Abstract
The tol-oprL region in Pseudomonas aeruginosa appears to be involved in pyocin uptake and required for cell viability. The complete nucleotide sequences of the tolQRA and oprL genes as well as the incomplete sequences of tolB and orf2 have been previously reported. In addition, the sequence of a P. aeruginosa iron-regulated gene (pig6) has been described and found to share homology with an open reading frame located upstream of the Escherichia coli tolQRA genes (U. A. Ochsner and M. L. Vasil, Proc. Natl. Acad. Sci. USA 93:4409–4414, 1996). In this study, we cloned the remainder of the P. aeruginosa tol-oprL gene cluster and determined its nucleotide sequence. This cluster was found to consist of seven genes in the order orf1 tolQ tolR tolA tolB oprL orf2. Transcriptional analysis of this gene cluster was performed by detecting the presence of mRNAs spanning adjacent genes as well as by using a promoterless lacZ reporter gene fused to each of the seven genes contained in the tol-oprL locus. The results show that there are three major transcriptional units or operons in this region, orf1-tolQRA, tolB, and oprL-orf2, in contrast to the E. coli tol-pal region, where there are only two operons, orf1-tolQRA and tolB-pal-orf2. Analysis of gene expression indicated that the tol-oprL genes of P. aeruginosa are both iron and growth phase modulated. The first operon, orf1-tolQRA, is iron regulated throughout growth, but iron-regulated expression of tolB and oprL fusions occurs only in late log phase. The expression of the three operons was significantly less repressed by iron in fur mutants than in the wild-type strain, suggesting the involvement of Fur in the iron regulation of all three operons. RegA is a positive yet nonessential regulator of tol-oprL expression.
The Tol system is one of two systems that are involved in macromolecule transport across the outer membrane of gram-negative bacteria. It has been shown that most group A colicins and filamentous phages gain entry into cells through this system in Escherichia coli (5, 48), and evidence has been obtained that tolQ, tolR, and tolA are involved in the transport of pyocin in Pseudomonas aeruginosa (9). Roles other than membrane transport, such as maintenance of outer membrane integrity, have also been assigned to the Tol-Pal complex. Mutations in the tol-pal genes cause the release of periplasmic contents (24, 49) and formation of vesicles (3). Tol-Pal proteins constitute one complex in the inner membrane and one near the outer membrane, and they have been proposed to form a contact site between outer and inner membranes which in turn may mediate interactions between the two membranes (4, 13). Both tolB and tolA interact with outer membrane porins, possibly affecting either porin assembly (39) or porin activity (24). Evidence suggests that tolA may also play a role in positioning the cell division sites since cell division in low- or high-osmolarity medium is impaired in tolA mutants (31). The Tol-Pal system in E. coli has recently been shown to consist of seven genes organized as two operons, orf1-tolQRA and tolB-pal-orf2 (47).
P. aeruginosa is an important human pathogen capable of causing a diverse range of infections in humans, especially in immunocompromised and cystic fibrosis patients (51). We have previously reported the cloning of the tolQRA genes from P. aeruginosa (9) and demonstrated that it was not possible to construct isogenic mutants in either tolQ or tolA, suggesting an essential role for these genes in P. aeruginosa. The oprL gene (pal in E. coli) has also been described in P. aeruginosa and P. putida (28, 40). The sequences of portions of tolB have previously been determined (9, 28). A DNA fragment encoding an iron-regulated gene (pig6) that exhibits high homology to E. coli orf1 in the orf1-tolQRA operon was isolated as a DNA fragment bound by the P. aeruginosa ferric uptake regulator (Fur) (33).
In E. coli, the expression of tolQRA is regulated by RcsC, a sensor protein in a two-component regulatory system controlling capsule synthesis, possibly through an unidentified mediator (7). The only environmental factor shown to affect tol-pal gene expression in E. coli was temperature (7). In contrast, we have shown that the expression of tolQ and tolA in P. aeruginosa is iron regulated and that growth temperature also affects expression of these genes (23). However, it was not clear whether the observed iron regulation of these genes in P. aeruginosa was dependent on interaction between orf1 promoter and Fur or other mediators. The effects of iron on other genes in the tol-pal cluster had not been determined. In this study, we further examined the genetic organization of the tol-oprL cluster in P. aeruginosa and determined that there are three major transcriptional units or operons in this region. All three operons were found to be iron regulated, and their expression was modulated during different phases of growth. In addition, we have shown that RegA, a transcriptional activator involved in exotoxin A production (16, 18), appears to positively regulate tol-oprL expression in P. aeruginosa.
MATERIALS AND METHODS
Strains, plasmids, primers, and culture conditions.
Bacterial strains, plasmids, and oligonucleotide primers used are described in Table 1. E. coli strains were routinely grown in Luria-Bertani (LB) broth or maintained on LB agar plates. P. aeruginosa strains were routinely maintained on M9-glucose agar plates or LB agar plates. Bacterial cultures were grown at 37°C with agitation at 220 rpm. Microaerobic conditions were achieved by incubating cultures statically in anaerobic jars with Anaerocult C packs from Merck & Co. (Whitehouse Station, N.J.). Antibiotics were added to the growth media at the following concentrations where appropriate: for E. coli, gentamicin at 15 μg/ml, ampicillin at 50 μg/ml, or tetracycline at 15 μg/ml; for P. aeruginosa gentamicin at 250 μg/ml or tetracycline at 200 μg/ml. All reagents and media were prepared with H2O purified by the Milli-Q system (Millipore, Bedford, Mass.).
TABLE 1.
Bacterial strains, plasmids, and primers
| Strain, plasmid, or primer | Relevant properties | Source or reference |
|---|---|---|
| E. coli DH5α | φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA recA1 hsdR17 deoR thi-1 supE44 gyrA96 relA1 | Life Technologies |
| P. aeruginosa | ||
| PAO1 | Prototroph | 21 |
| PAO1 C6 | fur mutant of PAO1 | 2 |
| PAO1 ΔpvdS | PAO1 with deletion of 460 bp of pvdS, Gmr cassette in pvdS gene | 32 |
| PAO6609 | met-9011 amiE200 rpsL pvd-9 | 20 |
| PAO6609 pchR | PAO6609 pchR::Ω-Tc | 17 |
| PA103 | Prototroph | 30 |
| PA103ΔregAB::Gm | PA103 with 1.8-kb EcoRV fragment containing regAB deleted and replaced with 1.6-kb EcoRV Gmr cassette | 38 |
| Plasmids | ||
| pHP45Ω | Source of Ω fragment (Smr/Spr with transcriptional termination signals in both orientations), Ampr | 36 |
| pNOT19 | Cloning vector, Ampr | 42 |
| pNOT8.0 | pNOT19 carrying 8.0-kb SphI chromosomal fragment (orf1-tolQRAB, oprL, and orf2), Ampr | This study |
| pPA3.5 | pUC19 carrying 3.5-kb SphI-PstI fragment (orf1-tolQRA and partial tolB) | 9 |
| pPA3.5RK | pRK415 carrying the 3.5-kb SphI-PstI fragment | 9 |
| pRK415 | Cloning vector; Tcr | 22 |
| pRKIz | orf1::lacZ fusion, pRK415 carrying PCR amplified orf1 P1 region oriented opposite Plac and lacZ-Gmr cassette from pZ1918 inserted in BamHI site; Tcr, Gmr | This study |
| pRKQzT | tolQ::lacZ fusion, pPA3.5RK with lacZ-Gmr cassette inserted in the BglII site of tolQ, HindIII fragment containing the terminator from pHP45Ω inserted in the HindIII site downstream of Plac; Tcr, Gmr | This study |
| pRKQz(ΔP1)T | tolQ (ΔP1)::lacZ fusion, pRK415 carrying 0.7-kb XbaI-BglII fragment (only coding region of orf1 and 5′ portion of tolQ) in XbaI-BamHI site with lacZ-Gmr cassette inserted in the KpnI site, HindIII Ω fragment in HindIII site downstream of Plac; Tcr, Gmr | This study |
| pRKRz | tolR::lacZ fusion, pRK415 carrying 1.9-kb SphI-SalI fragment (orf1-tolQ and partial tolR), lacZ-Gmr cassette in SalI site, HindIII Ω fragment downstream of Plac; Tcr, Gmr | This study |
| pRKRz (ΔP1) | tolR (ΔP1)::lacZ fusion, pRK415 carrying 0.6-kb BglII-SalI fragment (partial tolQ and partial tolR, but no P1 or orf1, oriented opposite Plac) in BamHI-SalI site with lacZ-Gmr cassette in SalI site; Tcr, Gmr | This study |
| pRKAzT | tolA::lacZ fusion, pPA3.5RK with lacZ-Gmr cassette in XhoI site of tolA, HindIII Ω fragment inserted downstream of Plac; Tcr, Gmr | This study |
| pRKAz (ΔP1)T | tolA (ΔP1)::lacZ fusion, same as pRKAzT with the SphI-SalI fragment (P1, orf1, and 5′ portion of tolQ) deleted; HindIII Ω fragment inserted in HindIII site downstream of Plac; Tcr, Gmr | This study |
| pRKBzT | tolB::lacZ fusion, pPA3.5RK with lacZ-Gmr cassette in PstI site, HindIII Ω fragment downstream of Plac; Tcr, Gmr | This study |
| pRKBz(ΔP1) | tolB (ΔP1)::lacZ fusion, pRK415 carrying 1.1-kb XhoI-PstI fragment (partial tolA and partial tolB, no P1, orf1 or tolQ, oriented opposite Plac) in SalI-PstI site, lacZ-Gmr cassette in KpnI site; Tcr, Gmr | This study |
| pRKPz (ΔPb) | oprL(ΔPb)::lacZ fusion, pRK415 carrying 1.2-kb PstI fragment (partial tolB and partial oprL, oriented opposite Plac) with lacZ-Gmr cassette in PstI site; Tcr, Gmr | This study |
| pRKIIz | orf2::lacZ fusion, pRK415 carrying 1.5-kb PstI-XhoI fragment (partial tolB, intact oprL, and partial orf2) with lacZ-Gmr cassette inserted in BamHI site, HindIII Ω fragment downstream of Plac; Tcr, Gmr | This study |
| pRKIIz (ΔPp) | orf2 (ΔPp)::lacZ fusion, pRK415 carrying 0.4-kb PstI fragment (3′ portion of oprL, 5′ portion of orf2) in PstI site with lacZ-Gmr cassette in SphI site; Tcr, Gmr | This study |
| pRKlacT | pRK415 with HindIII Ω fragment downstream of Plac and lacZ-Gmr cassette inserted in SalI site in the same orientation as Plac; Tcr, Gmr | This study |
| pRKlacO | pRK415 with the lacZ-Gmr cassette inserted in SalI site in orientation opposite Plac; Tcr, Gmr | This study |
| pUC19 | Cloning vector, Ampr | Life Technologies |
| pZ1918 | Vector, source of the cassette containing promoterless lacZ and Gmr (lacZ-Gmr) | 43 |
| Primers | ||
| RT-PCRa | ||
| orf1/tolQ-f | CGGACACCCTGAAGCCAC | |
| orf1/tolQ-r | GCCCTCCATCACCGCATC | |
| tolQ/tolR-f | ACAACCGCTTCTCCGCAC | |
| tolQ/tolR-r | CCAACCACCGCACCATAG | |
| tolR/tolA-f | ATGACCGATGCAGTCACC | |
| tolR/tolA-r | GCCTTCTTTTGTTCCGCC | |
| tolA/tolB-f | GCGGAAGCGGCGAAGAAG | |
| tolA/tolB-r | GCGATGGGAATGGCACGG | |
| tolB/oprL-f | CTAATCTACGCCACCCGCC | |
| tolB/oprL-r | CGCTGACCGCTGCCTTTC | |
| oprL/orf2-f | ACAGCTCCGACCTGAAGCC | |
| oprL/orf2-f | ACAGCTCCGACCTGAAGCC | |
| oprL/orf2-r | ACCTGCCGTGCCATACCC | |
| orf2drb | CGAAATCCCGGAAGGTCTC | |
| Amplification of orf1 P1 | ||
| Forward | GAGCGAGGAGCGGCACAC | |
| Reverse | TCCGAGCCCGTTCCATGAACTTG |
The primers used to test the region between two specific genes are represented by the names of the two genes; forward (upstream) primer and reverse (downstream) primer are indicated by “f” and “r,” respectively.
The reverse primer located downstream of orf2.
DNA manipulations.
Molecular biology techniques were generally performed as described by Sambrook et al. (41). Restriction enzymes, agarose, DNA size markers, and Taq DNA polymerase were purchased from Gibco-BRL (Burlington, Ontario, Canada). T4 DNA ligase was purchased from Promega (Madison, Wis.). DNA fragments were purified from agarose gels with Gene-Clean II (Bio/Can Scientific, Mississauga, Ontario, Canada). Plasmids were introduced into E. coli and P. aeruginosa by electroporation using a Gene Pulser electroporater (Bio-Rad, Richmond, Calif.) as previously described (10, 41). PCR products were cloned into pCR2.1-TOPO vector as recommended by the manufacturer (Invitrogen, Carlsbad, Calif.).
Isolation of tolA downstream region.
Chromosomal fragments that overlapped the 3.5-kb fragment containing orf1-tolQRA and partial tolB (9) were obtained from SphI or XhoI digests of chromosomal DNA of P. aeruginosa PAO, isolated as previously described (1), and fractionated on sucrose gradients (41). Fractions were hybridized with the 676-bp XhoI-KpnI fragment internal to tolA (Fig. 1). The probe was labeled with [32P]dCTP by random priming using an Oligolabelling kit from Amersham Pharmacia Biotech (Baie d'Urfé, Québec, Canada) according to the manufacturer's recommendations. The chromosomal DNA fragments that hybridized with the tolA probe were cloned into pUC19 or pNOT19 (42).
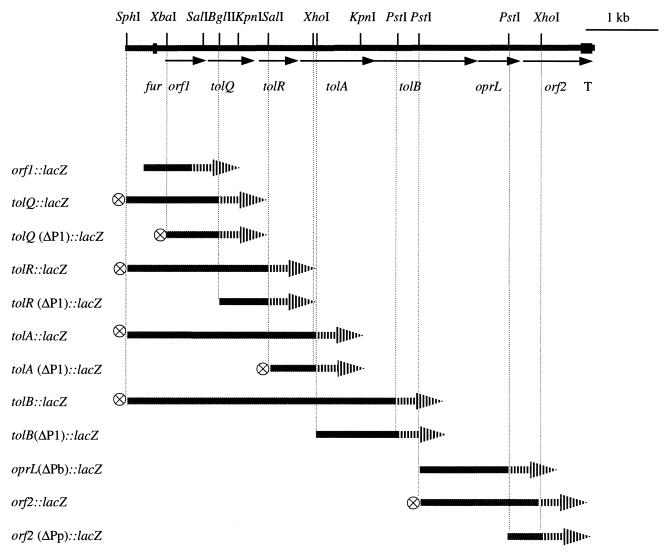
FIG. 1.
Genetic organization of tol-oprL cluster and lacZ fusion constructs of the tol-oprL genes. DNA fragments of the tol-oprL region are shown as solid bars. The relevant restriction sites, fur box, and putative terminator are indicated. The promoterless lacZ gene is shown by the arrowhead bars; the transcriptional terminator from pHP45Ω is represented by a circled cross. Only the fragments upstream of lacZ are shown in the diagram of the fusion constructs.
Nucleotide sequencing and sequence analysis.
The nucleotide sequences of orf1, tolB, and orf2 were determined by using the ABI Prism DyeDeoxy termination cycle sequencing system with AmpliTaq DNA polymerase (Perkin-Elmer, Norwalk, Conn.) and an ABI 1371A DNA sequencer by the University Core DNA Services (University of Calgary). Oligonucleotide primers were synthesized by Gibco-BRL. Analysis of the sequence was performed with PC/Gene software (Intelligenetics, Mountain View, Calif.).
Construction of lacZ transcriptional fusions.
Transcriptional fusions were constructed by cloning fragments of the tol-oprL cluster and a promoterless lacZ-Gmr cassette from pZ1918 (43) into pRK415 (22). The exact fragments and restriction sites used in constructing the fusions are described in Table 1 and Fig. 1. To construct the orf1::lacZ fusion, a fragment containing P1 was amplified with the P1 forward and reverse primers (Table 1) and cloned into the EcoRI site of pRK415. The lacZ-Gmr cassette was inserted into the downstream BamHI site. The tolQ::lacZ, tolA::lacZ, and tolB::lacZ fusions were constructed from pPA3.5RK by inserting the lacZ-Gmr cassette into the BglII, XhoI, and PstI sites, respectively. The tolR::lacZ fusion was constructed by cloning a 1.9-kb SphI-SalI fragment containing partial tolR and the upstream region into the SphI-SalI sites of pRK415. The lacZ-Gmr cassette was then cloned into the SalI site. A HindIII fragment containing transcription terminators from pHP45Ω (36) (HindIII Ω fragment) was inserted in the HindIII site downstream of the lacZ promoter (Plac) in pPA3.5RK. The tolQ(ΔP1)::lacZ fusion was constructed by cloning into pRK415 an XbaI-BglII fragment, containing only the coding region of orf1 and the 5′ portion of tolQ, into the XbaI-BamHI sites, inserting the lacZ-Gmr cassette in the KpnI site and the HindIII Ω fragment in the HindIII site. Similarly, tolR(ΔP1)::lacZ and tolB(ΔP1)::lacZ fusions were constructed by first cloning the BglII-SalI or XhoI-PstI fragment, respectively, lacking P1 and orf1 (Fig. 1) into pRK415 and then inserting the lacZ-Gmr cassette in the KpnI site. To construct the tolA(ΔP1)::lacZ fusions, pRKAz (23) was first digested with SalI and religated, deleting a SalI fragment (SalI-SphI fragment of the plasmid and SphI-SalI of the tol-oprL fragment containing P1 and orf1). The plasmid portion was restored by ligating a SalI fragment from pRK415 to the intermediate plasmid. The oprL(ΔPb)::lacZ fusion was made by inserting a PstI fragment containing the 3′ portion of tolB and the 5′ portion of oprL and the lacZ-Gmr cassette in the PstI site. The orf2::lacZ fusion in pRK415 contains a fragment with part of tolB, oprL, and part of orf2 in the PstI-SalI sites, the lacZ-Gmr cassette in the BamHI site, and the HindIII Ω fragment in the HindIII site. The orf2(ΔPp)::lacZ fusion contains a PstI fragment (3′ portion of oprL, 5′ portion of orf2, and a 6-bp SalI-PstI segment of pNOT19, without Pp, in the opposite orientation to Plac) in the PstI site with the lacZ-Gmr cassette in the SphI site.
β-Galactosidase assay.
β-Galactosidase assays were performed as previously described (35). Cultures of P. aeruginosa harboring the tol::lacZ fusions were grown in TSB-DC broth (34) at 37°C with aeration unless otherwise stated. The iron concentration of TSB-DC has previously been determined to be approximately 1.0 μM (34). Medium was supplemented with either ethylenediamine-di(o-hydroxyphenylacetic acid) (EDDHA; 400 μg/ml) or 50 μM FeCl3, and cells grown overnight in TSB-DC were used for inoculation of the cultures for assays. Samples were diluted 1/10 if the optical density at 600 nm of the culture exceeded 1.0. All assays were performed in triplicate. P. aeruginosa strains harboring pRKlacT or pRKlacO (Table 1) were used as negative controls.
Total RNA isolation and RT-PCR.
Total RNA from P. aeruginosa was isolated using a Qiagen RNA Midi kit (Qiagen, Mississauga, Ontario, Canada) from 5 ml of culture at 8 and 20 h of growth at 37°C in TSB-DC with EDDHA at 400 μg/ml. The RNA obtained was treated with amplification-grade DNase I (Gibco-BRL) before use. Reverse transcription-PCR (RT-PCR) was performed using a Titan one-tube RT-PCR kit (Boehringer Mannheim, Mississauga, Ontario, Canada), with minor modifications of the manufacturer's recommendations. Thirty nanograms of total RNA was used in each reaction. The cDNA was synthesized from the downstream primer by reverse transcriptase using RNA as the template. The double-stranded DNA was synthesized and amplified by PCR using both upstream and downstream primers. Reverse transcription was carried out at 50°C for 30 min, and 35 cycles of PCR were performed as follows: 10 cycles of denaturation at 95°C for 30 s, annealing at 60-64°C for 30 s, and elongation at 68°C for 45 s, followed by 25 cycles with increased elongation time in each cycle. An additional 5 s was added to each subsequent cycle; i.e., the 11th cycle has an elongation time of 50 s, and the 12th cycle has an elongation time of 55 s, etc. Amplification was stopped following a final elongation at 68°C for 10 min. The annealing temperature in each reaction was determined according to the composition of the primers used. RT-PCR products were examined by agarose gel electrophoresis. Negative controls used included RNA samples treated with RNase prior to reaction and heat inactivation of reverse transcriptase in Titan one-enzyme mixture before use. DNA contamination of the mRNA was determined by PCR using Taq polymerase without reverse transcriptase.
Nucleotide sequence accession numbers.
The nucleotide sequences reported here have been deposited in GenBank and assigned accession no. U39558 for orf1, tolQRAB, and AF177774 for orf2.
RESULTS
Organization of the tol-oprL genes in P. aeruginosa.
To complete the analysis of the gene organization of the tol-oprL region in P. aeruginosa, the regions upstream of tolQ, downstream of tolA, and downstream of oprL were sequenced. The previously cloned 3.5-kb SphI-PstI chromosomal DNA fragment in pPA3.5 (9) was used to sequence the upstream region of tolQ. To sequence the entire tolB and downstream of the tol-oprL cluster, an 8.0-kb SphI fragment and a 2.9-kb XhoI fragment, which hybridize with a 676-bp tolA probe (data not shown), were isolated from the P. aeruginosa chromosome. Sequence analysis confirmed that the tol-oprL region of P. aeruginosa contains seven genes in the order orf1 tolQ tolR tolA tolB oprL orf2 (Fig. 1). The stop codon of orf1 is separated from the start codon of tolQ only by 1 bp, suggesting possible translational coupling. The distance between tolQ and tolR is 22 bp, and that between tolR and tolA is only 2 bp. The distances present between tolA and tolB and between tolB and oprL are 38 and 49 bp, respectively. orf2 is separated from oprL by 9 bp. Fifty-two base pairs upstream of orf1 is a gene (ruvB) encoding Holliday junction-specific DNA helicase (19) and downstream of orf2 is a potential rho-independent transcriptional terminator, indicating that both boundaries of the gene cluster have been reached. As reported previously (33), the promoter region of orf1 has a fur box with 12 out 19 bp identical to the consensus sequence. The organization of the seven genes in the P. aeruginosa tol-oprL cluster is almost identical to that observed for the E. coli tol-pal gene cluster, although the latter has not been reported to contain a fur box.
orf1 potentially encodes a polypeptide of 148 amino acid residues with a molecular mass of 16.7 kDa that shares no homology with any protein of known function in GenBank. Directly following tolA is tolB, which encodes a predicted protein of 47.8 kDa. The first 21 amino acid residues at its N terminus form a potential secretory signal sequence. The predicted TolB protein is 78.7% identical to P. putida TolB and shares 44.2 and 40.3% identity with E. coli TolB (27) and Haemophilus influenzae TolB (44), respectively. orf2 potentially encodes a polypeptide of 275 amino acid residues with a molecular mass of 29.1 kDa, which is similar in size to the Orf2 of 262 amino acid residues in E. coli. A computer-predicted secretory signal sequence is also present at the N terminus of Orf2 with two alternative processing sites between residues 19 and 20 or between residues 21 and 22. The orf2 sequence of P. aeruginosa previously reported by Lim et al. suggests that orf2 encodes a protein of 107 residues and is followed by an insertion sequence (28). The orf2 sequence obtained from our PAO strain did not contain an insertion sequence.
Transcriptional analysis of the tol-oprL cluster.
To examine the operon structures of the tol-oprL cluster in P. aeruginosa, transcriptional fusion analysis was used. mRNA analysis has been shown to be difficult in studying the tol-oprL transcription in both P. aeruginosa (9, 23) and E. coli (47). Both Northern hybridization and primer extension proves to be problematic presumably due to the low abundance or instability of the transcripts. Therefore, a promoterless lacZ reporter gene was fused to the tol-oprL genes cloned on a low-copy-number plasmid, pRK415 (Fig. 1). The lacZ was fused in each of the seven genes containing only the individual gene's upstream region to test whether each gene has its own promoter. Fusions were also constructed with the presence of intact upstream genes and their potential promoter regions to test the expression directed by upstream promoters. A transcriptional terminator from pHP45Ω (36) was used in some of the constructs to eliminate the possible effect of the vector-encoded Plac when the gene under study was in the same orientation as Plac. No residual promoter activity was observed from the vector when the Plac promoter was in the opposite orientation to the fusion. The expression of these fusions was determined by measuring β-galactosidase activity.
As shown in Table 2, high levels of expression independent of the upstream genes were observed only with the orf1, tolB, and oprL fusions. When orf1 and its upstream region was present, tolQ, tolR, and tolA were also expressed at high levels. But when the orf1 region was deleted, tolR was expressed only at background levels and the expression of tolQ and tolA was greatly reduced to levels approximately 9- to 27-fold less than that in constructs containing orf1 and its promoter region. These data indicate the major transcriptional activity of orf1-tolQRA was from the orf1 promoter (P1). Low levels of expression of tolQ and tolA in the absence of P1 suggest that there could be also promoters present upstream of these two genes. In contrast to tolQ, tolR, and tolA fusions, the tolB fusion lacking the P1 promoter region exhibited a high level of expression as measured by β-galactosidase activity, suggesting that tolB itself has a strong promoter (designated as Pb) and that Pb in the absence of P1 is sufficient for strong expression of tolB. There was no difference between the expression of tolB(ΔP1)::lacZ and tolB::lacZ (including P1); however, pRKBzT was considerably less stable than pRKBz(ΔP1), which makes it difficult to compare the expression levels between fusions with both the P1 and Pb promoters to the fusion with only the Pb promoter. Despite growth in medium containing gentamicin, 65% of the cells containing pRKBz(ΔP1) lost the plasmid by 20 h of growth, as determined by comparing CFU counts on LB agar with and without gentamicin. In contrast, cultures harboring pRKBzT demonstrated a plasmid loss of only 9% during the same time period. This high level of plasmid instability was not detected with the other fusion constructs examined. The orf2(ΔPp)::lacZ fusion displayed a background level of expression, indicating that it is part of an operon that includes the upstream oprL. Strong expression of orf2, however, was observed in the absence of Pb, the tolB promoter. The orf2::lacZ fusion construct containing oprL and its upstream region, but lacking Pb, exhibited a high level of expression (Table 2), indicating the presence of the third major promoter upstream of oprL (designated Pp). The presence of Pp was confirmed by the high-level expression of the oprL(ΔPb)::lacZ fusion, which also lacks Pb. Therefore, there are three major operons in this cluster, consisting of orf1-tolQRA, tolB, and oprL-orf2. The organization of the tol-oprL region of P. aeruginosa is distinct from that in E. coli, where only two operons, orf1-tolQRA and tolB-pal-orf2, are present (47).
TABLE 2.
β-Galactosidase activity of PAO1 containing the tol::lacZ fusion constructs
| Fusion construct | β-Galactosidase activity (Miller units)a |
|---|---|
| orf1::lacZ | 10,560 ± 251 |
| tolQ::lacZ | 23,148 ± 358 |
| tolQ (ΔP1)::lacZ | 870 ± 35 |
| tolR::lacZ | 19,157 ± 296 |
| tolR (ΔP1)::lacZ | 353 ± 11 |
| tolA::lacZ | 14,739 ± 445 |
| tolA (ΔP1)::lacZ | 1,655 ± 34 |
| tolB::lacZ | 13,292 ± 649 |
| tolB(ΔP1)::lacZ | 14,115 ± 452 |
| oprL(ΔPb)::lacZ | 26,053 ± 1301 |
| orf2::lacZ | 23,021 ± 796 |
| orf2 (ΔPp)::lacZ | 158 ± 6 |
| pRKlacT | 345 ± 17 |
| pRKlacO | 198 ± 39 |
Means ± standard deviations of triplicate experiments. Samples were taken after 20 h of growth in TSB-DC medium supplemented with EDDHA at 400 μg/ml.
To examine the expression of the tol-oprL genes in P. aeruginosa, RT-PCR was also used to analyze mRNA isolated from strain PAO. Primers were designed to amplify the junction regions of each pair of adjacent genes from the mRNA template. If adjacent genes were cotranscribed, a PCR product would be generated following the synthesis of cDNA from RNA templates by reverse transcriptase. Otherwise no PCR products would be produced. The RT-PCR was performed on RNA isolated from both log-phase and stationary-phase cultures, and similar results were obtained. The results obtained correlate with the data obtained by analysis of the lacZ fusions. PCR products were generated from the primers amplifying the regions between orf1 and tolQ, tolQ and tolR, tolR and tolA, and oprL and orf2 (data not shown), demonstrating the members of each pair are in the same operon. No product was obtained from the primers amplifying the region from orf2 to downstream of the putative rho-independent terminator (primer orf2dr and oprL/orf2-f), showing that no detectable mRNA extended beyond the terminator was present in the RNA isolates. This result also suggests the functionality of the putative transcriptional terminator. PCR products were also obtained, however, from primers amplifying regions between tolA and tolB and between tolB and oprL. These data likely indicate the incomplete transcriptional termination between the major transcriptional units, resulting in transcripts containing the whole cluster and transcripts containing tolB, oprL, and orf2. Similar residual upstream promoter activity and possible presence of long transcripts have also been observed in the tol-pal region of E. coli (47).
Iron- and growth phase-modulated expression of the tol-oprL operons.
Previously tolQ and tolA fusions containing orf1 have been shown to be iron regulated during mid-log phase of growth (23). Therefore, experiments were conducted to test possible iron-regulated expression of the other two operons and to examine the iron regulation of all three operons during other phases of growth. Expression of the first operon was monitored by measuring the β-galactosidase activity of orf1::lacZ, tolQ::lacZ, and tolA::lacZ, while β-galactosidase activities of tolB(ΔP1)::lacZ and oprL(ΔPb)::lacZ were measured to represent expression of the second and third operons, respectively. Expression of tolQ(ΔP1)::lacZ and tolA(ΔP1)::lacZ was also measured to monitor activities of the putative independent promoters upstream tolQ and tolA, respectively. The expression of these fusions was monitored throughout growth.
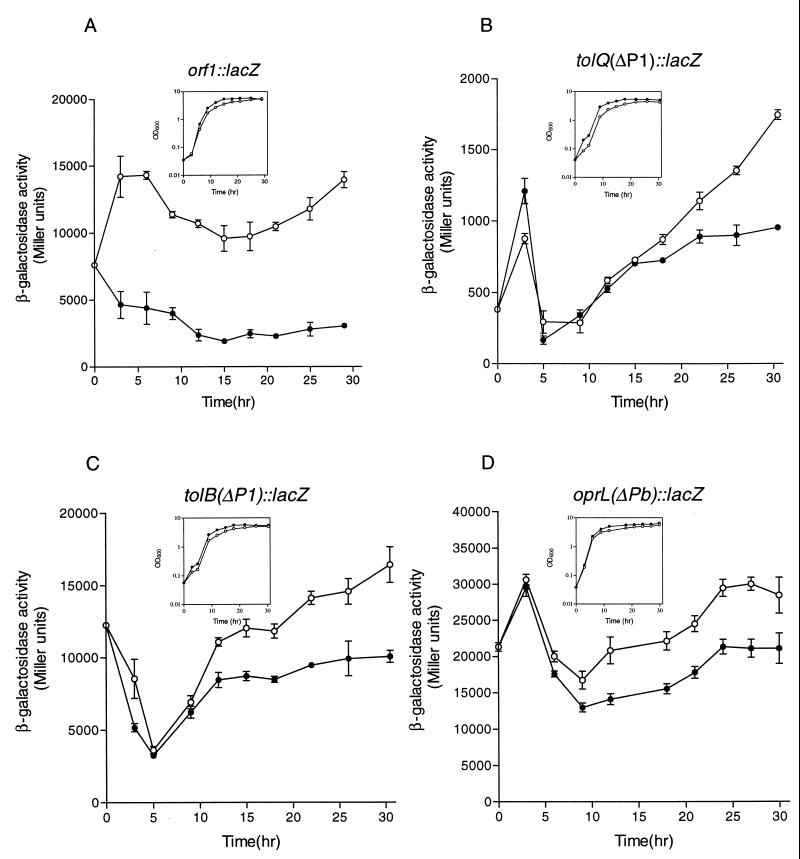
As shown in Fig. 2, the expression of all three operons appears to be iron regulated and growth phase dependent; however, differences in both gene expression and iron regulation profile were observed between the operons. In agreement with the observation above that orf1-tolQRA are in the same operon, the expression of orf1::lacZ (Fig. 2A), tolQ::lacZ, and tolA::lacZ (data not shown) was iron regulated in a similar manner throughout growth. In iron-rich conditions, expression of β-galactosidase activity was consistently lower than in iron-restricted conditions. In iron-restricted conditions, expression declined during log phase; however, in early stationary phase the expression again increased to maximum levels. The repression level (ratio of expression in iron-restricted to expression in iron-rich conditions) of orf1::lacZ was approximately 3.1-fold at 4 h and 4.8-fold at 30 h. The expression levels between iron-restricted and iron-rich medium were significantly different at all time points (P < 0.001) (Fig. 2A). In contrast, the expression of tolB(ΔP1)::lacZ and oprL(ΔPb)::lacZ (Fig. 2C and D) was also iron regulated, but only when the culture reached the late log phase of growth. β-Galactosidase activity was not iron regulated during the early stage of growth. The repression levels for tolB(ΔP1)::lacZ and oprL(ΔPb)::lacZ at 30 h were approximately 1.6- and 1.4-fold, respectively. These levels of repression were reproducible in that similar results were obtained in three experiments, and the differences in expression between iron-restricted and iron-rich conditions were significantly different (P < 0.01) at all time points between 12 and 30 h. The expression of tolQ(ΔP1)::lacZ (Fig. 2B) and tolA(ΔP1)::lacZ (data not shown) was similar to tolB(ΔP1)::lacZ and oprL(ΔPb)::lacZ in late log and stationary phase and was significantly different between iron-restricted and iron-rich conditions at time points between 18 and 30 h (P < 0.01). These results indicate that the expression of the operons in the tol-oprL region of P. aeruginosa is modulated by both iron and growth phase, but that there are differences in the expression of these operons.
FIG. 2.
Effect of iron and growth on expression of the tol-oprL genes. Cells were grown in TSB-DC medium supplemented with either 400 μg of EDDHA per ml (open circles) or 50 μM FeCl3 (solid circles). Bacterial growth is shown in the insets. β-Galactosidase activities are means ± standard deviations of triplicate cultures. Similar results were obtained in three independent experiments.
Involvement of Fur in iron regulation of tol-oprL expression.
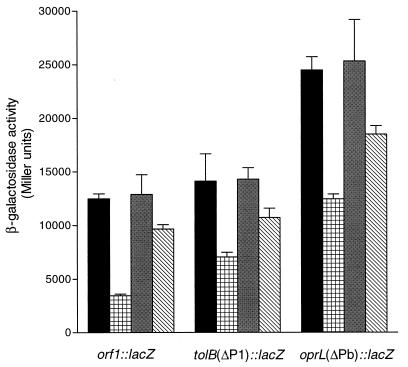
Fur has been shown to bind to the orf1 promoter region (33) and to affect expression of tolQ::lacZ or tolA::lacZ fusions containing the P1 promoter (23). As orf1-tolQRA form one operon, it is likely that Fur directly regulates the expression of these genes by binding to the fur box in P1. To further examine Fur involvement in the iron regulation of the orf1-tolQRA, tolB, and oprL-orf2 operons, the fusions orf1::lacZ, tolB(ΔP1)::lacZ and oprL(ΔPb)::lacZ were transferred to fur mutant C6, which has a point mutation causing a single amino acid residue change (A10→G) (2). Expression was tested in both iron-restricted and iron-rich conditions. The experiments were carried out in both aerobic and microaerobic conditions because it has been shown that Fur may affect gene expression differently in these conditions (2). The expression of the orf1, tolB, and oprL fusions in iron-rich medium in stationary phase was significantly higher in the fur mutant than in PAO, whereas the expression of these fusions in iron-restricted medium in PAO was similar to that in C6 (Fig. 3). Similar results were obtained under microaerobic conditions (data not shown). These results indicate that Fur is involved in the iron regulation of tolB and oprL-orf2 as well as orf1-tolQRA.
FIG. 3.
Effect of fur on expression of the three major tol-oprL transcriptional units under aerobic conditions. β-Galactosidase activities are means ± standard deviations from triplicate cultures. Samples were taken after 20 h of cultivation in TSB-DC with EDDHA at 400 μg/ml (PAO, black bars; C6, gray bars) or 50 μM FeCl3 (PAO, cross-hatched bars; C6, diagonal-hatched bars). The expression levels of each fusion in C6 were significantly different than in PAO in iron-rich medium (P < 0.01, analysis of variance).
RegA as a positive regulator in tol-oprL expression.
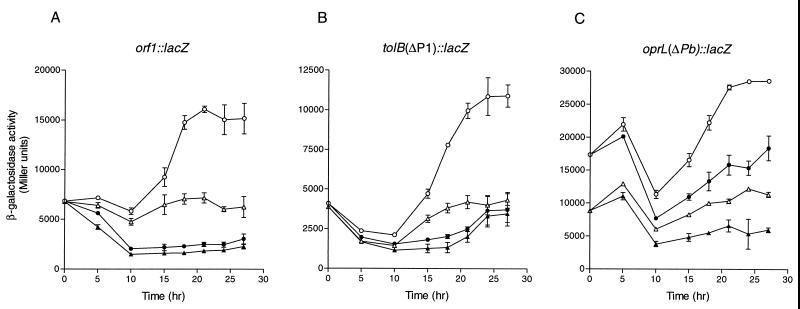
Although the data indicate the direct involvement of Fur in regulating orf1-tolQRA expression, no recognizable fur box motifs are present in either the Pb or Pp region. The involvement of Fur in tolB and oprL-orf2 expression is likely to be indirect, possibly through an iron-regulated mediator. The presence of expression peaks of the first operon (Fig. 2A) also suggests there are possibly regulators other than Fur in coordinating its expression. Several known iron-regulated transcriptional regulators including PchR, PvdS, and RegA were examined for their effects on the expression of tol genes. PchR is an AraC-type transcriptional activator involved in the synthesis of pyochelin and a ferripyochelin receptor (17). PvdS is an alternative sigma factor that is required for pyoverdine and exotoxin A production (8), and RegA is a transcriptional activator regulating the expression of toxA encoding exotoxin A (16, 18). Plasmids containing the orf1::lacZ, tolB(ΔP1)::lacZ, and oprL(ΔPb)::lacZ fusions were transferred to individual pchR, pvdS, and regAB mutants, and expression was compared to that of the parent strains in both iron-rich and iron-restricted conditions. No significant difference in expression of the three fusions was observed in the pchR and pvdS mutants compared to PAO (data not shown). The expression of all the three tol-oprL operon fusions, however, was affected by the regAB mutation (Fig. 4). Compared to the parent strain, the expression of these three operon fusions, and particularly oprL(ΔPb)::lacZ, was lower in the regAB mutant. In iron-restricted conditions, the expression levels of all three fusions in the regAB mutant remained relatively constant throughout growth, with only a slight increase in expression during stationary phase. In PA103, the parent strain, there was a significant increase in expression of all three lacZ fusions in iron-restricted conditions during stationary phase, similar to that observed with strain PAO (compare Fig. 2 and 4). There also appeared to be a decrease in the level of expression of the oprL(ΔPb)::lacZ fusion in PA103ΔregAB::Gm during log phase in both iron-rich and iron-restricted conditions compared to the parent strain (Fig. 4C). These data suggest that although regA is not required for expression of the orf1-tolQRA, tolB, or oprL-orf2 operon, it enhances expression of these genes in late log phase of growth under iron-restricted conditions and therefore serves as a positive regulator of tol-oprL gene expression.
FIG. 4.
Effect of regA on expression of the tol-oprL operons. PA103 carrying lacZ fusion constructs was grown in TSB-DC medium supplemented with either 400 μg of EDDHA per ml (open circles) or 50 μM FeCl3 (solid circles). PA103ΔregAB::Gm carrying lacZ fusion constructs grown in TSB-DC medium supplemented with either 400 μg of EDDHA per ml or 50 μM FeCl3 is represented by open or solid triangles, respectively. β-Galactosidase activities are means ± standard deviations from triplicate cultures. Similar results were obtained in three experiments.
DISCUSSION
The tol-oprL genes play important roles in gram-negative bacteria. The data from this and previous studies indicate that the tol-oprL region of P. aeruginosa consists of seven genes in the order orf1 tolQ tolR tolA tolB oprL orf2. An unrelated DNA helicase is located upstream of orf1, and the last gene, orf2, is followed by a putative rho-independent transcriptional terminator. This terminator appears to be functional as indicated by RT-PCR analysis in that no transcripts extended beyond the terminator were detectable. Previously, a terminator present within an insertion sequence has been reported to be located downstream of orf2 (28), but no insertion sequence could be identified in the cloned region in this study. It seems possible that the insertion sequence transposed into the orf2 region in the PAO isolate used in the study by Lim et al. (28).
Three operons appear to exist in the tol-oprL cluster in P. aeruginosa. lacZ fusion analysis indicates there are three major promoters, P1, Pb, and Pp, upstream of orf1, tolB, and oprL, respectively. It has been shown in E. coli that the TolQRA proteins form a complex in the inner membrane via their transmembrane domains (11, 26), and TolB, Pal, and Orf2 are positioned in the outer membrane or periplasm, possibly forming a complex with other outer membrane components (4, 6, 25, 26). In P. aeruginosa, it is likely that localization of the Tol-OprL proteins is similar to that in E. coli. By being arranged in operons, the major transcriptional activities of these genes may be coordinated but also differentially regulated, which may be important for their functions.
Interestingly, apart from the terminator downstream of orf2, no typical terminators could be identified downstream of the first two operons, tolA and tolB. The lack of obvious terminators makes it possible that the transcription from P1 could also read through tolB and orf2, and the transcription from Pb could read through oprL and orf2. The results from RT-PCR analysis support this possibility since mRNA spanning the regions between tolA and tolB and between tolB and oprL could be detected from the total RNA isolates. A similar phenomenon was also observed in E. coli, where the orf1 promoter is able to direct the transcription of the whole tol-pal cluster (47). In addition to the three major promoters in the P. aeruginosa tol-oprL region, weak promoter activity was also observed upstream of tolQ and tolA. This transcriptional organization may ensure a minimum expression of each gene product to perform essential functions.
Previously, using Northern hybridization analysis, we detected transcripts of approximately 1.5 kb using tolQ and tolR probes and an approximately 1.2-kb transcript using a tolA probe (9). A potential transcriptional start site was detected upstream of the tolA gene, using primer extension analysis (23). These data suggested that tolQ and tolR were cotranscribed and tolA was transcribed separately. In light of the current data obtained with lacZ reporter fusions and RT-PCR, it is likely that the transcripts detected in the previous studies (9, 23) were the result of endonucleolytic cleavage of the mRNA.
Iron-regulated expression of the tol-oprL genes in P. aeruginosa is somewhat unique. There are no reports suggesting that expression of the tol-pal genes of E. coli or H. influenzae is iron regulated, although this possibility may not have been investigated. All three operons in the tol-oprL cluster of P. aeruginosa displayed iron-regulated expression, although differences were observed during different stages of growth.
Expression of orf1-tolQRA was iron regulated throughout growth. The presence of 50 μM FeCl3 in the medium resulted in at least 50% reduction in expression compared to iron-restricted medium. Fur has been shown to play a central role in iron regulation in gram-negative bacteria (29, 33). In the presence of iron, Fur and cytoplasmic Fe2+ form a complex and bind to the fur box in the promoters; hence, transcription of the iron-regulated genes is repressed. In the absence of iron, Fur does not bind to the fur box and expression proceeds (29). Since binding of Fur-Fe2+ to the orf1 promoter has been demonstrated (33), it is clear that the iron regulation of these genes directly involves Fur. These data are confirmed by the observation that the repression of orf1 expression by iron is significantly decreased in fur mutants, where Fur-Fe2+ binding capacity is less efficient than for the wild-type complex (2).
Expression of orf1-tolQRA was iron regulated throughout growth; however, iron regulation of tolB and oprL-orf2 expression was detected only in late log to early stationary phase of growth. The decreased iron repression in the fur mutant indicated that Fur is also involved in the regulation of tolB and oprL-orf2 expression; however, such an involvement seems to be indirect. A search for fur boxes in the tolB and oprL promoter regions failed to identify any such motifs. Presence of an intermediate regulator, by which Fur may regulate the mediator and the mediator in turn would modulate the expression of the tol-oprL operons, was therefore postulated. This kind of hierarchy in iron regulation has been shown to be common in P. aeruginosa (46).
Expression of the orf1::lacZ, tolB(ΔP1)::lacZ, and oprL(ΔPb)::lacZ fusions was less iron regulated in the fur mutant C6 than in PAO (Fig. 3). Similar results were previously shown with tolQ::lacZ and tolA::lacZ fusions with the P1 promoter in two other fur mutants, A2 and A4 (2, 23). There was no difference in fur regulation between cultures grown in either aerobic or microaerobic conditions. Siderophore production, detected by chrome azurol S activity, was reported to be constitutive in C6 in high-iron medium regardless of the oxygen levels of the medium (2). Exotoxin A yields, however, were deregulated only in high-iron microaerobic conditions in the C6 mutant. RegA transcription was also shown to be constitutive in microaerobic but not aerobic conditions in C6 (2). Although RegA also enhances tol gene expression, Fur may regulate tol gene expression in a more similarly to siderophore biosynthesis gene expression than to exotoxin A or regA expression, since similar results were obtained with C6 in both aerobic and microaerobic conditions.
The transcriptional activator RegA was found to increase the expression of the tol-oprL genes in stationary phase in iron-restricted medium. In PA103, regA has been shown to have two promoters, P1 and P2, which direct the synthesis of the T1 and T2 transcripts, respectively. The T1 transcript encodes both regA and regB, while the T2 transcript encodes only regA (45). The regAB P1 promoter is not significantly affected by iron; however, the regA P2 promoter is iron regulated (45). P2 activity starts rising in late log to early stationary phase (45), which corresponds to the peak expression of orf1-tolQRA and the beginning of iron regulation of tolB and oprL-orf2. Although the mutation in PA103ΔregAB::Gm also eliminates the expression of regB, alterations in tol gene expression in this mutant are most likely due to the loss of regA since the iron-regulated expression of the tol genes more closely parallels the expression of the T2 transcript. RegA appears to be required for increased expression of the tol-oprL genes in iron-restricted medium, but it is not essential for the expression of these genes. In iron-rich medium, expression of the tol-oprL operons was also decreased in the regAB mutant compared to the parent. Although the decrease in expression observed with the orf1 and tolB fusions between the mutant and the parent was small, it was reproducible and significantly different between 5 and 21 h for orf1::lacZ and 5 and 24 h for tolB(ΔP1)::lacZ (P < 0.05). The difference observed in oprL(ΔPb)::lacZ expression between the two strains was significant throughout growth (P < 0.005). There is a level of iron-regulated expression of the tol-oprL genes in P. aeruginosa that is not due to either Fur-Fe2+ or RegA, suggesting that other potential regulators are involved in the expression of tol-oprL genes. It is not surprising to observe some iron regulation of the orf1-tolQRA operon in the regAB mutant because Fur would regulate orf1 P1 in this genetic background. But since Fur does not appear to regulate the Pb or Pp promoter directly, the iron regulation of these operons in the regAB mutant suggests the presence of other regulatory factors.
RegA is one of several factors that regulates the synthesis of exotoxin A. Other factors that influence exotoxin A expression include LasR (12), Fur (37), Vfr (50), PvdS (32), and PtxR (14). With the exception of lasR, these genes also regulate regA expression. PtxR and Vfr increase regA transcription through the P1 promoter (14, 50). PvdS activates expression of both the T1 and T2 transcripts in strain PAO (32). Although the effects of Vfr and PtxR on tol-oprL expression were not examined, it is possible that they would also enhance expression of the tol-oprL genes due to their roles in regA expression. Since pvdS was shown to be required for the detection of regA transcripts in PAO (32), it was somewhat surprising that there was no difference in the expression of the tol operon fusions in the pvdS mutant compared to PAO. Although neither regA nor toxA transcripts were detectable in this mutant, low levels of exotoxin A were detected in culture supernatants by immunoblotting (32). This suggests that low levels of regA were also expressed. The inability of these investigators to detect regA was likely due to the insensitivity of the assay and the short half-lives of the transcripts (32). The amount of RegA produced in the pvdS mutant may be sufficient for enhancement of tol-oprL gene expression.
The PA103 regAB mutant does not produce exotoxin A (38) yet still expresses the tol-oprL genes, although at lower levels than the parent strain. The mechanism by which RegA activates toxA expression, however, is not clear. RegA shares little homology with other known transcriptional regulators, and binding of RegA to the toxA promoter could not be demonstrated in mobility shift assays (15). Unlike other transcriptional regulators, RegA has been proposed to interact specifically with RNA polymerase prior to association with the promoter DNA (48). Further studies are needed to determine whether RegA activates toxA and tol-oprL gene expression in similar manners.
ACKNOWLEDGMENTS
This work was supported by a grant from the Canadian Bacterial Diseases Network of Centres of Excellence program.
We thank K. Poole, Queen's University, H. Schweizer, Colorado State University, D. Storey, University of Calgary, and M. Vasil, University of Colorado, for making available strains used in this study.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 2.Barton H A, Johnson Z, Cox C D, Vasil A I, Vasil M L. Ferric uptake regulator mutants of Pseudomonas aeruginosa with distinct alterations in the iron-dependent repression of exotoxin A and siderophores in aerobic and microaerobic environments. Mol Microbiol. 1996;21:1001–1017. doi: 10.1046/j.1365-2958.1996.381426.x. [DOI] [PubMed] [Google Scholar]
- 3.Bernadac A, Gavioli M, Lazzaroni J-C, Raina S, Lloubès R. Escherichia coli tol-pal mutants form outer membrane vesicles. J Bacteriol. 1998;180:4872–4878. doi: 10.1128/jb.180.18.4872-4878.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouveret E, Derouiche R, Rigal A, Lloubès R, Lazdunski C, Bénédetti H. Peptidoglycan-associated lipoprotein-TolB interaction. A possible key to explaining the formation of contact sites between the inner and outer membranes of Escherichia coli. J Biol Chem. 1995;270:11071–11077. doi: 10.1074/jbc.270.19.11071. [DOI] [PubMed] [Google Scholar]
- 5.Braun V, Herrmann C. Evolutionary relationship of uptake systems for biopolymers in Escherichia coli: cross-complementation between the TonB-ExbB-ExbD and the TolA-TolQ-TolR proteins. Mol Microbiol. 1993;8:261–268. doi: 10.1111/j.1365-2958.1993.tb01570.x. [DOI] [PubMed] [Google Scholar]
- 6.Clavel T, Germon P, Vianney A, Portalier R, Lazzaroni J C. TolB protein of Escherichia coli K-12 interacts with the outer membrane peptidoglycan-associated proteins Pal, Lpp and OmpA. Mol Microbiol. 1998;29:359–367. doi: 10.1046/j.1365-2958.1998.00945.x. [DOI] [PubMed] [Google Scholar]
- 7.Clavel T, Lazzaroni J C, Vianney A, Portalier R. Expression of the tolQRA genes of Escherichia coli K-12 is controlled by the RcsC sensor protein involved in capsule synthesis. Mol Microbiol. 1996;19:19–25. doi: 10.1046/j.1365-2958.1996.343880.x. [DOI] [PubMed] [Google Scholar]
- 8.Cunliffe H E, Merriman T R, Lamont I L. Cloning and characterization of pvdS, a gene required for pyoverdine synthesis in Pseudomonas aeruginosa: PvdS is probably an alternative sigma factor. J Bacteriol. 1995;177:2744–2750. doi: 10.1128/jb.177.10.2744-2750.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dennis J J, LaFontaine E R, Sokol P A. Identification and characterization of the tolQRA genes of Pseudomonas aeruginosa. J Bacteriol. 1996;178:7059–7068. doi: 10.1128/jb.178.24.7059-7068.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dennis J J, Sokol P A. Electrotransformation of Pseudomonas. In: Nickoloff J A, editor. Electroporation and electrofusion of microorganisms. Clifton, N.J: Humana Press; 1995. pp. 125–133. [DOI] [PubMed] [Google Scholar]
- 11.Derouiche R, Bénédetti H, Lazzaroni J C, Lazdunski C, Lloubès R. Protein complex within Escherichia coli inner membrane. TolA N-terminal domain interacts with TolQ and TolR proteins. J Biol Chem. 1995;270:11078–11084. doi: 10.1074/jbc.270.19.11078. [DOI] [PubMed] [Google Scholar]
- 12.Gambello M J, Kaye S, Iglewski B H. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect Immun. 1993;61:1180–1184. doi: 10.1128/iai.61.4.1180-1184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guihard G, Boulanger P, Bénédetti H, Lloubès R, Besnard M, Letellier L. Colicin A and the Tol proteins involved in its translocation are preferentially located in the contact sites between the inner and outer membranes of Escherichia coli cells. J Biol Chem. 1994;269:5874–5880. [PubMed] [Google Scholar]
- 14.Hamood A N, Colmer J A, Ochsner U A, Vasil M L. Isolation and characterization of Pseudomonas aeruginosa gene, ptxR, which positively regulates exotoxin A production. Mol Microbiol. 1996;21:87–110. doi: 10.1046/j.1365-2958.1996.6251337.x. [DOI] [PubMed] [Google Scholar]
- 15.Hamood A I, Iglewski B H. Expression of the Pseudomonas aeruginosa toxA positive regulatory gene (regA) in Escherichia coli. J Bacteriol. 1990;172:589–594. doi: 10.1128/jb.172.2.589-594.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hedstrom R C, Funk C R, Kaper J B, Pavlovskis O R, Galloway D R. Cloning of a gene involved in regulation of exotoxin A expression in Pseudomonas aeruginosa. Infect Immun. 1986;51:37–42. doi: 10.1128/iai.51.1.37-42.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinrichs D E, Poole K. Cloning and sequence analysis of a gene (pchR) encoding an AraC family activator of pyochelin and ferripyochelin receptor synthesis in Pseudomonas aeruginosa. J Bacteriol. 1993;175:5882–5889. doi: 10.1128/jb.175.18.5882-5889.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hindahl M S, Frank D W, Iglewski B H. Molecular studies of a positive regulator of toxin A synthesis in Pseudomonas aeruginosa. Antibiot Chemother. 1987;39:279–289. doi: 10.1159/000414353. [DOI] [PubMed] [Google Scholar]
- 19.Hishida T, Iwasaki H, Ishioka K, Shinagawa H. Molecular analysis of the Pseudomonas aeruginosa genes, ruvA, ruvB and ruvC, involved in processing of homologous recombination intermediates. Gene. 1996;182:63–70. doi: 10.1016/s0378-1119(96)00474-x. [DOI] [PubMed] [Google Scholar]
- 20.Hohnadel D, Haas D, Meyer J-M. Mapping of mutations affecting pyoverdine production in Pseudomonas aeruginosa. FEMS Microbiol Lett. 1986;36:195–199. [Google Scholar]
- 21.Holloway B W, Romling U, Tummler B. Genomic mapping of Pseudomonas aeruginosa PAO. Microbiology. 1994;140:2907–2929. doi: 10.1099/13500872-140-11-2907. [DOI] [PubMed] [Google Scholar]
- 22.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmid for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 23.LaFontaine E R, Sokol P A. Effects of iron and temperature on expression of the Pseudomonas aeruginosa tolQRA genes: role of the ferric uptake regulator. J Bacteriol. 1998;180:2836–2841. doi: 10.1128/jb.180.11.2836-2841.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lazdunski C J, Bouveret E, Rigal A, Journet L, Lloubès R, Bénédetti H. Colicin import into Escherichia coli cells. J Bacteriol. 1998;180:4993–5002. doi: 10.1128/jb.180.19.4993-5002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lazzaroni J C, Portalier R. The excC gene of Escherichia coli K-12 required for cell envelope integrity encodes the peptidoglycan-associated lipoprotein (PAL) Mol Microbiol. 1992;6:735–742. doi: 10.1111/j.1365-2958.1992.tb01523.x. [DOI] [PubMed] [Google Scholar]
- 26.Lazzaroni J C, Vianney A, Popot J L, Bénédetti H, Samatey F, Lazdunski C, Portalier R, Geli V. Transmembrane alpha-helix interactions are required for the functional assembly of the Escherichia coli Tol complex. J Mol Biol. 1995;246:1–7. doi: 10.1006/jmbi.1994.0058. [DOI] [PubMed] [Google Scholar]
- 27.Levengood S K, Webster R E. Nucleotide sequences of the tolA and tolB genes and localization of their products, components of a multistep translocation system in Escherichia coli. J Bacteriol. 1989;171:6600–6609. doi: 10.1128/jb.171.12.6600-6609.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim A, DeVos D, Brauns M, Mossialos D, Gaballa A, Qing D, Cornelis P. Molecular and immunological characterization of OprL, the 18kDa outer-membrane peptidoglycan-associated lipoprotein (PAL) of Pseudomonas aeruginosa. Microbiology. 1997;143:1709–1716. doi: 10.1099/00221287-143-5-1709. [DOI] [PubMed] [Google Scholar]
- 29.Litwin C M, Calderwood S B. Role of iron in regulation of virulence genes. Clin Microbiol Rev. 1993;6:137–149. doi: 10.1128/cmr.6.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu P V. The role of various fractions of Pseudomonas aeruginosa in its pathogenesis. III. Identification of the lethal toxins produced in vitro and in vivo. J Infect Dis. 1966;166:481–489. doi: 10.1093/infdis/116.4.481. [DOI] [PubMed] [Google Scholar]
- 31.Meury J, Devilliers G. Impairment of cell division in tolA mutant of Escherichia coli at low and high medium osmolarities. Biol Cell. 1999;91:67–75. [PubMed] [Google Scholar]
- 32.Ochsner U A, Johnson Z, Lamont I L, Cunliffe H E, Vasil M L. Exotoxin A production in Pseudomonas aeruginosa requires the iron-regulated pvdS gene encoding an alternative sigma factor. Mol Microbiol. 1996;21:1019–1028. doi: 10.1046/j.1365-2958.1996.481425.x. [DOI] [PubMed] [Google Scholar]
- 33.Ochsner U A, Vasil M L. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc Natl Acad Sci USA. 1996;93:4409–4414. doi: 10.1073/pnas.93.9.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohman D E, Sadoff J C, Iglewski B H. Toxin A-deficient mutants of Pseudomonas aeruginosa PA-103: isolation and characterization. Infect Immun. 1980;28:899–908. doi: 10.1128/iai.28.3.899-908.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Platt T, Muller-Hill B, Miller J H. Assays of the lac operon enzymes. In: Miller J H, editor. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 36.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 37.Prince R W, Storey D G, Vasil A I, Vasil M L. Regulation of toxA and regA by the Escherichia coli fur gene and identification of a Fur homologue in Pseudomonas aeruginosa PA103 and PAO1. Mol Microbiol. 1991;5:2823–2831. doi: 10.1111/j.1365-2958.1991.tb01991.x. [DOI] [PubMed] [Google Scholar]
- 38.Raivio T L, Hoffer D, Prince R W, Vasil M L, Storey D G. Linker insertion scanning of regA, an activator of exotoxin A production in Pseudomonas aeruginosa. Mol Microbiol. 1996;22:239–254. doi: 10.1046/j.1365-2958.1996.00102.x. [DOI] [PubMed] [Google Scholar]
- 39.Rigal A, Bouveret E, Lloubès R, Lazdunski C, Bénédetti H. The TolB protein interacts with the porins of Escherichia coli. J Bacteriol. 1997;178:7274–7279. doi: 10.1128/jb.179.23.7274-7279.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez-Herva J J, Ramos-Gonzalez M I, Ramos J L. The Pseudomonas putida peptidoglycan-associated outer membrane lipoprotein is involved in maintenance of the integrity of the cell envelope. J Bacteriol. 1996;178:1699–1706. doi: 10.1128/jb.178.6.1699-1706.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Schweizer H P. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol. 1992;6:1195–1204. doi: 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 43.Schweizer H P. Two plasmids, X1918 and Z1918, for easy recovery of the xylE and lacZ reporter genes. Gene. 1993;134:89–91. doi: 10.1016/0378-1119(93)90178-6. [DOI] [PubMed] [Google Scholar]
- 44.Sen K, Sikkema D J, Murphy T F. Isolation and characterization of the Haemophilus influenzae tolQ, tolR, tolA and tolB genes. Gene. 1996;178:75–81. doi: 10.1016/0378-1119(96)00338-1. [DOI] [PubMed] [Google Scholar]
- 45.Storey D G, Frank D W, Farinha M A, Kropinski A M, Iglewski B H. Multiple promoters control the regulation of the Pseudomonas aeruginosa regA gene. Mol Microbiol. 1990;4:499–503. doi: 10.1111/j.1365-2958.1990.tb00616.x. [DOI] [PubMed] [Google Scholar]
- 46.Verturi V, Weisbeek P, Koster M. Gene regulation of siderophore-mediated iron acquisition in Pseudomonas aeruginosa: not only the Fur repressor. Mol Microbiol. 1995;17:603–610. doi: 10.1111/j.1365-2958.1995.mmi_17040603.x. [DOI] [PubMed] [Google Scholar]
- 47.Vianney A, Muller M M, Clavel T, Lazzaroni J C, Portalier R, Webster R E. Characterization of the tol-pal region of Escherichia coli K-12: translational control of tolR expression by TolQ and identification of a new open reading frame downstream of pal encoding a periplasmic protein. J Bacteriol. 1996;178:4031–4038. doi: 10.1128/jb.178.14.4031-4038.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walker S L, Hiremath L S, Galloway D R. ToxR (RegA) activates Escherichia coli RNA polymerase to initiate transcription of Pseudomonas aeruginosa toxA. Gene. 1995;154:15–21. doi: 10.1016/0378-1119(94)00870-x. [DOI] [PubMed] [Google Scholar]
- 49.Webster R E. The tol gene products and the import of macromolecules into Escherichia coli. Mol Microbiol. 1991;5:1005–1011. doi: 10.1111/j.1365-2958.1991.tb01873.x. [DOI] [PubMed] [Google Scholar]
- 50.West S E H, Sample A K, Runyen-Janecky L J. The vfr gene product for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J Bacteriol. 1994;176:7532–7542. doi: 10.1128/jb.176.24.7532-7542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woods D E, Vasil M L. Pathogenesis of Pseudomonas aeruginosa infections. Infect Dis Ther. 1994;12:21–50. [Google Scholar]