Abstract
The iotaa component (ia) of Clostridium perfringens ADP ribosylates nonmuscle β/γ actin and skeletal muscle α-actin. Replacement of Arg-295 in ia with alanine led to a complete loss of NAD+-glycohydrolase (NADase) and ADP-ribosyltransferase (ARTase); that of the residue with lysine caused a drastic reduction in NADase and ARTase activities (<0.1% of the wild-type activities) but did not completely diminish them. Substitution of alanine for Glu-378 and Glu-380 caused a complete loss of NADase and ARTase. However, exchange of Glu-378 to aspartic acid or glutamine resulted in little effect on NADase activity but a drastic reduction in ARTase activity (<0.1% of the wild-type activity). Exchange of Glu-380 to aspartic acid caused a drastic reduction in NADase and ARTase activities (<0.1% of the wild-type activities) but did not completely diminish them; that of the residue to glutamine caused a complete loss of ARTase activity. Replacement of Ser-338 with alanine resulted in 0.7 to 2.3% wild-type activities, and that of Ser-340 and Thr-339 caused a reduction in these activities of 5 to 30% wild-type activities. The kinetic analysis showed that Arg-295 and Ser-338 also play an important role in the binding of NAD+ to ia, that Arg-295, Glu-380, and Ser-338 play a crucial role in the catalytic rate of NADase activity, and that these three amino acid residues and Glu-378 are essential for ARTase activity. The effect of amino acid replacement in ia on ARTase activity was similar to that on lethal and cytotoxic activities, suggesting that lethal and cytotoxic activities in ia are dependent on ARTase activity.
Clostridium perfringens type E produces iota-toxin, which is lethal and dermonecrotic (35, 37). Iota-toxin is a binary toxin which is composed of an enzymatic component called iotaa (ia) and a binding component called iotab (ib). ia causes ADP-ribosylation of skeletal muscle α-actin and nonmuscle β/γ actin, and ib is required for penetration of ia into the cytosol (1, 2, 3, 12, 34, 38, 40). It has been reported that iota-toxin may be associated with antibiotic-associated colitis caused by C. perfringens type E in rabbits (9, 26). In addition, C. spiroforme is known to cause antibiotic-associated enterotoxemia of rabbits and to produce iota-like-toxin (8), which shows partial antigenic identity with iota-toxin (32). Several workers have reported the presumptive involvement of iota-like-toxin in sudden outbreaks of enteritis in rabbit colonies (17, 20). It therefore appears that iota-toxin also is able to be a major agent in enterotoxemia (16, 37). Stiles and Wilkins (43) reported the purification of iota-toxin from cultures of C. perfringens type E. However, large amounts of iota-toxin were difficult to purify from the culture supernatant fluid of C. perfringens type E by to their method. To understand the mode of action of iota-toxin, large amounts of highly purified ia and ib are required. Bacillus subtilis is reported to produce and secrete secretory proteins of other gram-positive bacteria (39). We first tried to purify large amounts of ia from cultures of a B. subtilis transformant carrying the component gene.
Bacterial ADP-ribosylating toxins, such as diphtheria toxin (DT) (11), Pseudomonas exotoxin A (ETA) (45), cholera toxin (CT) (42), Escherichia coli heat-labile enterotoxin (LT) (42), pertussis toxin (22), Pseudomonas exoenzyme S (ExoS) (24), Clostridium botulinum C3 enzyme (C3) (5), C. botulinum C2 toxin (C2 toxin) (4), C. spiroforme toxin (2), and epidermal cell differentiation inhibitor (EDIN) from Staphylococcus aureus (44), have been studied as agents that contribute to the pathogenesis of bacteria. The Glu-14 and -226 residues in ia are included within the E-X-X-X-X-W sequence in the active sites of DT and ETA. ADP-ribosylating toxins such as CT, LT, and C3 are known to contain three conserved regions, aromatic residue-R/H, E-X-E, and hydrophobic residue-S-T-S-hydrophobic residue, in the cavity formed by the β/α motif (14). The analysis of the LT crystal suggested that the nicotinamide ring of NAD+ docks into the cavity (14). The role of these regions is thought to be as follows. The polar side chains of E-X-E extend toward the catalytic cavity and the consensus sequence involved in forming the NAD+ cleft; an aromatic residue-R/H located deep in the cavity binds NAD+; and the S-T-S consensus sequence is folded in a β strand representing the floor of the cavity. The Glu-378 and -380 residues in ia are included within the E-X-E sequences essential for the enzymatic activities of CT, LT, and ExoS. The Arg-295 residue in the component is present in the aromatic residue-R/H sequence. Ser-338, Ser-340, and Thr-339 are present in the S-T-S consensus sequence.
Recently, Perelle et al. (31) reported that Glu-378, Glu-380, and Arg-295 are involved in the ADP-ribosylation of ia. Damme et al. (13) reported the pivotal role of Glu-378 in ia which was photoaffinity labeled with [carbonyl-14C]NAD+. However, the reaction mechanism of ia has not been investigated by use of conservative substitutions and kinetic analysis. To study the functional roles of these conserved regions of ia in more detail, we replaced Arg-295, Glu-14, Glu-226, Glu-378, Glu-380, Ser-338, Ser-340, and Thr-339 with various amino acids by site-directed mutagenesis, determined the ADP-ribosyltransferase (ARTase) and NAD+-glycohydrolase (NADase) activities of these variant components, determined the cytotoxic and lethal activities of these variant components in the presence of ib, and analyzed the enzymatic properties of these components.
MATERIALS AND METHODS
Materials.
Restriction endonucleases and DNA-modifying enzymes were obtained from Takara Shuzo (Kyoto, Japan) and Toyobo (Osaka, Japan), respectively. pT-7 Blue was obtained from Novagen (Madison, Wis.). [adenylate-32P]NAD+ and [carbonyl-14C]NAD+ were obtained from New England Nuclear (Boston, Mass.) and Amersham (Tokyo, Japan), respectively. Purified rabbit muscle actin was purchased from Worthington Biochemical Corp., Lakewood, N.J. All other chemicals were of analytical grade.
Bacterial strains and plasmids.
C. perfringens type E strain NCIB 10748, kindly donated by M. Popoff (Institut Pasteur, Paris, France), was grown in brain heart infusion broth under anaerobic conditions. Plasmid DNA was extracted and purified as described by Perelle et al. (30). E. coli JM109 or C600 was the host for the plasmid used. B. subtilis ISW1214 was used for production of the toxin (28).
DNA cloning and expression of the ia gene.
C. perfringens type E strain NCIB 10748 carrying the entire iota-toxin gene was PCR amplified using a set of primers. A pair of primers for ia (5′-GAGAATTCAGAAAATACAATC-3′ and 5′-TCTTATCATAGCTGTAAGTG-3′) was designed from the published ia sequence (30). The PCR was carried out for 25 cycles under standard reaction conditions with a GeneAmp DNA amplification reagent kit (Perkin-Elmer Cetus, Norwalk, Conn.). After PCR amplification, the 1.7-kbp fragment obtained was gel purified and cloned into pT-7 Blue (pTIA). The 1.7-kbp EcoRI/XbaI fragment of the wild-type ia gene was subcloned from pTIA into the pHY300PLK (E. coli-B. subtilis shuttle vector) SmaI site and transformed into B. subtilis ISW1214.
Site-directed mutagenesis.
The 1.7-kbp EcoRI/XbaI fragment of the ia gene, encoding the entire reading frame for the 454 amino acids of ia, was introduced into the SmaI site of the pUC19 vector (pUIA) and used as a template for mutagenesis. Site-directed mutagenesis was carried out by the unique restriction enzyme site elimination technique with a Transformer mutagenesis kit (Clontech Laboratories, Inc., Palo Alto, Calif.) and synthetic oligonucleotide primers having mutations as described previously (28, 29). All mutants were obtained by this method and were identified by sequencing with a Dye Deoxy termination kit (Applied Biosystems), sequencing primers, and a DNA sequencer (374A; Applied Biosystems).
Purification of ia and ib from C. perfringens.
Native ia and ib were purified from culture supernatant fluid of C. perfringens NCTC 8084 as described previously (43).
Determination of lethal activity.
Serial twofold dilutions of ia and 5 μg of ib were mixed in 0.01 M phosphate buffer (pH 7.0) containing 0.9% NaCl (final volume, 1.0 ml). The mixture (0.1 ml) was injected intravenously into adult mice (about 25 g), and deaths occurring within 24 h were recorded.
Determination of cytotoxic activity.
Vero cells were cultivated in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum. For cytotoxicity assays, the cells were inoculated into 48-well tissue culture plates (Falcon, Oxnard, Calif.). Serial dilutions of various concentrations of ia and 200 ng of ib per ml were mixed in Dulbecco modified Eagle medium and inoculated onto a cell monolayer. The cells were observed for morphological alterations 8 h after inoculation.
NADase assay.
NAD+ glycohydrolysis due to ia was determined by the release of free radiolabeled nicotinamide into solution, resulting from the hydrolysis of [carbonyl-14C]NAD+ (41 mCi/mmol) (46). The assays were performed at various NAD+ concentrations in the presence of ia in a final volume of 50 μl of 40 mM Tris-HCl (pH 7.5) containing 10 mM EDTA, 10 mM dithiothreitol, and 100 μg of bovine serum albumin/ml at 37°C for 6 h. After incubation, hydrolyzed nicotinamide was separated from NAD+ by the addition of 200 μl of water-saturated ethyl acetate. The amount of nicotinamide in the ethyl acetate phase was determined by liquid scintillation counting. The data were corrected by subtraction of the radioactivity due to nonenzymatic hydrolysis of NAD+.
ARTase assay.
ARTase activity on globular (G)-actin due to ia was monitored by the incorporation of the radiolabeled ADP-ribose moiety of NAD+ into the trichloroacetic acid (TCA)-precipitable protein fraction of the reaction mixture. [adenylate-32P]NAD+ (800 Ci/mmol) was diluted to a final specific activity of 16 Ci/mmol with unlabeled NAD+ to give a final concentration of 5 μM. Reaction mixtures in a final volume of 0.1 ml of 20 mM Tris-HCl buffer (pH 7.5) containing 1 mM dithiothreitol, 40 μM ATP, 40 μM CaCl2, 50 μM MgCl2, and the specified concentrations of NAD+ and actin for the kinetic experiments (see below) were incubated with ia samples at 37°C for 60 min. After incubation, 0.5 ml of cold 7.5% TCA and 10 μg of bovine serum albumin were added, and the reaction mixtures were allowed to stand for 30 min on ice. Proteins were precipitated by centrifugation at 10,000 × g for 20 min. The precipitate was washed twice by centrifugation in 1 ml of ice-cold 7.5% TCA. The precipitated proteins were dissolved in 10 μl of 0.02 M Tris-HCl buffer (pH 7.5) containing 3% sodium dodecyl sulfate (SDS), 2% 2-mercaptoethanol, 5% glycerol, and 0.001% bromophenol blue and then subjected to SDS-polyacrylamide gel electrophoresis (PAGE) (12% polyacrylamide gel). The gel was stained with Coomassie blue, destained, and dried in a gel dryer; labeled actin was analyzed with a Fuji BAS 2000 system (Fuji Photo Film Co., Ltd., Tokyo, Japan). The radioactive bands were excised from the gel, and the incorporated radioactivity was counted in a liquid scintillation counter (Aloka Co., Ltd., Tokyo, Japan).
Kinetic experiments.
Initial rate data for the single-substrate NADase reaction were collected under conditions where NAD+ concentrations were varied from 0.2 to 2.5 Km. Initial rate data for the ARTase reaction were similarly determined for NAD+ binding at a fixed actin concentration (0.23 μM) and various NAD+ concentrations from 0.2 to 2.5 Km and for actin binding at a fixed NAD+ concentration (5 μM) and various actin concentrations from 0.2 to 2.5 Km. The kinetic parameters were obtained by analysis of a Hanes-Woolf plot (18).
Other procedures.
Protein was assayed by the method of Lowry et al. (25). Double-gel immunodiffusion and SDS-PAGE were carried out as described previously (23, 27). Anti-ia antiserum was prepared by immunizing a rabbit with 50 μg of purified ia as described previously (36). Determination of the N-terminal amino acid sequence was performed as described previously (21).
RESULTS
Purification of ia from culture supernatant fluid of the B. subtilis transformant carrying the ia gene.
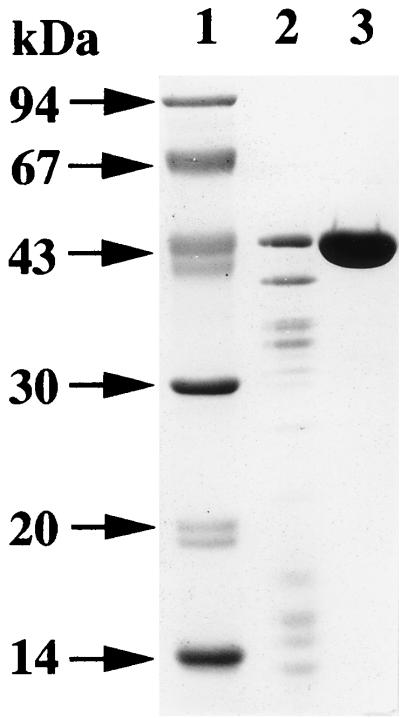
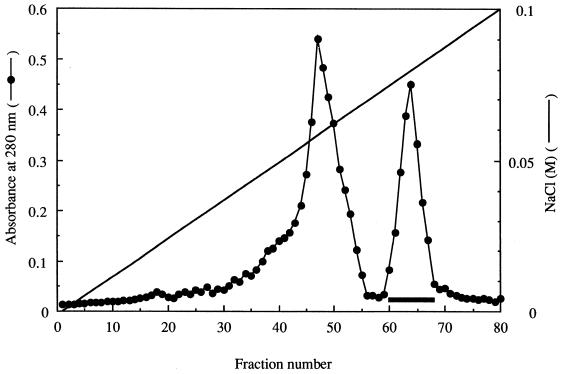
Growth of the B. subtilis transformant reached a maximum in Luria-Bertani broth after 7 h of incubation. The time course of ia production (ARTase activity) was roughly parallel to the growth of the transformant. ARTase activity remained constant in the culture within 5 h after growth stopped. Ammonium sulfate (313 g/liter) was added to the culture supernatant fluid and allowed to stand overnight at 4°C. SDS-PAGE of the ammonium sulfate fraction revealed that the corresponding band of ia (about 43 kDa) was dominant in the fraction (Fig. 1), suggesting that the B. subtilis transformant produces and secretes large amounts of the component. The ammonium sulfate fraction was dialyzed against 0.02 M Tris-HCl buffer (pH 7.5) and loaded onto a DEAE–Sepharose CL-6B column, previously equilibrated with the same buffer. Elution of the column was done with a 0 to 0.1 M NaCl linear gradient (300 ml total) in 0.02 M Tris-HCl buffer (pH 7.5). Figure 2 shows a typical elution profile of the ammonium sulfate fraction applied to the column. The eluted fractions were separated into two peaks, based on the absorbance at 280 nm. An immunodiffusion test of each fraction was performed with anti-ia antiserum. Only the second peak (fractions 61 to 68) reacted with the antiserum. Furthermore, the second peak showed ARTase activity, which was completely inhibited by anti-ia antiserum (data not shown). SDS-PAGE analysis of the preparation (50 μg of protein) revealed only one band of approximately 43 kDa, as shown in Fig. 1. An immunodiffusion test showed that the test preparation and the native component reacted with anti-ia antiserum, showing a single fused precipitin line (data not shown).
FIG. 1.
SDS-PAGE analysis of purified ia. Lanes: 1, molecular mass standards; 2, ammonium sulfate fraction; 3, purified r ia.
FIG. 2.
DEAE–Sepharose CL-6B column chromatography of the ammonium sulfate fraction. The ammonium sulfate fraction obtained from cultures of the B. subtilis transformant carrying the ia gene was applied to a DEAE–Sepharose CL-6B column (2 by 25 cm) equilibrated with 10 mM Tris-HCl buffer (pH 7.5). After application, the column was washed with 150 ml of the same buffer and then eluted with 300 ml of a linear gradient of 0 to 0.1 M NaCl in the same buffer. The fractions were collected and assayed for A280. The heavy horizontal bar indicates the peak containing ia.
Furthermore, the N-terminal sequence of the purified protein was identical to the amino acid sequence of ia expected from the gene and the native component. The purification steps for recombinant ia (r ia) are summarized in Table 1. The final step resulted in a homogeneous product that was purified approximately ninefold, starting from the ammonium sulfate fraction, with a yield of about 75% with respect to ARTase activity. The amount of purified r ia obtained from 1,000 ml of culture was about 35 mg. Stiles and Wilkins (43) reported that the dose required to kill 50% of mice injected intraperitoneally was 0.62 μg of ia in the presence of 0.94 μg of ib, but administration of 0.31 μg of ia and 0.47 μg of ib killed no mice. In the present work, approximately 2 ng of purified r ia was required to kill 50% of mice in the presence of approximately 500 ng of ib. However, 500 μg of purified r ia alone showed no lethal activity.
TABLE 1.
Summary of purification of r ia from culture supernatant fluid of the B. subtilis transformant
| Step | Total activity (mU)a | Total protein (mg) | Sp act (mU/mg) | Purification (fold) | Recovery of activity (%) |
|---|---|---|---|---|---|
| 70% (NH4)2SO4 precipitation | 790 | 183 | 4.32 | 1 | 100 |
| DEAE–Sepharose CL-6B | 598 | 35.2 | 39.6 | 9.2 | 74.6 |
One unit of activity (milliunit) was defined as the activity that catalyzed the transfer of 1 mmol of ADP-ribose from NAD+ to actin per min at 37°C.
Biological activities of ia with variations at Arg-295, Glu-14, Glu-226, Glu-378, Glu-380, Ser-338, Ser-340, and Thr-339.
The Arg-295 in ia was replaced with alanine, lysine, and histidine by site-directed mutagenesis; Glu-14 and -226 were replaced with alanine; Glu-378 and -380 were replaced with alanine, aspartic acid, and glutamine; Ser-338 and -340 were replaced with alanine, cysteine, threonine, and phenylalanine; and Thr-339 was replaced with alanine and phenylalanine. These variant components were purified from cultures of B. subtilis transformants carrying the corresponding genes by use of the purification procedure described for r ia. All of the purified variant components showed only one band of approximately 43 kDa on SDS-PAGE (data not shown).
R295A showed a complete loss of NADase, ARTase, cytotoxic, and lethal activities under our experimental conditions (Tables 2, 3, and 4). On the other hand, the NADase, ARTase, cytotoxic, and lethal activities of R295K were about 100- to 250-fold lower than those of the wild-type component (Tables 2, 3, and 4). The ARTase and cytotoxic activities of R295H were about 0.2% these activities in wild-type ia, but the lethal activity of R295H was about 6% the wild-type component activity; these results show that replacement of Arg-295 with histidine did not cause a severe reduction in lethal activity, compared with the reduction in enzymatic and cytotoxic activities. These observations suggest that Arg-295 is able to be partially substituted by basic amino acids, judging from the effect of the replacements on NADase, ARTase, cytotoxic, and lethal activities.
TABLE 2.
NADase and ARTase activities of wild-type ia and variant iaa
| Wild type or variant | NADase activity (ng/ml) | ARTase activity (μg/ml) |
|---|---|---|
| Wild type | 2.8 | 5.0 |
| E14A | — | 5.6 |
| E226A | — | 5.9 |
| R295A | ND | ND |
| R295K | 764 | 906 |
| R295H | — | 2,520 |
| S338A | 420 | 220 |
| S338C | 196 | 181 |
| S338T | — | ND |
| S338F | — | ND |
| T339A | 60 | 30.5 |
| T339F | — | 192 |
| S340A | 10 | 61 |
| S340C | — | 31 |
| S340T | — | 154 |
| E378A | ND | ND |
| E378D | 4.0 | 2,050 |
| E378Q | 5.0 | ND |
| E380A | ND | ND |
| E380D | 720 | 1,110 |
| E380Q | — | ND |
Wild-type ia or variant ia was incubated with 50 μM [carbonyl-14C]NAD+ at 37°C for 6 h, and NADase activity was determined by the release of radiolabeled nicotinamide. Wild-type ia or variant ia was incubated with 5 μM [32P]NAD+ in the presence of 2 μg of actin for 90 min at 37°C. Labeled actin was analyzed by SDS-PAGE and autoradiography. NADase and ARTase activities were expressed as the amount required to induce 50% maximal activity of each. ND, not detected (<5,000 μg/ml); —, not done.
TABLE 3.
Cytotoxic activities of wild-type ia and variant iaa
| Wild type or variant | Cytotoxic activity at the following dose of wild-type or variant ia (ng/ml):
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 1,000 | 500 | 100 | 50 | 10 | 5 | 1 | 0.5 | |
| Wild type | +++ | +++ | ++ | + | − | |||
| E14A | +++ | +++ | ++ | + | − | |||
| E226A | +++ | +++ | + | − | − | |||
| R295A | − | − | − | |||||
| R295K | +++ | ++ | + | − | ||||
| R265H | ++ | + | − | − | ||||
| S338A | +++ | +++ | ++ | + | − | |||
| S338C | +++ | ++ | + | − | ||||
| S338T | − | − | − | |||||
| S338F | − | − | − | |||||
| T339A | +++ | +++ | ++ | + | − | |||
| T339F | +++ | +++ | ++ | + | − | − | ||
| S340A | +++ | +++ | +++ | ++ | − | − | ||
| S340C | +++ | +++ | ++ | + | − | − | ||
| S340T | +++ | +++ | ++ | + | − | − | ||
| E378A | − | − | − | |||||
| E378D | − | − | − | |||||
| E378Q | − | − | − | |||||
| E380A | − | − | − | |||||
| E380D | +++ | ++ | − | − | ||||
| E380Q | − | − | − | |||||
Confluent monolayers of Vero cells (2 × 106 cells) were exposed to various doses of wild-type or variant ia in the presence of 200 ng of ib per ml at 37°C. Cell rounding was recorded after 8 h. Cell rounding in a Vero cell assay (cytotoxic activity) was scored as follows: +++, 100% rounding; ++, 50 to 80% rounding; +, 20 to 40% rounding; −, no rounding.
TABLE 4.
Lethal activities of wild-type ia and variant iaa
| Wild type or variant | No. of deaths at the following dose of wild-type or variant ia (ng/mouse):
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2,048 | 1,024 | 512 | 256 | 128 | 64 | 32 | 16 | 8 | 4 | 2 | 1 | |
| Wild type | 4 | 4 | 4 | 2 | 0 | |||||||
| E14A | 4 | 4 | 4 | 1 | 0 | |||||||
| E226A | 4 | 4 | 4 | 1 | 1 | |||||||
| R295A | 0 | 0 | 0 | |||||||||
| R295K | 4 | 4 | 0 | 0 | ||||||||
| R295H | 4 | 4 | 4 | 2 | 0 | |||||||
| S338A | 4 | 4 | 4 | 4 | 0 | |||||||
| S338C | 4 | 4 | 4 | 4 | 0 | |||||||
| S338T | 0 | 0 | 0 | |||||||||
| S338F | 0 | 0 | 0 | |||||||||
| T339A | 4 | 4 | 4 | 2 | 0 | |||||||
| T339F | 4 | 4 | 2 | 0 | 0 | |||||||
| S340A | 4 | 4 | 4 | 2 | 0 | |||||||
| S340C | 4 | 4 | 3 | 2 | 0 | |||||||
| S340T | 4 | 4 | 4 | 2 | 0 | |||||||
| E378A | 0 | 0 | 0 | |||||||||
| E378D | 0 | 0 | 0 | |||||||||
| E378Q | 0 | 0 | 0 | |||||||||
| E380A | 0 | 0 | 0 | |||||||||
| E380D | 2 | 0 | 0 | |||||||||
| E380Q | 0 | 0 | 0 | |||||||||
Various doses of wild-type or variant ia and ib (500 ng) were intravenously injected into 4 mice, and deaths occurring within 24 h were recorded.
Replacement of Glu-14 and -226 had little effect on the ARTase activity of ia (Table 2) and did not cause a significant reduction in the cytotoxic and lethal activities of ia in the presence of ib (Tables 3 and 4); these results indicate that these residues are not required for the biological activities of ia. Replacement of Glu-378 with alanine resulted in a complete loss of NADase, ARTase, cytotoxic, and lethal activities (Tables 2, 3, and 4), suggesting that Glu-378 is important for these activities. Conservative substitution of aspartic acid for Glu-378 caused a drastic decrease in ARTase, lethal, and cytotoxic activities, below our detection limit. That of glutamine reduced ARTase, lethal, and cytotoxic activities to below our detection limit. However, replacement of the residue with aspartic acid and glutamine had little effect on NADase activity. These results suggest that the carboxyl group of Glu-378 is essential for ARTase, lethal, and cytotoxic activities but not for NADase activity. As shown in Table 2, changing Glu-380 to alanine caused a complete loss of NADase and ARTase activities, changing it to aspartic acid resulted in over a 200-fold reduction in these enzymatic activities, and changing it to glutamine reduced ARTase activity to below our detection limit. Furthermore, the cytotoxic and lethal activities of E380D were about 500 and 1,000 times lower than those of r ia, respectively, but these activities of E380A and E380Q were below the detection limit (Tables 3 and 4). These observations show that Glu-380 is essential for any activities of ia.
Replacement of Ser-338 with alanine and cysteine resulted in about a 30- to 150-fold reduction in the NADase, ARTase, cytotoxic, and lethal activities of wild-type ia (Tables 2, 3, and 4). It is likely that Ser-338 is important but not essential for these activities. Replacement of Ser-338 with threonine and phenylalanine caused ARTase, cytotoxic, and lethal activities to fall below the detection limit, implying that replacement of Ser-338 with amino acids containing large side chains reduced these activities more severely than did that with amino acids containing small side chains. Replacement of Ser-340 with alanine, cysteine, and threonine caused only a 3- to 30-fold reduction in the NADase, ARTase, cytotoxic, and lethal activities of wild-type ia (Tables 2, 3, and 4). Replacement of Thr-339 with alanine or phenylalanine led to about a 6- to 30-fold reduction.
Kinetics of ADP-ribosylating activities of wild-type ia and variant ia.
Kinetic analyses of the six variant components (R295K, S338A, T339A, S340A, E378D, and E380D) were performed to help determine the mechanistic basis for the events caused by NADase and ARTase in the presence of increasing NAD+ or actin concentrations. The kinetic parameters were obtained by analysis of the Hanes-Woolf plots of initial velocities of the NADase reaction. Table 5 shows that the Km values for NAD+ binding to E380D, S338A, S340A, and T339A were similar to that of the wild type. The Km value for binding to R295K was significantly higher than that of the wild type (2.5-fold higher Km value), and the kcat values associated with R295K, S338A, and E380D were considerably lower than that of the wild type (200-, 67- and 250-fold lower kcat values, respectively); these results show that the kinetic effects on these residues almost exclusively involved kcat. The Km and kcat values for E378D were similar to those for the wild-type component.
TABLE 5.
Kinetic analysis of NADase activities of wild-type ia and variant iaa
| Wild type or variant | Km (μM) | Relative Km | kcat (min−1) | Relative kcat | kcat/Km | Relative kcat/Km |
|---|---|---|---|---|---|---|
| Wild type | 23 | 1 | 0.2 | 1 | 0.009 | 1 |
| R295K | 57 | 2.5 | 0.001 | 0.005 | 0.00002 | 0.002 |
| S338A | 24 | 1.0 | 0.003 | 0.015 | 0.0001 | 0.01 |
| T339A | 17.1 | 0.74 | 0.009 | 0.045 | 0.0005 | 0.06 |
| S340A | 13.1 | 0.57 | 0.034 | 0.17 | 0.0026 | 0.29 |
| E378D | 19 | 0.8 | 0.074 | 0.37 | 0.004 | 0.44 |
| E380D | 18.5 | 0.8 | 0.0008 | 0.004 | 0.00004 | 0.005 |
NADase activity was assayed with various amounts (5 to 25 μM) of NAD+ as described in Materials and Methods. The standard deviation was not greater than ±15% (n = 4).
The kinetic parameters obtained by analysis of the Hanes-Woolf plots of initial velocities for the ARTase reaction are summarized in Table 6. The Km values for NAD+ binding to the variant components for the ARTase reaction were determined from initial rate data obtained at a fixed actin concentration and various NAD+ concentrations. The results showed that the Km values of R295K and S338A were significantly higher than that of the wild-type and that the Km values for the ARTase activity of T339A, S340A, E378D, and E380D were identical to that of the wild type (Table 6); these results show that replacement of Arg-295 and Ser-338 resulted in a significant alteration of the Km for NAD+. Replacement of Arg-295, Glu-378, and Glu-380 drastically reduced kcat values. The kcat values associated with Arg-295, Glu-378, and Glu-380 were markedly reduced, by 1,000-, 250-, and 1,000-fold, respectively, compared to that of the wild type, as shown in Table 6. The kcat values associated with S338A, T339A, and S340A were about 5, 20, and 26% that of the wild type, respectively.
TABLE 6.
Kinetic analysis of ARTase activities of wild-type ia and variant iaa
| Varied ingredient | Wild type or variant | Kmb | Relative Km | kcat (min−1) | Relative kcat | kcat/Km | Relative kcat/Km |
|---|---|---|---|---|---|---|---|
| NAD+ | Wild type | 6.0 | 1 | 2,024 | 1 | 337 | 1 |
| R295K | 23 | 3.8 | 2.9 | 0.001 | 0.74 | 0.002 | |
| S338A | 20 | 3.3 | 104 | 0.05 | 5.1 | 0.015 | |
| T339A | 4.9 | 0.8 | 405 | 0.20 | 83 | 0.25 | |
| S340A | 11 | 1.8 | 518 | 0.26 | 48 | 0.14 | |
| E378D | 4.9 | 0.8 | 8.4 | 0.004 | 1.7 | 0.005 | |
| E380D | 5.5 | 0.9 | 2.0 | 0.001 | 0.4 | 0.001 | |
| Actin | Wild type | 152 | 1 | 441 | 1 | 2.9 | 1 |
| R295K | 96 | 0.6 | 3.6 | 0.008 | 0.04 | 0.014 | |
| S338A | 223 | 1.5 | 12 | 0.027 | 0.06 | 0.021 | |
| T339A | 265 | 1.7 | 344 | 0.78 | 1.3 | 0.45 | |
| S340A | 140 | 0.9 | 81 | 0.18 | 0.58 | 0.20 | |
| E378D | 126 | 0.8 | 5.1 | 0.012 | 0.04 | 0.014 | |
| E380D | 176 | 1.2 | 2.3 | 0.005 | 0.01 | 0.003 |
ARTase activity was measured with various NAD+ or actin concentrations as described in Materials and Methods. The standard deviation was not greater than ±15% (n = 4).
The Km concentration is given as micromolar for NAD+ and as picomolar for actin.
Next, the Km values for actin binding to the variant ia components for the ARTase reaction were determined from initial rate data obtained at a fixed NAD+ concentration and various actin concentrations. The Km values for actin binding to any variant components were in agreement with that determined for the wild type, within experimental error; none of the variants showed altered affinity for actin (Table 6). Replacement of Arg-295, Ser-338, Glu-378, and Glu-380 resulted in drastic reductions in kcat values. The kcat values associated with Arg-295, Ser-338, Glu-378, and Glu-380 were markedly reduced, by about 125-, 37-, 83-, and 200-fold, respectively, compared to that of the wild type as shown in Table 6. However, replacement of Thr-339 and Ser-340 reduced by about 78 and 18%, respectively, the kcat value associated with the wild type.
DISCUSSION
The yield of purified r ia was approximately 35 mg/liter from cultures of the B. subtilis transformant carrying the ia gene. The N-terminal sequence of r ia was identical to that of native ia and that deduced from the ia gene. The molecular mass and antigenicity of r ia also were coincident with those of native ia. Based upon the lethal activities of r ia purified from cultures of the B. subtilis transformant and of ia purified from C. perfringens cultures by Stiles and Wilkins (43), the specific activity of the former was calculated to be about 300 times higher than that of the latter; this result shows that our purification procedure is very useful for the isolation of large amounts of ia with high specific activity.
Replacement of Glu-14 and -226, which are included within the E-X-X-X-X-W sequence involved in the NAD+ binding sites of DT and ETA (15), had no effect on the activities of ia, confirming that ia belongs to the CT group of in the ADP-ribosylating enzyme family (14).
Replacement of Arg-295 with alanine in ia led to a complete loss of NADase, ARTase, lethal, and cytotoxic activities. Perelle et al. (31) also reported that this residue is essential for ARTase activity. Thus, it appears that Arg-295 of ia is equivalent to Arg-7 of LT and Arg-299 of C2 toxin, which are required for ARTase activity (6, 15). However, replacement of Arg-295 with histidine or lysine caused a drastic reduction in NADase and/or ARTase activities, although the activities were still detectable, showing that the charge of the basic side chain of Arg-295 is essential for the NADase and ARTase activities of ia. Substitution of lysine for Arg-295 in ia resulted in a significant reduction in the Km for NAD+, markedly reduced the kcat values for NADase and ARTase, but had little effect on the Km for actin. Accordingly, these observations suggest that Arg-295 plays an essential role in both the binding of ia to NAD+ and the catalytic actions of NADase and ARTase (Fig. 3).
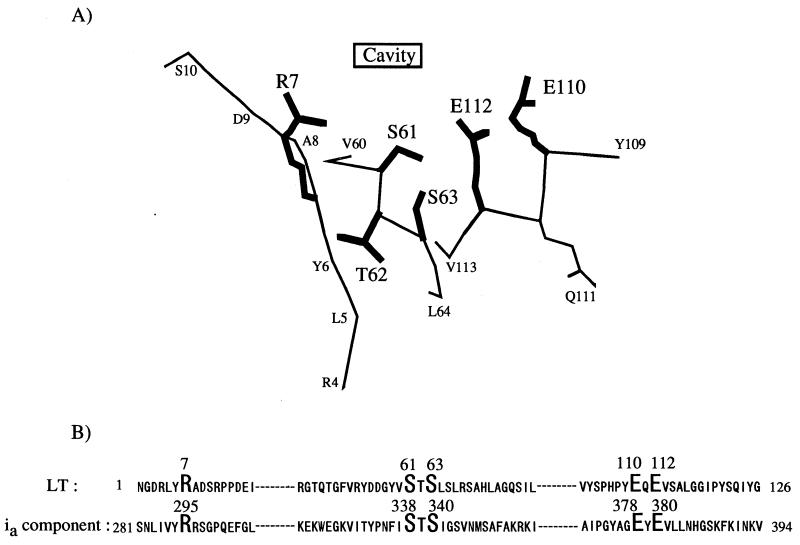
FIG. 3.
(A) Active-site structure of LT. This active-site configuration was drawn on the basis of the refined coordinates of LT (41). The Arg-7 (R7), Ser-61 (S61), Thr-62 (T62), Ser-63 (S63), Glu-110 (E110), and Glu-112 (E112) side chains are shown in bold. (B) Sequence alignments of LT and the ia component. Sequences were taken from porcine LT (SWISSProt accession no., P06717) and component ia (GenBank accession no., X73562). The residues in bold are those that are conserved in ADP-ribosylating enzymes.
Substitution of Glu-378 with alanine in ia resulted in a complete loss of NADase and ARTase activities, as reported by Perelle et al. (31); however, conservative substitution of Glu-378 with aspartic acid or glutamine in ia had little effect on NADase activity and drastically reduced but did not diminish ARTase activity. These results indicate that Glu-378 plays an important role in ARTase activity but not in NADase activity. It therefore is likely that the side chain of the amino acid at position 378 is important for maintenance of the active-site integrity in NADase activity and that the carboxyl group of the amino acid at position 378 is essential for ARTase activity. Glu-378 within the E-X-E motif (Glu378-X-Glu379) of C2 toxin and Glu-379 within the motif Glu379-X-Glu381 of ExoS were essential for ARTase activity but were not required for NADase activity (6, 33). Our results coincided with these results.
The kinetic analysis showed that replacement of Glu-378 with aspartic acid resulted in a severe reduction in the kcat values for ARTase activity but had little effect on the Km values for NAD+ and actin, suggesting that the residue plays an important role in catalytic mechanism but is not required for binding to NAD+ and actin. The glutamic acid at position 380 could be replaced with aspartic acid, although the replacement was not wholly effective, but could not be replaced with alanine and glutamine; these results show that conservative substitution, such as reduction of the carboxyl group at position 380 by one methylene unit or replacement of the carboxyl group by an uncharged amide, resulted in a significant reduction and a complete loss, respectively, of ARTase activity. Thus, the role of Glu-380 in ia appears to be equivalent to that of the corresponding residues in C2 toxin and LT.
Replacement of Glu-380 with aspartic acid markedly reduced the kcat values for ARTase and had little effect on the Km values for NAD+ and actin, suggesting that Glu-380 is essential for the catalytic mechanism of ARTase but not for binding to NAD+ and actin. Damme et al. (13) reported the Glu-378 in ia was photolabeled by NAD+ but that Glu-380 was not. Therefore, the Glu-378 and -380 residues seem to play different roles in the ARTase activity of ia.
Cieplak et al. (10) reported that substitution of Glu-112 (E110-X-E112) in LT resulted in a marked reduction in ARTase activity, suggesting that the residue plays a specific role in the mechanism of ADP-ribosylation and represents an essential catalytic residue. In addition, they suggested that Glu-110 is unlikely to play a specific role in the reaction mechanism. Hara et al. (19) reported that rat T-cell antigen RT 6.1 (Q207-X-E209) catalyzes NAD+ glycohydrolysis but not NAD+ ribosyltransfer and that mutant RT 6.1 in which Gln-207 was replaced with glutamic acid exhibited ARTase activity. Our result is coincident with their findings in that the first glutamic acid residue in the E-X-E motif is essential for ARTase activity but not for NADase activity. However, C3 and EDIN in the ADP-ribosyltransferase family, which ADP-ribosylate small GTP-binding proteins of the rho family, have a glutamine residue in the motif, suggesting that the residue in C3 and EDIN which corresponds to the residue at position 380 in ia is not required to be glutamic acid for ARTase activity. Thus, the residue in the motif may depend on the substrate.
Ser-338 could be replaced with alanine and cysteine, although the replacements were not wholly effective, but could not be replaced with threonine and phenylalanine; these results suggest that the hydroxyl group of Ser-338 is not essential for these activities and that the large side chain of the amino acid at position 338 completely disturbed ARTase activity. Thus, Ser-338 may be extremely close to the catalytic site, as shown in Fig. 3. Replacement of Ser-338 with alanine caused a significant reduction in the Km value of NAD+ for ARTase and drastically reduced the kcat values for NADase and ARTase activities. These observations imply that the side chain of the amino acid position at 338 is required for maintenance of the NAD+-binding site and catalytic site in these activities.
Bell and Eisenberg (7) reported that the S-T-S motif in DT binds to either the ribose or the phosphate of the AMP moiety of NAD+. Furthermore, Barth et al. (6) reported that Ser-348 in the motif S348-T-S350 in C2 toxin may play an essential role in NAD+ binding or catalysis. There is no contradiction between our results and their model. Replacement of Thr-339 or Ser-340 with alanine resulted in a significant but not severe reduction in NADase and ARTase activities. Replacement of Thr-339 and Ser-340 did not result in a drastic reduction in the Km and kcat values for NADase or ARTase, compared with that of Ser-338, suggesting that these residues do not play an important role in binding and catalytic reactions, as shown in Fig. 3.
NADase and ARTase activities catalyze the cleavage of the N-glycosidic bond of NAD+; the former transfers the ADP-ribose moiety to water, and the latter transfers the moiety to actin. Thus, it seems that Arg-298, Ser-338 and Glu-380, required for these activities in ia, play an important role in the cleavage of the N-glycosidic bond of NAD+ and that Glu-378, required for ARTase activity but not for NADase activity, is essential for the transfer of the ADP-ribose moiety to actin.
From these observations, changes in ARTase activity as a result of the substitution of various amino acid residues in ia were on the whole similar to those in cytotoxic and lethal activities, except for changes in these activities in R295H. It therefore is likely that the cytotoxicity and lethality of iota-toxin are closely related to the ARTase activity of ia.
ACKNOWLEDGMENTS
We thank Keiko Yamamoto and Akiko Maeda for competent technical assistance.
This research was supported in part by a grant from the Ministry of Education, Science, and Culture of Japan.
REFERENCES
- 1.Aktories K. Clostridial ADP-ribosyltransferases—modification of low molecular weight GTP-binding proteins and of actin by clostridial toxins. Med Microbiol Immunol. 1990;179:123–136. doi: 10.1007/BF00202390. [DOI] [PubMed] [Google Scholar]
- 2.Aktories K. Clostridial ADP-ribosylating toxins: effects on ATP and GTP-binding proteins. Mol Cell Biochem. 1994;138:167–176. doi: 10.1007/BF00928459. [DOI] [PubMed] [Google Scholar]
- 3.Aktories K, Wegner A. ADP-ribosylation of actin by clostridial toxins. J Cell Biol. 1989;109:1385–1387. doi: 10.1083/jcb.109.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aktories K, Barmann M, Ohishi I, Tsuyama S, Jakobs K H, Habermann E. Botulinum C2 toxin ADP-ribosylates actin. Nature. 1986;322:390–392. doi: 10.1038/322390a0. [DOI] [PubMed] [Google Scholar]
- 5.Aktories K, Rosener S, Blaschke U, Chhatwal G S. Botulinum ADP-ribosyltransferase C3. Purification of the enzyme and characterization of the ADP-ribosylation reaction in platelet membranes. Eur J Biochem. 1988;172:445–450. doi: 10.1111/j.1432-1033.1988.tb13908.x. [DOI] [PubMed] [Google Scholar]
- 6.Barth H, Preiss J C, Hofmann F, Aktories K. Characterization of the catalytic site of the ADP-ribosyltransferase Clostridium botulinum C2 toxin by site-directed mutagenesis. J Biol Chem. 1998;273:29506–29511. doi: 10.1074/jbc.273.45.29506. [DOI] [PubMed] [Google Scholar]
- 7.Bell C E, Eisenberg D. Crystal structure of nucleotide-free diphtheria toxin. Biochemistry. 1996;35:1137–1149. doi: 10.1021/bi962214s. [DOI] [PubMed] [Google Scholar]
- 8.Borriello S P, Carman R J. Association of iota-like toxin and Clostridium spiroforme with both spontaneous and antibiotic-associated diarrhea and colitis in rabbits. J Clin Microbiol. 1983;17:414–418. doi: 10.1128/jcm.17.3.414-418.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carman R J, Borriello S P. Observation on an association between Clostridium spiroforme and Clostridium perfringens type E iota enterotoxaemia in rabbits. Eur J Chemother Antibiol. 1982;2:143–144. [Google Scholar]
- 10.Cieplak W, Mead D J, Messer R J, Grant C C R. Site-directed mutagenic alteration of potential active-site residues of the A subunit of Escherichia coli heat-labile enterotoxin. Evidence for a catalytic role for glutamic acid 112. J Biol Chem. 1995;270:30545–30550. doi: 10.1074/jbc.270.51.30545. [DOI] [PubMed] [Google Scholar]
- 11.Collier R J. Diphtheria toxin: mode of action and structure. Bacteriol Rev. 1975;39:54–85. doi: 10.1128/br.39.1.54-85.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Considine R V, Simpson L L. Cellular and molecular actions of binary toxins possessing ADP-ribosyltransferase activity. Toxicon. 1991;29:913–936. doi: 10.1016/0041-0101(91)90076-4. [DOI] [PubMed] [Google Scholar]
- 13.Damme J, Jung M, Hofmann F, Just I, Vandekerckhove J, Aktories K. Analysis of the catalytic site of the actin ADP-ribosylating Clostridium perfringens iota toxin. FEBS Lett. 1996;380:291–295. doi: 10.1016/0014-5793(96)00052-x. [DOI] [PubMed] [Google Scholar]
- 14.Domenighini M, Rappuoli R. Three conserved consensus sequences identify the NAD-binding site of ADP-ribosylating enzymes, expressed by eukaryotes, bacteria and T-even bacteriophages. Mol Microbiol. 1996;21:667–674. doi: 10.1046/j.1365-2958.1996.321396.x. [DOI] [PubMed] [Google Scholar]
- 15.Domenighini M, Montecucco C, Ripka W C, Rappuoli R. Computer modeling of the NAD binding site of ADP-ribosylating toxins: active-site structure and mechanism of NAD binding. Mol Microbiol. 1991;5:23–31. doi: 10.1111/j.1365-2958.1991.tb01822.x. [DOI] [PubMed] [Google Scholar]
- 16.Eaton P, Fernie D S. Enterotoxaemia involving Clostridium perfringens iota toxin in a hysterectomy-derived rabbit colony. Lab Anim. 1980;14:347–351. doi: 10.1258/002367780781071049. [DOI] [PubMed] [Google Scholar]
- 17.Fernie D S, Knights J M, Thomson R O, Carman R J. Rabbit enterotoxaemia: purification and preliminary characterization of a toxin produced by Clostridium spiroforme. FEMS Microbiol Lett. 1984;21:207–211. [Google Scholar]
- 18.Hanes C S. CLXVII. Studies on plant amylases. I. The effect of starch concentration upon the velocity of hydrolysis by the amylase of germinated barley. Biochem J. 1932;26:1406–1421. doi: 10.1042/bj0261406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hara N, Tsuchiya M, Shimoyama M. Glutamic acid 207 in rodent T-cell RT6 antigens is essential for arginine-specific ADP-ribosylation. J Biol Chem. 1996;271:29552–29555. doi: 10.1074/jbc.271.47.29552. [DOI] [PubMed] [Google Scholar]
- 20.Holmes H T, Sonn R J, Patton N M. Isolation of Clostridium spiroforme from rabbits. Lab Anim Sci. 1988;38:167–168. [PubMed] [Google Scholar]
- 21.Hunter S E C, Brown J E, Oyston P C F, Sakurai J, Titball R W. Molecular genetic analysis of beta-toxin of Clostridium perfringens reveals sequence homology with alpha-toxin, gamma-toxin, and leukocidin of Staphylococcus aureus. Infect Immun. 1993;61:3958–3965. doi: 10.1128/iai.61.9.3958-3965.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaslow H R, Lim L K, Moss J, Lesikar D D. Structure-activity analysis of the activation of pertussis toxin. Biochemistry. 1987;26:123–127. doi: 10.1021/bi00375a018. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Liu S, Kulich S M, Barbieri J T. Identification of glutamic acid 381 as a candidate active site residue of Pseudomonas aeruginosa exoenzyme S. Biochemistry. 1996;35:2754–2758. doi: 10.1021/bi952340g. [DOI] [PubMed] [Google Scholar]
- 25.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 26.McDonel J L. Toxins of Clostridium perfringens types A, B, C, D, and E. In: Dorner F, Drews J, editors. Pharmacology of bacterial toxins. Oxford, England: Pergamon Press; 1986. pp. 477–517. [Google Scholar]
- 27.Nagahama M, Iida H, Nishioka E, Okamoto K, Sakurai J. Roles of the carboxy-terminal region of Clostridium perfringens alpha toxin. FEMS Microbiol Lett. 1994;120:297–302. doi: 10.1111/j.1574-6968.1994.tb07049.x. [DOI] [PubMed] [Google Scholar]
- 28.Nagahama M, Okagawa Y, Nakayama T, Nishioka E, Sakurai J. Site-directed mutagenesis of histidine residues in Clostridium perfringens alpha-toxin. J Bacteriol. 1995;177:1179–1185. doi: 10.1128/jb.177.5.1179-1185.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagahama M, Nakayama T, Michiue K, Sakurai J. Site-specific mutagenesis of Clostridium perfringens alpha-toxin: replacement of Asp-56, Asp-130, or Glu-152 causes loss of enzymatic and hemolytic activities. Infect Immun. 1997;65:3489–3492. doi: 10.1128/iai.65.8.3489-3492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perelle S, Gibert M, Boquet P, Popoff M R. Characterization of Clostridium perfringens iota-toxin genes and expression in Escherichia coli. Infect Immun. 1993;61:5147–5156. doi: 10.1128/iai.61.12.5147-5156.1993. . (Author's correction, 63:4967, 1995.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perelle S, Domenighini M, Popoff M R. Evidence that Arg-295, Glu-378, and Glu-380 are active-site residues of the ADP-ribosyltransferase activity of iota toxin. FEBS Lett. 1996;395:191–194. doi: 10.1016/0014-5793(96)01035-6. [DOI] [PubMed] [Google Scholar]
- 32.Popoff M R, Boquet P. Clostridium spiroforme toxin is a binary toxin which ADP-ribosylates cellular actin. Biochem Biophys Res Commun. 1988;152:1361–1368. doi: 10.1016/s0006-291x(88)80435-2. [DOI] [PubMed] [Google Scholar]
- 33.Radke J, Pederson K J, Barbieri J T. Pseudomonas aeruginosa exoenzyme S is a biglutamic acid ADP-ribosyltransferase. Infect Immun. 1999;67:1508–1510. doi: 10.1128/iai.67.3.1508-1510.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reuner K H, Presek P, Boschek C B, Aktories K. Botulinum C2 toxin ADP-ribosylates actin and disorganizes the microfilament network in intact cells. Eur J Cell Biol. 1987;43:134–140. [PubMed] [Google Scholar]
- 35.Sakurai J. Toxins of Clostridium perfringens. Rev Med Microbiol. 1995;6:175–185. [Google Scholar]
- 36.Sakurai J, Kobayashi K. Lethal and dermonecrotic activities of Clostridium perfringens iota toxin: biological activities induced by cooperation of two nonlinked components. Microbiol Immunol. 1995;39:249–253. doi: 10.1111/j.1348-0421.1995.tb02197.x. [DOI] [PubMed] [Google Scholar]
- 37.Sakurai J, Nagahama M, Ochi S. Major toxins of Clostridium perfringens. J Toxicol-Toxin Rev. 1997;16:195–214. [Google Scholar]
- 38.Schering B, Barmann M, Chhatwal G S, Geipel U, Aktories K. ADP-ribosylation of skeletal muscle and non-muscle actin by Clostridium perfringens iota toxin. Eur J Biochem. 1988;171:225–229. doi: 10.1111/j.1432-1033.1988.tb13780.x. [DOI] [PubMed] [Google Scholar]
- 39.Simonen M, Paya I. Protein secretion in Bacillus species. Microbiol Rev. 1993;57:109–137. doi: 10.1128/mr.57.1.109-137.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simpson L L, Stiles B G, Zepeda H H, Wilkins T D. Molecular basis for the pathological actions of Clostridium perfringens iota toxin. Infect Immun. 1987;55:118–122. doi: 10.1128/iai.55.1.118-122.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sixma T K, Kalk K H, van Santen B A, Dauter Z, Kingma J, Witholt B, Hol W G. Refined structure of Escherichia coli heat-labile enterotoxin, a close relative of cholera toxin. J Mol Biol. 1993;230:890–918. doi: 10.1006/jmbi.1993.1209. [DOI] [PubMed] [Google Scholar]
- 42.Spangler B D. Structure and function of cholera toxin and the related Escherichia coli heat-labile enterotoxin. Microbiol Rev. 1992;56:622–647. doi: 10.1128/mr.56.4.622-647.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stiles B G, Wilkins T D. Purification and characterization of Clostridium perfringens iota toxin: dependence on two nonlinked proteins for biological activity. Infect Immun. 1986;54:683–688. doi: 10.1128/iai.54.3.683-688.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugai M, Enomoto T, Hashimoto K, Matsumoto K, Matsuo Y, Ohgai H, Hong Y M, Inoue S, Yoshikawa K, Suginaka H. A novel epidermal cell differentiation inhibitor (EDIN): purification and characterization from Staphylococcus aureus. Biochem Biophys Res Commun. 1990;173:92–98. doi: 10.1016/s0006-291x(05)81026-5. [DOI] [PubMed] [Google Scholar]
- 45.Wick M J, Frank D W, Storey D G, Iglewski B H. Structure, function, and regulation of Pseudomonas aeruginosa exotoxin A. Annu Rev Microbiol. 1990;44:335–363. doi: 10.1146/annurev.mi.44.100190.002003. [DOI] [PubMed] [Google Scholar]
- 46.Xu Y, Barbarcon-Finck V, Barbieri J T. Role of histidine 35 of the S1 subunit of pertussis toxin in the ADP-ribosylation of transducin. J Biol Chem. 1994;269:9993–9999. [PubMed] [Google Scholar]