Abstract
In growing cells of the yeast Saccharomyces cerevisiae, proaminopeptidase I reaches the vacuole via the selective cytoplasm-to-vacuole targeting (cvt) pathway. During nutrient limitation, autophagy is also responsible for the transport of proaminopeptidase I. These two nonclassical protein transport pathways to the vacuole are distinct in their characteristics but in large part use identical components. We expanded our initial screen for aut− mutants and isolated aut9-1 cells, which show a defect in both pathways, the vacuolar targeting of proaminopeptidase I and autophagy. By complementation of the sporulation defect of homocygous diploid aut9-1 mutant cells with a genomic library, in this study we identified and characterized the AUT9 gene, which is allelic with CVT7. aut9-deficient cells have no obvious defects in growth on rich media, vacuolar biogenesis, and acidification, but like other mutant cells with a defect in autophagy, they exhibit a reduced survival rate and reduced total protein turnover during starvation. Aut9p is the first putative integral membrane protein essential for autophagy. A biologically active green fluorescent protein-Aut9 fusion protein was visualized at punctate structures in the cytosol of growing cells.
The vacuolar protein sorting pathway is the classical route for vacuolar proteins like carboxypeptidase Y (CPY) to reach the vacuole. In the yeast Saccharomyces cerevisiae, cytosolic proteins are delivered to the vacuole by two nonclassical pathways (10, 24). During vegetative growth, the cytoplasm-to-vacuole targeting (cvt) pathway selectively carries the cytosolic proform of aminopeptidase I with a half time of ∼40 min to the vacuole (11). The cvt pathway acts as a biogenetic pathway, delivering a resident vacuolar proteinase to its site of action.
To survive periods of nutrient limitation, eukaryotic cells break down significant amounts of their intracellular material inside the lysosome (vacuole) via autophagy, the second nonclassical route to the vacuole. In contrast to the cvt pathway, autophagy is an unselective, starvation-induced bulk flow process (for reviews, see references 3, 6, 13, 20, and 24). Under induced conditions, ∼2% of all proteins per h are transported to the vacuole via autophagy (22). Despite these differences, genetic studies demonstrated that the two processes use at least in part the same machinery. aut− (23) and apg− (25) mutants exhibiting defects in the autophagic process have been isolated in two independent screens. Most of the autophagy mutant cells are also impaired in the vacuolar uptake of proaminopeptidase I. Furthermore, there is a significant genetic overlap between these two sets of autophagy mutants and the cvt− mutants, isolated due to their defect in transporting proaminopeptidase I to the vacuole (8, 9, 19).
During autophagy, cytosolic material is enclosed in double-membrane layered vesicles called autophagosomes with a diameter of 300 to 900 nm (1, 12). The origin of the limiting membranes in yeast has not yet been determined. We detected a microtubule-associated protein complex consisting of Aut2p and Aut7p (12); in aut2− cells autophagosome-like vesicles accumulate in the cytosol during starvation. After reaching the vacuole, the outer membrane of the double membrane-layered autophagosomes fuses with the vacuolar membrane (2). The inner part of autophagosomes thus appears as monolayered autophagic vesicles inside the vacuole. In the vacuole, autophagic vesicles together with their cytosolic content are rapidly broken down dependent on active proteinase B. Therefore, addition of the proteinase B inhibitor phenylmethylsulfonyl fluoride (PMSF) leads to the accumulation of autophagic vesicles in the vacuole during periods of nutrient limitation. This provides an easy way to phenotypically monitor autophagy in S. cerevisiae.
The proform of aminopeptidase I was initially detected by electron microscopy in the cytosol clustered in a large complex termed the cvt complex (1). In growing cells, this complex is taken up in double-layered cvt vesicles which resemble autophagosomes but are smaller (140 to 160 nm) and exclude cytoplasmic material (1). Most likely by using the autophagic machinery, these cvt vesicles reach the vacuole in a way similar to autophagosomes. In starving cells, the cvt complex was found inside autophagosomes together with cytosolic material (1). We expanded our initial screen for aut− mutants and meanwhile isolated eight AUT genes (12, 18, 22; our unpublished results). Here we report the identification and characterization of AUT9, a gene essential for both the cvt and the autophagic protein transport pathways. AUT9 encodes a potential integral membrane protein; a biologically active fusion protein of Aut9p with the green fluorescent protein (GFP) is visualized at punctate structures in the cytoplasm of growing cells.
MATERIALS AND METHODS
Strains.
S. cerevisiae strains used for these studies are listed in Table 1. The aut9-1 mutant strain YBK367, obtained by ethyl methanesulfonate (EMS) mutagenesis, was backcrossed twice with YMTA and the wild-type strain WCG4. YTL1 was obtained by chromosomal deletion of the ADE2 gene in an aut9-1 mutant strain with a 2.3-kb BamHI fragment from pPL131 (P. Ljungdahl, Stockholm, Sweden). By using the oligonucleotides Δaut9 KAN1 and Δaut9 KAN2 (Table 2) and plasmid pUG6, a DNA fragment for the chromosomal replacement of AUT9 with a LoxP-Kanr-LoxP cassette was created by PCR (7, 26). The haploid null mutant strain YSR2 was made by transforming the single LoxP-Kanr-LoxP cassette into the WCG4 diploid strain followed by sporulation and tetrad dissection. Correct gene replacement was confirmed by Southern analysis (not shown).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| WCG4a | Mata his3-11,15 leu2-3,112 ura3 | 23 |
| YMTAa | Mata his3-11,15 leu2-3,112 ura3 pep4Δ::HIS3 | 23 |
| YBK367 | Mata leu2-3,112 ura3 his3-11,15 aut9-1 (backcrossed twice with YMTA and WCG4) | This study |
| YTL1 | Mata his3-11,15 leu2-3,112 ura3 ade2Δ1 aut9-1 | This study |
| YSR2 | Mata his3-11,15 leu2-3,112 ura3 aut9Δ::KAN | This study |
| YTL2 | Matα his3-11,15 leu2-3,112 ura3 ADE2 aut9-1 | |
| Mata his3-11,15 leu2-3,112 ura3 ade2 aut9-1 | This study |
TABLE 2.
Oligonucleotides used
| Name | Sequence |
|---|---|
| GFP1 AUT9 | 5′-CGG AAT TATCCG GGG ATG GAG AGA GAT GAA TAC CAG TTA CCC-3′ |
| GFP2 AUT9 | 5′-GAC ACA GTC GTC GAC ATC TTC CGA CGT CAG ACT TCT TGT AAT AC-3′ |
| GFP3 AUT9 | 5′-GAC ACA GTC AAG CTT ATC TTC CGA CGT CAG ACT TCT TGT AAT AC-3′ |
| Δaut9 KAN1 | 5′-TAA GAA CAG CCT GAA ATA TCA AAA TCA CGG AAT TAT TAG GTT CAG CTG AAG CTT CGT ACG-3′ |
| Δaut9 KAN2 | 5′-AGT TAT ATT GGA TGA TGT ACA CGA CAC AGT CTG CCT TAG CAT AGG CCA CTA GTG GAT CTG-3′ |
The cvt− mutant strains and corresponding wild-type strains are described in references 8 and 9.
Screening procedure.
The ade2 deletion allele from pPL131 was chromosomally introduced into an aut9-1 mutant strain, and the resulting strain YTL1 was mated with an aut9-1 ADE2 mutant strain. The resulting diploid YTL2 was transformed with yeast genomic libraries based on the centromeric shuttle vector YCp50 (17) and the 2 μm shuttle vector Yep24. Transformants were screened as described previously (22) for the ability to sporulate.
Plasmids. (i) Yeast genomic library plasmids and subclones.
Plasmid pTL1 was derived from Ycp50/6 after digestion with SalI and religation. Plasmid pTL2 was obtained by digesting pTL1 with EcoRI and subsequent religation. pTL3 was created by removing an NruI DNA fragment from pTL1 and religation.
Construction of GFP-Aut9 fusion proteins.
The yeast shuttle vectors pRN295 and pRN963 (CEN6/ARSH4, URA3, amp) both contain the GFP gene under the control of the inducible MET25 promoter and allow generation of C- and N-terminal GFP fusion proteins, respectively (15). DNA fragments containing the entire AUT9 gene were generated by PCR using pTL1 and oligonucleotides GFP1 AUT9, GFP2 AUT9, and GFP3 AUT9. These fragments contained a SmaI site before nucleotide 1 and a SalI or HindIII site starting at nucleotide 2991 of the AUT9 gene, respectively. pRN295 was digested with SmaI-SalI, and pRN963 was digested with SmaI-HindIII; ligation of the SmaI-SalI and SmaI-HindIII fragments, respectively, yielded pGFP-N/AUT9 and pGFP-C/AUT9.
Visualization of the fusion proteins with GFP was done with a Zeiss Axioscope.
Other procedures.
If not otherwise noted, the cells were incubated for starvation in 1% potassium acetate solution. Measurement of total protein turnover was done as described in reference 22. Cells were prepared for electron microscopy by permanganate fixation and embedding in Epon as described elsewhere (22).
RESULTS
Isolation of the AUT9 gene.
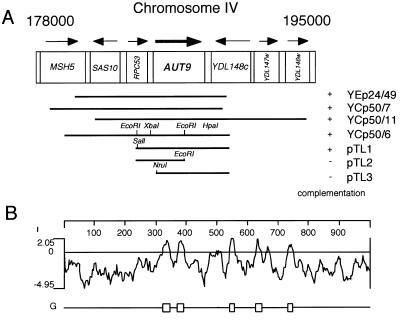
In a previous study, after EMS mutagenesis we isolated aut− mutant cells with a defect in autophagy (23) due to impairment in starvation-induced degradation of the cytosolic fatty acid synthase and an inability to accumulate autophagic vesicles in the vacuole during starvation in the presence of the proteinase B inhibitor PMSF. By expanding our initial screen, we isolated aut9-1 mutant cells (allelic with cvt7-1 mutant cells) as a member of a new complementation group. Like other aut− mutant cells, homozygous aut9-1 diploid cells are unable to undergo the cell differentiation process of sporulation (data not shown). After transformation with a YCp50 (17)-based yeast genomic library, we used this phenotype and a previously described procedure (18) to isolate three independent plasmids carrying complementing genomic fragments. Partial sequencing localized all genomic inserts to chromosome IV (Fig. 1A). Using an overexpressing YEp24-based genomic library, we identified an additional complementing genomic fragment, which also localized to chromosome IV (Fig. 1A). Subcloning of the genomic fragments identified open reading frame (ORF) YDL149w as the complementing ORF. Resequencing of AUT9 (YDL149w) revealed no differences to the sequence included in databases.
FIG. 1.
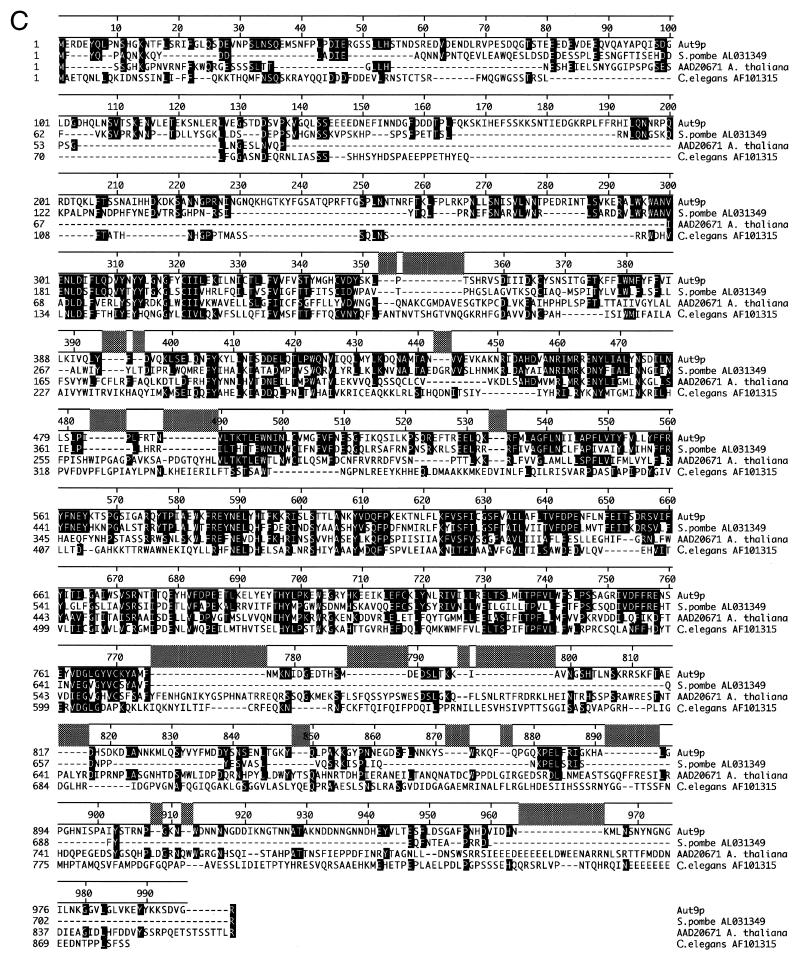
(A) Genomic fragments isolated after complementation of the sporulation defect of homocygous aut9Δ cells. (B) Hydrophobicity analysis of the Aut9p indicates five potential transmembrane domains. (C) Aut9p is the representative of a protein family of unknown function. Sequences were aligned by the Clustal method. Homologous residues (1 distance unit) are shaded.
For chromosomal deletion of YDL149w, a LoxP-Kanr-LoxP cassette (7, 26) was generated by PCR and integrated into the YDL149w locus of wild-type cells. Single integration of the construct was verified by tetrad dissection and correct gene replacement by Southern blotting (data not shown). The identity of YDL149w with AUT9 was confirmed by checking the inability of aut9-1 ydl149wΔ diploid cells to accumulate autophagic vesicles in the vacuole during starvation in the presence of PMSF (not shown).
AUT9 codes for a potential integral membrane protein.
AUT9 encodes a protein of 997 amino acids which shows homologies to proteins of unknown function from Schizosaccharomyces pombe, Arabidopsis thaliana, and Caenorhabditis elegans (Fig. 1C). Hydrophobicity analysis suggests five potential transmembrane domains (Fig. 1B). Prosite pattern search identified a potential bipartite nuclear localization signal (PS50079) between amino acids 795 and 812. The Proteome database (http://www.proteome.com/databases/YPD/reports/YDL149W.html) lists a potential mitochondrial transmembrane domain signature.
AUT9 is essential for autophagy and the cvt pathway but not for growth on rich media.
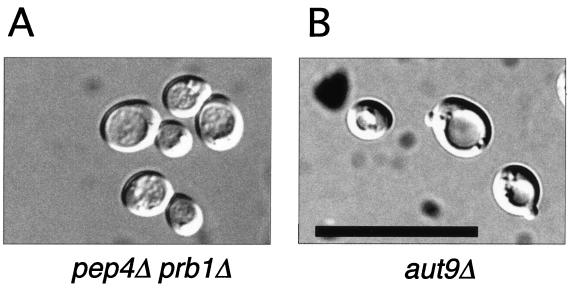
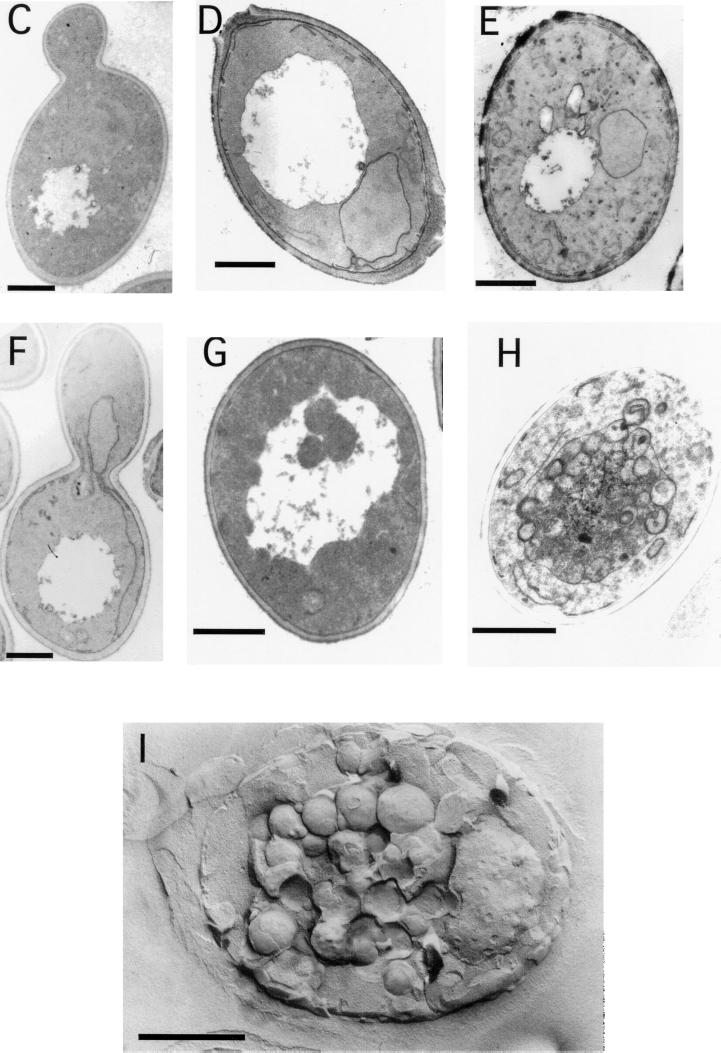
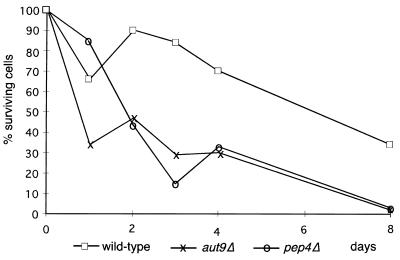
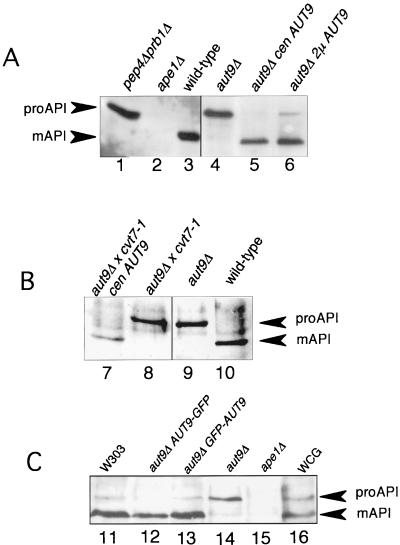
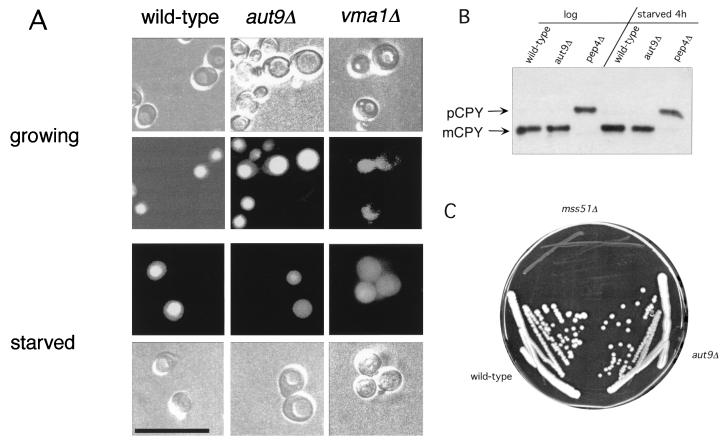
aut9Δ cells grew like wild-type cells on rich media at 18, 30, and 37°C (not shown); also, the morphology of their vacuoles appeared normal (Fig. 2A and B). As expected, aut9Δ cells exhibit phenotypes characteristic for mutant cells defective in autophagy. aut9Δ cells are unable to accumulate autophagic vesicles inside the vacuole during starvation for nitrogen in the presence of PMSF (Fig. 2B). We confirmed this phenotype by electron microscopy (Fig. 2C to I). aut9Δ cells further have a significantly reduced survival rate during starvation compared to wild-type cells (Fig. 3). aut9Δ cells are unable to mature proaminopeptidase I (Fig. 4A, lane 4); this demonstrates the essential function of Aut9p not only for autophagy but also for the cvt pathway.
FIG. 2.
aut9Δ cells (B to E) show a defect in the accumulation of autophagic vesicles in the vacuole during starvation in the presence of PMSF. As a control, we also checked cells with defects in the major vacuolar endoproteinases (pep4Δ prb1Δ) (A and F to I). Freeze etching nicely illustrates the accumulation of autophagic vesicles in starved pep4Δ prb1Δ cells (I). Cells were taken from the logarithmic growth phase (C and F), starved for 5 h (D and G), or starved for 24 h (E, H, and I). (A and B) Nomarski optics (bar, 20 μm). (C to H) Thin-section electron microscopy; I, freeze etching electron microscopy (bars, 1 μm).
FIG. 3.
aut9Δ cells, like pep4Δ cells, show a reduced survival rate during starvation compared to wild-type cells. Cells were starved in 1% potassium acetate solution, and aliquots were plated out to determine the proportion of surviving cells.
FIG. 4.
(A) Maturation of proaminopeptidase I is impaired in aut9Δ cells. (B) cvt7-1 cells are allelic with aut9Δ cells. (C) Fusion proteins of GFP with the amino and carboxy termini of Aut9p, respectively, are biologically active.
cvt7-1 mutant cells have been isolated due to their defect in vacuolar import and maturation of proaminopeptidase I (8). aut9Δ cvt7-1 diploid cells show mutant phenotypes (Fig. 4B, lane 8), which can be complemented by a plasmid-borne AUT9 gene (Fig. 4B, lane 7). This further confirms the allelism of aut9Δ and cvt7-1 cells.
Total protein turnover is reduced in starving aut9 (cvt7)-deficient cells.
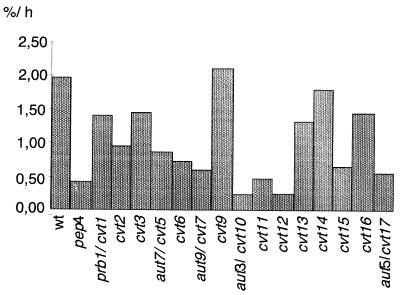
We next measured the significantly reduced total protein breakdown rate during starvation for nitrogen, another leading phenotype of mutants with autophagic defects by which to quantitatively follow the autophagic process. So far the autophagic defects of cvt− mutant cells have been checked only by following the accumulation of autophagic vesicles in the vacuole during starvation in the presence of the proteinase B inhibitor PMSF (8). Because autophagic vesicles are hard to visualize in the genetic background of the cvt− mutants, we included in our analysis not only cvt7 cells (allelic with aut9 cells) but all cvt− mutant strains isolated so far (8, 9) with the exception of cvt4-1 and cvt8-1, because these strains are allelic to vps39 and vps41 mutants, respectively. The cells were radiolabeled with [35S]methionine and then shifted to starvation medium. Aliquots were taken, and the amount of radiolabeled acid-soluble small peptides generated by proteolysis was measured. aut− mutant cells typically show only 20 to 30% of the breakdown rate of wild-type cells (12, 18, 22). aut− mutants in this respect resemble pep4Δ cells, which have a significantly reduced vacuolar protein degradation rate. As expected, cvt5-1 (allelic to aut7), cvt7-1 (allelic to aut9), and cvt10-1 (allelic to aut3) together with several other cvt− mutant strains exhibited significantly reduced turnover rates comparable with pep4Δ cells. Most interestingly, some cvt− mutant cells like cvt9-1 cells exhibit a wild-type-like or partially reduced total protein turnover rate during starvation (Fig. 5). A specific defect predominantly in the transport of proaminopeptidase I but not in bulk flow autophagy suggests a specific function of the respective gene products during the cvt pathway.
FIG. 5.
Total protein turnover during starvation for nitrogen was measured in cvt− mutants by labeling all proteins with [35S]methionine and determining the amount of acid-soluble small peptides generated by proteolysis. Data shown are the average from several independent experiments. Some cvt− mutants such as cvt9-1 cells show wild-type-like or partially reduced protein turnover, suggesting that these mutants are affected predominantly in the cvt pathway rather than in autophagy.
Proteinase A-deficient cells and cells deficient in vacuolar endoproteinase B both show an accumulation of autophagic vesicles inside the vacuole during starvation. pep4-deficient cells show as expected a ∼80% reduction in the starvation-induced protein breakdown. prb1-deficient cells, however, still have a residual proteolysis rate of about two-thirds of the wild-type level (Fig. 5) (14), a phenomenon which we do not fully understand. cvt17-1 mutant cells (allelic with aut5-1 cells) show a reduced total protein turnover rate. aut5-deficient cells exhibit a defect in lysing autophagic vesicles inside the vacuole (our unpublished results).
Biogenesis of the vacuole is not obviously disturbed in aut9Δ cells.
We were further interested in determining if the chromosomal deletion of AUT9 causes some defects in the biogenesis of the vacuole. The dye quinacrine is used to monitor the acidification of the vacuole (16). In growing and in starved aut9Δ cells, the pH-dependent accumulation of quinacrine inside the vacuole showed no significant differences compared to wild-type cells (Fig. 6A). As a control, we included vma1Δ cells, which exhibit a defect in vacuolar acidification.
FIG. 6.
(A) Accumulation of quinacrine in the vacuoles of growing and starved aut9Δ cells suggests wild-type-like acidification. Starved cells were incubated for 4 h in 1% potassium acetate. Bar, 20 μm. (B) In growing and in starved aut9Δ cells, only mature CPY is detectable in immunoblots. (C) Growth of aut9Δ cells is wild-type-like on YP medium containing ethanol as a carbon source. As a control, mss51Δ cells, known to exhibit a petite phenotype (5), are included.
Vacuolar protein sorting is the classical route for vacuolar proteins to reach the vacuole. Analyses of the steady-state levels of CPY in growing and in starved aut9-deleted cells led to the detection of only mature CPY; no accumulation of unprocessed precursor was found (Fig. 6B). This suggests that Aut9p is not involved in this vesicle-mediated protein transport pathway.
Since a potential mitochondrial transmembrane domain signature is reported in the Proteome database, we checked the ability of aut9Δ cells to grow on a nonfermentable carbon source such as ethanol. No difference compared to wild-type cells was detectable (Fig. 6C). As a control, we included mss51Δ cells, known to exhibit petite phenotypes (5).
Biologically active GFP-Aut9p in growing aut9Δ cells is located at punctate structures in the cytosol.
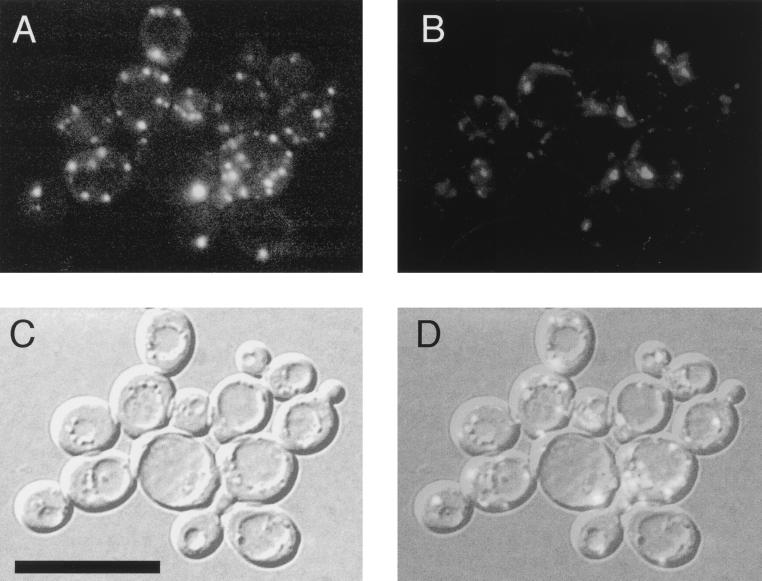
Using plasmids pRN295 and pRN963, we generated in-frame fusions of Aut9p with GFP from Aequoria victoria (4, 21) under control of the inducible MET25 promoter. GFP was fused to the amino and carboxy termini of Aut9p. Both fusion proteins were biologically active, as indicated by complementation of the maturation defect of proaminopeptidase I in aut9Δ cells (Fig. 4C, lanes 12 and 13). During growth on methionine-free medium to induce expression of the fusion proteins, only the amino-terminally fused GFP-Aut9p was detectable due to its green fluorescence. In aut9Δ cells growing on methionine-free medium, GFP-Aut9p was visualized at punctate structures in the cytosol, which were most prominent and numerous at an optical density (OD) of 2.2 (Fig. 7).
FIG. 7.
(A) Biologically active GFP-Aut9 fusion protein in aut9Δ cells grown to an OD of 2.2, visualized at punctate structures in the cytosol; (B) DAPI staining; (C) Nomarski optics; (D) overlay of panels A to C. Bar, 20 μm.
DISCUSSION
We expanded our initial screen for aut− mutants and, after EMS mutagenesis of wild-type cells, isolated aut9-1 mutant cells for their inability to degrade cytosolic fatty acid synthase during starvation for nitrogen. aut9-1 mutant cells are further unable to accumulate autophagic vesicles in the vacuole during starvation in the presence of PMSF. aut9-deficient cells are allelic with cvt7 mutant cells (Fig. 4B, lane 8) (8). This demonstrates the essential function of Aut9p for both autophagy and the cvt pathway.
Like other aut− mutant cells, homozygous aut9-1 diploid cells are severely impaired in sporulation. This phenotype allowed us to isolate the AUT9 gene by complementation with a centromeric and an overexpressing genomic library following a previously (22) described procedure. All four isolated complementing fragments contained ORF YDL149w (Fig. 1A), which we demonstrate is identical with AUT9. Aut9p has five potential transmembrane domains (Fig. 1B); it is a member of a protein family consisting of four proteins from different species such as S. cerevisiae, S. pombe, A. thaliana, and C. elegans (Fig. 1C). The biological function of this protein family is not known. Chromosomal deletion of the AUT9 gene did not affect growth on rich media at 18, 30, or 37°C (not shown). As expected, aut9-deficient cells showed the phenotypes characteristic for mutants with a defect in autophagy. During starvation for nitrogen in the presence of PMSF, no accumulation of autophagic vesicles was detectable using light (Fig. 2B) and electron (Fig. 2D and E) microscopy. The survival (Fig. 3) and total protein turnover rate (Fig. 5) during starvation are significantly reduced. Aut9p is also essential for the selective cvt pathway delivering proaminopeptidase I to the vacuole (Fig. 4A, lane 4); this finding is further supported by the allelism of aut9-deficient cells with cvt7-1 mutant cells (Fig. 4B, lanes 7 and 8). We further checked the relevance of AUT9 for vacuolar biogenesis and acidification. Analysis of the steady-state level of CPY in growing and starved cells did not show the accumulation of unmatured species in aut9Δ cells (Fig. 6B). The accumulation of the fluorescent dye quinacrine suggested wild-type like vacuolar acidification in growing and starved aut9Δ cells (Fig. 6A). In the Proteome database, a potential mitochondrial transmembrane signature domain is annotated. But the wild-type-like growth of aut9Δ cells (Fig. 6C) on rich media containing ethanol as a carbon source does not suggest an involvement of AUT9 in mitochondrial function. Hydrophobicity analysis of Aut9p points to the existence of about five transmembrane domains (Fig. 1B); Aut9p is therefore predicted to be the first integral membrane protein involved in autophagy. To learn more about the intracellular localization of Aut9p, we constructed fusion proteins of Aut9p with GFP. Fusion of GFP at the amino and carboxy termini of Aut9p led to biological activity, monitored by complementation of the maturation defect of proaminopeptidase I in aut9Δ cells (Fig. 4C, lanes 12 and 13). Only the amino-terminally fused GFP-Aut9p was visible in cells due to its green fluorescence. GFP-Aut9p was visualized at punctate structures, which were most prominent and numerous at an OD of 2.2 in the cytosol of aut9Δ cells (Fig. 7A).
The selective cytoplasm-to-vacuole targeting of proaminopeptidase I and the starvation-induced unspecific bulk flow autophagy are two nonclassical protein transport pathways to the vacuole. Although the characteristics of the two processes appear to be quite different, the mechanistic features and gene products involved are largely the same. It has been reported that almost all mutants with a defect in autophagy also show a block in vacuolar delivery and maturation of proaminopeptidase I (8, 19). This supports the idea that the autophagic machinery is also used for the transport of proaminopeptidase I to the vacuole. So far the autophagic capacity of the cvt− mutant strains has been checked only by their ability to accumulate autophagic vesicles inside the vacuole during starvation for nitrogen in the presence of PMSF. This phenotype has several limitations. First, in the cvt− mutant cells autophagic vesicles are hard to detect with Nomarski optics because the vacuole is not easily seen in this background after the cells are starved. Furthermore, the accumulation of autophagic vesicles is very difficult to quantitate. When checked by light microscopy, cells with a defect in the vacuolar endoproteinase A or B are phenotypically similar, and autophagic vesicles accumulate in the vacuoles of both types of mutant cells. As a more reliable phenotype to follow autophagy quantitatively, we measured the total intracellular protein breakdown rate and examined not only cvt7 cells, which are allelic with aut9 cells, but all cvt− mutant strains to obtain more precise data on the overlap between the two processes. Measurement of the total protein breakdown rate allows the detection of significant differences between proteinase A- and proteinase B-deficient cells. In proteinase A-deficient (pep4Δ) cells, vacuolar proteolysis is almost completely blocked (Fig. 5), whereas proteinase B-deficient (prb1Δ) cells still exhibit about two-thirds of the wild-type proteolysis rate (Fig. 5). The reason for the discrepancy between the accumulation of autophagic vesicles and the total protein breakdown rate of these two strains is not fully understood.
Among the cvt− mutants, several strains, especially cvt9-1, showed a wild-type-like or partially reduced turnover rate suggesting a mostly unaffected autophagy (Fig. 5). As possible components specific for the vacuolar targeting of proaminopeptidase I, we expect a receptor molecule which mediates the selective uptake of proaminopeptidase I in cvt vesicles or autophagosomes and components which are involved in the formation of the cvt complex itself.
Interestingly, cvt17-1 cells, which are allelic with aut5-1 mutant cells, also show a reduced protein turnover rate during starvation (Fig. 5). aut5-1/cvt17-1 mutant cells differ from other cvt− mutants in accumulating the proform of aminopeptidase I not in the cytosol but inside the vacuole (8). We could demonstrate that AUT5 is essential for the lysis of autophagic vesicles in the vacuole (our unpublished results). Components common to both pathways may include the autophagic machinery involved in formation of autophagosomes, their probable transport to and fusion with the vacuole, and finally the lysis of autophagic vesicles.
The visualization of the GFP-Aut9p at punctate structures in the cytosol of growing cells and the essential function of the putative integral membrane protein Aut9p for autophagy and the cvt pathway open the possibility to further analyze both pathways.
ACKNOWLEDGMENTS
This work was supported by DFG grant Wo 210/12-3.
We thank M. Schlumpberger for technical help and D. J. Klionsky for providing the cvt− mutant strains. We are grateful to Dieter H. Wolf for support and many helpful discussions.
REFERENCES
- 1.Baba M, Osumi M, Scott S V, Klionsky D J, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba M, Takeshige K, Baba N, Ohsumi Y. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J Cell Biol. 1994;124:903–913. doi: 10.1083/jcb.124.6.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Codogno P, Ogier-Denis E, Houri J J. Signal transduction pathways in macroautophagy. Cell Signal. 1997;9:125–130. doi: 10.1016/s0898-6568(96)00130-1. [DOI] [PubMed] [Google Scholar]
- 4.Cubitt A B, Heim R, Adams S R, Boyd A E, Gross L A, Tsien R Y. Understanding, improving and using green fluorescent proteins. Trends Biochem Sci. 1995;20:448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- 5.Decoster E, Simon M, Hatat D, Faye G. The MSS51 gene product is required for the translation of the COX1 mRNA in yeast mitochondria. Mol Gen Genet. 1990;224:111–118. doi: 10.1007/BF00259457. [DOI] [PubMed] [Google Scholar]
- 6.Dunn W J. Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol. 1994;4:139–143. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 7.Güldener U, Heck S, Fielder T, Beinhauer J, Hegemann J H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harding T M, Hefner-Gravink A, Thumm M, Klionsky D J. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole targeting pathway. J Biol Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- 9.Harding T M, Morano K A, Scott S V, Klionsky D J. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klionsky D J. Nonclassical protein sorting to the yeast vacuole. J Biol Chem. 1998;273:10807–10810. doi: 10.1074/jbc.273.18.10807. [DOI] [PubMed] [Google Scholar]
- 11.Klionsky D J, Cueva R, Yaver D S. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lang T, Schaeffeler E, Bernreuther D, Bredschneider M, Wolf D H, Thumm M. Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J. 1998;17:3597–3607. doi: 10.1093/emboj/17.13.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mortimore G E, Miotto G, Venerando R, Kadowaki M. Autophagy. Subcell Biochem. 1996;27:93–135. doi: 10.1007/978-1-4615-5833-0_4. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura N, Matsuura A, Wada Y, Ohsumi Y. Acidification of vacuoles is required for autophagic degradation in the yeast, Saccharomyces cerevisiae. J Biochem (Tokyo) 1997;121:338–344. doi: 10.1093/oxfordjournals.jbchem.a021592. [DOI] [PubMed] [Google Scholar]
- 15.Niedenthal R K, Riles L, Johnston M, Hegemann J H. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 16.Roberts C J, Raymond C K, Yamahiro C T, Stevens T H. Methods for studying the yeast vacuole. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- 17.Rose M D, Novick P, Thomas J H, Botstein D, Fink G R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- 18.Schlumpberger M, Schaeffeler E, Straub M, Bredschneider M, Wolf D H, Thumm M. AUT1, a gene essential for autophagocytosis in the yeast Saccharomyces cerevisiae. J Bacteriol. 1997;179:1068–1076. doi: 10.1128/jb.179.4.1068-1076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott S V, Hefner-Gravink A, Morano K A, Noda T, Ohsumi Y, Klionsky D J. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc Natl Acad Sci USA. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seglen P O, Berg T O, Blankson H, Fengsrud M, Holen I, Stromhaug P E. Structural aspects of autophagy. Adv Exp Med Biol. 1996;389:103–111. doi: 10.1007/978-1-4613-0335-0_12. [DOI] [PubMed] [Google Scholar]
- 21.Stearns T. Green fluorescent protein. The green revolution. Curr Biol. 1995;5:262–264. doi: 10.1016/s0960-9822(95)00056-x. [DOI] [PubMed] [Google Scholar]
- 22.Straub M, Bredschneider M, Thumm M. AUT3, a serine/threonine kinase gene, is essential for autophagocytosis in Saccharomyces cerevisiae. J Bacteriol. 1997;179:3875–3883. doi: 10.1128/jb.179.12.3875-3883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf D H. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- 24.Thumm M, Wolf D H. From proteasome to lysosome: studies on yeast demonstrate the principles of protein degradation in the eukaryote cell. In: Rivett A J, editor. Advances in molecular and cell biology. Greenwich, Conn: JAI Press; 1998. pp. 41–67. [Google Scholar]
- 25.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 26.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]