Abstract
Entry of herpes simplex virus 1 (HSV-1) into cells occurs by fusion with cell membranes; it requires gD as the receptor binding glycoprotein and the trigger of fusion, and the trio of the conserved glycoproteins gB, gH, and gL to execute fusion. Recently, we reported that the ectodomain of HSV-1 gH carries a hydrophobic α-helix (residues 377 to 397) with attributes of an internal fusion peptide (T. Gianni, P. L. Martelli, R. Casadio, and G. Campadelli-Fiume, J. Virol. 79:2931-2940, 2005). Downstream of this α-helix, a heptad repeat (HR) with a high propensity to form a coiled coil was predicted between residues 443 and 471 and was designated HR-1. The simultaneous substitution of two amino acids in HR-1 (E450G and L453A), predicted to abolish the coiled coil, abolished the ability of gH to complement the infectivity of a gH-null HSV mutant. When coexpressed with gB, gD, and gL, the mutant gH was unable to promote cell-cell fusion. These defects were not attributed to a defect in heterodimer formation with gL, the gH chaperone, or in trafficking to the plasma membrane. A 25-amino-acid synthetic peptide with the sequence of HR-1 (pep-gHwt25) inhibited HSV replication if present at the time of virus entry into the cell. A scrambled peptide had no effect. The effect was specific, as pep-gHwt25 did not reduce HSV-2 and pseudorabies virus infection. The presence of a functional HR in the HSV-1 gH ectodomain strengthens the view that gH has attributes typical of a viral fusion glycoprotein.
Key structural elements that characterize the viral fusion glycoproteins are the fusion peptide and the coiled-coil bundles of helices. The fusion peptide is a hydrophobic α-helix able to penetrate the membrane of the target cell; it may be located either at or close to the N terminus or in a fusion loop contained in the ectodomain, depending on whether the fusion glycoprotein undergoes a maturational cleavage or not. By means of the fusion peptide, the fusion glycoproteins form a bridge between the virion envelope and the cell membrane, an event that initiates pore formation and is critical for fusion of the viral and cellular membranes (13, 24). Heptad repeats (HRs)—usually two, termed HR1 and HR2—are located downstream of the fusion peptide. After binding to the receptor or induction by low pH, the trimeric fusion glycoproteins undergo a series of conformational changes. HR1 and HR2 form a structure of three α-helices termed a coiled coil, which is further modified to form a trimer of hairpins, also designated a six-helix bundle or fusion core. In this structure, three HR1 helices form a central coiled coil surrounded by three HR2 helices in an oblique antiparallel orientation. The formation of coiled-coil bundles is a key conformational change in the transition of the fusion glycoprotein from the fusion-inactive to the fusion-active state and likely contributes to bringing the viral and cellular membranes in close juxtaposition (13). Cellular fusion glycoproteins (e.g., the SNARE proteins) contain essentially the same structural elements, but located in different entities; typically, four distinct SNARE proteins are required for a fusion event (23, 42), one of which is located in the vesicle membrane and the other three in the target membrane. Bundles of coiled coils may therefore be formed between two regions of the same protein, e.g., the hairpin structure, or among the different proteins that participate in the fusion event, e.g., in the SNARE proteins (5, 6, 23, 42, 46).
Entry of most herpesviruses into the cell requires a minimum of three conserved glycoproteins—gB, gH, and gL—which are cumulatively involved in the execution of fusion of the virion envelope with the cell membrane and act downstream of the receptor recognition by the receptor-binding glycoprotein (7, 9, 12, 14, 20-22, 35, 43, 47, 49). The multigene fusion of herpesviruses is puzzling, because the specific role played by each member of the trio is still largely unknown and because no other viral fusion system requires such a high number of different entities (9, 47). Specifically, in HSV the receptor-binding glycoprotein is gD. The gD ectodomain comprises two separate domains with distinct functions. The region formed by the N terminus and the core recognizes two structurally unrelated alternative protein receptors, nectin1 and HVEM (herpesvirus entry mediator) (11, 16, 26, 27, 34). The second region (named the profusion domain) is located proximal to the membrane, is proline rich, and is most likely responsible for signaling receptor recognition and the triggering of fusion (10). Its role in fusion has been confirmed recently, and its location has been narrowed to amino acid residues 262 to 285 (52). In terms of fusion execution, while the role of gB remains to be elucidated, it is well established that gL forms a heterodimer with gH and serves as the chaperone for proper folding and transport of gH to the plasma membrane (8, 18, 22, 48, 51).
Recently, our laboratory showed that the ectodomain of HSV-1 gH contains a hydrophobic α-helix with attributes of an internal fusion peptide (17). Specifically, mutational disruption of the α-helix abolished virus infectivity and cell-cell fusion. Its replacement with the fusion peptide of human immunodeficiency virus gp41 or of vesicular stomatitis virus G partially rescued the infectivity and fusion activities of α-helix-deleted gH. Furthermore, an internal membrane α-helix is positionally conserved in the gH ectodomains of all herpesviruses examined, including human herpesviruses. Replacement of the natural α-helix in HSV-1 gH with the predicted membrane α-helix from varicella-zoster virus gH fully maintained the HSV-1 gH infectivity and fusion activities. We inferred from these results that the α-helix has attributes of an internal fusion peptide and, consequently, that HSV-1 gH has attributes typical of a viral fusion glycoprotein. Remarkably, gH appears to be part of a conserved mechanism of fusion in the Herpesviridae family.
To elucidate the molecular mechanism of HSV-mediated fusion, we asked whether gH carries HR motifs with the potential to form coiled coils, as predicted by the Lupas algorithm (29, 30). In accordance with the report of Lopper and Compton (28), which appeared while this paper was in preparation, we detected a motif (named HR-1) between residues 443 and 471, downstream of the sequence with attributes of a fusion peptide. A double amino acid substitution, which abrogated the predicted capacity to form the coiled coil, also abrogated infectivity and cell-cell fusion activity, indicating that the predicted coiled-coil motif represents a critical region in HSV-1 gH. A synthetic peptide corresponding to HR-1 inhibited HSV-1 infection.
MATERIALS AND METHODS
Cells and viruses.
BHK, COS, F6, and J-nectin 1 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% to 10% fetal calf serum. F6 is a stably transformed Vero cell line that expresses HSV-1 gH under the control of the HSV-1 gD promoter (14). The J1.1-2 cell line (J cells), a derivative of BHKtk− cells with high resistance to HSV infection, and its derivative J-nectin 1, stably expressing human nectin1, have been described previously (11). In the gH deletion mutant (ΔgH HSV) SCgHZ, the gH gene was replaced with a lacZ gene. SCgHZ was grown and titrated in the complementing F6 cells (14). R8102, a recombinant HSV-1 expressing lacZ under the control of the α27 promoter, pseudorabies virus (PrV) LacZ, and HSV-2(G) have been described previously (3, 25, 31).
Bioinformatic analysis.
The bioinformatic search for coiled coils was conducted by means of the optimized Lupas algorithm (29, 30), with window widths of 14, 21, and 28 and the MTIDK matrix (optimized for a better resolution between the scores of globular and coiled-coil proteins). A weighted and an unweighted scan were run in parallel, and the outputs were compared.
Plasmids.
Expression plasmids for HSV-1 gD, gB, gH, and gL have been described previously (2). EGFR2Δ (named Erb-2) carries the extracellular domain and transmembrane sequences of rat HER-2/neu (nucleotides 25 to 2096) (GenBank accession number NM_017003) and is deleted of the tyrosine kinase domain (44). Plasmid pCAGT7 contains the T7 RNA polymerase gene under the control of the CAG promoter, and the pT7EMCVLuc plasmid expresses firefly luciferase under the control of the T7 promoter (38, 41).
Constructs.
gHHR-EL, carrying the E450G and L453A substitutions in HR-1, was derived by site-directed mutagenesis with oligonucleotide 5′-GGC CCG CCT GCA GCT AGG AGC TCG GGC CCA GCA CCT GGT GGC CGA G-3′ containing a silent SacI site for ease of screening, as described previously (31).
Indirect immunofluorescence.
COS or BHK cells were grown on glass coverslips and transfected with the indicated plasmids by means of Polyfect (QIAGEN, Florence, Italy), according to the manufacturer's instructions. After 24 h, the cells were fixed with 4% paraformaldehyde for 10 min at room temperature, followed, when required, by 0.1% Triton X-100 in phosphate-buffered saline (PBS). Samples were incubated with the following monoclonal antibodies (MAbs): 52S and 53S against gH (39, 45) and a fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin G antibody (Jackson Immunoresearch). Samples were observed with a Zeiss microscope, and micrographs were taken with a Kodak DC290 digital camera.
Cell-cell fusion assay.
The luciferase-based cell-cell fusion assay was performed as described previously (33, 41), using the Luciferase Assay System from Promega (Florence, Italy). The total amounts of transfected plasmid DNA were made equal by addition of Erb-2 plasmid DNA. All samples were run three times and in triplicate.
Infectivity complementation assay.
The complementation assay was performed as described previously (10). Briefly, cells in T25 or T150 flasks were transfected with the appropriate gH plasmid. Four hours later, cells were infected with a gH−/+ stock of SCgHZ (7 PFU/cell). Unpenetrated virions were inactivated by two washes with PBS, followed by a 1-min rinse with 40 mM citric acid, 10 mM KCl, 135 mM NaCl, pH 3 (4). The monolayers were then rinsed twice with PBS and overlaid with medium containing 1% fetal calf serum. Cells were incubated overnight at 37°C, and the progeny virus was titrated in F6 cells. When appropriate, extracellular virions were pelleted and analyzed by Western blotting. Virions were pelleted by high-speed centrifugation from the medium of COS cells transfected with the appropriate plasmid, a wild-type (wt) gH or a gHHR-EL plasmid (15 μg of plasmid DNA/T150 flask), and superinfected with a stock of complemented (gH−/+) SCgHZ. Virions were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to enhanced chemiluminescence (ECL) nitrocellulose membranes (Amersham), and visualized by ECL with MAb H12 (40) against gH and MAb H170 against gD (Goodwin Institute, Plantation, FL).
Infection neutralization assays.
Lyophilized peptides pep-gHwt25, pep-gHwt30, pep-gHscr25, and pep-gD266-289 (synthesized by Primm, San Raffaele Biomedical Science Park, Milan, Italy, and by the Peptide Synthesis Facility of the University of Wisconsin Biotechnology Center) or bovine serum albumin (BSA) was dissolved in DMEM without serum at a 3 mM concentration; the solution was adjusted to a neutral pH by addition of 1 M Tris HCl, pH 10. The experiments were performed in 96-well plates with extracellular virions of R8102, or PrV LacZ, and HSV-2, at an input multiplicity of infection of 7 PFU/cell, except when otherwise stated. For cell coexposure, the cells were incubated with increasing concentrations of the peptides and the viral inoculum for 90 min at 37°C. After removal of the inoculum and rinsing of the cells with DMEM containing 1% fetal calf serum, the cells were incubated with the same concentrations of peptides for 6 or 16 h. For cell preexposure, cells were incubated with a single concentration of peptides or BSA (500 μM) for 30 min at 4°C, rinsed twice with PBS, and infected with R8102 for 90 min at 37°C. Following infection, unpenetrated virus was inactivated by means of an acid wash (40 mM citric acid, 10 mM KCl, 135 mM NaCl, pH 3) (4), and the cells were incubated for a further 16 h in the absence of the peptide. For postexposure, the cells were infected with R8102 for 45 min at 37°C; then peptides were added to the viral inoculum at a final concentration of 500 μM, and cells were incubated for a further 45 min. Unpenetrated virus was inactivated as described above. Virus preincubation was performed by incubating the virions with 500 μM peptides, BSA, or medium for 30 min at 37°C. Following infection for 90 min at 37°C, the viral inoculum was removed, and the extracellular virus was inactivated by an acid wash, as above. In all cases infectivity was measured as β-galactosidase activity using o-nitrophenyl-β-D-galactopyranoside (ONPG) as a substrate (11, 34). HSV-2(G) infection (5 PFU/cell) was detected on methanol-fixed cells by immunoreactivity, as detailed elsewhere (32). Cells were incubated first with 20% newborn calf serum in PBS for 30 min at 37°C and then with MAb LP1 against HSV α-trans-inducing factor (ascites fluid [1:400] in PBS containing 20% newborn calf serum) for 2 h at 37°C, followed by a biotinylated anti-mouse secondary antibody and streptavidin-β-galactosidase (Sigma), diluted 1:500 in PBS, for 90 min, at 37°C. ONPG (3 mg/ml in PBS) was used as a substrate. The absorbance at 405 nm was read in a Bio-Rad Microplate reader.
RESULTS
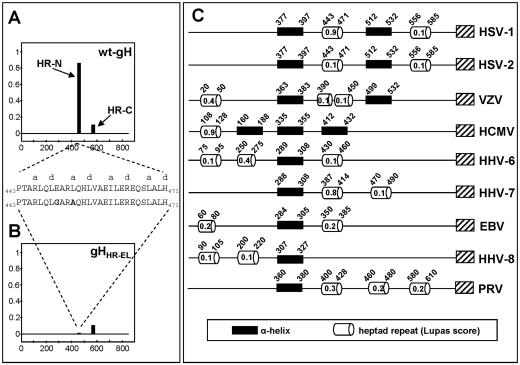
Bioinformatic prediction. In accordance with the prediction of Lopper and Compton (28), a bioinformatic search, performed by means of the optimized Lupas algorithm as detailed in Materials and Methods (29, 30), predicted an HR motif with a very high probability (score, 0.9 out of 1) of forming a coiled-coil structure, here designated HR-1, at residues 443 to 471 of HSV-1 gH. A second HR was predicted at residues 556 to 585, with a low probability of forming a coiled coil (score, 0.4 or 0.1, depending on whether the weighted or unweighted option was used). Because this score fell below the cutoff probability value (0.5) recommended by Lupas, we focused on HR-1. The results are summarized in Fig. 1A, which also shows the probability scores.
FIG. 1.
Identification of heptad repeat motifs in gHs of a number of herpesviruses. (A and B) Probability plots for heptad motif regions in HSV-1 gH were generated by means of the Lupas algorithm (29, 30) (http://www.ch.embnet.org/software/COILS_form.html) using the unweighted option, the MTIDK matrix, and window widths of 14, 21, and 28. The horizontal axes represent the primary sequence of gH protein, and the vertical axes represent the probability score to form coiled coils. (A) wt gH; (B) gHHR-EL. The amino acid sequences of HR-1 in wt gH and gHHR-EL are shown. “a” and “d” indicate the heptad repeat positions according to the Lupas algorithm output. (C) Bioinformatic analysis of the gHs from a number of herpesviruses. The ectodomain of all gHs shows a positionally conserved hydrophobic α-helix (black box) (17) and at least one heptad repeat motif (open cylinder), variously located relative to the hydrophobic α-helix. Additional heptad repeats and/or hydrophobic α-helices are present. Coiled-coil domains with probability scores lower than 0.1 were not represented. The figures within the open cylinders are the probability scores. A hatched box represents the transmembrane segment.
Remarkably, when the same search was performed on the gH sequences from the other human herpesviruses and PrV, HRs were predicted in all the gHs examined, although in some cases with probability scores below the recommended cutoff value, and with different locations in the ectodomain (Fig. 1C). Clearly, the significance of these predictions remains to be validated experimentally in each case, even more so because examples of viral fusion glycoproteins carrying functional HRs with low probability scores have been described, e.g., the F protein of Newcastle disease virus (36). Figure 1C further illustrates that all gHs examined also carry a positionally conserved membrane α-helix, as previously reported (17). Of note, in HSV-1, HSV-2, and human herpesvirus 7 (HHV-7) gHs, the HR motifs are located downstream of the α-helix, i.e., in the typical position of class 1 viral fusion glycoproteins. By contrast, in human cytomegalovirus (HCMV) and HHV-8 gHs, the HR motif is located upstream of the predicted α-helix, and in varicella-zoster virus, Epstein-Barr virus, and HHV-6 gHs, the HR motifs are located both upstream and downstream of the α-helix. The HR in HCMV gH was shown to constitute a critical region, as peptides with this sequence inhibit HCMV infection (28). Cumulatively, the bioinformatic analysis predicted that all gHs examined carry in their ectodomain a minimum complement of one HR and one hydrophobic α-helix, plus additional HRs and/or hydrophobic α-helices, variously located relative to the positionally conserved α-helix. The conservation of the motifs argues that they are critical to gH function throughout the Herpesviridae family, even though in some cases their relative position is noncanonical and the predicted probability falls below suggested cutoff values.
Functional analysis of HR-1.
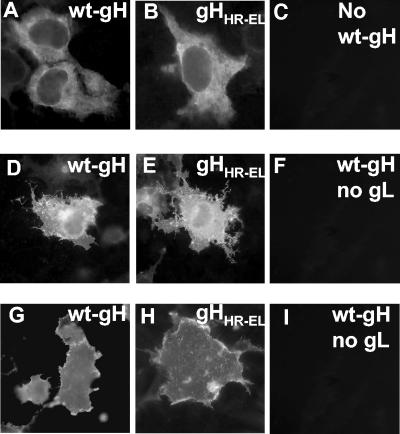
To investigate whether gH HR-1 plays a critical role in the fusion that leads to virus entry and to cell-cell fusion, we searched for substitutions that would abolish the probability of forming the coiled coil. The simultaneous E450G and L453A substitutions predicted an abolishment of the probability (see Fig. 1B). These substitutions were introduced into gH by site-directed mutagenesis, and the resulting construct, named gHHR-EL, was tested in two assays for proper folding (i.e., the ability to form a heterodimer with gL and to be trafficked to the plasma membrane) and in two functional assays (i.e., for the ability to complement the infectivity of the ΔgH HSV-1 mutant SCgHZ and the ability to induce cell-cell fusion when cotransfected with plasmids encoding gB, gD, and gL) (41, 48). In the first assay, the mutant or wt gH plasmid was transfected with or without a gL-encoding expression plasmid, and the immunofluorescence reactivity to two antibodies was measured. MAb 52S recognizes a gL-independent discontinuous epitope with critical residues at positions 536 to 537; MAb 53S recognizes a discontinuous epitope and strictly requires gL for reactivity (17, 40, 45). The results in Fig. 2 show that gHHR-EL maintained reactivity to MAb 52S (Fig. 2B), suggesting no major defect in proper folding. The reactivity of paraformaldehyde-fixed cells (Fig. 2H) indicates that gHHR-EL had no major defect in trafficking to the plasma membranes, while its reactivity to MAb 53S indicates that the ability to form a heterodimer with gL was also unaffected (Fig. 2E).
FIG. 2.
Expression of mutagenized gHHR-EL. (A-C) Paraformaldehyde-fixed, permeabilized COS cells transfected with a wt gH plasmid or a gHHR-EL plasmid and reacted with MAb 52S. In panel C, the wt gH plasmid was omitted. (D-F) Paraformaldehyde-fixed, permeabilized COS cells transfected with the indicated plasmid plus gL plasmid and stained with MAb 53S. In panel F, the gL plasmid was omitted. The positive reactivity in panels D and E indicates gH-gL heterodimer formation. (G-I) Paraformaldehyde-fixed COS cells transfected with a wt gH plasmid or a gHHR-EL plasmid, plus gL plasmid, and reacted with MAb 52S. In panel I, the gL plasmid was omitted. The reactivity denotes no major defect of gHHR-EL in trafficking to the plasma membrane.
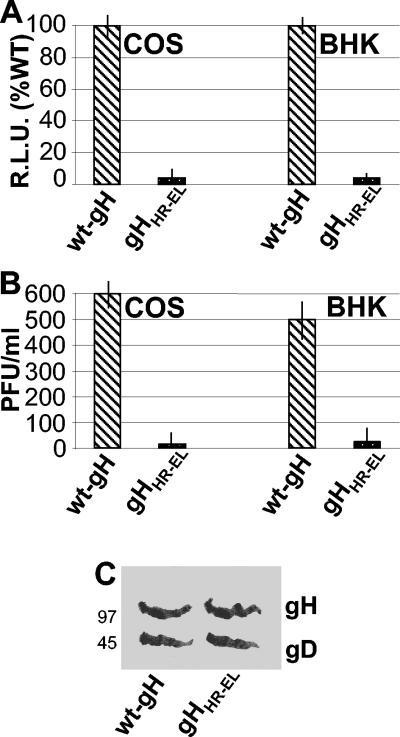
For the cell-cell fusion assay, the plasmid encoding gHHR-EL or wt gH was cotransfected into COS or in BHK cells together with expression plasmids encoding gB, gL, gD, and T7 RNA polymerase. The target cells were transfected with the luciferase reporter gene under the control of the T7 promoter (1, 33, 41, 48). The results in Fig. 3A show that gHHR-EL was completely unable to promote fusion of either COS or BHK cells, despite the fact that it exhibited no major defect in transport to the cell surface.
FIG. 3.
Cell fusion and infectivity complementation of gHHR-EL. (A) Quantification of luciferase-based cell-cell fusion assay in COS and BHK cells expressing the indicated gH plasmid plus gD, gB, and gL. Luciferase activity was expressed as relative light units (RLU). Each experiment was performed at least three times, and samples were run in triplicate; mean values are shown. Vertical bars, standard errors. (B) Infectivity complementation. The indicated cells were transfected with plasmids encoding wt gH or gHHR-EL and were infected 4 h later with a gH−/+ stock of SCgHZ (7 PFU/cell). Progeny virus was titrated at 24 h in gH-expressing cells. Each experiment was performed twice. The mean values of a typical experiment are shown. Vertical bars, standard errors. (C) gHHR-EL is incorporated into complemented virions. Extracellular virions from the experiment for which results are shown in panel B were pelleted by high-speed centrifugation, subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and visualized by Western blotting with MAb H12 against gH and MAb H170 against gD, followed by anti-mouse peroxidase and ECL. The numbers to the left are the migration positions of molecular mass markers (in kilodaltons).
In the infectivity complementation assay, COS or BHK cells were transfected with the mutant gHHR-EL or with wt-gH and superinfected 4 h later with SCgHZ; the progeny virus was titrated in complementing gH-expressing cells at 24 h after transfection (10, 14). When the ΔgH virus replicates in complementing cells, gH is supplied to virions in trans by transgenic gH, and the progeny virus is infectious for one cycle (gH−/+ stock). If gH is defective, the virus infectivity decreases or is abolished. Figure 3B shows that gHHR-EL completely failed to complement infectivity, in contrast to the wt gH, in both cell lines tested. Western blot detection of gH and, for comparison, of gD in the complemented virions ruled out the possibility that the defect in infectivity was due to a defect in the incorporation of mutant gH in virions (Fig. 3C).
Cumulatively, these data indicate that the substitution of two amino acids in HR-1, which disrupted the propensity to form the coiled coil, abolished the ability of gH to serve in infectivity and in cell-cell fusion.
HSV-1 infection is reduced by synthetic peptides with the sequence of gH HR-1.
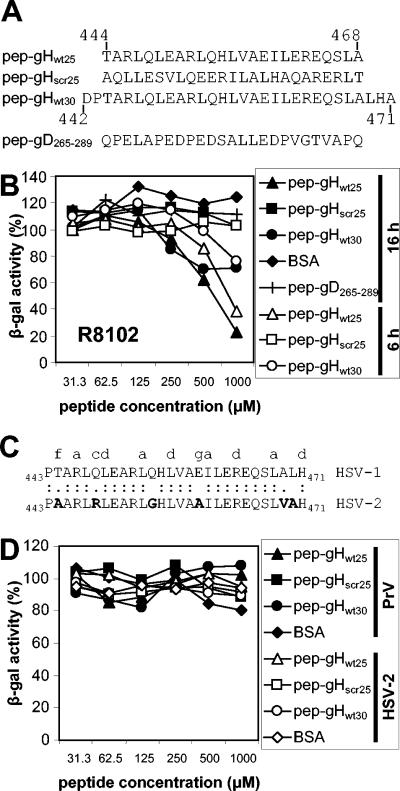
The objective of the following experiment was to verify the possibility that a synthetic peptide with the sequence of HR-1 can impair HSV-1 infectivity. Four peptides were synthesized (Fig. 4A). pep-gHwt25 and pep-gHwt30 were 25 and 30 residues long and encompassed 30 and 25 residues of HR-1, respectively. pep-gHscr25 was a scrambled peptide of gHwt25, in which the order of amino acids was modified so as to abolish the propensity to form a coiled coil. pep-gD265-289 was an unrelated 25-residue peptide containing the indicated sequence of the gD ectodomain. J-nectin 1 cells were infected with the HSV-1 recombinant R8102, which carries a lacZ reporter gene under the control of the α27 promoter (31). The recombinant virus allows one to readily quantify infection through the expression of the reporter gene. Cells were exposed to increasing concentrations of the peptides during and after virus infection. The results in Fig. 4B show that pep-gHwt25 inhibited HSV-1 infection in a dose-dependent manner. The extent of inhibition ranged from 50% to 80% at a 500 to 1,000 μM peptide concentration. pep-gHwt30 was less effective than pep-gHwt25, causing a maximum inhibition of 50%. The inhibitory effect of pep-gHwt25 was specific in that the scrambled pep-gHscr25 or an unrelated peptide with the gD sequence from residues 265 to 289 (pep-gD265-289) exerted almost no inhibition. Of note, two batches of pep-gHwt25 were independently synthesized by different facilities and gave overlapping results. Inhibition of R8102 infection by pep-gHwt25 was consistently observed, irrespective of whether the cells were harvested at 6 or 16 h after infection (Fig. 4B).
FIG. 4.
(A) Sequences of synthetic peptides corresponding to HSV-1 gH HR-1 and of a control peptide. Numbers indicate the positions of amino acid residues in the glycoprotein. pep-gHscr25 is the scrambled version of pep-gHwt25, and its sequence was designed to ablate the propensity to form a coiled coil. pep-gD265-289 encompasses residues 265 to 289 of mature HSV gD and was used as an unrelated 25-mer peptide. (B) Inhibition of HSV-1 infection by peptides corresponding to gH HR-1. J-nectin 1 cells were exposed to R8102, an HSV-1 recombinant carrying a reporter lacZ gene under the control of the α27 promoter, and increasing concentrations of the indicated peptides throughout the experiment. Infection was measured at 6 (open symbols) or 16 (closed symbols) h after infection as β-galactosidase activity, using ONPG as the substrate. (C) Alignment of gH HR-1 sequences from HSV-1 and HSV-2. Letters at the top indicate the positions in the heptad. Divergent residues are boldfaced. (D) The peptides corresponding to gH HR-1 fail to neutralize PrV and HSV-2(G) infections. J-nectin 1 cells were exposed to a PrV LacZ recombinant (closed symbols) or HSV-2(G) (open symbols) and increasing concentrations of the peptides. Following infection, nonpenetrated virus was inactivated by an acid wash. PrV infection was measured as for panel B. HSV-2(G) infection was measured by immunostaining, as detailed in Materials and Methods. The assays were run in triplicate. The standard error ranged between 2 and 8% of the respective average (not shown).
As shown in Fig. 1, HSV-2 gH carries a sequence predicted to form a coiled coil at low probability. It differs from the corresponding HSV-1 HR-1 sequence at positions “f” (which exhibits a nonpolar instead of a polar residue), “c,” and “g” (which exhibits a nonpolar residue instead of a charged residue) (Fig. 4C). We tested whether pep-gHwt25 could block HSV-2(G) infection. The results in Fig. 4D show that pep-gHwt25 and pep-gHwt30 had practically no effect on HSV-2 infection, indicating that the peptides were specific for the type 1 virus. These results rule out the possibility that the inhibitory effect of pep-gHwt25 on HSV-1 was due to a generalized cytotoxic effect, which could explain the observed inhibition. We further determined whether pep-gHwt25 exerted any effect on the closely related animal alphaherpesvirus PrV (3). As can be seen from Fig. 4D, pep-gHwt25 did not inhibit PrV LacZ infection, strengthening the conclusion that the peptide selectively targeted HSV-1 and ruling out the possibility that the effect on HSV was the consequence of nonspecific toxicity.
pep-gHwt25 inhibits the step of virus entry into the cells.
The objective of the experiments described below was to identify the step in replication inhibited by pep-gHwt25. We chose a peptide concentration (500 μM) that gave 50% inhibition in Fig. 4B, and we compared the effects of five different types of exposures.
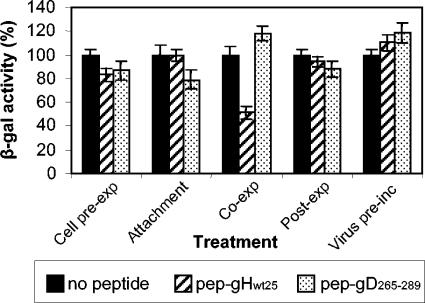
In cell preexposure, cells were exposed to the peptide prior to infection for 30 min at 4°C, the peptide was removed, and R8102 was added to cells. Alternatively, cells were simultaneously exposed to the peptide and the virus (coexposure), or the peptide was added after 45 min of virus absorption and penetration (postexposure). The results in Fig. 5 show that pep-gHwt25 inhibited R8102 replication only if present during the interval of virus attachment-entry into the cells. To determine whether pep-gHwt25 inhibited attachment or entry into cells, the virus was incubated with cells in the presence of the peptide at 4°C, the virus and peptide were then removed, and the cells were shifted to 37°C in the absence of the peptide. No inhibition was observed under these conditions (Fig. 5). Figure 5 further shows that the preincubation of virions with the peptide did not result in any inhibition of R8102 infection. Cumulatively, the results indicate that pep-gHwt25 inhibited infection only if present at the time of virus entry into the cells.
FIG. 5.
Inhibition of HSV infection by pep-gHwt25 occurs at the step of virus entry into the cell. J-nectin 1 cells were exposed to 500 μM pep-gHwt25 or pep-gD265-289 either prior to infection (Cell pre-exp), during virus attachment (Attachment), during attachment and entry (Co-exp), or after virus attachment for 45 min (Post-exp). Alternatively, the virus was preincubated with 500 μM pep-gHwt25 or BSA (Virus pre-inc) before addition to cells. Except in the “attachment” case, nonpenetrated virus was inactivated by an acid wash. Infectivity was measured at 16 h after infection as β-galactosidase activity, using ONPG as a substrate. The assays were run in triplicate. Vertical bars, standard errors.
DISCUSSION
We report that the ectodomain of HSV-1 gH carries a functional heptad repeat sequence (HR-1) with the potential to form a coiled coil, located 60 residues downstream of the hydrophobic α-helix with attributes of an internal fusion peptide. Evidence in support of this conclusion rests on the following.
(i) A bioinformatic search predicted the presence of two heptad repeats in the ectodomain of HSV-1 gH, both of which are located downstream of the potential fusion peptide (29, 30). HR-1, previously predicted by Lopper and Compton (28), exhibited a strong propensity to form a coiled coil, whereas the other HR exhibited a much weaker propensity, below the advised cutoff value. It is noteworthy that the best-characterized viral fusion glycoproteins, including orthomyxovirus hemagglutinin, paramyxovirus F, human immunodeficiency virus gp41, and Ebola virus GP2, all carry HR motifs downstream of the fusion peptide (5, 19, 36, 50). Importantly, HRs with the potential to form coiled coils were predicted in all examined gHs from human and animal herpesviruses, suggesting a conservation of these motifs. Intriguingly, in some cases they were located upstream of the putative fusion peptide, a location which is noncanonical, or the predicted probability was below the suggested cutoff value. In the case of HCMV the predictions were indeed validated by Lopper and Compton, who observed that peptides corresponding to coiled-coil sequences of HCMV gH and gB inhibited virus infectivity (28).
(ii) Substitution of two critical residues in HSV-1 gH HR-1, which abolished the predicted coiled-coil formation, abolished HSV-1 infectivity in the complementation assay and fusion in the cell-cell fusion assay. These defects were not due to defects in folding, heterodimer formation with gL, or trafficking to the plasma membrane, since the mutant gH was present at the cell surface and maintained reactivity to two MAbs that recognize discontinuous epitopes, one of which strictly requires gL for reactivity (8, 17, 45, 51). The presence of a functional domain in the 443-to-471 amino acid region was previously unrecognized. The only mutant described so far in this region was a mutant constructed by random mutagenesis and carrying a 5-residue insertion at residue 458 (15). This mutant exhibited only slightly reduced levels of gH cell surface expression, infectivity complementation, and cell-cell fusion activity (15). Interestingly, analysis of the insertion mutant by means of the Lupas algorithm reveals that the insertion only minimally modified the probability to form a coiled coil; the lack of a phenotype in this mutant is therefore fully consistent with the Lupas predictions of the insertion mutant.
(iii) A 25-residue peptide corresponding to HR-1 inhibited HSV-1 infection in a dose-dependent manner. The peptide was effective only if present at the time of virus entry into the cell; it was ineffective if present during virus attachment or after virus entry. The peptide selectively inhibited HSV-1 and not HSV-2 or PrV infection. The peptide concentrations required to inhibit HSV-1 entry were relatively high. However, concentrations of this order of magnitude were required in similar types of experiments with peptides corresponding to the C-terminal helix of Ebola virus GP2 or to HCMV gH and gB heptad repeat sequences (28, 50). Regardless of the concentrations required, the findings that coiled-coil motifs are conserved in the gHs from different herpesviruses and represent a critical region in HSV-1 and HCMV gHs reinforce the importance of this type of structural motif. Of note, the peptide corresponding to HSV-1 gH HR-1 had no effect if administered to virions or cells but needed to be present at the time of virus entry into the cells, suggesting that the target of the peptide is not gH in its native conformation but rather a gH undergoing conformational modifications toward a fusion-active conformation. HR motifs have been predicted also in bovine herpesvirus 1 gB, and peptides with the HR sequences decreased virus replication at 48 h, at a postentry step (37). Because the algorithm employed for the prediction was not described, and because the HRs do not correspond to those predicted by the Lupas algorithm, it is difficult to compare the significance of the present findings and Lopper and Compton's findings with those on bovine herpesvirus 1.
(iv) As mentioned in the introduction, coiled coils are bundles of α-helices that are wound into a superhelix; the superhelix may be intramolecular or intermolecular (23, 29, 30, 46). In most viral fusion glycoproteins, HR1 and HR2 form an intramolecular hairpin (6, 13, 19, 36, 50). By contrast, in the v-SNARE-t-SNARE complexes, four-helix bundles are formed; most frequently, one helix is supplied by v-SNARE and three helices are supplied by t-SNARE (23, 46). At present it remains to be determined whether HSV-1 gH is a trimer during fusion and whether HR-1 trimers form six-helix bundles with the putative HR-2. Because the role of gB in the execution of HSV-1 fusion remains to be elucidated, and because coiled coils are functional domains in HCMV gB, we do not dismiss the possibility that gH may form helix bundles with gB.
Altogether, the concurrent presence in HSV-1 gH of a hydrophobic α-helix with attributes of a fusion peptide and an HR potentially capable of forming a coiled coil emphasizes that HSV-1 gH has some of the critical attributes typical of class 1 viral fusion glycoproteins. Clearly, the requirement for a minimum of three to four glycoproteins differentiates the fusion induced by herpesviruses from the fusion executed by type 1 fusion glycoproteins.
ADDENDUM IN PROOF
The HSV-1 gH candidate fusion peptide is locased in a loop made by cysteines 2 and 4 (T.M. Cairns, D. J. Landsburg, J. C. Whitbeck, R. J. Eisenberg, and G. H. Cohen, Virology 332:550-562, 2005).
Acknowledgments
We thank T. Minson, T. Mettenleiter, B. Roizman, and P. L. Lollini for gifts of viruses and plasmids. We are indebted to Elisabetta Romagnoli for invaluable assistance.
This work was supported by the FIRB autonomous and coordinated project, Cofin-MIUR 40%, CNR-Functional Genomics, University of Bologna 60%, and Fondo Pallotti.
REFERENCES
- 1.Avitabile, E., G. Lombardi, and G. Campadelli-Fiume. 2003. Herpes simplex virus glycoprotein K, but not its syncytial allele, inhibits cell-cell fusion mediated by the four fusogenic glycoproteins, gD, gB, gH, and gL. J. Virol. 77:6836-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avitabile, E., G. Lombardi, T. Gianni, M. Capri, and G. Campadelli-Fiume. 2004. Coexpression of UL20p and gK inhibits cell-cell fusion mediated by herpes simplex virus glycoproteins gD, gH-gL, and wild-type gB or an endocytosis-defective gB mutant and downmodulates their cell surface expression. J. Virol. 78:8015-8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babic, N., B. G. Klupp, B. Makoschey, A. Karger, A. Flamand, and T. C. Mettenleiter. 1996. Glycoprotein gH of pseudorabies virus is essential for penetration and propagation in cell culture and in the nervous system of mice. J. Gen. Virol. 77:2277-2285. [DOI] [PubMed] [Google Scholar]
- 4.Brunetti, C. R., R. L. Burke, B. Hoflack, T. Ludwig, K. S. Dingwell, and D. C. Johnson. 1995. Role of mannose-6-phosphate receptors in herpes simplex virus entry into cells and cell-to-cell transmission. J. Virol. 69:3517-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37-43. [DOI] [PubMed] [Google Scholar]
- 6.Bullough, P. A., F. M. Hughson, A. C. Treharne, R. W. Ruigrok, J. J. Skehel, and D. C. Wiley. 1994. Crystals of a fragment of influenza haemagglutinin in the low pH induced conformation. J. Mol. Biol. 236:1262-1265. [DOI] [PubMed] [Google Scholar]
- 7.Cai, W. H., B. Gu, and S. Person. 1988. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J. Virol. 62:2596-2604. Erratum, 62:4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cairns, T. M., R. S. Milne, M. Ponce-de-Leon, D. K. Tobin, G. H. Cohen, and R. J. Eisenberg. 2003. Structure-function analysis of herpes simplex virus type 1 gD and gH-gL: clues from gDgH chimeras. J. Virol. 77:6731-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campadelli-Fiume, G., F. Cocchi, L. Menotti, and M. Lopez. 2000. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev. Med. Virol. 10:305-319. [DOI] [PubMed] [Google Scholar]
- 10.Cocchi, F., D. Fusco, L. Menotti, T. Gianni, R. J. Eisenberg, G. H. Cohen, and G. Campadelli-Fiume. 2004. The soluble ectodomain of herpes simplex virus gD contains a membrane-proximal pro-fusion domain and suffices to mediate virus entry. Proc. Natl. Acad. Sci. USA 101:7445-7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cocchi, F., L. Menotti, P. Mirandola, M. Lopez, and G. Campadelli-Fiume. 1998. The ectodomain of a novel member of the immunoglobulin superfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex viruses 1 and 2 in human cells. J. Virol. 72:9992-10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole, N. L., and C. Grose. 2003. Membrane fusion mediated by herpesvirus glycoproteins: the paradigm of varicella-zoster virus. Rev. Med. Virol. 13:207-222. [DOI] [PubMed] [Google Scholar]
- 13.Colman, P. M., and M. C. Lawrence. 2003. The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell Biol. 4:309-319. [DOI] [PubMed] [Google Scholar]
- 14.Forrester, A., H. Farrell, G. Wilkinson, J. Kaye, N. Davis Poynter, and T. Minson. 1992. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J. Virol. 66:341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galdiero, M., A. Whiteley, B. Bruun, S. Bell, T. Minson, and H. Browne. 1997. Site-directed and linker insertion mutagenesis of herpes simplex virus type 1 glycoprotein H. J. Virol. 71:2163-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 17.Gianni, T., P. L. Martelli, R. Casadio, and G. Campadelli-Fiume. 2005. The ectodomain of herpes simplex virus glycoprotein H contains a membrane α-helix with attributes of an internal fusion peptide, positionally conserved in the Herpesviridae family. J. Virol. 79:2931-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gompels, U., and A. Minson. 1986. The properties and sequence of glycoprotein H of herpes simplex virus type 1. Virology 153:230-247. [DOI] [PubMed] [Google Scholar]
- 19.Greenberg, M., N. Cammack, M. Salgo, and L. Smiley. 2004. HIV fusion and its inhibition in antiretroviral therapy. Rev. Med. Virol. 14:321-337. [DOI] [PubMed] [Google Scholar]
- 20.Haan, K. M., S. K. Lee, and R. Longnecker. 2001. Different functional domains in the cytoplasmic tail of glycoprotein B are involved in Epstein-Barr virus-induced membrane fusion. Virology 290:106-114. [DOI] [PubMed] [Google Scholar]
- 21.Huber, M. T., and T. Compton. 1998. The human cytomegalovirus UL74 gene encodes the third component of the glycoprotein H-glycoprotein L-containing envelope complex. J. Virol. 72:8191-8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutchinson, L., H. Browne, V. Wargent, N. Davis-Poynter, S. Primorac, K. Goldsmith, A. C. Minson, and D. C. Johnson. 1992. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J. Virol. 66:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jahn, R., T. Lang, and T. C. Sudhof. 2003. Membrane fusion. Cell 112:519-533. [DOI] [PubMed] [Google Scholar]
- 24.Jardetzky, T. S., and R. A. Lamb. 2004. Virology: a class act. Nature 427:307-308. [DOI] [PubMed] [Google Scholar]
- 25.Kieff, E. D., S. L. Bachenheimer, and B. Roizman. 1971. Size, composition, and structure of the deoxyribonucleic acid of herpes simplex virus subtypes 1 and 2. J. Virol. 8:125-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krummenacher, C., A. V. Nicola, J. C. Whitbeck, H. Lou, W. Hou, J. D. Lambris, R. J. Geraghty, P. G. Spear, G. H. Cohen, and R. J. Eisenberg. 1998. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J. Virol. 72:7064-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ligas, M. W., and D. C. Johnson. 1988. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by beta-galactosidase sequences binds to but is unable to penetrate into cells. J. Virol. 62:1486-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopper, M., and T. Compton. 2004. Coiled-coil domains in glycoproteins B and H are involved in human cytomegalovirus membrane fusion. J. Virol. 78:8333-8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lupas, A. 1997. Predicting coiled-coil regions in proteins. Curr. Opin. Struct. Biol. 7:388-393. [DOI] [PubMed] [Google Scholar]
- 30.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 31.Menotti, L., R. Casadio, C. Bertucci, M. Lopez, and G. Campadelli-Fiume. 2002. Substitution in the murine nectin1 receptor of a single conserved amino acid at a position distal from the herpes simplex virus gD binding site confers high affinity binding to gD. J. Virol. 76:5463-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menotti, L., F. Cocchi, and G. Campadelli-Fiume. 2002. Critical residues in the CC′ ridge of the human nectin1 receptor V domain enable herpes simplex virus entry into the cell and act synergistically with the downstream region. Virology 301:6-12. [DOI] [PubMed] [Google Scholar]
- 33.Milne, R. S., S. L. Hanna, A. H. Rux, S. H. Willis, G. H. Cohen, and R. J. Eisenberg. 2003. Function of herpes simplex virus type 1 gD mutants with different receptor-binding affinities in virus entry and fusion. J. Virol. 77:8962-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 35.Mori, Y., X. Yang, P. Akkapaiboon, T. Okuno, and K. Yamanishi. 2003. Human herpesvirus 6 variant A glycoprotein H-glycoprotein L-glycoprotein Q complex associates with human CD46. J. Virol. 77:4992-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrison, T. G. 2003. Structure and function of a paramyxovirus fusion protein. Biochim. Biophys. Acta 1614:73-84. [DOI] [PubMed] [Google Scholar]
- 37.Okazaki, K., and H. Kida. 2004. A synthetic peptide from a heptad repeat region of herpesvirus glycoprotein B inhibits virus replication. J. Gen. Virol. 85:2131-2137. [DOI] [PubMed] [Google Scholar]
- 38.Okuma, K., M. Nakamura, S. Nakano, Y. Niho, and Y. Matsuura. 1999. Host range of human T-cell leukemia virus type I analyzed by a cell fusion-dependent reporter gene activation assay. Virology 254:235-244. [DOI] [PubMed] [Google Scholar]
- 39.Peng, T., M. Ponce de Leon, M. J. Novotny, H. Jiang, J. D. Lambris, G. Dubin, P. G. Spear, G. H. Cohen, and R. J. Eisenberg. 1998. Structural and antigenic analysis of a truncated form of the herpes simplex virus glycoprotein gH-gL complex. J. Virol. 72:6092-6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng, T., M. Ponce-de-Leon, H. Jiang, G. Dubin, J. M. Lubinski, R. J. Eisenberg, and G. H. Cohen. 1998. The gH-gL complex of herpes simplex virus (HSV) stimulates neutralizing antibody and protects mice against HSV type 1 challenge. J. Virol. 72:65-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pertel, P. E., A. Fridberg, M. L. Parish, and P. G. Spear. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313-324. [DOI] [PubMed] [Google Scholar]
- 42.Rizo, J., and T. C. Sudhof. 2002. Snares and Munc18 in synaptic vesicle fusion. Nat. Rev. Neurosci. 3:641-653. [DOI] [PubMed] [Google Scholar]
- 43.Roop, C., L. Hutchinson, and D. C. Johnson. 1993. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J. Virol. 67:2285-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rovero, S., A. Amici, E. D. Carlo, R. Bei, P. Nanni, E. Quaglino, P. Porcedda, K. Boggio, A. Smorlesi, P. L. Lollini, L. Landuzzi, M. P. Colombo, M. Giovarelli, P. Musiani, and G. Forni. 2000. DNA vaccination against rat her-2/Neu p185 more effectively inhibits carcinogenesis than transplantable carcinomas in transgenic BALB/c mice. J. Immunol. 165:5133-5142. [DOI] [PubMed] [Google Scholar]
- 45.Showalter, S. D., M. Zweig, and B. Hampar. 1981. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect. Immun. 34:684-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skehel, J. J., and D. C. Wiley. 1998. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell 95:871-874. [DOI] [PubMed] [Google Scholar]
- 47.Spear, P. G., and R. Longnecker. 2003. Herpesvirus entry: an update. J. Virol. 77:10179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turner, A., B. Bruun, T. Minson, and H. Browne. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72:873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, X., W. J. Kenyon, Q. Li, J. Mullberg, and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus uses different complexes of glycoproteins gH and gL to infect B lymphocytes and epithelial cells. J. Virol. 72:5552-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watanabe, S., A. Takada, T. Watanabe, H. Ito, H. Kida, and Y. Kawaoka. 2000. Functional importance of the coiled-coil of the Ebola virus glycoprotein. J. Virol. 74:10194-10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Westra, D. F., H. B. Kuiperij, G. W. Welling, A. J. Scheffer, T. H. The, and S. Welling-Wester. 1999. Domains of glycoprotein H of herpes simplex virus type 1 involved in complex formation with glycoprotein L. Virology 261:96-105. [DOI] [PubMed] [Google Scholar]
- 52.Zago, A., C. R. Jogger, and P. G. Spear. 2004. Use of herpes simplex virus and pseudorabies virus chimeric glycoprotein D molecules to identify regions critical for membrane fusion. Proc. Natl. Acad. Sci. USA 101:17498-17503. [DOI] [PMC free article] [PubMed] [Google Scholar]