Summary
Rice blast, caused by Magnaporthe oryzae, significantly impacts grain yield, necessitating the identification of broad‐spectrum resistance genes and their functional mechanisms for disease‐resistant crop breeding. Here, we report that rice with knockdown OsHDAC1 gene expression displays enhanced broad‐spectrum blast resistance without effects on plant height and tiller numbers compared to wild‐type rice, while rice overexpressing OsHDAC1 is more susceptible to M. oryzae. We identify a novel blast resistance transcription factor, OsGRAS30, which genetically acts upstream of OsHDAC1 and interacts with OsHDAC1 to suppress its enzymatic activity. This inhibition increases the histone H3K27ac level, thereby boosting broad‐spectrum blast resistance. Integrating genome‐wide mapping of OsHDAC1 and H3K27ac targets with RNA sequencing analysis unveils how OsHDAC1 mediates the expression of OsSSI2, OsF3H, OsRLR1 and OsRGA5 to regulate blast resistance. Our findings reveal that the OsGRAS30–OsHDAC1 module is critical to rice blast control. Therefore, targeting either OsHDAC1 or OsGRAS30 offers a promising approach for enhancing crop blast resistance.
Keywords: OsHDAC1, OsGRAS30, ChIP‐Seq, transcriptome, H3K27ac, rice broad‐spectrum blast resistance
Introduction
Rice (Oryza sativa L.) is one of the world's most important cereal crops, feeding half of the global population (Zhang et al., 2020). Rice blast is caused by the pathogenic fungus Magnaporthe oryzae and is one of the most serious diseases affecting rice, often causing serious decreases in rice production (Chakraborty et al., 2021). Plants have developed two layers of defence against pathogens, including pathogen‐associated molecular pattern‐triggered immunity (PTI) and effector‐triggered immunity (ETI) (Jones and Dangl, 2006). PTI occurs at the plasma membrane surface and is controlled by pattern recognition receptors (Couto and Zipfel, 2016). Pathogens secrete effector molecules encoded by virulence genes into plant cells to induce pathogen virulence and attenuate plant PTI (Xin et al., 2018). Such pathogen effectors are detected by intracellular plant receptors known as nucleotide‐binding leucine‐rich repeat resistance (R) proteins, activating ETI (Thordal‐Christensen, 2020). To date, over 100 blast R genes in the rice genome and 24 virulence genes in M. oryzae have been identified (Li et al., 2019). Blast R genes interact directly or indirectly with pathogenic effectors following M. oryzae infection, to trigger changes in the expression of defence response‐related genes, triggering diverse physiological events including hormone signalling activation, hypersensitivity reactions, biosynthesis of chemical molecules that degrade pathogen cells and production of reactive oxygen species (Liu et al., 2021). However, such resistance is race‐specific and is easily lost due to the rapid evolution of pathogen virulence genes (Deng and Naqvi, 2019). In contrast, non‐race‐specific resistance is broad‐spectrum and may fight crop diseases more effectively. To date, a few genes conferring broad‐spectrum resistance to pathogens have been identified in plants, including RPW8.1 and RPW8.2 for broad‐spectrum resistance to powdery mildew in Arabidopsis (Xiao et al., 2001; Yang et al., 2023; Zhao et al., 2021, 2023), STV11 for durable resistance to rice stripe virus (Wang et al., 2014), bsr‐d1, OsUBC45 and RBL1 for broad‐spectrum blast resistance in rice (Li et al., 2017; Sha et al., 2023; Wang et al., 2023), Lr34 and TapsIPK1 for broad‐spectrum resistance to rust fungi in wheat (Fukuoka et al., 2009; Wang et al., 2022) and ZmGLK36 for broad‐spectrum resistance to maize rough dwarf disease (Xu et al., 2023). In plants, a growth–defence tradeoff exists due to resource constraints, which requires plants to prioritize either growth or defence according to changes in external and internal factors (Huot et al., 2014). This tradeoff means that most defence genes also affect agricultural traits (He et al., 2022). Thus, broad‐spectrum genes with no or little effect on crop growth should be identified and integrated into breeding strategies to optimize the growth–defence balance, maximize crop yields and meet rising global food demands.
Recent reports have shown that epigenetic modification‐mediated transcription regulation is involved in many biological processes in plants (Chang et al., 2020; Chen et al., 2020; Liang et al., 2020; Yang et al., 2022). Histone acetylation is one of the most extensively characterized epigenetic modifications and is generally associated with active transcription (Shvedunova and Akhtar, 2022). Histone deacetylases (HDACs) regulate histone acetylation levels and gene transcription by removing acetyl moieties from the acetylated lysines of histones, which are added by histone acetyltransferases (Kumar et al., 2021), crucial players in plant growth and development as well as responses to environmental stimuli (Chen et al., 2020). In addition, HDACs are involved in disease resistance in some plant species. For example, Arabidopsis HDA6 suppresses the expression of defence‐related genes (Wang et al., 2017; Wu et al., 2021). HDA19 is required for salicylic acid (SA)‐mediated defence responses (Choi et al., 2012), and HDA19 can interact with WRKY38 or WRKY62 to regulate plant basal defence responses (Kim et al., 2008). HD2B is involved in the reprogramming of defence gene expression and innate immunity (Latrasse et al., 2017). In rice, HDA701 negatively regulates broad‐spectrum resistance to rice pathogens by modulating histone H3K9 acetylation of defence‐related genes (Chen et al., 2022a). SRT702 negatively regulates resistance to rice blast, false smut and bacterial blight by modulating histone H4K5 and H4K8 acetylation to affect the expression of disease‐resistance genes (Chen et al., 2024). HDT701 negatively regulates plant innate immunity by modulating histone H4 acetylation of defence‐related genes (Ding et al., 2012), and the RNase P protein subunit Rpp30 can target HDT701, conferring resistance to fungal and bacterial pathogens (Li et al., 2021). In wheat, HOS15 can interact with HDA6 to regulate defence responses to powdery mildew (Zhi et al., 2020). However, the molecular mechanisms of HDAC activity in plant immunity remain largely unknown.
Seventeen HDAC genes have been identified in the rice genome (Hou et al., 2021). Here, we demonstrate OsHDAC1 as a negative regulator of immunity against rice blast with no effects on plant height and tiller numbers and identify a novel GRAS transcription factor OsGRAS30, which can interact with OsHDAC1 and suppress its activity, thereby enhancing rice broad‐spectrum blast resistance. Further, we have found that OsHDAC1 suppresses the transcription of blast resistance‐related genes by binding their promoters and reducing histone H3K27 acetylation. Moreover, we show that chemical HDAC inhibitors confer rice blast resistance, providing a novel strategy for controlling rice blast. Overall, our data reveal the functional characteristics of the OsGRAS30–OsHDAC1 module in mediating broad‐spectrum blast resistance in rice.
Results
OsHDAC1 negatively regulates broad‐spectrum resistance to M. Oryzae in rice
Recently, chromatin histone modifications have been reported to play a role in the regulation of plant disease defences (Chen et al., 2022a; Li et al., 2021; Niu et al., 2019). In our previous work, to examine the function of OsHDAC1, we constructed OsHDAC1 RNA interference (RNAi) lines in rice, as homozygous OsHDAC1 mutant seeds could not be obtained, and also generated overexpression (OE) transgenic plants (Chung et al., 2009; Hou et al., 2022). In this study, to explore whether OsHDAC1 is involved in rice blast resistance, we first challenged rice plants with the virulent M. oryzae Guy11. To detect changes in OsHDAC1 protein levels in these rice plants, we produced an OsHDAC1‐specific antibody (Figure S1). Analysis of changes in OsHDAC1 expression after M. oryzae infection showed that the transcript (Figure 1a) and protein (Figure 1b) levels of OsHDAC1 were downregulated. To investigate the possible involvement of OsHDAC1 in blast resistance, two OsHDAC1 RNAi lines and two OsHDAC1 OE lines, along with wild‐type (WT) Nipponbare rice, were inoculated with M. oryzae Guy11 by spraying with conidia. The two OsHDAC1 RNAi lines (Ri2 and Ri3) developed only small, scattered lesions, while WT Nipponbare showed typical blast lesions (Figure 1c), indicating that silencing of OsHDAC1 enhances resistance to blast. Fungal DNA quantification showed that the two OsHDAC1 RNAi lines harboured only 10% and 30% as much M. oryzae as WT, respectively (Figure 1d). In contrast, the OsHDAC1 OE5 and OE6 lines had larger lesions compared with WT (Figure 1e). The two OE lines had significantly greater fungal DNA content, which was increased by approximately 3‐ to 4‐fold (Figure 1f). We then inoculated the OsHDAC1 RNAi and OE lines and WT by spraying conidia from mixed M. oryzae isolates, including AH4, Sc09‐153‐07, TM3‐2, RB1, RB3, NC1, HLJ1‐3, HLJ09‐17‐1 and HLJ5‐3 from different regions of China (Figure S2). The OsHDAC1 RNAi lines showed enhanced resistance to multiple virulent isolates of M. oryzae, whereas the OE lines showed higher susceptibility, compared with WT (Figure 1g). Relative fungal biomass was reduced by approximately 3‐fold in the RNAi lines and increased by 1.7‐ and 1.5‐fold in the OE lines (Figure 1h). In addition, field tests demonstrated that the OsHDAC1 OE and RNAi lines had no effects on plant height, tiller numbers, 1000 seed weight and seed setting rate (Figure S3). These results indicate that OsHDAC1 negatively regulates rice broad‐spectrum blast resistance.
Figure 1.
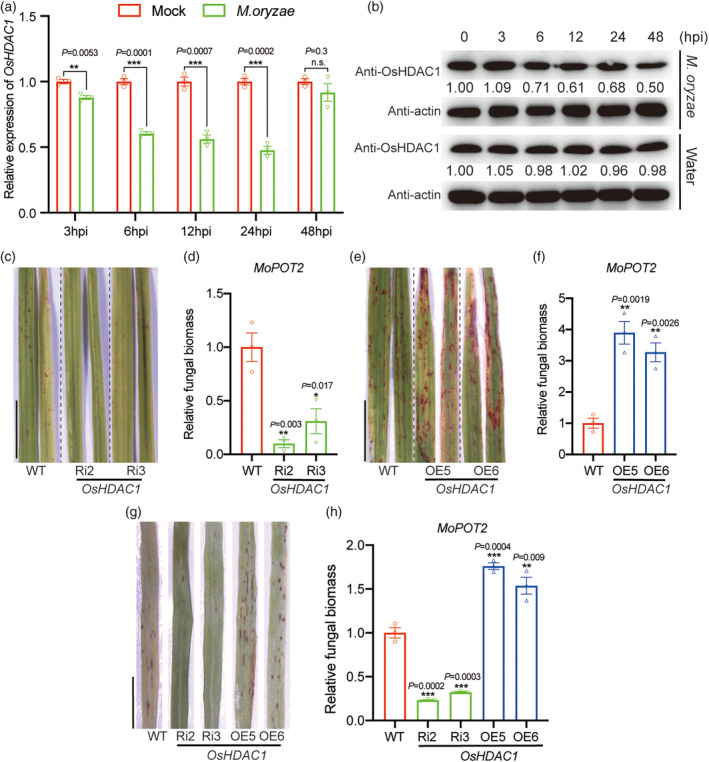
OsHDAC1 regulates blast resistance in rice. (a) Transcript variation of OsHDAC1 in response to M. oryzae Guy11 treatment was analysed by RT–qPCR using total RNA extracted from leaves at 3, 6, 12, 24 or 48‐h post‐infection (hpi). Gene expression levels in the WT without M. oryzae treatment were set to 1.00. Values are means ± SEM (n = 3). Experiments were repeated three times with similar results. (b) Protein level of OsHDAC1was reduced in response to M. oryzae Guy11 treatment, as shown by immunoblotting analysis. Water treatment was used as a control, and the actin protein was applied as an equal loading control. (c) Disease symptoms after spray‐inoculation with a spore suspension of M. oryzae Guy11 for 5 days post inoculation (dpi) in OsHDAC1 RNAi lines and WT. Scale bar = 1 cm. Representative data from three independent experiments. (d) Relative fungal biomass in OsHDAC1 RNAi lines compared with WT determined by qPCR for the M. oryzae Pot2 (MoPot2) gene normalized to rice Ubiquitin. Fungal biomass in the WT was set to 1.00. Values are means ± SEM (n = 3). Experiments were repeated three times with similar results. (e) Disease symptoms after spray‐inoculation with a spore suspension of M. oryzae Guy11 for 5 dpi in OsHDAC1 OE lines and WT. Scale bar = 1 cm. Representative data from three independent experiments. (f) Relative fungal biomass in OsHDAC1 OE lines compared with WT determined using qPCR for the MoPot2 gene normalized to rice Ubiquitin. Fungal biomass in the WT was set to 1.00. Values are means ± SEM (n = 3). Experiments were repeated three times with similar results. (g) Disease symptoms of the WT, OsHDAC1 OE and RNAi rice leaves inoculated with mixed isolates including AH4, Sc09‐153‐07, TM3‐2, RB1, RB3, NC1, HLJ1‐3, HLJ09‐17‐1 and HLJ5‐3. Scale bar = 1 cm. Representative data from three independent experiments. (h) Relative fungal biomass in OsHDAC1 RNAi and OE lines compared with WT determined using qPCR for the MoPot2 gene normalized to rice Ubiquitin. Fungal biomass in the WT was set to 1.00. Values are means ± SEM (n = 3). Experiments were repeated three times with similar results. Asterisks represent significant differences determined by Student's t‐test (*P < 0.05; **P < 0.01; ***P < 0.001). WT, wild‐type; Ri, RNA interference; OE, overexpression.
OsGRAS30 interacts with OsHDAC1
To gain mechanistic insights into OsHDAC1 function, we attempted to identify proteins interacting with OsHDAC1 in a yeast screening experiment. We identified the gene LOC_Os05g49930, named OsGRAS30, which belongs to the GRAS transcription factor family and interacts with OsHDAC1 (Figure 2a). Previous studies indicated that overexpression of the GRAS transcription factor family proteins GIGR2 and OsSLR1 promotes resistance to M. oryzae (De‐Vleesschauwer et al., 2016; Tanabe et al., 2016). Therefore, we speculated that OsGRAS30 might be involved in OsHDAC1‐mediated M. oryzae resistance. To confirm the OsHDAC1–OsGRAS30 interaction in vivo, OsHDAC1‐HA and OsGRAS30‐Myc were co‐expressed in rice protoplasts and subjected to coimmunoprecipitation (Co‐IP) analysis. OsGRAS30‐Myc could be co‐purified with OsHDAC1‐HA using an anti‐HA antibody, but no OsGRAS30‐Myc was obtained from the control (Figure 2b). We conclude that OsHDAC1 interacts with OsGRAS30 in vivo. Further, in a split‐luciferase (Split‐LUC) complementation imaging assay, we transiently co‐infiltrated Nicotiana benthamiana leaves with OsHDAC1‐nLUC and cLUC‐OsGRAS30 fusion constructs, and LUC fluorescence was apparent in N. benthamiana leaves, demonstrating OsHDAC1–OsGRAS30 interaction in planta (Figure 2c). To confirm the OsHDAC1–OsGRAS30 interaction in vitro, we expressed and purified recombinant GST‐OsHDAC1 and MBP‐OsGRAS30‐His protein from Escherichia coli and performed an in vitro pull‐down assay. MBP‐OsGRAS30‐His was pulled down by GST‐OsHDAC1 but not by GST itself (Figure 2d), suggesting that MBP‐OsGRAS30‐His interacts with GST‐OsHDAC1 in vitro. In addition, laser confocal microscopy showed that OsHDAC1‐sGFP and OsGRAS30‐mCherry were co‐localized in the nucleus of rice protoplasts (Figure 2e).
Figure 2.
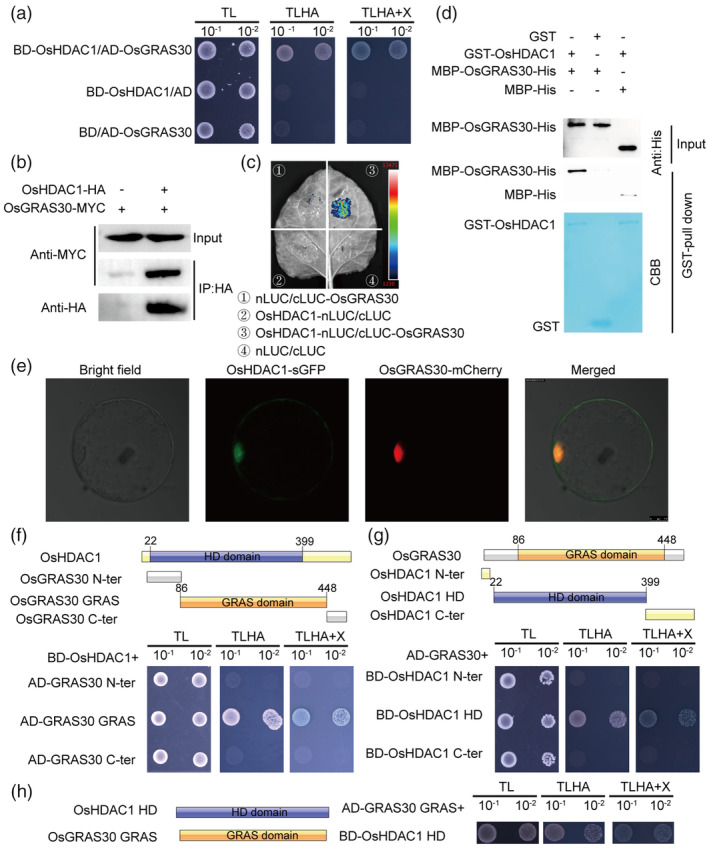
OsHDAC1 interacts with OsGRAS30. (a) Yeast two‐hybrid (Y2H) assay showing OsHDAC1 and OsGRAS30 interaction in yeast. Empty BD and AD were used as the negative controls. (b) Coimmunoprecipitation assay showed that OsHDAC1 interacted with OsGRAS30 in rice protoplast. + or ‐, presence or absence of protein. (c) Split‐luciferase complementation assay showing the interaction between OsHDAC1 and OsGRAS30 in N. benthamiana leaves. Empty nLUC and cLUC were used as negative controls. (d) GST pull‐down assay showing the interaction between OsHDAC1 and OsGRAS30 in vitro. GST‐OsHDAC1 and GST were stained with CBB as loading controls. GST and MBP tags were used as the negative control. + or –, presence or absence of protein. (e) Confocal microscopy result of OsHDAC1‐sGFP co‐localizing with OsGRAS30‐mCherry in rice protoplasts. Scale bar = 7.5 μm. (f–h) Y2H assays showing the interaction between different regions of OsHDAC1 and different regions of OsGRAS30 in yeast. Schematic of the domain structure of OsHDAC1 (f) and OsGRAS30 (g) are indicated. SD, synthetic dropout medium; BD, binding domain; AD, activation domain; TL, SD medium lacking leucine and tryptophan; TLHA, SD medium lacking leucine, tryptophan, histidine and adenine; X, X‐α‐gal. HA, HA epitope tag sequence; MYC, MYC epitope tag sequence; GST, GST epitope tag sequence; His, His epitope tag sequence; MBP, MBP epitope tag sequence; CBB, Coomassie brilliant blue; LUC, luciferase gene; N‐ter, N‐terminal region; C‐ter, C‐terminal region; HD, histone deacetylase domain; GRAS, GRAS domain.
We attempted to delineate the interaction interface by testing three OsHDAC1 fragments for interaction with three OsGRAS30 fragments in a yeast two‐hybrid (Y2H) assay. First, we divided OsGRAS30 into three parts: the N‐terminal region (amino acids 1–85), GRAS domain (amino acids 86–448) and C‐terminal region (amino acids 449–501). The results showed that the GRAS domain region of OsGRAS30 was important for its interaction with OsHDAC1 (Figure 2f). Then, we divided OsHDAC1 into three parts: the N‐terminal region (amino acids 1–22), histone deacetylase domain (amino acids 22–399) and C‐terminal region (amino acids 400–518). As shown in Figure 2g, the histone deacetylase domain of OsHDAC1 interacted with OsGRAS30. We further found that the histone deacetylase domain of OsHDAC1 interacted with the GRAS domain region of OsGRAS30 (Figure 2h). These results indicate that the histone deacetylase domain of OsHDAC1 and the GRAS domain region of OsGRAS30 are essential for their interaction.
OsGRAS30 functions as a positive regulator of rice blast resistance
To investigate the role of OsGRAS30 in the response to M. oryzae, we first assessed the change in its expression levels after blast infection. The RNA level began to decrease at 3 h post‐infection (hpi) with blast and was further reduced at 12 hpi, reaching 40% of the level in plants with no blast infection (Figure 3a). We then generated loss‐of‐function and gain‐of‐function transgenic plants. We used clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 technology to create OsGRAS30‐knockout mutants, with one site in the exon selected for guide RNA (gRNA) design, as shown in Figure 3b. DNA sequencing revealed that among the mutant lines isolated in this study, osgras30 #1 has a single nucleotide deletion, and osgras30 #2 has a two‐nucleotide deletion at the gRNA site (Figure 3b). Each mutation causes a frameshift in the OsGRAS30‐coding sequence. We generated rice transgenic rice plants overexpressing OsGRAS30‐MYC using the pCXUN vector containing OsGRAS30 cDNA fused to the MYC tag driven by the Ubiquitin promoter in the WT background (Figure S4a). Of 10 OE lines, 4 exhibited increased OsGRAS30 RNA levels to varying degrees (Figure S4b). We selected lines OE4 and OE8, which displayed the highest OsGRAS30 RNA levels, for further study.
Figure 3.
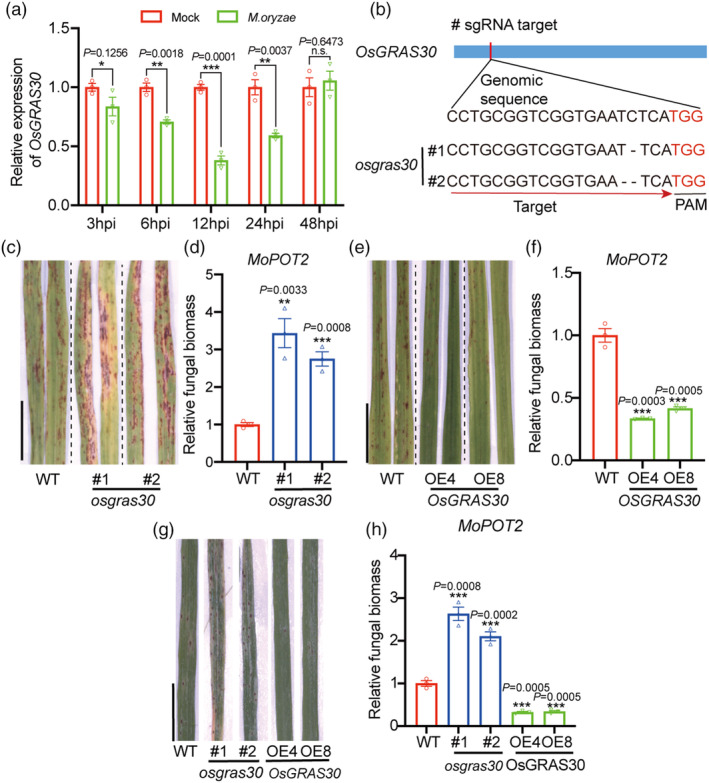
OsGRAS30 is correlated with blast resistance in rice. (a) Transcript variation of OsGRAS30 in response to M. oryzae. Total RNA was extracted from leaves after 3, 6, 12, 24 or 48‐h M. oryzae infection and subjected to RT–qPCR. Expression levels of genes in the WT without M. oryzae treatment were set to 1.00. Values are mean ± SEM (n = 3). Experiments were repeated three times with similar results. (b) Gene structure and mutation sites of OsGRAS30. sgRNA, small guide RNA; PAM, protospacer adjacent motif. (c) Disease symptoms after spray‐inoculation with a spore suspension of M. oryzae for 5 dpi in OsGRAS30 mutants and WT. Scale bar = 1 cm. Representative data from three independent experiments. (d) Relative fungal biomass in OsGRAS30 mutants compared with WT determined using qPCR for the MoPot2 normalized to rice Ubiquitin. Fungal biomass in the WT treated with M. oryzae was set to 1.00. Values are means ± SEM (n = 3). Experiments were repeated three times with similar results. (e) Disease symptoms after spray‐inoculation with a spore suspension of M. oryzae for 5 dpi in OsGRAS30 OE lines and WT. Scale bar = 1 cm. Representative data from three independent experiments. (f) Relative fungal biomass in OsGRAS30 OE lines compared with WT determined by qPCR for the MoPot2 normalized to rice Ubiquitin. Fungal biomass in the WT treated with M. oryzae was set to 1.00. Values are means ± SEM (n = 3). Experiments were repeated three times with similar results. (g) Disease symptoms of the OsGRAS30 mutant and OE lines in rice leaves inoculated with mixed isolates including AH4, Sc09‐153‐07, TM3‐2, RB1, RB3, NC1, HLJ1‐3, HLJ09‐17‐1 and HLJ5‐3 compared with WT. Scale bar = 1 cm. Representative data from three independent experiments. (h) Relative fungal biomass determined in OsGRAS30 mutant and OE lines compared with WT determined using qPCR for the MoPot2 gene normalized to rice Ubiquitin. Values are means ± SEM (n = 3). Experiments were repeated three times with similar results. Asterisks mark significant changes compared with WT without M. oryzae treatment (a) or WT with M. oryzae treatment (d, f and h) based on Student's t‐test: *P < 0.05, **P < 0.01, ***P < 0.001. WT, wild‐type; OE, overexpression.
Next, two OsGRAS30 mutant lines (osgras30 #1 and #2), the OsGRAS30 OE lines, and WT were inoculated with blast by spraying Guy11 conidia. The results showed that the osgras30 #1 and osgras30 #2 lines exhibited higher susceptibility to blast treatment compared with WT plants (Figure 3c), and the fungal DNA contents in the OsGRAS30 mutant lines were increased by 2.7‐ and 3.3‐fold compared with WT (Figure 3d). In contrast, we observed enhanced resistance to blast after inoculating the OsGRAS30 OE4 and OE8 lines and WT (Figure 3e). These two OE lines contained lower fungal DNA content than that of WT (Figure 3f). We further inoculated the OsGRAS30 mutant and OE lines along with WT by spraying conidia from mixed M. oryzae isolates including AH4, Sc09‐153‐07, TM3‐2, RB1, RB3, NC1, HLJ1‐3, HLJ09‐17‐1 and HLJ5‐3 (Figure S2). The OsGRAS30 OE lines showed enhanced resistance to multiple virulent isolates of M. oryzae, whereas the mutant lines showed higher susceptibility, compared with WT (Figure 3g). Relative fungal biomass increased by 2‐ and 2.5‐fold in the mutant lines and decreased by 3‐fold in the OE lines (Figure 3h). Taken together, these data from the mutant and OE lines suggest that the OsGRAS30 gene positively regulates broad‐spectrum resistance to rice blast.
OsHDAC1 acts genetically downstream of OsGRAS30
To analyse the genetic interactions between OsGRAS30 and OsHDAC1, we used Agrobacterium tumefaciens strain EHA105 containing OsHDAC1‐RNAi constructs (Hou et al., 2022) to transform the background of osgras30 #1 plants and thereby generated homozygous double OsHDAC1 RNAi/osgras30 lines. Through this process, we obtained 10 putative OsHDAC1 RNAi lines of osgras30 #1 rice with reduced OsHDAC1 expression; OsHDAC1 RNA levels were reduced by approximately 90% in the OsHDAC1 RNAi #5 and #9 lines compared with the osgras30 #1 line (Figure 4a). Then, two OsHDAC1 RNAi lines with the osgras30 #1 background, the osgras30 mutants and WT were inoculated with conidia from mixed M. oryzae isolates. The results indicated that the OsHDAC1 RNAi #5 and #9 lines in the osgras30 #1 background showed enhanced resistance to multiple virulent isolates of M. oryzae compared with OsGRAS30 mutants, whereas OsHDAC1 RNAi #5 and #9 lines in the osgras30 #1 background showed similar blast resistance phenotype observed in the WT (Figure 4b). Measurement of M. oryzae DNA content confirmed the phenotype results (Figure 4c). This demonstrates that OsGRAS30 acts, at least in part, genetically upstream of OsHDAC1 to positively regulate blast resistance.
Figure 4.
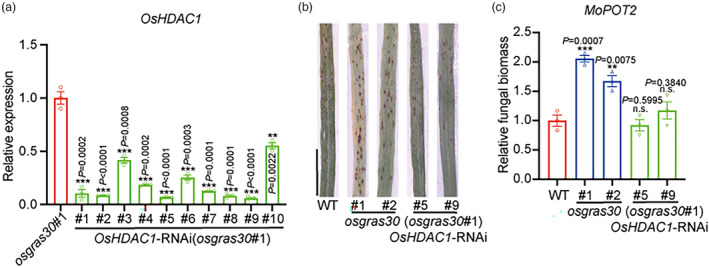
OsGRAS30 promotes blast resistance in an OsHDAC1‐dependent manner. (a) Expression levels of OsHDAC1 in OsHDAC1 RNAi lines (from #1 to #10) with the osgras30 #1 background. Total RNA was extracted from leaves and subjected to RT–qPCR. Expression levels of OsHDAC1 in the osgras30 #1 plants were set to 1.00. Values are mean ± SEM (n = 3). Experiments were repeated three times with similar results. (b) Disease symptoms after spray‐inoculation with a spore suspension of M. oryzae for 5 dpi in OsHDAC1 RNAi lines with the osgras30 #1 background compared with WT and OsGRAS30 mutants. Scale bar = 1 cm. Representative data from three independent experiments. (c) Relative fungal biomass in OsHDAC1 RNAi lines with the osgras30 #1 background compared with WT and OsGRAS30 mutants determined using qPCR for the MoPot2 gene normalized to rice Ubiquitin. Fungal biomass in the WT plants infected with M. oryzae was set to 1.00. Values are means ± SEM (n = 3). Experiments were repeated three times with similar results. Asterisks mark significant changes compared with osgras30 #1(a and c) based on Student's t‐test: **P < 0.01, ***P < 0.001.
OsGRAS30 represses OsHDAC1 enzyme activity to induce H3K27ac hypoacetylation
To further investigate how OsGRAS30 functions in OsHDAC1‐mediated immunity in response to M. oryzae. We first detected the abundance of OsHDAC1 proteins in WT, OsGRAS30 mutant and OsGRAS30 OE lines by immunoblotting using an antibody against OsHDAC1. The results indicated that the OsHDAC1 protein level remained unchanged in the OsGRAS30 mutant and OsGRAS30 OE lines (Figure S5a), and the transcript level of OsHDAC1 was also unchanged in OsGRAS30 mutant (Figure S5b,c), suggesting that OsGRAS30 did not affect OsHDAC1 abundance or stability. Thus, we speculated that OsGRAS30 interferes with the deacetylation activity of OsHDAC1. To confirm this hypothesis, we analysed HDAC activity in WT, OsGRAS30 mutants, and OE plants, and the results showed that HDAC activity was significantly elevated in osgras30 #1 and #2 lines and markedly reduced in OsGRAS30 OE4 and OE8 lines compared with WT (Figure 5a). In vitro activity analysis revealed that OsGRAS30 inhibited the deacetylation activity of OsHDAC1 by binding to OsHDAC1 (Figure 5b).
Figure 5.
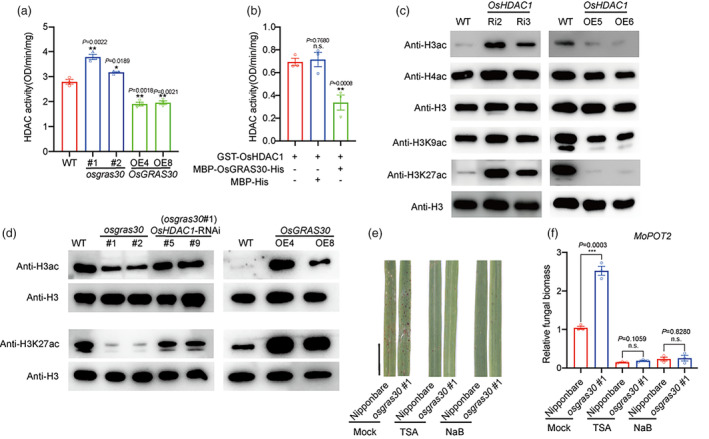
OsGRAS30 represses OsHDAC1 to induce H3K27ac hypoacetylation. (a) In vivo HDAC activity assays in WT, OsGRAS30 mutant and OE lines. Values are mean ± SEM (n = 3). Experiments were repeated three times with similar results. (b) In vitro analysis of the effect of OsGRAS30 on OsHDAC1 activity. Values are mean ± SEM (n = 3). + or –, presence or absence of proteins. Experiments were repeated three times with similar results. (c) Overall H3ac, H4ac, H3K9ac and H3K27ac levels in WT, OsHDAC1 RNAi and OE lines using immunoblotting. Histone H3 was applied as an equal loading control. Representative data from three independent experiments. (d) Immunoblotting analysis of the overall H3ac, H4ac, H3K9ac and H3K27ac levels in WT, OsGRAS30 mutant lines, and OsHDAC1 RNAi lines in the background of osgras30 #1 and OsGRAS30 OE lines (line numbers are given above the graph). H3 protein was applied as an equal loading control. Representative data from three independent experiments. (e) Disease symptoms after spray‐inoculation with a spore suspension of M. oryzae for 5 dpi in osgras30#1 and WT treated with HDAC inhibitors including TSA and NaB. Untreated was used as a control. Scale bar = 1 cm. Representative data from three independent experiments. (f) Relative fungal biomass was determined using qPCR for the MoPot2 gene normalized to rice Ubiquitin. Values are means ± SEM (n = 3). Experiments were repeated three times with similar results. Asterisks mark significant changes compared with WT (a and b) or WT with M. oryzae treatment (f) based on Student's t test: *P < 0.05, **P < 0.01, ***P < 0.001. n. s., not significant change. WT, wild‐type; Ri, RNA interference; OE, overexpression.
HDACs function to remove the acetyl group from acetylated lysines in histones (Kumar et al., 2021). Thus, we assessed overall histone acetylation in WT, OsHDAC1 RNAi and OsHDAC1 OE lines by immunoblotting, and the results showed that the overall histone 3 acetylation (H3ac) level was dramatically elevated in OsHDAC1 RNAi lines and dramatically reduced in OsHDAC1 OE lines compared with WT (Figure 5c). Previous reports have indicated that H3K27me3 and H3K27ac regulate functional genes in M. oryzae (Lin et al., 2022; Zhang et al., 2021), and H3K9ac plays an essential role in plant defence responses in rice (Chen et al., 2022a). H3K9ac and H3K27ac are the most characterized epigenetic marks invariably associated with transcriptional activation and have been shown to affect many developmental and biological processes in higher plants (Dasgupta et al., 2022; Zhou et al., 2010). This prompts us to further assess the level of histone H3 acetylation at lysines 9 and 27 by immunoblotting using specific antibodies. The results indicated that the H3K27ac levels were higher in the OsHDAC1 RNAi lines and lower in the OsHDAC1 OE lines compared with WT rice plants (Figure 5c), suggesting that H3K27ac is a critical regulatory site of OsHDAC1. We then challenged rice plants with the virulent M. oryzae Guy11, and observed that H3K27ac levels were upregulated after M. oryzae infection (Figure S6). These results confirmed that H3K27ac was involved in blast defence responses in rice. Further, we detected total acetylation levels of histone 3 and 4 in OsGRAS30 mutant lines, OsGRAS30 OE lines, OsHDAC1 RNAi /osgras30 #1 lines, and WT plants to assess whether OsGRAS30 impacts OsHDAC1‐mediated H3 acetylation level. The results showed that overall H3ac levels were decreased in OsGRAS30 mutant lines, but OsHDAC1 RNAi lines in the osgras30 #1 background restored this decrease, while H3ac levels were elevated in the OsGRAS30 OE lines (Figure 5d). We specifically examined H3K9ac and H3K27ac levels in these lines and found that H3K27ac shows a similar trend to H3ac (Figure 5d). These results demonstrate that OsGRAS30 regulates H3K27ac levels by suppressing the deacetylated activity of OsHDAC1. In addition, we applied a series of different concentrations of HDAC inhibitors including trichostatin A (TSA) and sodium butyrate (NaB) to WT plants inoculated with spore suspensions for 5 days and found that the resistance of rice plants to blast infection increased with the HDAC inhibitor concentration (Figure S7). Furthermore, treatment of WT and osgras30 #1 rice plants with HDAC inhibitors greatly improved their symptoms, and osgras30 #1 treated with HDAC inhibitors showed greater blast resistance compared with untreated WT plants (Figure 5e,f). Taken together, these observations demonstrate that OsGRAS30 enhances blast resistance by suppressing OsHDAC1 in rice.
OsHDAC1 and H3K27ac co‐occupy targets in rice genome
To elucidate the molecular determinants and targets of OsHDAC1, we performed chromatin immunoprecipitation sequencing (ChIP–Seq) to detect OsHDAC1 binding sites in the genome of the OsHDAC1 OE5 line using an antibody against the HA tag (Figure S8) because the HA antibody performed better than did our customized OsHDAC1 antibody in ChIP and two biological replicates showed a strong correlation (Figure S9). A total of 21, 471 OsHDAC1‐binding sites, corresponding to 15, 420 genes, were identified (Dataset S1). Analysis of the OsHDAC1 distribution in the rice genome showed that among the OsHDAC1 targets, 60.19% of binding peaks occurred in gene regions and OsHDAC1 enrichment was mostly detected near transcription start site (TSS), indicating that OsHDAC1 were highly enriched in the promoter region surrounding the TSS to regulate transcriptional initiation (Figure 6a,b). Examination of the correlation between OsHDAC1 binding and gene expression levels revealed that OsHDAC1 was more enriched in genes with high‐transcription levels than in genes with low‐transcription levels, and which was located mainly around the TSS of these genes (Figure 6c). We further identified potential DNA‐binding motifs in OsHDAC1‐binding sites using DREME, and a significantly enriched consensus G‐box motif was identified (Figure 6d), which is similar to the report that HAD9 in Arabidopsis could bind to the G‐box (Chen et al., 2016). Therefore, OsHDAC1 is associated mainly with active genes and is preferentially enriched in their promoter regions.
Figure 6.
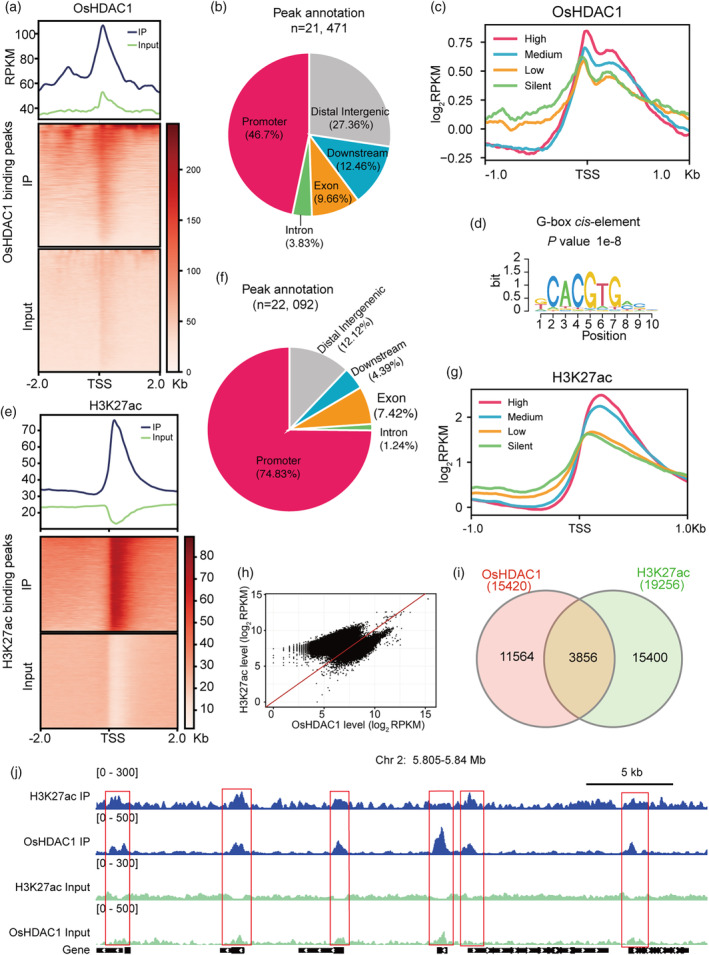
OsHDAC1 co‐occupies with H3K27ac at genome wide. (a) Metagene plot and heatmap showing OsHDAC1 binding levels on the 2.0‐kb flanking genomic region surrounding TSS in rice. (b) Pie chart showing the distribution of OsHDAC1 binding peaks at annotated genic and intergenic regions in the rice genome. (c) Metagene plot showing OsHDAC1 binding profiles on the 1.0‐kb flanking genomic region surrounding TSS of genes with high, medium, low and silent expression. (d) DREME identified representative DNA motifs in OsHDAC1 binding sites. (e) Metagene plot and heatmap showing H3K27ac enrichment levels on the 2.0‐kb flanking genomic region surrounding TSS in rice. (f) Pie chart shows the distribution of H3K27ac enrichment peaks at annotated genic and intergenic regions in the rice genome (g) Metagene plot showing OsHDAC1 binding profiles on the 1.0‐kb flanking genomic region surrounding TSS of genes with high, medium, low and silent expression. (h) Correlation between OsHDAC1 binding and H3K27ac level in OsHDAC1 target genes. (i) Venn diagram showing overlap of OsHDAC1 binding genes and H3K27 acetylated genes. (j) IGV screen shoot showing the co‐localization between OsHDAC1 and H3K27ac at a representative region in chromosome 2. TSS, the transcription start site.
Next, we investigated genome‐wide targets of H3K27ac by ChIP‐Seq using an antibody against H3K27ac in WT plants with two biological replicates, whose results showed a strong correlation (Figure S10). A total of 22, 092 H3K27ac peaks, corresponding to 19, 256 genes, were identified (Dataset S2). Analysis of the H3K27ac distribution in the rice genome showed that H3K27ac peaks were located mainly in gene regions (83.49%), especially in promoter regions surrounding the TSS, and H3K27ac enrichment was mostly detected around the TSS (Figure 6e,f). In addition, we observed that H3K27ac was enriched in genes with high‐transcription levels and occurred mainly around the TSS of these genes, whereas silent genes showed a low level of H3K27ac enrichment (Figure 6g). Interestingly, analysis of the relationship between OsHDAC1 binding and acetylation levels showed that OsHDAC1 tends to be positively correlated with the H3H27ac level at the target sites (Figure 6h), and 3, 856 genes were co‐bound by OsHDAC1 and H3K27ac (Figure 6i). In humans, HDAC levels have also been shown to positively correlate with histone acetylation levels, and it is suggested that HDACs function to remove the acetyl group added by HATs in active genes and to reset chromatin modification after gene activation (Wang et al., 2002, 2009). Finally, a randomly selected region on chromosome 2 confirmed that OsHDAC1‐occupied peaks indeed colocalized with H3K27ac (Figure 6j), which further supports the correlation between OsHDAC1 and H3K27ac. Overall, we discovered that H3K27ac is associated with expressed genes in the rice genome and that OsHDAC1 functions to remove the acetyl moieties from H3K27ac to control the H3K27ac level in co‐occupied target genes.
OsHDAC1 modulate blast resistance by directly suppressing blast resistance‐related genes
To further investigate the effect of OsHDAC1 on gene expression associated with rice resistance to blast. We first profiled mRNA from 2‐week‐old rice leaves of WT and OsHDAC1 Ri2 plants via RNA‐Seq. There are three biological replicates, whose results showed a strong correlation (Figure S11). In total, 2, 398 upregulated and 2, 953 downregulated genes (expression level fold change >2 and FDR <0.05) were identified in the OsHDAC1 RNAi lines compared with corresponding WT plants (Figures 7a and S12; Dataset S3). Next, we investigated the association between OsHDAC1 and H3K27ac binding and altered gene expression in OsHDAC1 Ri2 plants. The results showed that the expression of 220 genes bound by OsHDAC1 and enriched by H3K27ac was significantly increased in the OsHDAC1 Ri2 lines, while the expression of 223 genes bound by OsHDAC1 and enriched by H3K27ac was significantly decreased in the OsHDAC1 Ri2 lines (Figure 7b).
Figure 7.
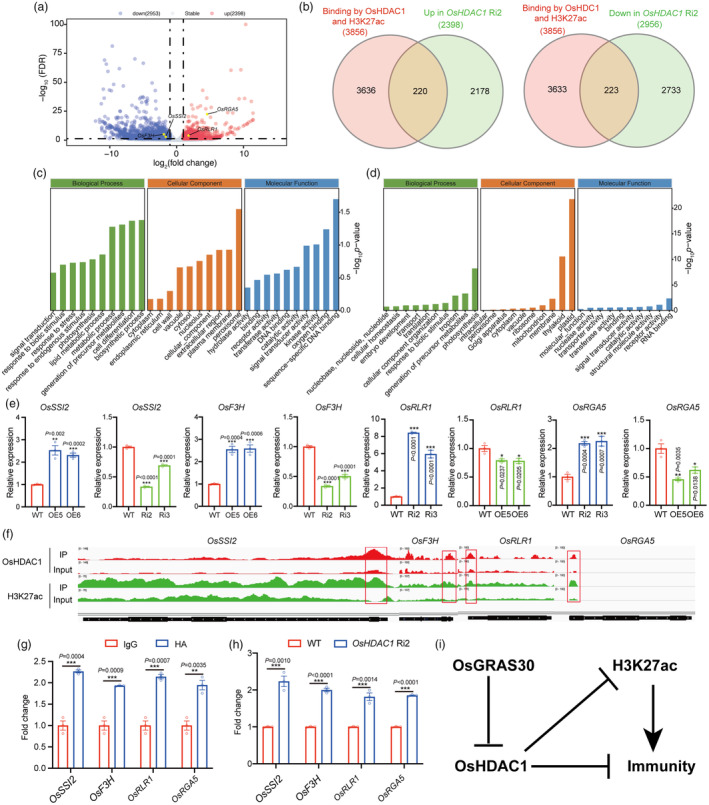
OsHDAC1 regulates the expression of blast resistance‐related genes. (a) Volcano plot showing differentially expressed genes (DEGs) associated with OsHDAC1 in the OsHDAC1 Ri2 line compared with the WT. (b) Venn diagram showing overlap of DEGs in the OsHDAC1 Ri2 line, OsHDAC1 binding genes and H3K27ac enrichment genes. (c) Go results showing the related biological pathway enriched from downregulated DEGs in OsHDAC1 Ri2 line compared with WT. (d) Go results showing the related biological pathway enriched from upregulated DEGs in OsHDAC1 Ri2 line compared with WT. (e) The transcript level of blast resistance‐related genes in OsHDAC1 OE and RNAi lines compared with WT. Total RNA was extracted from two‐week‐old rice leaves and subjected to RT–qPCR. Expression levels of genes in the WT were set to 1.00. Values are mean ± SEM (n = 3). Experiments were repeated three times with similar results. (f) IGV views showing OsHDAC1 and H3K27ac enrichment in blast resistance‐related genes. Input serves as the negative control. (g) ChIP‐qPCR results of OsHDAC1 binding levels at blast resistance‐related genes in OsHDAC1 OE plants. ChIP‐qPCR results of IgG binding levels of genes were set to 1.00. Values are mean ± SEM (n = 3). Experiments were repeated three times with similar results. (h) ChIP‐qPCR results of H3K27ac levels at blast resistance‐related genes in OsHDAC1 Ri2 line compared with WT plants. ChIP‐qPCR results of H3K27ac levels of genes in the WT were set to 1.00. Values are mean ± SEM (n = 3). Experiments were repeated three times with similar results. Asterisks mark significant changes compared with WT (e and h) or IgG (g) based on Student's t test: *P < 0.05, **P < 0.01, ***P < 0.001. (i) A proposed working model showing the role of OsGRAS30 in manipulating the OsHDAC1‐mediated broad‐spectrum blast resistance in rice. WT, wild‐type; Ri, RNA interference; OE, overexpression.
The differentially expressed genes (DEGs) were functionally characterized based on Gene Ontology (GO) analysis. The main GO terms assigned to downregulated DEGs affected by OsHDAC1 were the biosynthetic process and metabolic process (Figure 7c; Dataset S4). The KEGG analysis revealed that these downregulated DEGs enriched in fatty acid degradation and biosynthesis (Figure S13a; Dataset S5). Prior study revealed that fatty acid was involved in broad‐spectrum blast resistance in rice (He et al., 2020), suggesting that OsHDAC1 negatively regulates blast resistance by modulating fatty acid contents. Suppression of the fatty acid desaturase gene OsSSI2 has been reported to enhance resistance to blast by regulating fatty acid content (Jiang et al., 2009), and we verified that OsHDAC1 could increase the expression of OsSSI2 by binding its promoter region and altering the H3K27ac levels (Figure 7e–h). Similarly, OsF3H has been reported to involve in blast resistance by mediating the biosynthesis, signalling and transport of SA (Chen et al., 2022b; Wu et al., 2022). Our result showed that OsF3H is the direct target of OsHDAC1 by RT–qPCR and ChIP–PCR experiments (Figure 7e–h). Conversely, we found that upregulated DEGs involved in biotic stimulus were enriched (Figures 7d and S13b, Dataset S6 and S7), and genes related to blast resistance (OsRLR1 and OsRGA5) were upregulated in OsHDAC1 Ri2 plants and downregulated in OsHDAC1 OE5 plants (Figure 7e). Among, OsRLR1 confers broad‐spectrum resistance in rice by increasing hydrogen peroxide (H2O2) content (Du et al., 2021); OsRGA5 recognizes the M. oryzae effector AVR1‐CO39 to relieve OsRGA4 to promote blast resistance (Césari et al., 2013, 2014; Okuyama et al., 2011), supporting the role of these genes in blast resistance. Further, we found that OsHDAC1 was significantly enriched in OsRLR1 and OsRGA5 via ChIP–qPCR, and the H3K27ac levels in these target genes were significantly elevated in the OsHDAC1 Ri2 lines compared with WT plants (Figure 7f–h). Together, these results suggest that OsHDAC1 binds to blast resistance‐related genes and alters their H3K27ac levels to regulate their expression, revealing a link between blast resistance and OsHDAC1.
Discussion
Rice feeds half of the world's population, and rice blast is a very serious disease that affects rice production (Chakraborty et al., 2021). Functional characterization of non‐race specific resistance‐related genes is particularly important for molecular breeding to improve broad‐spectrum disease resistance and the development of strategies for effective control of this disease in rice. In this study, we report the functional identification of the OsGRAS30–OsHDAC1 module in rice blast resistance. OsGRAS30 acts genetically upstream of OsHDAC1 and interacts with OsHDAC1, repressing its deacetylation activity; inactivated OsHDAC1 induces the H3K27ac hyperacetylation and regulates the expression of blast resistance‐related genes, leading to broad‐spectrum blast resistance (Figure 7i).
Silencing of OHDAC1 confers broad‐spectrum resistance to M. Oryzae in rice
In plants, histone deacetylases are involved in the regulation of both growth and stress response functions (Chakraborty et al., 2021). A few HDAC members have been reported to regulate plant defence pathways, including HDA6 (Wang et al., 2017), HDA19 (Choi et al., 2012; Kim et al., 2008), HD2B (Latrasse et al., 2017) and SRT2 (Wang et al., 2010) in Arabidopsis; HDA701 (Chen et al., 2022a) and HDT701 (Ding et al., 2012; Li et al., 2021) in rice; and HDA6 in wheat (Zhi et al., 2020). Although HDA701 and HDT701 have been demonstrated to confer disease resistance in rice, the downstream factors related to their role in plant immunity remain unclear (Chen et al., 2022a; Li et al., 2021). Previous studies showed that homozygous OsHDAC1 mutant seeds could not be obtained and that partial inhibition of OsHDAC1 repressed root growth (Hou et al., 2022). In this work, the protein abundance of OsHDAC1 was downregulated at the early stage of blast infection (Figure 1b) and we further demonstrate that partial inhibition of OsHDAC1 contributes to plant defence against multiple isolates of M. oryzae without affecting plant height and tiller numbers (Figures 1 and S3). Similarly, mutations in a few genes are reported to constitutively promote the nucleotide‐binding leucine‐rich repeat immune response, as well as to cause plant growth retardation (Palma et al., 2010; Shirano et al., 2002). These findings suggest that OsHDAC1 is important for rice growth but knockdown of OsHDAC1 enhances broad‐spectrum resistance to rice blast fungus M. oryzae, making it a prime candidate gene for manipulation in breeding programs.
OsGRAS30 suppresses OsHDAC1 activity, leading to enhanced blast resistance in rice
OsGRAS30 is a member of the GRAS family, and previous studies have revealed that GRAS transcription factors function in plant growth, development, and resistance to various biotic and abiotic stresses (Jaiswal et al., 2022). Our data suggest that OsGRAS30 positively regulates blast resistance in rice, which is supported by our finding that loss of function and overexpression of OsGRAS30 decreased and increased blast resistance, respectively (Figure 3). Previous reports showed that EAR domain‐containing transcription factors trigger PRC2‐mediated chromatin marking in Arabidopsis (Baile et al., 2021). HDA9 and WRKY53 transcription factors are mutual antagonists (Zheng et al., 2020), and MADS‐domain transcription factor AGL15 involves in the recruitment of histone deacetylase complex components (Hill et al., 2008). Thus, the interaction of transcription factor and chromatin remodelling components is valuable for understanding the transcription factor as well as the chromatin remodelling mechanism. Our in vitro and in vivo assays both suggest that interaction between OsGRAS30 and OsHDAC1 (Figure 2), and overexpression of OsGRAS30 led to reduced OsHDAC1 deacetylation capacity and enhanced blast resistance, while mutation of OsGRAS30 increased blast sensitivity (Figures 3 and 5b,c), indicating that OsGRSA30 functions in blast resistance by inhibiting OsHDAC1 activity. This revealed that a GRAS transcription factor is involved in plant resistance responses by modulating the activity of an epigenetic modification enzyme. Additionally, the GRAS domain of OsGRAS30 binds to the histone deacetylase domain of OsHDAC1 (Figure 2f–h), and GRAS family DELLA proteins were found to interact with the DNA‐binding domains of EIN3/EIL1, which blocks DNA binding domain of EIN3/EIL1 to inhibit its transcriptional regulation of downstream genes in Arabidopsis (An et al., 2012). This implies that OsGRAS30 could block histone deacetylase domain of OsHDAC1 to inhibit its deacetylation capacity. Meanwhile, we did not detect that MBP‐OsGRAS30‐His was acetylated in vitro, suggesting OsGRAS30 is not a substrate for OsHDAC1. Further, the blast resistance phenotype of the OsGRAS30 mutant lines was reversed by OsHDAC1 RNAi (Figure 4b,c), validating that OsGRAS30 can directly target OsHDAC1 to decrease OsHDAC1 enzyme activity. HDACs function to remove the acetyl group from acetylated lysines in histones (Kumar et al., 2021), and we revealed that H3K27ac is a favoured substrate of OsHDAC1 (Figure 5c), while OsGRAS30 induces H3K27ac hyperacetylation by regulating the activity of OsHDAC1 (Figure 5d).
It has revealed that acetylation plays an essential role in crop defences during plant–pathogen interactions. For example, infection of maize by the fungus Cochliobolus carbonum increases the acetylation level of histones H3 and H4, which affects the expression of defence‐related genes (Walley et al., 2018). In rice, the HD2‐type HDAC HDT701 regulates plant innate immunity by modulating H4K5 acetylation in defence‐related genes (Ding et al., 2012), and another report revealed that OsRpp30 confers bacterial, fungal and viral resistance by targeting HDT701 to regulate the H4K5 acetylation level (Li et al., 2021). Similarly, the RPD3‐type HDAC HDA701 affects disease resistance by regulating H3K9 acetylation (Chen et al., 2022a). Our data reveal that the RPD3‐type HDAC OsHDAC1 is associated with the removal of acetyl groups from H3K27, but not H3K9 (Figure 5c), suggesting that OsHDAC1 may use a different mechanism than HDA701 to regulate blast resistance.
Genome‐wide mapping of OsHDAC1 targets reveals its regulatory mechanism
HDACs are generally associated with transcriptional repression (Berger, 2007; Robert et al., 2004). However, recent genome‐wide occupancy studies in plants including Arabidopsis and maize (Chen et al., 2016; Yang et al., 2016), as well as humans (Wang et al., 2009), indicate that HDACs mainly target transcriptionally active genes. In accordance with this conclusion, our data provide evidence that the rice RPD3‐type HDAC OsHDAC1 is preferentially enriched in promoter regions and positively related to gene expression (Figure 6a–c). In addition, OsHDAC1 can bind to the G‐box (Figure 6d), which is consistent with the result obtained for HAD9 in Arabidopsis (Chen et al., 2016). These findings suggest that the interaction of HDACs with active genes may be conserved across plants and mammalian cells. Interestingly, our data show that the level of OsHDAC1 binding tends to be positively correlated with the enrichment of H3K27ac (Figure 6h,j). Similarly, the level of HDA101 binding also appears to be positively correlated with the amount of H4K5ac in maize (Yang et al., 2016). The probable explanation is that HDACs function to remove acetyl groups at active genes added by HATs and to reset chromatin modification after gene activation (Wang et al., 2009). 25.01% of OsHDAC1‐bound genes happen H3K27ac enrichment (Figure 6i), implying that OsHDAC1 bind to these target genes and controls the H3K27ac level. In addition, the more direct targets of OsHDAC1 show no H3K27ac enrichment (Figure 6i). A possible reason for this finding is that other target genes that bind to OsHDAC1 undergo acetylation at sites other than H3K27. Furthermore, our RNA‐Seq data indicated that the knockdown of OsHDAC1 did not affect the transcript levels of most genes bound by OsHDAC1 (Figures 6i and 7b), in accordance with HDACs identified in mammalian cells, maize and Arabidopsis (Chen et al., 2016; Wang et al., 2009; Yang et al., 2016).
Plant disease resistance responses involve transcriptional reprogramming and expression changes in many genes (Tsuda and Somssich, 2015). Rice blast resistance is a complex process in which the expression of numerous genes must be fine‐tuned (Tsuda and Somssich, 2015). Blast R gene‐mediated resistance depends on interactions between R proteins and corresponding pathogenic Avr proteins (Lazar et al., 2022), such as OsRGA5 positively regulates rice blast resistance by affecting the recognition of pathogenic Avr‐CO39 protein (Cesari et al., 2013). In addition, OsRGA4 and OsRGA5 are a pair of R genes for blast resistance, and both are required for blast resistance (Césari et al., 2013; Okuyama et al., 2011), which upon recognition of the pathogen effector AVR‐Pia by direct binding to OsRGA5, OsRGA4 is relieved to induce cell death (Césari et al., 2014). Upregulation of OsRGA5 due to OsHDAC1 knockdown may possibly contribute to the recognition of the M. oryzae effectors. OsRLR1 improve broad‐spectrum blast resistance by regulating H2O2 accumulation (Du et al., 2021). OsRGA5, along with OsRLR1 were identified downstream of OsHDAC1. Our results indicate that OsHDAC1 bind to the promoter region of these genes and alters the H3K27ac levels, inhibiting their expression to suppress immune activation (Figure 7). Thus, an important finding of our study is that positive regulators of blast resistance‐related genes are associated with OsHDAC1 binding and are inhibited by OsHDAC1 (Figure 7). Conversely, negative regulators of blast resistance‐related genes associated with OsHDAC1 binding are upregulated by OsHDAC1 (Figure 7). These genes including OsF3H and OsSSI2 caused the decreased production of SA and fatty acid, and their inhibition could achieve broad‐spectrum resistance to M. oryzae in rice (Chen et al., 2022b; Jiang et al., 2009). Overall, our findings revealed that OsHDAC1 directly affect these targets and thus alters their expression to regulate blast resistance in rice.
A chemical intervention strategy reduces HDAC activity to increase blast resistance in rice
Rice blast, caused by M. oryzae, is an important disease affecting rice production, and thus it threatens global food security (Chakraborty et al., 2021). The symptoms of rice blast can occur throughout the rice growth period (Chakraborty et al., 2021). Genetic breeding of durably resistant plants is considered an effective strategy for controlling plant diseases, although it is difficult and time consuming to accomplish without affecting plant growth due to the limited set of currently available broad‐spectrum resistance genes without detrimental effects (He et al., 2022). Some genes have been engineered to increase disease resistance directly. The present study showed that knockdown of OsHDAC1 via RNAi conferred rice blast disease resistance, and our preliminary field test results showed no difference in plant height and tiller numbers, between OsHDAC1‐knockdown and WT rice (Figures 1 and S3), revealing that the development of resistant rice can be achieved by decreasing the expression, rather than inducing loss of function, of a target gene. Another strategy for disease resistance is chemical intervention, i.e., application of small‐molecule chemicals or drugs to enhance resistance via interference with the functions of target genes. HDAC inhibitor treatment enhanced rice blast resistance (Figure S7) provides a foundation for further application of such chemicals to disease intervention in vitro.
Materials and methods
Plasmid construction and generation of transgenic plants
For the purification of OsHDAC1‐His and MBP‐OsGRAS30‐His recombinant proteins, a fragment containing the OsHDAC1 coding sequence (CDS) was amplified and cloned into the EcoRI site of the pET32a vector, and the OsGRAS30 CDS fragment was amplified and inserted into the EcoRI site of the pMalC2x vector. To generate gene constructs fused to the DNA binding domain or activation fragment for Y2H assays, the OsHDAC1 CDS fragments, as well as the N‐terminal region (nucleotides 1–63), histone deacetylase domain region (nucleotides 64–1197), and C‐terminal region (nucleotides 1198–1557) of OsHDAC1 were amplified and cloned into the EcoRI site of the pGBKT7 vector; the OsGRAS30 CDS fragments, as well as the N‐terminal region (nucleotides 1–255), GRAS domain region (nucleotides 256–1344) and C‐terminal region (nucleotides 1345–1503) of OsGRAS30 were amplified and cloned into the EcoRI site of the pGADT7 vector. To generate the OsHDAC1‐nLUC and cLUC‐OsGRAS30 constructs, the OsHDAC1 CDS fragments were amplified and cloned into the KpnI and SacI sites of the pCambia1300‐nLUC vector, while OsGRAS30 CDS fragments were amplified and cloned into the KpnI and SalI sites of the pCambia1300‐cLUC vector. To generate the OsGRAS30‐mCherry constructs, the amplified OsGRAS30 CDS fragments were fused to a mCherry tag to generate fusion genes and cloned into the BamH1 and SalI sites of the pBI121 vector. To generate OsGRAS30 OE plants, the amplified OsGRAS30 CDS fragment was fused to a MYC tag to generate fusion genes and cloned into the XcmI site of the pCXUN vector. To generate OsGRAS30 mutant plants, guide sequences were designed using CRISPR‐P (http://cbi.hzau.edu.cn/crispr) and inserted into the sgRNA expression vector backbone pYLsgRNA‐OsU6 using annealed oligonucleotides. Then, the sgRNAs were inserted into the pYLCRISPR/Cas9Pubi‐H vector as described previously (Hou et al., 2022). The OsGRAS30‐MYC and CRISPR/Cas9‐OsGRAS30 constructs were transfected into A. tumefaciens strain EHA105 for transformation of WT (Nipponbare) and OsHDAC1‐RNAi constructs (Hou et al., 2022) were transfected into EHA105 for transformation in the background of osgras30 #1. These transformations were performed by Wuhan BioRun Biosciences Co., Ltd. (Wuhan, China).
Plant materials and growth conditions
All rice plants used in this study, including Nipponbare (Oryza sativa L. ssp. japonica), the OsHDAC1 RNAi and OE lines in the background of Nipponbare (Hou et al., 2022), the OsGRAS30 mutants and OE lines, and the OsHDAC1 RNAi lines in the background of osgras30 #1 were grown in hydroponic cultures in Kimura B nutrient salts under a photoperiod of 14 h light at 28 °C and 10 h dark at 26 °C with 70% humidity. For growth in the field, germinated rice seedlings were transplanted into the Huashan rice experimental paddy fields at the Wuhan University in Wuhan, Hubei province, China in mid‐May; grains were harvested in October. Urea was used as the nitrogen source, applied at 2 kg N/100 m2. The rice plants were transplanted at a density of three rows of 38 plants with spacing between rice plants of 20 cm. The N. benthamiana plants used in this study were grown in soil under a photoperiod of 14 h light at 22 °C and 10 h dark at 22 °C with 70% humidity.
Inoculation of rice leaves with M. Oryzae strains
The M. oryzae strains Guy11, AH4, Sc09‐153‐07, TM3‐2, RB1, RB3, NC1, HLJ1‐3, HLJ09‐17‐1 and HLJ5‐3 were used in this study. M. oryzae cultivation and inoculation assay were performed using methods described previously with modifications (Li et al., 2017). Briefly, M. oryzae strains were gown on complete agar medium in a growth chamber at 25 °C under a 12‐h light/12‐h dark photoperiod to produce spores, which were collected with sterile water at a final concentration of 5 × 105 spores/mL in suspension. For spray inoculation, 2‐week‐old rice seedlings were sprayed in a dew growth chamber with spore suspensions from various M. oryzae strains. At 5 days after inoculation, the lesion area was measured using Photoshop according to a previously reported method (Li et al., 2017), and relative fungal biomass was determined by qPCR analysis of the M. oryzae POT2 gene, the expression of which was normalized to that of the rice Ubiquitin gene, as described previously (Wang et al., 2018). All inoculation experiments were repeated three times independently.
HDAC inhibitor treatments
TSA (Selleck Chemicals, Cat. # S1045, Houston, TX, USA) was dissolved in dimethyl sulfoxide. NaB (Beyotime, Cat. # ST1636, Shanghai, China) was dissolved in double‐distilled water. Two‐week‐old rice seedlings were spray‐inoculated with M. oryzae spore suspensions and transferred to Kimura B nutrient salts containing the indicated concentration of TSA or NaB for 5 days. Control plants were grown in an identical volume of dimethyl sulfoxide or double‐distilled water as a mock treatment.
RNA extraction and RT–qPCR
RNA was extracted from the leaves of 2‐week‐old seedlings with Trizol reagent (ThermoFisher, Cat. # 15596026, Waltham, MA, USA) and then reverse‐transcribed into cDNA using the SuperScript III transcriptase kit (ThermoFisher, Cat. # 18080093). RT–qPCR analysis was conducted using SYBR green master mix (Bio‐Rad, Cat. # 1725150, Hercules, CA, USA) on the CFX96 real‐time detection system 690 (Bio‐Rad, USA). The rice Ubiquitin (LOC_Os03g13170) gene was used as an internal control (Wang et al., 2018). The mRNA levels relative to the Ubiquitin level were calculated according to the 2−∆∆Ct method.
In vitro purification of recombinant proteins
The GST‐OsHDAC1, OsHDAC1‐His and MBP‐OsGRAS30‐His proteins were purified as described previously (Hou et al., 2022). These constructs were transformed into E. coli strain BL21(DE3); expression was induced by adding 0.5 mM isopropyl ß‐D‐1‐thiogalactopyranoside (Coolaber, Cat. # CI6621, Beijing, China) to the culture, followed by incubation for 12 h at 16 °C. The fusion proteins were purified using glutathione resin (GeneScript, Cat. # L00206, Nanjing, China) or high‐affinity Ni‐NTA resin (GeneScript, Cat. # L00250) in accordance with the manufacturer's instructions.
Antibodies
To generate an antibody against rice OsHDAC1, purified OsHDAC1‐His recombinant protein was used as an antigen to immunize rabbits. Immunization of rabbits and serum collection were performed by Dai‐An Biotech (Wuhan, China). The antibodies used for immunoblotting and ChIP were as follows: anti‐OsHDAC1 (1:1000; Dai‐An), anti‐HA (1:5000 for immunoblot, 1:100 for ChIP; Abcam, Cat. # ab9110, Cambridgeshire, UK), anti‐MYC (1:5000; Proteintech, Cat. # 60003‐2‐Ig) anti‐IgG (1:100; Abcam, Cat. # ab172730), anti‐H3 (1:2000; Abcam, Cat. # ab1791), anti‐H3ac (1:2000; Millipore, Cat. # 17‐625, MA, USA), anti‐H4ac (1:2000; PTMbio, Cat. # PTM‐189, Hangzhou, China), anti‐H3K9ac (1:2000; Millipore, Cat. # 07–352), anti‐H3K27ac (1:5000 for immunoblot, 1:100 for ChIP; Abcam, Cat. # ab4729), anti‐His (1:5000, Proteintech, Cat. # 1B7G5, Wuhan, China), anti‐actin (1:2000, ABclonal, Cat. # AC009, Wuhan, China), goat anti‐rabbit IgG‐horseradish peroxidase (HRP) (1:5000; Beyotime, Cat. # A0277) and goat anti‐mouse IgG‐HRP (1:5000; Beyotime, Cat. # A0216).
Protein extraction and immunoblotting
The leaves of 2‐week‐old seedlings were ground into powder in liquid nitrogen. Then, histones were extracted using a protein extraction kit (Bestbio, Cat. # BB‐31171, Shanghai, China), and total protein was extracted using another protein extraction kit (Bestbio, Cat. # BB‐319815). Protein concentrations were measured using a bicinchoninic acid protein assay kit (Bio‐sharp, Cat. # BL521A, Hefei, China). Subsequently, proteins were heated for 10 min at 95 °C, separated using a 15% or 12% sodium dodecyl sulfate–polyacrylamide electrophoresis gel, transferred to polyvinylidene fluoride membranes, and detected as described previously (Hou et al., 2017). All western blots were developed using the ECL Plus immunoblotting detection system (GE Healthcare, Little Chalfont, UK).
Y2H assay
Y2H assays were performed using the Matchmaker GAL4 Two‐hybrid System 3 (Clontech, Mountain View, CA, USA) according to the manufacturer's instructions. Briefly, the bait and prey constructed in this work were co‐transformed into S. cerevisiae AH109 cells. After cultivation on SD medium lacking Leu and Trp (Takara, Cat. # 630316, Kyoto, Japan) at 30 °C for 3 days, yeast transformants were transferred onto SD medium lacking Leu, Trp, His and Ade (Takara, Cat. # 630322) or medium supplemented with 5‐bromo‐4‐chloro‐3‐indoxyl‐α‐d‐galactopyranoside (Takara, Cat. # 630463) and grown at 30 °C for 3 days.
Co‐IP assay
Co‐IP assays were performed as described previously (Zhang et al., 2017). The OsHDAC1‐HA and OsGRAS30‐MYC constructs or HA and OsGRAS30‐MYC constructs were transformed into rice protoplasts using a plant protoplast preparation and transformation kit (Real‐Times, Cat. # RTU4052, Beijing, China) in accordance with the manufacturer's instructions. Then, total proteins were extracted from rice protoplasts and incubated with rProtein A sepharose (GE Healthcare, USA) and an anti‐HA antibody. Proteins bound to sepharose were detected using an anti‐MYC antibody.
Split‐LUC assay
Split‐LUC assays were performed according to a previously described method (Hou et al., 2022). The OsHDAC1‐nLUC and cLUC‐OsGRAS30 constructs were transformed into A. tumefaciens strain GV3101(pSoup‐p19). Equal amounts of bacterial suspension were infiltrated into the leaves of 5‐week‐old N. benthamiana plants. The inoculated leaves were sprayed with 0.5 mM luciferin (Coolaber, Cat. # CL6928) after 3 days and then kept in the dark for 5 min. Luminescence images were taken using the Tanon 5200 imaging apparatus (Shanghai, China).
In vitro pull‐down assay
Pull‐down assays were performed as described previously (Hou et al., 2022). Glutathione resin containing GST or GST‐OsHDAC1 was incubated with MBP‐OsGRAS30‐His in phosphate‐buffered saline at 4 °C for 2 h. The proteins eluted from the resin were detected by immunoblotting using an anti‐His antibody.
Subcellular localization assay
To assess the co‐localization of OsHDAC1 with OsGRAS30 in rice protoplasts, the OsHDAC1‐sGFP (Hou et al., 2022) and OsGRAS30‐mCherry constructs were transformed into rice protoplasts using a plant protoplast preparation and transformation kit in accordance with the manufacturer's instructions. The transformed protoplasts were observed using a confocal laser scanning microscope (Leica SP8, Wetzlar, Germany). Images were analysed using LAS‐AF software. The settings used for confocal microscopy were as follows: for sGFP, excitation 488 nm, emission 500–550 nm; for mCherry, excitation 552 nm, emission 575–625 nm; fluorescence intensity, 1%–10%; gain value of the HyD detector, 100–200.
HDAC activity assay
To assess HDAC activity in vivo, leaves of 2‐week‐old seedlings from WT, OsGRAS30 mutant and OE lines were ground into powder in liquid nitrogen and extracted using a protein extraction kit (Bestbio, Cat. # BB‐319815). Protein concentrations in each sample were measured using a bicinchoninic acid protein assay kit (Bio‐sharp, Cat. # BL521A), and approximately 4 μg total protein was used to assess in vivo HDAC activity. For the in vitro HDAC activity assay, purified recombinant GST‐OsHDAC1 (~2 μg) and MBP‐OsGRAS30‐His (~2 μg) or MBP‐His (~2 μg) proteins were incubated at 37 °C for 1 h, and the mixed proteins were used to assess in vitro HDAC activity. Subsequently, the mixed proteins were subjected to in vivo and in vitro HDAC activity assays using the Epigenase HDAC activity/inhibition direct assay kit (Epigentek, Cat. # P‐4034, Farmingdale, NY, USA) in accordance with the manufacturer's instructions. After the reactions were complete, the optical density at 450 nm was measured using a SpectraMax iD5 microplate reader (Molecular Devices, Sunnyvale, CA, USA). The activity of the HDAC enzyme in proportion to optical density (measured as intensity in relative fluorescence units [RFU]) was calculated using the formula:
ChIP‐Seq or ‐PCR
Two‐week‐old seedlings were ground into a fine powder and suspended in a nuclear isolation buffer (10 mM HEPES [pH 8.0], 1 M sucrose, 5 mM KCl, 5 mM MgCl2, 0.6% Triton X‐100, 0.4 mM phenylmethylsulfonyl fluoride, 1× protease inhibitor cocktail, 1% formaldehyde) for 20 min for crosslinking. ChIP assays were performed using a ChIP kit (Bersinbio, Cat. # Bes5002, Guangzhou, China) in accordance with the manufacturer's instructions. Finally, the eluted DNA samples were digested with proteinase K (Invitrogen, Cat. # 25530049, Waltham, MA, USA) and RNase A and purified for subsequent sequencing or qPCR.
ChIP‐Seq for analysis of OsHDAC1 occupancy was performed using an antibody against the HA tag in OsHDAC1 OE lines, with two replicates. The input was used as a negative control, and IgG was used as an experimental control for the HA antibody. ChIP‐Seq for genome‐wide H3K27ac analysis was performed using an antibody against H3K27ac in WT plants, with two replicates. Immunoprecipitated and purified DNA was used for library construction and sequencing, which were performed on the Illumina Novaseq 6000 sequencing system by Seqhealth Biotech (Wuhan, China).
ChIP–qPCR was performed as described previously (Ke et al., 2020). In brief, the immunoprecipitated and purified DNA was subjected to qPCR to amplify the target sequences. The Ubiquitin (LOC_Os03g13170) promoter was used as an internal reference control.
Analysis of ChIP‐Seq data
A total of 10 Gb high‐quality 150‐bp paired‐end reads were generated from each sample. Raw data obtained from Illumina sequencing were processed and filtered using FastP to generate clean reads. These reads were mapped to the Nipponbare genome (MSU 7.0) using Bowtie2 with the default parameters, and only unique alignments were retained. The SAM‐formatted output files generated by Bowtie2 were transformed to BAM format, followed by sorting and indexing using Samtools. Peaks were called using MACS2, and all duplicate reads were retained for OsHDAC1 and H3K27ac ChIP‐Seq analysis. The q‐value cutoff applied to calculate statistical significance was <0.01. Other parameters were set to their default values. The abundance of OsHDAC1 and H3K27ac ChIP‐Seq reads was normalized to the corresponding input level. The genomic regions enriched in OsHDAC1 and H3K27ac were determined by comparison of the corresponding ChIP library with its input DNA library. All statistical analyses and figure construction were conducted using R.
RNA‐Seq and data analysis
Total RNA was isolated from 2‐week‐old seedlings of WT and OsHDAC1 Ri2 plants in three biological replicates using a TRIzol kit according to previously reported methods (Hou et al., 2021). Subsequently, mRNA enrichment, cDNA synthesis, library construction and sequencing were performed using the Illumina Novaseq 6000 system by Seqhealth Biotech (Wuhan, China).
Finally, 6 Gb high‐quality 150‐bp paired‐end reads were generated from each sample. Raw data obtained from Illumina sequencing were processed and filtered using Fastp to generate clean reads. These reads were aligned to the Nipponbare reference genome (MSU 7.0) using the splice junction mapper TopHat2. HTSeq was used to obtain the read counts mapped to each gene. The read counts were normalized as reads per kilobase per million reads to assess relative expression levels. EdgeR was used for differential expression analysis. Genes showing an absolute value of log2 fold change (OsHDAC1 Ri2/WT) > 1 and adjusted P‐value <0.05 were considered differentially expressed. All the statistical analyses and figure construction were conducted using R.
Statistical analysis
All the values are presented as the mean ± standard error of the mean, and the number (n) of samples is indicated. Grey values were determined using ImageJ software. Statistically significant differences between control and experimental groups were determined using Student's t‐test, with values of P < 0.05 indicating statistical significance.
Primer sequences
The sequences of primers used in this study are listed in Table S1.
Conflict of interest
The authors declare no competing interests.
Author contributions
J.H. and L.L. conceived the ideas and designed the experiments. J.H. performed most experiments and data analysis. H.X., and Q.S. performed part experiments, including LUC, Pull‐down, RT–qPCR and material identification. P.Y., performed bioinformatics analysis, J.Y., and X.M. helped to perform part experiments, including Y2H, and agronomic traits analysis. H.H. performed part data analysis and discussion. J.H. and L.L. supervised the research and wrote the article.
Supporting information
Figure S1 Specificity testing results for the anti‐OsHDAC1 polyclonal antibody in immunoblotting analysis in WT and OsHDAC1 OE5 plants.
Figure S2 The standard M. oryzae strain Guy11 and M. oryzae strains AH4, Sc09‐153‐07, TM3‐2, RB1, RB3, NC1, HLJ1‐3, HLJ09‐17‐1 and HLJ5‐3 isolated in various regions of China were used for rice leaf infection analyses.
Figure S3 Phenotypes and statistics of key agronomic traits of OsHDAC1 RNAi and OE plants in the field.
Figure S4 Diagrammatic representation of vector construction and expression analysis of OsGRAS30 in transgenic plants.
Figure S5 Protein abundance and transcript levels of OsHDAC1 in OsGRAS30 overexpression lines, mutants and osgras30 #1/OsHDAC1 RNAi lines.
Figure S6 H3K27ac level was increased in response to M. oryzae Guy11 treatment, as shown by immunoblotting analysis.
Figure S7 Rice blast disease symptoms after treatment with HDAC inhibitors.
Figure S8 Specificity testing of the anti‐HA antibody for chromatin immunoprecipitation sequencing (ChIP‐Seq) analysis in OsHDAC1 OE5 plants.
Figure S9 The correlation analysis of the two biological replicates of OsHDAC1 in ChIP‐seq.
Figure S10 The correlation analysis of the two biological replicates of H3K27ac in ChIP‐seq.
Figure S11 The correlation analysis of the three biological replicates of OsHDAC1 in RNA‐seq.
Figure S12 Heatmap showing different expression of numerous DEGs in rice seedlings between the OsHDAC1 Ri2 line and WT.
Figure S13 Significantly enriched KEGG terms among OsHDAC1‐regulated DEGs.
Table S1 Primers used in this study.
Dataset S1 OsHDAC1 binding sites identified by ChIP‐Seq.
Dataset S2 H3K27 distribution sites identified by ChIP‐Seq.
Dataset S3 List of DEGS in OsHDAC1 Ri2 plants vs WT plants.
Dataset S4 GO pathways of downregulated DEGS in OsHDAC1 Ri2 plants vs WT plants.
Dataset S5 KEGG pathways of downregulated DEGS in OsHDAC1 Ri2 plants vs WT plants.
Dataset S6 GO pathways of upregulated DEGS in OsHDAC1 Ri2 plants vs WT plants.
Dataset S7 KEGG pathways of upregulated DEGS in OsHDAC1 Ri2 plants vs WT plants.
Acknowledgements
We thank Prof. Yun‐Yuan Xu (Institute of Botany, Chinese Academy of Sciences) for kindly providing us with the pBI121 vector, Prof. Qiang Cai and Prof. Zhi‐Hong Zhang (Wuhan University) for kindly providing us with M. oryzae strains, Prof. Hong‐Chun Yang (Wuhan University) for kindly providing us with the rice yeast two hybrid library. This work was funded by grants from the National Natural Science Foundation of China (NSFC No. 31871238).
Data availability statement
RNA‐ and ChIP‐sequencing raw data are available at the National Genomics Data Center, Beijing Institute of Genomics (China National Center for Bioinformation), Chinese Academy of Sciences, under the Genome Sequence Archive (GSA) accession number CRA012210. All data supporting the findings of this study are available within the paper and within its supplemental data.
References
- An, F. , Zhang, X. , Zhu, Z. , Ji, Y. , He, W. , Jiang, Z. , Li, M. et al. (2012) Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 22, 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baile, F. , Merini, W. , Hidalgo, I. and Calonje, M. (2021) EAR domain‐containing transcription factors trigger PRC2‐mediated chromatin marking in Arabidopsis. Plant Cell 33, 2701–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, S.L. (2007) The complex language of chromatin regulation during transcription. Nature 447, 407–412. [DOI] [PubMed] [Google Scholar]
- Césari, S. , Thilliez, G. , Ribot, C. , Chalvon, V. , Michel, C. , Jauneau, A. , Rivas, S. et al. (2013) The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR‐Pia and AVR1‐CO39 by direct binding. Plant Cell 25, 1463–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Césari, S. , Kanzaki, H. , Fujiwara, T. , Bernoux, M. , Chalvon, V. , Kawano, Y. , Shimamoto, K. et al. (2014) The NB‐LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 33, 1941–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty, M. , Mahmud, N.U. , Ullah, C. , Rahman, M. and Islam, T. (2021) Biological and biorational management of blast diseases in cereals caused by Magnaporthe oryzae . Crit. Rev. Biotechnol. 41, 994–1022. [DOI] [PubMed] [Google Scholar]
- Chang, Y.N. , Zhu, C. , Jiang, J. , Zhang, H. , Zhu, J.K. and Duan, C.G. (2020) Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 62, 563–580. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Lu, L. , Mayer, K.S. , Scalf, M. , Qian, S. , Lomax, A. , Smith, L.M. et al. (2016) POWERDRESS interacts with HISTONE DEACETYLASE 9 to promote aging in Arabidopsis . Elife 5, e17214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Ding, A.B. and Zhong, X. (2020) Functions and mechanisms of plant histone deacetylases. Sci. China Life Sci. 63, 206–216. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Duan, Y. , Qiao, F. , Liu, H. , Huang, J. , Luo, C. , Chen, X. et al. (2022a) A secreted fungal effector suppresses rice immunity through host histone hypoacetylation. New Phytol. 235, 1977–1994. [DOI] [PubMed] [Google Scholar]
- Chen, S. , Sun, B. , Shi, Z. , Miao, X. and Li, H. (2022b) Identification of the rice genes and metabolites involved in dual resistance against brown planthopper and rice blast fungus. Plant Cell Environ. 45, 1914–1929. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Liu, C. , Wang, H. , Liu, Q. , Yue, Y. , Duan, Y. , Wang, Z. et al. (2024) Ustilaginoidea Virens‐Secreted Effector Uv1809 Suppresses Rice Immunity by Enhancing OsSRT2‐Mediated Histone Deacetylation. Plant Biotechnol. J. 22, 148–164. 10.1111/pbi.14174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, S.M. , Song, H.R. , Han, S.K. , Han, M. , Kim, C.Y. , Park, J. , Lee, Y.H. et al. (2012) HDA19 is required for the repression of salicylic acid biosynthesis and salicylic acid‐mediated defense responses in Arabidopsis . Plant J. 71, 135–146. [DOI] [PubMed] [Google Scholar]
- Chung, P.J. , Kim, Y.S. , Jeong, J.S. , Park, S.H. , Nahm, B.H. and Kim, J.K. (2009) The histone deacetylase OsHDAC1 epigenetically regulates the OsNAC6 gene that controls seedling root growth in rice. Plant J. 59, 764–776. [DOI] [PubMed] [Google Scholar]
- Couto, D. and Zipfel, C. (2016) Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552. [DOI] [PubMed] [Google Scholar]
- Dasgupta, P. , Prasad, P. , Bag, S.K. and Chaudhuri, S. (2022) Dynamicity of histone H3K27ac and H3K27me3 modifications regulate the cold‐responsive gene expression in Oryza sativa L. ssp. indica. Genomics 114, 110433. [DOI] [PubMed] [Google Scholar]
- Deng, Y.Z. and Naqvi, N.I. (2019) Metabolic basis of pathogenesis and host adaptation in rice blast. Annu. Rev. Microbiol. 73, 601–619. [DOI] [PubMed] [Google Scholar]
- De‐Vleesschauwer, D. , Seifi, H.S. , Filipe, O. , Haeck, A. , Huu, S.N. , Demeestere, K. and Höfte, M. (2016) The DELLA protein SLR1 integrates and amplifies salicylic acid‐ and jasmonic acid‐dependent innate immunity in rice. Plant Physiol. 170, 1831–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, B. , Bellizzi, M.dR. , Ning, Y. , Meyers, B.C. and Wang, G.L. (2012) HDT701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histone H4 acetylation of defense‐related genes in rice. Plant Cell 24, 3783–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, D. , Zhang, C. , Xing, Y. , Lu, X. , Cai, L. , Yun, H. , Zhang, Q. et al. (2021) The CC‐NB‐LRR OsRLR1 mediates rice disease resistance through interaction with OsWRKY19. Plant Biotechnol. J. 19, 1052–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka, S. , Saka, N. , Koga, H. , Ono, K. , Shimizu, T. , Ebana, K. , Hayashi, N. et al. (2009) Loss of function of a proline‐containing protein confers durable disease resistance in rice. Science 325, 998–1001. [DOI] [PubMed] [Google Scholar]
- He, M. , Su, J. , Xu, Y. , Chen, J. , Chern, M. , Lei, M. , Qi, T. et al. (2020) Discovery of broad‐spectrum fungicides that block septin‐dependent infection processes of pathogenic fungi. Nat. Microbiol. 5, 1565–1575. [DOI] [PubMed] [Google Scholar]
- He, Z. , Webster, S. and He, S.Y. (2022) Growth‐defense trade‐offs in plants. Curr. Biol. 32, R634–R639. [DOI] [PubMed] [Google Scholar]
- Hill, K. , Wang, H. and Perry, S.E. (2008) A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J. 53, 172–185. [DOI] [PubMed] [Google Scholar]
- Hou, H. , Zheng, X. , Zhang, H. , Yue, M. , Hu, Y. , Zhou, H. , Wang, Q. et al. (2017) Histone deacetylase is required for GA‐induced programmed cell death in maize aleurone layers. Plant Physiol. 175, 1484–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, J. , Ren, R. , Xiao, H. , Chen, Z. , Yu, J. , Zhang, H. , Shi, Q. et al. (2021) Characteristic and evolution of HAT and HDAC genes in Gramineae genomes and their expression analysis under diverse stress in Oryza sativa . Planta 253, 72. [DOI] [PubMed] [Google Scholar]
- Hou, J. , Zheng, X. , Ren, R. , Shi, Q. , Xiao, H. , Chen, Z. , Yue, M. et al. (2022) The histone deacetylase 1/GSK3/SHAGGY‐like kinase 2/BRASSINAZOLE‐RESISTANT 1 module controls lateral root formation in rice. Plant Physiol. 189, 858–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot, B. , Yao, J. , Montgomery, B.L. and He, S.Y. (2014) Growth‐defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant 7, 1267–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal, V. , Kakkar, M. , Kumari, P. , Zinta, G. , Gahlaut, V. and Kumar, S. (2022) Multifaceted roles of GRAS transcription factors in growth and stress responses in plants. iScience 25, 105026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C.J. , Shimono, M. , Maeda, S. , Inoue, H. , Mori, M. , Hasegawa, M. , Sugano, S. et al. (2009) Suppression of the rice fatty‐acid desaturase gene OsSSI2 enhances resistance to blast and leaf blight diseases in rice. Mol. Plant Microbe Interact. 22, 820–829. [DOI] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Ke, Y. , Yuan, M. , Liu, H. , Hui, S. , Qin, X. , Chen, J. , Zhang, Q. et al. (2020) The versatile functions of OsALDH2B1 provide a genic basis for growth‐defense trade‐offs in rice. Proc. Natl. Acad. Sci. U. S. A. 117, 3867–3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K.C. , Lai, Z. , Fan, B. and Chen, Z. (2008) Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20, 2357–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, V. , Thakur, J.K. and Prasad, M. (2021) Histone acetylation dynamics regulating plant development and stress responses. Cell. Mol. Life Sci. 78, 4467–4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrasse, D. , Jégu, T. , Li, H. , de Zelicourt, A. , Raynaud, C. , Legras, S. , Gust, A. et al. (2017) MAPK‐triggered chromatin reprogramming by histone deacetylase in plant innate immunity. Genome Biol. 18, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar, N. , Mesarich, C.H. , Petit‐Houdenot, Y. , Talbi, N. , Li de la Sierra‐Gallay, I. , Zélie, E. , Blondeau, K. et al. (2022) A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins. PLoS Pathog. 18, e1010664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Zhu, Z. , Chern, M. , Yin, J. , Yang, C. , Ran, L. , Cheng, M. et al. (2017) A natural allele of a transcription factor in rice confers broad‐spectrum blast resistance. Cell 170, 114–126. [DOI] [PubMed] [Google Scholar]
- Li, W. , Chern, M. , Yin, J. , Wang, J. and Chen, X. (2019) Recent advances in broad‐spectrum resistance to the rice blast disease. Curr. Opin. Plant Biol. 50, 114–120. [DOI] [PubMed] [Google Scholar]
- Li, W. , Xiong, Y. , Lai, L.B. , Zhang, K. , Li, Z. , Kang, H. , Dai, L. et al. (2021) The rice RNase P protein subunit Rpp30 confers broad‐spectrum resistance to fungal and bacterial pathogens. Plant Biotechnol. J. 19, 1988–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Z. , Riaz, A. , Chachar, S. , Ding, Y. , Du, H. and Gu, X. (2020) Epigenetic modifications of mRNA and DNA in plants. Mol. Plant 13, 14–30. [DOI] [PubMed] [Google Scholar]
- Lin, C. , Wu, Z. , Shi, H. , Yu, J. , Xu, M. , Lin, F. , Kou, Y. et al. (2022) The additional PRC2 subunit and Sin3 histone deacetylase complex are required for the normal distribution of H3K27me3 occupancy and transcriptional silencing in Magnaporthe oryzae . New Phytol. 236, 576–589. [DOI] [PubMed] [Google Scholar]
- Liu, Z. , Zhu, Y. , Shi, H. , Qiu, J. , Ding, X. and Kou, Y. (2021) Recent progress in rice broad‐spectrum disease resistance. Int. J. Mol. Sci. 22, 11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, D. , Lin, X.L. , Kong, X. , Qu, G.P. , Cai, B. , Lee, J. and Jin, J.B. (2019) SIZ1‐mediated SUMOylation of TPR1 suppresses plant immunity in Arabidopsis . Mol. Plant 12, 215–228. [DOI] [PubMed] [Google Scholar]
- Okuyama, Y. , Kanzaki, H. , Abe, A. , Yoshida, K. , Tamiru, M. , Saitoh, H. , Fujibe, T. et al. (2011) A multifaceted genomics approach allows the isolation of the rice Pia‐blast resistance gene consisting of two adjacent NBS‐LRR protein genes. Plant J. 66, 467–479. [DOI] [PubMed] [Google Scholar]
- Palma, K. , Thorgrimsen, S. , Malinovsky, F.G. , Fiil, B.K. , Nielsen, H.B. , Brodersen, P. , Hofius, D. et al. (2010) Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog. 6, e1001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, F. , Pokholok, D.K. , Hannett, N.M. , Rinaldi, N.J. , Chandy, M. , Rolfe, A. , Workman, J.L. et al. (2004) Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell 16, 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha, G. , Sun, P. , Kong, X. , Han, X. , Sun, Q. , Fouillen, L. , Zhao, J. et al. (2023) Genome editing of a rice CDP‐DAG synthase confers multipathogen resistance. Nature 618, 1017–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirano, Y. , Kachroo, P. , Shah, J. and Klessig, D.F. (2002) A gain‐of‐function mutation in an Arabidopsis Toll Interleukin1 receptor‐nucleotide binding site‐leucine‐rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14, 3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvedunova, M. and Akhtar, A. (2022) Modulation of cellular processes by histone and non‐histone protein acetylation. Nat. Rev. Mol. Cell Biol. 23, 329–349. [DOI] [PubMed] [Google Scholar]
- Tanabe, S. , Onodera, H. , Hara, N. , Ishii‐Minami, N. , Day, B. , Fujisawa, Y. , Hagio, T. et al. (2016) The elicitor‐responsive gene for a GRAS family protein, CIGR2, suppresses cell death in rice inoculated with rice blast fungus via activation of a heat shock transcription factor, OsHsf23. Biosci. Biotechnol. Biochem. 80, 145–151. [DOI] [PubMed] [Google Scholar]
- Thordal‐Christensen, H. (2020) A holistic view on plant effector‐triggered immunity presented as an iceberg model. Cell. Mol. Life Sci. 77, 3963–3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda, K. and Somssich, I.E. (2015) Transcriptional networks in plant immunity. New Phytol. 206, 932–947. [DOI] [PubMed] [Google Scholar]
- Walley, J.W. , Shen, Z. , McReynolds, M.R. , Schmelz, E.A. and Briggs, S.P. (2018) Fungal‐induced protein hyperacetylation in maize identified by acetylome profiling. Proc. Natl. Acad. Sci. U. S. A. 115, 210–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, A. , Kurdistani, S.K. and Grunstein, M. (2002) Requirement of Hos2 histone deacetylase for gene activity in yeast. Science 298, 1412–1414. [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Zang, C. , Cui, K. , Schones, D.E. , Barski, A. , Peng, W. and Zhao, K. (2009) Genome‐wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138, 1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Gao, F. , Wu, J. , Dai, J. , Wei, C. and Li, Y. (2010) Arabidopsis putative deacetylase AtSRT2 regulates basal defense by suppressing PAD4, EDS5 and SID2 expression. Plant Cell Physiol. 51, 1291–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. , Liu, Y. , He, J. , Zheng, X. , Hu, J. , Liu, Y. , Dai, H. et al. (2014) STV11 encodes a sulphotransferase and confers durable resistance to rice stripe virus. Nat. Commun. 5, 4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Hu, Q. , Wu, Z. , Wang, H. , Han, S. , Jin, Y. , Zhou, J. et al. (2017) Histone deacetylase 6 represses pathogen defense responses in Arabidopsis thaliana . Plant Cell Environ. 40, 2972–2986. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Zhou, L. , Shi, H. , Chern, M. , Yu, H. , Yi, H. , He, M. et al. (2018) A single transcription factor promotes both yield and immunity in rice. Science 361, 1026–1028. [DOI] [PubMed] [Google Scholar]
- Wang, N. , Tang, C. , Fan, X. , He, M. , Gan, P. , Zhang, S. , Hu, Z. et al. (2022) Inactivation of a wheat protein kinase gene confers broad‐spectrum resistance to rust fungi. Cell 185, 2961–2974.e19. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Yue, J. , Yang, N. , Zheng, C. , Zheng, Y. , Wu, X. , Yang, J. et al. (2023) An ERAD‐related ubiquitin‐conjugating enzyme boosts broad‐spectrum disease resistance and yields in rice. Nat. Food 4, 774–787. [DOI] [PubMed] [Google Scholar]
- Wu, Z. , He, L. , Jin, Y. , Chen, J. , Shi, H. , Wang, Y. and Yang, W. (2021) HISTONE DEACETYLASE 6 suppresses salicylic acid biosynthesis to repress autoimmunity. Plant Physiol. 187, 2592–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, T. , Zhang, H. , Yuan, B. , Liu, H. , Kong, L. , Chu, Z. and Ding, X. (2022) Tal2b targets and activates the expression of OsF3H03g to hijack OsUGT74H4 and synergistically interfere with rice immunity. New Phytol. 233, 1864–1880. [DOI] [PubMed] [Google Scholar]
- Xiao, S. , Ellwood, S. , Calis, O. , Patrick, E. , Li, T. , Coleman, M. and Turner, J.G. (2001) Broad‐spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291, 118–120. [DOI] [PubMed] [Google Scholar]
- Xin, X.‐F. , Kvitko, B. and He, S.Y. (2018) Pseudomonas syringae: what it takes to be a pathogen. Nat. Rev. Microbiol. 16, 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z. , Zhou, Z. , Cheng, Z. , Zhou, Y. , Wang, F. , Li, M. , Li, G. et al. (2023) A transcription factor ZmGLK36 confers broad resistance to maize rough dwarf disease in cereal crops. Nat. Plants 9, 1720–1733. [DOI] [PubMed] [Google Scholar]
- Yang, H. , Liu, X. , Xin, M. , Du, J. , Hu, Z. , Peng, H. , Rossi, V. et al. (2016) Genome‐wide mapping of targets of maize histone deacetylase HDA101 reveals its function and regulatory mechanism during seed development. Plant Cell 28, 629–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L. , Zhang, P. , Wang, Y. , Hu, G. , Guo, W. , Gu, X. and Pu, L. (2022) Plant synthetic epigenomic engineering for crop improvement. Sci. China Life Sci. 65, 2191–2204. [DOI] [PubMed] [Google Scholar]
- Yang, X.M. , Zhao, J.H. , Xiong, X.Y. , Hu, Z.W. , Sun, J.F. , Su, H. , Liu, Y.J. et al. (2023) Broad‐spectrum resistance gene RPW8.1 balances immunity and growth via feedback regulation of WRKYs. Plant Biotechnol J. 22, 116–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Li, J. , Li, F. , Liu, H. , Yang, W. , Chong, K. and Xu, Y. (2017) OsMAPK3 phosphorylates OsbHLH002/OsICE1 and inhibits its ubiquitination to activate OsTPP1 and enhances rice chilling tolerance. Dev. Cell 43, 731–743. [DOI] [PubMed] [Google Scholar]
- Zhang, Z. , Gao, S. and Chu, C. (2020) Improvement of nutrient use efficiency in rice: current toolbox and future perspectives. Theor. Appl. Genet. 133, 1365–1384. [DOI] [PubMed] [Google Scholar]
- Zhang, W. , Huang, J. and Cook, D.E. (2021) Histone modification dynamics at H3K27 are associated with altered transcription of in planta induced genes in Magnaporthe oryzae . PLoS Genet. 17, e1009376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Z.X. , Xu, Y.J. , Lei, Y. , Li, Q. , Zhao, J.Q. , Li, Y. , Fan, J. et al. (2021) ANNEXIN 8 negatively regulates RPW8.1‐mediated cell death and disease resistance in Arabidopsis . J. Integr. Plant Biol. 63, 378–392. [DOI] [PubMed] [Google Scholar]
- Zhao, J.H. , Huang, Y.Y. , Wang, H. , Yang, X.M. , Li, Y. , Pu, M. , Zhou, S.X. et al. (2023) Golovinomyces cichoracearum effector‐associated nuclear localization of RPW8.2 amplifies its expression to boost immunity in Arabidopsis . New Phytol. 238, 367–382. [DOI] [PubMed] [Google Scholar]
- Zheng, Y. , Ge, J. , Bao, C. , Chang, W. , Liu, J. , Shao, J. , Liu, X. et al. (2020) Histone deacetylase HDA9 and WRKY53 transcription factor are mutual antagonists in regulation of plant stress response. Mol. Plant 13, 598–611. [DOI] [PubMed] [Google Scholar]
- Zhi, P. , Kong, L. , Liu, J. , Zhang, X. , Wang, X. , Li, H. , Sun, M. et al. (2020) Histone deacetylase TaHDT701 functions in TaHDA6‐TaHOS15 complex to regulate wheat defense responses to Blumeria graminis f.sp. tritici. Int. J. Mol. Sci. 21, 2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Wang, X. , He, K. , Charron, J.B. , Elling, A.A. and Deng, X.W. (2010) Genome‐wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Mol. Biol. 72, 585–595. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Specificity testing results for the anti‐OsHDAC1 polyclonal antibody in immunoblotting analysis in WT and OsHDAC1 OE5 plants.
Figure S2 The standard M. oryzae strain Guy11 and M. oryzae strains AH4, Sc09‐153‐07, TM3‐2, RB1, RB3, NC1, HLJ1‐3, HLJ09‐17‐1 and HLJ5‐3 isolated in various regions of China were used for rice leaf infection analyses.
Figure S3 Phenotypes and statistics of key agronomic traits of OsHDAC1 RNAi and OE plants in the field.
Figure S4 Diagrammatic representation of vector construction and expression analysis of OsGRAS30 in transgenic plants.
Figure S5 Protein abundance and transcript levels of OsHDAC1 in OsGRAS30 overexpression lines, mutants and osgras30 #1/OsHDAC1 RNAi lines.
Figure S6 H3K27ac level was increased in response to M. oryzae Guy11 treatment, as shown by immunoblotting analysis.
Figure S7 Rice blast disease symptoms after treatment with HDAC inhibitors.
Figure S8 Specificity testing of the anti‐HA antibody for chromatin immunoprecipitation sequencing (ChIP‐Seq) analysis in OsHDAC1 OE5 plants.
Figure S9 The correlation analysis of the two biological replicates of OsHDAC1 in ChIP‐seq.
Figure S10 The correlation analysis of the two biological replicates of H3K27ac in ChIP‐seq.
Figure S11 The correlation analysis of the three biological replicates of OsHDAC1 in RNA‐seq.
Figure S12 Heatmap showing different expression of numerous DEGs in rice seedlings between the OsHDAC1 Ri2 line and WT.
Figure S13 Significantly enriched KEGG terms among OsHDAC1‐regulated DEGs.
Table S1 Primers used in this study.
Dataset S1 OsHDAC1 binding sites identified by ChIP‐Seq.
Dataset S2 H3K27 distribution sites identified by ChIP‐Seq.
Dataset S3 List of DEGS in OsHDAC1 Ri2 plants vs WT plants.
Dataset S4 GO pathways of downregulated DEGS in OsHDAC1 Ri2 plants vs WT plants.
Dataset S5 KEGG pathways of downregulated DEGS in OsHDAC1 Ri2 plants vs WT plants.
Dataset S6 GO pathways of upregulated DEGS in OsHDAC1 Ri2 plants vs WT plants.
Dataset S7 KEGG pathways of upregulated DEGS in OsHDAC1 Ri2 plants vs WT plants.
Data Availability Statement
RNA‐ and ChIP‐sequencing raw data are available at the National Genomics Data Center, Beijing Institute of Genomics (China National Center for Bioinformation), Chinese Academy of Sciences, under the Genome Sequence Archive (GSA) accession number CRA012210. All data supporting the findings of this study are available within the paper and within its supplemental data.


