amount of AJ18P5 culture was used for reverse transcription (see Materials and Methods). Although the RT-PCR amplifications of AJ18P5 (Fig. 3D) and of the CVB3/TDs at pass 5 were not matched for template concentration, a strong amplification with S3 was detected for the AJ18P5 culture and for all the CVB3/TD cultures. The sensitivity of RT-PCR using E1 and S, S2, or S3 is similar: approximately 400 molecules of CVB3 RNA can be detected. The fact that no increase in concentration was needed to detect viral RNA with S3 (nt 33 to 61) suggests that genomes with deletions of 30 and 49 nucleotides are well represented in the AJ18P5 viral RNA population.
FIG. 6.
Replication of recombinant CVB3/TDs in HeLa cell culture and RT-PCR. (A) Each deleted 5′-NTR sequence was cloned into pCMVCVB3/28 (CVB3/TD8, CVB3/TD13, CVB3/TD18, CVB3/TD31, and CVB3/TD50). Transfection of cDNA in HeLa cell cultures maintained a noncytopathic phenotype and the respective deletions through five passages of HeLa cells by supernatants. Monolayers were fixed and stained with crystal violet. (B) Viral RNA from each CVB3/TD (pass 5) was analyzed by RT-PCR with primers specific for sequences at or near the 5′ end. Primers used are indicated. The arrow indicates a 600-bp marker. Lane M, 100-bp DNA ladder; lane +, CVB3/28; lane 1, CVB3/TD8; lane 2, CVB3/TD13; lane 3, CVB3/TD18; lane 4, CVB3/TD31; lane 5, CVB3/TD50. (C) Map of primer positions on CVB3 genome.
CVB3/TD strains are encapsidated.
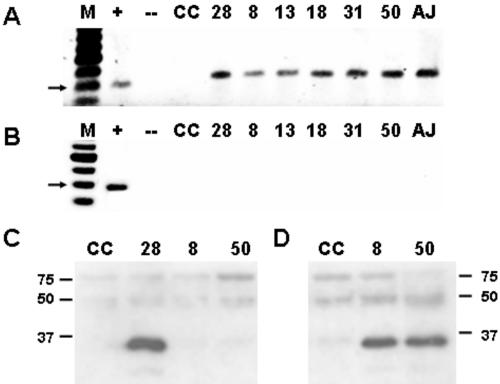
Our preliminary results suggested that CVB3/TD infection of HeLa cells in culture was due either to transmission of residual cDNA genomes from the transfected cells to fresh cells nor to free viral RNA, as CVB3/TD strains were transmitted to fresh HeLa cultures following DNase I and RNase A/T1 treatment. We repeated this work using progeny CVB3/TD strains from each of the cloned CVB3/TD genomes. Following incubation of CVB3/TD8-50 in the presence of RNase A/T1 and DNase 1, virus preparations were inoculated onto HeLa cell cultures. After 72 h, total RNA was prepared from cultures for RT-PCR assay of viral genomes. When all TDs were passaged in HeLa culture, CVB3 RNA was detected in each of the cultures. However, if the viral inocula were first preincubated in the presence of anti-CVB3 neutralizing serum prior to inoculation onto HeLa cells, infection and replication were completely inhibited, and no viral RNA was detectable after 72 h (Fig. 7, compare panels B and A). The detection of the largest capsid protein, VP1, by Western blot analysis in cells inoculated with CVB3/TD8 and TD50 (Fig. 7C), together with the ablation of detectable CVB3/TD RNA in cells infected with virus in the presence of anti-CVB3 neutralizing antibody, strongly suggests that CVB3/TD strains are normally encapsidated and transmitted.
FIG. 7.
CVB3/TD strains are neutralized by anti-CVB3 polyclonal serum and infect cells to produce viral proteins. (A) Cultures of 105 HeLa cells were inoculated with 103 TCID50 units of CVB3/28 (lane 28), 107 rTCID50 TD8 (lane 8), 104 rTCID50 TD13 (lane 13), 105 rTCID50 TD18 (lane 18), 105 rTCID50 TD31 (lane 31), 106 rTCID50 TD50 (lane 50), or 0.1 ml of heart homogenate taken 18 days p.i. (lane AJ). Total RNA was prepared after 72 h for RT-PCRs with primers KS1 and KS2 (amplimer size, 390 bp). Lane M, 100-bp marker; lane +, CVB3/28 viral RNA RT-PCR; lane -, negative control RT-PCR; lane CC, uninfected cell control. The arrow indicates a 400-bp marker. (B) Same as in panel A, except CVB3 antiserum (1:500) was added to virus stocks in infections, washes, and medium. (C) Cultures of 105 HeLa cells were inoculated with 106 TCID50 units of CVB3/28 (lane 28), 107 rTCID50 TD8 (lane 8), 107 rTCID50 TD50 (lane 50), or uninfected cell control (lane CC). Cells were harvested at 7 h, resuspended in Laemmli buffer, electrophoresed, and analyzed by Western blotting, and viral proteins were detected as described in Materials and Methods. Mobility of molecular weight markers of 150, 75, 50, and 37 is noted. (D) Same as in panel C, but cells were harvested at 26 h p.i.
Negative-strand viral RNA is encapsidated in CVB3/TD virions.
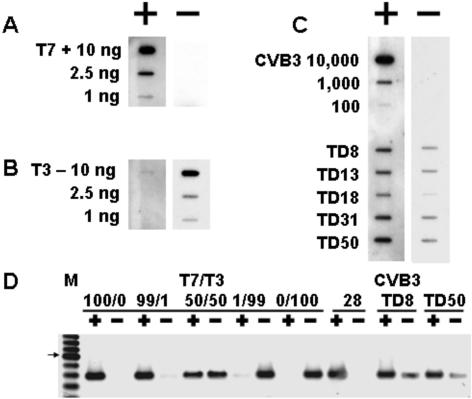
The 5′-terminal region of the enteroviral genome is intimately involved with RNA replication and translational events (2, 36, 64, 84, 118). Thus, deletion of the terminal 5′ sequence of the CVB3 genome could be expected to have an impact upon virus replication. Enterovirus-infected cells have been determined to have approximately 40 to 70 strands of positive-strand RNA for every viral molecule of negative polarity (78). As a first step toward assessing impact, we assayed populations of CVB3/TD RNA by slot blot strand-specific hybridization. The specificity of the oligomeric probes was tested and verified using in vitro-transcribed positive- and negative-strand CVB3 RNAs (Fig. 8A and B). Parental CVB3 and different CVB3/TD strains were concentrated by ultracentrifugation, resuspended in saline, and treated with RNase A/T1 to remove contaminating RNA. Viral RNA was then extracted with Trizol. Negative-strand viral RNA was not detected in total RNA from parental CVB3/28 stocks using 1 × 104 TCID50 per slot (Fig. 8C). The CVB3/TD RNA stocks were also tested for the presence of host cellular RNA by RT-PCR for GADPH mRNA (using 3-GADPH to prime cDNA and amplification with 5- and 3-GADPH); in each case, GADPH RNA was not detectable (data not shown). That parental CVB3 stocks did not contain detectable negative-strand RNA is consistent with others' findings using approaches with greater sensitivity (78) and indicates that the partial purification through sucrose, combined with RNase treatment, removed residual contaminating, nonencapsidated viral or cellular RNA. However, negative-strand viral RNA in purified CVB3/TD stocks was readily detectable (Fig. 8C). We normalized the pixel intensities of the viral RNA versus the intensity of 1 ng of T7 transcript (for the positive strand [Fig. 8A]) or 1 ng of T3 transcript (for the negative strand [Fig. 8B]) to compensate for differences in hybridization efficiency of the probe oligonucleotide. This analysis determined that ratios of positive- to negative-strand CVB3/TD RNA in these preparations (Fig. 8C) ranged between 3 and 12.
FIG. 8.
CVB3/TD strains package negative-strand RNA. (A) Dilutions of T7 RNA polymerase-transcribed positive-strand RNA and (B) T3 RNA polymerase-transcribed negative-strand RNA from a linearized CVB3 subclone were slot blotted. (C) RNA was prepared from virus stocks pretreated with RNase as described in Materials and Methods using 104, 103 and 102 TCID50 units of CVB3/28 (CVB3 10,000, 1,000, and 100, respectively) or 107 rTCID50 TD8, 107 rTCID50 TD13, 105 rTCID50 TD18, 108 rTCID50 TD31, or 108 rTCID50 TD50. Blots were probed with 32P-labeled E1 (positive strand) or 5Puff (negative strand). (D) Strand-specific RNAs (+, positive; -, negative) were purified from T7 and T3 RNA polymerase transcripts of a subclone of CVB3/28 cDNA, mixtures of transcripts, and RNA of CsCl gradient-purified CVB3/28, CVB3/TD8, and CVB3/TD50 as described in Materials and Methods. RNA (0.3 μg) was used for each preparation. cDNAs prepared from these RNAs were amplified with KS1 and KS2, electrophoresed, and visualized as described in Materials and Methods. Lane M, 100-bp ladder. The arrow indicates a 600-bp band.
To further test the hypothesis that negative-strand CVB3/TD RNA is encapsidated, we centrifuged CVB3/28, TD8, and TD50 to equilibrium in CsCl gradients. RNA was purified from the fraction of the gradients closest in density to 1.34 g/ml, the density at which CVB3/28 contained the peak of infectious titer (data not shown). Strand-specific cDNA was synthesized with biotinylated primers (KS1 and KS2 [Table 1]) and isolated using streptavidin magnetic beads. T7 (positive-strand) and T3 (negative-strand) RNA transcripts from the CVB3/28 cloned cDNA and mixtures of both served as control templates for the RT reactions (Fig. 8D). CsCl-purified CVB3/28 had no detectable negative-strand RNA, although CsCl-purified CVB3/TD8 and TD50 (Fig. 8D) contained a mixture of positive and negative strands. Amplification of control transcript mixtures that contained a 99:1 mix of positive and negative strands detected both strands, but the minor component was only barely detectable (Fig. 8D). In 50:50 mixes of the transcripts, both strands were amplified to an equivalent level (Fig. 8D). Comparison of the amplification of CVB3/TD8 and TD50 fractions to the amplified 99:1 and 50:50 mixtures of T7 and T3 transcripts indicates that the ratio of strands in the CVB3/TD RNAs is less than 50:50 but contains more negative-strand RNA than the 99:1 mixture. The ratio of positive to negative strands detected by slot blot assay was 3:1 for CVB3/TD8 and TD50, results that were consistent with observations from the slot blot data (Fig. 8C). Although more positive- than negative-strand RNA is packaged, a significant proportion of the viral RNA is of the negative-strand type.
VPg is covalently attached to the CVB3/TD genomic RNA.
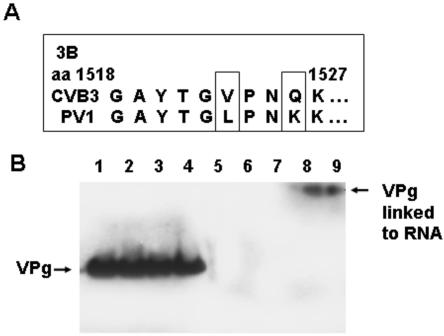
As part of the process of picornavirus replication, the viral protein, VPg, is uridylated (103). Uridylated VPg is thought to serve as a protein primer (85) for transcription either by an interaction with residues in the poly(A) tract of the positive strand to initiate negative-strand RNA replication or with the two 3′-terminal adenosine residues of the negative strand to initiate positive-strand RNA replication, resulting in covalent attachment of VPg protein to the 5′ end of the viral RNA (1, 97). However, we had observed that three of the five CVB3/TD genomes do not have a 5′-terminal uridine residue (Fig. 4). To determine whether the virion RNA of CVB3/TDs had VPg attached, we employed an antibody generated from immunization with a peptide containing the 10 N-terminal amino acid sequences of protein 3B from PV1 (73). Alignments of this region of 3B in PV1 and CVB3 revealed two changes in CVB3 relative to the PV sequence (Fig. 9A) (73). Virion RNA of PV1 (Fig. 9B, lane 1), CVB3/28 (lane 2), CVB3/TD8 (lane 3), and CVB3/TD50 virions (lane 4) was RNase treated, electrophoresed, and blotted. The N10 anti-VPg antibody detected a protein on Western blots of CVB3 RNA which migrated at the same size as that from PV (Fig. 9B). VPg was detected with N10 from the two CVB3/TD samples (Fig. 9B, lanes 3 and 4), at the same mobility as the PV and CVB3 control VPg proteins (lanes 1 and 2). Samples were digested with proteinase K to remove protein and then electrophoresed as before; no signal was detected with samples from CVB3/28, CVB3/TD8, and TD50 when probed with the N10 antibody (Fig. 9B, lanes 5 to 7). In the absence of both RNase and proteinase treatment, N10 detected a high-mobility band in both CVB3/TD8 and TD50 virion RNA (Fig. 9B, lanes 8 and 9) as expected for VPg with the attached virion RNA. As a further test of the protein's identity, protein covalently linked to RNA from CVB3/TD8 and CVB3/TD50 virion preparations was isolated from gels and sequenced. The first five amino terminal amino acids for each sample were determined as NH2-GAYTG, consistent with the VPg from CVB3 (Fig. 9A). These results indicate that the CVB3/TD virion RNAs are VPg linked. As equivalent amounts of viral RNA were used for PV1 Sabin, CVB3/28, and the two TD viruses, the similar intensity of the signal for VPg for the four RNase-treated samples (Fig. 9B, lanes 1 to 4) indicates that a similar amount of viral RNA has VPg attached. This observation suggests that VPg is linked to both the positive- and negative-strand encapsidated RNA, since CsCl-purified TD8 and TD50 have a positive- to negative-strand ratio of 3:1 (Fig. 8C) and attachment to the negative strand alone would diminish the signal visibly. If VPg is attached to the positive strand of TD8 and TD50 (which have 5′-terminal AG and GC, respectively [Fig. 4]), VPg attachment can occur independently of a 5′-terminal diuridine sequence.
FIG. 9.
CVB3/TD viruses have VPg covalently attached to genomic RNA. (A) Alignment of amino-terminal sequences of VPg between CVB3/28 and PV1. Differences are boxed. (B) Viral RNA was purified from stocks of the CVB3/28, TD8, and TD50 viruses (105 TCID50 units) and analyzed by Western blotting. Blots were probed with antibody to PV1 VPg (N10) as described in Materials and Methods. RNA treated with RNase A/T1: PV1 Sabin (lane 1), CVB3/28 (lane 2), CVB3/TD8 (lane 3), and CVB3/TD50 (lane 4); RNA treated with RNase A/T1 and proteinase K: CVB3/28 (lane 5), CVB3/TD8 (lane 6), and CVB3/TD50 (lane 7); untreated RNA: CVB3/TD8 (lane 8) and CVB3/TD50 (lane 9).
CVB3/TD8 and CVB3/TD50 replicate and persist in the murine heart.
Despite the inability to detect intact 5′ termini from hearts of infected mice late (such as AJ18 [Fig. 3]), we could not discount the possibility that helper viruses remained in the wild-type population of terminally deleted CVB genomes. Having demonstrated that the clonal CVB3/TD strains were capable of replication in cell culture, we tested replication of the molecularly cloned progeny CVB3/TD strains in vivo. We used scid mice, which lack an adaptive immune response (15), and C3H/HeJ mice, which are immunologically normal and produce anti-CVB immune responses subsequent to experimental CVB infection (12, 18, 44). scid mice lack functional T and B cells and so do not clear CVB infections (22, 46, 112), whereas immunologically competent mice establish B- and T-cell responses against experimental CVB infections within 7 to 14 days of exposure to virus (17), which usually results in the eradication of infectious virus from the mouse within 10 to 14 days (21, 108). Mice were inoculated intraperitoneally with CVB3/TD8 or CVB3/TD50. Aliquots of heart homogenates from CVB3/TD-inoculated scid mice on days 4, 8, and 38 p.i. or C3H/HeJ mice on days 14, 18, 28, and 175 p.i. were inoculated onto HeLa cell monolayers. No CPE was noted in these cultures (data not shown). RNA from 72-h cultures was assayed for CVB3 RNA using RT-PCR and primers specific for a conserved sequence in the interior of the 5′ NTR (KS1 and KS2 [Table 1]). Viral RNA was detected in HeLa cultures of homogenates of all scid mouse hearts through day 38 p.i. and in cultures of homogenates of all C3H/HeJ mouse hearts through day 175 p.i. (data not shown). Viral RNA was detected by RT-PCR using internal primers (KS1 and KS2) from day 175 p.i. heart homogenates of C3H/HeJ mice inoculated with CVB3/TD8 or CVB3/TD50 (Fig. 10A). The amounts of amplified cDNA varied between individual homogenate cultures, despite the relatively even signal generated by amplification of GADPH cDNA from each culture (Fig. 10B). This may be due to the relatively focal nature of CVB3 infection of murine hearts resulting in variability of sampling. RT-PCR with 5′-specific primers using RNA from HeLa cultures inoculated with day 175 heart homogenates indicated that the 5′ terminus had not regenerated to the CVB3/28 wild type (Fig. 10C and D). There was a lack of amplification with the S primer in all three heart homogenate cultures from mice inoculated with CVB3/TD8 and in one case, with the S1 primer (Fig. 10C). RNA from HeLa cultures of clonal CVB3/TD8 did amplify in RT-PCR with S (Fig. 6B) due to a 13-nt overlap of the S primer with the 5′ end of this genome. The S1 primer amplified RNA poorly from one heart homogenate culture and not at all from another (Fig. 10C) despite the 21-nt overlap of this primer with the CVB3/TD8 genome. These results suggest that the CVB3/TD8 is evolving during replication in the murine heart, perhaps to a more extensive deletion. No similar alteration in amplification with 5′-specific primers is seen with RNA from CVB3/TD50-inoculated heart homogenate cultures (Fig. 10D). Necrosis and inflammation were not observed in the scid or C3H/HeJ hearts by light microscopic examination of hematoxylin and eosin-stained heart sections (data not shown). No inflammation or fatty replacement was observed in pancreas sections (data not shown). These results demonstrate that clonal CVB3/TD strains can replicate in vivo in the absence or in the presence of an adaptive immune system following i.p. inoculation and can persist for weeks postinoculation.
FIG. 10.
Inoculation of mice with CVB3/TD results in persistent cardiac virus infections. (A) Total RNA from homogenates of day 175 p.i. hearts of C3H/HeJ mice inoculated with CVB3/TD8 or TD50 was assayed by RT-PCR for the presence of CVB3 RNA with KS1 and KS2. Southern blot analysis (lower panel) with primer E3 confirms that amplified cDNA is CVB3. (B) Heart homogenates were assayed for cellular mRNA with 5-GADPH and 3-GADPH. Southern blot analysis (lower panel) with primer 5-GADPH. (C, D, E) HeLa cells (105) were inoculated with homogenates of day 175 p.i. hearts of C3H/HeJ mice inoculated with CVB3/TD8 (C) or TD50 (D) or with 106 TCID50 units of CVB3/28 (E). Total RNA from 72-h (C, D) or 24-h (E) cultures was assayed by RT-PCR for the presence of CVB3 RNA with KS1 and S, S1, S2, S3, or S4; electrophoresed; and visualized as described in Materials and Methods. Southern blot analysis (lower panels) with primer E3 confirms that amplified cDNA is CVB3. Lane M, 100-bp ladder; lane +, CVB3/28 viral RNA; He, HeLa cell total RNA; lane -, no RNA. The arrow indicates a 600-bp band.
DISCUSSION
These results represent the first report of an in vivo evolution of a wild-type enterovirus population into a quasispecies (30) that has 5′-terminal genomic deletions ranging from 7 to 49 nucleotides. Progeny virus populations from infectious cDNA copies of CVB3 genomes containing these naturally modified 5′ genomic termini replicate in cell cultures and in mice, persist in culture or in mice for weeks to months, do not repair the 5′-terminal genomic deletions, package negative-strand RNA in virions, and possess viral RNA terminated with VPg despite the genomic RNA terminating in bases other than U. These results provide a new basis by which previous detection of enteroviral RNA in cardiomyopathic heart tissue in the absence of infectious virus (4, 60, 92) may be evaluated.
Picornavirus infections are generally considered to be acute and cleared rapidly by the host adaptive immune response, yet long-term persistence in animals and humans has been demonstrated (4, 54, 60, 87, 92, 95, 104, 116). The mechanism by which long-term picornavirus persistence occurs in the presence of an intact immune system remains unclear. It has been shown that DI virus populations, described for many virus families (28, 30, 98, 99) can persist for long periods of time in cell cultures. Spontaneously occurring picornavirus DI populations from cell culture systems demonstrate variously sized deletions in the capsid protein (P1) coding region of the viral genome (57), although defective viruses in foot-and-mouth disease have been shown to have deletions in the leader protein-encoding region (37). Further, defective interfering picornavirus populations have not been observed in animals or humans (99). In this study of coxsackievirus replication in cardiac cells in culture and in mice, the virus evolves to a population of partially deleted genomes.
The relatively rapid movement from a cytolytic to a noncytolytic phenotype, accompanied by the disappearance of detectable wild-type 5′ genomic termini and their replacement by diverse TD 5′ genomic termini, is consistent with predicted rapid changes in quasispecies populations when environmental conditions are altered (30, 66). An alternative possibility is that a slower accumulation of the deleted genomes occurred over the time between inoculation of the parental virus and the appearance of the CVB3/TD genomes. However, given the low yield of virus shown by the clonal CVB3/TD strains, we believe it is unlikely that a parental CVB3 population coexists with a minor population of TD-like genomes, since the latter should be at a significant replicative disadvantage and would soon be eliminated from the virus population. In vivo, the adaptive immune response to the CVB3 infection which clears the productive (cytolytic) wild-type CVB3 may confer a selective advantage to the more slowly replicating CVB3/TD variants. In vitro, the selection for these defective viruses in ACM may be related to the short period of viability of these primary cultures (48 to 72 h) (113): the protocol for serial passage of these viruses may have favored genomes which were packaged but still intracellular when the freeze-thaw lysis was performed.
A striking degree of structural conservation exists in domain I of TD genomes, despite differences in the deletion lengths. The results of chemical probing suggest that folding of structural elements within the domain I cloverleaf is determined locally; those genomes with stem-loop b sequences adopt the same structure regardless of the presence of stem a; the same is true for stem-loop c and stem-loop d. The stability of RNA structure in stem-loop d is quite remarkable, particularly in CVB3/TD50, and includes the non-Watson-Crick pyrimidine-pyrimidine pairs in the internal loop first indicated by the nuclear magnetic resonance analysis of Ohlenschläger and colleagues (80). In our structural analysis of the entire 5′ NTR, we found no indications of structural perturbation downstream of the cloverleaf in any of the TD genomes. Thus, as suggested by others (11, 14, 42), elements of the 5′ NTR responsible for IRES function and replication are largely independent.
Artificially created deletions of 5′ termini of enteroviral genomes have been shown to be deleterious for viral replication (11, 106). Enterovirus cloverleaf domain I is an essential structured region for viral RNA replication (11, 84, 106). Mutations in stem-loop b of domain I have been demonstrated to prevent cytopathic virus replication (2, 3) as well as RNA replication (3, 84), while insertions and deletions in stem-loop d can prevent binding of PV 3CD protein (2). Domain I appears to be critical for initiation of negative-strand RNA replication (11, 42, 106). We have observed that all clonal CVB3/TD strains replicate so slowly that uninfected cells soon predominate in infected cultures, thereby preventing apparent CPE (data not shown). Thus, just the deletion of the 5′-terminal 7 nt (which occurs in each of the CVB3/TD strains characterized here) can attenuate replication. It is noteworthy that although stem-loop b contains a PCBP binding site (84), the absence of this region in CVB3/TD31 and TD50 does not further accentuate the replication-defective phenotype of these viruses. As the residual stem-loop d in the CVB3/TD domain I can bind the CVB3 3C protein (119), we speculate that this structure can form a replication complex with 3CD to permit sufficient negative-strand replication. The absence of a cloverleaf PCBP binding site may decrease viral RNA circularization via the cloverleaf ternary complex, believed to be important for RNA replication (42). However, interactions of the CVB3 IRES with host proteins such as PCBP may allow sufficient circularization of genomes for replication: engineered PV genomes lacking the IRES do not efficiently replicate RNA (75). It is clear that the cloverleaf domain is an important region for viral RNA replication since the CVB3/TD genomes have a more extreme phenotype than even engineered PV genomes with complete deletions of the 3′ NTR (16): PV genomes in which the 3′ NTR has been deleted nonetheless replicate in culture to titers within 10% of those of wild-type (parental) virus and induce widespread CPE in infected cultures.
Alterations of positive/negative viral RNA strand ratios have been linked with CVB persistence, as shown by in situ hybridization in murine heart muscle (54) or by strand-specific RT-PCR in skeletal muscle (104) and in persistence of PV following experimental infection of the murine central nervous system (38). Deletions of 1 to 3 nt at the 5′ terminus or mutations in the first 6 nt of PV1 have been shown to alter positive- to negative-strand ratios in in vitro replication (100). We observed negative-strand RNA, as well as positive-strand RNA, in populations of CVB3/TD virions that had been incubated with RNase. The ratios of positive to negative strands in the preparations ranged between 3:1 and 12:1; this was a remarkable finding in that negative-strand RNA was not detectable in parental CVB3 virions prepared in the same manner. Assay of CsCl-banded CVB3/TD virions confirmed that negative-strand RNA was packaged. The logic of specific viral RNA packaging has dictated a packaging signal, a sequence or RNA structure which initiates the assembly of the capsid pentamers into a complete virion. No such specific signal has yet been identified in the human enteroviruses, although the generation of replicons with substitutions of foreign proteins in the P1 region which can be encapsidated in trans indicates that such a signal cannot be present in the capsid encoding region (47, 70, 90, 91). In PV, only newly synthesized genomes are packaged (79). It seems likely that the high packaging specificity for the positive strand in wild-type CVB3 and in PV1 (78) is linked to the efficiency of positive-strand RNA replication, which is decreased by the 5′-end deletions in the CVB3/TD strains. If, as has been suggested previously (79), the specificity of packaging is via viral replication proteins bound to positive-strand RNA, perhaps through interactions with the 5′ terminus of the genome, only newly replicated viral RNA can be packaged. It is interesting that one PV replication protein, 2C, has been shown to bind to a double-stranded sequence at the 3′ end of the negative-strand viral RNA (8) and stem-loop b (9), sequences which are partially or completely deleted in the CVB3/TDs. If the replication complex binding to these deleted genomes is altered, it may make negative- and positive-strand replication almost equivalent in efficiency. One consequence of this might be promiscuous packaging of both strands of viral RNA in the virions of CVB3/TDs.
Enteroviral RNA is covalently bound at the 5′ terminus to the viral protein, VPg (1). Because uridylated VPg acts as a protein primer in the replication of enterovirus RNA by the RNA-dependent RNA polymerase 3D, VPg remains covalently attached at the 5′ terminus. Encapsidated enteroviral RNA contains VPg, while cellular enteroviral positive-strand RNA used as mRNA is not (77). The CVB3/TD8, TD13, and TD50 genomes have 5′ termini that are not U. Only the CVB3/TD18 genome begins with 5′-UU, while CVB3/TD31 begins with a single U residue (5′-UA). Thus, the 3′-terminal AA of the negative strand, which would interact in priming of enteroviral RNA synthesis by uridylated VPg (93), is not represented as 5′-UU in 4/5 of the CVB3/TD genomes. Despite these findings, VPg was easily detected by Western blot analysis in CVB3/TD8 and TD50 RNA, two genomes without 5′-terminal uridine residues. Since a quarter of the encapsidated RNA is negative-strand RNA, the signal might be reduced if only negative-strand RNA had attached VPg despite the presence of negative-strand viral RNA. However, the level of VPg detected in relation to the amount of encapsidated viral RNA was not decreased in the CVB3/TDs, indicating that the encapsidated positive-strand RNA is likely to have VPg attached despite the lack of the 5′-terminally repeated U residues. Sequence analysis of the VPg encoding region (3B) as well as the cis-acting replication element (CRE) region in the virus population from cultures of day 18 A/J heart homogenate (defined as nt 4364 to 4422, by analogy to PV RNA) (39) revealed no differences between the CVB3/TD RNAs and the parental CVB3/28 RNA (data not shown). The CRE, a stem-loop RNA structure in the coding sequence of enterovirus nonstructural protein 2C (39, 68), has been shown to function in VPg uridylylation. A slide-back model has been proposed for the CRE-based uridylylation of VPg (86) in PV1 in which interactions between VPg and 3D polymerase prime the linking of uridine to VPg through interactions with adenosine residues in the loop of the CRE. Thus, sites at which mutations might alter VPg uridylylation were not mutated in the CVB3/TDs. A 5′-terminal U residue is not conserved in the TDs (Fig. 4B). As poliovirus VPg can be linked to any of the nucleotides (although VPgpU is greatly favored in concentration [86]), VPg linked to nonuridine nucleotides may be used as a primer for synthesis of positive-strand RNA when the negative-strand template lacks 3′-terminal uridine residues. However, as the TD population does not favor U as the 5′-terminal residue as would be expected if positive-strand replication were primed by CRE-generated VPgpUpU, it can be assumed that the process of addition of nucleotides to the protein primer must occur at a site other than CRE in these mutants due to the lack of the complete 5′ cloverleaf, possibly at the 3′ end of the negative strand.
It is assumed that such deletions may constantly be occurring at some level in infected cells, due to pausing or premature termination of nascent RNAs. Are the TD genomes synthesized randomly, so that any number of different deletions occur, or is there a selection for relatively few classes of deletions which can promote successful replication? Do short deletions give rise to longer deletions? We may speculate on the mechanism based on an interpretation of the current results in which enzymatic amplification of intact 5′ termini was not possible with the mixed population present in the AJ day 18 heart cultures but was possible when using RNA extracted from clonal populations of CVB3/TD8 and TD13. Failure to amplify the TD8 and TD13 genomes from a mixed, naturally occurring population suggests that these may be in much lower abundance than other genomic ends. If the CVB3/TD8 and TD13 genomes were, in fact, present in low abundance in the AJ18 population, this might indicate movement of the population toward the genomes containing greater deletions. We have established that the 5′ deletions are not repaired during passage of clonal CVB3/TD strains in cell culture; therefore, we assume that for these strains, there is little or no evolutionary movement back to the wild type under these conditions. Inoculation of CVB3/TD strains into mice similarly did not result in reversion to a wild-type cytopathic phenotype. However, inoculation of mice with CVB3/TD8 produced cardiac virus which could not be amplified by primers specific for the first 20 nt of the CVB3 genome by RT-PCR amplification, unlike CVB3/TD8. Although the numbers of mice were too small to determine a consistent trend, this result, which suggests further evolution of the 5′ end, can occur, probably by further deletion. Consequently, we suggest that the TD genomes are proceeding through sequence space in directions away from the wild-type virus. As the portion of stem-loop d preserved in the largest deletion that we have characterized (CVB3/TD50, a deletion of 49 nucleotides from the 5′ genomic terminus) is the minimum for forming a complex of 3CD with RNA of the 5′ end of the positive strand (32, 119), larger deletions may well be nonviable. Consistent with this, our structural analysis shows that the stem-loop d portion of TD50 is structurally indistinguishable from the stem-loop d in the full-length molecule.
While the viability in the natural environment of enteroviruses with significant replication deficits is expected to be quite low due to the inability of such virus populations to compete successfully with wild-type virus, defective replication may nonetheless provide an avenue for prolonged viral genetic survival under specific circumstances. We did not observe inflammatory disease in any hearts from mice inoculated with a CVB3/TD strain. Persistence of CVB in the heart muscle by this mechanism following infection by a cardiovirulent virus might, however, also encourage long-term persistence of the inflammatory disease initially induced by the wild-type virus infection. The concept of viral persistence through selection of variants from the viral quasispecies in the face of active immunity is known (reviewed in reference 30). The results presented here document the emergence of a unique enterovirus quasispecies either in murine cell culture or in experimentally infected mice. It remains to be shown whether these genomic deletions can be detected in enteroviral RNA isolated from human cases of myocarditis and dilated cardiomyopathy.
Acknowledgments
This work was supported by U.S. Public Health Service grants R03AI053196 (N.M.C.), R21AI49540-01A1 (S.T.) from the National Institutes of Health, P20 RR16469 (W.T.) from the INBRE Program of the National Center for Research Resources, and 0225561Z (K.-S. K.) from the American Heart Association.
We thank J. Butler, P. Karki, and J. S. Leser for excellent technical assistance. We also thank F. Schiff of the ERACE Foundation, the Stein family, M. Guthrie, and E. Barnett for their generosity in supporting this work in memory of loved ones.
REFERENCES
- 1.Ambros, V., and D. Baltimore. 1978. Protein is linked to the 5′ end of poliovirus RNA by a phosphodiester linkage to tyrosine. J. Biol. Chem. 253:5263-5266. [PubMed] [Google Scholar]
- 2.Andino, R., G. E. Rieckhof, P. L. Achacoso, and D. Baltimore. 1993. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 12:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andino, R., G. E. Rieckhof, and D. Baltimore. 1990. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63:369-380. [DOI] [PubMed] [Google Scholar]
- 4.Andreoletti, L., T. Bourlet, D. Moukassa, L. Rey, D. Hot, Y. Li, V. Lambert, B. Gosselin, J. F. Mosnier, C. Stankowiak, and P. Wattre. 2000. Enteroviruses can persist with or without active viral replication in cardiac tissue of patients with end-stage ischemic or dilated cardiomyopathy. J Infect. Dis. 182:1222-1227. [DOI] [PubMed] [Google Scholar]
- 5.Baboonian, C., M. J. Davies, J. C. Booth, and W. J. McKenna. 1997. Coxsackie B viruses and human heart disease. Curr. Top. Microbiol. Immunol. 223:31-52. [DOI] [PubMed] [Google Scholar]
- 6.Baboonian, C., and W. McKenna. 2003. Eradication of viral myocarditis: is there hope? J. Am. Coll. Cardiol. 42:473-476. [DOI] [PubMed] [Google Scholar]
- 7.Baboonian, C., and T. Treasure. 1997. Meta-analysis of the association of enteroviruses with human heart disease. Heart 78:539-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banerjee, R., and A. Dasgupta. 2001. Interaction of picornavirus 2C polypeptide with the viral negative-strand RNA. J. Gen. Virol. 82:2621-2627. [DOI] [PubMed] [Google Scholar]
- 9.Banerjee, R., W. Tsai, W. Kim, and A. Dasgupta. 2001. Interaction of poliovirus-encoded 2C/2BC polypeptides with the 3′ terminus negative-strand cloverleaf requires an intact stem-loop b. Virology 280:41-51. [DOI] [PubMed] [Google Scholar]
- 10.Barton, D. J., B. J. Morasco, and J. B. Flanegan. 1999. Translating ribosomes inhibit poliovirus negative-strand RNA synthesis. J. Virol. 73:10104-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ Cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beck, M. A., and S. M. Tracy. 1989. Murine cell-mediated immune response recognizes an enterovirus group-specific antigen(s). J. Virol. 63:4148-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bedard, K. M., and B. L. Semler. 2004. Regulation of picornavirus gene expression. Microbes Infect. 6:702-713. [DOI] [PubMed] [Google Scholar]
- 14.Borman, A. M., P. Le Mercier, M. Girard, and K. M. Kean. 1997. Comparison of picornaviral IRES-driven internal initiation of translation in cultured cells of different origins. Nucleic Acids Res. 25:925-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosma, G. C., R. P. Custer, and M. J. Bosma. 1983. A severe combined immunodeficiency mutation in the mouse. Nature 301:527-530. [DOI] [PubMed] [Google Scholar]
- 16.Brown, D. M., S. E. Kauder, C. T. Cornell, G. M. Jang, V. R. Racaniello, and B. L. Semler. 2004. Cell-dependent role for the poliovirus 3′ noncoding region in positive-strand RNA synthesis. J. Virol. 78:1344-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chapman, N. M., C. J. Gauntt, and S. Tracy. 2002. Immunology of the coxsackieviruses, p. 391-403. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 18.Chapman, N. M., K. S. Kim, S. Tracy, J. Jackson, K. Hofling, J. S. Leser, J. Malone, and P. Kolbeck. 2000. Coxsackievirus expression of the murine secretory protein interleukin-4 induces increased synthesis of immunoglobulin G1 in mice. J. Virol. 74:7952-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman, N. M., A. Ragland, J. S. Leser, K. Hofling, S. Willian, B. L. Semler, and S. Tracy. 2000. A group B coxsackievirus/poliovirus 5′ nontranslated region chimera can act as an attenuated vaccine strain in mice. J. Virol. 74:4047-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chapman, N. M., A. I. Ramsingh, and S. Tracy. 1997. Genetics of coxsackievirus virulence. Curr. Top. Microbiol. Immunol. 223:227-258. [DOI] [PubMed] [Google Scholar]
- 21.Chapman, N. M., Z. Tu, S. Tracy, and C. J. Gauntt. 1994. An infectious cDNA copy of the genome of a non-cardiovirulent coxsackievirus B3 strain: its complete sequence analysis and comparison to the genomes of cardiovirulent coxsackieviruses. Arch. Virol. 135:115-130. [DOI] [PubMed] [Google Scholar]
- 22.Chow, L. H., K. W. Beisel, and B. M. McManus. 1992. Enteroviral infection of mice with severe combined immunodeficiency. Evidence for direct viral pathogenesis of myocardial injury. Lab. Investig. 66:24-31. [PubMed] [Google Scholar]
- 23.Cole, C. N., and D. Baltimore. 1973. Defective interfering particles of poliovirus. II. Nature of the defect. J. Mol. Biol. 76:325-343. [DOI] [PubMed] [Google Scholar]
- 24.Cole, C. N., D. Smoler, E. Wimmer, and D. Baltimore. 1971. Defective interfering particles of poliovirus. I. Isolation and physical properties. J. Virol. 7:478-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coller, B. A., N. M. Chapman, M. A. Beck, M. A. Pallansch, C. J. Gauntt, and S. M. Tracy. 1990. Echovirus 22 is an atypical enterovirus. J. Virol. 64:2692-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dalldorf, G. 1955. The coxsackie viruses. Annu. Rev. Microbiol. 9:277-296. [DOI] [PubMed] [Google Scholar]
- 27.De Jager, H., and S. Van Creveld. 1956. Myocarditis in newborns, caused by coxsackie virus: clinical and pathological data. Ann. Paediatr. 187:100-118. [PubMed] [Google Scholar]
- 28.DePolo, N. J., and J. J. Holland. 1986. Very rapid generation/amplification of defective interfering particles by vesicular stomatitis virus variants isolated from persistent infection. J. Gen. Virol. 67:1195-1198. [DOI] [PubMed] [Google Scholar]
- 29.DiLella, A. G., and S. L. Woo. 1987. Hybridization of genomic DNA to oligonucleotide probes in the presence of tetramethylammonium chloride. Methods Enzymol. 152:447-451. [DOI] [PubMed] [Google Scholar]
- 30.Domingo, E., C. Escarmis, L. Menendez-Arias, and J. J. Holland. 1999. Viral quasispecies and fitness variations, p. 141-161. In E. Domingo and J. J. Holland (ed.), Origin and evolution of viruses. Academic Press, London, England.
- 31.Drescher, K., K. Kono, S. Bopegamage, S. D. Carson, and S. Tracy. 2004. Coxsackievirus B3 infection and type 1 diabetes development in NOD mice: insulitis determines susceptibility of pancreatic islets to virus infection. Virology 329:381-394. [DOI] [PubMed] [Google Scholar]
- 32.Du, Z., J. Yu, N. B. Ulyanov, R. Andino, and T. L. James. 2004. Solution structure of a consensus stem-loop D RNA domain that plays important roles in regulating translation and replication in enteroviruses and rhinoviruses. Biochemistry 43:11959-11972. [DOI] [PubMed] [Google Scholar]
- 33.Dunn, J. J., S. S. Bradrick, N. M. Chapman, S. M. Tracy, and J. R. Romero. 2003. The stem loop II within the 5′ nontranslated region of clinical coxsackievirus B3 genomes determines cardiovirulence phenotype in a murine model. J. Infect. Dis. 187:1552-1561. [DOI] [PubMed] [Google Scholar]
- 33a.Ehresmann, C., F. Baudin, M. Mougel, P. Romby, J. P. Ebel, and B. Ehresmann. 1987. Probing the structure of RNAs in solution. Nucleic Acids Res. 15:9109-9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Hagrassy, M. M., J. E. Banatvala, and D. J. Coltart. 1980. Coxsackie-B-virus-specific IgM responses in patients with cardiac and other diseases. Lancet 2:1160-1162. [DOI] [PubMed] [Google Scholar]
- 35.Gamarnik, A. V., and R. Andino. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 74:2219-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Arriaza, J., S. C. Manrubia, M. Toja, E. Domingo, and C. Escarmis. 2004. Evolutionary transition toward defective RNAs that are infectious by complementation. J. Virol. 78:11678-11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Girard, S., A. S. Gosselin, I. Pelletier, F. Colbere-Garapin, T. Couderc, and B. Blondel. 2002. Restriction of poliovirus RNA replication in persistently infected nerve cells. J. Gen. Virol. 83:1087-1093. [DOI] [PubMed] [Google Scholar]
- 39.Goodfellow, I., Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hagino-Yamagishi, K., and A. Nomoto. 1989. In vitro construction of poliovirus defective interfering particles. J. Virol. 63:5386-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herold, J., and R. Andino. 2000. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol. 74:6394-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Higuchi, R., C. Fockler, G. Dollinger, and R. Watson. 1993. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (N.Y.) 11:1026-1030. [DOI] [PubMed] [Google Scholar]
- 44.Hofling, K., S. Tracy, N. Chapman, K. S. Kim, and J. Smith Leser. 2000. Expression of an antigenic adenovirus epitope in a group B coxsackievirus. J. Virol. 74:4570-4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huber, S. A. 1997. Autoimmunity in myocarditis: relevance of animal models. Clin. Immunol. Immunopathol. 83:93-102. [DOI] [PubMed] [Google Scholar]
- 46.Hufnagel, G., N. Chapman, and S. Tracy. 1995. A non-cardiovirulent strain of coxsackievirus B3 causes myocarditis in mice with severe combined immunodeficiency syndrome. Eur. Heart J. 16(Suppl. O):18-19. [DOI] [PubMed] [Google Scholar]
- 47.Jia, X. Y., M. Van Eden, M. G. Busch, E. Ehrenfeld, and D. F. Summers. 1998. trans-encapsidation of a poliovirus replicon by different picornavirus capsid proteins. J. Virol. 72:7972-7977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson, J. P., R. H. Yolken, D. Goodman, J. A. Winkelstein, and J. E. Nagel. 1982. Prolonged excretion of group A coxsackievirus in an infant with agammaglobulinemia. J. Infect. Dis. 146:712. [DOI] [PubMed] [Google Scholar]
- 49.Kaplan, G., A. Levy, and V. R. Racaniello. 1989. Isolation and characterization of HeLa cell lines blocked at different steps in the poliovirus life cycle. J. Virol. 63:43-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaplan, G., and V. R. Racaniello. 1988. Construction and characterization of poliovirus subgenomic replicons. J. Virol. 62:1687-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khetsuriani, N., D. R. Prevots, L. Quick, M. E. Elder, M. Pallansch, O. Kew, and R. W. Sutter. 2003. Persistence of vaccine-derived polioviruses among immunodeficient persons with vaccine-associated paralytic poliomyelitis. J. Infect. Dis. 188:1845-1852. [DOI] [PubMed] [Google Scholar]
- 52.Kim, K. S., K. Hofling, S. D. Carson, N. M. Chapman, and S. Tracy. 2002. The primary viruses of myocarditis, p. 23-54. In L. T. Cooper, Jr. (ed.), Myocarditis: from bench to bedside. Humana Press, Totowa, N.J.
- 53.King, A. M. Q., F. Brown, P. Christian, et al. 2000. Picornaviridae, p. 657-678. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 54.Klingel, K., C. Hohenadl, A. Canu, M. Albrecht, M. Seemann, G. Mall, and R. Kandolf. 1992. Ongoing enterovirus-induced myocarditis is associated with persistent heart muscle infection: quantitative analysis of virus replication, tissue damage, and inflammation. Proc. Natl. Acad. Sci. USA 89:314-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klingel, K., B. M. McManus, and R. Kandolf. 1995. Enterovirus-infected immune cells of spleen and lymph nodes in the murine model of chronic myocarditis: a role in pathogenesis? Eur. Heart J. 16(Suppl. O):42-45. [DOI] [PubMed] [Google Scholar]
- 56.Klump, W. M., I. Bergmann, B. C. Muller, D. Ameis, and R. Kandolf. 1990. Complete nucleotide sequence of infectious coxsackievirus B3 cDNA: two initial 5′ uridine residues are regained during plus-strand RNA synthesis. J. Virol. 64:1573-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuge, S., I. Saito, and A. Nomoto. 1986. Primary structure of poliovirus defective-interfering particle genomes and possible generation mechanisms of the particles. J. Mol. Biol. 192:473-487. [DOI] [PubMed] [Google Scholar]
- 58.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 59.Le, S. Y., and M. Zuker. 1990. Common structures of the 5′ non-coding RNA in enteroviruses and rhinoviruses. Thermodynamical stability and statistical significance. J. Mol. Biol. 216:729-741. [DOI] [PubMed] [Google Scholar]
- 60.Li, Y., T. Bourlet, L. Andreoletti, J. F. Mosnier, T. Peng, Y. Yang, L. C. Archard, B. Pozzetto, and H. Zhang. 2000. Enteroviral capsid protein VP1 is present in myocardial tissues from some patients with myocarditis or dilated cardiomyopathy. Circulation 101:231-234. [DOI] [PubMed] [Google Scholar]
- 61.Liu, P. P., and J. W. Mason. 2001. Advances in the understanding of myocarditis. Circulation 104:1076-1082. [DOI] [PubMed] [Google Scholar]
- 62.Longson, M., F. M. Cole, and D. Davies. 1969. Isolation of a coxsackie virus group B, type 5, from the heart of a fatal case of myocarditis in an adult. J. Clin. Pathol. 22:654-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lundquist, R. E., M. Sullivan, and J. V. Maizel, Jr. 1979. Characterization of a new isolate of poliovirus defective interfering particles. Cell 18:759-769. [DOI] [PubMed] [Google Scholar]
- 64.Lyons, T., K. E. Murray, A. W. Roberts, and D. J. Barton. 2001. Poliovirus 5′-terminal cloverleaf RNA is required in cis for VPg uridylylation and the initiation of negative-strand RNA synthesis. J. Virol. 75:10696-10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.MacLennan, C., G. Dunn, A. P. Huissoon, D. S. Kumararatne, J. Martin, P. O'Leary, R. A. Thompson, H. Osman, P. Wood, P. Minor, D. J. Wood, and D. Pillay. 2004. Failure to clear persistent vaccine-derived neurovirulent poliovirus infection in an immunodeficient man. Lancet 363:1509-1513. [DOI] [PubMed] [Google Scholar]
- 66.Martinez, M. A., C. Carrillo, F. Gonzalez-Candelas, A. Moya, E. Domingo, and F. Sobrino. 1991. Fitness alteration of foot-and-mouth disease virus mutants: measurement of adaptability of viral quasispecies. J. Virol. 65:3954-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 68.McKnight, K. L., and S. M. Lemon. 1998. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA 4:1569-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mena, I., C. M. Perry, S. Harkins, F. Rodriguez, J. Gebhard, and J. L. Whitton. 1999. The role of B lymphocytes in coxsackievirus B3 infection. Am. J. Pathol. 155:1205-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meyer, R. G., M. L. Meyer-Ficca, H. Kaiser, H. C. Selinka, R. Kandolf, and J. H. Kupper. 2004. Plasmid-based generation of recombinant coxsackievirus B3 particles carrying capsid gene replacement replicons. Virus Res. 104:17-26. [DOI] [PubMed] [Google Scholar]
- 71.Misbah, S. A., G. P. Spickett, P. C. Ryba, J. M. Hockaday, J. S. Kroll, C. Sherwood, J. B. Kurtz, E. R. Moxon, and H. M. Chapel. 1992. Chronic enteroviral meningoencephalitis in agammaglobulinemia: case report and literature review. J. Clin. Immunol. 12:266-270. [DOI] [PubMed] [Google Scholar]
- 72.Modlin, J. F., and H. A. Rotbart. 1997. Group B coxsackie disease in children. Curr. Top. Microbiol. Immunol. 223:53-80. [DOI] [PubMed] [Google Scholar]
- 72a.Moine, H., B. Ehresmann, C. Ehresmann, and P. Romby. 1998. Probing RNA structure and function in solution, p. 77-116. In R. W. Simons and M. Grunberg-Manago (ed.), RNA structure and function. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 73.Molla, A., K. S. Harris, A. V. Paul, S. H. Shin, J. Mugavero, and E. Wimmer. 1994. Stimulation of poliovirus proteinase 3Cpro-related proteolysis by the genome-linked protein VPg and its precursor 3AB. J. Biol. Chem. 269:27015-27020. [PubMed] [Google Scholar]
- 74.Muckelbauer, J. K., M. Kremer, I. Minor, G. Diana, F. J. Dutko, J. Groarke, D. C. Pevear, and M. G. Rossmann. 1995. The structure of coxsackievirus B3 at 3.5 A resolution. Structure 3:653-667. [DOI] [PubMed] [Google Scholar]
- 75.Murray, K. E., B. P. Steil, A. W. Roberts, and D. J. Barton. 2004. Replication of poliovirus RNA with complete internal ribosome entry site deletions. J. Virol. 78:1393-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nicholson, R., J. Pelletier, S. Y. Le, and N. Sonenberg. 1991. Structural and functional analysis of the ribosome landing pad of poliovirus type 2: in vivo translation studies. J. Virol. 65:5886-5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nomoto, A., N. Kitamura, F. Golini, and E. Wimmer. 1977. The 5′-terminal structures of poliovirion RNA and poliovirus mRNA differ only in the genome-linked protein VPg. Proc. Natl. Acad. Sci. USA 74:5345-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Novak, J. E., and K. Kirkegaard. 1991. Improved method for detecting poliovirus negative strands used to demonstrate specificity of positive-strand encapsidation and the ratio of positive to negative strands in infected cells. J. Virol. 65:3384-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nugent, C. I., K. L. Johnson, P. Sarnow, and K. Kirkegaard. 1999. Functional coupling between replication and packaging of poliovirus replicon RNA. J. Virol. 73:427-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ohlenschläger, O., J. Wohnert, E. Bucci, S. Seitz, S. Hafner, R. Ramachandran, R. Zell, and M. Gorlach. 2004. The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf RNA and its interaction with the proteinase 3C. Structure (Camb.) 12:237-248. [DOI] [PubMed] [Google Scholar]
- 81.Pallansch, M. A. 1997. Coxsackievirus B epidemiology and public health concerns. Curr. Top. Microbiol. Immunol. 223:13-30. [DOI] [PubMed] [Google Scholar]
- 82.Pallansch, M. A., and R. P. Roos. 2001. Enteroviruses: polioviruses, coxsackieviruses, echoviruses and newer enteroviruses, p. 723-775. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 83.Palmenberg, A. C. 1990. Proteolytic processing of picornaviral polyprotein. Annu. Rev. Microbiol. 44:603-623. [DOI] [PubMed] [Google Scholar]
- 84.Parsley, T. B., J. S. Towner, L. B. Blyn, E. Ehrenfeld, and B. L. Semler. 1997. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124-1134. [PMC free article] [PubMed] [Google Scholar]
- 85.Paul, A. V., J. H. van Boom, D. Filippov, and E. Wimmer. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280-284. [DOI] [PubMed] [Google Scholar]
- 86.Paul, A. V., J. Yin, J. Mugavero, E. Rieder, Y. Liu, and E. Wimmer. 2003. A “slide-back” mechanism for the initiation of protein-primed RNA synthesis by the RNA polymerase of poliovirus. J. Biol. Chem. 278:43951-43960. [DOI] [PubMed] [Google Scholar]
- 87.Pauschinger, M., A. Doerner, U. Kuehl, P. L. Schwimmbeck, W. Poller, R. Kandolf, and H. P. Schultheiss. 1999. Enteroviral RNA replication in the myocardium of patients with left ventricular dysfunction and clinically suspected myocarditis. Circulation 99:889-895. [DOI] [PubMed] [Google Scholar]
- 88.Peng, T., Y. Li, Y. Yang, C. Niu, P. Morgan-Capner, L. C. Archard, and H. Zhang. 2000. Characterization of enterovirus isolates from patients with heart muscle disease in a selenium-deficient area of China. J. Clin. Microbiol. 38:3538-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pestova, T. V., C. U. Hellen, and E. Wimmer. 1991. Translation of poliovirus RNA: role of an essential cis-acting oligopyrimidine element within the 5′ nontranslated region and involvement of a cellular 57-kilodalton protein. J. Virol. 65:6194-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Porter, D. C., D. C. Ansardi, and C. D. Morrow. 1995. Encapsidation of poliovirus replicons encoding the complete human immunodeficiency virus type 1 gag gene by using a complementation system which provides the P1 capsid protein in trans. J. Virol. 69:1548-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Porter, D. C., D. C. Ansardi, J. Wang, S. McPherson, Z. Moldoveanu, and C. D. Morrow. 1998. Demonstration of the specificity of poliovirus encapsidation using a novel replicon which encodes enzymatically active firefly luciferase. Virology 243:1-11. [DOI] [PubMed] [Google Scholar]
- 92.Rabausch-Starz, I., A. Schwaiger, K. Grunewald, H. K. Muller-Hermelink, and N. Neu. 1994. Persistence of virus and viral genome in myocardium after coxsackievirus B3-induced murine myocarditis. Clin. Exp. Immunol. 96:69-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Racaniello, V. R. 2001. Picornaviridae: the viruses and their replication. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 94.Ramsingh, A. I. 1997. Coxsackieviruses and pancreatitis. Front. Biosci. 2:e53-e62. [DOI] [PubMed] [Google Scholar]
- 95.Reetoo, K. N., S. A. Osman, S. J. Illavia, C. L. Cameron-Wilson, J. E. Banatvala, and P. Muir. 2000. Quantitative analysis of viral RNA kinetics in coxsackievirus B3-induced murine myocarditis: biphasic pattern of clearance following acute infection, with persistence of residual viral RNA throughout and beyond the inflammatory phase of disease. J. Gen. Virol. 81:2755-2762. [DOI] [PubMed] [Google Scholar]
- 96.Rivera, V. M., J. D. Welsh, and J. V. Maizel, Jr. 1988. Comparative sequence analysis of the 5′ noncoding region of the enteroviruses and rhinoviruses. Virology 165:42-50. [DOI] [PubMed] [Google Scholar]
- 97.Rothberg, P. G., T. J. Harris, A. Nomoto, and E. Wimmer. 1978. O4-(5′-uridylyl)tyrosine is the bond between the genome-linked protein and the RNA of poliovirus. Proc. Natl. Acad. Sci. USA 75:4868-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Roux, L., and J. J. Holland. 1979. Role of defective interfering particles of Sendai virus in persistent infections. Virology 93:91-103. [DOI] [PubMed] [Google Scholar]
- 99.Roux, L., A. E. Simon, and J. J. Holland. 1991. Effects of defective interfering viruses on virus replication and pathogenesis in vitro and in vivo. Adv. Virus Res. 40:181-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sharma, N., B. J. O'Donnell, and J. B. Flanegan. 2005. 3′-Terminal sequence in poliovirus negative-strand templates is the primary cis-acting element required for VPgpUpU-primed positive-strand initiation. J. Virol. 79:3565-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Skinner, M. A., V. R. Racaniello, G. Dunn, J. Cooper, P. D. Minor, and J. W. Almond. 1989. New model for the secondary structure of the 5′ non-coding RNA of poliovirus is supported by biochemical and genetic data that also show that RNA secondary structure is important in neurovirulence. J. Mol. Biol. 207:379-392. [DOI] [PubMed] [Google Scholar]
- 102.Stern, S., D. Moazed, and H. F. Noller. 1988. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 164:481-489. [DOI] [PubMed] [Google Scholar]
- 103.Takegami, T., R. J. Kuhn, C. W. Anderson, and E. Wimmer. 1983. Membrane-dependent uridylylation of the genome-linked protein VPg of poliovirus. Proc. Natl. Acad. Sci. USA 80:7447-7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tam, P. E., and R. P. Messner. 1999. Molecular mechanisms of coxsackievirus persistence in chronic inflammatory myopathy: viral RNA persists through formation of a double-stranded complex without associated genomic mutations or evolution. J. Virol. 73:10113-10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tam, P. E., M. L. Weber-Sanders, and R. P. Messner. 2003. Multiple viral determinants mediate myopathogenicity in coxsackievirus B1-induced chronic inflammatory myopathy. J. Virol. 77:11849-11854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Teterina, N. L., D. Egger, K. Bienz, D. M. Brown, B. L. Semler, and E. Ehrenfeld. 2001. Requirements for assembly of poliovirus replication complexes and negative-strand RNA synthesis. J. Virol. 75:3841-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tracy, S. 1984. A comparison of genomic homologies among the coxsackievirus B group: use of fragments of the cloned coxsackievirus B3 genome as probes. J. Gen. Virol. 65:2167-2172. [DOI] [PubMed] [Google Scholar]
- 108.Tracy, S., N. M. Chapman, and Z. Tu. 1992. Coxsackievirus B3 from an infectious cDNA copy of the genome is cardiovirulent in mice. Arch. Virol. 122:399-409. [DOI] [PubMed] [Google Scholar]
- 109.Tracy, S., K. M. Drescher, N. M. Chapman, K. S. Kim, S. D. Carson, S. Pirruccello, P. H. Lane, J. R. Romero, and J. S. Leser. 2002. Toward testing the hypothesis that group B coxsackieviruses (CVB) trigger insulin-dependent diabetes: inoculating nonobese diabetic mice with CVB markedly lowers diabetes incidence. J. Virol. 76:12097-12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tracy, S., K. Hofling, S. Pirruccello, P. H. Lane, S. M. Reyna, and C. J. Gauntt. 2000. Group B coxsackievirus myocarditis and pancreatitis: connection between viral virulence phenotypes in mice. J. Med. Virol. 62:70-81. [DOI] [PubMed] [Google Scholar]
- 111.Trono, D., R. Andino, and D. Baltimore. 1988. An RNA sequence of hundreds of nucleotides at the 5′ end of poliovirus RNA is involved in allowing viral protein synthesis. J. Virol. 62:2291-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tu, Z., N. M. Chapman, G. Hufnagel, S. Tracy, J. R. Romero, W. H. Barry, L. Zhao, K. Currey, and B. Shapiro. 1995. The cardiovirulent phenotype of coxsackievirus B3 is determined at a single site in the genomic 5′ nontranslated region. J. Virol. 69:4607-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wagoner, L. E., L. Zhao, D. K. Bishop, S. Chan, S. Xu, and W. H. Barry. 1996. Lysis of adult ventricular myocytes by cells infiltrating rejecting murine cardiac allografts. Circulation 93:111-119. [DOI] [PubMed] [Google Scholar]
- 114.Wessely, R., A. Henke, R. Zell, R. Kandolf, and K. U. Knowlton. 1998. Low-level expression of a mutant coxsackieviral cDNA induces a myocytopathic effect in culture: an approach to the study of enteroviral persistence in cardiac myocytes. Circulation 98:450-457. [DOI] [PubMed] [Google Scholar]
- 115.Wessely, R., K. Klingel, L. F. Santana, N. Dalton, M. Hongo, W. Jonathan Lederer, R. Kandolf, and K. U. Knowlton. 1998. Transgenic expression of replication-restricted enteroviral genomes in heart muscle induces defective excitation-contraction coupling and dilated cardiomyopathy. J. Clin. Investig. 102:1444-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Why, H. J., B. T. Meany, P. J. Richardson, E. G. Olsen, N. E. Bowles, L. Cunningham, C. A. Freeke, and L. C. Archard. 1994. Clinical and prognostic significance of detection of enteroviral RNA in the myocardium of patients with myocarditis or dilated cardiomyopathy. Circulation 89:2582-2589. [DOI] [PubMed] [Google Scholar]
- 117.Willian, S., S. Tracy, N. Chapman, S. Leser, J. Romero, B. Shapiro, and K. Currey. 2000. Mutations in a conserved enteroviral RNA oligonucleotide sequence affect positive strand viral RNA synthesis. Arch. Virol. 145:2061-2086. [DOI] [PubMed] [Google Scholar]
- 118.Xiang, W., K. S. Harris, L. Alexander, and E. Wimmer. 1995. Interaction between the 5′-terminal cloverleaf and 3AB/3CDpro of poliovirus is essential for RNA replication. J. Virol. 69:3658-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zell, R., K. Sidigi, E. Bucci, A. Stelzner, and M. Gorlach. 2002. Determinants of the recognition of enteroviral cloverleaf RNA by coxsackievirus B3 proteinase 3C. RNA 8:188-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zell, R., K. Sidigi, A. Henke, J. Schmidt-Brauns, E. Hoey, S. Martin, and A. Stelzner. 1999. Functional features of the bovine enterovirus 5′-non-translated region. J. Gen. Virol. 80:2299-2309. [DOI] [PubMed] [Google Scholar]
- 121.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]