Abstract
The most highly expressed protein in the productive life cycle of human papillomaviruses (HPVs) is E1∧E4, but its function is not well understood. To investigate the role of E1∧E4, we undertook a genetic analysis in the context of the complete HPV type 31 (HPV31) genome. A mutant HPV31 genome (E4M9) was constructed that contained a stop codon in the E4 open reading frame at amino acid 9 and was silent in the overlapping E2 coding sequence. Wild-type and mutant genomes were transfected into normal human foreskin keratinocytes (HFKs) and selected for drug resistance, and pooled cultures were examined for effects of E1∧E4 on viral functions. Southern blot analyses of transfected HFKs demonstrated that cells carrying the E4M9 mutant genomes were maintained as episomes at copy numbers similar to those in keratinocytes transfected with wild-type HPV31. Both sets of cells grew at similar rates, exhibited comparable extensions of life spans, and had equivalent levels of early transcripts. Following suspension of the cells in a semisolid medium, differentiation-dependent genome amplification and late gene expression were significantly decreased in cells maintaining the E4M9 mutant genome compared to those with wild-type HPV31. One explanation for these effects could be a reduction in the number of cells harboring mutant genomes that enter S phase upon differentiation. An analysis of cells containing E4M9 mutant genomes in organotypic raft cultures indicated a reduction in bromodeoxyuridine incorporation in differentiated suprabasal cells compared to that seen in wild-type rafts. Our results indicate that the HPV31 E1∧E4 protein plays a significant role in promoting HPV genome amplification and S phase maintenance during differentiation.
Human papillomaviruses (HPVs) are small double-stranded DNA viruses that infect cutaneous and mucosal epithelial tissues (22). Infection by HPVs induces a range of responses, from benign papillomas to invasive carcinoma. The HPV types that infect the genital epithelia are among the best characterized and are categorized as either high- or low-risk viruses based on their association with cervical cancer. The high-risk HPV types include types 16, 18, 31, 33, and 45, and these viral sequences are found in 99% of cervical carcinomas (33, 53). Low-risk HPVs are not associated with the development of malignancy but rather with benign genital warts and include HPV viral types 6 and 11 (5, 52, 53).
Human papillomaviruses infect cells in the basal layer of stratified epithelium. Infection occurs through microlesions of the skin that expose basal cells to viral entry. Following entry, viral genomes are maintained as nuclear episomes at approximately 50 to 100 copies/cell (22). As infected basal cells divide, HPV episomes are coordinately replicated with cellular DNA and distributed to daughter cells. Following cell division, one daughter cell detaches from the basement membrane and migrates towards the suprabasal layers of the epithelium. This process occurs coordinately with cellular differentiation and induces the amplification of viral genomes to several thousand copies per cell. The expression of HPV late genes is coincident with DNA amplification and is followed by the assembly and release of progeny virions (19, 24).
During the productive life cycle, HPV open reading frames are expressed from polycistronic messages that are initiated predominantly from two promoters. For HPV31, early transcripts, which encode the viral proteins E1, E2, E6, E7, E1∧E4, and E5, are expressed from the early promoter p97 (24). The viral proteins E1 and E2 are involved in viral replication as well as the regulation of early transcription. E1 binds to the origin of replication and exhibits ATPase as well as helicase activity (23, 49). E2 acts by forming a complex with E1 and facilitating its binding to the origin of viral replication (18, 32, 49). In addition, the E2 protein acts as a transcription factor that positively and negatively regulates early gene expression by binding to specific E2 recognition sites within the upstream regulatory region (URR) (2, 21). For the high-risk HPVs, E6 and E7 function as oncoproteins that target the cell cycle regulators p53 and Rb, respectively. The E6 protein facilitates binding of the tumor suppressor p53 to the E3 ubiquitin ligase, E6-AP, resulting in polyubiquitination of p53 and its degradation by proteasomes (45, 46, 51). In addition, E6 has been shown to activate the transcription of hTERT, the catalytic subunit of telomerase (25). The E7 protein binds the tumor suppressor Rb and blocks its binding to E2F transcription factors, resulting in the constitutive activation of S-phase genes (1, 13, 29, 34).
Upon epithelial differentiation, additional HPV genes are expressed from transcripts initiated at the late promoter, which for HPV type 31 (HPV31) is designated p742 (24). These differentiation-specific late genes include E5 and E1∧E4 as well as the viral capsid proteins L1 and L2. The E5 protein has been shown to have weak transforming capabilities in vitro (27, 47) and to play a role in supporting HPV late functions (16, 20). Transcripts encoding E1∧E4 are generated as a result of RNA splicing and consist of the first five codons for E1 fused to the open reading frame of E4 (6, 36). The E1∧E4 open reading frame is present in most early and late transcripts, but high-level synthesis is restricted to differentiated suprabasal cells (10, 31, 39). Initial studies indicated that E1∧E4 induces the collapse of the cytokeratin network when it is overexpressed in transient assays, and they suggested a role for the protein in facilitating viral egress (6, 9, 41). However, no such collapse is seen in lesions induced by many high-risk HPV types (10, 11, 40). In HPV lesions, the expression of E1∧E4 occurs in the upper layers of stratified epithelia, coordinating with the onset of genome amplification but preceding the expression of L1 (10, 37). This pattern of E1∧E4 expression suggests a possible role in regulating late viral functions. Many studies of E1∧E4 have relied on heterologous expression systems, and few have examined the effects of E1∧E4 in the context of the entire HPV genome. In this study, we have investigated the role of E1∧E4 in the life cycle of high-risk HPVs by use of a genetic analysis. Our studies indicate that E1∧E4 proteins play important roles in promoting the differentiation-dependent productive phase of the viral life cycle.
MATERIALS AND METHODS
Plasmids.
The plasmid pBRmod-HPV31 contains the HPV31 genome inserted into the EcoRI site of a pBR322-derived plasmid (19). The mutant plasmids E4M4 and E4M9 were constructed in the pBRmod background by site-directed PCR mutagenesis. The E4M4 mutant contains a nucleotide substitution in the E4 open reading frame at nucleotide 39 (T→A) which generates a stop codon (UAA) after the first four amino acids of the E4 coding sequence. The E4M9 mutant contains a nucleotide substitution in the E4 open reading frame at nucleotide 53 (T→A) and places a stop codon (UAG) after the first nine amino acids of the E4 sequence. All wild-type and mutant genomes were sequenced prior to and posttransfection to confirm the wild-type and E1∧E4 mutant sequences.
Cell culture.
Normal human foreskin keratinocytes (HFKs) were isolated from neonatal foreskin as described previously (44) and were maintained in keratinocyte growth medium supplemented with bovine pituitary extract (Clonetics). HPV31-transfected HFKs were grown in serum-containing medium (E medium) supplemented with 5 ng/ml mouse epidermal growth factor (Collaborative Biomedics) in the presence of mitomycin C-treated J2 3T3 fibroblast feeders. Prior to Southern and Northern analyses, monolayer keratinocytes were treated with 0.5 mM EDTA in phosphate-buffered saline (PBS) to remove J2 fibroblasts.
Transfection of HFKs.
Ten micrograms of pBRmod-HPV31 wild-type and pBRmod-HPV31 E1∧E4 mutant plasmids were digested with EcoRI to isolate HPV31 genomes. Restriction digest mixtures were heat inactivated, and fragments were religated with T4 DNA ligase (10 U/900 μl) at 14°C for approximately 15 h. DNAs were then precipitated with isopropyl alcohol and resuspended in 10 mM Tris-1 mM EDTA (pH 7.5). One microgram of the religated DNA was cotransfected with one microgram of the neomycin resistance plasmid pSV2Neo by use of the FuGene reagent (Roche) as described by the manufacturer. At approximately 24 h posttransfection, transfected keratinocytes were trypsinized and replated in E medium in the presence of J2 fibroblasts. A green fluorescent protein reporter plasmid, pEGFPN.1, and the pSV2Neo plasmid were transfected into separate sets of HFKs to determine their transfection efficiencies. At 2 days posttransfection, transfected keratinocytes were selected for 8 days with G418 (Invitrogen) as previously described (16).
Differentiation of keratinocytes in organotypic raft cultures.
Untransfected HFKs, along with HFKs carrying wild-type HPV31 and HPV31 E1∧E4 mutant genomes, were grown in organotypic raft cultures as previously described (30). Briefly, cells were plated onto a collagen matrix containing J2 fibroblasts, allowed to grow to confluence, and transferred to a metal grid placed on an air-liquid interface in the presence of E medium for differentiation. Raft cultures were harvested after 13 days, fixed in 4% paraformaldehyde, paraffin embedded, sectioned, and stained with hematoxylin and eosin for morphological analysis or examined by immunohistochemistry. For the identification of cells that were actively synthesizing DNA, 20 μM bromodeoxyuridine (BrdU) was added to the raft culture medium 12 h prior to the harvest of rafts.
Differentiation of keratinocytes in semisolid medium.
Cells containing wild-type HPV31 and HPV31 E1∧E4 mutant genomes were grown in monolayer cultures before being suspended in methylcellulose. Prior to suspension, fibroblasts were removed with 0.5 mM EDTA in PBS, and keratinocytes were then trypsinized and suspended in semisolid medium containing 1.5% methylcellulose as previously described (16). Approximately 2 × 106 keratinocytes were trypsinized, plated onto a 10-cm petri dish containing 25 ml of 1.5% methylcellulose, and incubated at 37°C in a CO2-containing humidified incubator for 24 and 48 h. Prior to harvest, the cells were washed by centrifugation four times with PBS. Harvested cells were used for Southern and Northern analyses.
Immunofluorescence with organotypic raft cultures.
E1∧E4 expression in organotypic raft cultures was determined by immunohistochemistry using paraffin-embedded raft sections. The paraffin-embedded tissue sections were placed on silanized slides and incubated at 50°C for 30 min. After incubation, the slides were washed three times in xylene to remove the paraffin and then rehydrated with ethanol. The slides were then heated in 10 mM sodium citrate, pH 6.0, at 95°C for 20 min followed by a 20-minute incubation at room temperature. A previously described E1∧E4 rabbit polyclonal antibody (40) was used to detect the expression of E1∧E4. Staining with 300 mM 4′,6′-diamidino-2-phenylindole dihydrochloride (DAPI) was used to identify nuclei. BrdU incorporation was detected with a monoclonal BrdU antibody (Roche).
FISH.
Fluorescence in situ hybridization (FISH) analysis was performed as previously described (12). Briefly, slides were baked and deparaffinized by three xylene washes followed by two ethanol washes. The slides were then heated in 100 mM Tris base-50 mM EDTA, pH 7.0, and proteins were digested with Digest All −3 pepsin (Zymed) with a 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) wash before and after digestion. The slides were dehydrated with a series of ethanol washes at 75%, 85%, 95%, and 100% and then air dried. HPV31 DNA was labeled with digoxigenin by use of the Bioprime DNA labeling system (Invitrogen), added to slides, denatured for 3 min at 85°C, and incubated overnight at 37°C. Labeled DNAs were detected with a fluorescein isothiocyanate-anti-digoxigenin antibody at 1:400 (Zymed), and nuclei were identified following labeling with 300 mM DAPI.
Southern blot analysis.
Prior to the cell harvest for DNA isolation, J2 fibroblasts were removed by EDTA treatment. Monolayer and methylcellulose-treated keratinocytes containing wild-type HPV31 and HPV31 E1∧E4 mutant genomes were lysed in lysis buffer (400 mM NaCl, 10 mM Tris-HCl, and 10 mM EDTA). Cell extracts were incubated with 50 μg/ml RNase A as well as proteinase K sequentially to remove residual RNAs and proteins, followed by phenol-chloroform extraction. DNAs were quantitated by spectrophotometry, and 5 μg of total DNA was digested with restriction enzymes. DpnI was added to all samples to digest any residual input DNA. Samples were run in a 0.8% agarose gel at 70 V for approximately 14 h. DNA transfer was performed with DuPont Gene Screen Plus nylon membranes (NEN Research Products) using vacuum transfer according to the manufacturer's protocols. The HPV31 genome was isolated from the pBRmod-HPV31 wild-type plasmid by restriction digestion with the enzyme EcoRI for use as a probe. The probe was labeled with [α-32P]dCTP by the use of Ready-To-Go DNA labeling beads (Amersham Biosciences). After being labeled, the probe was purified through Probe Quant G-50 microcolumns (Amersham Biosciences) prior to use. Nylon membranes were placed in prehybridization/hybridization solution (4× SSC, 5× Denhardt's solution, 10% dextran sulfate, 1% sodium dodecyl sulfate [SDS], 50% formamide, and 0.1 mg/ml denatured salmon sperm DNA) for approximately 1 h at 42°C and hybridized to the labeled probe at 42°C overnight. After hybridization, the membranes were washed twice for 15 min each in 2× SSC-0.1% SDS, 0.5× SSC-0.1% SDS, and 0.1× SSC-1% SDS and once for 30 min at 50°C in 0.1× SSC-1% SDS. The bands were visualized by autoradiography, and quantitative analysis was performed with a PhosphorImager (Molecular Dynamics).
Northern blot analysis.
The isolation of total RNAs from cells containing wild-type HPV31 and HPV31 E1∧E4 mutant genomes grown in monolayer or methylcellulose-treated keratinocytes was performed by use of the TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Ten micrograms of total RNA was analyzed in a 1.0% agarose gel containing 2.2 M formaldehyde and 1× morpholinepropanesulfonic acid (MOPS) buffer (10× MOPS [0.2 M], 50 mM sodium acetate, 10 mM EDTA). RNAs were quantitated with a spectrophotometer, and gels were stained for rRNA to confirm equal loading. RNAs were then transferred onto a Zeta-Probe nylon membrane (Bio-Rad). Following UV cross-linking, nylon membranes were placed in a prehybridization/hybridization solution (1 mM EDTA, 0.5 M Na2HPO4, and 7% SDS) for 10 min at 65°C. The HPV31 genome was used as a probe in the same manner as that described for Southern blot analysis. Purified labeled HPV31 probes were denatured and added to prehybridized nylon membranes in the fresh prehybridization/hybridization solution described above and incubated at 65°C overnight. After hybridization, the nylon membranes were washed twice with 2× SSC-10% SDS for 5 min at room temperature and once with 0.2× SSC-1% SDS for 15 min at 55°C. Hybridization was visualized by autoradiography, and quantitative analysis was performed with a PhosphorImager (Molecular Dynamics).
RESULTS
Generation of HPV31 E1∧E4 mutant genomes.
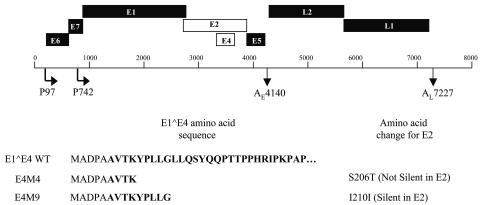
In order to study the role of the E1∧E4 protein in the productive life cycle of human papillomaviruses, we constructed two mutant HPV31 genomes (Fig. 1). The sites where a translation termination codon could be inserted in E4 were limited because the E4 open reading frame (ORF) overlaps that of E2. Given these limitations, we created two mutations, one of which was silent in E2 and one of which disturbed the E2 amino acid sequence. The first mutant, E4M4, contained a translation termination codon inserted after the first four amino acid codons of the E4 open reading frame. This mutation also altered a serine to a threonine in the E2 amino acid sequence (S206T) (Fig. 1). The second mutant genome, called E4M9, contained a translation termination codon after nine codons of the E4 coding sequence. This mutation is silent in E2 (I210I). Normal HFKs were transfected with wild-type HPV31 as well as the two HPV31 E1∧E4 mutant genomes along with a drug-selectable marker. A total of four different isolates of HFKs from different donors were used for this study. Following transfection and selection, pooled cultures were expanded. DNAs were extracted from transfected cells, and the presence of the mutations was confirmed by sequencing.
FIG. 1.
Diagram of HPV31 genome showing viral open reading frames (ORF) with early and late promoters and polyadenylation sites. The E4 ORF and the overlapping E2 ORF are shown as open boxes. The N-terminal amino acid sequences of wild-type E1∧E4 as well as of the E4M4 and E4M9 mutant forms are indicated at the bottom of the diagram. For E4M4, a translation termination codon was inserted after the first four amino acids of the E4 ORF, while for E4M9, a stop codon was inserted after nine amino acids. The E4M4 mutation introduced a change in the E2 coding sequence from a serine to a threonine, while the E4M9 mutation was silent in E2. The E1∧E4 protein consists of the first five amino acids of E1 fused to the E4 open reading frame (bold letters).
Keratinocytes transfected with HPV31 E1∧E4 mutant genomes maintain episomes.
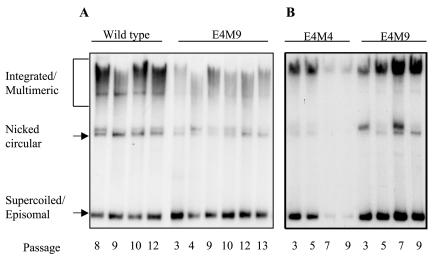
We first sought to investigate if the abrogation of E1∧E4 expression had any effect on episomal maintenance of the viral genome or on the ability of undifferentiated keratinocytes to proliferate. To determine if the HPV31 E1∧E4 mutant genomes could be stably maintained as episomes, we isolated DNAs and performed Southern blot analysis as a function of the passage number. Equal amounts of total DNA isolated from monolayer cultures of keratinocytes containing wild-type HPV31 and HPV31 E1∧E4 mutant genomes were digested with the restriction enzyme DpnI to degrade any residual input DNA. As shown in Fig. 2, Southern analysis revealed three different migrating forms of HPV genomes, namely, integrated/multimeric, nicked circular, and supercoiled or episomal forms. HFKs containing E4M9 mutant genomes were found to maintain viral DNA as episomes at levels similar to that of cells containing wild-type HPV31 for both early and late passages (Fig. 2A and B). Keratinocytes harboring E4M4 mutant genomes maintained viral episomes at levels comparable to that of cells harboring the wild type for early passages, but upon further passaging, a majority of integrated copies were observed (Fig. 2B). The loss of episomal copies of E4M4 mutant genomes at later passages was consistently observed in multiple independent experiments using keratinocytes isolated from four different hosts. Since the E2 coding sequences were altered in the E4M4 mutant genome, we cannot exclude the possibility that these effects may have been due to the alteration of the E2 amino acid sequence and not that of E1∧E4. However, this alteration in the E2 coding sequence did not result in the loss of any previously documented activity of E2, such as its effects on gene expression and short-term replication. Given the phenotype with E4M4, we focused our subsequent studies primarily on the E4M9 mutant genome. We observed no differences in proliferation rates among wild-type, E4M9 mutant, and early-passage E4M4 mutant cell lines or in the expression of early transcripts (data not shown). In a small number of experiments, cells transfected with the E4M9 mutant genome exhibited reduced levels of viral DNA, but overall we believe that there were no significant differences with respect to wild-type copy numbers in undifferentiated cells.
FIG. 2.
Southern analysis of keratinocytes stably transfected with wild-type HPV31, E4M4 mutant, or E4M9 mutant genomes. Equal amounts of total DNA were isolated from keratinocytes harboring wild-type, E4M4, and E4M9 genomes at various passages and were examined by Southern analysis using a linearized HPV31 DNA genome as a probe. Two representative Southern blots from two different HFK cell lines are shown. (A) Wild type- and E4M9-transfected keratinocytes were able to stably replicate HPV genomes as episomes at various passages. (B) In contrast, E4M4-transfected keratinocytes were unable to be stably maintained as episomes at late passages. Three different migrating forms of HPV are indicated (integrated/multimeric, nicked circular, and supercoiled/episomal).
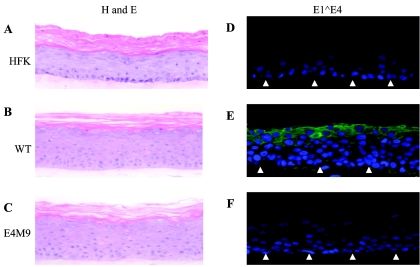
Raft cultures of cells with E1∧E4 mutant genomes exhibit similar morphologies to cells with wild-type HPV31.
Since the E4M9 mutant genomes were stably maintained as episomes in undifferentiated cells, we next investigated if there were any significant differences in the differentiation-dependent phase of the life cycle. For these studies, we first analyzed the ability of cells to differentiate in organotypic raft cultures. Keratinocytes that maintained HPV31 wild-type and E4M9 genomes and normal HFKs were grown in raft cultures for 13 days and then harvested, and tissue cross sections were stained with hematoxylin and eosin. Untransfected HFKs exhibited a typical pattern of morphological differentiation, including a reduction in the number of nuclei upon stratification (Fig. 3A) (19). In contrast, cell lines carrying wild-type HPV31 and the E4M9 mutant maintained their nuclei in the upper stratified layers and exhibited a thickening of the stratified layers compared to untransfected HFKs (Fig. 3B and C). In addition, we observed no significant differences in the expression of cellular markers of differentiation such as involucrin and keratin 10 between the two sets of HPV-positive cells (data not shown). Our studies suggest that E1∧E4 does not exert a significant effect on the differentiation program of HPV-containing keratinocytes. Interestingly, we consistently observed a reduction in the number of suprabasal nuclei present in raft cultures of cells with E4M9 mutant genomes compared to cells with wild-type genomes (Fig. 3E and F).
FIG. 3.
Differentiation of keratinocytes containing wild-type HPV 31 or E4M9 mutant genomes in organotypic rafts. Raft cultures were grown for 13 days, harvested, and fixed in paraformaldehyde. Tissue cross sections were stained with hematoxylin and eosin (A, B, and C) or examined by immunohistochemistry using a polyclonal antibody to HPV31 E1∧E4 (green staining; D, E, and F). The blue staining in panels D, E, and F corresponds to nuclear staining with DAPI. The closed arrows represent the basal layer.
E1∧E4 is a late protein that is primarily expressed in differentiated cells. To confirm that E4M9 mutant cell lines did not express the E1∧E4 protein, we performed immunohistochemistry on cross sections of raft cultures by using an E1∧E4-specific rabbit polyclonal antibody. As expected, no staining was observed in sections from untransfected HFK or E4M9 raft cultures (Fig. 3D and F). In contrast, organotypic rafts of wild-type HPV31-infected cells exhibited E1∧E4 staining in the upper stratified layers (Fig. 3E), in agreement with the results of other studies examining E1∧E4 expression in HPV-induced lesions and raft cultures (10, 40).
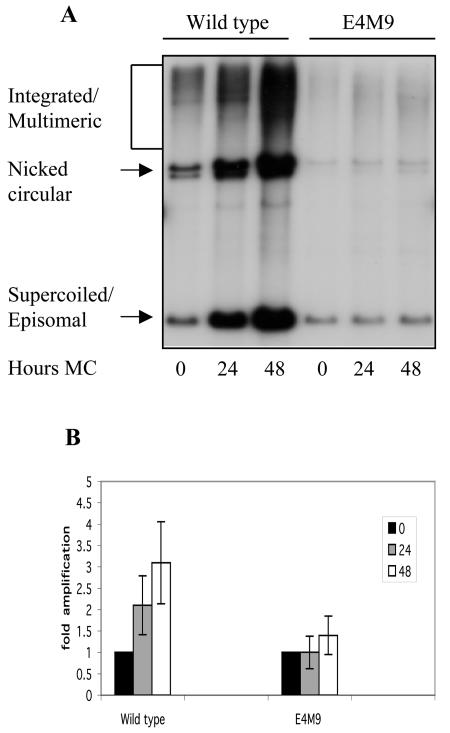
Keratinocytes that maintain E1∧E4 mutant genomes are impaired in differentiation-dependent amplification.
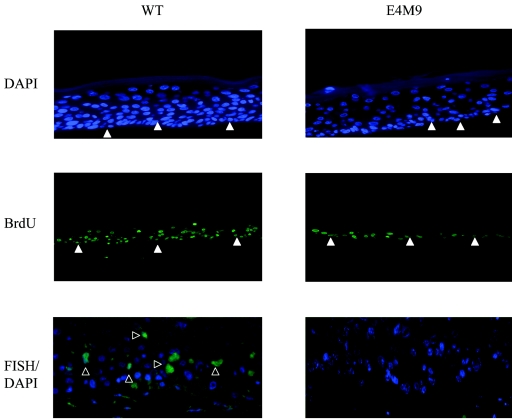
We next examined if the loss of E1∧E4 function had an effect on the differentiation-dependent amplification of viral DNA. Since it is difficult to utilize raft cultures to quantitate levels of amplification, we used suspension in 1.5% methylcellulose to induce differentiation. Previous studies have demonstrated that the resuspension of HPV-positive cells in methylcellulose induces differentiation-dependent late viral functions such as DNA amplification and late gene expression, including the synthesis of E1∧E4 proteins (16, 24, 43, 44). For these studies, wild-type HPV31- and E4M9 mutant-infected keratinocytes were suspended in 1.5% methylcellulose for 24 and 48 h and DNAs were harvested. Southern blot analysis was performed on the total DNAs isolated from these cells to determine the degree of amplification of viral genomes. The E4M9 mutant-infected keratinocytes were found to exhibit significantly less genome amplification than cells with wild-type genomes at both the 24- and 48-h time points (Fig. 4A). We analyzed data from 12 different experiments using four sets of cells derived from different host keratinocytes and observed similar results. Keratinocytes harboring wild-type genomes exhibited 2.1 ± 0.7-fold amplification at 24 h and approximately 3.1 ± 1-fold amplification at 48 h when the data were normalized to those for monolayer cells (Fig. 4B). These levels of amplification are consistent with those observed in previous studies (16, 19, 44). In contrast, keratinocytes harboring E4M9 mutant genomes exhibited 1.0 ± 0.4-fold amplification at 24 h and approximately 1.4 ± 0.5-fold amplification at 48 h. These results indicate that HPV31 E1∧E4 mutant genomes in differentiating keratinocytes are impaired in the ability to amplify viral genomes and that E1∧E4 has a significant role in promoting late viral functions. We also observed similar reductions in viral DNA amplification with early-passage cells that maintained E4M4 mutant genomes (data not shown). This indicates that the effects seen with the E4 mutant genomes were not the result of cis effects due to the mutations. In support of our methylcellulose analyses, we performed a FISH analysis of raft cultures of untransfected, wild type-transfected, and E4M9 mutant-transfected cells and found that only wild-type rafts exhibited significant levels of viral DNA amplification (Fig. 5). This indicates that the quantitative reduction in amplification seen for cells suspended in methylcellulose is seen with other systems for differentiation.
FIG. 4.
Differentiation-dependent amplification of HPV genomes in keratinocytes containing wild-type or E4M9 mutant genomes. Equal amounts of total DNA were isolated from keratinocytes that were grown in monolayers or suspended in methylcellulose for 24 and 48 h, and Southern blot analysis was performed by using a linearized HPV31 genome as a probe. (A) The levels of episomal forms of HPV31 DNA were found to be significantly less in E4M9 keratinocytes than in cells with wild-type genomes. (B) Bar graph showing amplification, as determined from phosphorimaging analysis of 12 different experiments.
FIG. 5.
Analysis of wild-type and E1∧E4 mutant organotypic raft cultures. Wild-type and E4M9 organotypic raft cultures were stained with DAPI to determine which cells had positive nuclei. In addition, the rafts were incubated with BrdU for 12 h before being harvested to measure DNA synthesis. Immunofluorescence was performed on cross sections of stratified rafts following incubation with antibodies to BrdU. Cross sections were also analyzed by FISH analysis to determine HPV DNA amplification (open arrows). The closed arrows represent the basal layer.
Late transcripts are decreased in differentiated keratinocytes harboring the E1∧E4 mutant.
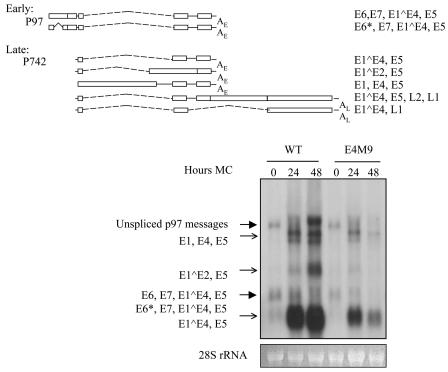
Since the studies described above indicated that the mutation of E1∧E4 had a significant effect on the amplification of viral genomes, we next investigated whether late viral gene expression was similarly altered. Equal amounts of total RNAs were extracted from keratinocytes harboring wild-type HPV31 and the E4M9 mutant which were grown in monolayers or differentiated for 24 or 48 h in methylcellulose. In undifferentiated keratinocytes, unspliced p97 messages and messages encoding E6, E7, E1∧E4, and E5 and E6*, E7, E1∧E4, and E5 were found to be expressed in cell lines carrying both the wild type and the E1∧E4 mutant (Fig. 6, large arrows). Upon differentiation, keratinocytes that maintained wild-type HPV31 expressed late transcripts that encoded E1, E4, and E5; E1∧E2 and E5; and E1∧E4 and E5, as previously described (Fig. 6, small arrows) (19, 24). While the E4M9 mutant cell lines expressed similar amounts of early transcripts (Fig. 6, large arrows) as wild-type HPV31-transfected keratinocytes, significant differences were seen in the levels of late transcripts upon differentiation. The E4M9 mutant-transfected keratinocytes expressed significantly smaller amounts of late transcripts at both the 24- and 48-h time points (Fig. 6, small arrows). The level of late transcripts for E4M9 was slightly higher at 24 h than that at 48 h, but this was due to a greater amount of RNA being loaded onto the gel at 24 h than at 48 h. This is evident if the levels of 28S rRNA are compared. Similar effects of the reduction of late transcripts in the E4M9 mutant were observed in multiple experiments using four different sets of wild-type and E1∧E4 mutant cell lines. These data suggest that E1∧E4 has a role not only in promoting the amplification of viral genomes but also in the activation of expression of late viral transcripts.
FIG. 6.
Late transcription is reduced upon differentiation of keratinocytes containing E1∧E4 mutant genomes. In the upper panel, the most abundant early and late transcripts containing E1∧E4 are shown. For the lower panel, Northern blot analysis was performed using equal amounts of total RNA isolated from keratinocytes grown in monolayers or following suspension in methylcellulose for 24 or 48 h. Linearized HPV31 DNA was used as a probe. Keratinocytes with E4M9 mutant genomes were found to express lower levels of late transcripts (open arrows) than those seen for wild-type cells. 28S rRNA was used as a loading control and is shown below the Northern analysis.
DNA synthesis is decreased in organotypic raft cultures of cells with E1∧E4 mutant genomes.
One way in which the loss of E1∧E4 could act to reduce viral genome amplification and late gene expression would be to decrease the number of cells that reenter S phase upon differentiation. To determine the number of differentiating cells that are actively synthesizing DNA, we performed BrdU incorporation analysis on wild-type and E4M9 mutant raft cultures 12 h prior to cell harvest. BrdU incorporation was then detected in histological cross sections of these rafts by use of a BrdU monoclonal antibody. As shown in Fig. 5, the E4M9 mutant rafts exhibited low levels of BrdU incorporation in the upper stratified layers, and staining was primarily localized to the basal layer. In contrast, raft cultures of cells containing wild-type genomes incorporated BrdU throughout both the suprabasal and basal layers (Fig. 5). Similar results were seen for raft cultures in which BrdU was added for 24 h before the cell harvest (data not shown). Consistent with the results shown in Fig. 3, an examination of DAPI-stained cross sections of organotypic raft cultures identified reduced numbers of suprabasal nuclei in raft cultures of cells with E1∧E4 mutant genomes (E4M9) compared to those with wild-type DNAs (Fig. 5).
DISCUSSION
Our genetic analyses of HPV31 E1∧E4 function in the life cycle of high-risk human papillomaviruses suggest that this protein acts to regulate viral DNA amplification and activation of late gene expression in highly differentiated suprabasal epithelial cells. Since we observed minimal differences in episomal copy numbers, early viral transcripts, or the growth rates of cells grown in monolayer cultures that maintained either wild-type or E1∧E4 mutant genomes, we conclude that E1∧E4 has little effect on undifferentiated cells. Upon the differentiation of cells in methylcellulose, we observed significant reductions in the ability of cells containing E1∧E4 mutant genomes to activate viral DNA amplification and late gene expression. This suggested a role for E1∧E4 in activating late viral functions and is consistent with published observations that high-level expression of the E1∧E4 protein closely correlates with viral DNA amplification (10, 40, 44). These results are in agreement with a recent study that examined cottontail rabbit papillomaviruses, in which E4 was shown to be required for viral DNA amplification and the expression of L1 proteins (38).
One way in which E1∧E4 could act in differentiating cells would be by altering the ability of suprabasal keratinocytes to reenter or remain in S phase. In our studies, BrdU incorporation into suprabasal cells was reduced in E4M9 mutant rafts compared to that in rafts of cells with wild-type genomes. The mutant rafts exhibited BrdU incorporation largely in the basal layer, while DNA synthesis was detected in both the basal and upper stratified layers of rafts with cells that contained wild-type genomes. We also observed a reduction in the number of suprabasal nuclei in rafts of cells with E4M9 mutant genomes from that seen in rafts of keratinocytes containing wild-type genomes, but the number was still larger than that seen in stratified HFKs. This reduction may contribute in part to the reduction in amplification that we observed. High-risk E7 proteins have been shown to play major roles in maintaining cells that are active in the cell cycle upon differentiation (17). Furthermore, we have shown previously that viral DNA amplification and late gene expression occur in differentiated cells that have reentered S phase (44). E1∧E4 could thus act together with E7 to further activate DNA synthesis in suprabasal cells.
Previous studies indicated that E1∧E4 proteins are posttranslationally modified through either proteolysis, phosphorylation, or the formation of multimeric complexes. Proteolysis occurs at various times in the life cycle, and the processed forms are present at different cellular locations. For HPV1 and HPV16, the full-length E1∧E4 protein is present in the lower differentiated layers of the epithelium while the cleaved products are found in the upper differentiated layers (7, 10). In a recent study of HPV1 E1∧E4 proteins, a 16-kDa proteolytically cleaved form of the protein induced rereplication of the cellular DNA. When the full-length 17-kDa protein was expressed, no rereplication of cellular DNA was evident. However, when both the full-length 17-kDa E1∧E4 protein and the 16-kDa protein were coexpressed, cellular proliferation and cellular DNA replication were inhibited. This suggests that E1∧E4 proteins could have different functions that are dependent on processing and multimerization effects (26). A further analysis of the importance of these posttranslational modifications will require a detailed mutational analysis in the context of the viral life cycle. It is formally possible that short E1∧E4 peptides are generated in our mutants and that they still retain some functionality. However, these peptides are quite short and may not be stable.
In transient transfection assays, the overexpression of HPV16 and HPV18 E1∧E4 proteins and the truncated 16-kDa form of the HPV1 E1∧E4 protein from heterologous promoters was found to result in arrest at G2/M (3, 26, 35). In addition, this phenotype could be rescued by the addition of cyclin B1 (26). Therefore, E1∧E4 proteins could function by altering the activity or cellular localization of cell cycle regulators such as cyclin A or B. The 16-kDa truncated form of the full-length 17-kDa E1∧E4 protein of HPV1 has been shown to increase cyclin A expression with a corresponding decrease in the level of cyclin B1. This loss of cyclin B1 may contribute to the G2/M arrest discussed earlier (26). Since there are no reports of direct binding of the E1∧E4 protein to cell cycle regulators, any interaction, if it occurs, would possibly be mediated indirectly, perhaps through keratin filaments, which associate specifically with HPV16 E1∧E4 (50).
A second way in which E1∧E4 could act would be to interfere with late gene expression at a posttranscriptional level. Two of the late transcripts are those that encode E1 and E2, and their high-level expression likely contributes to amplification. E1∧E4 could act to modulate the stability of these transcripts. Such a mechanism would be consistent with the results of yeast two-hybrid studies that used HPV16 E1∧E4 as bait and identified an association with a putative RNA helicase, E4-DBP (8). E4-DBP is thought to be involved in regulating mRNA stability, and its RNA-independent ATPase activity can be partially inhibited by E1∧E4 (8). In addition, E4-DBP was found to bind HPV16 late transcripts. By altering the activity of this helicase, E1∧E4 may regulate the turnover of cellular and viral RNAs, including those encoding E1 and E2. A reduction in E1 and E2 expression would result in decreased viral genome amplification.
A third possible mode of action for E1∧E4 is the binding of cellular proteins that directly regulate viral gene expression and replication. While E1∧E4 proteins are primarily localized to the cytoplasm, a subset of these proteins have been shown to be present in nuclear or perinuclear regions (10, 11, 40). Recently, it was shown that HPV1 E1∧E4 can redistribute the ND10 protein PML from ND10 to inclusion bodies (42). Nuclear domains, or ND10 bodies, are clusters of protein found throughout the nuclei that are reported to be sites of viral replication and transcription for DNA viruses such as herpes simplex virus type 1 (HSV-1) and adenovirus type 5 (Ad5) (14). In addition, these domains are thought to be potential sites for papillomaviral replication (4, 48). Both Ad5 and HSV-1 viral proteins redistribute the ND10 proteins from ND10 bodies in a manner similar to that reported for E1∧E4 (15, 28, 42). Therefore, it is possible that E1∧E4 is involved in redirecting replication as well as transcription to ND10 bodies. Unfortunately, the significance of this redistribution is not yet fully understood. Since E1∧E4 is found in both nuclear and cytoplasmic fractions, it could also encode a shuttling activity that contributes to the effects we observed.
Our findings regarding the function of the E1∧E4 protein share some similarities with a recent report examining the effects of a loss of E5 function from complete HPV31 genomes (16). In that study, HPV31 E5, like E1∧E4, was found to contribute to the activation of differentiation-dependent functions such as viral genome amplification and late gene transcription. One possible explanation for these similarities in phenotypes could be that the insertion of a translation termination codon in the E4 ORF had a destabilizing effect on late transcripts that also encode E5. The majority of HPV31 early transcripts contain E1∧E4/E5 coding sequences, and we observed no differences in the levels of these messages in cells that contained wild-type genomes or genomes with translation termination codons inserted in E4. This demonstrates that the insertion of a translation termination codon in E4 does not lead to alterations in RNA stability. Furthermore, preliminary studies examining the ability of keratinocytes that maintain genomes with mutations in either E1∧E4 or E5 to remain competent for proliferation after being induced to differentiate in methylcellulose suggest that there are differences in effects between the two viral gene products (F. Fehrmann, L. Laimins, and R. Wilson, unpublished). Finally, it is possible that E5 acts together with E1∧E4 to mediate effects on late viral functions and that the loss of either partner alters this ability. In summary, our study has identified an important role for the E1∧E4 protein in the late phase of the productive life cycle of oncogenic human papillomaviruses. Further mutational analyses of E1∧E4 in the context of the complete viral genome are required to elucidate the particular mechanism by which this important viral protein acts.
Acknowledgments
We thank members of the Laimins laboratory for helpful discussions.
R.W. was supported by an Institutional NRSA in support of the Carcinogenesis Training Program at Northwestern University (T32CA09560-17) and by a Public Health Service grant from the National Cancer Institute (1F31CA103648-01). Initial studies were supported by a grant from the National Cancer Institute (CA74202) to L.A.L., and subsequent work was supported by a grant from the Illinois Department of Public Heath.
The contents of this work are solely the responsibility of the authors and do not necessary reflect the official views of the Illinois Department of Public Health.
REFERENCES
- 1.Cheng, S., D. C. Schmidt-Grimminger, T. Murant, T. R. Broker, and L. T. Chow. 1995. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 9:2335-2349. [DOI] [PubMed] [Google Scholar]
- 2.Cripe, T. P., T. H. Haugen, J. P. Turk, F. Tabatabai, P. G. Schmid III, M. Durst, L. Gissmann, A. Roman, and L. P. Turek. 1987. Transcriptional regulation of the human papillomavirus-16 E6-E7 promoter by a keratinocyte-dependent enhancer, and by viral E2 trans-activator and repressor gene products: implications for cervical carcinogenesis. EMBO J. 6:3745-3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davy, C. E., D. J. Jackson, Q. Wang, K. Raj, P. J. Masterson, N. F. Fenner, S. Southern, S. Cuthill, J. B. Millar, and J. Doorbar. 2002. Identification of a G(2) arrest domain in the E1 wedge E4 protein of human papillomavirus type 16. J. Virol. 76:9806-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Day, P. M., R. B. Roden, D. R. Lowy, and J. T. Schiller. 1998. The papillomavirus minor capsid protein, L2, induces localization of the major capsid protein, L1, and the viral transcription/replication protein, E2, to PML oncogenic domains. J. Virol. 72:142-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Villiers, E. M. 1994. Human pathogenic papillomavirus types: an update. Curr. Top. Microbiol. Immunol. 186:1-12. [DOI] [PubMed] [Google Scholar]
- 6.Doorbar, J., D. Campbell, R. J. Grand, and P. H. Gallimore. 1986. Identification of the human papilloma virus-1a E4 gene products. EMBO J. 5:355-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doorbar, J., I. Coneron, and P. H. Gallimore. 1989. Sequence divergence yet conserved physical characteristics among the E4 proteins of cutaneous human papillomaviruses. Virology 172:51-62. [DOI] [PubMed] [Google Scholar]
- 8.Doorbar, J., R. C. Elston, S. Napthine, K. Raj, E. Medcalf, D. Jackson, N. Coleman, H. M. Griffin, P. Masterson, S. Stacey, Y. Mengistu, and J. Dunlop. 2000. The E1E4 protein of human papillomavirus type 16 associates with a putative RNA helicase through sequences in its C terminus. J. Virol. 74:10081-10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doorbar, J., S. Ely, J. Sterling, C. McLean, and L. Crawford. 1991. Specific interaction between HPV-16 E1-E4 and cytokeratins results in collapse of the epithelial cell intermediate filament network. Nature 352:824-827. [DOI] [PubMed] [Google Scholar]
- 10.Doorbar, J., C. Foo, N. Coleman, L. Medcalf, O. Hartley, T. Prospero, S. Napthine, J. Sterling, G. Winter, and H. Griffin. 1997. Characterization of events during the late stages of HPV16 infection in vivo using high-affinity synthetic Fabs to E4. Virology 238:40-52. [DOI] [PubMed] [Google Scholar]
- 11.Doorbar, J., E. Medcalf, and S. Napthine. 1996. Analysis of HPV1 E4 complexes and their association with keratins in vivo. Virology 218:114-126. [DOI] [PubMed] [Google Scholar]
- 12.Duensing, S., A. Duensing, E. R. Flores, A. Do, P. F. Lambert, and K. Munger. 2001. Centrosome abnormalities and genomic instability by episomal expression of human papillomavirus type 16 in raft cultures of human keratinocytes. J. Virol. 75:7712-7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dyson, N., P. M. Howley, K. Munger, and E. Harlow. 1989. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934-937. [DOI] [PubMed] [Google Scholar]
- 14.Everett, R. D. 2001. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 20:7266-7273. [DOI] [PubMed] [Google Scholar]
- 15.Everett, R. D., and G. G. Maul. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fehrmann, F., D. J. Klumpp, and L. A. Laimins. 2003. Human papillomavirus type 31 E5 protein supports cell cycle progression and activates late viral functions upon epithelial differentiation. J. Virol. 77:2819-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flores, E. R., B. L. Allen-Hoffmann, D. Lee, and P. F. Lambert. 2000. The human papillomavirus type 16 E7 oncogene is required for the productive stage of the viral life cycle. J. Virol. 74:6622-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frattini, M. G., and L. A. Laimins. 1994. Binding of the human papillomavirus E1 origin-recognition protein is regulated through complex formation with the E2 enhancer-binding protein. Proc. Natl. Acad. Sci. USA 91:12398-12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frattini, M. G., H. B. Lim, and L. A. Laimins. 1996. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc. Natl. Acad. Sci. USA 93:3062-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Genther, S. M., S. Sterling, S. Duensing, K. Munger, C. Sattler, and P. F. Lambert. 2003. Quantitative role of the human papillomavirus type 16 E5 gene during the productive stage of the viral life cycle. J. Virol. 77:2832-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gloss, B., H. U. Bernard, K. Seedorf, and G. Klock. 1987. The upstream regulatory region of the human papilloma virus-16 contains an E2 protein-independent enhancer which is specific for cervical carcinoma cells and regulated by glucocorticoid hormones. EMBO J. 6:3735-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howley, P. M. 1996. Papillomaviridae: the viruses and their replication, p. 947-978. In P. M. Howley (ed.), Fundamental virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 23.Hughes, F. J., and M. A. Romanos. 1993. E1 protein of human papillomavirus is a DNA helicase/ATPase. Nucleic Acids Res. 21:5817-5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hummel, M., J. B. Hudson, and L. A. Laimins. 1992. Differentiation-induced and constitutive transcription of human papillomavirus type 31b in cell lines containing viral episomes. J. Virol. 66:6070-6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klingelhutz, A. J., S. A. Foster, and J. K. McDougall. 1996. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature 380:79-82. [DOI] [PubMed] [Google Scholar]
- 26.Knight, G. L., J. R. Grainger, P. H. Gallimore, and S. Roberts. 2004. Cooperation between different forms of the human papillomavirus type 1 E4 protein to block cell cycle progression and cellular DNA synthesis. J. Virol. 78:13920-13933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leechanachai, P., L. Banks, F. Moreau, and G. Matlashewski. 1992. The E5 gene from human papillomavirus type 16 is an oncogene which enhances growth factor-mediated signal transduction to the nucleus. Oncogene 7:19-25. [PubMed] [Google Scholar]
- 28.Leppard, K. N., and R. D. Everett. 1999. The adenovirus type 5 E1b 55K and E4 Orf3 proteins associate in infected cells and affect ND10 components. J. Gen. Virol. 80:997-1008. [DOI] [PubMed] [Google Scholar]
- 29.Martin, L. G., G. W. Demers, and D. A. Galloway. 1998. Disruption of the G1/S transition in human papillomavirus type 16 E7-expressing human cells is associated with altered regulation of cyclin E. J. Virol. 72:975-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyers, C., and L. A. Laimins. 1994. In vitro systems for the study and propagation of human papillomaviruses. Curr. Top. Microbiol. Immunol. 186:199-215. [DOI] [PubMed] [Google Scholar]
- 31.Middleton, K., W. Peh, S. A. Southern, H. M. Griffin, K. Sotlar, T. Nakahara, A. El-Sherif, L. Morris, R. Seth, M. Hibma, D. Jenkins, and P. F. Lambert. 2003. Organization of human papillomavirus productive cycle during neoplastic progression provides a basis for selection of diagnostic markers. J. Virol. 77:10186-10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohr, I., R. Clark, S. Sun, E. Androphy, P. MacPherson, and M. Botchan. 1990. Targeting the E1 replication factor to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science 250:1694-1699. [DOI] [PubMed] [Google Scholar]
- 33.Monsonego, J., F. X. Bosch, P. Coursaget, J. T. Cox, E. Franco, I. Frazer, R. Sankaranarayanan, J. Schiller, A. Singer, T. Wright, W. Kinney, C. Meijer, and J. Linder. 2004. Cervical cancer control, priorities and new directions. Int. J. Cancer 108:329-333. [DOI] [PubMed] [Google Scholar]
- 34.Munger, K., B. A. Werness, N. Dyson, W. C. Phelps, E. Harlow, and P. M. Howley. 1989. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 8:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakahara, T., A. Nishimura, M. Tanaka, T. Ueno, A. Ishimoto, and H. Sakai. 2002. Modulation of the cell division cycle by human papillomavirus type 18 E4. J. Virol. 76:10914-10920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nasseri, M., R. Hirochika, T. R. Broker, and L. T. Chow. 1987. A human papilloma virus type 11 transcript encoding an E1-E4 protein. Virology 159:433-439. [DOI] [PubMed] [Google Scholar]
- 37.Palefsky, J. M., B. Winkler, J. P. Rabanus, C. Clark, S. Chan, V. Nizet, and G. K. Schoolnik. 1991. Characterization of in vivo expression of the human papillomavirus type 16 E4 protein in cervical biopsy tissues. J. Clin. Investig. 87:2132-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peh, W. L., J. L. Brandsma, N. D. Christensen, N. M. Cladel, X. Wu, and J. Doorbar. 2004. The viral E4 protein is required for the completion of the cottontail rabbit papillomavirus productive cycle in vivo. J. Virol. 78:2142-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peh, W. L., K. Middleton, N. Christensen, P. Nicholls, K. Egawa, K. Sotlar, J. Brandsma, A. Percival, J. Lewis, W. J. Liu, and J. Doorbar. 2002. Life cycle heterogeneity in animal models of human papillomavirus-associated disease. J. Virol. 76:10401-10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pray, T. R., and L. A. Laimins. 1995. Differentiation-dependent expression of E1-E4 proteins in cell lines maintaining episomes of human papillomavirus type 31b. Virology 206:679-685. [DOI] [PubMed] [Google Scholar]
- 41.Roberts, S., I. Ashmole, G. D. Johnson, J. W. Kreider, and P. H. Gallimore. 1993. Cutaneous and mucosal human papillomavirus E4 proteins form intermediate filament-like structures in epithelial cells. Virology 197:176-187. [DOI] [PubMed] [Google Scholar]
- 42.Roberts, S., M. L. Hillman, G. L. Knight, and P. H. Gallimore. 2003. The ND10 component promyelocytic leukemia protein relocates to human papillomavirus type 1 E4 intranuclear inclusion bodies in cultured keratinocytes and in warts. J. Virol. 77:673-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruesch, M. N., and L. A. Laimins. 1998. Human papillomavirus oncoproteins alter differentiation-dependent cell cycle exit on suspension in semisolid medium. Virology 250:19-29. [DOI] [PubMed] [Google Scholar]
- 44.Ruesch, M. N., F. Stubenrauch, and L. A. Laimins. 1998. Activation of papillomavirus late gene transcription and genome amplification upon differentiation in semisolid medium is coincident with expression of involucrin and transglutaminase but not keratin-10. J. Virol. 72:5016-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75:495-505. [DOI] [PubMed] [Google Scholar]
- 46.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 47.Straight, S. W., P. M. Hinkle, R. J. Jewers, and D. J. McCance. 1993. The E5 oncoprotein of human papillomavirus type 16 transforms fibroblasts and effects the downregulation of the epidermal growth factor receptor in keratinocytes. J. Virol. 67:4521-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swindle, C. S., N. Zou, B. A. Van Tine, G. M. Shaw, J. A. Engler, and L. T. Chow. 1999. Human papillomavirus DNA replication compartments in a transient DNA replication system. J. Virol. 73:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ustav, M., and A. Stenlund. 1991. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 10:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, Q., H. Griffin, S. Southern, D. Jackson, A. Martin, P. McIntosh, C. Davy, P. J. Masterson, P. A. Walker, P. Laskey, M. B. Omary, and J. Doorbar. 2004. Functional analysis of the human papillomavirus type 16 E1-E4 protein provides a mechanism for in vivo and in vitro keratin filament reorganization. J. Virol. 78:821-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Werness, B. A., A. J. Levine, and P. M. Howley. 1990. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 248:76-79. [DOI] [PubMed] [Google Scholar]
- 52.Zur Hausen, H. 1994. Human papillomaviruses. Annu. Rev. Microbiol. 48:427-447. [DOI] [PubMed] [Google Scholar]
- 53.Zur Hausen, H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2:342-350. [DOI] [PubMed] [Google Scholar]