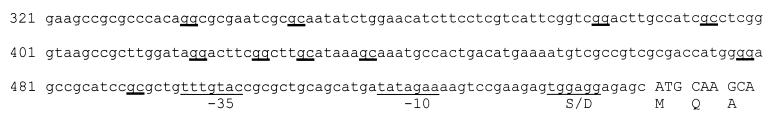Abstract
We have identified two Sinorhizobium meliloti chromosomal loci affecting the poly-3-hydroxybutyrate degradation pathway. One locus was identified as the gene acsA, encoding acetoacetyl coenzyme A (acetoacetyl-CoA) synthetase. Analysis of the acsA nucleotide sequence revealed that this gene encodes a putative protein with a molecular weight of 72,000 that shows similarity to acetyl-CoA synthetase in other organisms. Acetyl-CoA synthetase activity was not affected in cell extracts of glucose-grown acsA::Tn5 mutants; instead, acetoacetyl-CoA synthetase activity was drastically reduced. These findings suggest that acetoacetyl-CoA synthetase, rather than CoA transferase, activates acetoacetate to acetoacetyl-CoA in the S. meliloti poly-3-hydroxybutyrate cycle. The second locus was identified as phbC, encoding poly-3-hydroxybutyrate synthase, and was found to be required for synthesis of poly-3-hydroxybutyrate deposits.
The catabolism of intracellular carbon stores is a strategy employed by many bacterial species to survive nutritionally suboptimal conditions. Polyhydroxyalkanoates (PHA), such as poly-3-hydroxybutyrate (PHB), accumulate in cells when growth is limited but carbon availability is not (2). This stored carbon can then be utilized during conditions of otherwise limiting carbon availability (45). PHB synthesis and degradation are collectively referred to as the PHB cycle. The extent of PHB accumulation is dependent on the relative rates of synthesis and degradation, which in turn are controlled by growth conditions (36, 41).
PHA have attracted substantial industrial interest for their use as high-quality, biodegradable plastics (2). This interest has driven research efforts directed at PHA biosynthesis. The genes encoding the enzymes responsible for PHA synthesis have been isolated and characterized from a number of bacterial species (33, 43), including Sinorhizobium meliloti strains Rm41 (49) and Rm1021 (51). The degradative portion of the cycle has not been subjected to similar attention, and it is less well defined both genetically and biochemically.
We have recently isolated a number of S. meliloti mutants that are affected in the ability to utilize PHB cycle intermediates as sole carbon sources to support growth (11). The mutations mapped to loci on both the chromosome and the pRmeSU47b (pEXO) megaplasmid. Two loci on the megaplasmid have been identified via enzymatic and nucleotide sequence analyses. One encodes the enzyme 3-hydroxybutyrate dehydrogenase (3), and the other encodes methylmalonyl coenzyme A (methylmalonyl-CoA) mutase (10). In this paper, we further characterize and identify two of the chromosomal loci.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
Bacterial strains and plasmids are listed in Table 1. The construction of new strains is described in the text. Bacterial culture in Luria-Bertani (LB) and TY complex media and modified M9 minimal salts medium and antibiotic selection were carried out as previously described (12). Modified M9 minimal salts medium was supplemented with glucose, dl-3-hydroxybutyrate (sodium salt), acetoacetate (lithium salt), or acetate (potassium salt) at 15 mM. PHB-accumulating medium YM was described previously (48).
TABLE 1.
Bacterial strains and plasmids useda
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Sinorhizobium meliloti | ||
| Rm1021 | SU47 str-21 (Smr) | 35 |
| Rm5320 | Rm1021 pRmeSU47aΩ30::Tn5-11 (mobilizable pRmeSU47a) | 20 |
| Rm11105 | phbC1::Tn5 (=aau-1::Tn5) | 11 |
| Rm11134 | acsA7::Tn5 (=aau-7::Tn5) | 11 |
| Rm11135 | acsA8::Tn5 (=aau-8::Tn5) | 11 |
| Rm11144 | phbC1::Tn5-233 (=aau-1::Tn5-233) | 11 |
| Rm11149 | acsA49::Tn5-B20 | This study |
| Rm11153 | phbC1::TnV (=aau-1::TnV) | This study |
| Rm11160 | acsA7::Tn5-233 (=aau-7::Tn5-233) | 11 |
| Rm11368 | phbC1::Tn5, pRmeSu47aΩ30::Tn5-11 | This study |
| Escherichia coli | ||
| DH5α | F−endA1 hsdR17 (rK− mK−) supE44 thi-1 recA1 gyrA96 relA1 Δ(argF-lacZYA)U169 φ80dlacZΔM15, λ− | BRL Inc. |
| MT607 | pro-82 thi-1 hsdR17 supE44 recA56 | 20 |
| MT614 | MT607ΩTn5 | T. Finan, unpublished |
| MT616 | MT607 (pRK600) | 20 |
| G312 | MT607ΩTn5-B20 | 17 |
| Agrobacterium strain GMI9023 | C58 Rfr Smr, cured of pTiC58 and pATC58 | 40 |
| Plasmids | ||
| pLAFR1 | IncP cosmid cloning vector, Tcr | 21 |
| pSP329 | IncP cloning vector, Tcr | 8 |
| pVK101 | IncP cloning vector, Kmr Tcr | 29 |
| pUC18 | Cloning vector, ColE1 oriV, bla | 52 |
| pTC338 | PLAFR1 clone, Tcr, complements aau-1 mutant | This study |
| pTC350 | 7-kb EcoRI fragment from pTC338 in pUC18, contains acsA | This study |
| pTC355 | 7-kb EcoRI fragment from pTC338 in pVK101, contains acsA | This study |
| pTC381 | aau-1::TnV EcoRI self-ligation product, Kmr | This study |
| pGQ105 | aau-1 and aau-7 complementing 4-kb KpnI fragment from pTC350 in pSP329, Tcr | This study |
| pGQ109 | HindIII fragment from pTC381 in pUC18 | This study |
Abbreviations for antibiotics: Km, kanamycin; Rf, rifampin; Sm, streptomycin; Tc, tetracycline.
Genetic and molecular biology techniques.
Replacement of Tn5 insertions with Tn5-233 (16) (encoding Gmr Spr [see the footnote to Table 1 for abbreviations for antibiotics]), replacement of Tn5-233 with TnV (23) (encoding Nmr), identification of complementing recombinant plasmids from the Rm1021 pLAFR1 cosmid library (21), isolation of Tn5 and Tn5-B20 (42) insertions on IncP plasmids, homogenotization of these insertions using incompatibility of plasmid pPH1JI, conjugation of IncP plasmids between S. meliloti and Escherichia coli using the mobilizing strain MT616, and transduction between S. meliloti strains using phage φM12 (19) were carried out as described previously (11–13). Molecular biology was carried out using standard methods (4). Automated DNA sequence analysis was performed at the MOBIX DNA sequencing facility (McMaster University, Hamilton, Ontario, Canada), using an ABI 373A instrument. Manual DNA sequence analysis was performed with a dsDNA Cycle Sequencing System kit (GIBCO BRL) and [33P]dATP. An IS50 primer (14) was used to prime sequencing reactions from cloned Tn5 and TnV insertions. Sequences were analyzed using DNA Strider, version 1.2 (34), MacVector 6.0.1 (Oxford Molecular Group), Clustal W (46), and BLAST (1).
Biochemical assay.
Cell cultures were grown to stationary phase in 1 liter of M9-glucose in 2.8-liter Fernbach flasks at 30°C, with shaking at 200 rpm. Culture purity was ascertained by streaking on TY agar plates. Cells were collected by centrifugation; the pellets were washed twice with ice-cold washing buffer (20 mM Tris-Cl [pH 8], 1 mM MgCl2) and suspended in 4 ml of ice-cold sonication buffer (20 mM Tris-HCl [pH 8], 1 mM MgCl2, 10% [wt/vol] glycerol, 10 mM β-mercaptoethanol) per g (wet weight) of cells. The cells were disrupted by sonication on ice (Branson Sonifier cell disruptor model 200). Cell debris was removed by centrifugation (SS34 rotor, 12,000 rpm, 20 min), and the cell extracts were stored at −70°C. The extracts were centrifuged again in a microcentrifuge for 15 min at 4°C to reduce membrane-associated NADH oxidase background activity. Protein concentration was determined by the method of Bradford (6), using bovine serum albumin as the standard. All enzyme assays were carried out by continuous coupled assay at room temperature. Values presented are the means of at least three assays ± standard deviations.
Acetyl-CoA synthetase activity was determined by a modification of the method of Brown et al. (7). The reaction mixture (1 ml) contained 100 μmol of Tris-Cl (pH 8.0), 5 μmol of MgCl2, 5 μmol of NAD, 0.1 μmol of CoA, 5 μmol of l-malate, 150 U of malate dehydrogenase, 2.75 U of citrate synthase, 10 μmol of potassium acetate, 10 μmol of ATP, and cell extract. The reaction was initiated by addition of ATP. The formation of acetyl-CoA from acetate and CoA was measured by coupling the reaction to the rate of reduction of NAD+ via malate dehydrogenase and citrate synthase, determined at 340 nm.
Acetoacetyl-CoA synthetase activity was determined in the same way as acetyl-CoA synthetase activity except that 10 μmol of lithium acetoacetate was substituted for potassium acetate. The acetoacetyl-CoA produced by the synthetase was quickly broken down to acetyl-CoA by endogenous thiolase present in crude extract (5). Thus, acetoacetyl-CoA synthetase activity was measured via determination of the rate of acetyl-CoA production as above.
Acetoacetate:succinyl-CoA transferase activity was measured by determining the rate of acetyl-CoA production, in a coupled assay manner similar to that used to measure acetoacetyl-CoA synthetase activity. The reaction mixture (1 ml) contained 100 μmol of Tris-Cl (pH 8.0), 5 μmol of MgCl2, 5 μmol of NAD+, 0.1 μmol of CoA, 5 μmol of l-malate, 150 U of malate dehydrogenase, 2.75 U of citrate synthase, 10 μmol of lithium acetoacetate, 0.05 μmol of succinyl-CoA, and cell extract. The reaction was initiated by the addition of succinyl-CoA.
Thiolase activity was measured in both directions. The cleavage of 1 mol of acetoacetyl-CoA to 2 mol of acetyl-CoA by thiolase was determined by measuring the rate of acetyl-CoA production, in a coupled assay similar to that used to measure acetyl-CoA synthetase activity. The reaction mixture (1 ml) contained 100 μmol of Tris-Cl (pH 8.0), 5 μmol of MgCl2, 5 μmol of NAD+, 0.1 μmol of CoA, 5 μmol of l-malate, 150 U of malate dehydrogenase, 2.75 U of citrate synthase, 0.05 μmol of acetoacetyl-CoA, 10 μmol of ATP, and cell extract. The reaction was initiated by addition of acetoacetyl-CoA. The condensation of 2 mol of acetyl-CoA to 1 mol of acetoacetyl-CoA by thiolase was determined by measuring the rate of formation of acetoacetyl-CoA via a coupled reaction with 3-hydroxyacetyl-CoA dehydrogenase, in which the rate of oxidation of NADH to NAD+ by 3-hydroxyacetyl-CoA dehydrogenase was determined at 340 nm. The reaction mixture (1 ml) contained 250 μmol of Tris-HCl, 250 nmol of NADH, 2 μmol of EDTA, 1 mg of bovine serum albumin, 2 U of 3-hydroxyacetyl-CoA dehydrogenase, 0.6 μmol of acetyl-CoA, and cell extract. The reaction was initiated by adding acetyl-CoA.
PHB was determined by the spectrophotometric method of Law and Slepecky (31), using strains grown in YM medium for 48 h.
Symbiotic assay.
Assay for symbiotic performance on alfalfa plants was carried out as described before (3). The plants were harvested for shoot dry weight determination 5 weeks after inoculation.
Starvation assay.
Saturated TY broth cultures were subcultured 1:50 to YM and cultured for 48 h. The cells were washed twice in saline before subculture 1:50 to carbon-free M9 medium. Cell titer was monitored by plating for viable cells on LB agar. Values are the means from triplicate cultures.
Nucleotide sequence accession number.
The GenBank accession number for the sequence reported in this paper is AF080217.
RESULTS
Isolation of complementing clones.
The pLAFR1 cosmid clone bank was introduced into the mutant strains Rm11105 and Rm11134, harboring mutations in the aau-1 and aau-7 loci, respectively. Both of these mutations render strains carrying them unable to utilize the PHB cycle intermediates 3-hydroxybutyrate and acetoacetate as sole carbon sources. Following selection on M9 supplemented with 3-hydroxybutyrate as the sole carbon source, we obtained two cosmid clones, designated pTC338 (from the Rm11105 complementation) and pGQ2 (from the Rm11134 complementation). Complementation was confirmed by reintroduction of the clones into the mutant strains by conjugation. Surprisingly, both pTC338 and pGQ2 complemented each of the two mutants. Restriction analysis confirmed that these two clones shared common-sized EcoRI fragments (data not shown). A common 7-kb EcoRI fragment was subcloned from pTC338 into the unique EcoRI site of pVK101. The resulting plasmid, pTC355, was able to complement both mutants. Further subcloning experiments determined that a 4-kb KpnI subfragment of the 7-kb EcoRI fragment (pGQ105) was also capable of complementation.
Since aau-1 and aau-7 were previously mapped to two distinct regions on the S. meliloti chromosome (11), the complementation could not be homologous in the case of both loci. At least one of the two loci must have been unlinked to the 7-kb EcoRI fragment. To resolve this, a Tn5-B20 insertion which abolished the ability to complement strain Rm11105 for 3-hydroxybutyrate utilization was isolated in pTC355. This insertion was then recombined into the genome by homogenotization, resulting in strain Rm11149. Transduction mapping indicated the insertion to be 100% (60 of 60) linked to the aau-7::Tn5-233 insertion in strain Rm11160 and unlinked (none of 50) to the aau-1::Tn5-233 insertion in strain Rm11144. Southern blot analysis (data not presented) confirmed that the aau-7::Tn5 insertion was located in the 7-kb EcoRI fragment. Therefore, pTC355 appears to reverse the aau-7 phenotype by homologous complementation and the aau-1 phenotype by nonhomologous complementation.
Genetic and sequence analysis of aau-7 complementing region.
To better define the region of cosmid clone pGQ2 responsible for complementation of the aau-7 phenotype, 14 independent Tn5 insertions which abolished aau-7 complementation ability were isolated in that clone. All of these insertions were located in the 4-kb KpnI fragment. EcoRI fragments, containing the Tn5 insertions, were subcloned from each of the 14 pGQ2::Tn5 plasmids into pUC18. Subclones of each of the two possible orientations for each insertion were retained. One side of each subclone was deleted by cleavage with HindIII followed by self-ligation, and the sequence of the DNA flanking each insertion was obtained using the IS50 primer. In this way, the precise site of each Tn5 insertion was determined. Combined with the sequence obtained using custom-designed primers, the sequence assembled into a bidirectional contig of 3,295 bp, extending to the distal KpnI site. Analysis of the sequence revealed a single open reading frame (ORF) of 1,950 bp (650 amino acids) encoding a predicted gene product with a molecular weight of 72,000 (Fig. 1). The presumptive ATG start codon at position 548 was preceded by a putative ribosome binding site 5 bp upstream and a ς70-type (−35 −10) promoter motif 24 bp upstream. There are also several ς54-type promoter motifs (47) in the upstream region (Fig. 2), although ς54 (rpoN) mutants are not defective in growth on 3-hydroxybutyrate (data not presented). The G+C content of 64.4% for this ORF is comparable to the average G+C content of 61.6% in the S. meliloti genome, and the codon usage is similar to the preference in S. meliloti (data not shown). No ORF longer than 25 amino acid residues was observed between the end of this ORF and the distal KpnI site. Clustal W (46) analysis shows that the 650-amino-acid sequence exhibits homology to acetyl-CoA synthetase sequences from several organisms (15, 18, 24, 25, 30, 39, 50) (Table 2), although the other sequences are more similar to each other than they are to the S. meliloti sequence.
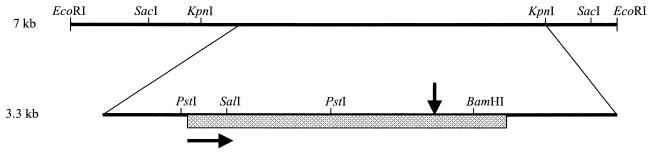
FIG. 1.
Physical map of the 7-kb EcoRI fragment and the 3,295-bp sequenced region containing S. meliloti acsA. The hatched bar indicates the ORF of 1,950 bp. The horizontal arrow indicates the direction of transcription. The vertical arrow shows the site of acsA7::Tn5 (=aau-7::Tn5) insertion.
FIG. 2.
Promoter region of S. meliloti acsA. The putative ribosome binding site is underlined and labeled “S/D”; the putative promoter is underlined and indicated by “−10” and “−35”; The gg and gc rpoN-like recognition motifs are marked with heavy underlines.
TABLE 2.
Amino acid identities and similarities between acetoacetyl-CoA synthetase and acetyl-CoA synthetasesa
| Source | % Identity (% similarity)b
|
|||||||
|---|---|---|---|---|---|---|---|---|
| S.m. | M.s. | P.b. | S.c.-2 | R.e. | E.c. | S.c. | B.s. | |
| S.m. | — | |||||||
| M.s. | 24 (40) | — | ||||||
| P.b. | 24 (42) | 43 (59) | — | |||||
| S.c.-2 | 24 (39) | 41 (59) | 56 (74) | — | ||||
| R.e. | 25 (41) | 46 (62) | 47 (62) | 43 (59) | — | |||
| E.c. | 23 (39) | 43 (59) | 50 (66) | 45 (61) | 50 (66) | — | ||
| S.c. | 22 (38) | 37 (56) | 55 (70) | 55 (70) | 46 (62) | 45 (63) | — | |
| B.s. | 17 (31) | 33 (46) | 31 (45) | 30 (46) | 31 (47) | 32 (46) | 27 (41) | — |
GenBank accession numbers for the analyzed sequences: S.m. (Sinorhizobium meliloti), AF080217; M.s. (Methanothrix soehngenii), P27095; P.b. (Phycomyces blakesleeanus), S46276; B.s. (Bacillus subtilis), P39062; S.c. (Saccharomyces cerevisiae), S30019; S.c.-2 (S. cerevisiae acs2), P52910; R.e. (Ralstonia eutropha), P31683; E.c. (Escherichia coli), P27550.
Values were derived using MacVector 6.0.1 (Clustal W algorithm). —, not applicable.
To determine the exact site of insertion of the aau-7::Tn5 insertion in strain Rm11134, this mutation was transferred by recombination from the S. meliloti genome onto plasmid pTC338, which carries DNA homologous to the insertion site. First, plasmid pTC338 was introduced into strain Rm11134. Next, pTC338 was conjugally transferred into E. coli MT607, and transconjugants were selected for growth on LB-Km-Tc at 37°C after 24 h. The Kmr EcoRI fragment was subcloned into pUC18, BamHI deletions were generated, and sequence was obtained using the IS50 primer. The site of insertion is defined by a 9-bp repeat of nucleotides 2056 to 2065 (Fig. 1), corresponding to an interruption of sequence at amino acid residue 503.
Identification of aau-1.
Since a homologously complementing clone of aau-1 was not obtained, we decided to identify the exact site of insertion of the aau-1::Tn5 insertion. The aau-1::Tn5 was first replaced with aau-1::Tn5-233 (Gmr Spr), which was in turn replaced by TnV to make strain Rm11153. TnV contains the replication origin derived from pSC101 and is devoid of EcoRI sites (23). By self-ligation of EcoRIdigested genomic DNA from strain Rm11153, followed by transformation of E. coli DH5α to Kmr, a clone consisting of TnV flanked by the aau-1 EcoRI fragment was isolated and designated pTC381. A fragment containing the TnV-flanking regions of pTC381, along with the ends of the IS50 elements, was released by HindIII digestion and subcloned into HindIII-digested pUC18. One side of the flanking region was deleted by cleavage with EcoRI followed by self-ligation, and the sequence of the TnV flanking region was obtained using the IS50 primer. The sequence (185 bp) is identical to the segment of the phbC gene from 2532 to 2717 in S. meliloti 1021 (51) and exhibits 98% (181/185) identity to the corresponding segment of the phbC gene in S. meliloti 41 (49).
We had previously reported the chromosomal location of aau-1 (11), while phbC was more recently reported to map to the megaplasmid pRmeSU47a (51). To resolve the incongruity between the two reports, we attempted to mobilize aau-1::Tn5 by using the pRmeSU47a-located Tn5-11 insertion (=Tn5 containing oriT, Gmr Smr) in strain Rm5320. The Tn5-11 insertion in strain Rm5320 was therefore transduced into strain Rm11105, and the resulting transductant was designated Rm11368. Rm11368 was mated with Agrobacterium strain GMI9023, followed by selection for transconjugants on TY-Rf-Gm-Sp and TY-Rf-Nm. Transconjugants arose only on TY-Rf-Gm-Sp, and 50 of these transconjugant colonies were screened for Nmr; all were found to be Nms. This result clearly excludes a pRmeSU47a location for phbC and is consistent with our earlier chromosome mapping data (11) and recently reported genomic sequence data (9).
Biochemical characterization of the mutants.
A series of enzyme assays was carried out with cell extracts of representative mutants (Table 3). The level of acetyl-CoA synthetase was not reduced in any mutant strain, including the aau-7 mutant. In an attempt to rationalize the aau-7 mutant phenotype with biochemical function, acetoacetyl-CoA synthetase activity was assayed and found to be drastically reduced in the aau-7 mutant strain. In addition, homologous complementation of the aau-7 mutant strain with plasmid pGQ105 restored acetoacetyl-CoA synthetase activity and actually increased it to a level greater than four times higher than the wild-type level. This confirms that aau-7 encodes acetoacetyl-CoA synthetase (acetoacetyl-CoA ligase; EC 6.2.1.16), which activates acetoacetate to acetoacetyl-CoA by the single reaction ATP + CoA + acetoacetate =>acetoacetyl-CoA + AMP + PPi. We have thus designated the gene acsA. Although the aau-7 strain exhibited lower than wild-type succinyl-CoA transferase activity, the presence of acsA-bearing pGQ105 did not increase the activity to a level significantly greater than that in the wild-type strain. Although the aau-1 mutant exhibited slightly reduced levels of acetoacetyl-CoA synthetase and ketothiolase activities, this perhaps reflects physiological effects related to reduced provision of acetoacetate substrate in the absence of accumulated PHB. The slightly reduced levels of 3-ketothiolase activities in each of the other mutant strains perhaps reflect lower levels of available acetoacetyl-CoA substrate in these backgrounds. The aau-1 mutant did not accumulate PHB after growth in YM, while all other strains accumulated PHB to 60 to 70% of cell dry mass, thus further confirming the synonymity of aau-1 and phbC, encoding PHB synthase.
TABLE 3.
Effects of PHB metabolism mutations on enzyme activities
| Strain | Sp act (nmol/min/mg of protein; mean ± SD [n ≥ 3])
|
||||
|---|---|---|---|---|---|
| Acetyl-CoA synthetase | Acetoacetyl-CoA synthetase | 3-Ketothiolase
|
Succinyl-CoA transferase | ||
| Thiolytic | Condensation | ||||
| Rm1021 | 25.04 ± 4.04 | 26.48 ± 0.22 | 1,686.34 ± 57.68 | 69.73 ± 16.36 | 9.92 ± 1.39 |
| Rm11105 (aau-1) | 25.27 ± 3.95 | 21.26 ± 1.33 | 1,095.35 ± 56.17 | 48.18 ± 7.84 | 11.62 ± 0.67 |
| Rm11134 (aau-7) | 24.48 ± 0.67 | 4.24 ± 3.48 | 1,465.06 ± 44.17 | 46.46 ± 7.87 | 5.55 ± 0.76 |
| Rm11134 (pGQ105) | 33.37 ± 3.90 | 111.75 ± 9.45 | 1,318.62 ± 41.89 | 71.30 ± 10.35 | 11.41 ± 1.73 |
Physiological traits.
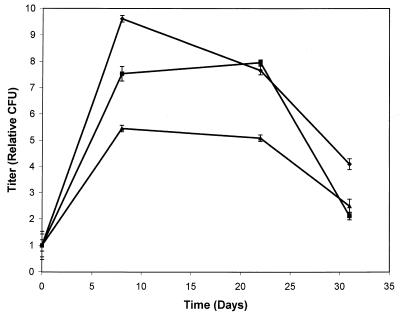
To determine whether the ability to accumulate or metabolize PHB deposits affects cell survival ability, we designed a carbon starvation assay to investigate the starvation survival of the PHB-negative phbC mutant and the PHB degradation-deficient acsA mutant. Strains were cultivated under PHB-accumulating conditions in YM to stationary phase, transferred to carbon nutrient-free M9 medium, and incubated. Viable counting after the transfer to carbon nutrient-free medium indicates that the mutant strains do not propagate as well as the wild-type strain (Fig. 3) upon initial subculture. This is presumably because the wild-type strain is utilizing the accumulated PHB stores, while the mutant strains cannot store or utilize PHB. After 1 month of incubation, however, none of the cultures had dropped below the initial titer.
FIG. 3.
Population change during incubation in growth medium lacking nutrient carbon. ⧫, wild-type strain Rm1021; ■, PHB synthesis mutant strain Rm11105; ▴, PHB degradation-deficient strain Rm11134. The CFU present in each of the cultures immediately after subculture to carbon nutrient-free growth medium (ca. 5 × 106 per ml) was given a relative value of 1, and values are presented as the means of three samples ± standard deviation. The standard deviation values immediately after inoculation (time = 0 days) were 0.21, 0.53, and 0.43 for Rm1021, Rm11105, and Rm11134, respectively.
The symbiotic properties of the mutants were investigated by inoculation of axenic alfalfa seedlings cultivated in the absence of fixed nitrogen. All mutants formed root nodules which fixed N2, as evidenced by the green shoots and shoot dry weight similar to wild type (data not shown).
DISCUSSION
We have demonstrated that acsA, encoding acetoacetyl-CoA synthetase, is required for acetoacetate metabolism in S. meliloti. Unlike the acetyl-CoA synthetase enzymes of other organisms which are rarely able to use C4 fatty acids as substrates (28, 37, 39), it appears that acetoacetate is the substrate for the S. meliloti AcsA. The absence of any significant ORFs downstream of acsA on the smallest complementing subclone indicates that the carbon utilization phenotype is not caused by polar effects on a downstream gene. This work establishes that in S. meliloti, acetoacetyl-CoA synthetase is responsible for activation of acetoacetate, even in the presence of considerable levels of acetoacetate:succinyl-CoA transferase activity. Therefore, we propose that acetoacetyl-CoA synthetase activity is an integral part of the degradation portion of the PHB cycle in S. meliloti. To our knowledge, this is the first report of the mutation and sequence determination of a gene encoding acetoacetyl-CoA synthetase.
Three distinct mechanisms for activation of acetoacetate to acetoacetyl-CoA have been found in bacteria. In E. coli, acetoacetate is activated by a CoA transferase (encoded by atoD and atoA) and then converted to two molecules of acetyl-CoA by ketothiolase (encoded by atoB) (27). It has been suggested that in other bacteria, such as Azotobacter beijerinckii, acetoacetate is activated by an acetoacetate:succinyl-CoA transferase (41). In Zoogloea ramigera, however, no CoA transferase activity was detected. It was reasoned that in this organism, acetoacetyl-CoA synthetase might be responsible for acetoacetate activation, and an enzyme with a molecular weight of 70,000 having this activity was purified from this organism (22). We are not aware of any report on characterization of the Z. ramigera gene encoding this enzyme.
Although acsA is homologous to acetyl-CoA synthetase-encoding genes, mutation of acsA did not affect the ability to use acetate as a sole carbon source (11). This is consistent with the finding that acetyl-CoA synthetase activity was not affected in the acsA mutant, suggesting the presence of an acetate-specific acetyl-CoA synthetase activity in S. meliloti. S. meliloti also possesses the acetate kinase-phosphotransacetylase pathway for acetate activation (44), which is considered to be the primary low-affinity acetate catabolic pathway in bacteria, with a certain amount of contribution of the higher-affinity acetyl-CoA synthetase pathway (7). Complete abolition of ability to grow on acetate requires disruption of both pathways in E. coli (30). In contrast, disruption of the acetate kinase-phosphotransacetylase pathway alone is sufficient to block the ability of Salmonella enterica serotype Typhimurium to use acetate as a sole carbon source (32), while some other bacteria, such as Ralstonia eutropha and Bacillus subtilis, use acetyl-CoA synthetase but not acetate kinase-phosphotransacetylase for growth on acetate (25, 39). Investigation of the acetate growth phenotype of S. meliloti mutants containing lesions in both acetate activation pathways may be required to understand the relative contributions of the alternate pathways to acetate metabolism in this organism.
It is intriguing that disruption of phbC affects growth on acetoacetate, and without substantial decrease in the in vitro-measured acetoacetyl-CoA synthetase activity in cell extracts. We reason that disruption of phbC will result in increased intracellular levels of 3-hydroxybutyryl-CoA and acetoacetyl-CoA. The increased concentration of acetoacetyl-CoA might inhibit the activity of acetoacetyl-CoA synthetase activity in vivo while not affecting acetoacetyl-CoA synthetase activity as measured in vitro. This might also explain the ability of plasmid-encoded (multiple-copy) acsA to suppress the phbC growth phenotype and is also consistent with our recent observation that disruption of phbB, encoding acetoacetyl-CoA reductase, also affects growth on acetoacetate (P. Aneja et al., unpublished data).
The demonstration that symbiotic N2 fixation ability is not affected in any of the PHB cycle mutant strains is consistent with reports indicating that neither PHB synthesis nor degradation is required for effective symbiosis (3, 38, 51), although PHB synthesis has been shown to be important for symbiotic competition (51). PHB is accumulated as an endogenous source of carbon and energy, and fluorescence microscopy provided visual evidence of PHB utilization during carbon-free starvation in Legionella pneumophila (26). This is consistent with our finding that during carbon-free starvation of S. meliloti, PHB synthesis and PHB degradation mutants showed reduced ability to proliferate during the first 30 days of incubation. The mutants were not, however, deficient in the ability to survive prolonged cultivation in the absence of an external nutrient carbon source.
ACKNOWLEDGMENTS
This work was supported by operating grants to T.C.C. from the Natural Sciences and Engineering Research Council of Canada and Fonds pour la Formation de Chercheurs et l'Aide à la Recherche (Québec).
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson A J, Dawes E A. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev. 1990;54:450–472. doi: 10.1128/mr.54.4.450-472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aneja P, Charles T C. Poly-3-hydroxybutyrate degradation in Rhizobium (Sinorhizobium) meliloti: isolation and characterization of a gene encoding 3-hydroxybutyrate dehydrogenase. J Bacteriol. 1999;181:849–857. doi: 10.1128/jb.181.3.849-857.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1997. [Google Scholar]
- 5.Bergstrom J D, Edmond J. Rat liver acetoacetyl-CoA synthetase. Methods Enzymol. 1985;110:3–9. doi: 10.1016/s0076-6879(85)10054-6. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 7.Brown T D K, Jones-Mortimer M C, Kornberg H L. The enzymatic interconversion of acetate and acetyl-coenzyme A in Escherichia coli. J Gen Microbiol. 1977;102:327–336. doi: 10.1099/00221287-102-2-327. [DOI] [PubMed] [Google Scholar]
- 8.Cangelosi G A, Best E A, Martinetti G, Nester E W. Genetic analysis of Agrobacterium. Methods Enzymol. 1991;204:384–397. doi: 10.1016/0076-6879(91)04020-o. [DOI] [PubMed] [Google Scholar]
- 9.Capela D, Barloy-Hubler F, Gatius M-T, Gouzy J, Galibert F. A high-density physical map of Sinorhizobium meliloti 1021 chromosome derived from bacterial artificial chromosome library. Proc Natl Acad Sci USA. 1999;96:9357–9362. doi: 10.1073/pnas.96.16.9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charles T C, Aneja P A. Methylmalonyl-CoA mutase encoding gene of Sinorhizobium meliloti. Gene. 1999;226:121–127. doi: 10.1016/s0378-1119(98)00555-1. [DOI] [PubMed] [Google Scholar]
- 11.Charles T C, Cai G-Q, Aneja P. Megaplasmid and chromosomal loci for the PHB degradation pathway in Rhizobium (Sinorhizobium) meliloti. Genetics. 1997;146:1211–1220. doi: 10.1093/genetics/146.4.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charles T C, Finan T M. Analysis of a 1600-kilobase Rhizobium meliloti megaplasmid using defined deletions generated in vivo. Genetics. 1991;127:5–20. doi: 10.1093/genetics/127.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charles T C, Finan T M. Genetic map of Rhizobium meliloti megaplasmid pRmeSU47b. J Bacteriol. 1990;172:2469–2476. doi: 10.1128/jb.172.5.2469-2476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charles T C, Nester E W. A chromosomally encoded two-component sensory transduction system is required for virulence of Agrobacterium tumefaciens. J Bacteriol. 1993;175:6614–6625. doi: 10.1128/jb.175.20.6614-6625.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Virgilio C, Burckert N, Barth G, Neuhaus J M, Boller T, Wiemken A. Cloning and disruption of a gene required for growth on acetate but not on ethanol: the acetyl-coenzyme A synthetase gene of Saccharomyces cerevisiae. Yeast. 1992;8:1043–1051. doi: 10.1002/yea.320081207. [DOI] [PubMed] [Google Scholar]
- 16.De Vos G F, Walker G C, Signer E R. Genetic manipulations in Rhizobium meliloti utilizing two new transposon Tn5 derivatives. Mol Gen Genet. 1986;204:485–491. doi: 10.1007/BF00331029. [DOI] [PubMed] [Google Scholar]
- 17.Driscoll B T, Finan T M. NADP+-dependent malic enzyme of Rhizobium meliloti. J Bacteriol. 1996;178:2224–2231. doi: 10.1128/jb.178.8.2224-2231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eggen R I L, Geerling A C M, Boshoven A B P, de Vos W M. Cloning, sequence analysis, and functional expression of acetyl coenzyme A synthetase gene from Methanothrix soehngenii in Escherichia coli. J Bacteriol. 1991;173:6383–6389. doi: 10.1128/jb.173.20.6383-6389.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finan T M, Hartwieg E K, LeMieux K, Bergman K, Walker G C, Signer E R. General transduction in Rhizobium meliloti. J Bacteriol. 1984;159:120–124. doi: 10.1128/jb.159.1.120-124.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finan T M, Kunkel B, DeVos G F, Signer E R. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986;167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedman A M, Long S R, Brown S E, Buikema W J, Ausubel F M. Construction of a broad host range cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 22.Fukui T, Ito M, Tomita K. Purification and characterization of acetoacetyl-CoA synthetase from Zoogloea ramigera I-16-M. Eur J Biochem. 1982;127:423–428. doi: 10.1111/j.1432-1033.1982.tb06889.x. [DOI] [PubMed] [Google Scholar]
- 23.Furuichi T, Inouye M, Inouye S. Novel one-step cloning vector with a transposable element: application to the Myxococcus xanthus genome. J Bacteriol. 1985;164:270–275. doi: 10.1128/jb.164.1.270-275.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garre V, Murillo F J, Torres-Martinez S. Isolation of the facA (acetyl-CoA synthetase) gene of Phycomyces blakesleeanus. Mol Gen Genet. 1994;244:278–286. doi: 10.1007/BF00285455. [DOI] [PubMed] [Google Scholar]
- 25.Grundy F J, Waters D A, Takova T Y, Henkin T M. Identification of genes involved in utilization of acetate and acetoin in Bacillus subtilis. Mol Microbiol. 1993;10:259–271. doi: 10.1111/j.1365-2958.1993.tb01952.x. [DOI] [PubMed] [Google Scholar]
- 26.James B W, Mauchline W S, Dennis P J, Keevil C W, Wait R. Poly-3-hydroxybutyrate in Legionella pneumophila, an energy source for survival in low-nutrient environments. Appl Environ Microbiol. 1999;65:822–827. doi: 10.1128/aem.65.2.822-827.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jenkins L S, Nunn W D. Genetic and molecular characterization of the genes involved in short-chain fatty acid degradation in Escherichia coli: the ato system. J Bacteriol. 1987;169:42–52. doi: 10.1128/jb.169.1.42-52.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jetten M S M, Stams A J M, Zehnder A J B. Isolation and characterization of acetyl-coenzyme A synthetase from Methanothrix soehngenii. J Bacteriol. 1989;171:5430–5435. doi: 10.1128/jb.171.10.5430-5435.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knauf V C, Nester E W. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982;8:45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- 30.Kumari S, Tishel R, Eisenbach M, Wolfe A J. Cloning, characterization, and functional expression of acs, the gene which encodes acetyl coenzyme A synthetase in Escherichia coli. J Bacteriol. 1995;177:2878–2886. doi: 10.1128/jb.177.10.2878-2886.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Law J H, Slepecky R A. Assay of poly-β-hydroxybutyric acid. J Bacteriol. 1961;82:33–36. doi: 10.1128/jb.82.1.33-36.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LeVine S M, Ardeshir F, Ames G F-L. Isolation and characterization of acetate kinase and phosphotransacetylase mutants of Escherichia coli and Salmonella typhimurium. J Bacteriol. 1980;143:1081–1085. doi: 10.1128/jb.143.2.1081-1085.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madison L L, Huisman G W. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol Mol Biol Rev. 1999;63:21–53. doi: 10.1128/mmbr.63.1.21-53.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marck C. DNA Strider: a “C” program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 1988;16:1829–1836. doi: 10.1093/nar/16.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meade H M, Long S R, Ruvkun G B, Brown S E, Ausubel F M. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon mutagenesis. J Bacteriol. 1982;149:114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oeding V, Schlegel H G. β-Ketothiolase from Hydrogenomonas eutropha H16 and its significance in the regulation of poly-β-hydroxybutyrate metabolism. Biochem J. 1973;134:239–248. doi: 10.1042/bj1340239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel J J, Gerson T. Formation and utilisation of carbon reserves by Rhizobium. Arch Microbiol. 1974;101:211–220. doi: 10.1007/BF00455939. [DOI] [PubMed] [Google Scholar]
- 38.Povolo S, Tombolini R, Morea A, Anderson A J, Casella S, Nuti M P. Isolation and characterization of mutants of Rhizobium meliloti unable to synthesize poly-β-hydroxybutyrate (PHB) Can J Microbiol. 1994;40:823–829. [Google Scholar]
- 39.Priefert H, Steinbüchel A. Identification and molecular characterization of the acetyl coenzyme A synthetase gene (acoE) of Alcaligenes eutrophus. J Bacteriol. 1992;174:6590–6599. doi: 10.1128/jb.174.20.6590-6599.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenberg C, Huguet T. The pAtC58 plasmid is not essential for tumour induction. Mol Gen Genet. 1984;196:533–536. [Google Scholar]
- 41.Senior P J, Dawes E A. The regulation of poly-β-hydroxybutyrate metabolism in Azotobacter beijerinckii. Biochem J. 1973;134:225–248. doi: 10.1042/bj1340225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon R, Quandt J, Klipp W. New derivatives of transposon Tn5 suitable for mobilization of replicons, generation of operon fusions, and induction of genes in Gram-negative bacteria. Gene. 1989;80:161–169. doi: 10.1016/0378-1119(89)90262-x. [DOI] [PubMed] [Google Scholar]
- 43.Steinbüchel A, Hustede E, Libergesell M, Pieper U, Timm A, Valentin H. Molecular basis for biosynthesis and accumulation of polyhydroxyalkanoic acids in bacteria. FEMS Microbiol Rev. 1992;103:217–230. doi: 10.1111/j.1574-6968.1992.tb05841.x. [DOI] [PubMed] [Google Scholar]
- 44.Summers M L, Denton M C, McDermott T R. Genes coding for phosphotransacetylase and acetate kinase in Sinorhizobium meliloti are in an operon that is inducible by phosphate stress and controlled by PhoB. J Bacteriol. 1999;181:2217–2224. doi: 10.1128/jb.181.7.2217-2224.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tal S, Okon Y. Production of the reserve material poly-β-hydroxybutyrate and its function in Azospirillum brasilense Cd. Can J Microbiol. 1985;31:608–613. [Google Scholar]
- 46.Thompson J D, Higgins D G, Gibson T J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thöny B, Hennecke H. The −24/−12 promoter comes of age. FEMS Microbiol Rev. 1989;5:341–357. doi: 10.1016/0168-6445(89)90028-4. [DOI] [PubMed] [Google Scholar]
- 48.Tombolini R, Nuti M P. Poly (β-hydoxyalkanoate) biosynthesis and accumulation by different Rhizobium species. FEMS Microbiol Lett. 1989;60:299–304. [Google Scholar]
- 49.Tombolini R, Povolo S, Buson A, Squartini A, Nuti M P. Poly-β-hydroxybutyrate (PHB) biosynthetic genes in Rhizobium meliloti 41. Microbiology. 1995;141:2553–2559. doi: 10.1099/13500872-141-10-2553. [DOI] [PubMed] [Google Scholar]
- 50.Van den Berg M A, Steensma H Y. ACS2, a Saccharomyces cerevisiae gene encoding acetyl-coenzyme A synthetase, essential for growth on glucose. Eur J Biochem. 1995;231:704–713. doi: 10.1111/j.1432-1033.1995.tb20751.x. [DOI] [PubMed] [Google Scholar]
- 51.Willis L B, Walker G C. The phbC (poly-β-hydroxybutyrate synthase) gene of Rhizobium (Sinorhizobium) meliloti and characterization of phbC mutants. Can J Microbiol. 1998;44:554–564. doi: 10.1139/w98-033. [DOI] [PubMed] [Google Scholar]
- 52.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]