Abstract
DNA damage is unavoidable, and organisms across the evolutionary spectrum possess DNA repair pathways that are critical for cell viability and genomic stability. To understand the role of base excision repair (BER) in protecting eukaryotic cells against alkylating agents, we generated Schizosaccharomyces pombe strains mutant for the mag1 3-methyladenine DNA glycosylase gene. We report that S. pombe mag1 mutants have only a slightly increased sensitivity to methylation damage, suggesting that Mag1-initiated BER plays a surprisingly minor role in alkylation resistance in this organism. We go on to show that other DNA repair pathways play a larger role than BER in alkylation resistance. Mutations in genes involved in nucleotide excision repair (rad13) and recombinational repair (rhp51) are much more alkylation sensitive than mag1 mutants. In addition, S. pombe mutant for the flap endonuclease rad2 gene, whose precise function in DNA repair is unclear, were also more alkylation sensitive than mag1 mutants. Further, mag1 and rad13 interact synergistically for alkylation resistance, and mag1 and rhp51 display a surprisingly complex genetic interaction. A model for the role of BER in the generation of alkylation-induced DNA strand breaks in S. pombe is discussed.
DNA damage emanates from the inherent chemical instability of nucleic acids, from errors made by DNA polymerase during DNA replication, and from exposure to DNA-damaging agents present in the environment or produced by certain endogenous cellular processes (reviewed in reference 15). All organisms possess a panel of DNA repair mechanisms to repair damaged DNA. DNA excision repair pathways recognize and remove damaged segments from one DNA strand and then resynthesize new DNA, using the opposing undamaged strand as a template. Excision repair includes base excision repair (BER) and nucleotide excision repair (NER). An alternative approach to handling damaged DNA is by recombinational repair. These DNA repair pathways have been best characterized in Escherichia coli, but analogous pathways have been found in all organisms examined to date (15).
BER initiation occurs by the action of DNA glycosylases that recognize specific types of damaged or abnormal DNA bases and cleave the glycosylic bond linking the base to the sugar-phosphate backbone. DNA glycosylases recognize bases such as uracil, deaminated adenine (hypoxanthine), and certain alkylated and oxidized purines and pyrimidines (reviewed in references 10, 15, 21, 31, and 47). Releasing a damaged base from the DNA produces an apurinic/apyrimidinic (AP) site, and it is worth noting that AP sites are themselves a form of DNA damage. In addition to being generated by DNA glycosylases, AP sites can be formed spontaneously. AP endonucleases (which cleave 5′ to the AP site) or AP lyases (which cleave 3′ to the AP site) cleave the DNA backbone adjacent to the AP site. AP endonuclease produces a 5′-deoxyribose phosphate moiety, which must be removed to allow subsequent DNA ligation; removal occurs by the action of deoxyribose phosphodiesterase or Flap endonuclease 1 (FEN1). Cleavage by AP lyase produces a 3′ phosphate that cannot prime the new DNA synthesis required for repair. 3′-Phosphodiesterase enzymes remove the 3′-phosphate, generating a 3′-hydroxyl primer. Following modification of the appropriate DNA terminus, DNA polymerase synthesizes a patch of new DNA that can be as small as 1 base or as large as 13 bases; such DNA repair synthesis is followed by DNA ligation (reviewed in references 21, 31, and 47).
Like BER, NER requires several enzymatic steps. Although the general strategy for NER has been highly conserved, the reaction mechanism is more complex in eukaryotes than in prokaryotes (reviewed in references 15, 31, and 47). To initiate NER, the DNA sugar-phosphate backbone is incised both 3′ and 5′ to the damaged base(s). In E. coli this reaction requires 3 proteins (UvrA, -B, and -C), whereas in Saccharomyces cerevisiae and mammalian cells it requires the concerted action of at least 13 proteins (15, 40). NER was originally thought to repair exclusively bulky DNA lesions and DNA cross-links, known to distort the DNA helix. However, in vivo studies revealed a role for NER in the repair of methylated DNA base lesions that do not cause major helical distortions and in providing cellular resistance to simple methylating agents (38, 48, 49). Indeed, biochemical studies confirmed that subtle types of DNA damage can be substrates for NER, including thymine glycols, 8-oxoguanine, O4-ethylthymine, O4-methylthymine, O6-methylguanine, AP sites, and N6-methyladenine (17, 22, 26, 36, 38, 46). Thus, NER may play a role in alkylation resistance larger than was originally thought.
In addition to BER and NER, Schizosaccharomyces pombe has an additional DNA excision repair pathway initiated by the enzyme UV damage endonuclease (UVDE) (reviewed in reference 51). UVDE cleaves 5′ to UV photoproducts as well as to other aberrant DNA bases, including cisplatin-cross-linked diadducts, uracil, dihydrouracil, AP sites, and a variety of mismatched normal bases (3, 24, 25). Thus, UVDE-mediated excision repair has a wide substrate range and is likely to be important for resistance to a number of DNA-damaging agents. Although the downstream enzymatic components of the S. pombe UVDE-mediated excision repair are unidentified, genetic epistasis analysis suggests the involvement of the products of rad2 (encoding the S. pombe FEN1 homologue), rad18 (an essential gene that plays a role in UV and γ-ray resistance), and rhp51 (encoding a RecA homologue that plays an essential role in recombination) (reviewed in reference 51).
Strand breaks in DNA are repaired by recombinational repair (RR) mechanisms. Less is known about RR enzymatic mechanisms than about to the excision repair pathways described above, although recent advances have increased our understanding at the molecular genetic level. RR genes have been identified in S. cerevisiae, and their homologues have been identified in both S. pombe and mammals (reviewed in references 15 and 23). RR repair of DNA strand breaks proceeds by either homologous recombination or illegitimate recombination. In S. cerevisiae, homologous recombination predominates for the repair of DNA strand breaks and requires the products of the RAD52 epistasis group, which includes the RAD50, -51, -52, -54, -55, and -57 genes. Mutations in any one of these S. cerevisiae genes produces a severe defect in homologous recombination accompanied by sensitivity to the lethal effects of γ rays (an agent which produces both single- and double-strand breaks in DNA), reduced mitotic and meiotic recombination, and defects in mating-type switching (16). In addition to γ rays, S. cerevisiae mutant in genes belonging to the RAD52 group are very sensitive to the killing effects of the simple methylating agent methyl methanesulfonate (MMS). This observation lead to dubbing MMS a radiomimetic, and it has been shown that MMS can induce DNA strand breaks, in addition to alkylated bases (14, 39, 42, 45).
One of the central components of S. cerevisiae RR is the Rad51 protein (reviewed in reference 4). S. cerevisiae Rad51 homologues are found in S. pombe, chickens, and mammals, and these proteins, together with S. cerevisiae Rad51, are all homologues of the E. coli recombination protein RecA. Biochemical studies show that like RecA, S. cerevisiae and human Rad51 form nucleoprotein filaments in the presence of DNA and promote DNA strand transfer and annealing of cDNA. For S. cerevisiae and possibly other eukaryotes, the Rad52, Rad55, and Rad57 proteins stimulate such Rad51 activities.
In an effort to develop S. pombe as a model organism for the study of cellular responses to alkylating agents, we cloned an S. pombe cDNA encoding a 3-methyladenine (3MeA) DNA glycosylase, Mag1 (32). This cDNA was cloned by its ability to suppress the alkylation-sensitive phenotype of 3MeA DNA glycosylase-deficient E. coli. The S. pombe Mag1 3MeA DNA glycosylase turned out to be homologous to a certain group of 3MeA DNA glycosylases, namely, E. coli AlkA, S. cerevisiae Mag, and Bacillus subtilis AlkA. Structural studies indicate that E. coli AlkA (and most likely its homologues) is a member of the helix-hairpin-helix family of DNA glycosylases (28, 50). Members of this family share a common three-dimensional structure and catalytic mechanism. Although S. pombe mag1 has several features in common with the E. coli and S. cerevisiae 3MeA DNA glycosylase genes, in contrast to E. coli alkA and S. cerevisiae MAG, S. pombe mag1 expression is not inducible by MMS treatment (32).
Here we set out to determine the biological role of S. pombe Mag1 and its contribution to alkylation resistance. This led us to determine the relative roles of BER, NER, and RR in providing S. pombe with resistance to alkylating agents. We determined the alkylation sensitivity of S. pombe mag1 mutants compared to strains deficient in NER, RR, or FEN1. We further determined the interaction between BER, NER, and RR repair pathways for providing alkylation sensitivity.
MATERIALS AND METHODS
S. pombe strains and growth conditions.
The genotypes of strains used in this study are listed in Table 1. S. pombe were routinely grown aerobically in rich yeast extract medium supplemented with adenine (2, 33). S. pombe strains disrupted at the rhp51, rad13, or rad2 locus by ura4 insertion were generated in the laboratory of A. M. Carr (Sussex University, Sussex, United Kingdom) and kindly given to us by T. Enoch (Harvard Medical School, Boston, Mass.) (1, 35); all three strains were disrupted with the ura4 gene and able to grow in the absence of uracil. ura4-D18 leu1-32 S. pombe (designated strain TE236) obtained from T. Enoch) was used as the wild type for DNA repair. S. pombe was cultured using standard techniques (2, 33).
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| TE236 | leu1-32 ura4-D18 | Derived from 972 h− |
| AM006 | leu1-32 ura4-D18 mag1::ura4+ | This study |
| TE793 | leu1-32 ura4-D18 ade6-704 rad2::ura4+ | 35 |
| TE570 | leu1-32 ura4-D18 ade6-704 rad3::ura4+ | 1 |
| TE791 | leu1-32 ura4-D18 ade6-704 rad13::ura4+ | 35 |
| TE792 | leu1-32 ura4-D18 ade6-704 rhp51::ura4+ | 35 |
| AM008 | leu1-32 ura4-D18 ade6-704 mag1::ura4+ rad13::ura4+ | This study |
| AM013 | leu1-32 ura4-D18 ade6-704 mag1::ura4+ rhp51::ura4+ | This study |
| AM019 | leu1-32 ura4-D18 ade6-704 rad13::ura4+ rhp51::ura4+ | This study |
| AM022 | leu1-32 ura4-D18 ade6-704 mag1::ura4+ rad13::ura4+ rhp51::ura4+ | This study |
Construction of mutant S. pombe strains.
A 1.9-kb HindIII fragment containing the S. pombe ura4 gene was isolated from the plasmid pUC8-ura4 (generously provided by C. Hoffman, Boston College, Newton, Mass.), blunt ended with the Klenow fragment of E. coli polymerase I and inserted into the unique EcoRV site within the mag1 cDNA (32). The mag1::ura4 DNA fragment was excised from plasmid by BcgI and BclI digestion, separated from vector by gel electrophoresis, purified using a Qiax DNA purification kit (Qiagen), and used to transform uraD-18 leu1-32 S. pombe. Resulting clones were selected for growth in the absence of uracil; the Ura+ phenotype of individual clones was confirmed by serially plating twice onto nonselective rich medium and then plating onto medium lacking uracil and medium containing 5-fluoroorotic acid (5FOA). Clones that were Ura+ and 5FOA sensitive were selected for Southern analysis (Fig. 1). One mag1::ura4 clone (clone 4 in Fig. 1B) was backcrossed three times with ura4-D18 leu1-32 S. pombe, and the resulting mag1 ura4D-18 leu1-32 S. pombe cells were characterized for sensitivity to the methylating agent MMS. S. pombe strains with ura4 disruptions in multiple DNA repair genes were generated by mating strains of opposite mating type on malt medium agar plates. Resulting progeny were analyzed for growth in the absence of uracil and genotypes were confirmed by Southern analysis.
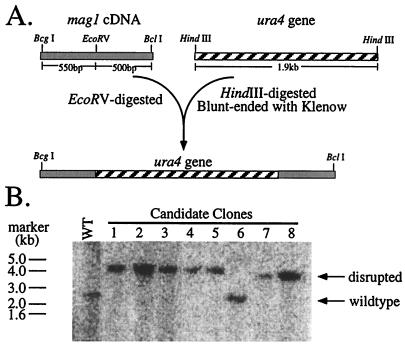
FIG. 1.
Disruption of mag1 in S. pombe. (A) The mag1::ura4 disruption construct was made as follows. A blunt-ended DNA fragment containing the S. pombe ura4 gene (hatched box) was ligated into a unique EcoRV site in the mag1 cDNA (shaded box) as diagrammed. The BcgI/BclI fragment was isolated, purified, and used to transform ura4D-18 leu1-32 S. pombe. (B) Genomic DNA from 5FOA-resistant S. pombe clones was isolated, digested with HindIII, and analyzed by Southern using radiolabeled mag1 cDNA as a probe. The positions and molecular sizes of marker DNA are indicated.
DNA isolation and Southern hybridization.
Genomic DNA isolations were performed as described elsewhere (2). Genomic DNA was digested with the indicated restriction enzymes and size fractionated on 1% agarose gels, transferred to a nylon membrane (Nytran; Schleicher & Schuell), and hybridized to mag1 cDNA labeled with 32P by the random primer method (NEBlot; New England Biolabs).
Generation of plasmid for expression of S. cerevisiae APN1 in S. pombe.
Two primers (5′-CGC GCT CGA GCC TTC GAC ACC TAG CTT T-3′ and 5′-CGC GGG GCC CTA CGT ACG TTG AGA TAA T-3′) were used to amplify the S. cerevisiae APN1 open reading frame (ORF) and introduce ApaI and XhoI restriction sites 5′ and 3′, respectively, to the gene. Insertion into the ApaI and XhoI sites within the S. pombe expression vector pREP3-HA (a gift from Dieter Wolf, Harvard School of Public Health, Boston, Mass., and described in reference 13) resulted in a plasmid (pREP-APN1sc) that would express the S. cerevisiae APN1 gene as a hemagglutinin fusion protein under the control of the thiamine-repressible nmt1 promoter in S. pombe. pREP-APN1sc and or the empty vector were introduced into S. pombe strains by using electroporation as described elsewhere (2). Transformants were isolated and maintained in minimal medium containing uracil, adenine, and thiamine (5 μg/ml).
S. pombe colony formation in MMS.
Colony-forming ability of S. pombe was determined after treatment with MMS, either as a chronic dose in solid medium or as an acute dose in liquid culture. For chronic exposure, serial dilutions of a logarithmically growing S. pombe (5 × 106 to 2 × 107 cells/ml) were plated on MMS-containing solid media, and colonies were scored after 3 to 4 days at 30°C. For acute exposure, the indicated doses of MMS were added to logarithmically growing S. pombe. Aliquots were removed at the indicated time, serially diluted, and spread on solid media. Colony formation was determined after 3 to 5 days of growth at 30°C. In general, MMS doses were selected such that colonies could form in the most sensitive strain and toxicity could be detected in the most resistant strain.
MMS gradient survival assay.
Gradient plates, which contain an increasing concentration of MMS across the width of a square petri dish, were made by adding S. pombe medium containing agar in two steps in a manner previously described (11). The medium used was either rich yeast extract medium (for S. pombe strains not harboring a plasmid) or essential minimal medium supplemented with uracil and adenine as described elsewhere (2) (for strains harboring a plasmid). In the initial step, molten MMS-containing agar was allowed to solidify as a wedge by propping up one edge of the square petri dish approximately 0.5 cm. Following solidification of the first layer, the petri dish was laid flat, and a second layer of molten agar was overlaid and allowed to solidify. Following this second solidification, plates were dried for 5 to 10 min at 55°C with the lids off. The edge of a sterile glass slide was used to transfer stationary-phase S. pombe mixed with molten agar from a sterile surface to the MMS gradient plate. In this manner, cells were spread uniformly across the gradient and between samples. MMS sensitivity was determined after allowing 3 to 4 days growth at 30°C.
RESULTS
Disruption of the S. pombe mag1 locus.
To study the role of BER in protecting S. pombe against the simple methylating agent MMS, we disrupted the mag1 3MeA DNA glycosylase gene by insertion of the ura4 gene. The ura4 disruption was positioned within the coding region at a predicted cleft-like structure containing the putative Mag1 active site. Amino acids encoding the cleft are highly conserved among E. coli AlkA and its homologues; furthermore, in AlkA, structural and biochemical studies have confirmed the existence of the cleft and the importance of this region for catalytic activity (28, 50). Due to Mag1's homology to AlkA, it is very likely that the two enzymes share the same biochemical determinants. In support of this hypothesis, site-directed mutagenesis of a codon encoding an aspartate residue (Asp170) predicted to be essential for catalysis abolished the ability of S. pombe mag1 to complement the MMS sensitivity of 3MeA DNA glycosylase-deficient S. cerevisiae (data not shown); the equivalent aspartate in AlkA (Asp238) is critical for catalytic activity (28, 50). Further, unlike the parental plasmid containing S. pombe mag1, a plasmid containing the ura4-disrupted mag1 cDNA did not provide MMS resistance to 3MeA DNA glycosylase-deficient E. coli (data not shown). Thus, the biological activity of mag1 was indeed disrupted by the ura4 insertion.
Following transformation with the disruption construct, S. pombe clones that consistently exhibited the Ura+ phenotype were screened for the presence of the mag1::ura4 allele by Southern analysis. The mag1 cDNA hybridized to a HindIII fragment of approximately 2.5 kb in size in wild-type S. pombe. Disruption of mag1 with ura4 is predicted to result in a HindIII fragment approximately 4.5 kb in size. Seven of eight clones tested had the mutated mag1 allele and had lost the wild-type allele (Fig. 1). In an effort to determine the effect of the mag1 disruption of 3MeA and 7-methylguanine DNA glycosylase activity in S. pombe, repeated attempts were made to measure glycosylase activity in wild-type and mag1 S. pombe cell extracts. However, such DNA glycosylase activity was undetectable even in wild-type S. pombe extracts, preventing us from determining the effect of mag1 disruption on activity. We previously showed that the gene transcript is constitutively expressed in S. pombe, suggesting that our failure to detect Mag1 activity reflects a problem with in vitro reaction conditions; note that S. pombe Mag1 activity can be measured in extracts from mag1-expressing E. coli, indicating that this gene does indeed encode a 3MeA DNA glycosylase (32).
Sensitivity of mag1-deficient S. pombe to MMS.
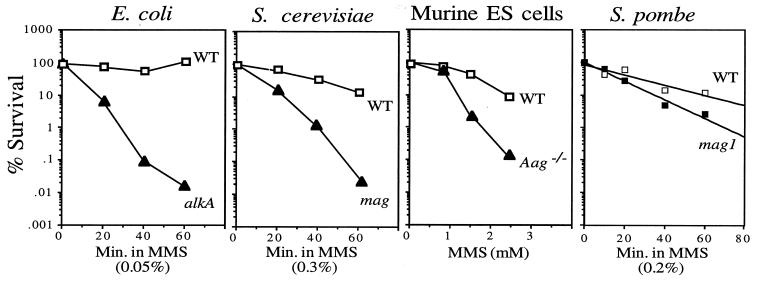
In all organisms that we previously tested, inactivation of 3MeA DNA glycosylase genes dramatically increased MMS sensitivity (Fig. 2). However, to our surprise, mag1 S. pombe tested for sensitivity to the methylating agent MMS displayed very little MMS sensitivity (Fig. 2). Indeed, when tested under chronic exposure to MMS in gradient plates (11), mag1 S. pombe appeared no more MMS sensitive than wild-type cells (Fig. 3B). To reconcile our findings, we reasoned that DNA repair pathways other than BER might play a more prominent role in S. pombe alkylation resistance.
FIG. 2.
MMS sensitivity of 3MeA DNA glycosylase-deficient cells. The colony-forming ability of cells from various organisms following MMS treatment was determined. For E. coli, S. cerevisiae, and murine embryonic stem cells, doses and times are as indicated and data shown are adapted from references 8, 12, and 32. For S. pombe, 0.2% MMS was added to logarithmically growing wild-type (WT; TE236) or mag1 (AM006) S. pombe. Aliquots were collected at the indicated times and scored for colony-forming ability as described in Materials and Methods. The data shown for S. pombe are the mean from five independent experiments.
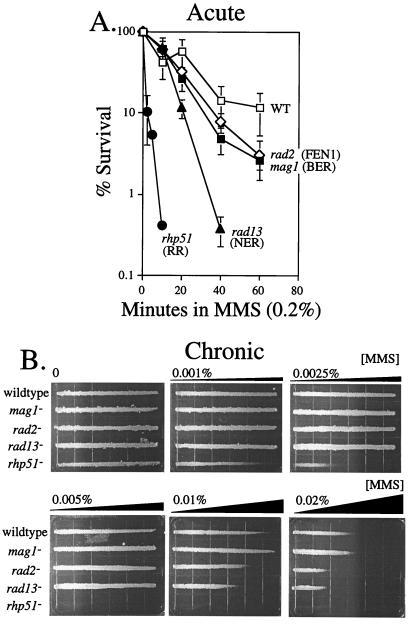
FIG. 3.
MMS sensitivity of S. pombe deficient in DNA repair. (A) MMS (0.2%) was added to logarithmically growing S. pombe. Aliquots were collected at the indicated times and scored for colony-forming ability as described in the Materials and Methods. The strains analyzed were wild-type (TE236), mag1 (AM006), rad2 (TE793), rad13 (TE791), and rhp51 (TE792) S. pombe. The data shown are the mean and standard error from three experiments for rad2, rad13, and rhp51 and five experiments for wild type and mag1. (B) MMS gradient plate analysis was performed on the indicated strains as described in Materials and Methods. The concentration of MMS indicated reflects the concentration of the bottom layer of agar and corresponds to the highest concentration of the gradient.
MMS sensitivity of S. pombe deficient in RR, NER, or FEN1.
Since Mag1-initiated BER did not appear to play a major role in alkylation resistance, we set out to determine which DNA repair pathways do contribute to alkylation resistance in S. pombe. The MMS sensitivity of various DNA repair-deficient S. pombe strains was determined. rad13, rad2, and rhp51 strains were chosen based on their known participation in pathways other than BER. The rad13 gene was originally cloned by virtue of its ability to reverse the UV-sensitive phenotype of rad13 S. pombe (7); its product is homologous to mammalian XPG/ERCC5 and to S. cerevisiae Rad2 (unrelated to S. pombe Rad2), both of which are required for the initiation of NER and have structure-specific 3′-exinuclease activities (18, 19). Similarly S. pombe rad2 was cloned by its ability to reverse the UV sensitivity of rad2 S. pombe and encodes a homologue of S. cerevisiae RAD27 and mammalian FEN1 (35). FEN1 is another structure-specific DNA endonuclease, and its substrates include “flaps” of DNA that result from DNA polymerase-mediated displacement of single-stranded DNA 3′ to the newly synthesized DNA. Such structures are thought to occur during lagging-strand DNA synthesis and possibly during BER and UVDE-mediated excision repair (30, 51). In support of a role for S. pombe Rad2 in DNA repair, rad2 strains have a reduced ability to repair cyclobutane pyrimidine dimers and 6-4 photoproducts and presumably accounts for the UV-sensitive phenotype (35). The S. pombe rhp51 gene was cloned by low-stringency hybridization to S. cerevisiae RAD51 (34); as mentioned, Rad51 from both yeasts are RecA homologues and are thus involved in DNA strand exchange during RR (4).
The MMS sensitivity of wild-type, mag1, rhp51 (RR-deficient), rad13 (NER-deficient), and rad2 (FEN1-deficient) S. pombe single mutants was determined by two different methods. The first method measured colony-forming ability following MMS exposure for up to 1 h (acute exposure) (Fig. 3A), and the second method measured growth of S. pombe along an MMS gradient plate (chronic exposure) (Fig. 3B). As previously indicated, 3MeA DNA glycosylase deficiency alone (mag1) had a small but measurable effect on the colony-forming ability of S. pombe given an acute dose of MMS. In contrast, the survival of mag1 S. pombe given a chronic dose of MMS from an MMS gradient plate was the same as for the wild type (Fig. 3). In contrast, FEN1-deficient (rad2) S. pombe was sensitive to MMS in both assays, although sensitivity was greater in the MMS gradient plates (Fig. 3). NER-deficient (rad13) S. pombe was considerably more sensitive to MMS in both assays and, surprisingly, much more sensitive to MMS than mag1 S. pombe. NER and FEN1 deficiencies had quantitatively similar effects on sensitivity to chronic MMS exposure, although they are most likely involved in different DNA repair pathways. RR-deficient (rhp51) S. pombe was profoundly sensitive to both acute and chronic MMS exposures (Fig. 3).
MMS sensitivity of S. pombe deficient in both Mag1 and NER.
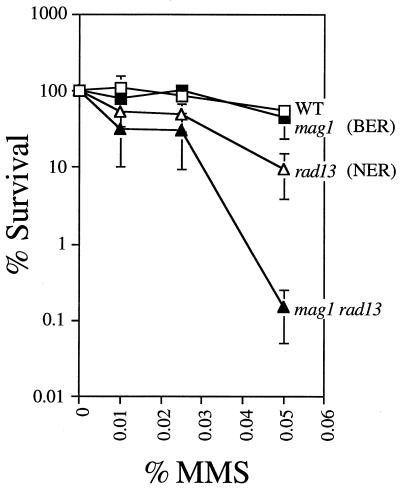
To determine whether Mag1-initiated BER and Rad13-initiated NER play redundant roles in DNA alkylation repair, mag1 rad13 S. pombe double mutants were made. Candidate double-mutant clones were genotyped by Southern analysis (data not shown). Using ura4 as a probe, a characteristic ura4-containing restriction fragment corresponding to each mutant allele was detected; in the double mutant, both ura4 insertion alleles were present. In the absence of NER, the mag1 mutation had a much more profound effect on MMS sensitivity than in the presence of NER (i.e., in wild-type S. pombe) (Fig. 4). Thus, rad13 mag1 S. pombe were much more sensitive to MMS compared to S. pombe mutated in rad13 alone (Fig. 4). The synergistic interaction of rad13 and mag1 for the MMS-sensitive phenotype was observed in several independent clones (data not shown), and such interaction indicates that the two repair pathways do indeed play redundant roles.
FIG. 4.
MMS sensitivity of S. pombe mutant for mag1 and NER. Logarithmically growing wild-type (WT; TE236), mag1 (AM006), rad13 (TE791), and mag1 rad13 (AM008) S. pombe strains were treated with the indicated dose of MMS for 1 h as described in Materials and Methods. Aliquots were collected and analyzed for colony-forming ability. The data shown are the mean and standard error of three independent experiments.
MMS sensitivity of S. pombe deficient in both Mag1 and RR.
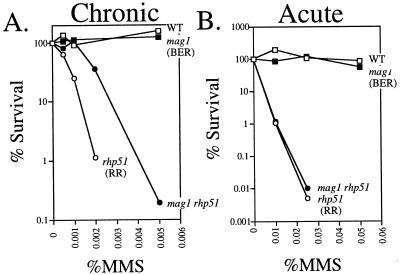
To determine whether Mag1-initiated BER and Rhp51-initiated RR play redundant roles in DNA alkylation repair, we generated a mag1 rhp51 S. pombe double mutant that turned out to display a quite unexpected phenotype. mag1 rhp51 cells were actually less MMS sensitive than the rhp51 single mutant (Fig. 5A), and this phenotype was consistent for several clones obtained from independent crosses (data not shown). However, it is worth noting that this unexpected genetic interaction between mag1 and rhp51 was observed upon chronic exposure of S. pombe to MMS (Fig. 5A) and was not observed upon acute MMS exposure; i.e., mag1 rhp51 and rhp51 S. pombe strains were similarly MMS sensitive when treated with various doses of MMS for 1 h (Fig. 5B).
FIG. 5.
MMS sensitivity of S. pombe mutant for mag1 and recombinational repair. (A) Logarithmically growing S. pombe was diluted and plated onto solid agar medium containing the indicated doses of MMS. Colony-forming ability was determined. The data are from a representative experiment. Because RR-deficient strains are hypersensitive to MMS, slight variations in dose due to the reactivity of MMS have large effects on viability. Thus, there was significant between-experiment variability in all experiments using rhp51 S. pombe and its derivative strains. The between-experiment variability made it impossible to pool results and calculate values for the mean and standard error for each data point. However, the relationship between the strains was always the same, and similar results were observed in three independent experiments. (B) Logarithmically growing S. pombe was treated with the indicated dose of MMS for 1 h as described in Materials and Methods. Aliquots were collected and analyzed for colony-forming ability. The data shown are from a representative experiment; similar results were obtained with three independent experiments. Strains analyzed were wild-type (WT; TE 236), mag1 (AM006), rhp51 (TE792), and mag1 rhp51 (AM013) S. pombe.
During BER, a single-stranded DNA break is formed by AP endonuclease at the abasic site produced by DNA glycosylase. Classic studies have shown that the number of MMS-induced DNA strand breaks increases for some time after MMS removal (37, 45); it has been suggested that glycosylases and AP endonucleases are responsible for some fraction of these MMS-induced DNA strand breaks. If the BER enzymes required for the repair of such single-strand DNA breaks were limiting, an accumulation of DNA strand breaks could result. Alternatively, AP sites could give rise to DNA strand breaks due to their inherent chemical instability or their ability to block DNA replication and thus indirectly result in strand breaks. Since, the major role of RR is to repair DNA strand breaks, we hypothesized that DNA strand breaks generated during BER could be substrates for RR, and that in the absence of RR these BER-induced DNA strand breaks could contribute to MMS-induced cytotoxicity. Thus, without Mag1-initiated BER, fewer MMS-induced DNA strand breaks may accumulate, rendering RR-deficient cells more MMS resistant as in Fig. 5A. However, why such MMS resistance is apparent only during chronic exposure remains unclear.
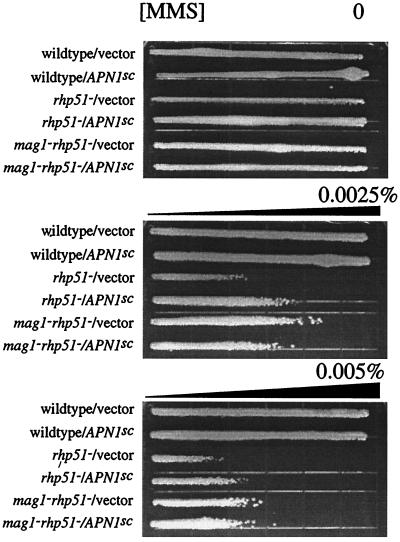
To determine whether Mag1-induced cytotoxicity (in RR-deficient cells) is due to excessive AP sites or other downstream intermediates of BER, we expressed the S. cerevisiae AP endonuclease gene, APN1 in S. pombe. Expression of APN1 in rhp51 S. pombe cells partially reversed their MMS sensitivity (Fig. 6). It is worth noting that the level of MMS resistance observed in rhp51 S. pombe expressing APN1 is similar to that observed in mag1 rhp51 S. pombe with vector. Thus, expression of APN1 appears to reverse the negative contribution of Mag1 to MMS sensitivity, suggesting that Mag1-induced cytotoxicity is due to excessive AP sites. Note that expression of APN1 in the absence of Mag1 activity had no effect on MMS sensitivity (Fig. 6).
FIG. 6.
MMS sensitivity of S. pombe overexpressing S. cerevisiae AP endonuclease. MMS gradient plate analysis was performed on wild-type, rhp51, and mag1 rhp51 S. pombe strains expressing either the pREP vector plasmid or the pREP-APNsc plasmid expressing the S. cerevisiae AP endonuclease gene APN1 as described in Materials and Methods. The MMS concentration (0.005%) reflects the concentration of the bottom layer of agar and corresponds to the highest concentration of the gradient. The strains and plasmids are as indicated. Similar results were obtained from three independent experiments using two independent transformants for each strain.
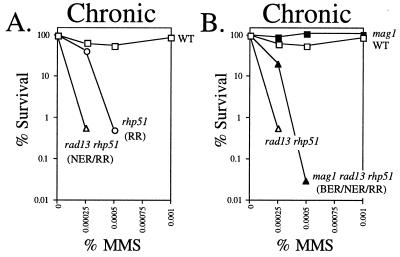
Since NER also plays a role in the repair of MMS-induced DNA damage, we postulated that MMS-induced NER intermediates such as single-strand DNA gaps might also generate RR substrates. To test this, we determined the MMS sensitivity of rad13 rhp51 double-mutant (NER and RR-deficient) and mag1 rad13 rhp51 triple-mutant (Mag1-, NER-, and RR-deficient) S. pombe. All genotypes were confirmed by Southern analysis (data not shown). It turned out that rad13 rhp51 S. pombe was in fact more MMS sensitive than the rhp51 single mutant, suggesting that RR substrates do not accumulate during NER and that the interaction between BER and RR is quite different from that between NER and RR (Fig. 7A). The additive nature of the rhp51 and rad13 MMS sensitivity confirms that rhp51 and rad13 are involved in different DNA repair pathways and suggests that NER intermediates do not normally become RR substrates. Finally, the mag1 mutation still conferred MMS resistance upon rad13 rhp51 double mutants, just at it had upon the rhp51 single mutant; thus, even in NER-deficient cells mag1 and rhp51 display their unexpected genetic interaction (Fig. 7B).
FIG. 7.
MMS sensitivity of S. pombe mutant for mag1, NER, and RR. Logarithmically growing S. pombe was diluted and plated onto solid agar medium containing the indicated doses of MMS. Colony-forming ability was determined. The data shown are from a representative experiment. Similar results were observed in three independent experiments. Strains analyzed were wild-type (WT; TE236), rhp51 (TE792), and rad13 rhp51 (AM019) (A) and wild-type (TE236), mag1 (AM006), rad13 rhp51 (AM019) and mag1 rad13 rhp51 (AM022) (B) S. pombe.
DISCUSSION
Here we demonstrate that although S. pombe employs the same range of DNA alkylation repair pathways as several other organisms, the relative importance of each pathway for providing resistance to each organism varies dramatically. In wild-type S. pombe, the Mag1 3MeA DNA glycosylase was not a major determinant of MMS sensitivity. In contrast to S. pombe, 3MeA DNA glycosylase-deficient E. coli was profoundly sensitive to MMS. MMS doses that had no effect on survival of the wild type allowed less than 0.01% survival of 3MeA DNA glycosylase-deficient E. coli (8). Similarly, in S. cerevisiae, doses of MMS that allowed greater than 10% survival in the wild type allowed less than 0.0001% survival in mag1 S. cerevisiae (9). The contribution of 3MeA DNA glycosylase activity to MMS sensitivity of mammalian cells was more modest but nevertheless significant; MMS doses that allowed approximately 10% survival of the wild type caused less than 1% survival of 3MeA DNA glycosylase-deficient Aag−/− null cells (12). Note that there was no detectable 3MeA DNA glycosylase activity in cell extracts made from Aag−/− embryonic stem cells, suggesting that the presence of another 3MeA DNA glycosylase with redundant activity is unlikely (12).
NER and RR were shown to play major roles in S. pombe MMS resistance. NER-deficient rad13 S. pombe strains are significantly MMS sensitive and RR-deficient rhp51 strains are profoundly MMS sensitive compared to the wild type. Similarly, S. cerevisiae strains mutant for NER genes are modestly MMS sensitive, whereas S. cerevisiae mutant for genes involved in RR are extremely MMS sensitive (49). In contrast, NER-deficient mammalian cells are not sensitive to MMS (20, 43). Moreover, it appears in S. pombe that BER plays a secondary role in DNA methylation repair, which is revealed in the absence of NER.
We have also shown a role for S. pombe FEN1 (Rad2) in MMS resistance. rad2 S. pombe strains were identified by virtue of their UV sensitivity. In contrast, mutants in the S. cerevisiae rad2 homologue, rth1 (also known as rad27), are not sensitive to either UV or γ irradiation, but they do display an elevated mitotic recombination, elevated spontaneous mutation, temperature sensitivity, and MMS sensitivity (41, 44). Here we show that rad2 S. pombe is moderately sensitive to MMS. The nuclease activity of FEN1 is known to be important in mammalian cells for processing of Okazaki fragments during DNA lagging-strand synthesis, for UVDE-mediated excision repair, and also for the long-patch subpathway of BER. Our data do not indicate which function of FEN1 (Rad2) is important for MMS resistance.
It is still formally possible that BER plays a significant role in providing alkylation resistance to S. pombe, initiated by glycosylases other than Mag1. In support of this notion, a predicted ORF for a second enzyme with sequence similarity to E. coli AlkA and S. pombe Mag1 has been identified in the S. pombe genome, representing a putative second S. pombe 3MeA DNA glycosylase. Additionally, two separate S. pombe ORFs bearing sequence similarity to the major AP endonucleases in E. coli, ExoIII and EndoIV, have been identified (5). Although the structural genes for BER enzymes exist in S. pombe, 3MeA DNA glycosylase activity and AP endonuclease activity were undetectable in crude S. pombe extracts, suggesting that these genes may not be expressed at very high levels (reference 29 and data not shown).
The interaction between S. pombe BER and NER was shown to be synergistic; i.e., a deficiency in both BER and NER produced much more MMS sensitivity than that predicted from an additive contribution of each repair pathways. Synergism is observed between DNA repair genes when neither DNA repair pathway is operating to its full capacity in wild-type cells. Thus, when one pathway is absent, the other can at least partially compensate, and in some cases a DNA damage-sensitive phenotype is avoided altogether (20). Synergism between S. pombe BER and NER indicates several points. First, it confirms that Mag1-initiated BER does indeed function in S. pombe, albeit in a supportive role, for the repair of DNA methylation damage. Furthermore, these data indicate that insertion of ura4 into the mag1 ORF did disrupt the biological activity of Mag1, as predicted. The synergism also indicates that while the S. pombe BER and NER pathways are distinct, they can act on the same methylated DNA lesions, most likely 3MeA (20), and that at least one of these lesions (again, most likely 3MeA) is lethal in S. pombe if left unrepaired.
Although a similar synergistic interaction between BER and NER was observed for S. cerevisiae, for this organism, BER predominates over NER (48). Further, NER does not seem to play any role in MMS resistance in mammalian fibroblasts (43). While the list of known in vitro substrates for NER has expanded from just DNA helix-distorting lesions to include O6-methylguanine, AP sites, N6-methyladenine, and some mismatched bases, it is not yet known whether 3MeA is repaired by NER (17, 22). Given the biological evidence presented here and similar studies in S. cerevisiae, it seems highly likely that 3MeA can be repaired by NER, at least in S. pombe and S. cerevisiae (48). It is worth noting that XPG/ERCC5, the human homologue of the S. pombe rad13 gene studied here, has been reported to stimulate the excision of thymine glycol DNA lesions by the human thymine glycol DNA glycosylase and can thus act in an accessory role for BER (6, 27). It is not known whether S. pombe rad13 has a similar stimulatory effect on thymine glycol DNA glycosylase or whether XPG and its homologues stimulate other DNA glycosylases.
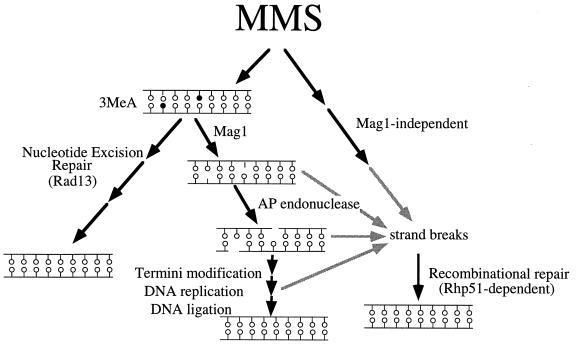
Analysis of the MMS sensitivity of single and double mutants for mag1 and rhp51 revealed an unexpected genetic relationship. The observation that rhp51 S. pombe is more sensitive to MMS than mag1 rhp51 S. pombe is consistent with a model for MMS-induced single-strand DNA breaks accumulating as BER intermediates. As diagrammed in Fig. 8, the successive action of 3MeA DNA glycosylase and AP endonuclease creates a DNA break in one strand whose repair is completed by termini modification (by either deoxyribose phosphodiesterase or FEN1), DNA replication, and DNA ligation. If any of the latter three steps of BER were rate limiting, DNA strand breaks would accumulate in the genome and BER intermediates (either singly or closely opposed to one another), could act as substrates for RR involving S. pombe Rhp51. Alternatively, AP sites could give rise to DNA strand breaks due to their inherent chemical instability or their ability to block DNA replication. A stalled replication fork on the leading strand with continued DNA synthesis on the lagging strand could produce extended regions of single-stranded DNA that are potential substrates for RR. Indeed, heterologous expression of an S. cerevisiae AP endonuclease gene, APN1, in mag1 rhp51 S. pombe reverses the contribution of mag1 to MMS sensitivity, suggesting that Mag1-induced DNA strand breaks are due to unrepaired AP sites (Fig. 6). The absence of Mag1-initiated BER may reduce the number of RR substrates, making RR-deficient cells more MMS resistant. Both of these models indicate that 3MeA DNA lesions are less lethal to S. pombe than DNA strand breaks. It is worth noting that the absence of Mag1 only partially reverses the MMS-sensitive phenotype of rhp51 S. pombe, suggesting that a small but significant portion of MMS-induced DNA strand breaks are attributable to Mag1-generated BER intermediates. It is unclear how the remaining MMS-induced DNA strand breaks are produced.
FIG. 8.
Model for MMS-induced strand breaks and DNA repair in S. pombe. As diagrammed, BER and NER can repair potentially lethal 3MeA lesions. BER generates DNA strand breaks during the repair process, and these can be substrates for Rhp51-dependent recombinational repair. Alternatively, MMS can induce DNA strand breaks by a mechanism whose molecular details are unknown.
The genetic interaction between mag1 and rhp51 was observed only when S. pombe was treated chronically with MMS, not when S. pombe was treated acutely. Although rhp51 S. pombe was still extremely sensitive to acute MMS exposure, there was no difference in the MMS sensitivity of rhp51 and mag1 rhp51 S. pombe. In the model presented, the contribution of BER to DNA strand breaks requires that at least one of the downstream components of BER be limiting, such that BER intermediates accumulate. Perhaps these components are induced under acute but not chronic exposure to MMS, such that BER intermediates are processed more efficiently under acute than under chronic MMS exposure. Our previous studies showed that acute exposure of S. pombe to MMS did not affect mag1 transcript levels (32). However, preliminary results show that chronic MMS exposure may moderately induce mag1 transcript levels. Whether other components of the BER pathway are differentially expressed under various treatment conditions remains to be determined.
The studies described here illustrate an important point: a single DNA repair enzyme or a single DNA repair pathway does not solely determine sensitivity to DNA-damaging agents. Rather, the interactions between different DNA repair pathways are clearly very important for cell survival and thus can have both positive and negative outcomes. The data presented here demonstrate that a single type of DNA repair defect can have no effect, a positive effect, or a negative effect on cell survival of DNA damage, depending on the constellation of other repair pathways in the cell. We have also demonstrated that DNA repair intermediates from one DNA repair pathway can be substrates for a second DNA repair pathway. It is highly likely that the balance and interactions between DNA repair pathways differ among organisms and even among different tissues within the same organism. DNA-damaging agents are used in the clinic to treat cancers, and several gene therapy approaches are being developed to alter DNA repair capacity such that the efficacy of cancer chemotherapy is increased. It is therefore very important to explore potential interactions among DNA repair pathways in order to understand their influences on DNA damage susceptibility and to manipulate DNA repair pathways as effectively as possible.
ACKNOWLEDGMENTS
This research was supported by grants NCI CA55042 and NIEHS P01-ES03926. A.M. was supported by a fellowship from the Pharmaceutical Research and Manufacturers of America Foundation. L.S. is a Burroughs Wellcome Toxicology Scholar.
We thank Charles Hoffman for the S. pombe ura4 plasmid, Tamar Enoch for S. pombe strains, and Dieter Wolf for the S. pombe expression plasmid. We are also grateful to Tamar Enoch and Bruce Demple for their thoughtful and constructive advice during this research and Brian Glassner and Lauren Posnick for helpful comments on the manuscript.
REFERENCES
- 1.al-Khodairy F, Carr A M. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 1992;11:1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfa C, Fantes P, Hyams M, Warbrick E. Experiments with fission yeast. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1993. [Google Scholar]
- 3.Avery A M, Kaur B, Taylor J, Mello J A, Essigmann J M, Doetsch P W. Substrate specificity of ultraviolet DNA endonuclease (UVDE/Uve1p) from Schizosaccharomyces pombe. Nucleic Acids Res. 1999;27:2256–2264. doi: 10.1093/nar/27.11.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumann P, West S C. Role of human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem Sci. 1998;23:247–251. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- 5.Bennett R A O. The Saccharomyces cerevisiae ETH1 gene, an inducible homolog of exonuclease III that provides resistance to DNA-damaging agents and limits spontaneous mutagenesis. Mol Cell Biol. 1999;19:1800–1809. doi: 10.1128/mcb.19.3.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bessho T. Nucleotide excision repair 3′ endonuclease XPG stimulates the activity of base excision repair enzyme thymine glycol DNA glycosylase. Nucleic Acids Res. 1999;27:979–983. doi: 10.1093/nar/27.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carr A M, Sheldrick K S, Murray J M, al-Harithy R, Watts F Z, Lehmann A R. Evolutionary conservation of excision repair in Schizosaccharomyces pombe: evidence for a family of sequences related to the Saccharomyces cerevisiae RAD2 gene. Nucleic Acids Res. 1993;21:1345–1349. doi: 10.1093/nar/21.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Derfler B, Maskati A, Samson L. Cloning a eukaryotic DNA glycosylase repair gene by the suppression of a DNA repair defect in Escherichia coli. Proc Natl Acad Sci USA. 1989;86:7961–7965. doi: 10.1073/pnas.86.20.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, Derfler B, Samson L. Saccharomyces cerevisiae 3-methyladenine DNA glycosylase has homology to the AlkA glycosylase of E. coli and is induced in response to DNA alkylation damage. EMBO J. 1990;9:4569–4575. doi: 10.1002/j.1460-2075.1990.tb07910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunningham R P. DNA glycosylases. Mutat Res. 1997;383:189–196. doi: 10.1016/s0921-8777(97)00008-6. [DOI] [PubMed] [Google Scholar]
- 11.Cunningham R P, Saporito S M, Spitzer S G, Weiss B. Endonuclease IV (nfo) mutant of Escherichia coli. J Bacteriol. 1986;168:1120–1127. doi: 10.1128/jb.168.3.1120-1127.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engelward B P, Dreslin A, Christensen J, Huszar D, Kurahara C, Samson L. Repair-deficient 3-methyladenine DNA glycosylase homozygous mutant mouse cells have increased sensitivity to alkylation-induced chromosome damage and cell killing. EMBO J. 1996;15:945–952. [PMC free article] [PubMed] [Google Scholar]
- 13.Forsburg S L. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 1993;21:2955–2956. doi: 10.1093/nar/21.12.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox B W, Fox M. Sensitivity of the newly synthesized and template DNA of lymphoma cells to damage by methyl methanesulfonate, and the nature of associated “repair” processes. Mutat Res. 1969;8:629–638. doi: 10.1016/0027-5107(69)90081-5. [DOI] [PubMed] [Google Scholar]
- 15.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: American Society for Microbiology; 1995. [Google Scholar]
- 16.Game J C. DNA double-strand breaks and the RAD50-RAD57 genes in Saccharomyces. Semin Cancer Biol. 1993;4:73–83. [PubMed] [Google Scholar]
- 17.Grossman L, Lin C, Ahn Y. Nucleotide excision repair in Escherichia coli. In: Nickoloff J A, Hoekstra M F, editors. DNA damage and repair in lower eukaryotes. Totowa, N.J: Humana Press; 1998. pp. 11–27. [Google Scholar]
- 18.Habraken Y, Sung P, Prakash L, Prakash S. A conserved 5′ to 3′ exonuclease activity in the yeast and human nucleotide excision repair proteins RAD2 and XPG. J Biol Chem. 1994;269:31342–31345. [PubMed] [Google Scholar]
- 19.Habraken Y, Sung P, Prakash L, Prakash S. Human xeroderma pigmentosum group G gene encodes a DNA endonuclease. Nucleic Acids Res. 1994;22:3312–3316. doi: 10.1093/nar/22.16.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haynes R H, Kunz B A. DNA repair and mutagenesis in yeast. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces cerevisiae: life cycle and inheritance. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1981. pp. 371–414. [Google Scholar]
- 21.Hickson I D. Base excision repair of DNA damage. Austin, Tex: Landes Bioscience and Chapman & Hall; 1997. [Google Scholar]
- 22.Huang J C, Hsu D S, Kazantsev A, Sancar A. Substrate spectrum of human excinuclease: repair of abasic sites, methylated bases, mismatches and bulky adducts. Proc Natl Acad Sci USA. 1994;91:12213–12217. doi: 10.1073/pnas.91.25.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanaar R, Hoeijmakers J H J, van Gent D C. Molecular mechanisms of DNA double-strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- 24.Kanno S, Iwai S, Takao M, Yasui A. Repair of apurinic/apyrimidinic sites by UV damage endonuclease; a repair protein for UV and oxidative damage. Nucleic Acids Res. 1999;27:3096–3103. doi: 10.1093/nar/27.15.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaur B, Fraser J L A, Freyer G A, Davey S, Doetsch P W. A Uve1p-mediated mismatch repair pathway in Schizosaccharomyces pombe. Mol Cell Biol. 1999;19:4703–4710. doi: 10.1128/mcb.19.7.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein J C, Bleeker M J, Roelen H C, Rafferty J A, Margison G P, Brugghe H F, van den Elst H, van der Marel G A, van Boom J H, Kreik E, Berns A J. Role of nucleotide excision repair in processing of O4-alkylthymine in human cells. J Biol Chem. 1994;269:25521–25528. [PubMed] [Google Scholar]
- 27.Klungland A, Hoss M, Ganz D, Canstatinou A, Clarkson S G, Doetsch P W, Bolton P H, Wood R D, Lindahl T. Base excision repair of oxidative DNA damage activated by XPG protein. Mol Cell. 1999;3:33–42. doi: 10.1016/s1097-2765(00)80172-0. [DOI] [PubMed] [Google Scholar]
- 28.Labahn J, Scharer O D, Long A, Ezaz-Nikpay, Khosro, Verdine G L, Ellenberger T E. Structural basis for the excision repair of alkylation-damaged DNA. Cell. 1996;86:321–329. doi: 10.1016/s0092-8674(00)80103-8. [DOI] [PubMed] [Google Scholar]
- 29.Levin J, Demple B. Analysis of class II (hydrolytic) and class I (β-lyase) apurinic/apyrimidinic endonucleases with a synthetic DNA substrate. Nucleic Acids Res. 1990;18:5069–5075. doi: 10.1093/nar/18.17.5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lieber M R. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 31.Lindahl T, Karran P, Wood R D. DNA excision repair pathways. Curr Opin Genet Dev. 1997;7:158–169. doi: 10.1016/s0959-437x(97)80124-4. [DOI] [PubMed] [Google Scholar]
- 32.Memisoglu A, Samson L. Cloning and characterization of a cDNA encoding a 3-methyladenine DNA glycosylase from the fission yeast Escherichia coli. Gene. 1996;177:229–235. doi: 10.1016/0378-1119(96)00308-3. [DOI] [PubMed] [Google Scholar]
- 33.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 34.Muris D F, Vreeken K, Carr A M, Broughton B C, Lehmann A R, Lohman P H, Pastink A. Cloning the RAD51 homologue of Schizosaccharomyces pombe. Nucleic Acids Res. 1993;21:4586–4591. doi: 10.1093/nar/21.19.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray J M, Tavossoli M, Al-Harithy R, Sheldrick K S, Lehman A R, Carr A M, Watts F Z. Structural and functional conservation of the human homolog of the Schizosaccharomyces pombe rad2 gene, which is required for chromosome segregation and recovery from DNA damage. Mol Cell Biol. 1994;14:4878–4888. doi: 10.1128/mcb.14.7.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reardon J T, Bessho T, Kung H C, Bolton P H, Sancar A. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc Natl Acad Sci USA. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reiter H, Strauss B, Robbins M, Marone R. Nature of the repair of methyl methanesulfonate-induced damage in Bacillus subtilis. J Bacteriol. 1967;93:1056–1062. doi: 10.1128/jb.93.3.1056-1062.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samson L, Thomale J, Rajewsky M F. Alternative pathways for the in vivo repair of O6-alkylguanine and O4-alkythymine in Escherichia coli: the adaptive response and nucleotide excision repair. EMBO J. 1988;7:2261–2267. doi: 10.1002/j.1460-2075.1988.tb03066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartz J L. Monofunctional alkylating agent-induced S-phase-dependent DNA damage. Mutat Res. 1989;216:111–118. doi: 10.1016/0165-1161(89)90011-3. [DOI] [PubMed] [Google Scholar]
- 40.Siede W. The genetics and biochemistry of the repair of UV-induced DNA damage in Saccharomyces cerevisiae. In: Nickoloff J A, Hoekstra M F, editors. DNA damage and repair in prokaryotes and lower eukaryotes. Totowa, N.J: Humana Press; 1998. pp. 307–334. [Google Scholar]
- 41.Sommers C H, Miller E J, Dujon B, Prakash S, Prakash L. Conditional lethality of null mutations in RTH1 that encoded the yeast counterpart of a mammalian 5′-to 3′-exonuclease required for lagging strand DNA synthesis in reconstituted systems. J Biol Chem. 1995;270:4193–4196. doi: 10.1074/jbc.270.9.4193. [DOI] [PubMed] [Google Scholar]
- 42.Strauss B S, Wahl R. The presence of breaks in the deoxyribonucleic acid of Bacillus subtilis treated in vivo with the alkylating agent, methylmethanesulfonate. Biochim Biophys Acta. 1964;80:116–126. [Google Scholar]
- 43.Thielmann H W, Edler L, Friemel S. Xeroderma pigmentosum patients from Germany: repair capacity of 45 XP fibroblast strains of the Mannheim XP Collection as measured by colony-forming ability and unscheduled DNA synthesis following treatment with methyl methanesulfonate and N-methyl-N-nitrosourea. J Cancer Res Clin Oncol. 1986;112:245–57. doi: 10.1007/BF00395919. [DOI] [PubMed] [Google Scholar]
- 44.Tishkoff D X, Filosi N, Gaida G M, Kolodner R D. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 45.Van den Berg H W. Alkaline sucrose gradient sedimentation studies of DNA from HeLa S3 cells exposed to methyl methanesulfonate or methylazoxymethanol acetate. Biochim Biophys Acta. 1974;353:215–226. doi: 10.1016/0005-2787(74)90186-5. [DOI] [PubMed] [Google Scholar]
- 46.Voigt J M, Van Houten B, Sancar A, Topal M D. Repair of O6-methylguanine by ABC excinuclease of Escherichia coli in vitro. J Biol Chem. 1989;264:5172–5176. [PubMed] [Google Scholar]
- 47.Wood R D. DNA repair in eukaryotes. Annu Rev Biochem. 1996;65:135–167. doi: 10.1146/annurev.bi.65.070196.001031. [DOI] [PubMed] [Google Scholar]
- 48.Xiao W, Chow B L. Synergism between yeast nucleotide and base excision repair. Curr Genet. 1998;33:92–99. doi: 10.1007/s002940050313. [DOI] [PubMed] [Google Scholar]
- 49.Xiao W, Chow B L, Rathgeber L. The repair of DNA methylation damage in Saccharomyces cerevisiae. Curr Genet. 1996;30:461–468. doi: 10.1007/s002940050157. [DOI] [PubMed] [Google Scholar]
- 50.Yamagata Y, Kato M, Odawara K, Tokuno Y, Nakashima Y, Matsushima N, Yasumura K, Tomita K, Ihara K, Fujii Y, Nakabeppu Y, Sekiguchi M, Fujii S. Three-dimensional crystal structure of a DNA repair enzyme, 3-methyladenine DNA glycosylase II, from Escherichia coli. Cell. 1996;86:311–319. doi: 10.1016/s0092-8674(00)80102-6. [DOI] [PubMed] [Google Scholar]
- 51.Yasui A, McCready S J. Alternative repair pathways for UV-induced DNA damage. Bioessays. 1998;20:291–297. doi: 10.1002/(SICI)1521-1878(199804)20:4<291::AID-BIES5>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]