Summary
The exploitation of heterosis to integrate parental advantages is one of the fastest and most efficient ways of rice breeding. The genomic architecture of heterosis suggests that the grain yield is strongly correlated with the accumulation of numerous rare superior alleles with positive dominance. However, the improvements in yield of hybrid rice have shown a slowdown or even plateaued due to the limited availability of complementary superior alleles. In this study, we achieved a considerable increase in grain yield of restorer lines by inducing an alternative splicing event in a heterosis gene OsMADS1 through CRISPR‐Cas9, which accounted for approximately 34.1%–47.5% of yield advantage over their corresponding inbred rice cultivars. To achieve a higher yield in hybrid rice, we crossed the gene‐edited restorer parents harbouring OsMADS1 GW3p6 with the sterile lines to develop new rice hybrids. In two‐line hybrid rice Guang‐liang‐you 676 (GLY676), the yield of modified hybrids carrying the homozygous heterosis gene OsMADS1 GW3p6 significantly exceeded that of the original hybrids with heterozygous OsMADS1. Similarly, the gene‐modified F1 hybrids with heterozygous OsMADS1 GW3p6 increased grain yield by over 3.4% compared to the three‐line hybrid rice Quan‐you‐si‐miao (QYSM) with the homozygous genotype of OsMADS1. Our study highlighted the great potential in increasing the grain yield of hybrid rice by pyramiding a single heterosis gene via CRISPR‐Cas9. Furthermore, these results demonstrated that the incomplete dominance of heterosis genes played a major role in yield‐related heterosis and provided a promising strategy for breeding higher‐yielding rice varieties above what is currently achievable.
Keywords: heterosis, crispr/cas9, rice, breeding, grian yield, grain size
Introduction
Heterosis, or hybrid vigour, describes the situation in which hybrid offspring exhibit superior phenotypic performance than that of their parents. Hybrid rice often shows a 10%–20% yield advantage over their corresponding inbred rice cultivars. The exploitation of heterosis has been widely applied in hybrid rice breeding and has achieved remarkable achievements in grain yield. Several classical hypotheses to explain heterosis were proposed soon after plant breeders began to purposefully exploit heterosis, for example, three non‐mutually exclusive hypotheses: dominance, overdominance and epistasis. Among them, dominance and overdominance are the most prominent genetic models for the genetic basis of heterosis. And extensive studies in multiple plants have provided support for these genetic models (Krieger et al., 2010; Liu et al., 2020a; Seymour et al., 2016; Torgeman and Zamir, 2023; Wang et al., 2023; Xiao et al., 1995; Zhou et al., 2012). In search of the genetic basis of heterosis, a direct approach is to evaluate the contributions of heterosis quantitative trait loci (hQTL) to the heterotic advantage through the analysis of their heterotic effects (Lippman and Zamir, 2007; Schnable and Springer, 2013). Recently, the genomic architecture of rice heterosis for yield‐related traits revealed that the grain yield was strongly correlated with the accumulation of numerous rare superior alleles with positive dominance, and a small number of heterosis loci from female parents explained a large proportion of the yield advantage of hybrids over their male parents (Huang et al., 2015, 2016). Although the genetic basis of rice heterosis has well been elucidated, it is still labour‐intensive and time‐consuming to exploit the heterotic effect from a crossing hybridization in crop hybrid breeding.
CRISPR‐Cas‐based genome editing is a powerful and transformative method to introduce predictable and precise genome modifications into plants to obtain desired traits, which holds great promise for crop improvement (Gao, 2021). To date, genome editing has been successfully applied to improve multiple agronomic traits in various crops, such as grain yield (Song et al., 2022), grain quality (Xu et al., 2021), and disease‐resistance traits (Wei et al., 2021b). Therefore, based on a better understanding of heterosis, there is still significant potential for the development of hybrid breeding improvements through gene editing technologies. Alternative splicing (AS) is pervasive to increase transcriptomic and proteomic complexity, which contributed potentially to genomic diversity to regulate complex traits (Wang and Brendel, 2006). Accumulating evidence in rice supports that AS induced by variation at either intron donor or acceptor sites could modulate important agronomic traits, such as Waxy controlling grain quality (Cai et al., 1998) and RLI1 controlling Pi starvation signalling and growth (Guo et al., 2022). Recently, it was found that mutations at splice sites could cause messenger RNA (mRNA) missplicing and gene disruption through CRISPR‐Cas9 systems (Li et al., 2019; Tang et al., 2021).
In this study, we achieved an alternative 3′ splicing event of the heterosis gene OsMADS1 in the male parent by disrupting the 3′ splice site through the CRISPR‐Cas9 system. The gene‐edited restore line with the AS of OsMADS1 displayed a higher yield and better grain quality than wild‐type plants. Notably, the modified hybrids harbouring a homozygous or heterozygous genotype of the heterosis gene (OsMADS1 GW3p6 ) under the same hybrid genetic background have a better performance in grain yield compared to the original hybrid variety with heterozygous OsMADS1 GW3p6 or homozygous OsMADS1, respectively. Our results implied that the dominance effect underlying functional complementation was an indispensable contributor to yield heterosis and provided a valuable breeding target and an efficient method for hybrid rice breeding without compromising heterotic performance.
Results
A strategy to improve hybrid breeding based on the genetic basis of rice heterosis
Relying solely on random hybridization for hybrid breeding is a labour‐intensive and time‐consuming task. A comprehensive understanding of the genetic mechanisms of heterosis could efficiently guide hybrid breeding. To this end, as shown in Figure 1a, various genetic populations were designed to uncover the genetic underpinnings underlying heterosis through the identification of hQTLs and estimation of their effects (Liu et al., 2020b; Ouyang et al., 2022). Based on different heterotic populations, we have summarized the research on the genetic mechanisms of heterosis in plants over the past few decades. As shown in Table S1, dominance, overdominance, and epistasis contribute to the heterosis, with the dominance effect remaining the primary genetic mechanism.
Figure 1.
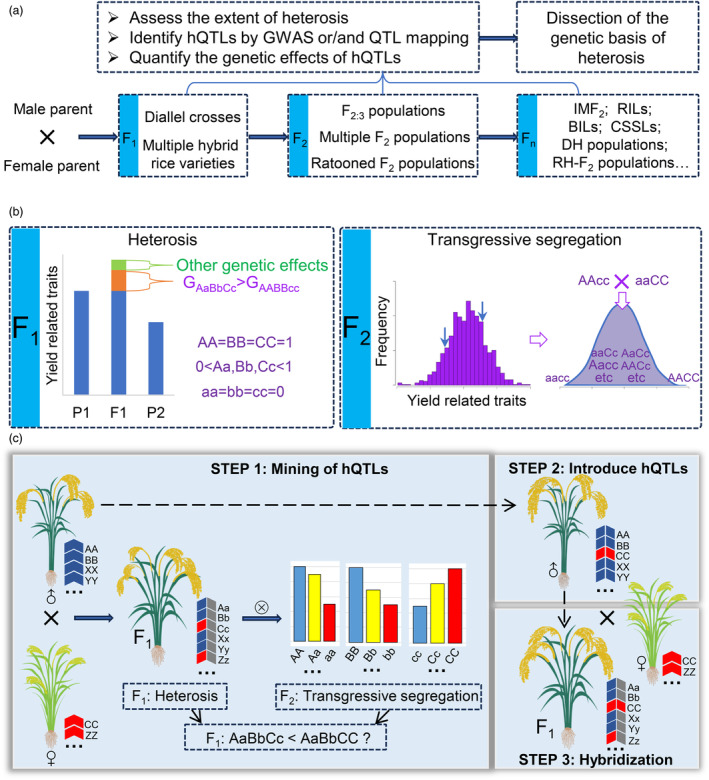
The theoretical basis and roadmap of hybrid breeding strategy. (a) The commonly used genetic populations and approaches for exploring the genetic basis of heterosis. F1, first filial generation; F2, second filial generation; F2:3, F3 families derived from a single F2 plant; IMF2, immortalized F2 population; RILs, recombinant inbred lines; BILs, backcross inbred lines; CSSLs, chromosome segment substitution lines; DH, doubled haploid. RH‐F2, residual heterozygous − second filial generation. (b) Two schematic diagrams explaining heterosis and transgressive segregation. Left dashed box: The schematic diagram describes the possible contributions of the yield advantage over their corresponding paternal parents. The green part in the bar chart represents yield advantage formed by other genetic effects, such as epistasis and overdominance. The orange part indicates yield advantage formed by dominance effect. The letters A, a, B, b, C, and c indicate favourable alleles at three loci. Upper case letters increase the value of the trait, lower case letters decrease it. The letters A, a, B, and b indicate the alleles at two loci in paternal parent, and C and c represent the allele at one locus in maternal parent. GAaBbCc and GAABBcc indicate the genetic values of the genotypes of ‘AaBbCc’ and ‘AABBcc’ respectively. P1, P2, and F1 indicate the male, female parents and their first filial generation separately. Right dashed box: The histogram on the left represents the common phenomenon of transgressive segregation. The two blue arrows represent the range of parental yield production. Right schematic illustrating a simple model for transgressive segregation. If the beneficial alleles are dispersed among the parents in a cross, it is possible to observe offspring with trait values that are higher or lower than those of the parents. (c) The roadmap for improving breeding strategies through hybridization. ‘F1: AaBbCc<AaBbCC?’ indicate the hypothesis being tested in this study, which is whether F1 plants with genotype ‘AaBbCC’ (maternal homozygous genotype at one favourable allele) have higher yields compared to those with genotype ‘AaBbCc’ (maternal heterozygous genotype). The letters XX, YY and ZZ represent three additional favourable genes derived from the male and female parents. Similarly, upper case letters increase the value of the trait, lower case letters decrease it.
In our previous work, we employed an integrated quantitative genetics approach to study a large number of hybrid rice varieties and their F2 populations, including many time‐honoured elite hybrids with strong heterosis. Our research also revealed that yield‐related heterosis in hybrid rice was primarily attributed to the accumulation of favourable alleles with incomplete dominance, and rare favourable alleles from the female parents contributed significantly to the superiority of the hybrid over their male parents (Gu et al., 2023; Huang et al., 2015, 2016). Furthermore, heterosis and transgressive segregation are the reasons why plant breeding works (Mackay et al., 2021). Although the genetic basis of transgressive segregation appears to be largely distinct from that underlying heterosis, numerous quantitative genetic studies consistently support that the primary cause of both phenomena is the action of complementary genes (deVicente and Tanksley, 1993; Rieseberg et al., 1999). We have drawn a simple diagram to explain the genetic basis and underlying connection of heterosis and transgressive segregation. Based on the information depicted in Figure 1b, we consider ‘AA’ and ‘BB’ as favourable alleles from the paternal parent, while ‘CC’ represents superior alleles from the maternal parent, and their heterozygous genotypes exhibit incomplete dominance. Compared to other genetic effects such as overdominance and epistasis, a larger portion of the advantage of hybrids over their male parents is likely caused by dominant effects. Furthermore, the contribution of heterozygous favourable alleles from the maternal parent to the advantage over the male parents, minus the contribution reduced by the paternal favourable alleles' heterozygosity, remains significantly greater than the phenotypic value of the paternal genotype. In other words, for the manifestation of a superior advantage over the paternal parent, the genetic value of ‘AaBbCc’ should exceed that of ‘AABBcc’ (Figure 1b). Certainly, in the F2 segregating progeny, there are still many individuals that surpass their parents in certain traits (directional transgressive segregation) and exhibit a phenomenon similar to hybrid vigour observed in F1 hybrids (Figure 1b). This ubiquitous phenomenon can also be explained by the dispersion of favourable genes (Mackay et al., 2021; Wang et al., 2015).
Given all the information above, can we manipulate the maternal heterotic genes to maximize their beneficial properties for yield‐related heterosis in a more directed manner, based on our understanding of their genetic effects? For this purpose, we propose a hybrid breeding improvement strategy. This strategy consists of three main steps (Figure 1c). The first step is to identify the heterotic genes from the female parents. In the F2 segregating population of hybrid varieties exhibiting heterotic performance, the incomplete dominant loci in the maternal parent are identified through QTL mapping. The second step involves genetic manipulation through hybrid backcrossing or gene editing to introduce the favourable allele genotypes from the maternal parent into the paternal parent. This manipulation aims to incorporate the advantageous heterotic genes from the maternal parent into the genetic background of the paternal parent. The third step is to cross the improved paternal parent, which now contains the favourable heterotic genes from the maternal parent, with the original maternal parent to obtain improved hybrid rice varieties. This cross will combine the desirable traits of both parents, potentially leading to increased yield or other favourable traits without sacrificing heterotic performance. The subsequent results will validate how this strategy can further enhance yield without compromising heterotic performance.
Alternative splicing induced by CRISPR‐Cas9 in a heterosis gene OsMADS1 GW3p6 improves the grain yield and quality in rice
We reviewed the mapping results of the F2 hybrid rice population of GLY676, which includes both parents, and found that the majority of the QTLs exhibited incomplete dominance (Table S2). Among these loci, the heterotic gene GW3p6 from the maternal parent not only showed incomplete dominance but also made a significant contribution to yield‐related traits (LOD = 46.25, phenotypic variance = 14.98%). Therefore, GW3p6 appears to be a promising candidate gene for validating this breeding strategy.
qLGY3/OsLG3b/GW3p6, encoding a MIKC‐type MADS‐box transcription factor OsMADS1, strikingly increases the grain length and thus the grain yield in hybrid rice or inbred rice cultivars (Liu et al., 2018; Wang et al., 2019; Yu et al., 2018). A 15‐bp non‐homologous genomic fragment spanning the intron‐exon junction of OsMADS1 resulted in an alternatively spliced protein, which is associated with the increase in grain length and weight. Moreover, a vast repertoire of mRNA isoforms could be produced through AS of multiexon genes' precursor mRNAs (pre‐mRNAs). Most pre‐mRNAs with GT‐AG splice sites are processed by spliceosomes to generate mature mRNAs by removing introns and joining of exons based on canonical GU‐AG rule (Reddy et al., 2013). According to the GU‐AG rule and the genomic position of 15‐bp non‐homologous fragment, we designed a sgRNA targeting the splice acceptor site (dinucleotides AG) at the seventh intron of OsMADS1 (Figure 2a). The vector pYLCRISPR/Cas9‐OsU3:sgRNA‐OsMADS1 was introduced into Fuhui676 (FH676, male parent of two‐line elite hybrid rice GLY676) through Agrobacterium‐mediated transformation.
Figure 2.
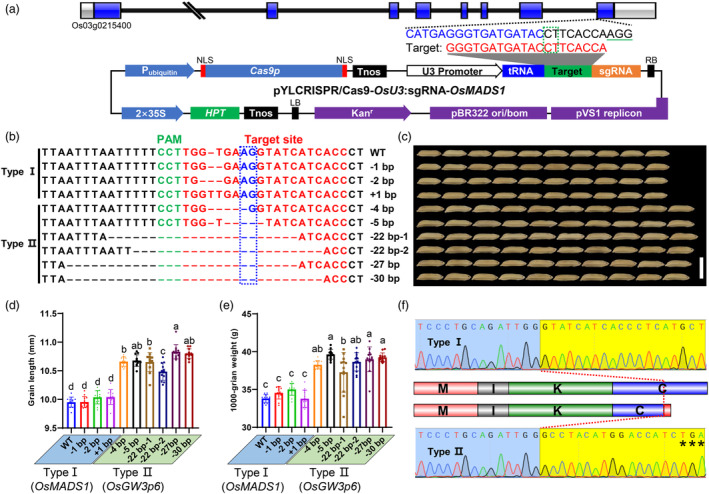
Alternative splicing induced by CRISPR‐Cas9 in OsMADS1 GW3p6 and the grain phenotypes of transgenic plants. (a) Structure of OsMADS1 and CRISPR‐Cas9 vector. The green dotted box and green line indicate the positions of 3′‐splice site and PAM site, respectively. HPT stands for the hygromycin B phosphotransferase gene in the CRISPR‐Cas9 vector. (b) Genomic sequence of T2 lines of FH676 at the target site. The Arabic numerals and their signs represented the number of inserted or deleted nucleotides. (c) Grain phenotypes of T2 lines. Scale bar, 1 cm. (d–e) Statistical analysis of grain length (d) and TGW (e) of T2 lines. The letters in parentheses represented the respective mRNAs of OsMADS1 and OsMADS1 GW3p6 . Data shown as mean ± s.d. (n = 12 plants). Statistical analyses were performed by Duncan's multiple range tests. The presence of the same lowercase letter denotes a non‐significant difference between means (P > 0.05). (f) cDNA sequencing of Type I & II plants. The red dotted line indicates the junction between exon 7 and 8 of OsMADS1. The asterisks show the position of the stop codon. The schematic illustration in the middle indicates the functional domains of OsMADS1 (upper) and OsMADS1GW3p6 (lower).
By sequencing the PCR amplicons of the non‐homologous genomic fragment and selecting hygromycin B (hyg)‐resistant plants, we obtained six T0 transgenic plants comprising uniform bi‐allelic and heterozygous mutations. Ultimately, over 40 individual T2 transgenic lines were examined to screen for homozygous mutations and investigate the phenotypic effect of the mutants. A total of 10 genotypes exist in all T2 lines, including one with a 1 bp insertion and the others with various deletions (Figure 2b). Based on whether the 3′ acceptor site (AG) was disrupted or not, we divided these genotypes into two types: Type I and Type II. Type I plants, harbouring the original splice site, exhibited similar grain length and thousand‐grain weight (TGW) to the transgenic negative lines (WT). But compared to Type I plants, the grain length and TGW of Type II plants were increased by 6.9% and 12.8%, respectively (Figure 2c–e). Unlike Type I with original mature mRNA of OsMADS1, 3′RACE and cDNA sequencing revealed that Type II plants produced an alternative 3′ splicing event: the splice site shifted to the 32nd nucleotide (AG/GC) of exon 8 of OsMADS1, causing a premature termination codon and a truncated exon 8 (Figure 2f and Figure S1). This AS isoform is identical to that of qLGY3/OsLG3b/GW3p6, indicating that the alternatively spliced protein of OsMADS1 is truncated by 32 amino acid residues to influence its C‐terminal domain (Liu et al., 2018). All these results suggested that AS of OsMADS1 induced by CRISPR‐Cas9 system can significantly increase grain length and grain weight by disrupting the 3′ splice site.
The Type II genotype could simultaneously improve grain yield and quality with no apparent trade‐off in other agronomic traits
As the genome editing vectors can be segregated out from mutant genomes through crossing or selfing, we crossed T2 plants with FH676 to screen for transgene‐free mutant plants, and thus obtained transgene‐free plants in the BC2F2 progeny (Figure 3a). Neither the genomic fragment of hpt (hygromycin B phototransferase) nor cas9 protein was detected in these transgene‐free plants (Figure 3b and Figure S2). Phenotypic comparison of the two types of plants indicated that Type II plants had remarkable increases in grain length (8.5%), TGW (13%), and grain yield per plant (9.2%) compared to FH676 and Type I plants (Figure 3c–g and Figure S3), whereas other important agronomically traits did not change significantly (Figure 3a and Figure S4). OsMADS1 GW3p6 , a heterosis gene derived from the female parent, explained a large proportion in the yield related better‐parent heterosis in the two‐line hybrid rice GLY676, and its heterozygous state displayed a positive partial dominance effect (Huang et al., 2016; Wang et al., 2019). We therefore crossed BC2F2 plants with FH676 to evaluate the genetic effects of heterozygous OsMADS1. As shown in Figure 3c–g, the genetic effects of heterozygous Type II alleles showed a partial positive dominance effect, which is consistent with previous findings (Huang et al., 2016; Wang et al., 2019). This result further supported the notion that the heterozygous state of the heterotic gene OsMADS1 GW3p6 could greatly enhance grain yield. OsMADS1 lgy3 is responsible for better appearance quality in terms of grain length‐to‐width ratio and grain chalkiness (Liu et al., 2018), and recent study has shown that OsMADS1 affects grain quality by regulating gene expressions and regulatory networks of starch and storage protein metabolisms in rice grains (Liu et al., 2023). Therefore, we used image analysis systems and scanners to analyse the grain appearance quality of the gene‐edited BC2F2 plants. We observed a slight difference between Type I and Type II plants in grain chalkiness except for grain length‐to‐width ratio (Figure S5). The percentage of endosperms with chalkiness and the degree of endosperm chalkiness of Type II plants are approximately 4.4% (about 30% decrease) and 0.95% (about 31% decrease) lower than that of Type I plants, respectively (Figure 3h,i and Figure S5). Intriguingly, the plants with a heterozygous state of OsMADS1 GW3p6 also exhibit partial positive dominance for the phenotype of chalky grain and chalkiness area (Figure 3h,i). Further comparisons of endosperm transparency showed no significance difference between the two types of OsMADS1 (Figure S5). Collectively, the CRISPR‐Cas9‐mediated AS isoform of OsMADS1 played dual roles in increasing grain yield and quality. More importantly, these transgene‐free restore lines with type II genotypes, coupled with this gene‐editing target site that effectively caused AS of OsMADS1, hold tremendous promise in rice breeding, especially in improving the male parent in hybrid rice breeding.
Figure 3.
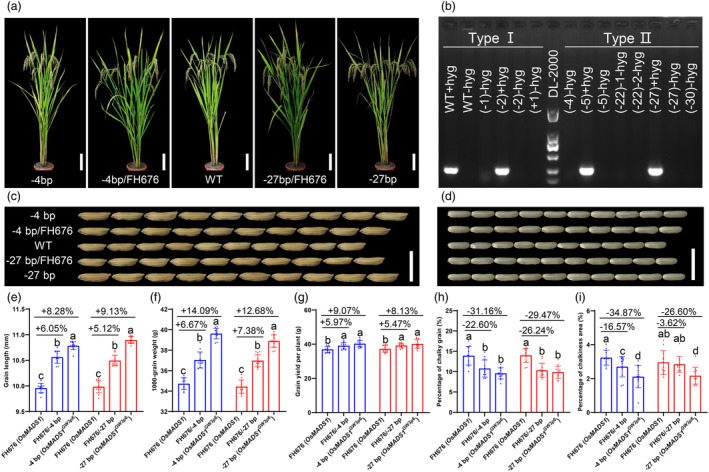
The phenotype of grain size and quality in FH676 transgene‐free plants. (a) Comparison of plant morphology in BC2F2 plants. Scale bars = 20 cm. (b) PCR amplicons of hyg resistance gene in BC2F2 and T2 plants. All positive plants with the hyg‐B‐resistance gene are conducted in T2 lines. (c–i) Grain morphology (c), brown rice (d), grain length (e), TGW (f), grain yield per plant (g), percentage of chalky grain (h), and chalkiness area (i) in FH676 and BC2F2 plants with the heterozygous and homozygous genotypes of Type II. The letters in parentheses represent the same mRNA as OsMADS1 and OsMADS1 GW3p6 . Data shown as mean ± s.d. (n = 12 plants). Statistical analyses were performed by Duncan's multiple range tests. The presence of the same lowercase letter denotes a non‐significant difference between means (P > 0.05).
Introducing Type II genotype into hybrid rice GLY676 could further boost grain yield
Extensive genomic analyses of hybrid rice varieties have revealed that the heterozygosity of whole‐genome genotypes has a limited contribution to yield‐related heterosis through in‐depth analyses of the effects of heterozygous genotypes, but a strong correlation is observed between the yield and the accumulation of superior alleles (Huang et al., 2015; Yang et al., 2017a, 2017b). Since the genetic effect of OsMADS1/OsMADS1 GW3p6 is partial positive dominance in grain yield (Figure 3c–g), it is very promising to manipulate heterosis genes to break the yield ceiling of current hybrid rice varieties.
Therefore, we crossed Type II plants of FH676 with the male sterile line Guangzhan63‐4S (GZ) to produce new hybrids (Figure 4a). Multiple molecular markers distributed on each chromosome are used to check the heterozygosity of new rice hybrids (Figure S6a). Genotyping analysis indicated the purity of new hybrid seeds is 100%, which ensures the same genetic background as the GLY676 (Figure S6b–d). Although the grain length and TGW of the new F1 hybrids exhibited mid‐parent heterosis (less than FH676), their grain length and TGW are still higher than GLY676 and the plants of WT (Type I) × GZ (Figure 4b,c). Compared to FH676, the plants of WT (Type I) × GZ, GLY676, and the new hybrids (homologous genotype of OsMADS1 GW3p6 ) exhibited higher grain yield per plant, accounting for an average of 16.5%, 19.8%, and 22.1%, respectively (Figure 4d). We found no obvious differences in grain appearance quality among these hybrid combinations (Figure 4e), indicating that increasing the dosage of OsMADS1 GW3p6 (from heterozygous to homozygous state) could improve the yield of hybrid rice without an apparent trade‐off in grain quality. Intriguingly, over three successive years of field trialling, GLY676 and the new hybrids out‐yielded FH676 by an average of 11.24% and 13.16%, respectively (Figure 4f, Figure S7). Similarity, a new hybrid generated by crossing a near‐isogenic line of OsMADS1 GW3p6 in FH676 genetic background (NIL) with GZ had comparable grain yield performance with other new modified hybrids carrying the homologous OsMADS1 GW3p6 genotype (Figure 4d,f). Thus, we produced the improved hybrids with practical potential in rice yield. These results also prove that our hybrid breeding strategy is effective and lend credence to the view that dominance effect underlying functional complementation is an indispensable contributor to yield heterosis.
Figure 4.
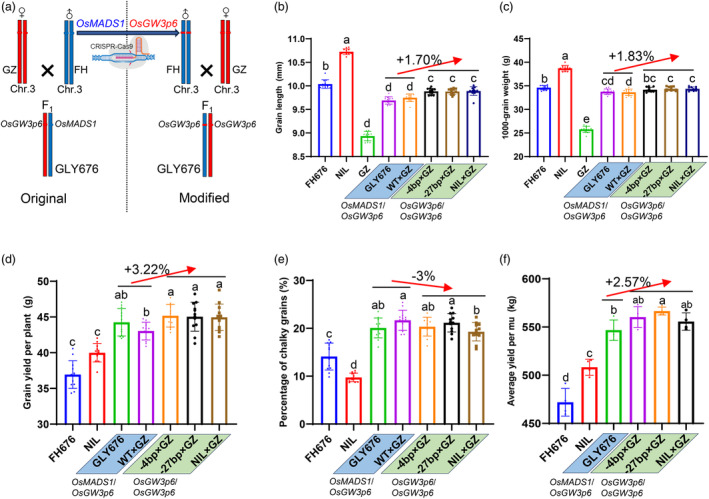
The performance of grain yield and grain quality among various hybrid combinations of GLY676. (a) The schematic diagram shows the genotype of chromosome 3 of the new hybrids. FH and GZ indicate the male and female parents of GLY676, respectively. The colours of blue and red represent the genotypes of the male parent and female parent, respectively. The heterozygous genotype of the F1 progeny (GLY676) of FH and GZ is represented by red and blue boxes arranged side by side. The arrowhead indicates using CRISPR‐Cas9 to change OsMADS1 to OsMADS1 GW3p6 in FH676 plants. (b–f) Statistical analyses of grain length (b), TGW (c), grain yield per plant (d), percentage of chalky grain (e), and average yield per mu (f) among different hybrid combinations of GLY676. The genotypes below represent the same haplotypes present in these genetic materials. Values indicate means ± s.d. Exact P values are performed by Duncan's multiple range tests. (b–e) n = 12 plants. (f) n = 4 paddies; each paddy contained 480 plants in the 20.6‐m2 field. mu, 666.66 m2 or a fifteenth hectare.
The increased yield in three‐line hybrid rice by assimilating a heterosis gene demonstrated an effective and time‐saving strategy for hybrid breeding
To test if this approach that introduces a heterosis gene OsMADS1 GW3p6 into the parental plants to enhance grain yield has great potential in hybrid rice breeding, we first perform haplotype analysis to find the proportion of the haplotype of OsMADS1 GW3p6 in 2932 commercial hybrid rice varieties. The results showed that approximately 5.25% of the 2932 hybrid rice varieties carried the haplotype of OsMADS1 GW3p6 (Dataset S1). Subsequently, we collected 566 hybrid rice parents, consisting of 407 male parents and 159 female parents, to investigate the frequency of OsMADS1 GW3p6 application in all parents of hybrid rice (Dataset S2). About 3.36% of the parent materials possessed the haplotype of OsMADS1 GW3p6 , and the ratio in female parents (8.18%) far outstrips that in male parents (1.47%). Intriguingly, only three varieties with the homozygous genotype of OsMADS1 GW3p6 occur in 2932 hybrid rice varieties, which may be associated with a lower possibility for the homozygous genotype of OsMADS1 GW3p6 in the progeny formed by crossing their parents harbouring the haplotype of OsMADS1 GW3p6 . In addition, diverse hybrid rice varieties containing geographic and chronological information enable us to examine the dynamic utilization process of OsMADS1 GW3p6 . We found that the haplotype of OsMADS1 GW3p6 mainly existed in three planting areas: the middle and lower reaches of the Yangtze River, southwestern China, and Southern China (Figure 5a). Among these areas, the highest proportion of the haplotype in southwestern China may be relevant to the breeding concept − heavy panicle and large grain size of hybrid rice − pursued by breeders in this region. According to the release time of these hybrid rice, we divided them into three periods: 1976–2000, 2001–2010, and 2011–2020. Haplotype analysis showed a noticeable increase with time in the proportion of OsMADS1 GW3p6 (Figure 5b). All these data indicated that OsMADS1 GW3p6 is a rare superior allele that has not yet been widely used and has excellent potential in hybrid rice breeding.
Figure 5.
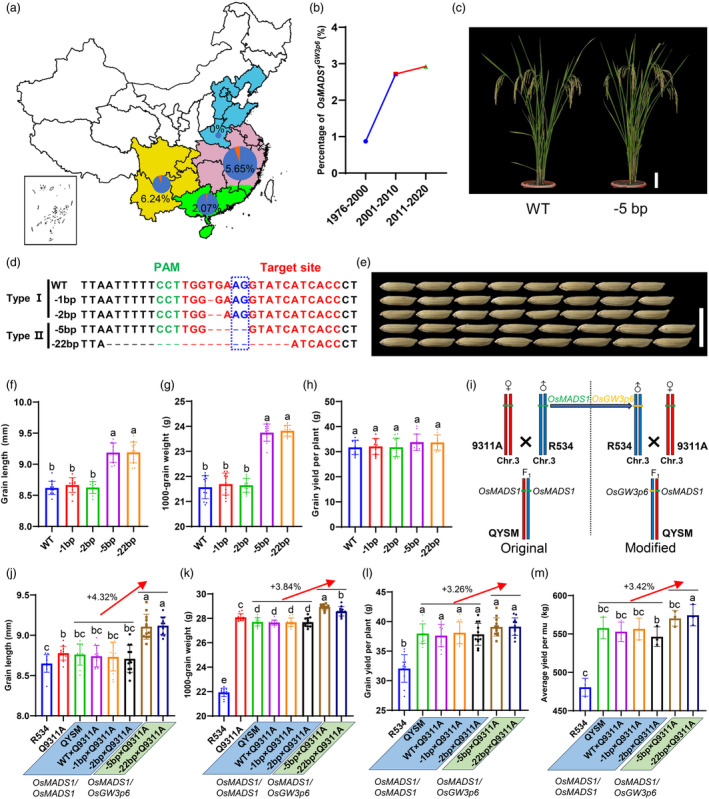
Introducing a heterosis gene into hybrid rice QYSM showed great potential for yield increase. (a) The geographic distribution of the hybrid rice accessions containing OsMADS1 and OsMADS1 GW3p6 alleles. The colour of blue and orange in the pie graph represent the haplotypes of OsMADS1 and OsMADS1 GW3p6 , respectively. The values closed to the pie graph represent the proportion of OsMADS1 GW3p6 haplotype in this area. The size of the pie graph indicates the number of hybrid rice varieties. (b) The proportion of the haplotype of OsMADS1 GW3p6 in different breeding periods. (c) Gross morphologies of transgenic plants with the genotypes of Type I and Type II. (d) The genotypes of T2 plants of R534 at the target site of OsMADS1. (e–h) Grain size and shape (e), grain length (f), TGW (g), and grain yield per plant (h) in transgenic T2 plants of R534. n = 12 plants. (i) The illustration represents the genotypes of the hybrid rice QYSM and its parents in chromosome 3 and exhibits the general process of the strategy for hybrid breeding by introducing a heterosis gene. Blue and red indicated the genotypes of male parents and female parents, respectively. The green and orange boxes indicate the genotypes of OsMADS1 and Type II (or OsMADS1 GW3p6 ), respectively. The heterozygous genotypes of OsMADS1 (OsMADS1/Type II or OsMADS1/OsMADS1 GW3p6 ) in modified QYSM are represented by green and orange boxes arranged side by side. (j–m) Statistical analyses of grain length (j), TGW (k), grain yield per plant (l), and average yield per mu (m) in various hybrid combinations of QYSM. The genotypes below represent the same haplotypes present in these genetic materials. (j–k) Data shown as mean ± s.d. (n = 12 plants). (m) n = 3 paddies, each paddy contained 240 plants in the 10.66‐m2 field. mu, 666.66 m2 or a fifteenth hectare. Statistical analyses were performed by Duncan's multiple range tests.
To further validate the feasibility of this approach, we tried to modify OsMADS1 in other representative hybrid rice through the CRISPR‐Cas9 system. Wu‐shan‐si‐miao (R534) is an inbred rice variety featuring high yield and grain quality and is widely used as a restorer line for two‐line and three‐line hybrid rice due to its high combining ability. Therefore, R534 carrying the haplotype of OsMADS1 was selected as the candidate plant for genome editing. In proof‐of‐concept experiments, we obtained a total of 17 T0 transgenic lines, and four genotypes were generated in all transgenic lines of R534 through Sanger sequencing (Figure 5c,d). Comparison of plant phenotype showed that the genome‐edited plants with Type II genotype increased the grain length, TGW, and grain yield per plant by 6.40%, 9.94%, and 5.89%, respectively, without an apparent negative impact on other agronomic traits (Figure 5e–h, Figure S8). These data also indicated that the genotype of Type II created by CRISPR‐Cas9 technology had great potential for yield improvement in R534.
Quan‐you‐si‐miao (QYSM), a three‐line high‐yield hybrid rice variety, is bred by crossing R534 and the male‐sterile line Quan‐9311A (Q9311A). The genetic background of QYSM is heterozygous, but we found it had a homozygous genotype of OsMADS1 through Sanger sequencing. For the purpose of crop improvement, we made multiple hybrids to test their yield performance (Figure 5i). In terms of grain length and TGW, we found QYSM and new hybrid combinations with homozygous haplotype of OsMADS1 (WT × Q9311A, −1 bp × Q9311A, and −2 bp × Q9311A) exhibited mid‐parent heterosis, but other new combinations with heterozygous haplotype of OsMADS1 GW3p6 (−5 bp × Q9311A, and −22 bp × Q9311A) showed better‐parent heterosis (Figure 5j,k). Compared with the hybrids with the homozygous haplotype of OsMADS1, the F1 hybrids harbouring the heterozygous haplotype of OsMADS1 GW3p6 increased the grain length and TGW by 4.32% and 3.84%, respectively (Figure 5j,k). Besides, the new hybrids with Type II genotype produced higher grain weight than QYSM and Type I hybrids, thus resulting in a 3.26% increase in grain yield per plant compared with the QYSM and the hybrids with Type I genotype (Figure 5l). More importantly, the field test indicated that the plants with Type II genotype, on average, had an 3.42% increase compared with QYSM and Type I hybrids (Figure 5m). All these data suggested that this approach could quickly improve grain yield in QYSM, implying great potential in hybrid rice breeding.
Discussion
Despite the emergence about various genetic models to explain the genetic basis of heterosis, little consensus has yet been reached. More studies provided empirical support for the dominance hypothesis compared to other genetic models (Li et al., 2016; Shen et al., 2022; Yang et al., 2017a). For example, the hybrids pyramiding multiple heterotic loci in the female or combined parental genome background showed comparable yield performance to that of the hybrid rice variety Shanyou63 (Shen et al., 2022). This result demonstrated that the dominance effect played an important role in yield heterosis, and yield‐related heterosis could be achieved by manipulating several major dominant heterotic loci. Previous research has revealed that OsMADS1 GW3p6 is a typical heterosis locus that can significantly increase grain yield (Wang et al., 2019). Moreover, the genetic effect of heterozygous OsMADS1/OsMADS GW3p6 exhibited incomplete dominance (Figure 3c–g), and introducing the heterosis gene OsMADS1 GW3p6 into restorer lines greatly enhanced the grain yield without an apparent negative impact on other agronomic traits (Figure 3g, Figure S4). The above characters of OsMADS1 GW3p6 ensure a higher yield by pyramiding a dominant heterotic gene in hybrid rice. Additionally, this study provided solid evidence that the grain yield of the hybrids with the homozygous genotype of OsMADS1 GW3p6 (or Type II) was higher than that of the hybrids with the heterozygous genotype of OsMADS1 GW3p6 in hybrid rice GLY676, implying dominance model played a leading role in yield‐related heterosis. In the current scenario of similar and saturated combinations of complementary genotypes in hybrid rice, these results demonstrate the feasibility of the breeding strategies that aim to maximize the contribution of heterosis genes in the female parent to further enhance yield. Because it is challenging to obtain genetically modified seeds of Q9311A owing to extremely low fertility, we did not achieve homozygous genotype of OsMADS1 GW3p6 (or Type II) in QYSM. Nevertheless, the results of increased yield in the modified F1 of QYSM carrying heterozygous OsMADS1 GW3p6 , demonstrated that accumulating heterozygous heterosis gene with incomplete dominance can still improve hybrid rice. It is two sides of the same coin of our hybrid breeding strategy. In the future, we will introduce the Type II genotype into the maintainer line Q9311B, which will help us obtain a new QYSM with a homozygous Type II genotype to further investigate the extent of increase in grain yield.
Our study also showed the potential of single heterosis gene to increase rice production in hybrid rice. For the trait of grain yield per plant, OsMADS1 GW3p6 (or Type II genotype) in hybrid combinations of GLY676 and QYSM explained over 40% and over 30% of the yield advantage of hybrids over their male parents, respectively (Data S1). This data proved the genomic loci from the female parents contributed a lot to better‐parent heterosis of rice yield. As genomic insights into rice heterosis have suggested that the heterozygous state of heterotic loci mostly acted through the way of positive partial dominance, we observed a remarkable growth in grain yield by introducing Type II genotype into QYSM even though the genotype of OsMADS1 GW3p6 in QYSM is heterozygous. Meanwhile, we found that the proportion of the haplotype of OsMADS1 GW3p6 is low in 2932 hybrid rice varieties, which is because OsMADS1 GW3p6 may have no effect in other genetic backgrounds. These results also inspire us that hQTLs from male parents, including OsMADS1 GW3p6 , should be introduced into more hybrid rice varieties to verify whether this approach can further improve grain yield. In this study, we have achieved an increase in grain yield in two representative two‐line and three‐line hybrid rice varieties (Figure 4d,f; Figure 5l,m). Numerous published studies on the genome analysis of hybrid rice have shown that the breeders introduced different exogenous genomes of other rice subpopulations to construct male and female parents of hybrid rice, thereby shaped the genome landscape of heterosis in hybrid rice (Gu et al., 2023; Lin et al., 2020). Therefore, we inferred from these results that the approach of introducing OsMADS1 GW3p6 into hybrid rice would be effective in most hybrid rice varieties.
Yield‐related heterosis is characterized by a substantial increase in yield compared to the parental lines, which is attributed to the rearrangement of genotypes (or hQTLs) inherited from the parents. Among these hQTLs, many heterosis genes exhibit pleiotropy in important agronomic traits, such as the major hQTL DTH8 with pleiotropic effects on grain yield, heading date, and plant height (Yan et al., 2011). And previous studies also shown that several major heading date genes determined strong heterosis of commercial rice hybrids in diverse ecological regions (Huang et al., 2016; Zhou et al., 2021). In this study, both FH676 and R534 carrying the Type II genotype showed a slight increase in heading date compared to the wild type (Figures S4b and S8b). However, we did not observe significant differences in heading date among the improved F1 hybrids (Figure S9). Considering the potential inconvenience in agricultural production and the adaptation to ecological regions caused by significant changes in heading date, OsMADS1 GW3p6 , which exhibits less pronounced phenotypic variations in heading date, remains highly valuable in breeding.
Genome editing is a powerful and prevalent method in crop improvement and is set to revolutionize plant breeding. Our study reflected several advantages in harnessing CRISPR‐Cas9 to edit a heterosis gene for cross breeding. First, our approach is time‐ and labour‐saving. Compared to the tedious work of conventional cross breeding, we estimated that this approach could save at least 50% of time and labour costs for hybrid rice breeding. The extensive use of DNA‐free genome editing methods to obtain transgene‐free mutant parents will further accelerate the process of hybrid rice breeding (Liang et al., 2017; Svitashev et al., 2016; Zhang et al., 2016). Second, introducing hQTLs into hybrid rice could effectively prevent the genetic drag between heterosis genes and deleterious genes. Precise gene editing enables the optimization of agronomic traits in hybrid rice without damaging their unique genetic backgrounds. In addition, previous studies have revealed that OsMADS1 GW3p6 originated from tropical japonica (Liu et al., 2018; Wang et al., 2019; Yu et al., 2018). So, it may be difficult to effectively utilize this gene due to linkage drag, resulting in a low proportion of OsMADS1 GW3p6 in 2932 hybrid rice varieties. This study showed massive potential for crop improvement by fine‐tuning the genome of hybrid rice. Third, our breeding strategy with good versatility will be applicable to combine desirable traits in hybrid rice breeding. For example, a three‐line hybrid variety with increased grain aroma could be produced by introducing new alleles of BADH2, demonstrating a similar strategy to our approach (Hui et al., 2022). In view of this, some essential QTLs that control certain agronomic traits without a trade‐off effect in other traits will be suitable targets for precisely modifying traits in hybrid rice. Additionally, the ever‐increasing known quantitative trait nucleotides integrating with various genome editing tools will further expand the scope of rapid genetic improvement in hybrid breeding (Wei et al., 2021a). Further research should quantify the genetic effect of more hQTLs and still need to verify the increasing effect in agronomic traits by editing the candidate hQTLs in hybrid rice (Mackay et al., 2021; Wang and Han, 2022). We believed that the total grain output of the genome‐edited parental inbred lines could exceed that of their hybrid rice by introducing multiple heterosis loci in the future.
In conclusion, we achieved an alternative 3′ splicing event of OsMADS1 by disrupting the 3′ splice site using the CRISPR‐Cas9 system. We also observed that AS of OsMADS1 could simultaneously enhance rice yield and grain quality. Also, the new hybrids harbouring homozygous Type II genotypes or OsMADS1 GW3p6 under the same hybrid genetic backgrounds have a better performance in grain yield compared to the hybrid variety GLY676, which demonstrates that our hybrid breeding strategy is a promising and time‐saving strategy for breeding higher‐yielding rice varieties beyond what is currently achievable. Our research also showed great potential of single heterosis gene from the female parents in increasing grain yield of hybrid rice. Continuous improvements in both rice yield and grain quality are long‐term goals to cope with the challenges of population growth and climate change. With the development of rice functional genomics and powerful genome editing technologies, increasing superior alleles could be harnessed in rice breeding (Wang and Han, 2022). Overall, our study provided a valuable breeding target and an efficient method for hybrid rice breeding.
Materials and methods
Plant materials and growth conditions
The indica varieties FH676, GZ63‐4S, NIL‐GW3p6, GLY676, R534, Q9311A, and QYSM were used in this study. All these plants, including transgenic and non‐transgenic plants, were planted in the experimental fields of the CAS Center for Excellence in Molecular Plant Sciences in Shanghai and Hainan, China, from 2018 to 2023. Twelve 30‐day‐old seedlings of each line were transplanted with 17 cm spacing in single row plots in the rice paddy field, and rows were 30 cm apart.
Phenotypic evaluation
Fully filled grains on each plant were used to measure grain length, grain width, thousand‐grain weight, and yield per plant. And the grain size of rice was measured by an automatic digital grain scanner, and the statistics of grain length and width were conducted from the image analysis software (Wseen SC‐G). Ten plants from the middle of each row were harvested individually for trait measurement. The phenotypic data of grain number per panicle, panicle length, and seed setting rate was obtained from the three longest panicles of each plant.
The percentage of chalky grains was scored by the visual assessment of the percentage of white‐core grains in random samples of more than 100 dehulled grains from each plant. The degree of endosperm chalkiness was obtained from image processing software (Wseen SC‐E) by scanning the milled rice.
To obtain the total yields of field‐grown rice, we planted different types of rice varieties in the same paddy fields in parallel. For all rice varieties of GLY676 and QYSM, the average planting density of rice is about 15 152 plants per mu. 480 and 240 plants of every rice variety grown in the 20.6‐m2 and 10.65‐m2 paddies were harvested, respectively. Threshed seeds from each plant were air dried and stored at room temperature for 2 months before further weigh. The seed purity of F1 hybrids was confirmed by multiple molecular markers.
Plasmid construction and plant transformation
According to the genomic position of the 15‐bp non‐homologous fragment, we designed a sgRNA targeting the splice acceptor site at the seventh intron of OsMADS1. The sgRNA target of CRISPR‐Cas9 was designed on the website: http://crispr.hzau.edu.cn/CRISPR2. A 20‐bp target region was inserted into the intermediate vector pYLgRNA‐OsU3/LacZ, and the integrated U3‐target‐gRNA region was cloned and subsequently inserted into the vector pYLCRISPR/Cas9‐MH.
The constructs were introduced into Agrobacterium tumefaciens strain EHA105 and transferred into the FH676 and R534 by Agrobacterium‐mediated transformation. The primers used in this study are listed in Table S3.
Screening of transgene‐free plants
The coding sequence of hpt and Cas9 protein were selected as the indicator of exogenous genomic fragments. We designed one and two pairs of primers targeting hpt and cas9, respectively. After PCR analysis, the plants without hpt and cas9 genome fragments could be identified as transgene‐free plants. In T2, BC1F1, BC2F1, and BC2F2 lines of FH676, we screened transgene‐free plants through PCR analysis of three molecular markers. Since T2 generation, the BC2F2 plants of FH676 that have been negative for three genomic fragments in all their generations could be further confirmed as transgene‐free plants.
RACE and cDNA analysis
The leaves from 2‐week‐old seedlings were harvested and grinded, and the total RNA of rice leaves was extracted with Trizol reagent (Thermo Fisher Scientific, 15 596 026). The genomic DNA removal and first‐strand cDNA synthesis were conducted using ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo, FSQ‐301). The genomic fragments containing the seventh and eighth exons of OsMADS1 were amplified and sequenced to identify the AS. The full‐length transcripts of OsMADS1 and OsMADS1 GW3p6 were determined by rapid amplication of cDNA ends (RACE). The cDNA was synthesized following the protocol provided in SMARTer RACE 5′/3′ kit (Takara, 634 858/634859). The primers used for RACE AND cDNA analysis are listed in Table S3.
Statistical analysis
Statistical tests were performed by GraphPad Prism 8 and SPSS Statistics 23. All values are reported as mean ± s.d.
Competing of interests
The authors declare no competing interest.
Author contributions
C.W. performed most of the experiments, Z.W. and C.W. contributed the generation of the hybrid combinations. C.W., Z.W., Y.C., L.H., Y.W., D.Y., W.Lv., and K.L. characterized the genotypes and phenotypes of the gene‐edited plants. C.W., Z.Z., Q.Z., and L.S. analysed the data. C.W., and B.H. conceived and designed experiments and wrote the manuscript. B.H. directed the research.
Supporting information
Dataset S1 The haplotype of OsMADS1 in hybrid rice.
Dataset S2 The haplotype of OsMADS1 in the parents of hybrid rice.
Data S1 Phenotypic source data in this study.
Figure S1 The electropherograms of cDNA sequencing in T2 transgenic plants.
Figure S2 PCR amplicons of Cas9 in BC2F2 and T2 plants.
Figure S3 Statistical analyses of important agronomic traits in BC2F2 lines of FH676 with Type I genotypes.
Figure S4 Comparisons of agricultural traits in BC2F2 plants.
Figure S5 Grain appearance in BC2F2 plants.
Figure S6 Purity identification of hybrid rice seeds.
Figure S7 The average yield per mu among different hybrid combinations of GLY676 in Hainan and Shanghai.
Figure S8 Comparisons of agronomic traits in transgenic plants of R534.
Figure S9 Heading date in multiple hybrid combinations of GLY676 and QYSM.
Table S1 Progress of the genetic basis of heterosis in plants.
Table S2 QTL mapping results in GLY676 F2 population.
Table S3 Primers and probes used in this study.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (31788103) and the Chinese Academy of Sciences (XDB27010301) to B. H., National Natural Science Foundation of China (32300503), Shanghai Post‐doctoral Excellence Program (2020460) and the China Postdoctoral Science Foundation (2020M681413) to C. W., the National Key Research and Development Program of China (2020YFE0202300 to Z.Q.). We thank Dr. Ahong Wang and Dr. Yu Fang provided the seeds of R534, Q9311A, and QYSM.
Data availability statement
All data supporting the findings of this study are available in the article, supplementary information, and Data S1. Source data are provided in this paper.
References
- Cai, X.‐L. , Wang, Z.‐Y. , Xing, Y.‐Y. , Zhang, J.‐L. and Hong, M.‐M. (1998) Aberrant splicing of intron 1 leads to the heterogeneous 5′ UTR and decreased expression of waxy gene in rice cultivars of intermediate amylose content. Plant J. 14, 459–465. [DOI] [PubMed] [Google Scholar]
- Gao, C. (2021) Genome engineering for crop improvement and future agriculture. Cell 184, 1621–1635. [DOI] [PubMed] [Google Scholar]
- Gu, Z. , Gong, J. , Zhu, Z. , Li, Z. , Feng, Q. , Wang, C. , Zhao, Y. et al. (2023) Structure and function of rice hybrid genomes reveal genetic basis and optimal performance of heterosis. Nat. Genet. 55, 1745–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, M. , Zhang, Y. , Jia, X. , Wang, X. , Zhang, Y. , Liu, J. , Yang, Q. et al. (2022) Alternative splicing of REGULATOR OF LEAF INCLINATION 1 modulates phosphate starvation signaling and growth in plants. Plant Cell 34, 3319–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Yang, S. , Gong, J. , Zhao, Y. , Feng, Q. , Gong, H. , Li, W. et al. (2015) Genomic analysis of hybrid rice varieties reveals numerous superior alleles that contribute to heterosis. Nat. Commun. 6, 6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X. , Yang, S. , Gong, J. , Zhao, Q. , Feng, Q. , Zhan, Q. , Zhao, Y. et al. (2016) Genomic architecture of heterosis for yield traits in rice. Nature 537, 629–633. [DOI] [PubMed] [Google Scholar]
- Hui, S. , Li, H. , Mawia, A.M. , Zhou, L. , Cai, J. , Ahmad, S. , Lai, C. et al. (2022) Production of aromatic three‐line hybrid rice using novel alleles of BADH2 . Plant Biotechnol. J. 20, 59–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger, U. , Lippman, Z.B. and Zamir, D. (2010) The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat. Genet. 42, 459–463. [DOI] [PubMed] [Google Scholar]
- Li, D. , Huang, Z. , Song, S. , Xin, Y. , Mao, D. , Lv, Q. , Zhou, M. et al. (2016) Integrated analysis of phenome, genome, and transcriptome of hybrid rice uncovered multiple heterosis‐related loci for yield increase. Proc. Natl. Acad. Sci. USA 113, E6026–E6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. , Xiong, X. , Wang, F. , Liang, J. and Li, J.‐F. (2019) Gene disruption through base editing‐induced messenger RNA missplicing in plants. New Phytol. 222, 1139–1148. [DOI] [PubMed] [Google Scholar]
- Liang, Z. , Chen, K. , Li, T. , Zhang, Y. , Wang, Y. , Zhao, Q. , Liu, J. et al. (2017) Efficient DNA‐free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 8, 14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Z. , Qin, P. , Zhang, X. , Fu, C. , Deng, H. , Fu, X. , Huang, Z. et al. (2020) Divergent selection and genetic introgression shape the genome landscape of heterosis in hybrid rice. Proc. Natl. Acad. Sci. USA 117, 4623–4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman, Z.B. and Zamir, D. (2007) Heterosis: revisiting the magic. Trends Genet. 23, 60–66. [DOI] [PubMed] [Google Scholar]
- Liu, Q. , Han, R. , Wu, K. , Zhang, J. , Ye, Y. , Wang, S. , Chen, J. et al. (2018) G‐protein βγ subunits determine grain size through interaction with MADS‐domain transcription factors in rice. Nat. Commun. 9, 852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Wang, Q. , Chen, M. , Ding, Y. , Yang, X. , Liu, J. , Li, X. et al. (2020a) Genome‐wide identification and analysis of heterotic loci in three maize hybrids. Plant Biotechnol. J. 18, 185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Li, M. , Zhang, Q. , Wei, X. and Huang, X. (2020b) Exploring the molecular basis of heterosis for plant breeding. J. Integr. Plant Biol. 62, 287–298. [DOI] [PubMed] [Google Scholar]
- Liu, Z. , Li, P. , Yu, L. , Hu, Y. , Du, A. , Fu, X. , Wu, C. et al. (2023) OsMADS1 regulates grain quality, gene expressions, and regulatory networks of starch and storage protein metabolisms in rice. Int. J. Mol. Sci. 24, 8017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay, I.J. , Cockram, J. , Howell, P. and Powell, W. (2021) Understanding the classics: the unifying concepts of transgressive segregation, inbreeding depression and heterosis and their central relevance for crop breeding. Plant Biotechnol. J. 19, 26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang, Y. , Li, X. and Zhang, Q. (2022) Understanding the genetic and molecular constitutions of heterosis for developing hybrid rice. J. Genet. Genomics 49, 385–393. [DOI] [PubMed] [Google Scholar]
- Reddy, A.S.N. , Marquez, Y. , Kalyna, M. and Barta, A. (2013) Complexity of the alternative splicing landscape in plants. Plant Cell 25, 3657–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg, L.H. , Archer, M.A. and Wayne, R.K. (1999) Transgressive segregation, adaptation and speciation. Heredity 83, 363–372. [DOI] [PubMed] [Google Scholar]
- Schnable, P.S. and Springer, N.M. (2013) Progress toward understanding heterosis in crop plants. Annu. Rev. Plant Biol. 64, 71–88. [DOI] [PubMed] [Google Scholar]
- Seymour, D.K. , Chae, E. , Grimm, D.G. , Martín Pizarro, C. , Habring‐Müller, A. , Vasseur, F. , Rakitsch, B. et al. (2016) Genetic architecture of nonadditive inheritance in Arabidopsis thaliana hybrids. Proc. Natl. Acad. Sci. USA 113, E7317–E7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, G. , Hu, W. , Wang, X. , Zhou, X. , Han, Z. , Sherif, A. , Ayaad, M. et al. (2022) Assembly of yield heterosis of an elite rice hybrid is promising by manipulating dominant quantitative trait loci. J. Integr. Plant Biol. 64, 688–701. [DOI] [PubMed] [Google Scholar]
- Song, X. , Meng, X. , Guo, H. , Cheng, Q. , Jing, Y. , Chen, M. , Liu, G. et al. (2022) Targeting a gene regulatory element enhances rice grain yield by decoupling panicle number and size. Nat. Biotechnol. 40, 1403–1411. [DOI] [PubMed] [Google Scholar]
- Svitashev, S. , Schwartz, C. , Lenderts, B. , Young, J.K. and Mark Cigan, A. (2016) Genome editing in maize directed by CRISPR‐Cas9 ribonucleoprotein complexes. Nat. Commun. 7, 13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Y. , Abdelrahman, M. , Li, J. , Wang, F. , Ji, Z. , Qi, H. , Wang, C. et al. (2021) CRISPR/Cas9 induces exon skipping that facilitates development of fragrant rice. Plant Biotechnol. J. 19, 642–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgeman, S. and Zamir, D. (2023) Epistatic QTLs for yield heterosis in tomato. Proc. Natl. Acad. Sci. USA 120, e2205787119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vicente, M.C. and Tanksley, S.D. (1993) QTL analysis of transgressive segregation in an interspecific tomato cross. Genetics 134, 585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B.‐B. and Brendel, V. (2006) Genomewide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. USA 103, 7175–7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. and Han, B. (2022) Twenty years of rice genomics research: From sequencing and functional genomics to quantitative genomics. Mol. Plant 15, 593–619. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Greaves, I.K. , Groszmann, M. , Wu, L.M. , Dennis, E.S. and Peacock, W.J. (2015) Hybrid mimics and hybrid vigor in Arabidopsis. Proc. Natl. Acad. Sci. USA 112, E4959–E4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Tang, S. , Zhan, Q. , Hou, Q. , Zhao, Y. , Zhao, Q. , Feng, Q. et al. (2019) Dissecting a heterotic gene through GradedPool‐Seq mapping informs a rice‐improvement strategy. Nat. Commun. 10, 2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B. , Hou, M. , Shi, J. , Ku, L. , Song, W. , Li, C. , Ning, Q. et al. (2023) De novo genome assembly and analyses of 12 founder inbred lines provide insights into maize heterosis. Nat. Genet. 55, 312–323. [DOI] [PubMed] [Google Scholar]
- Wei, X. , Qiu, J. , Yong, K. , Fan, J. , Zhang, Q. , Hua, H. , Liu, J. et al. (2021a) A quantitative genomics map of rice provides genetic insights and guides breeding. Nat. Genet. 53, 243–253. [DOI] [PubMed] [Google Scholar]
- Wei, Z. , Abdelrahman, M. , Gao, Y. , Ji, Z. , Mishra, R. , Sun, H. , Sui, Y. et al. (2021b) Engineering broad‐spectrum resistance to bacterial blight by CRISPR‐Cas9‐mediated precise homology directed repair in rice. Mol. Plant 14, 1215–1218. [DOI] [PubMed] [Google Scholar]
- Xiao, J. , Li, J. , Yuan, L. and Tanksley, S.D. (1995) Dominance is the major genetic basis of heterosis in rice as revealed by QTL analysis using molecular markers. Genetics 140, 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y. , Lin, Q. , Li, X. , Wang, F. , Chen, Z. , Wang, J. , Li, W. et al. (2021) Fine‐tuning the amylose content of rice by precise base editing of the Wx gene. Plant Biotechnol. J. 19, 11–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, W.H. , Wang, P. , Chen, H.X. , Zhou, H.J. , Li, Q.P. , Wang, C.R. , Ding, Z.H. et al. (2011) A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 4, 319–330. [DOI] [PubMed] [Google Scholar]
- Yang, J. , Mezmouk, S. , Baumgarten, A. , Buckler, E.S. , Guill, K.E. , McMullen, M.D. , Mumm, R.H. et al. (2017a) Incomplete dominance of deleterious alleles contributes substantially to trait variation and heterosis in maize. PLoS Genet. 13, e1007019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, M. , Wang, X. , Ren, D. , Huang, H. , Xu, M. , He, G. and Deng, X.W. (2017b) Genomic architecture of biomass heterosis in Arabidopsis. Proc. Natl Acad. Sci. USA 114, 8101–8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J. , Miao, J. , Zhang, Z. , Xiong, H. , Zhu, X. , Sun, X. , Pan, Y. et al. (2018) Alternative splicing of OsLG3b controls grain length and yield in japonica rice. Plant Biotechnol. J. 16, 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Liang, Z. , Zong, Y. , Wang, Y. , Liu, J. , Chen, K. , Qiu, J.‐L. et al. (2016) Efficient and transgene‐free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7, 12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, G. , Chen, Y. , Yao, W. , Zhang, C. , Xie, W. , Hua, J. , Xing, Y. et al. (2012) Genetic composition of yield heterosis in an elite rice hybrid. Proc. Natl. Acad. Sci. USA 109, 15847–15852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, X. , Nong, C. , Wu, B. , Zhou, T. , Zhang, B. , Liu, X. , Gao, G. et al. (2021) Combinations of Ghd7, Ghd8, and Hd1 determine strong heterosis of commercial rice hybrids in diverse ecological regions. J. Exp. Bot. 72, 6963–6976. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dataset S1 The haplotype of OsMADS1 in hybrid rice.
Dataset S2 The haplotype of OsMADS1 in the parents of hybrid rice.
Data S1 Phenotypic source data in this study.
Figure S1 The electropherograms of cDNA sequencing in T2 transgenic plants.
Figure S2 PCR amplicons of Cas9 in BC2F2 and T2 plants.
Figure S3 Statistical analyses of important agronomic traits in BC2F2 lines of FH676 with Type I genotypes.
Figure S4 Comparisons of agricultural traits in BC2F2 plants.
Figure S5 Grain appearance in BC2F2 plants.
Figure S6 Purity identification of hybrid rice seeds.
Figure S7 The average yield per mu among different hybrid combinations of GLY676 in Hainan and Shanghai.
Figure S8 Comparisons of agronomic traits in transgenic plants of R534.
Figure S9 Heading date in multiple hybrid combinations of GLY676 and QYSM.
Table S1 Progress of the genetic basis of heterosis in plants.
Table S2 QTL mapping results in GLY676 F2 population.
Table S3 Primers and probes used in this study.
Data Availability Statement
All data supporting the findings of this study are available in the article, supplementary information, and Data S1. Source data are provided in this paper.


