Abstract
The papillomavirus capsid mediates binding to the cell surface and passage of the virion to the perinuclear region during infection. To better understand how the virus traffics across the cell, we sought to identify cellular proteins that bind to the minor capsid protein L2. We have identified syntaxin 18 as a protein that interacts with bovine papillomavirus type 1 (BPV1) L2. Syntaxin 18 is a target membrane-associated soluble N-ethylmaleimide-sensitive factor-attachment protein receptor (tSNARE) that resides in the endoplasmic reticulum (ER). The ectopic expression of FLAG-tagged syntaxin 18, which disrupts ER trafficking, blocked BPV1 pseudovirion infection. Furthermore, the expression of FLAG-syntaxin 18 prevented the passage of BPV1 pseudovirions to the perinuclear region that is consistent with the ER. Genetic studies identified a highly conserved L2 domain, DKILK, comprising residues 40 to 44 that mediated BPV1 trafficking through the ER during infection via an interaction with the tSNARE syntaxin 18. Mutations within the DKILK motif of L2 that did not significantly impact virion morphogenesis or binding at the cell surface prevented the L2 interaction with syntaxin 18 and disrupted BPV1 infection.
Papillomaviruses (PVs) have been identified as the causative agents of lesions in squamous epithelial cells of many organisms, primarily higher vertebrates (4). These species-specific viruses infect cutaneous and mucosal surface squamous epithelial cells. Bovine papillomavirus type 1 (BPV1) has been studied extensively since the early 1980s and has served as the prototype for many virus-host cell interaction studies involving the papillomaviruses (5, 12, 14, 39, 41, 51). Infection begins with binding of the virus to the surface of the cell, and the α2β4 integrin complex has been suggested as a potential receptor for the virus (27). A recent study demonstrated the colocalization of papillomavirus virus-like particles with the clathrin adaptor molecule AP-2 and with the transferrin receptor, a marker of late endosomes and lysosomes, indicating that papillomaviruses internalize via clathrin-coated vesicles (8). The events that are necessary for uncoating of the virus and for viral DNA entry into the nucleus have not been well defined. The kinetics of infection are such that internalization occurs with a half-life of 4 h, and the transcription of viral packaged DNA occurs after 12 h (8).
Two structural viral proteins form the nonenveloped icosahedral capsid at a ratio of approximately 30:1 L1 to L2, and they mediate the delivery of the histone-bound ∼8-kb double-stranded circular papillomavirus genome to the nucleus during infection (11, 45). The L1/L2 ratio suggests that there is one L2 at each of 12 vertices of the 55-nm-diameter particles (45). The major capsid protein L1 is sufficient to form virus-like particles that morphologically and immunologically resemble the native virion (10, 23, 24, 40). The coassembly of L2 into virus-like particles enhances both the ability to package DNA and the ability to mediate infection (38, 39, 51). Conversely, specific mutations in L2 result in noninfectious particles without affecting viral packaging (38).
L2 interacts with the viral coat protein L1 as well as with the E2 nonstructural regulatory protein, which is involved in viral DNA replication, viral transcription, and viral genome maintenance (9, 34). These interactions result in the relocation of L1 and E2 to nuclear domains 10 (ND10s) (9). L2 sequences at residues 390 to 420 are needed to localize L2 at ND10 (2). The significance of the ND10 relocation is currently uncertain, but data show that ND10s enhance papillomavirus transcription and infection (7, 9, 49). Additionally, L2 has two nuclear localization signals and a DNA binding domain that may have roles in translocating the genome to the nucleus during infection and/or in the recruitment of PV DNA to the viral capsid during assembly (7, 13, 28, 43, 52).
Syntaxin 18 is a member of the SNAP (soluble N-ethylmaleimide-sensitive factor-attachment protein) receptor family whose role is to traffic vesicles within the cell into or out of the endoplasmic reticulum (ER) (18, 44). Syntaxin 18 is primarily found with an immunofluorescent pattern overlapping that of calnexin (ER marker) rather than that of ERGIC53 (ER-Golgi intermediate compartment marker) or GM130 (cis-Golgi protein). Syntaxin 18 was discovered in a yeast two-hybrid screen designed to identify binding partners of α-SNAP (18). Syntaxin 18 has a C-terminal transmembrane domain responsible for anchoring it within the ER and a SNARE (SNAP receptor) domain that binds SNARE domains on other membrane proteins to form a core fusion complex. Although the presence of an N-terminal bundle comprising three helices is well known for syntaxin 1, and perhaps for syntaxin 5, there are no obvious coiled coil structures in the N-terminal region of syntaxin 18 (44). Fractionation experiments have determined that the majority of syntaxin 18 is found in the ER (18). The expression of FLAG-syntaxin 18 causes aggregation of the ER and disrupts vesicle trafficking into or out of the ER with no obvious changes to the mitochondria, lysosomes, or peroxisomes (18). Indeed, FLAG-syntaxin 18 overexpression prevents the export of vesicular stomatitis virus glycoprotein-green fluorescent protein (GFP) from the ER to the Golgi yet has no effect upon the localization of the endosomal marker EEA1 or upon the uptake of transferrin (18).
In this study, we describe the interaction of BPV1 L2 with syntaxin 18 and report the L2-mediated translocation of BPV1 pseudovirions to the ER following infection. We have identified a conserved motif in the L2 protein that is required for binding syntaxin 18. We show that L2 mutant proteins defective in syntaxin 18 binding and ER vesicle trafficking generate infection-deficient pseudovirions.
MATERIALS AND METHODS
Cells, antibodies, and DNA constructs.
COS-7 cells (American Type Culture Collection) and 293T cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Anti-hemagglutinin (HA)-conjugated beads, anti-FLAG-conjugated beads, the mouse anti-FLAG M2 antibody, and the FLAG peptide were purchased from Sigma (St. Louis, Mo.). The mouse anti-promyelocytic leukemia protein (PML) antibody PG-M3 was purchased from Santa Cruz (Santa Cruz, Calif.). The rabbit anti-L2 antibody BL2, mouse anti-L2 antibody C6, mouse anti-L1 antibody 5B6, mouse anti-syntaxin 18 antibody 1E1, rabbit anti-syntaxin 18, pFLAG-syntaxin 18 DNA, and human BPV L2 (hBPV L2) have been previously described (3, 18, 20, 26, 41, 50). The ECTAP plasmid used for tandem affinity purification was made by inserting the Kozak sequence, the FLAG epitope, and two HA epitopes into Ad-TrakCMV at BglII and SalI sites (19). ECTAP BPV1 L2 was made by cloning L2 from hBPVL2 into ECTAP, downstream and in frame with the HA and FLAG epitopes at KpnI and SalI sites. Single-residue mutants were generated by PCR according to standard techniques (42). The deletion mutants were cloned by using standard techniques; an EcoRI site was introduced at the newly formed junction and cloned into vector pcDNA3.1 (Invitrogen, Carlsbad, Calif.).
Transfections and infections.
Transfections were performed by the use of Fugene 6 per the manufacturer's suggestions (Roche, Basel, Switzerland). Briefly, the ratio of Fugene 6 to DNA was 11 μl:5 μg in 250 μl of serum-free Dulbecco's modified Eagle's medium. Fugene 6 was incubated with the medium for 15 min at room temperature prior to the addition of DNA and then incubated at room temperature for an additional 15 min. The transfection mix was added to cells dropwise, and cells were incubated overnight at 37°C with 5% CO2. Pseudovirion infections were performed by incubating cells grown on coverslips to 60% confluence with pseudovirions at a multiplicity of infection of >10 for 2 h at 4°C followed by incubation at 37°C with 5% CO2 for the described length of time.
Identification of interacting proteins.
Tandem affinity purification (TAP) was performed as previously described (33, 48), with the following changes. COS-7 cells at 60% confluence were transfected at >70% efficiency with ECTAP BPV1L2 or the ECTAP control plasmid. Transfection was visualized with fluorescein isothiocyanate fluorescence to detect the expression of GFP. ECTAP expresses only GFP, while ECTAP BPV1 L2 coexpresses HA/FLAG-tagged L2 and GFP from different cassettes. The cells were harvested after 24 h, resuspended in hypotonic buffer (10 mM Tris-HCl, pH 7.3, 10 mM KCl, 1.5 mM MgCl2), and incubated on ice for 5 min. The cells were then pelleted, resuspended in 12 ml of immunoprecipitation (IP) buffer (20 mM Tris-HCl, pH 8.0, 10% glycerol, 5 mM MgCl2, 0.1% Tween 20, 0.1 M KCl, 1× protease inhibitor cocktail [Amersham Biosciences], and 0.5 mM dithiothreitol), and sonicated for 30 s at 40% output and 50% interval with a Branson Sonifier 250 instrument. Lysates were clarified by centrifugation (12,000 × g, 20 min, 4°C), and 10 mg of extract was run through one round of FLAG epitope immunoprecipitation with 100 μl of anti-FLAG-conjugated beads and eluted with the FLAG peptide at 0.25 mg/ml, followed by one round of anti-HA immunoprecipitation with 7.5 μl of anti-HA-conjugated beads. Samples were eluted from anti-HA-conjugated beads with 30 μl of 100 mM glycine-HCl (pH 2.5) neutralized with 5 μl Tris-HCl, pH 7.5, mixed 1:1 with 2× Laemmli buffer, boiled for 3 min, and subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% SDS-PAGE). Proteins in the gel were visualized with freshly made Coomassie stain. Bands of interest were analyzed by matrix-assisted laser desorption ionization-time of flight protein fingerprinting (Proteomics Core Facility of the Genomics Institute and the Abramson Cancer Center, University of Pennsylvania, Philadelphia, Pa.).
Immunoprecipitation.
Transfected COS-7 cells from a 10-cm2 dish were harvested in 200 μl RIPA buffer (1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 160 mM NaCl, 10 mM Tris-HCl, pH 7.4, 5 mM EDTA), incubated on ice for 20 min, and clarified by centrifugation (16,000 × g, 5 min, 4°C). Coimmunoprecipitation was performed by incubating 100 μl of FLAG-syntaxin 18-transfected extract, 100 μl of L2 (or vector)-transfected extract, 25 μl of protein A/G (50/50) beads (Amersham Biosciences, Piscataway, N.J.), the corresponding antibody, and 375 μl of IP buffer (20 mM Tris-HCl, pH 8.0, 10% glycerol, 5 mM MgCl2, 0.1% Tween 20, 0.1 M KCl, 1× protease inhibitor cocktail [Amersham Biosciences], and 0.5 mM dithiothreitol) for a total volume of 600 μl. An antibody to L2, syntaxin 18, or FLAG was used at 1:200, and the protein A/G beads were equilibrated by three washes with IP buffer. The samples were incubated overnight at 4°C with gentle rocking. The next morning, the samples were washed three times with IP buffer, resuspended in 40 μl of 2× SDS-Laemmli buffer, and subjected to 10% SDS-PAGE.
Western blots.
After transfer to nitrocellulose, blots were blocked in 5% Carnation milk. The primary antibodies were incubated for 3 h in TNET (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100), followed by 30 min of incubation with horseradish peroxidase-conjugated secondary antibodies (Amersham). The blot was then visualized by enhanced chemiluminescence (Amersham). The blots were washed three times in TNET for 5 min between the different steps. Primary antibodies were used at a 1:1,000 dilution.
Immunofluorescence.
Cells that were transfected or infected on coverslips were fixed in 100% methanol for 5 min at −20°C at 24 h posttransfection. Antibody working dilutions were as follows: mouse anti-syntaxin 18 1E1, 1:50; mouse-anti L2 C6, 1:100; mouse anti-FLAG M2, 1:100; mouse anti-PML PG-M3, 1:100; rabbit anti-syntaxin 18, 1:50; and rabbit anti-L2 BL2, 1:500. Cells were permeabilized in immunofluorescence buffer (0.2% fish skin gelatin, 0.2% Triton X-100 in phosphate-buffered saline [PBS]) for 5 min, followed by a 1-h incubation in immunofluorescence buffer with primary antibodies. Cells were washed three times in PBS. Fluorescently labeled Alexa fluor anti-rabbit 594, anti-mouse 488, and anti-mouse 647 (Molecular Probes, Eugene, Oreg.) were used as secondary antibodies in a 30-min incubation. Coverslips were washed in PBS and mounted on glass slides by the use of Prolong antifade mounting medium (Molecular Probes) ± 0.5 μM DAPI (4′,6′-diamidino-2-phenylindole) for nuclear staining (Pierce, Rockford, Ill.). All steps were performed at room temperature. Fluorescence microscopy was performed with a Zeiss Axioskop 2 microscope with Zeiss Axiovision software (Zeiss, Thornwood, N.Y.), and confocal microscopy was performed with a Leica TCSNT microscope with Leica TCSNT software (Leica, Northvale, N.J.).
Transmission electron microscopy.
Cell entry and intracellular trafficking of BPV pseudovirions were assessed with infected cells as described above by transmission electron microscopy (TEM) as described previously (47).
Pseudovirion production and titers.
Titer analysis and the production of BPV pseudovirions containing BPV1 packaging enhancing sequences and the GFP marker were performed as previously described (6). The generation of secreted alkaline phosphatase (SEAP) pseudovirions was done as described above, and an assay for infection was performed as described previously (35).
RESULTS
TAP identification of syntaxin 18 as an L2-interacting protein.
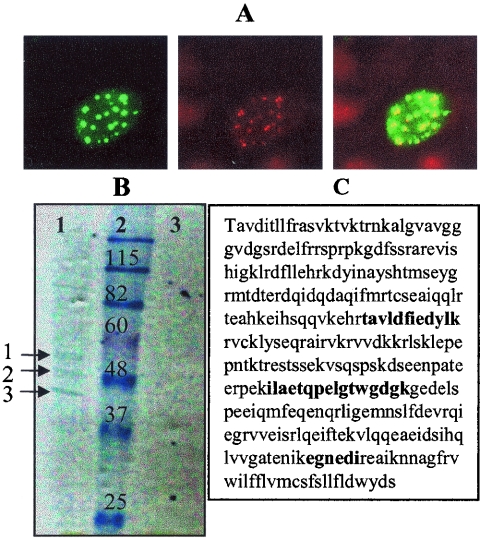
To identify cellular proteins that interact with hBPV1 L2, we cloned a codon-modified version with improved expression in mammalian cells in frame 3′ of one copy of the FLAG epitope and two copies of the HA epitope to generate ECTAP BPV1L2. The addition of the tags to L2 did not affect its ability to colocalize with PML (an ND10 marker) (7, 9, 49). Figure 1A shows a COS-7 cell transfected with ECTAP BPV1L2. The left panel shows staining for BPV1 L2, the middle panel shows staining for PML (clone PG-M3), and the merged image (right panel) shows the colocalization of both signals (Fig. 1A). We were unable to establish cell lines that maintained stable expression of the tagged L2 protein. Therefore, COS-7 cells were transfected with the ECTAP BPV1L2 plasmid and incubated overnight, and lysates were prepared for TAP. TAP was performed on 10 mg of protein lysate from ECTAP BPV1L2-transfected COS-7 cells and from control ECTAP transfections. Immunoprecipitates were separated by SDS-PAGE, and the proteins were visualized by Coomassie blue staining (Fig. 1B). Several protein bands were identified from the ECTAP BPV1L2-transfected cells, whereas none were visualized in immunoprecipitates from extracts of ECTAP vector-transfected cells (Fig. 1B, lanes 1 and 3). Three proteins were identified by matrix-assisted laser desorption ionization-time of flight protein fingerprinting, namely, BPV1 L2, actin, and syntaxin 18 (Fig. 1B, lane 1, arrows 1, 2, and 3, respectively). The interaction with actin has been previously described and shown to be critical for infection (47). The peptides identifying syntaxin 18 corresponded to three distinct regions of the protein, as shown in bold in Fig. 1C.
FIG. 1.
Identifying the interaction of BPV1 L2 with syntaxin 18. (A) ECTAP BPV1L2 is properly distributed within cells. COS-7 cells were transfected with ECTAP BPV1L2 for 24 h. The left panel shows staining for BPV1 L2, the middle panel shows staining for PML, and the right panel shows a merged image. (B) Coomassie blue-stained gel of TAP extracts from ECTAP BPV1L2-transfected COS-7 cells (lane 1) and ECTAP-transfected control cells (lane 3). Molecular weight markers are shown in lane 2. The proteins labeled 1, 2, and 3 in lane 1 were identified as L2, actin, and syntaxin 18, respectively. No proteins were seen in lane 3. (C) Peptide matches to the syntaxin 18 sequence are shown in bold.
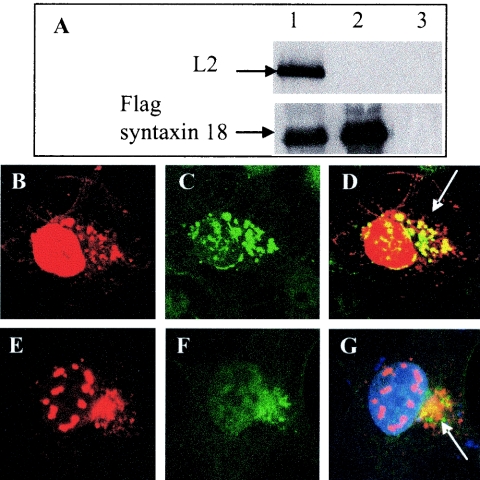
BPV1 L2 coimmunoprecipitates with syntaxin 18 and colocalizes with syntaxin 18 in the ER.
L2 contains two nuclear localization signals, and its C-terminal domain is needed to localize with PML in ND10s (1, 2). Since syntaxin 18 is an ER-resident protein, its coimmunoprecipitation with L2 was unexpected. We confirmed the interaction by coimmunoprecipitation experiments and by immunofluorescence colocalization. COS-7 cells were transfected for 24 h with untagged full-length L2 (hBPV1L2) and FLAG-syntaxin 18, either together or alone (Fig. 2A). RIPA extracts were immunoprecipitated with mouse anti-FLAG M2 as described above and then analyzed by SDS-PAGE. Western blotting was performed with an anti-BPV1 L2 monoclonal antibody (clone C6) (Fig. 2A, upper panel) and an anti-syntaxin 18 rabbit antibody (Fig. 2A, lower panel). The M2 antibody coimmunoprecipitated FLAG-syntaxin 18 and L2 when cells were cotransfected (Fig. 2A, lane 1), and as expected, it immunoprecipitated FLAG-syntaxin 18 when cells were transfected with this protein alone (Fig. 2A, lane 2) and was unable to immunoprecipitate L2 in the absence of FLAG-syntaxin 18 (Fig. 2A, lane 3). Confocal microscopy analysis of COS-7 cells cotransfected with hBPV1 L2 and FLAG-syntaxin 18 confirmed their interaction at 24 h posttransfection (Fig. 2B to D). Untagged BPV1 L2 stained with rabbit anti-BPV1 L2 (Fig. 2B) and FLAG-syntaxin 18 stained with the mouse anti-FLAG M2 antibody (Fig. 2C) had overlapping immunofluorescence (Fig. 2D, merged image). Additionally, transfected untagged BPV1 L2 stained with the rabbit anti-BPV1 L2 antibody (Fig. 2E) colocalized with endogenous syntaxin 18 stained with the anti-syntaxin 18 mouse 1E1 antibody (Fig. 2F) in the perinuclear region of transfected COS-7 cells (Fig. 2G, merged image).
FIG. 2.
Syntaxin 18 and BPV1 L2 coimmunoprecipitate and colocalize. (A) COS-7 cells were cotransfected with FLAG-syntaxin 18 and hBPV L2 (lane 1), transfected with FLAG-syntaxin 18 (lane 2), or transfected with hBPV1 L2 (lane 3). Cells were harvested after 24 h in RIPA buffer, and samples were immunoprecipitated with a FLAG antibody. The top panel was blotted with mouse monoclonal anti-L2 (clone C6), and the bottom panel was blotted with a rabbit polyclonal anti-syntaxin 18 antibody. L2 coimmunoprecipitated with FLAG-syntaxin 18 (lane 1) and did not immunoprecipitate with the FLAG antibody (lane 3). (B to D) COS-7 cells cotransfected with FLAG-syntaxin 18 and hBPV1 L2 were fixed and stained after 24 h for BPV1 L2 (B) and for FLAG (C). Overlapping fluorescence is indicated by an arrow (D). (E to G) COS-7 cells transfected with hBPV1 L2 were fixed and stained after 24 h for BPV1 L2 (E) and for endogenous syntaxin 18 (F). Overlapping fluorescence is indicated by an arrow (G).
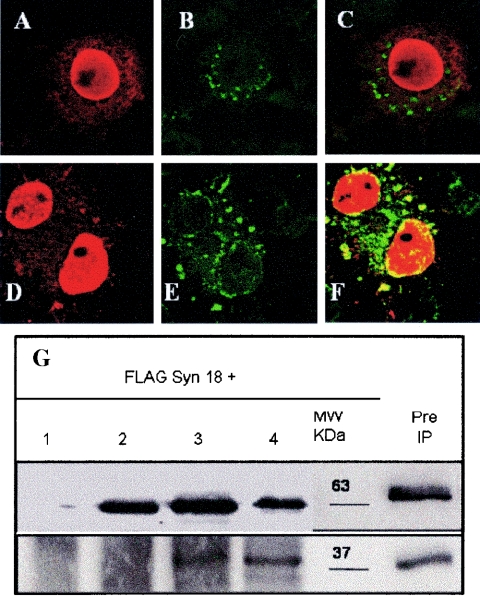
In previous studies, the first 88 residues of L2 were shown to play a critical role in BPV1 infection (46, 47). To determine if this region is involved in binding to syntaxin 18, we constructed the hBPV1 L2 deletion mutants L2 Δ30-90 and L2 Δ60-90. L2 deletion mutant constructs were cotransfected with FLAG-syntaxin 18 into COS-7 cells and analyzed by confocal microscopy. L2 constructs were stained with the C6 antibody, and FLAG-syntaxin 18 was stained with the M2 antibody as described above. L2 Δ30-90 (Fig. 3A) entered the nucleus but did not overlap with FLAG-syntaxin 18 (Fig. 3B and C). L2 Δ60-90 (Fig. 3D) entered the nucleus and overlapped with FLAG-syntaxin 18 (Fig. 3E and F). We confirmed the lack of interaction of L2 Δ30-90 and the interaction of L2 Δ60-90 with syntaxin 18 by coimmunoprecipitation experiments. RIPA extracts from COS-7 cells transfected for 24 h with FLAG-syntaxin 18 alone or with the various L2s were immunoprecipitated with the anti-BPV1 L2 C6 antibody (Fig. 3G). Western analysis of L2 with rabbit anti-BPV1 L2 (Fig. 3G, upper panel) showed that the mutant proteins were expressed and immunoprecipitated the same as the wild type. Consecutive blotting of the membrane with the anti-FLAG M2 antibody confirmed that L2 Δ30-90 did not interact with FLAG-syntaxin 18 (Fig. 3G, lane 2), whereas L2 Δ60-90 (Fig. 3G, lane 3) interacted with FLAG-syntaxin 18, although less well than hBPV1 L2 (Fig. 3G, lane 4).
FIG. 3.
BPV1 L2 residues 30 to 60 mediate the interaction with syntaxin 18. COS-7 cells were transfected with FLAG-syntaxin 18 and either L2 Δ30-90 (A to C) or L2 Δ60-90 (D to F). The cells were fixed and stained after 24 h with rabbit anti-BPV1 L2 (A and D) or a mouse anti-FLAG M2 antibody (B and E). The merged images show no overlapping of syntaxin 18 with L2 Δ30-90 (C) and do show overlapping with L2 Δ60-90 (F). (G) FLAG-syntaxin 18 was cotransfected with the pcDNA3 vector (lane 1), L2 Δ30-90 (lane 2), L2 Δ60-90 (lane 3), or hBPV1 L2 (lane 4). The molecular size marker shows the protein sizes in kilodaltons (kDa), and preimmunoprecipitated full-length L2 and FLAG-syntaxin 18 are also shown. Immunoprecipitation was performed with the anti-L2 monoclonal C6 antibody and Western blotting was performed with a rabbit anti-BPV1 L2 antibody (top), with a mouse anti-FLAG antibody (bottom) used on the same blot. The results showed that L2 Δ60-90 and full-length BPV1 L2 coimmunoprecipitated with FLAG-syntaxin 18, whereas L2 Δ30-90 did not. The L2 C6 antibody did not immunoprecipitate FLAG-syntaxin 18 (lane 1, bottom panel).
L2 residues 30 to 90 are necessary for infection but not viral assembly.
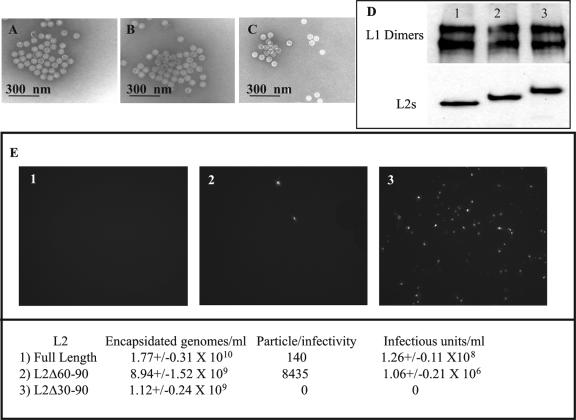
L2 plays a role in both DNA encapsidation and infectivity, and thus we addressed the possible functional roles of L2 residues 30 to 90 in virion assembly or infection. We attempted to generate BPV1 pseudovirions with the L2 Δ30-90 and L2 Δ60-90 deletion mutants (38, 39, 51). We cotransfected codon-modified BPV1 L1, hBPV1 L2 (or the L2 Δ30-90 or L2 Δ60-90 mutant), and a reporter construct (green fluorescent protein or the enzymatic reporter gene SEAP) and tested them for reporter encapsidation and infection as described previously (6). Transmission electron microscopy confirmed the presence of well-formed, apparently full, 55-nm virion particles generated with L2 Δ30-90 (Fig. 4A) and L2 Δ60-90 (Fig. 4B) compared to full-length L2 (Fig. 4C). Each of the mutant L2 proteins copurified with L1 on a Nycodenz step gradient, as did the wild-type L2 protein from the pseudovirion preparations (Fig. 4D, lane 1 [L2 Δ30-90], lane 2 [L2 Δ60-90], and lane 3 [hBPV1 L2]).
FIG. 4.
Role of hBPV1 L2 residues 30 to 90 in viral production and infection. The electron microscopic images of virions generated with L2 Δ30-90 (A), L2 Δ60-90 (B), and hBPV1 L2 (C) seem indistinguishable. (D) Denatured Nycodenz-purified virions (lane 1, Δ30-90; lane 2, Δ60-90; lane 3, BPV1 L2) run in a 10% SDS-PAGE gel were blotted for L1 (upper panel) and L2 (lower panel). (E) COS-7 cells (3 × 106) were incubated with pseudovirions generated with L2 Δ30-90 (panel 1), L2 Δ60-90 (panel 2), and BPV1 L2 (panel 3). Infected cells fluoresced green due to GFP transduction. The levels of genome encapsidation, particle-to-infectivity ratios, and infectious units per ml were determined and are summarized in the bottom panel.
We next analyzed the infectious titers of the pseudovirions generated with L2 Δ30-90 and L2 Δ60-90 compared to that with wild-type L2 by GFP transduction (Fig. 4E). To test for the presence of encapsidated reporter DNA within the Benzonase-treated pseudovirions, we performed quantitative PCRs on DNA extracts from each preparation. Nearly equivalent reporter DNA packaging was observed upon coexpression of BPV1 L1 with all L2 constructs. Although the levels of encapsidated minigenomes were not statistically significant compared to the wild-type level, pseudovirions generated with L2 Δ30-90 (Fig. 4E, panel 1) were completely noninfectious, whereas pseudovirions generated with L2 Δ60-90 (Fig. 4E, panel 2) had a 60-fold reduction compared to wild-type L2 (Fig. 4E, panel 3 and bottom panel).
Fine mapping of L2 residues necessary for syntaxin 18 interaction and infectivity.
The difference in infectious titers was so dramatic between mutants that we constructed L2 mutants with deletions along residues 30 to 90 as well as single-residue mutants with conserved residues (Table 1). We found a direct correlation between infection and coimmunoprecipitation with syntaxin 18 (Table 1). Not surprisingly, the pseudovirions generated with the various L2 mutants contained similar numbers of genomes. Our first observation was that L2 mutants with a deletion of residues 31 to 44 or 41 to 54 were unable to coimmunoprecipitate with syntaxin 18, and pseudovirions generated with these constructs were noninfectious (Table 1, constructs Δ31-44 and Δ41-54). Wild-type levels of infectivity were observed with pseudovirions generated from the deletion mutant L2 Δ51-64 and the L2 point mutants E34A and D36A. BPV1 pseudovirions generated with the L2 Δ61-74 mutant had a 60-fold increase in the particle-to-infectivity ratio. Pseudovirions generated with the L2 Δ74-90 mutant had a 22-fold reduction in infectivity. BPV1 pseudovirions generated with the L2 point mutant D40A exhibited a 2,285-fold decrease in infectivity compared to the wild type.
TABLE 1.
Summary of L2 mutant constructs, their corresponding infectivity rates, and their coimmunoprecipitation with syntaxin 18 (Syn18)
| Construct | No. of Benzonase-resistant genomes per ml | Infectious units per ml | Particle/infectivity ratio | Coimmunoprecipitation with Syn18 |
|---|---|---|---|---|
| Δ31-44 | 8.67 × 109 ± 1.37 × 109 | 0 | ∞ | No |
| Δ41-54 | 1.03 × 1010 ± 0.16 × 1010 | 0 | ∞ | No |
| Δ51-64 | 1.50 × 1010 ± 0.25 × 1010 | 0.56 × 108 ± 0.08 ×108 | 270 | Yes |
| Δ61-74 | 1.16 × 1010 ± 0.17 × 1010 | 1.48 × 106 ± 0.27 × 106 | 7,830 | Yes |
| Δ74-90 | 1.48 × 1010 ± 0.20 × 1010 | 4.71 × 106 ± 0.69 × 106 | 3,140 | Yes |
| E34A | 1.61 × 1010 ± 0.34 × 1010 | 0.94 × 108 ± 0.12 × 108 | 170 | Yes |
| D36A | 1.93 × 1010 ± 0.31 × 1010 | 0.73 × 108 ± 0.10 × 108 | 265 | Yes |
| D40A | 2.56 × 1010 ± 0.46 × 1010 | 0.80 × 105 ± 0.14 × 105 | 320,000 | No |
| ANS | 1.39 × 1010 ± 0.18 × 1010 | 0 | ∞ | No |
The loss of infectivity by pseudovirions generated with the L2 mutants Δ31-44, Δ41-54, and D40A led us to focus on a highly conserved stretch from residues 40 to 44 among PV L2s. We replaced L2 residues 41-KILK-44 with ANS and saw a loss of the syntaxin 18 interaction, and pseudovirions generated with L2ANS were noninfectious (Table 1, ANS).
Noninfectious pseudovirions attach to and enter the cell.
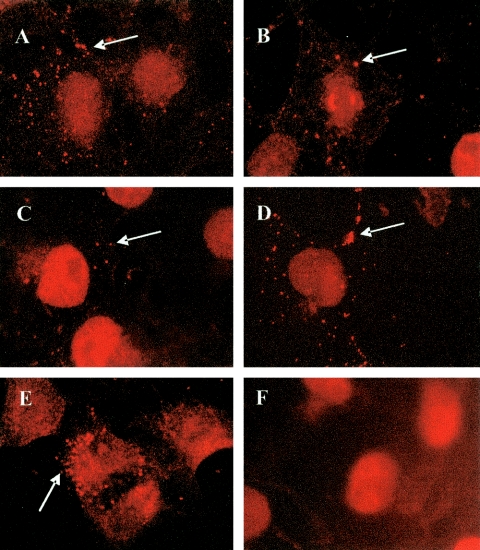
To determine the subcellular localization of the block to pseudovirion infectivity, we examined cells infected with pseudovirions generated with the various L2 mutants by indirect fluorescence microscopy. Although the incubation of COS-7 cells with the noninfectious pseudovirions L2 Δ30-90 (Fig. 5A ), L2 Δ31-44 (Fig. 5B), L2 Δ41-54 (Fig. 5C), L2 D40A (Fig. 5D), and L2ANS (Fig. 5E) did not show any GFP-transduced cells, staining with the 5B6 antibody did show that these pseudovirions were able to attach and remain attached to cells for up to 24 h (arrows in Fig. 5A to E). COS-7 cells in the absence of pseudovirus lacked the punctate pseudovirion fluorescence seen with 5B6 (Fig. 5F).
FIG. 5.
BPV1 pseudovirion infection. COS-7 cells were infected with BPV pseudovirions generated with the following L2s: Δ30-90 (A), Δ31-44 (B), Δ41-54 (C), D40A (D), and ANS (E). Virions are shown in orange after staining with the 5B6 antibody (arrows). (F) Control, uninfected COS-7 cells stained with 5B6 show no punctate fluorescence indicative of pseudovirions.
Disruption of ER trafficking with FLAG-syntaxin 18 prevents BPV1 infection.
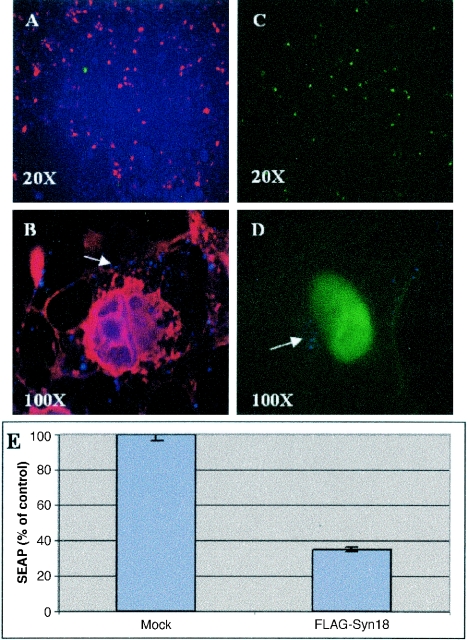
The ectopic expression of FLAG-syntaxin 18 results in its acting as a dominant negative inhibitor of trafficking into and out of the ER (18). We therefore examined whether FLAG-syntaxin 18 expression would disrupt BPV1 infection. The FLAG-syntaxin 18 expression construct was transfected into cells 12 h prior to infection with BPV1 hBPV1 L2-generated pseudovirions carrying minigenomes that expressed either GFP or secreted alkaline phosphatase. The infections were compared to similar infections on vector-transfected cells (Fig. 6). FLAG-syntaxin 18 expression (Fig. 6A, red) resulted in a >60% drop in GFP transduction efficiency compared to that in vector-transfected cells (compare green cells in Fig. 6A and C). In addition, we found no evidence of coexpression of FLAG-syntaxin 18 and GFP in the same cells. BPV1 pseudovirions, as visualized with the mouse antibody 5B6 (Fig. 6B and D, blue dots and arrow), remained trapped within the cell periphery in the FLAG-syntaxin 18 (Fig. 6B, red)-transfected cells 24 h after virus addition, whereas the virions were in the perinuclear region in vector-transfected cells (Fig. 6D). Also, the expression of FLAG-syntaxin 18 resulted in a 70% reduction in infection, as determined by a bulk culture analysis of SEAP production (Fig. 6E).
FIG. 6.
FLAG-syntaxin 18 prevents BPV1 pseudovirion infection. (A and B) COS-7 cells transfected with FLAG-syntaxin 18 12 h before being infected with particles containing wild-type L2. Cells were immunostained for FLAG-syntaxin 18 (red) and GFP was visualized (green). Over 40% of cells were transfected with syntaxin 18, and few cells were infected. (C and D) Vector-transfected COS-7 cells infected with BPV1 L2 pseudovirions had 50% GFP transduction at 24 h. Panels B and D show merged images of GFP fluorescence, pseudovirion staining with the 5B6 antibody (blue dots; arrows), and FLAG-syntaxin 18 staining with the M2 antibody (red). (E) Summary of three experiments showing the level of SEAP expression on cells infected with SEAP-containing wild-type L2 pseudoviruses. FLAG-syntaxin 18-transfected cells had 30% ± 3.4% expression compared to mock-transfected cells after infection.
Visualization of infection pathway leading to the ER by TEM.
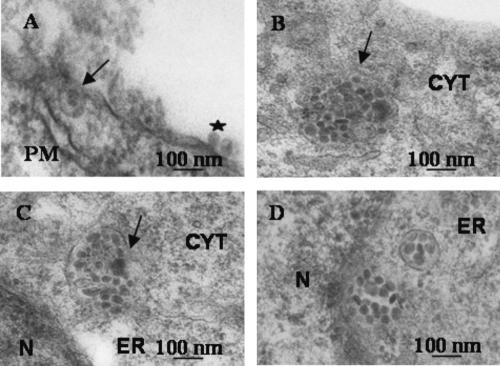
The interaction of L2 with the ER resident syntaxin 18 suggested that PVs pass through the ER during infection. We therefore wanted to see whether we would find pseudovirion particles in the ER in infected cells. 293T cells grown on coverslips were incubated with BPV1 pseudovirions generated with hBPV1 L2 at 4°C for 2 h. The cells were warmed to 37°C (time zero), coverslips were fixed 5 min, 2 h, or 7 h later, and sections were prepared for TEM. Virions bound to the plasma membrane (Fig. 7A, star) and internalization were evident after 5 min (Fig. 7A, arrow). Virions were seen within the cytoplasm in vesicles at 2 h (Fig. 7B, arrow), and at 7 h, the membranes of vesicles filled with virions were observed fusing to the ER (Fig. 7C, arrow). At the same time, virion-filled vesicles were found in the ER and some vesicles had dissolved, releasing the virions into the perinuclear region (Fig. 7D).
FIG. 7.
Ultrastructural analysis of BPV1 pseudovirion infection. Cells were preincubated at 4°C for 2 h with virions before being warmed to 37°C. Samples were obtained 5 min (A), 2 h (B), and 7 h (C and D) after the warming step. (A) A virion is bound at the plasma membrane (PM; star), and internalization into a vesicle is occurring (arrow). (B) A virion-filled vesicle has pinched off from the plasma membrane into the cytoplasm (arrow). (C) The ER and a virus-filled vesicle membrane are fusing (arrow). (D) Viruses are found free floating in the ER or perinuclear (N, nucleus) region and in a formed vesicle.
DISCUSSION
The endocytosis of nonenveloped DNA viruses can be considered a two-step process involving penetration of the cell by virions, followed by their sorting into the appropriate cellular organelles. The primary goal of these viruses during endocytosis is to reach an environment where viral disassembly can occur and the genome is free to gain access to the nucleus. Nuclear translocation of the viral DNA is needed for replication and transcription of the viral genome. Most endocytosed material, including viruses, is engulfed in vesicles and sorted from the cell membrane into the endosomal-lysosomal compartments (30). The low pH of these compartments often leads to viral disassembly and genome release. Recently, a pathway has been identified that shows the sorting of virions into the ER, where disassembly occurs (31, 36). We have investigated the route of infection of papillomaviruses (PVs) and showed that the minor capsid L2 is involved in the sorting of viral particles into the ER, a step that is necessary for infection.
The PVs and polyomaviruses are grouped within one family based on criteria that include their size, their double-stranded DNA content, and their ability to replicate and assemble their virions in the nucleus (22). Indeed, the family name “papovavirus” is a result of the grouping of the initial three members, i.e., rabbit papillomaviruses, mouse polyomavirus, and simian vacuolating virus (SV40). The penetration of cells by papovaviruses during early infection seems to be different: SV40 was shown to enter cells via caveolin-rich vesicles (37); polyomaviruses enter cells in nonclathrin- and noncaveola-derived vesicles (16, 17); and papillomaviruses enter cells in clathrin-coated vesicles (8). Yet regardless of these differences in initial vesicular entry, with the addition of the present results, trafficking of papovaviruses through the ER is emerging as a common step that is necessary for infection (15, 37).
SV40 was shown to disassemble in the ER (31), and it has been suggested that polyomaviruses do the same (15), but the mechanisms of virion disassembly and DNA translocation remain unclear. Polyomaviruses reach the ER by 3 h after infection (29), SV40 reaches the ER at 5 h (31), and we show that PV virions reach the ER at 7 h. The time variance may be dependent on the different initial steps of entry. Our electron microscopic analysis suggests that the membranes of PV virion-filled vesicles fuse with the ER membrane, releasing the viral particles into the ER.
It is reasonable to believe that the trafficking of virions within cells is mediated by corresponding capsid proteins. The role of the L2 minor capsid protein in PV during viral infection and viral production has remained elusive. With this study, we demonstrate that the minor capsid L2 has the ability to mediate the ER entry of BPV1 virions by its association with the tSNARE syntaxin 18. The observation that the tSNARE syntaxin 18 interacts with L2 was unexpected since PVs have not previously been shown to interact with the ER. We identified the highly conserved residues 40DKILK44 as being responsible for the interaction with syntaxin 18, and the only substitutions found in this region were replacements of lysine (K) with glutamine (Q) (i.e., DQ/KILQ/K), which are conservative changes. This motif was shown to be crucial for infection but not for viral production or binding of the virions to the cell membrane. Our hypothesis is that the L2 region encompassing residues 40 to 44 is on the surface of the virion, allowing for binding to syntaxin 18 during trafficking. In support of this, we were unable to coimmunoprecipitate L2 with syntaxin 18 containing a C-terminal deletion leading to a lack of the transmembrane domain (data not shown) (18), suggesting that L2 may bind to the luminal portion of syntaxin 18, although more work needs to be done on the topology of this interaction.
The lack of infection of pseudovirions generated with L2 mutants unable to interact with syntaxin 18 and the observation of pseudovirions in the ER during infection led us to conclude that if virions are prevented from reaching the ER, then infection does not occur. In support of this conclusion, we were able to prevent the infection of wild-type L2 pseudovirions by the expression of dominant negative syntaxin 18 (FLAG-syntaxin 18). The expression of FLAG-syntaxin 18 disrupted vesicle trafficking into or out of the ER without affecting clathrin-mediated endocytosis (18). In these experiments, the trafficking of the virion was blocked in some as yet unidentified organelle in the cytoplasm outside the ER.
In summary, the data described in this study demonstrate that BPV1 L2 residues 40DKILK44 are critical for its association with syntaxin 18 and for the trafficking of BPV1 to the ER. The importance of this sequence in mediating infection and its high level of conservation among all L2s, including the oncogenic human papillomaviruses, suggest that this motif may represent an epitope for a broadly neutralizing antibody. Our data were obtained using pseudovirions generated in vitro, and although studies have shown similarities in viral infections with pseudovirions and wild-type virions, our studies need to be verified with wild-type virions. The finding of this identical sequence in numerous, diverse capsid/envelope proteins (e.g., picornavirus VP1 and VP2, blue tongue virus VP5, and human immunodeficiency virus Env) in membrane-exposed regions (21, 25, 32) leads to the hypothesis that it may be an important signal in infections by other nonenveloped and enveloped viruses.
Acknowledgments
We thank John Schiller and Christopher Buck (National Cancer Institute, Bethesda, Md.) for providing the BPV1 pseudovirion constructs, Eric Cooper for the ECTAP plasmid, and Ian Fraser (University of Queensland, Australia) and the late Jian Zhou for providing codon-modified BPV1 L2 and the monoclonal antibody C6.
This research was funded in part by grants from the American Cancer Society (RSG MBC-103111) and the NIH (P50 CA098252) to R.R. and by grants from the NIH (R01 CA77385 and an S1 minority supplement) to P.H. and P.M.
REFERENCES
- 1.Becker, K. A., L. Florin, C. Sapp, G. G. Maul, and M. Sapp. 2004. Nuclear localization but not PML protein is required for incorporation of the papillomavirus minor capsid protein L2 into virus-like particles. J. Virol. 78:1121-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, K. A., L. Florin, C. Sapp, and M. Sapp. 2003. Dissection of human papillomavirus type 33 L2 domains involved in nuclear domains (ND) 10 homing and reorganization. Virology 314:161-167. [DOI] [PubMed] [Google Scholar]
- 3.Booy, F. P., R. B. Roden, H. L. Greenstone, J. T. Schiller, and B. L. Trus. 1998. Two antibodies that neutralize papillomavirus by different mechanisms show distinct binding patterns at 13 A resolution. J. Mol. Biol. 281:95-106. [DOI] [PubMed] [Google Scholar]
- 4.Bosch, F. X., A. Lorincz, N. Munoz, C. J. Meijer, and K. V. Shah. 2002. The causal relation between human papillomavirus and cervical cancer. J. Clin. Pathol. 55:244-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brobst, D. F., and G. C. Dulac. 1969. Meningeal tumors induced in calves with the bovine cutaneous papilloma virus. Pathol. Vet. 6:135-145. [DOI] [PubMed] [Google Scholar]
- 6.Buck, C. B., D. V. Pastrana, D. R. Lowy, and J. T. Schiller. 2004. Efficient intracellular assembly of papillomaviral vectors. J. Virol. 78:751-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day, P. M., C. C. Baker, D. R. Lowy, and J. T. Schiller. 2004. Establishment of papillomavirus infection is enhanced by promyelocytic leukemia protein (PML) expression. Proc. Natl. Acad. Sci. USA 101:14252-14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Day, P. M., D. R. Lowy, and J. T. Schiller. 2003. Papillomaviruses infect cells via a clathrin-dependent pathway. Virology 307:1-11. [DOI] [PubMed] [Google Scholar]
- 9.Day, P. M., R. B. Roden, D. R. Lowy, and J. T. Schiller. 1998. The papillomavirus minor capsid protein, L2, induces localization of the major capsid protein, L1, and the viral transcription/replication protein, E2, to PML oncogenic domains. J. Virol. 72:142-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Bruijn, M. L., H. L. Greenstone, H. Vermeulen, C. J. Melief, D. R. Lowy, J. T. Schiller, and W. M. Kast. 1998. L1-specific protection from tumor challenge elicited by HPV16 virus-like particles. Virology 250:371-376. [DOI] [PubMed] [Google Scholar]
- 11.Doorbar, J., and P. H. Gallimore. 1987. Identification of proteins encoded by the L1 and L2 open reading frames of human papillomavirus 1a. J. Virol. 61:2793-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dvoretsky, P. M., E. Woodard, T. A. Bonfiglio, L. H. Hempelmann, and I. P. Morse. 1980. The pathology of breast cancer in women irradiated for acute postpartum mastitis. Cancer 46:2257-2262. [DOI] [PubMed] [Google Scholar]
- 13.Fay, A., W. H. T. Yutzy, R. B. Roden, and J. Moroianu. 2004. The positively charged termini of L2 minor capsid protein required for bovine papillomavirus infection function separately in nuclear import and DNA binding. J. Virol. 78:13447-13454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geraldes, A. 1969. Malignant transformation of hamster cells by cell-free extracts of bovine papillomas (in vitro). Nature 222:1283-1284. [DOI] [PubMed] [Google Scholar]
- 15.Gilbert, J., and T. Benjamin. 2004. Uptake pathway of polyomavirus via ganglioside GD1a. J. Virol. 78:12259-12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbert, J. M., and T. L. Benjamin. 2000. Early steps of polyomavirus entry into cells. J. Virol. 74:8582-8588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbert, J. M., I. G. Goldberg, and T. L. Benjamin. 2003. Cell penetration and trafficking of polyomavirus. J. Virol. 77:2615-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatsuzawa, K., H. Hirose, K. Tani, A. Yamamoto, R. H. Scheller, and M. Tagaya. 2000. Syntaxin 18, a SNAP receptor that functions in the endoplasmic reticulum, intermediate compartment, and cis-Golgi vesicle trafficking. J. Biol. Chem. 275:13713-13720. [DOI] [PubMed] [Google Scholar]
- 19.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirose, H., K. Arasaki, N. Dohmae, K. Takio, K. Hatsuzawa, M. Nagahama, K. Tani, A. Yamamoto, M. Tohyama, and M. Tagaya. 2004. Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi. EMBO J. 23:1267-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hogle, J. M., M. Chow, and D. J. Filman. 1985. Three-dimensional structure of poliovirus at 2.9 A resolution. Science 229:1358-1365. [DOI] [PubMed] [Google Scholar]
- 22.Howley, P., and P. Lowy. 2001. Papillomaviruses and their replication, p. 1019-1052. In D. M. Knipe and P. M. Howley (ed.), Fundamental virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 23.Kirnbauer, R., F. Booy, N. Cheng, D. R. Lowy, and J. T. Schiller. 1992. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc. Natl. Acad. Sci. USA 89:12180-12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirnbauer, R., J. Taub, H. Greenstone, R. Roden, M. Durst, L. Gissmann, D. R. Lowy, and J. T. Schiller. 1993. Efficient self-assembly of human papillomavirus type 16 L1 and L1-L2 into virus-like particles. J. Virol. 67:6929-6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, W. J., L. Gissmann, X. Y. Sun, A. Kanjanahaluethai, M. Muller, J. Doorbar, and J. Zhou. 1997. Sequence close to the N-terminus of L2 protein is displayed on the surface of bovine papillomavirus type 1 virions. Virology 227:474-483. [DOI] [PubMed] [Google Scholar]
- 27.Liu, X., A. Mayeda, M. Tao, and Z. M. Zheng. 2003. Exonic splicing enhancer-dependent selection of the bovine papillomavirus type 1 nucleotide 3225 3′ splice site can be rescued in a cell lacking splicing factor ASF/SF2 through activation of the phosphatidylinositol 3-kinase/Akt pathway. J. Virol. 77:2105-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mallon, R. G., D. Wojciechowicz, and V. Defendi. 1987. DNA-binding activity of papillomavirus proteins. J. Virol. 61:1655-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mannova, P., and J. Forstova. 2003. Mouse polyomavirus utilizes recycling endosomes for a traffic pathway independent of COPI vesicle transport. J. Virol. 77:1672-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marsh, M., and A. Helenius. 1989. Virus entry into animal cells. Adv. Virus Res. 36:107-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Norkin, L. C., H. A. Anderson, S. A. Wolfrom, and A. Oppenheim. 2002. Caveolar endocytosis of simian virus 40 is followed by brefeldin A-sensitive transport to the endoplasmic reticulum, where the virus disassembles. J. Virol. 76:5156-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oberste, M. S., K. Maher, and M. A. Pallansch. 2002. Molecular phylogeny and proposed classification of the simian picornaviruses. J. Virol. 76:1244-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogawa, H., K. Ishiguro, S. Gaubatz, D. M. Livingston, and Y. Nakatani. 2002. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296:1132-1136. [DOI] [PubMed] [Google Scholar]
- 34.Okun, M. M., P. M. Day, H. L. Greenstone, F. P. Booy, D. R. Lowy, J. T. Schiller, and R. B. Roden. 2001. L1 interaction domains of papillomavirus L2 necessary for viral genome encapsidation. J. Virol. 75:4332-4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pastrana, D. V., C. B. Buck, Y. Y. Pang, C. D. Thompson, P. E. Castle, P. C. FitzGerald, S. Kruger Kjaer, D. R. Lowy, and J. T. Schiller. 2004. Reactivity of human sera in a sensitive, high-throughput pseudovirus-based papillomavirus neutralization assay for HPV16 and HPV18. Virology 321:205-216. [DOI] [PubMed] [Google Scholar]
- 36.Pelkmans, L., and A. Helenius. 2003. Insider information: what viruses tell us about endocytosis. Curr. Opin. Cell Biol. 15:414-422. [DOI] [PubMed] [Google Scholar]
- 37.Pelkmans, L., J. Kartenbeck, and A. Helenius. 2001. Caveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ER. Nat. Cell Biol. 3:473-483. [DOI] [PubMed] [Google Scholar]
- 38.Roden, R. B., P. M. Day, B. K. Bronzo, W. H. T. Yutzy, Y. Yang, D. R. Lowy, and J. T. Schiller. 2001. Positively charged termini of the L2 minor capsid protein are necessary for papillomavirus infection. J. Virol. 75:10493-10497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roden, R. B., H. L. Greenstone, R. Kirnbauer, F. P. Booy, J. Jessie, D. R. Lowy, and J. T. Schiller. 1996. In vitro generation and type-specific neutralization of a human papillomavirus type 16 virion pseudotype. J. Virol. 70:5875-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roden, R. B., R. Kirnbauer, A. B. Jenson, D. R. Lowy, and J. T. Schiller. 1994. Interaction of papillomaviruses with the cell surface. J. Virol. 68:7260-7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roden, R. B., E. M. Weissinger, D. W. Henderson, F. Booy, R. Kirnbauer, J. F. Mushinski, D. R. Lowy, and J. T. Schiller. 1994. Neutralization of bovine papillomavirus by antibodies to L1 and L2 capsid proteins. J. Virol. 68:7570-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J. R., and D. W. Russell (ed.). 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Sun, X. Y., I. Frazer, M. Muller, L. Gissmann, and J. Zhou. 1995. Sequences required for the nuclear targeting and accumulation of human papillomavirus type 6B L2 protein. Virology 213:321-327. [DOI] [PubMed] [Google Scholar]
- 44.Teng, F. Y., Y. Wang, and B. L. Tang. 2001. The syntaxins. Genome Biol. 2:REVIEWS3012. [DOI] [PMC free article] [PubMed]
- 45.Trus, B. L., R. B. Roden, H. L. Greenstone, M. Vrhel, J. T. Schiller, and F. P. Booy. 1997. Novel structural features of bovine papillomavirus capsid revealed by a three-dimensional reconstruction to 9 A resolution. Nat. Struct. Biol. 4:413-420. [DOI] [PubMed] [Google Scholar]
- 46.Yang, R., P. M. Day, W. H. T. Yutzy, K. Y. Lin, C. F. Hung, and R. B. Roden. 2003. Cell surface-binding motifs of L2 that facilitate papillomavirus infection. J. Virol. 77:3531-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang, R., W. H. T. Yutzy, R. P. Viscidi, and R. B. Roden. 2003. Interaction of L2 with beta-actin directs intracellular transport of papillomavirus and infection. J. Biol. Chem. 278:12546-12553. [DOI] [PubMed] [Google Scholar]
- 48.You, J., J. L. Croyle, A. Nishimura, K. Ozato, and P. M. Howley. 2004. Interaction of the bovine papillomavirus E2 protein with Brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117:349-360. [DOI] [PubMed] [Google Scholar]
- 49.Zhao, K. N., K. Hengst, W. J. Liu, Y. H. Liu, X. S. Liu, N. A. McMillan, and I. H. Frazer. 2000. BPV1 E2 protein enhances packaging of full-length plasmid DNA in BPV1 pseudovirions. Virology 272:382-393. [DOI] [PubMed] [Google Scholar]
- 50.Zhou, J., W. J. Liu, S. W. Peng, X. Y. Sun, and I. Frazer. 1999. Papillomavirus capsid protein expression level depends on the match between codon usage and tRNA availability. J. Virol. 73:4972-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou, J., D. J. Stenzel, X. Y. Sun, and I. H. Frazer. 1993. Synthesis and assembly of infectious bovine papillomavirus particles in vitro. J. Gen. Virol. 74:763-768. [DOI] [PubMed] [Google Scholar]
- 52.Zhou, J., X. Y. Sun, K. Louis, and I. H. Frazer. 1994. Interaction of human papillomavirus (HPV) type 16 capsid proteins with HPV DNA requires an intact L2 N-terminal sequence. J. Virol. 68:619-625. [DOI] [PMC free article] [PubMed] [Google Scholar]