Abstract
Primordial germ cell (PGC) specification is the first step in the development of the germline. Recent work has elucidated human-mouse differences in PGC differentiation and identified cell states with enhanced competency for PGC-like cell (PGCLC) differentiation in vitro in both species. However, it remains a subject of debate how different PGC competent states in vitro relate to each other, to embryonic development, and to the origin of PGCs in vivo. Here we review recent literature on human PGCLC differentiation in the context of mouse and non-human primate models. In contrast to what was previously thought, recent work suggests human pluripotent stem cells (hPSCs) are highly germline competent. We argue that para-doxical observations regarding the origin and signaling requirements of hPGCLCs may be due to local cell interactions. These confound assays of competence by generating endogenous signaling gradients and spatially modulating the ability to receive exogenous inductive signals. Furthermore, combinatorial signaling suggests that there is no unique germline competent state: rather than a one-dimensional spectrum of developmental progression, competence should be considered in a higher dimensional landscape of cell states.
Introduction
Primordial germ cell (PGC) specification is the first step in the development of the germline that occurs around the onset of gastrulation in mammals. Efficient differentiation of human PGCs in vitro is required for in vitro gametogenesis: a technology with the potential to transform reproductive medicine [1]. In addition, understanding PGC differentiation is essential for understanding pluripotency, as germline competence is a defining feature of pluripotent stem cells [2]. Difficulties with efficient differentiation of human pluripotent stem cell (hPSCs) into PGC-like cells (hPGCLCs) have been attributed to the limited germline competence of hPSCs [3–5]. This prompted a search for pluripotent states with increased germ cell competence. Here we provide a concise review of the generation of PGCLCs from pluripotent cells. We first discuss signals required for PGC differentiation followed by PGC-competent states. We then describe recent work achieving efficient PGCLC differentiation from hPSCs, suggesting that there is no inherent limitation in germline competence of hPSCs. We propose two explanations for discrepancies in the literature describing germline competent states.
First, to address apparent discrepancies in hPSC to hPGCLC differentiation efficiency, we argue that assays of PGCLC differentiation efficiency in response to growth factors in the media do not measure competence as often presumed. Competence has been defined as “the ability [of a tissue] to respond to a specific inductive signal” [6,7]. At face value, this definition encompasses issues with the inductive signal reaching its receptors [8]. However, what is often not explicitly stated but implicitly assumed is that competence is a cell-intrinsic, epigenetic property. Here, we define competence as “the ability to differentiate upon appropriate activation of the inductive signaling pathways,” where “appropriate” means that signaling is activated at the right level, for the right duration, etc. By this definition, PGC differentiation efficiency in response to an inductive signal (in this case exogenous BMP) does not actually reflect competence of individual cells. Instead, it reflects tissue level properties due to local cell-cell interactions. Cell interactions spatially modulate hPSC responses to exogenous BMPs and give rise to additional endogenous cell signaling gradients that play an essential but underappreciated role in PGC differentiation. Thus, PGCLC differentiation is limited, not because cells are not competent, but instead because most cells do not receive the appropriate inductive signals to begin with.
As a second cause for a lack of a consensus regarding PGC(LC) competence we propose that there is not a unique competent state. Combinatorial action of exogenous BMPs with multiple endogenous signals in controlling germ cell fate suggests PGCLC progenitor states cannot be simply placed on a one-dimensional spectrum from less to more developmentally advanced. Instead, germline competency may be multidimensional, forming a plateau in the Waddington landscape through which multiple paths can lead to PGCLCs. For example, Wnt signaling followed by BMP signaling may take cells along one path, while BMP followed by Wnt takes a different path. From this perspective there is neither a unique competent state, nor a unique inductive signal: different states may give rise to identical PGCLCs in response to different signals. Our discussion focuses on the relationship between cell signaling and germline competence; for a detailed discussion of the transcription factor network and epigenetic changes underlying PGC differentiation, we refer to several excellent recent reviews [9–13].
Signals for germline differentiation in the mouse
PGCs originate in the posterior embryonic epiblast of the pre-primitive streak and early primitive streak mouse embryo [14] where they are marked by expression of the PGC marker PRDM1 starting ~E6.25 [15]. Early gene knockout studies revealed that BMP4 and BMP8b from the extra-embryonic ectoderm (ExE; placenta precursor) are required for PGC induction, while loss of BMP2 from the visceral endoderm (VE; yolk-sac precursor) leads to a reduction in PGCs. Furthermore, grafting experiments suggested the entire embryonic epiblast is germline competent [16–21]. A landmark study by Ohinata et al. [22] further dissected PGC induction. It found that BMP4 from the ExE acts directly on the epiblast and that exogenous BMP4 is sufficient to induce PGC fate across the entire dissected epiblast between E5.5 and E6.5. In contrast, BMP8b from the ExE restricts induction of the anterior visceral endoderm, which in turn secretes inhibitors that restrict the primitive streak and PGC induction to the posterior epiblast. Exogenous BMP8b therefore has little impact on dissected epiblasts to generate PGCs. Exogenous BMP2 is also sufficient to induce PRDM1 expressing cells in epiblast explants, but PRDM1 expression is lower than with BMP4. Importantly, BMPs could not induce PRDM1 expression in Wnt3−/− epiblasts, which could be rescued by supplying exogenous WNT3A, showing that BMPs and WNTs may act combinatorially. WNT3 in the epiblast induces Nodal expression in the mouse epiblast during mouse gastrulation [23–25]. However, a Nodal signaling inhibitor had little impact on PRDM1 expression in BMP-treated epiblast explants. In summary, BMP4 and WNT3 are required for mouse PGC differentiation.
While exogenous BMP4 is sufficient for PGC induction in the epiblast, several factors that do not impact induction efficiency were found to positively impact survival and proliferation [22,26,27]. The protocol for PGCLC induction from mouse epiblasts thus included BMP4, BMP8b, SCF, LIF, and EGF and has remained largely unchanged for in vitro induction of PGCLCs from PSCs from various species, including human and mouse.
The roles of Nodal and BMP signaling in mouse PGCs have been re-investigated recently. BMP signaling in PGCs was found to be reduced relative to the neighboring extraembryonic mesoderm [28,29], indicating that PGCs may downregulate BMP signaling after they are induced. Surprisingly, BMPR1A−/− mESCs could be differentiated to PGCLCs, and BMP activation or inhibition on day 2 in 3D mouse gastruloids, respectively decreased or increased the number of TFAP2C positive cells on day 5 [28,30]. Therefore, it was suggested that BMP signaling may not be directly required for mPGCLC induction. However, a quantitative evaluation of signaling levels and a full panel of PGC markers over time was not performed, nor were BMP manipulations at different times. This leaves open the possibility that PGCLC induction is in fact compromised, or that BMP signaling is required at lower levels and a different time course than assumed. Recent reinvestigation also found that although Nodal−/− embryos generate PRDM1-expressing cells, these do not express other PGC markers like TFAP2C, SOX2, and DPPA3, which were not evaluated in earlier work. This suggests that PGC induction may have also been compromised by Nodal inhibition in the earlier studies. However, genetic experiments with Nodal are difficult to interpret because Nodal is part of multiple feedback loops within and between the epiblast and extraembryonic tissues. Moreover, Nodal is involved in both maintenance of pluripotency in the epiblast and its differentiation into a diverse spectrum of fates [25] (Figure 1). Importantly, BMP4 expression is also lost in Nodal-mutants as Nodal is both downstream of BMP signaling and part of a feedback loop that maintains BMP4 expression. Thus, the lack of sustained BMP signaling may be responsible for the loss of PGC markers in the absence of Nodal, although reduced PGCLC differentiation from Nodal−/− ESCs suggests a direct requirement for Nodal signaling in the generation of PGCLCs [31].
Figure 1.
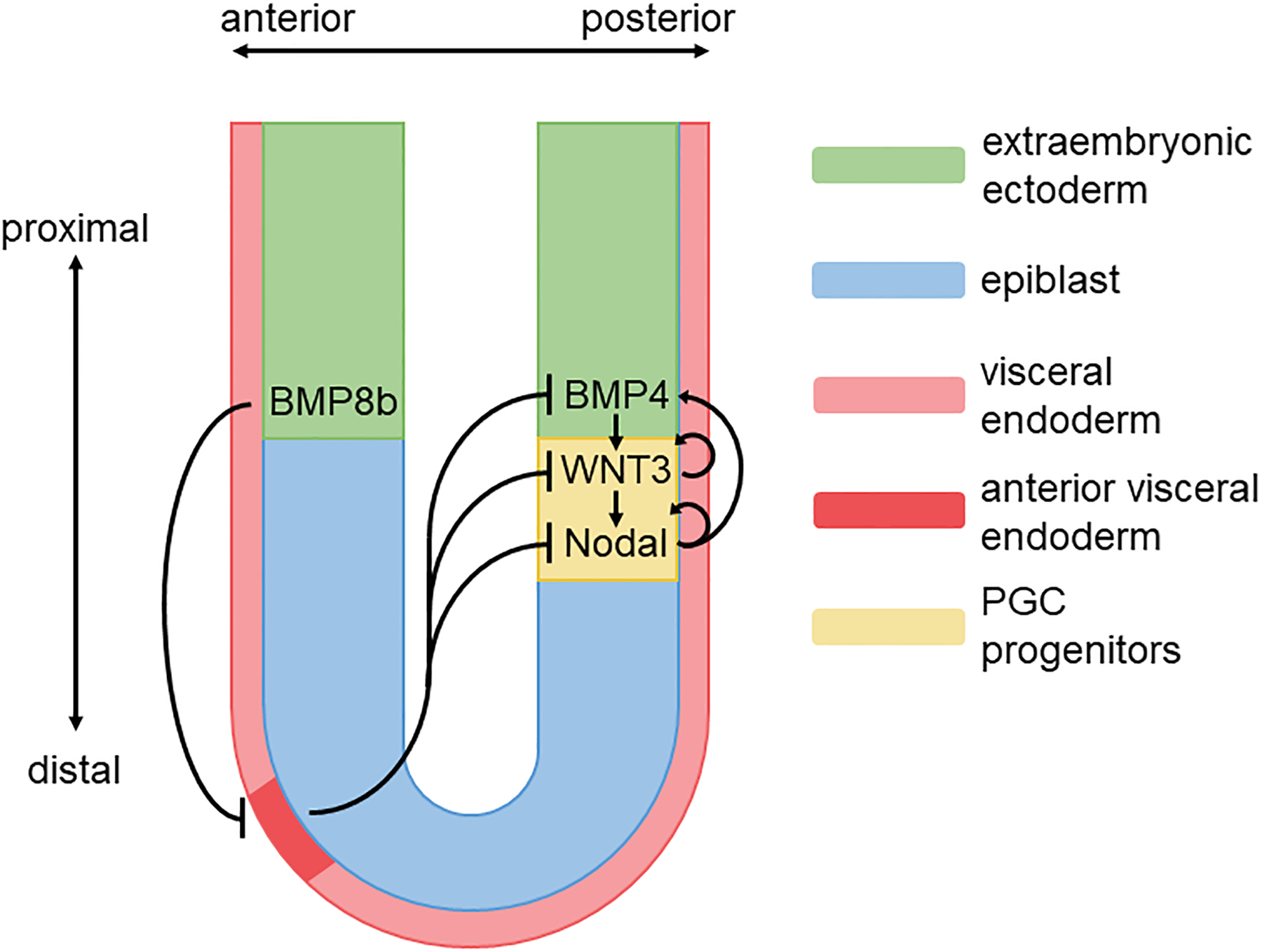
Signals for PGC specification in the peri-gastrulation mouse embryo. BMP4 from the extraembryonic ectoderm (ExE) induces Wnt3 in the epiblast, which in turn induces Nodal. Nodal signaling forms a positive feedback to maintain BMP expression in the ExE. The ExE also secretes BMP8b, which restricts the anterior visceral endoderm (AVE) to its initial position in the distal part of the visceral endoderm after which it migrates to the anterior border of the epiblast and ExE. Inhibitors from the AVE restrict the signals controlling PGC differentiation to the posterior proximal epiblast.
Mouse germline competence and pluripotency in vitro
When the protocol to generate PGCLCs from epiblast cells was first developed [22], two types of mouse pluripotent stem cells had been established in culture: embryonic stem cells (mESCs) and epiblast stem cells (mEpiSCs) which represent distinct “naïve” and “primed” pluripotent epiblast states [32]. mESCs are derived from and correspond to the pre-implantation epiblast progenitor cells in the ~E4 embryo [33,34]. mESC colonies have a dome shaped morphology and are maintained in culture media supplemented with MEK and GSK3 inhibitors and leukemia inhibitory factor (LIF) (2i + LIF) [3,35]. In contrast, EpiSCs are derived from embryos up to E7.5 and represent post-implantation epiblast, likely E6.5-E7.5, although variations in the derivation and culture conditions confound precise staging [36–38]. They are maintained in media supplemented with Activin and FGF2, and form an epithelial monolayer. mESCs and EpiSCs also differ in their ability to contribute to chimeras, X-inactivation status, gene expression, and chromatin modification profiles, matching the pre- and post-implantation embryonic epiblasts they represent.
Although WNT3 and BMP4 act combinatorially in PGC differentiation in vivo [22,39], the extent to which germline competence in vitro is mediated by endogenous Wnt is unknown and competence is evaluated as the fraction of cells expressing PGC marker genes after treatment with only BMP4 and maintenance factors. By this measure, it was found that neither mESCs nor EpiSCs efficiently differentiate into PGC-like cells [4]. It was reasoned that since these correspond to the E4-E4.5 pre-implantation and E6.5-E7.5 post-implantation epiblast, both are outside the epiblast competence window for PGC induction of E5.5-E6.5. By switching mESCs to EpiSC maintenance conditions, cells were found to pass through a transient intermediate state resembling the E5.5 epiblast named mouse epiblast-like cells (mEpiLCs) from which they efficiently differentiate into PGCLCs [4].
The discovery of intermediate but transient cell types like EpiLCs motivated a search for protocols to stabilize these distinct intermediate or “formative” pluripotent states, which can be functionally defined by germline competence [3,5]. Formative pluripotent cells (FPCs) have a flattened epithelial morphology and, unlike primed pluripotent cells, maintain the ability to contribute to chimeras and express intermediate levels of naïve and primed pluripotency genes. Several recent studies have identified conditions to stably maintain FPCs in culture [40–45]. A common theme in these protocols is an emphasis on the importance of controlling Wnt, FGF, and TGF-beta signaling. However, these FPCs differ in their culture conditions and properties, and consequently all go by different names (Table 1). To earn the designation of FPCs, cells should efficiently differentiate into PGCLCs upon BMP treatment, although this property has not been demonstrated in each case.
Table 1.
Formative pluripotent states.
| Name | FGF pathway modulation | Wnt pathway modulation | TGF-b pathway modulation | other characteristics | species examined | PGCLC differentiation? |
|---|---|---|---|---|---|---|
| RSC (rosette-like) [40] | MEKi | Inhibit: IWP2 | – | FBS, LIF | Mouse | In chimeras |
| fPSCs (formative) [41] | FGF2 | Inhibit: XAV939 | Activin A | spheroids | Mouse | Yes |
| FSCs (formative) [44] | – | Inhibit: XAV939 | Low Activin A | RARi | Mouse, human | Yes |
| FTW-ESCs/XPSC (X = chimera) [42] | FGF2 | Activate: GSK3i or Wnt3a | TGFb1 or Activin A | MEF feeders | Mouse, human, horse | Yes |
| INTPSCs [45] | FGF2 | Activate: GSK3i | Activin A | KSR,BSA | mouse | Yes |
| PiCs [46] | – | – | – | L-proline, FBS, LIF, feeders | mouse | Yes [47] |
MEKi inhibits the MAPK pathway downstream of Fibroblast Growth Factor (FGF). IWP2 is a porcupine inhibitor which blocks Wnt secretion. GSK3 inhibitor (most commonly CHIR99021) acts as a canonical Wnt agonist. RARi: retinoic acid receptor inhibitor. XAV939 is a tankyrase inhibitor, inhibiting canonical Wnt signaling. FBS: fetal bovine serum. LIF: Leukemia Inhibitory Factor. MEF: mouse embryonic fibroblast. BSA: bovine serum albumin. KSR: knockout serum replacement.
PGC differentiation of human cells
hPSCs resemble EpiSCs more closely than mESCs in many aspects including maintenance conditions, morphology, X-inactivation status, and gene expression profiles. After the discovery of EpiSCs, hPSCs were quickly equated with mEpiSCs [48]. Subsequently, conditions that capture varying degrees of naïve pluripotency in human cells were discovered [49]. Accordingly, the limited success in differentiating hPSCs to hPGCLCs [50–53] with exogenous BMPs was interpreted as low germline competence of primed cells. Attempts to improve hPGCLC induction efficiency have therefore focused on first achieving a competent state, followed by PGC induction with BMPs.
In one approach mimicking the mouse EpiLC protocol, human pluripotent cells maintained under conditions which induce naïve-like features (“4i” or NHSM [54]) were converted to an epi-like state with FGF and Activin after which they differentiated to hPGCLCs more efficiently (from <5% to 27%) [52]. More recently, a similar strategy was pursued with improved naïve human pluripotency protocols, leading to “reset” hPGCLCs with comparable efficiency [55]. A different approach [53,56] exposed cells to primitive streak inducing signals to create “incipient mesoderm-like cells” (iMeLC) or “precursors of mesendoderm” (PreME). PreME differentiation also enhanced subsequent hPGCLC induction by BMP treatment, in line with the earlier finding that mPGC induction requires the gene brachyury (TBXT) which marks mesoderm precursors [39]. Several of the conditions that maintain mouse FPC states were also found suitable for human cells but hPGCLC competence was not compared to other approaches.
It is counter-intuitive that both developmental progression towards mesoderm (by iMeLC differentiation) and developmental reversal (by resetting to formative or naïve pluripotency) would each enhance hPGCLC competence. One attempt to reconcile this discrepancy defined iMeLCs as an intermediate between primed and naïve pluripotency; however, this approach projected a high-dimensional gene expression profile on two points without evaluating the alternative hypothesis that iMeLC is an intermediate state between pluripotency and primitive streak [57]. Further work showed 4i-derived precursors and iMeLC to be similar to each other and distinct from the resetting naïve cells [55].
To begin to understand this, it is important to consider several differences between human and mouse in both the germline competent progenitors and signaling requirements. PGCs are first observed in the amnion [58–60] in monkey embryos, in contrast to mouse, rabbit, and pig PGCs, which all originate from the pre-streak posterior epiblast. This raises the question if monkey/human PGCs derive from amnion. However, the fact that amnion and PGC differentiation occur simultaneously suggest that rather than PGCs deriving from the amnion, both the PGCs and the amnion derive from pluripotent cells in overlapping signaling environments. Consistent with this, and in contrast to mouse cells, hPSCs treated with BMP4 in the absence of Wnt differentiate to what are thought to be amnionic ectoderm-like cells (hAELCs), although this remains subject to debate [61–66]. Furthermore, TFAP2A-positive progenitors give rise to both hAELCs and hPGCLCs [57,67]. On the other hand, in spatially organized stem cell models for the human embryo [68–70], hPGCLCs appear scattered among both the amnion-like and primitive streak-like cells [63,71,72]. Moreover, PGCs appear transcriptionally between the epiblast and primitive streak in rare single cell RNA-sequencing data of a gastrulating human embryo [73]. Altogether, this suggests that primate PGCs arise in signaling conditions that are intermediate between those giving rise to primitive streak and amnion, in line with the fact that iMeLC differentiation followed by BMP4, i.e., a combination of primitive streak and amnion induction signals, improves PGCLC differentiation.
Furthermore, a comparison between hPSCs, mouse embryos, cynomolgus monkey embryos, and cynomolgus monkey PSCs (cmPSCs) revealed that hPSCs and cmPSCs are transcriptionally very similar to each other and the post-implantation monkey epiblast, but that the latter is transcriptionally stable for a week and resembles the E5.5 mouse epiblast [74]. This is consistent with earlier work transcriptionally placing hPSCs between mESCs and EpiLCs which led to characterization of hPSCs as having an “extended primed pluripotency”, competent to generate all cell types that emerge after implantation including amnion and germ cells [1]. Corroborating this, enhancers for PGC genes are active in hPCSs [75]. Therefore, it is unclear whether hPSCs can be called primed by mouse standards.
PGCLC differentiation efficiency from hPSCs reflects tissue organization, not competence
Consistent with the idea that hPSCs are more like mouse formative PSCs than primed PSCs and therefore highly germline competent, two recent approaches obtained over 50% hPGCLCs from hPSCs using BMP4 without first inducing a competent state.
The first approach [63], by the current authors, used micropatterning to control colony geometry. Disc-shaped colonies treated with exogenous BMP4 form self-organized cell fate patterns that model human gastrulation in vitro [76]. With a colony diameter of 700um, PGCLCs constitute 5–10% of the cells and are localized on the interface between the amnion-like and primitive streak-like regions [63] (Figure 2). Crucially, response to exogenous BMP in this system is restricted to the colony edge due to BMP receptors localizing basolaterally [77]. Furthermore, like in mouse gastrulation, the self-organized patterning in this human model depends on a BMP-Wnt-Nodal signaling hierarchy: exogenous BMP4 induces endogenous Wnt3, which in turn induces endogenous Nodal, leading to dynamic and overlapping gradients of these signals [78–80]. Both Wnt inhibition and Nodal knockout block PGCLC differentiation but either can be rescued by exogenous activation of the Nodal pathway, demonstrating a key role for Nodal in hPGCLC induction from hPSCs. Importantly, Nodal signaling duration is a critical parameter and despite the requirement for Nodal, prolonged Nodal signaling inhibits PGCLC differentiation, instead inducing endoderm. There is minimal cell rearrangement in micropatterned colonies [78], suggesting PGCLCs are induced at a fixed distance from the colony edge by the right combination of BMP, Wnt, and Nodal (and possibly other signals). Because in smaller colonies all cells are close enough to the edge to respond to BMP, optimizing micropatterned colony size increased the efficiency of PGCLC differentiation using BMP alone to 50%, with the remaining 50% becoming amniotic ectoderm-like due to higher BMP response on the colony edge.
Figure 2. Effect of tissue geometry and endogenous signaling on PGCLC differentiation.
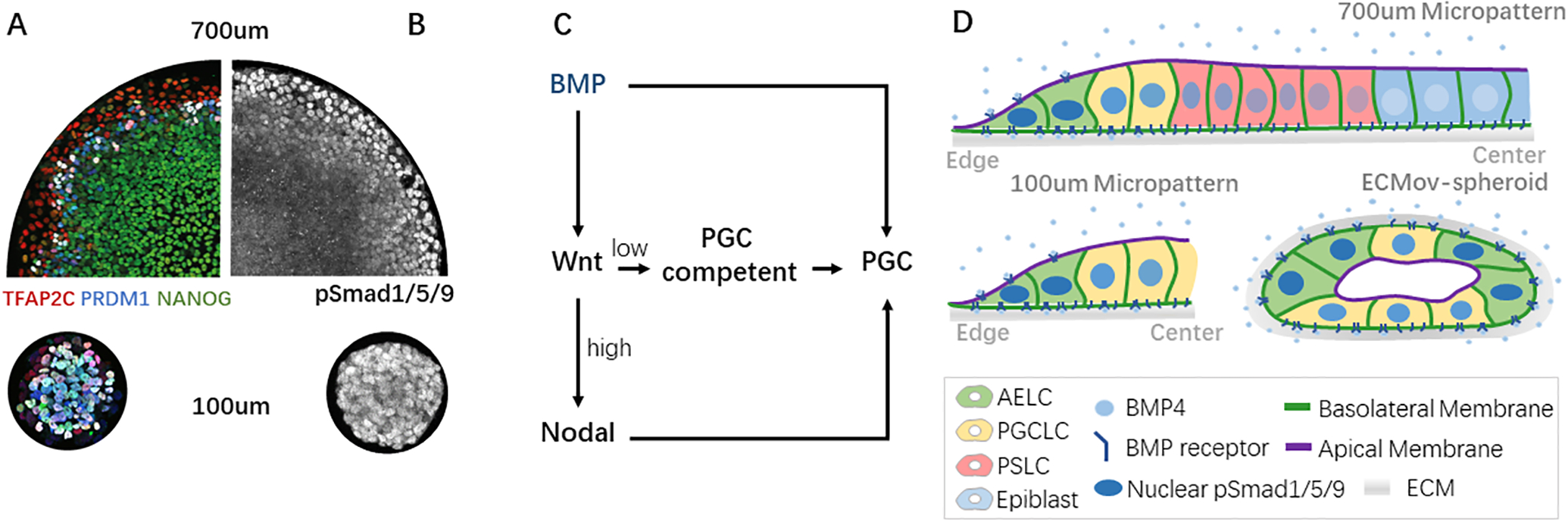
A, B) Immunofluorescence for (a) early hPGC marker genes, and b) pSmad1/5/9 staining (maximal intensity projection along z) in 700um and 100um micropatterned hPSC colonies treated with BMP4 for 2 days. c) Simplified diagram of signaling hierarchy controlling hPGCLC induction in hPSCs. d) Cartoon showing how receptor localization and tissue organization impact BMP response in micropatterned colonies and hypothetically in ECMov-spheroids to enable efficient hPGCLC differentiation. AELC: Amniotic Ectoderm-like cell; PGCLC: Primordial Germ Cell-Like cell; PSLC: Primitive Streak-Like cell; ECM: Extracellular Matrix.
In a different approach, Overeem [81] et al. added a solution of extracellular matrix (ECM) to form hPSCs spheroids, which upon BMP treatment formed hPGCLCs with over 50% efficiency. They showed that ECM overlay increases BMP response after which PGCLC differentiation requires a much lower BMP4 concentration than in the standard protocol. Although this remains to be tested, their results could be explained by the basal localization of BMP receptors and tissue geometry. In contrast to standard tissue culture where the basal surface faces the substrate and is shielded from exogenous BMP, with ECM overlay the cells form spheroids with the basement membrane facing the media so that the receptors are more uniformly exposed to exogenous BMP. Too high a BMP response leads to amnion differentiation, explaining the need to reduce the BMP dose with this approach. It will be important to investigate whether the 50% efficiency can be accounted for by remaining non-uniformity in BMP response and determine how required endogenous Wnt and Nodal signaling downstream of BMP are affected by the ECM overlay. Other signaling pathways may also play important roles, e.g., YAP is likely affected by the tissue organization and has recently been indicated as important for hPGCLC differentiation, in part by modulating Wnt [82].
Overall, a picture emerges wherein differences in PGCLC differentiation efficiency between approaches can be explained by tissue organization, which modulates both the response to exogenous BMP and endogenous cell signaling interactions downstream. Although cell intrinsic differences in competence may also contribute to differentiation efficiency, they cannot account for the recent results. To begin to unravel the relative contributions of tissue organization and competence in determining fate, it will be necessary to both measure how signal reception is affected by tissue organization and directly relate cell signaling activity to fate.
If hPSCs are highly germline competent, it is not clear why strategies using naïve cells improve differentiation. It is worth considering the possibility that these cells are more BMP-responsive due to changes in receptor polarization. Similarly, iMeLC differentiation could improve hPGCLC induction in part by modulating endogenous Wnt and Nodal expression through Wnt and Nodal autoregulation, thereby creating the right combinatorial signaling for PGCLC induction. More generally, some differences in competence in the literature may be accounted for by tissue geometry and endogenous signaling. For example, differences between hPSC lines [57] previously interpreted as a consequence of epigenetic lineage priming, could instead reflect differences that directly modulate responses to inductive signals like receptor polarization or cell junction integrity. In support of this notion, mouse epiblasts efficiently make PGCLCs in floating culture but not on a 2D substrate, where their receptors are likely shielded from the media [22].
Multiple paths through the Waddington landscape
Induction has been defined as involving separate inducing and responding tissues [7]. BMP is produced by the extra-embryonic cells while Wnt and Nodal are produced by the epiblast itself. Nevertheless, it is illuminating to conceptually separate signal interpretation from the signal source and to consider how combinatorial signals are interpreted.
Individually, these signals induce distinct non-PGC fates. For human, BMP alone induces amnion while Wnt and Nodal induce primitive streak. Only the right combination of these signals produces PGCs. Rather than separate them into permissive and instructive signals, we propose to consider their combinatorial action as moving cells around a higher-dimensional Waddington landscape. BMP moves cells in the amniotic ectoderm (AELC) direction in this landscape, while Wnt and Nodal move cells towards mesendoderm (a.k.a. primitive-streak, PSLC). PGCs lie in between and to get there, cells can take multiple paths. If they are exposed to BMP first, they will move in the AELC direction, but timely exposure to Nodal and Wnt will turn them in PSLC direction and land them in the PGC basin. This is what happens when hPSCs are differentiated with BMP directly, as endogenous Wnt and Nodal are activated downstream with a delay. Conversely, if cells are first exposed to Wnt and Nodal they move in the PSLC direction, but BMP exposure before they commit may redirect them to become PGCs, which resembles protocols with an iMeLC step.
From this perspective there is no “true” competent state or inductive signal. Nodal and Wnt induce a state that is competent for PGC differentiation in response to BMP, while BMP induces a different state that is competent for PGC differentiation in response to Nodal and Wnt. The right inductive signal, i.e. the right direction in which to move, depends on the current state. Competence does not lie on a line that reflects developmental stage and on which cells can only move forward or backward, but on a plateau on which can move in multiple directions, as Waddington himself described it [6]. Different parts of the tissue at the same developmental time may explore different parts of this plateau. They may also arrive in the same place at different times through different paths, i.e., differentiation is asynchronous if signaling is heterogeneous.
More than a metaphor, this picture can be made precise and falsifiable with mathematical modeling [63,83,84], as illustrated in Figure 3a. It also serves as a reminder that PGCs cannot be understood in isolation: PGC competence is lost when cells commit to alternate fates, therefore understanding commitment to alternate fates is as important as understanding PGC fate itself for improving PGC differentiation [85,86].
Figure 3. Different routes through the Waddington landscape.
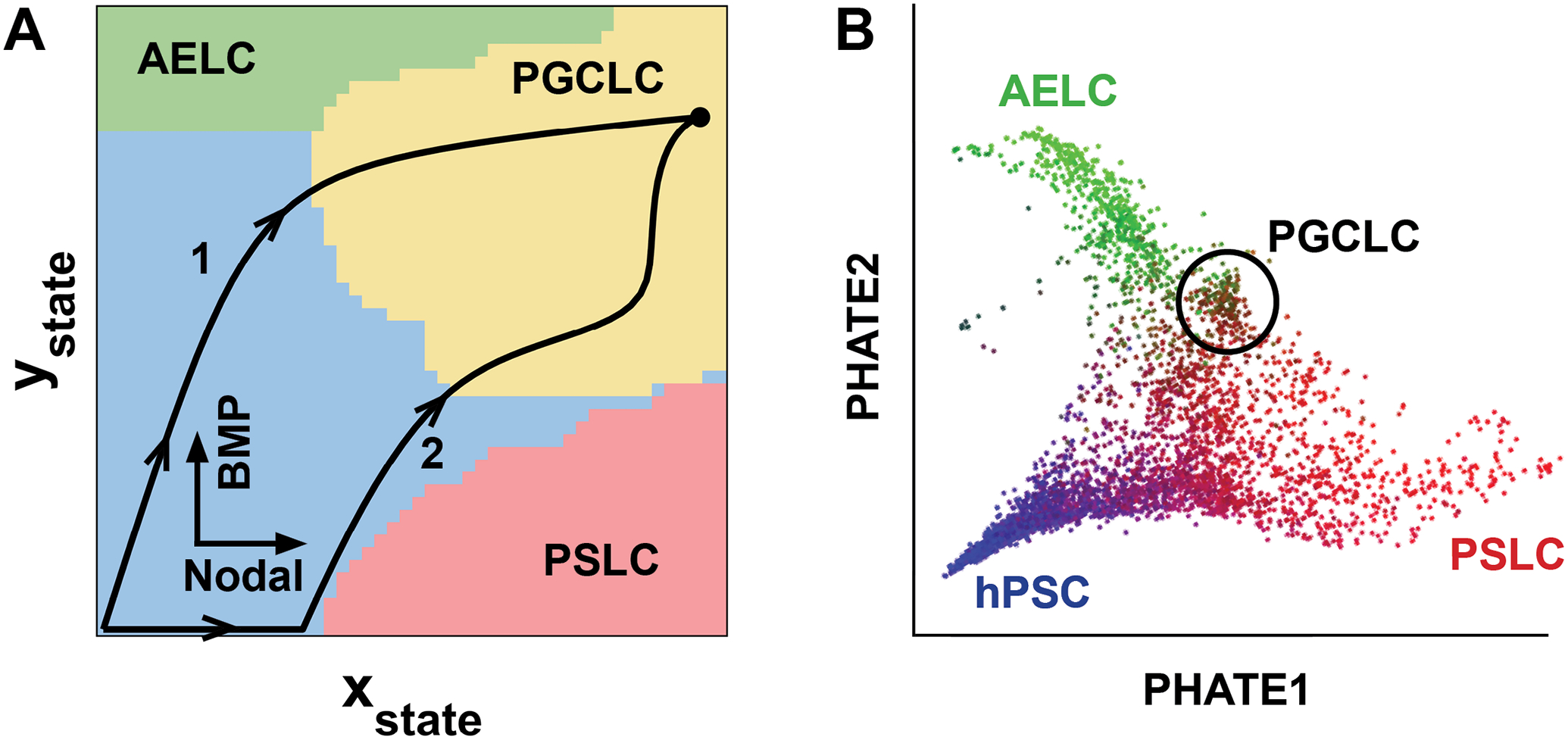
a) A landscape of cell states is shown with basins for amniotic ectoderm-like cells (AELC), primitive streak-like cells (PSLC), and primordial germ cell-like (PGCLC) cells derived from a mathematical model for hPGCLC differentiation [63]. The size of the basins does not reflect the probability of reaching it. Two paths through the landscape driven by distinct signaling dynamics are shown. Path 1 is continuous stimulation with a high level of BMP and low level of Activin/Nodal signaling. Path 2 represents initial treatment with CHIR + a high dose of Activin followed by treatment with a high dose of BMP4. x- and y-axes, respectively correspond to PSLC + SOX17 and AELC + TFAP2C as defined in Jo et al. [63]. The model is a simplification that does not capture many known complications, e.g., it does not separately model the roles of Nodal and Wnt. Nevertheless, it explains the effect of many signaling perturbations on PGCLC differentiation in micropatterned colonies and could be refined without fundamentally changing the picture above. b) PHATE visualization [88] of scRNA-sequencing data of BMP treated colonies is structured like the model landscape with primitive streak and amnion branches going in two directions and PGCLCs in the middle. Colors: SOX2 (blue), TBXT (red), ISL1 (green).
This model resolves apparent contradictions between an amnion-like or mesoderm-like origin for PGCLCs: it can be both. That explains the description in the literature of different hPGCLC-competent populations marked by combinations of amnion markers induced by BMP like TFAP2A or mesendoderm markers induced by Nodal and Wnt like TBXT [57,67,87]. It also explains the appearance of PGCLCs in both the amnion-like and PS-like regions of human embryoids [63,71] and the placement of hPGCLCs between those fates in scRNA-sequencing data of BMP treated hPSCs (Figure 3b).
Conclusion and outlook
In contrast to what was previously thought, hPSCs appear broadly competent to differentiate to hPGCLCs in response to BMP4. hPGCLC differentiation efficiency was previously found to be limited not because cells are not competent but because most cells do not receive the appropriate inductive signals: response to exogenous BMPs strongly depends on tissue organization and endogenous signaling gradients. Although cell state and tissue organization are linked – e.g., the polarization of the epithelium reflects gene expression, and conversely modulating epithelial polarization with matrix proteins will affect its state – significant insight can be gained from disentangling their effects. Local cell interactions that organize the tissue may therefore be key to understanding human germline differentiation in vitro. In this regard, an important open question is whether endogenous signaling in hPGCLC differentiation can be treated as purely autocrine, or whether interactions between distinct populations are required (e.g. amnion-like cells and hPGCLCs).
Germline competence can be considered higher dimensional when endogenous signals are placed on equal footing with exogenous BMPs. There may be distinct competent states along different trajectories through the landscape that are equal in developmental advancement but require distinct signals to reach the same endpoint. This framework may enable a unified understanding of different competent in vitro states described in the literature. If multiple trajectories exist, it will be important to determine which trajectories are realized in vivo, and whether PGCLCs retain any memory of their trajectory, i.e., whether the result of different protocols is truly equivalent. Transcriptional analysis indicates that the expression profile of hPGCLCs derived with BMP followed by endogenous Wnt and Nodal is equivalent to that of hPGCLCs derived with exogenous Wnt and Nodal activation followed by BMP [63]. On the other hand, hPGCLCs derived from naïve human pluripotent cells may retain epigenetic memory of their origin and mature faster than those derived from standard hPSCs [55]. It therefore merits further investigation to determine if and how different PGCLC differentiation protocols affect the resulting state and subsequent maturation of the cells.
It is interesting to ask why there would be multiple paths towards the same state. One possibility is that it is simply a consequence of controlling three fates with two (sets of) signals. For example, in an oversimplified model for differentiation consisting of two independent switches, where off–off is the pluripotent state, on-off is amnion, off-on is primitive streak, and on–on is PGCLC, one can imagine that it is immaterial which switch is flipped first to get to the on–on state. Another possibility is that flexibility in the relative timing of signals is required by evolution to generate PGCs from divergent sources of BMP across mammalian species. For example, the amnion in primates appears to substitute for the BMP4-producing role of the extraembryonic ectoderm in mice, while in rabbits the margin of the epiblast expresses BMP4, all possibly with different timing relative to Wnt and Nodal expression [65,89]. Insight into how and why multiple progenitor states can give rise to the same final state could also be gained by comparing with other examples, like definitive endoderm, which can arise from both visceral endoderm and epiblast [90,91].
In conclusion, there are many dimensions to germline competence. By breaking down what germline competence means, we will gain deeper insight into human pluripotency and accelerate progress toward in vitro gametogenesis.
Acknowledgments
We thank Ben Allen, Sundeep Kalantry, and Aryeh Warmflash for discussions and feedback on the manuscript. This work was funded by the National Institute of General Medical Sciences (NIGMS R35GM138346) and the National Science Foundation(2033654).
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.
Data availability
No data was used for the research described in the article.
References
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
- 1.Saitou M, Hayashi K: Mammalian in vitro gametogenesis. Science 2021, 374:eaaz6830, 10.1126/science.aaz6830. [DOI] [PubMed] [Google Scholar]
- 2.De Los Angeles A, Ferrari F, Xi R, Fujiwara Y, Benvenisty N, Deng H, Hochedlinger K, Jaenisch R, Lee S, Leitch HG, et al. : Hallmarks of pluripotency. Nature 2015, 525:469–478, 10.1038/nature15515. [DOI] [PubMed] [Google Scholar]
- 3.Kinoshita M, Smith A: Pluripotency deconstructed. Dev Growth Differ 2018, 60:44–52, 10.1111/dgd.12419. [DOI] [PubMed] [Google Scholar]
- 4.Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M: Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 2011, 146:519–532, 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 5.Pera MF, Rossant J: The exploration of pluripotency space: charting cell state transitions in peri-implantation development. Cell Stem Cell 2021, 28:1896–1906, 10.1016/j.stem.2021.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Waddington CH: How animals develop (routledge). 2015.
- 7.Gilbert SF, Barresi MJ: Developmental biology (sinauer associates). 2019.
- 8.Sagner A, Briscoe J. In Morphogen interpretation: concentration, time, competence, and signaling dynamics, vol. 6. Wiley Interdiscip Rev Dev Biol; 2017. e271, 10.1002/wdev.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sybirna A, Wong FCK, Surani MA: Genetic basis for primordial germ cells specification in mouse and human: conserved and divergent roles of PRDM and SOX transcription factors. Curr Top Dev Biol 2019, 135:35–89, 10.1016/bs.ctdb.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 10.Tang WWC, Kobayashi T, Irie N, Dietmann S, Surani MA: Specification and epigenetic programming of the human germ line. Nat Rev Genet 2016, 17:585–600, 10.1038/nrg.2016.88. [DOI] [PubMed] [Google Scholar]
- 11.Saitou M: Mammalian germ cell development: from mechanism to in vitro reconstitution. Stem Cell Rep 2021, 16: 669–680, 10.1016/j.stemcr.2021.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramakrishna NB, Murison K, Miska EA, Leitch HG: Epigenetic regulation during primordial germ cell development and differentiation. Sex Dev 2021, 15:411–431, 10.1159/000520412. [DOI] [PubMed] [Google Scholar]
- 13.Hancock GV, Wamaitha SE, Peretz L, Clark AT: Mammalian primordial germ cell specification. Development 2021, 148, 10.1242/dev.189217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawson KA, Hage WJ: Clonal analysis of the origin of primordial germ cells in the mouse. Ciba Found Symp 1994, 182: 68–84. discussion 84–91. 10.1002/9780470514573.ch5. [DOI] [PubMed] [Google Scholar]
- 15.Ohinata Y, Payer B, O’Carroll D, Ancelin K, Ono Y, Sano M, Barton SC, Obukhanych T, Nussenzweig M, Tarakhovsky A,et al. : Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 2005, 436:207–213, 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- 16.Tam PP, Zhou SX: The allocation of epiblast cells to ectodermal and germ-line lineages is influenced by the position of the cells in the gastrulating mouse embryo. Dev Biol 1996, 178:124–132, 10.1006/dbio.1996.0203. [DOI] [PubMed] [Google Scholar]
- 17.Ying Y, Liu XM, Marble A, Lawson KA, Zhao GQ: Requirement of Bmp8b for the generation of primordial germ cells in the mouse. Mol Endocrinol 2000, 14:1053–1063, 10.1210/mend.14.7.0479. [DOI] [PubMed] [Google Scholar]
- 18.Fujiwara T, Dunn NR, Hogan BL: Bone morphogenetic protein 4 in the extraembryonic mesoderm is required for allantois development and the localization and survival of primordial germ cells in the mouse. Proc Natl Acad Sci U S A 2001, 98: 13739–13744, 10.1073/pnas.241508898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawson KA, Dunn NR, Roelen BA, Zeinstra LM, Davis AM, Wright CV, Korving JP, Hogan BL: Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev 1999, 13:424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ying Y, Zhao GQ: Cooperation of endoderm-derived BMP2 and extraembryonic ectoderm-derived BMP4 in primordial germ cell generation in the mouse. Dev Biol 2001, 232: 484–492, 10.1006/dbio.2001.0173. [DOI] [PubMed] [Google Scholar]
- 21.Saitou M: Specification of the germ cell lineage in mice. Front Biosci (Landmark Ed) 2009, 14:1068–1087, 10.2741/3294. [DOI] [PubMed] [Google Scholar]
- 22.Ohinata Y, Ohta H, Shigeta M, Yamanaka K, Wakayama T, Saitou M: A signaling principle for the specification of the germ cell lineage in mice. Cell 2009, 137:571–584, 10.1016/j.cell.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 23.Liu P, Wakamiya M, Shea MJ, Albrecht U, Behringer RR, Bradley A: Requirement for Wnt3 in vertebrate axis formation. Nat Genet 1999, 22:361–365, 10.1038/11932. [DOI] [PubMed] [Google Scholar]
- 24.Ben-Haim N, Lu C, Guzman-Ayala M, Pescatore L, Mesnard D, Bischofberger M, Naef F, Robertson EJ, Constam DB: The nodal precursor acting via activin receptors induces mesoderm by maintaining a source of its convertases and BMP4. Dev Cell 2006, 11:313–323, 10.1016/j.devcel.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Arnold SJ, Robertson EJ: Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol 2009, 10:91–103, 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- 26.Dolci S, Williams DE, Ernst MK, Resnick JL, Brannan CI, Lock LF, Lyman SD, Boswell HS, Donovan PJ: Requirement for mast cell growth factor for primordial germ cell survival in culture. Nature 1991, 352:809–811, 10.1038/352809a0. [DOI] [PubMed] [Google Scholar]
- 27.Matsui Y, Toksoz D, Nishikawa S, Nishikawa S, Williams D, Zsebo K, Hogan BL: Effect of Steel factor and leukaemia inhibitory factor on murine primordial germ cells in culture. Nature 1991, 353:750–752, 10.1038/353750a0. [DOI] [PubMed] [Google Scholar]
- 28.Morgani SM, Hadjantonakis A-K: Quantitative analysis of signaling responses during mouse primordial germ cell specification. Biol Open 2021, 10, bio058741, 10.1242/bio.058741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Senft AD, Bikoff EK, Robertson EJ, Costello I: Genetic dissection of Nodal and Bmp signalling requirements during primordial germ cell development in mouse. Nat Commun 2019, 10:1089, 10.1038/s41467-019-09052-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooke CB, Barrington C, Baillie-Benson P, Nichols J, Moris N: Gastruloid-derived primordial germ cell-like cells (Gld-PGCLCs) develop dynamically within integrated tissues. Development, dev 2023, 201790, 10.1242/dev.201790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulas C, Kalkan T, Smith A: NODAL secures pluripotency upon embryonic stem cell progression from the ground state. Stem Cell Rep 2017, 9:77–91, 10.1016/j.stemcr.2017.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nichols J, Smith A: Naive and primed pluripotent states. Cell Stem Cell 2009, 4:487–492, 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 33.Evans MJ, Kaufman MH: Establishment in culture of pluripotential cells from mouse embryos. Nature 1981, 292: 154–156, 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 34.Martin GR: Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA 1981, 78:7634–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wray J, Kalkan T, Smith AG: The ground state of pluripotency. Biochem Soc Trans 2010, 38:1027–1032, 10.1042/BST0381027. [DOI] [PubMed] [Google Scholar]
- 36.Brons IGM, Smithers LE, Trotter MWB, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. : Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 2007, 448:191–195, 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 37.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RDG: New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 2007, 448:196–199, 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 38.Kojima Y, Kaufman-Francis K, Studdert JB, Steiner KA,Power MD, Loebel DAF, Jones V, Hor A, de Alencastro G, Logan GJ, et al. : The transcriptional and functional properties of mouse epiblast stem cells resemble the anterior primitive streak. Cell Stem Cell 2014, 14:107–120, 10.1016/j.stem.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 39.Aramaki S, Hayashi K, Kurimoto K, Ohta H, Yabuta Y, Iwanari H, Mochizuki Y, Hamakubo T, Kato Y, Shirahige K, et al. :A mesodermal factor, T, specifies mouse germ cell fate by directly activating germline determinants. Dev Cell 2013, 27: 516–529, 10.1016/j.devcel.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 40.Neagu A, van Genderen E, Escudero I, Verwegen L, Kurek D, Lehmann J, Stel J, Dirks RAM, van Mierlo G, Maas A, et al. :In vitro capture and characterization of embryonic rosettestage pluripotency between naive and primed states. Nat Cell Biol 2020, 22:534–545, 10.1038/s41556-020-0508-x. [DOI] [PubMed] [Google Scholar]
- 41.Wang X, Xiang Y, Yu Y, Wang R, Zhang Y, Xu Q, Sun H, Zhao Z-A, Jiang X, Wang X, et al. : Formative pluripotent stem cells show features of epiblast cells poised for gastrulation. Cell Res 2021, 31:526–541, 10.1038/s41422-021-00477-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.*.Yu L, Wei Y, Sun H-X, Mahdi AK, Pinzon Arteaga CA, Sakurai M, Schmitz DA, Zheng C, Ballard ED, Li J, et al. : Derivation of intermediate pluripotent stem cells amenable to primordial germ cell specification. Cell Stem Cell 2021, 28:550–567.e12, 10.1016/j.stem.2020.11.003. [DOI] [PubMed] [Google Scholar]; One of two papers stabilizing formative pluripotency in both mouse and human cells.
- 43.Shyh-Chang N, Li L: Stabilizing formative pluripotent states with germ cell competency. Cell Stem Cell 2021, 28:361–363, 10.1016/j.stem.2021.02.021. [DOI] [PubMed] [Google Scholar]
- 44.*.Kinoshita M, Barber M, Mansfield W, Cui Y, Spindlow D, Stirparo GG, Dietmann S, Nichols J, Smith A: Capture of mouse and human stem cells with features of formative pluripotency. Cell Stem Cell 2021, 28:453–471.e8, 10.1016/j.stem.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; One of two papers stabilizing formative pluripotency in both mouse and human cells.
- 45.Tsukiyama T, Ohinata Y: A modified EpiSC culture condition containing a GSK3 inhibitor can support germline-competent pluripotency in mice. PLoS One 2014, 9, e95329, 10.1371/journal.pone.0095329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.D’Aniello C, Habibi E, Cermola F, Paris D, Russo F, Fiorenzano A, Di Napoli G, Melck DJ, Cobellis G, Angelini C,et al. : Vitamin C and l-proline antagonistic effects capture alternative states in the pluripotency continuum. Stem Cell Rep 2017, 8:1–10, 10.1016/j.stemcr.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cermola F, D’Aniello C, Tatè R, De Cesare D, Martinez-Arias A, Minchiotti G, Patriarca EJ: Gastruloid development competence discriminates different states of pluripotency. Stem Cell Rep 2021, 16:354–369, 10.1016/j.stemcr.2020.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Los Angeles A, Loh Y-H, Tesar PJ, Daley GQ: Accessing naïve human pluripotency. Curr Opin Genet Dev 2012, 22: 272–282, 10.1016/j.gde.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou J, Hu J, Wang Y, Gao S: Induction and application of human naive pluripotency. Cell Rep 2023, 42, 112379, 10.1016/j.celrep.2023.112379. [DOI] [PubMed] [Google Scholar]
- 50.Kee K, Gonsalves JM, Clark AT, Pera RAR: Bone morphogenetic proteins induce germ cell differentiation from human embryonic stem cells. Stem Cell Dev 2006, 15:831–837, 10.1089/scd.2006.15.831. [DOI] [PubMed] [Google Scholar]
- 51.Clark AT, Bodnar MS, Fox M, Rodriquez RT, Abeyta MJ, Firpo MT, Pera RAR: Spontaneous differentiation of germ cells from human embryonic stem cells in vitro. Hum Mol Genet 2004, 13:727–739, 10.1093/hmg/ddh088. [DOI] [PubMed] [Google Scholar]
- 52.Irie N, Weinberger L, Tang WWC, Kobayashi T, Viukov S, Manor YS, Dietmann S, Hanna JH, Surani MA: SOX17 is a critical specifier of human primordial germ cell fate. Cell 2015, 160:253–268, 10.1016/j.cell.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sasaki K, Yokobayashi S, Nakamura T, Okamoto I, Yabuta Y, Kurimoto K, Ohta H, Moritoki Y, Iwatani C, Tsuchiya H, et al. : Robust in vitro induction of human germ cell fate from pluripotent stem cells. Cell Stem Cell 2015, 17:178–194, 10.1016/j.stem.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 54.Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, et al. : Derivation of novel human ground state naive pluripotent stem cells. Nature 2013, 504:282–286, 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 55.*.Alves-Lopes JP, Wong FCK, Tang WWC, Gruhn WH, Ramakrishna NB, Jowett GM, Jahnukainen K, Surani MA: Specification of human germ cell fate with enhanced progression capability supported by hindgut organoids. Cell Rep 2023, 42, 111907, 10.1016/j.celrep.2022.111907. [DOI] [PubMed] [Google Scholar]; Compares different human germline-competent states.
- 56.Sugawa F, Araúzo-Bravo MJ, Yoon J, Kim K-P, Aramaki S, Wu G, Stehling M, Psathaki OE, Hübner K, Schöler HR: Human primordial germ cell commitment in vitro associates with a unique PRDM14 expression profile. EMBO J 2015, 34: 1009–1024, 10.15252/embj.201488049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen D, Sun N, Hou L, Kim R, Faith J, Aslanyan M, Tao Y, Zheng Y, Fu J, Liu W, et al. : Human primordial germ cells are specified from lineage-primed progenitors. Cell Rep 2019, 29: 4568–4582.e5, 10.1016/j.celrep.2019.11.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sasaki K, Nakamura T, Okamoto I, Yabuta Y, Iwatani C, Tsuchiya H, Seita Y, Nakamura S, Shiraki N, Takakuwa T, et al. : The germ cell fate of cynomolgus monkeys is specified in the nascent amnion. Dev Cell 2016, 39:169–185, 10.1016/j.devcel.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 59.Li J, Zhu Q, Cao J, Liu Y, Lu Y, Sun Y, Li Q, Huang Y, Shang S, Bian X, et al. : Cynomolgus monkey embryo model captures gastrulation and early pregnancy. Cell Stem Cell 2023, 30: 362–377.e7, 10.1016/j.stem.2023.03.009. [DOI] [PubMed] [Google Scholar]
- 60.Bergmann S, Penfold CA, Slatery E, Siriwardena D, Drummer C, Clark S, Strawbridge SE, Kishimoto K, Vickers A, Tewary M,et al. : Spatial profiling of early primate gastrulation in utero. Nature 2022, 609:136–143, 10.1038/s41586-022-04953-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chhabra S, Warmflash A: BMP-treated human embryonic stem cells transcriptionally resemble amnion cells in the monkey embryo. Biol Open 2021, 10, bio058617, 10.1242/bio.058617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shao Y, Taniguchi K, Gurdziel K, Townshend RF, Xue X, Yong KMA, Sang J, Spence JR, Gumucio DL, Fu J: Self-organized amniogenesis by human pluripotent stem cells in a biomimetic implantation-like niche. Nat Mater 2017, 16: 419–425, 10.1038/nmat4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.*.Jo K, Teague S, Chen B, Khan HA, Freeburne E, Li H, Li B, Ran R, Spence JR, Heemskerk I: Efficient differentiation of human primordial germ cells through geometric control reveals a key role for Nodal signaling. Elife 2022, 11, e72811, 10.7554/eLife.72811. [DOI] [PMC free article] [PubMed] [Google Scholar]; Achieves efficient differentiation of hPCGCLCs from hPSCs without first inducing a mesoderm-like or naïve-like competent state using small micropatterns.
- 64.Rostovskaya M, Andrews S, Reik W, Rugg-Gunn PJ: Amniogenesis occurs in two independent waves in primates. Cell Stem Cell 2022, 29:744–759.e6, 10.1016/j.stem.2022.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.*.Yang R, Goedel A, Kang Y, Si C, Chu C, Zheng Y, Chen Z, Gruber PJ, Xiao Y, Zhou C, et al. : Amnion signals are essential for mesoderm formation in primates. Nat Commun 2021, 12: 5126, 10.1038/s41467-021-25186-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; Elucidates differentiation of amnion and its signaling interactions with neighboring tissues in monkey embryos and human embryoids.
- 66.Shao Y, Taniguchi K, Townshend RF, Miki T, Gumucio DL, Fu J: A pluripotent stem cell-based model for post-implantation human amniotic sac development. Nat Commun 2017, 8:208, 10.1038/s41467-017-00236-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Castillo-Venzor A, Penfold CA, Morgan MD, Tang WW, Kobayashi T, Wong FC, Bergmann S, Slatery E, Boroviak TE, Marioni JC, et al. : Origin and segregation of the human germline. Life Sci Alliance 2023, 6, e202201706, 10.26508/lsa.202201706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fu J, Warmflash A, Lutolf MP: Stem-cell-based embryo models for fundamental research and translation. Nat Mater 2021, 20: 132–144, 10.1038/s41563-020-00829-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heemskerk I: Full of potential: pluripotent stem cells for the systems biology of embryonic patterning. Dev Biol 2019, 10.1016/j.ydbio.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 70.Bao M, Cornwall-Scoones J, Zernicka-Goetz M: Stem-cell-based human and mouse embryo models. Curr Opin Genet Dev 2022, 76, 101970, 10.1016/j.gde.2022.101970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zheng Y, Xue X, Shao Y, Wang S, Esfahani SN, Li Z, Muncie JM, Lakins JN, Weaver VM, Gumucio DL, et al. : Controlled modelling of human epiblast and amnion development using stem cells. Nature 2019, 573:421–425, 10.1038/s41586-019-1535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Minn KT, Fu YC, He S, Dietmann S, George SC, Anastasio MA, Morris SA, Solnica-Krezel L: High-resolution transcriptional and morphogenetic profiling of cells from micropatterned human ESC gastruloid cultures. Elife 2020, 9, 10.7554/eLife.59445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.*.Tyser RCV, Mahammadov E, Nakanoh S, Vallier L, Scialdone A, Srinivas S: Single-cell transcriptomic characterization of a gastrulating human embryo. Nature 2021, 600:285–289, 10.1038/s41586-021-04158-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; Important transcriptional reference for PGCs and surrounding cell types in a CS7 human embryo.
- 74.Nakamura T, Okamoto I, Sasaki K, Yabuta Y, Iwatani C, Tsuchiya H, Seita Y, Nakamura S, Yamamoto T, Saitou M:A developmental coordinate of pluripotency among mice, monkeys and humans. Nature 2016, 537:57–62, 10.1038/nature19096. [DOI] [PubMed] [Google Scholar]
- 75.Tang WWC, Castillo-Venzor A, Gruhn WH, Kobayashi T, Penfold CA, Morgan MD, Sun D, Irie N, Surani MA: Sequential enhancer state remodelling defines human germline competence and specification. Nat Cell Biol 2022, 24:448–460, 10.1038/s41556-022-00878-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Warmflash A, Sorre B, Etoc F, Siggia ED, Brivanlou AH:A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat Methods 2014, 11: 847–854, 10.1038/nmeth.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Etoc F, Metzger J, Ruzo A, Kirst C, Yoney A, Ozair MZ, Brivanlou AH, Siggia ED: A balance between secreted inhibitors and edge sensing controls gastruloid self-organization. Dev Cell 2016, 39:302–315, 10.1016/j.devcel.2016.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chhabra S, Liu L, Goh R, Kong X, Warmflash A: Dissecting the dynamics of signaling events in the BMP, WNT, and NODAL cascade during self-organized fate patterning in human gastruloids. PLoS Biol 2019, 17, e3000498, 10.1371/journal.pbio.3000498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heemskerk I, Burt K, Miller M, Chhabra S, Guerra MC, Liu L, Warmflash A: Rapid changes in morphogen concentration control self-organized patterning in human embryonic stem cells. Elife 2019, 8, 10.7554/eLife.40526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martyn I, Kanno TY, Ruzo A, Siggia ED, Brivanlou AH: Self-organization of a human organizer by combined Wnt and Nodal signalling. Nature 2018, 10.1038/s41586-018-0150-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.*.Overeem AW, Chang YW, Moustakas I, Roelse CM, Hillenius, Helm TVD, Schrier VFVD, Gonçalves MAFV, Mei H, Freund C, et al. : Efficient and scalable generation of primordial germ cells in 2D culture using basement membrane extract overlay. Cell Reports Methods 2023, 10.1016/j.crmeth.2023.100488. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]; Achieves efficient differentiation of hPCGCLCs from hPSCs without first inducing a mesoderm-like or naïve-like competent state using ECM overlay.
- 82.Kagiwada S, Aramaki S, Wu G, Shin B, Kutejova E, Obridge D, Adachi K, Wrana JL, Hübner K, Schöler HR: YAP establishes epiblast responsiveness to inductive signals for germ cell fate. Development 2021, 148:dev199732, 10.1242/dev.199732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Raju A, Siggia ED: A geometrical perspective on development. Dev Growth Differ 2023, 10.1111/dgd.12855. [DOI] [PubMed] [Google Scholar]
- 84.*.Sáez M, Briscoe J, Rand DA: Dynamical landscapes of cell fate decisions. Interface Focus 2022, 12, 20220002, 10.1098/rsfs.2022.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Reviews recent progress in mathematically formalizing the Waddington landscape.
- 85.Camacho-Aguilar E, Yoon S, Ortiz-Salazar MA, Warmflash A: Combinatorial interpretation of BMP and WNT allows BMP to act as a morphogen in time but not in concentration. Dev Biol 2022, 10.1101/2022.11.11.516212. [DOI] [Google Scholar]
- 86.Teague S, Primavera G, Chen B, Freeburne E, Khan H, Jo K, Johnson C, Heemskerk I: The time integral of BMP signaling determines fate in a stem cell model for early human development. bioRxiv 2023, 10.1101/2023.04.10.536068. 2023.04.10.536068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yokobayashi S, Okita K, Nakagawa M, Nakamura T, Yabuta Y, Yamamoto T, Saitou M: Clonal variation of human induced pluripotent stem cells for induction into the germ cell fate. Biol Reprod 2017, 96:1154–1166, 10.1093/biolre/iox038. [DOI] [PubMed] [Google Scholar]
- 88.Moon KR, van Dijk D, Wang Z, Gigante S, Burkhardt DB,Chen WS, Yim K, Elzen A van den, Hirn MJ, Coifman RR, et al. : Visualizing structure and transitions in high-dimensional biological data. Nat Biotechnol 2019, 37:1482–1492, 10.1038/s41587-019-0336-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hopf C, Viebahn C, Püschel B: BMP signals and the transcriptional repressor BLIMP1 during germline segregation in the mammalian embryo. Dev Gene Evol 2011, 221:209–223, 10.1007/s00427-011-0373-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kwon GS, Viotti M, Hadjantonakis A-K: The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Dev Cell 2008, 15:509–520, 10.1016/j.devcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nowotschin S, Setty M, Kuo Y-Y, Liu V, Garg V, Sharma R, Simon CS, Saiz N, Gardner R, Boutet SC, et al. : The emergent landscape of the mouse gut endoderm at single-cell resolution. Nature 2019, 569:361–367, 10.1038/s41586-019-1127-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.


