Abstract
Nocardioides sp. strain KP7 grows on phenanthrene but not on naphthalene. This organism degrades phenanthrene via 1-hydroxy-2-naphthoate, o-phthalate, and protocatechuate. The genes responsible for the degradation of phenanthrene to o-phthalate (phd) were found by Southern hybridization to reside on the chromosome. A 10.6-kb DNA fragment containing eight phd genes was cloned and sequenced. The phdA, phdB, phdC, and phdD genes, which encode the α and β subunits of the oxygenase component, a ferredoxin, and a ferredoxin reductase, respectively, of phenanthrene dioxygenase were identified. The gene cluster, phdAB, was located 8.3 kb downstream of the previously characterized phdK gene, which encodes 2-carboxybenzaldehyde dehydrogenase. The phdCD gene cluster was located 2.9 kb downstream of the phdB gene. PhdA and PhdB exhibited moderate (less than 60%) sequence identity to the α and β subunits of other ring-hydroxylating dioxygenases. The PhdC sequence showed features of a [3Fe-4S] or [4Fe-4S] type of ferredoxin, not of the [2Fe-2S] type of ferredoxin that has been found in most of the reported ring-hydroxylating dioxygenases. PhdD also showed moderate (less than 40%) sequence identity to known reductases. The phdABCD genes were expressed poorly in Escherichia coli, even when placed under the control of strong promoters. The introduction of a Shine-Dalgarno sequence upstream of each initiation codon of the phdABCD genes improved their expression in E. coli. E. coli cells carrying phdBCD or phdACD exhibited no phenanthrene-degrading activity, and those carrying phdABD or phdABC exhibited phenanthrene-degrading activity which was significantly less than that in cells carrying the phdABCD genes. It was thus concluded that all of the phdABCD genes are necessary for the efficient expression of phenanthrene-degrading activity. The genetic organization of the phd genes, the phylogenetically diverged positions of these genes, and an unusual type of ferredoxin component suggest phenanthrene dioxygenase in Nocardioides sp. strain KP7 to be a new class of aromatic ring-hydroxylating dioxygenases.
The aerobic degradation of polycyclic aromatic hydrocarbons (PAHs), especially of low-molecular-weight ones such as naphthalene and phenanthrene, has been extensively studied (8, 19, 20). Bacteria degrade phenanthrene by one of two distinct routes. Bacteria capable of growing on both phenanthrene and naphthalene mineralize these substrates via salicylate and catechol (the salicylate pathway), while those capable of growing on phenanthrene but not naphthalene degrade phenanthrene via protocatechuate (the protocatechuate pathway) (29).
On the salicylate pathway, phenanthrene and naphthalene are converted to 1-hydroxy-2-naphthoate and salicylate, respectively, by the same set of enzymes (30, 54). The amino acid sequences of these enzymes (48, 54) were >90% identical to those of naphthalene-degrading enzymes encoded by the NAH7 plasmid in Pseudomonas putida G7 (11, 13, 42), by the pDTG1 plasmid in P. putida NCIB 9816-4 (31), and by the C18 plasmid in Pseudomonas sp. strain C18 (10). This observation has led to the expectation that the sequence diversity in the genes for the degradation of naphthalene and phenanthrene in bacteria would be limited. However, this view has recently been modified. Pseudomonas sp. strain U2 isolated by selective enrichment on naphthalene converts this substrate to salicylate, but salicylate is transformed differently from other naphthalene-degrading bacteria to gentisate. The organization of genes for naphthalene dioxygenase in strain U2 was different from that in other naphthalene-degrading bacteria, the genes for salicylate 5-hydroxylase being inserted in the gene cluster for naphthalene dioxygenase (18). Similarly, the genes for the degradation of naphthalene and phenanthrene in Burkholderia sp. strain RP007 have been shown to be significantly different in sequence and gene order from the above-mentioned naphthalene- or phenanthrene-degrading genes (32).
The catabolic genes from bacteria capable of growing on phenanthrene but not naphthalene have not previously been characterized. Nocardioides sp. strain KP7 was isolated from a Kuwait beach on the basis of its ability to degrade phenanthrene 2 years after the oil pollution occurred in 1991 (27). This organism transforms phenanthrene via the protocatechuate pathway. The genes and enzymes involved in the transformation of 1-hydroxy-2-naphthoate to o-phthalate in this organism have recently been characterized (24–26). In the present study, we characterize the genes for phenanthrene dioxygenase, the enzyme involved in the first step of phenanthrene catabolism, in this organism.
(A preliminary account of this work was presented at the May 1998 IUPAC International Conference. “Degradation Processes in the Environment” [39].)
MATERIALS AND METHODS
Bacterial strains and culture conditions.
A gene library of Nocardioides sp. KP7 (27), a phenanthrene-degrading bacterium, was constructed in Escherichia coli S17-1 (43). E. coli HB101 (40), JM109 (55), and BL21(DE3) (45) were used as the hosts for plasmid construction or gene expression. E. coli strains were grown at 37°C on Luria-Bertani medium (40) or M9-glucose medium (40) supplemented with 100 μg of ampicillin per ml. Nocardioides sp. KP7 was grown at 30°C in 5 ml of marine broth (Difco Laboratories) containing 10 mg of phenanthrene. The stationary-phase culture exhibited a tinge of pink.
PFGE and hybridization.
The cells of Nocardioides sp. strain KP7 (2 × 107 to 5 × 107) were embedded in a gel plug made of 1.0% (wt/vol) CleanCut agarose (Bio-Rad Laboratories) and treated with restriction enzymes by using a CHEF genomic DNA plug kit (Bio-Rad). Plugged samples were placed in slots of a running gel made of 1.0% (wt/vol) pulsed-field certified agarose (Bio-Rad) and fixed by pouring molten 1.0% CleanCut agarose in the slots. Pulsed-field gel electrophoresis (PFGE) was performed with CHEF model DR II apparatus (Bio-Rad) under the following conditions: electrophoresis buffer, 0.5× Tris-borate-EDTA; voltage, 200 V (6 V/cm); reorientation angle, 120°; pulse time, linearly increased from 60 to 120 s; run times, 22 h; temperature, 15°C. Chromosome DNAs of Saccharomyces cerevisiae YNN295 and concatemers of lambda 50-kb DNA (Bio-Rad) were used as size standards. The DNA bands were visualized by ethidium bromide. DNAs in a pulsed-field gel were blotted onto a nylon membrane of Hybond-N+ (Amersham). The 2.5-kb HindIII-BamHI DNA fragment (Fig. 1; nucleotides 6776 to 9312 of entry AB000735 in the DDBJ/GenBank/EMBL DNA databases), which contained almost the entire phdI gene (the structural gene for 1-hydroxy-2-naphthoate dioxygenase) (26) and the entire phdJ gene (the structural gene for trans-2′-carboxybenzalpyruvate hydratase-aldolase) (25), was used as a probe. Labeling of probe DNA, hybridization, and signal development were carried out by using the Amersham ECL (enhanced chemiluminescence) direct nucleic acid labeling and detection system.
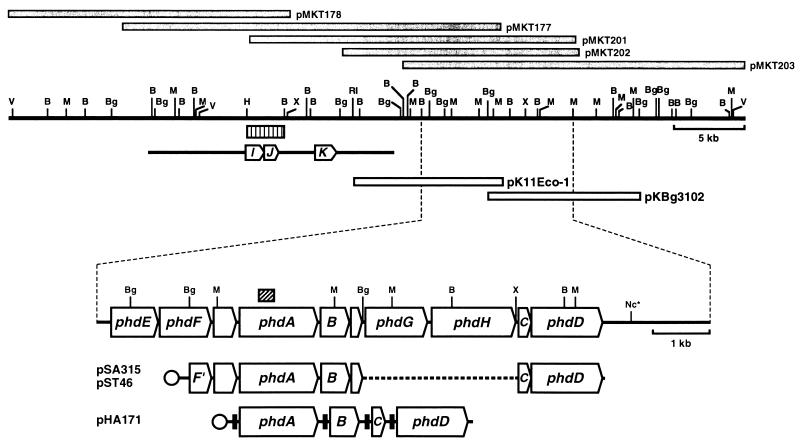
FIG. 1.
Physical map and genetic organization of phenanthrene-degrading genes. Gray bars indicate the DNA inserts in the library clones. Open bars show inserts in the two clones used in this study. Arrows on the enlarged physical map show the locations and orientations of the phenanthrene-degrading genes: phdA, the α subunit of the oxygenase component; B, phdB, the β subunit of the oxygenase component; C, phdC, the ferredoxin component; phdD, the ferredoxin reductase component; phdE, dihydrodiol dehydrogenase; phdF, extradiol dioxygenase; phdG, hydratase-aldolase; phdH, aldehyde dehydrogenase. The box with oblique indicates the 302-bp PCR-amplified fragment that was used for probing the library clones. The typical expression plasmids are shown at the bottom. Circles indicate promoters. Transcription of DNA segments containing phdABCD genes cloned in pSA315 and pST46 is under the control of the lac and T7 promoters, respectively. The transcription in pHA171 of the phdABCD genes preceded by the SD sequence that is effective in E. coli (filled boxes) is under the control of the T7 promoter. The bar with three arrows indicates the previously reported sequence (AB000735 in DDBJ/GenBank/EMBL databases), arrows marked with I, J, and K showing the locations and orientations of the phenanthrene-degrading genes phdI (1-hydroxy-2-naphthoate dioxygenase), phdJ (trans-2′-carboxybenzalpyruvate hydratase-aldolase), and phdK (2-carboxybenzaldehyde dehydrogenase). The striped bar shows the 2.5-kb HindIII-BamHI DNA fragment that was used as probe in the hybridization following PFGE (Fig. 2). Restriction sites: B, BamHI; Bg, BglII; H, HindIII, M, MluI; Nc, NcoI; RI, EcoRI; V, EcoRV; X, XbaI. The asterisk indicates that the other NcoI restriction sites are not shown.
Cloning and sequencing.
The gene library of Nocardioides sp. strain KP7 based on pLAFR3 (44) was constructed as reported by Iwabuchi et al. (26). To produce a probe against the gene for the ring-hydroxylating dioxygenase of Nocardioides sp. strain KP7, the degenerate primers DO-1s [5′-TG(TC)AG(TC)T(AT)(TC)CA(TC)GG(GATC)TGG-3′] and DO-1a [5′-TC(GATC)(GA)C(GATC)GC(AG)AA(TC)TTCCA(AG)TT-3′] were designed from conserved amino acid sequences of the known α subunits of the ring-hydroxylating dioxygenase (10, 15, 23, 31, 34, 42, 47, 48, 58). A PCR with this primer pair and KP7 total DNA as a template amplified a 0.3-kb fragment. This fragment was cloned in the SrfI site of pCR-Script SK(+) (Stratagene Cloning Systems), giving pK11-300-5, and the 302-bp-long sequence was determined. The genomic library was then screened by colony hybridization, using the 302-bp fragment as a probe. Labeling of the probe DNA, hybridization, and signal development were carried out by using the Amersham ECL direct nucleic acid labeling and detection system. The four clones, pMKT177 (24, 26), pMKT201, pMKT202, and pMKT203, gave a positive signal.
The 10.5-kb EcoRI fragment of pMKT177 and the 10.5-kb BglII fragment of pMKT203 were cloned into the EcoRI site and BglII site, respectively, of pSL301 (7) to produce pK11Eco-1 and pKBg3102 (Fig. 1). Using pK11Eco-1, pKBg3102, and their subclone derivatives, the nucleotide sequence of the 10.6-kb segment encompassing the genes for phenanthrene dioxygenase (phdABCD) was determined using a Taq DyeDeoxy terminator cycle sequencing kit and a 373A DNA sequencer (Perkin-Elmer Applied Biosystems).
Homology search and phylogenetic analysis.
The nucleotide sequence was processed by SeqEd version 1.0.3 and Genetyx version 8.0 software, and a homology search was performed with BLAST 2.0 (gapped BLAST) (1). Protein sequences were aligned by using CLUSTAL W version 1.7 (49), phylogenetic analysis was performed by the PHYLIP version 3.572c package (16), and a neighbor-joining tree was constructed by the NEIGHBOR program. The resulting unrooted tree was depicted by using the TreeView version 1.5 package (48).
Construction of expression plasmids.
The 3-kb BglII fragment from pKEco-1 containing phdE, phdA, and phdB was cloned into the BglII site of pSL301 to give pKG1021. The 2-kb XbaI-NcoI fragment from pKBg3102 containing phdC and phdD was cloned between the XbaI and NcoI sites of pSL301 to give pKXN382. A HindIII site was introduced immediately after the termination codon of phdD by using a PCR primer, 5′-CCCCAAGCTTGGAATTCTTCCTCGGCTCATGCCGTCGGT-3′ (the underlined sequences are the HindIII and EcoRI sites), and the XbaI-HindIII fragment containing phdC and phdD was cloned between the XbaI and HindIII sites of pKG1021 to give pSA315 (Fig. 1). pT7-7(BglIIX) is a derivative of pT7-7 (3) in which the BglII site had been disrupted for the convenience of DNA cloning (39). The 4.6-kb EcoRI fragment from pSA315 including the phdABCD genes was cloned into the EcoRI site of pT7-7 (BglIIX), giving pST46 (Fig. 1). The phd genes in pSA315 and pST46 were arranged in the order phdA-phdB-phdC-phdD under the control of the lac and T7 promoters, respectively. The 1.36-kb fragment containing phdA was prepared by PCR using pKG1021 as the template and primers 5′-GGGAATTCCATATGTCGGTAGTCAGCGGGGAT-3′ (the underlined sequence is the NdeI site) and 5′-CCGGAATTCGGTCGCAACTCATAAGACAGC-3′ (the underlined sequence is the EcoRI site). The amplified product was cloned between the NdeI and EcoRI sites of pT7-7(BglIIX) to give pHA102. The entire phdA gene in pHA102 was located under the control of the T7 promoter and an efficient Shine-Dalgarno (SD) sequence derived from the pT7-7(BglIIX) vector. The 0.56-kb fragment containing phdB was prepared by PCR using pKG1021 as the template and primers 5′-CCGGAAT TCAAGGAGATATACATATGC TGAC TAC TG T TGACGAGAATC-3′ (the underlined sequences are the EcoRI and NdeI sites) and 5′-CGCGGATCCAGATCTGCCTGCGGGCTAGAAG AAGAACGC-3′ (the underlined sequences are the BamHI and BglII sites). The PCR product was cloned between the EcoRI and BamHI sites of pT7-7(BglIIX) to give pHB103. The phdB gene in pHB103 was preceded by the efficient SD sequence, AAGGAG, derived from one of the primers used, and transcribed under the control of the T7 promoter. The 0.25-kb DNA fragments containing AAGGAG-phdC was PCR amplified by using pKXN382 as the template and the two primers 5′-CGCGGATCCAGATCTAAGGAGATATACATATGCGTGTGGATGTTGACCCACAGCGG-3′ (the underlined sequences are the BamHI, BglII, and NdeI sites) and 5′-GCCCAAGCTTGGCCTCCCGTCACTGAGCGGG-3′ (the underlined sequence is the HindIII site) and then cloned between the BamHI and HindIII sites of pT7-7(BglIIX) to give pHC102. The 1.26-kb fragment containing phdD was prepared by PCR using pKXN382 as the template and the two primers 5′-GGGAATTCCATATGACGGGAGGCCAGGTGGCGGCGC-3′ (the underlined sequence is the NdeI site) and 5′-CCCCAAGCTTGGAATTCTTCCTCGGCTCATGCCGTCGGT-3′ (the underlined sequence is the HindIII site), and the amplified fragment was cloned between the NdeI and HindIII sites of pT7-7(BglIIX) to give pHD115. To introduce the SD sequence upstream of phdD, two oligonucleotides, 5′-CTAGAAAGCTTAGATCTAAGGAGATATACA-3′ (the underlined sequences are the HindIII and BglII sites) and 5′-TATGTATATCTCCTTAGATCTAAGCTTT-3′ (the underlined sequences are the BglII and HindIII sites), were annealed each other, and the double-stranded oligonucleotides were inserted between the XbaI-NdeI sites of pHD115 to give pHD101. The 0.56-kb EcoRI-BamHI fragment from pHB103 containing AAGGAG-phdB was cloned between the EcoRI and BamHI sites of pHA102 to give pHA111. E. coli BL21(DE3) carrying pHA111 overproduced three products: PhdA, PhdB, and a PhdB derivative most likely translated from the Met codon within the 3′ end of the phdA gene. To eliminate this by-product, pHA111 was digested with EcoRI, filled in with T4 DNA polymerase, and recircularized. Production of the undesired product was eliminated in the resulting plasmid, pHA111eV. pHB103 was digested with NdeI and recircularized to give pHB111, in which the phdB gene was located under the control of the T7 promoter and the efficient SD sequence derived from the pT7-7(BglIIX) vector. The 0.25-kb BamHI-HindIII fragment from pHC102 containing AAGGAG-phdC was cloned between the BamHI and HindIII sites of pHA102, pHA111eV, and pHB111, giving pHA121 (phdA-phdC), pHA141eV (phdA-phdB-phdC), and pHB121 (phdB-phdC), respectively. The 1.3-kb HindIII fragment from pHD101 containing AAGGAG-phdD was cloned into the HindIII sites of pHA121, pHA141eV, and pHB121, giving pHA162 (phdA-phdC-phdD), pHA171 (phdA-phdB-phdC-phdD), and pHB141 (phdB-phdC-phdD), respectively. pHA171 was digested with BglII (eliminating the phdC gene) and recircularized to give pHA151 (phdA-phdB-phdD).
Biotransformation of phenanthrene.
E. coli BL21(DE3) or JM109 cells carrying one of the phd-expressing plasmids were grown overnight at 37°C in 100 ml of M9-glucose medium containing 100 μg of ampicillin per ml. The cells were collected by centrifugation, washed, and resuspended in a fresh M9-glucose medium, after which adjusting the cell densities were adjusted to an optical density at 600 nm of 1.0. An aliquot (10 ml) of the cell suspension was distributed into sealed vials (100 ml), and 40 μl of 50 mM phenanthrene dissolved in N,N-dimethylformamide was added to make the final concentration of phenanthrene 200 μM. When required isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 100 μM was added. The cells were incubated at 37°C for the time indicated. The volume of each culture was adjusted with methanol to 20 ml to completely dissolve the substrate and metabolites, mixed well, and centrifuged at 12,000 × g for 10 min. The resulting supernatant was loaded into a reverse-phase column (TSKgel ODS-80Ts; 4.6 by 250 mm; Tosoh) fitted to a high-pressure liquid chromatography (HPLC) system (Tosoh). Phenanthrene and its hydroxylation product were separated by using a methanol gradient (from 60% [vol/vol] to 90% [vol/vol] methanol in 5 min followed by 90% [vol/vol] methanol for 15 min) at a flow rate of 1 ml/min. Electron impact mass spectrometry (EI-MS) of the collected hydroxylation product was carried out with a JMS-SX102 mass spectrometer (JEOL, Tokyo, Japan).
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been submitted to the DDBJ/GenBank/EMBL DNA databases under accession no. AB017794, AB017795, and AB031319.
RESULTS
Localization of the phd gene cluster on the chromosome DNA.
To examine whether the phd gene cluster lies on the chromosome or on a plasmid, the total cellular DNA of Nocardioides sp. strain KP7 was subjected to PFGE and then transferred to a membrane which was probed with the 2.5-kb HindIII-BamHI DNA fragment containing the phdI and phdJ sequences (Fig. 1). Undigested KP7 DNA barely moved from the slot and hybridized with the probe (Fig. 2, lanes 3 and 9). No DNA band with a size between 50 and 2,200 kb was apparent. The DraI digestion (lanes 4 and 10) gave a result similar to that with the nondigested DNA. In the cases of digestion with XbaI and HindIII, the probe hybridized to the 350- and 80-kb bands, respectively (lanes 11 and 12). These results indicate that the phd gene cluster resides on a very large DNA, most likely the chromosome DNA.
FIG. 2.
Localization of phenanthrene-degrading genes on the chromosome DNA. Total DNA of Nocardioides sp. strain KP7 was subjected to PFGE (lanes 1 to 8) and hybridization with the 2.5-kb HindIII-BamHI DNA fragment shown in Fig. 1 (lanes 9 to 12). Lanes 1 and 7, S. cerevisiae YNN295 chromosomal DNA as molecular mass markers; lanes 2 and 8, ladder of phage lambda DNA concatemers as molecular mass markers; lanes 3 and 9, undigested total DNA of KP7; lanes 4 and 10, DraI-digested total DNA of KP7; lanes 5 and 11, XbaI-digested total DNA of KP7; lanes 6 and 12, HindIII-digested total DNA of KP7.
Cloning and sequencing of phd genes.
The degenerate primers DO-1s and DO-1a, which were designed from the conserved amino acid sequences of the α subunits of ring-hydroxylating dioxygenases, were used to amplify a 302-bp fragment from the genomic DNA of Nocardioides sp. strain KP7 (Fig. 1). The amino acid sequence encoded by the fragment showed significant homology to the α subunits of known ring-hydroxylating dioxygenases. The gene library of strain KP7 was then screened by using the 302-bp fragment as a probe. Four clones gave a positive signal. One of them, pMKT177, has previously been characterized as a clone containing the phdIJK gene cluster, which encodes the enzymes responsible for the transformation of 1-hydroxy-2-naphthoate to o-phthalate (24–26). The other positive clones are designated pMKT201, pMKT202, and pMKT203. The physical map of 52-kb-long DNA encompassing pMKT178, pMKT177, and pMKT203 was constructed, and pK11Eco-1 and pKBg3102 were used for DNA sequencing of the 10.6-kb-long BamHI-MluI fragment (Fig. 1). The gene cluster phdEFABGHCD plus two small open reading frames (ORFs) was identified downstream of the previously characterized phdIJK genes. In this study, the phdA, phdB, phdC, and phdD genes were further characterized.
PhdA showed significant but moderate sequence homology to the α subunits of known ring-hydroxylating dioxygenases. The amino acid sequences of the α subunits of PAH dioxygenases from Rhodococcus sp. strain NCIMB 12038 (NarAa, AF082663) and strain I24 (NidA) (50) showed 55 to 56% identity with the PhdA sequence, whereas the other known α subunits exhibited a sequence identity lower than 40%. The amino acid sequence alignment of PhdA to the known α subunits revealed a limited number of conserved amino acid residues, the highly conserved ones corresponding to those involved in the coordination to the [2Fe-2S] Rieske-type cluster and to the catalytic nonheme iron (Fig. 3) (28).
FIG. 3.
Alignment of well-conserved sequence regions in the N-terminal and central part of the α subunits of ring-hydroxylating dioxygenases. The three-dimensional structure of the NdoBC oxygenase component from Pseudomonas sp. strain NCIB 9816-4 has recently been determined (28). Highlighted characters represent the residues involved in binding to the Rieske-type [2Fe-2S] cluster and to the mononuclear iron atom. Shadowed characters indicate the Asp residues hydrogen bonded to active-site ligand His on the same molecule and to [2Fe-2S] Rieske center ligand His on the neighboring α subunit. The Rieske-type [2Fe-2S] cluster-binding sequence, CXHX17CX2H, in PhdA was located at Cys91 to His114. The residues involved in coordinating the catalytic iron were also conserved as His219 and His224 (and Asp376). Asterisks indicate residues conserved in all proteins. Definition of abbreviations are given in the legend to Fig. 5.
The phdB gene was found downstream of the phdA gene, and PhdB exhibited significant sequence homology to the β subunits of the ring-hydroxylating dioxygenases. The β subunits of PAH dioxygenases from Rhodococcus sp. strain I24 (NidB) (50) and strain NCIMB 12038 (NarAb, AF082663) manifested relatively high sequence identity to PhdB (55 and 52%, respectively). The other β subunits exhibited sequence identity lower than 40%.
The phdC gene was located 2.9 kb downstream of the phdB gene. PhdC, which is predicted to be 69 amino acids long, did not show significant homology to any of the ferredoxin components of known ring-hydroxylating dioxygenases, except for DitA3 (33), but showed significant homology to the [3Fe-4S] and [4Fe-4S] ferredoxins.
The phdD gene was found just downstream of the phdC gene, and the encoded gene product showed significant sequence similarity to the proteins of the reductase family. PhdD contained amino acid sequences typical of ADP-binding βαβ folds (53) which may constitute putative flavin adenine dinucleotide (Val6 to Ala37)- and NAD (Ser148 to Glu176)-binding sites (35) with the completely conserved consensus motif GXGX2GX3A.
Involvement of the phdABCD genes in enzyme activity.
To determine whether the phdABCD genes actually encode the components of the ring-hydroxylating dioxygenase, the genes were introduced into E. coli. Plasmids pSA315, containing the phdABCD genes under the control of the lac promoter, and pST46, containing the phdABCD genes under the control of the T7 promoter, were introduced into E. coli JM109 and BL21(DE3), respectively. The phenanthrene transformation activity in these cells was then examined in the presence or absence of IPTG. Both E. coli cultures showed very low phenanthrene transformation activity. One possible reason for this was inefficient translation from the phdABCD messages in the E. coli cells. Another possibility was inefficient transcription prevented by extra DNA sequences (e.g., the 3′-half region of phdF and two small ORFs) (Fig. 1).
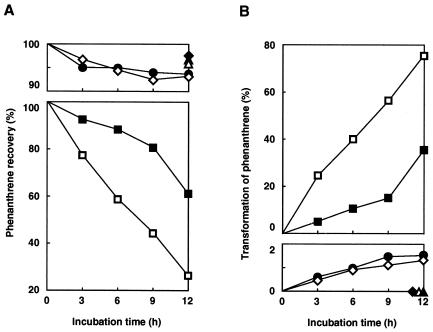
To improve the expression of the phdABCD genes, an SD sequence whose effectiveness in E. coli had previously been demonstrated was introduced upstream of each of the phdABCD genes as described in Materials and Methods. The constructed plasmids were subsequently introduced into BL21(DE3), and the phenanthrene-transforming activity in the absence of IPTG was then examined in BL21(DE3) cells carrying one of these plasmids. The cells carrying pHA171 (phdABCD) showed the highest activity to transform phenanthrene to a more hydrophilic product (Fig. 4). The EI-MS spectrum of the product showed the molecular ion at m/z 212 (M+), indicating the formation of phenanthrene dihydrodiol. Phenanthrene decreased linearly to 26% of the input value up to 12 h, while the concentration of dihydrodiol increased linearly during this period. After extension of the incubation to 24 h, phenanthrene had completely disappeared (data not shown). Cells carrying pHA141eV (phdABC) lacking the reductase gene, phdD, showed one-third of the transforming activity of the pHA171 (phdABCD). Cells carrying pHA151 (phdABD) lacking the ferredoxin gene, phdC, and those carrying pHA111eV (phdAB) showed much less activity than cells carrying pHA171 (phdABCD). These results indicated that both electron transport proteins, PhdC and PhdD, were involved in the enzyme activity. Cells carrying pHA162 (phdACD), pHB141 (phdBCD), and pT7-7(BglIIX) showed no transformation activity, demonstrating the indispensability of the α and β subunits for the enzyme activity.
FIG. 4.
Requirements of the phdABCD genes for phenanthrene transformation activity. The removal of phenanthrene (A) and formation of the dihydrodiol (B) were monitored by HPLC at 254 nm in a suspension of BL21(DE3) cells carrying expression plasmids pHA171 (phdABCD) (□), pHA141eV (phdABC) (■), pHA151 (phdABD) (●), pHA111eV (phdAB) (◊), pHB141 (phdBCD) (⧫), pHA162 (phdACD) (▵), and pT7-7 (BglIIX) (▴). Prolonged incubation (24 h) of the cells carrying pHA171 resulted in the complete removal of phenanthrene. Therefore, the rate of dihydrodiol formation was calculated by taking the amount of dihydrodiol at 24 h as 100%. Values are averages from two independent experiments.
PhdA and PhdB expressed in E. coli cells from pHA171 (phdABCD) and pHA141eV (phdABC) were detected as major bands by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) even without the addition of IPTG, and its addition at a final concentration of 0.1 mM increased the level of their expression. However, the IPTG addition markedly reduced the activity of the phenanthrene transformation in these cells (data not shown), suggesting that the high-level expression of the phd genes caused the formation of inclusion bodies. In contrast to the PhdA and PhdB expression, PhdC and PhdD could not be identified in the E. coli cells by SDS-PAGE in the presence or absence of IPTG (data not shown).
DISCUSSION
We report in this study the cloning, sequencing, and functional expression in E. coli of a novel PAH ring-hydroxylating dioxygenase which is composed of the PhdABCD proteins. The gene clusters responsible for PAH degradation have been localized on both plasmids (nah, P. putida strain [31]; ndo, P. putida strain [42]; dox, Pseudomonas sp. strain C18 [10]; nag, Pseudomonas sp. strain U2 [18]; phn, Burkholderia sp. strain RP007 [32]) and chromosomes (pah, P. putida OUS82 [48]; nah, P. stutzeri AN10 [6]). The results of this study show that the phd gene cluster was localized on the chromosome (Fig. 2) and that the order of the phd genes (Fig. 1) was quite different from that of analogous gene sets so far reported. Each of the subunits of the PhdABCD dioxygenase showed a modest (<60%) sequence identity to all of the known dioxygenase subunits, although the basic sequence features of each protein family were conserved.
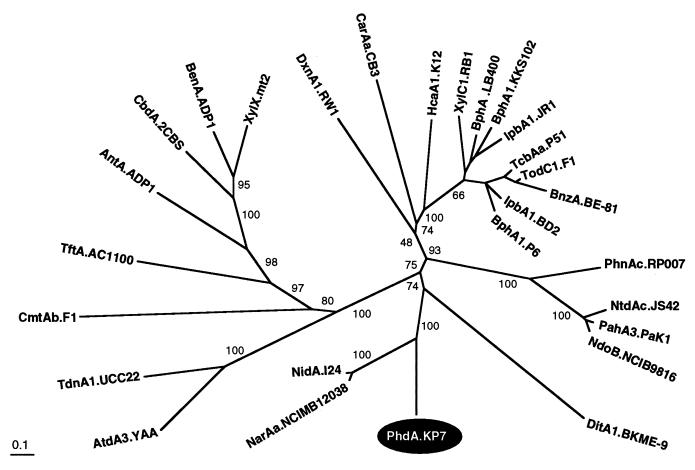
By phylogenetic analysis, PhdA did not form a cluster with most of known α-subunit sequences but formed a deep branch with two newly described α subunits from Rhodococcus sp., NarAa (AF082663) and NidA (50) (Fig. 5). Phylogenetic analysis of the β subunits gave an unrooted tree similar to that for the α subunits, indicating that PhdB formed a distinct cluster with the two β subunits, NarAb (AF082663) and NidB (50) (data not shown). In both unrooted trees, the branches of PhdA and PhdB were supported by low bootstrap values which might be due to the distant relationships of PhdA and PhdB to other counterparts.
FIG. 5.
Phylogenetic tree obtained from the alignment of PhdA with related proteins. The protein sequences of the 28 α subunits of ring-hydroxylating dioxygenases including PhdA are classified. The multiple-alignment analysis was performed with the PHYLIP software package, and the phylogenetic unrooted tree was drawn by using TreeView. The numbers on some branches refer to the percentage confidence estimated by a bootstrap analysis with 1,000 replications. The scale bar indicates the percentage divergence. The sequence abbreviations, enzyme substrates, species, and DDBJ/GenBank/EMBL references are as follows: NdoB.NCIB9816, naphthalene, P. putida NCIB 9816, M23914; PahA3.PaK1, naphthalene, P. aeruginosa PaK1, D84146; NtdAc.JS42, 2-nitrotoluene, Pseudomonas sp. strain JS42, U49504; PhnAc.RP007, phenanthrene, Burkholderia sp. strain RP007, AF061751; BnzA.BE-81, benzene, P. putida BE-81, M17904; TodC1.F1, toluene, P. putida F1, J04996; TcbAa.P51, chlorobenzene, Pseudomonas sp. strain P51, U15298; IbpA1.BD2, isopropylbenzene, Rhodococcus erythropolis BD2, U24277; BphA1.P6, biphenyl, R. globerulus P6, X80041; IbpA1.JR1, isopropylbenzene, Pseudomonas sp. strain JR1, U53507; BphA1.KKS102, biphenyl, Pseudomonas sp. strain KKS102, D17319; BphA.LB400, biphenyl, Burkholderia cepacia LB400, M86348; XylC1.RB1, substrate unknown, Cycloclasticus oligotrophus RB1, U51165; HcaA1.K12, phenylpropionate, E. coli K-12, AE000340; DxnA1.RW1, dibenzo-p-dioxin, Sphingomonas sp. strain RW1, X72850; CarAa.CB3, carbazole, Sphingomonas sp. strain CB3, AF060489; XylX.mt2, toluate, P. putida mt2, M64747; BenA.ADP1, benzoate, Acinetobacter calcoaceticus ADP1, AF009224; CbdA.2CBS, 2-halobenzoate, B. cepacia 2CBS, X79076; AntA.ADP1, anthranilate, A. calcoaceticus ADP1, AF071556; TftA.AC1100, 2,4,5-trichlorophenoxyacetic acid, B. cepacia AC1100, U11420; CmtAb.F1, p-cumate, P. putida F1, U24215; TdnA1.UCC22, aniline, P. putida UCC22, D85415; AtdA3.YAA, aniline, Acinetobacter sp. strain YAA, AB008831; NidA.I24, indene, Rhodococcus sp. strain I24, AF121905; NarAa.NCIMB12038, naphthalene, Rhodococcus sp. strain NCIMB 12038, AF082663; DitA1.BKME-9, diterpenoid, P. abietaniphila BKME-9, AF119621; PhdA.KP7, phenanthrene, Nocardioides sp. strain KP7, AB017794.
The three-dimensional structure of the oxygenase component (α3 [NdoB], β3 [NdoC]) of naphthalene dioxygenase from Pseudomonas sp. NCIB 9816-4 was determined (28). A long narrow gorge which may provide access for substrates to catalytic iron was found in NdoB. The five residues constituting the narrowest part of the channel near the catalytic iron in NdoB were completely conserved in PhdA as Asn212, Phe213, His219, His 224, and Phe366. However, the residues lining the substrate-binding pocket below the catalytic iron and those covering the upper part of the catalytic iron were diverged. This sequence diversity may contribute to the substrate specificity difference between NdoB and PhdA.
The Lys97, Gly98, Val100, Gln115, Ser116, Pro118, and Trp211 residues in NdoB and the Ser75, Arg77, Arg78, Pro105, and Trp108 residues in NdoC form a possible interaction domain with the [2Fe-2S] ferredoxin. As discussed below, the ferredoxin component, PhdC, of the PhdABCD dioxygenase is a [3Fe-4S] or [4Fe-4S] ferredoxin, different from the [2Fe-2S] ferredoxins adopted in other ring-hydroxylating dioxygenases. Therefore, the binding site for PhdC on PhdA and PhdB is thought to be distinct from the binding site on NdoB and NdoC. As expected, the residues forming the putative ferredoxin-binding site in NdoB and NdoC were not conserved in PhdA and PhdB, except for Pro128 in PhdA (corresponding to Pro118 in NdoB) and Trp88 in PhdB (corresponding to Trp108 in NdoC).
Twenty-four residues at the N terminus of NdoC are involved in formation of the NdoC trimer (28). Alignment of the PhdB sequence with the NdoC sequence indicates that PhdB was 19 residues shorter than NdoC in the N-terminal region, and the 10 N-terminal residues of PhdB did not show any homology with NdoC. The long loop formed by residues 68 to 85 in NdoC is also involved in trimer formation and in the interaction with the Rieske domain of NdoB. The corresponding sequence in PhdB exhibited only 11% (2/18) amino acid identity and 55% (10/18) similarity. It is thus expected that the mode of interaction between the PhdB subunits would be quite different from that of NdoC.
The ferredoxin component of the PhdABCD dioxygenase, PhdC, was the first example of the [3Fe-4S] or [4Fe-4S] type of ferredoxin (39), the other ferredoxin components being the [2Fe-2S] type (4). More recently, another example of the [3Fe-4S] or [4Fe-4S] type of ferredoxin, DitA3, has been reported as a component of the diterpenoid dioxygenase of Pseudomonas abietaniphila BKME-9 (33). A homology search showed the wide and diverse distribution of the PhdC-like bacterial ferredoxin family among the high-G+C (9, 37) and low-G+C (14) gram-positive bacteria and members of the taxa Proteobacteria (17, 33), Thermotogales (36), and Archaea (22, 52). The ferredoxins closely related to PhdC are the electron transport proteins in the multicomponent P-450 systems in Streptomyces griseolus (37, 51). The ferredoxins from the hyperthermophilic archaea Pyrococcus furiosus and Thermococcus litoralis have been shown to bind a [4Fe-4S] cluster which can readily be converted to a stable [3Fe-4S] form (56). The three Cys residues which serve as ligands for the iron-sulfur cluster are conserved in all [3Fe-4S] and [4Fe-4S] types of ferredoxin, while substitution of the Cys residue second from the N termini has been observed in some proteins in this family. The Asp residue at this position can act as a ligands for the [4Fe-4S] cluster, as Cys does (57), while the Ala residue in ferredoxin-2 (SubB) from S. griseolus coordinates a [3Fe-4S] cluster in place of the second Cys (37). SubB was most similar to PhdC (51% sequence identity), whereas Tyr was found at the second Cys position of PhdC as in the case of DitA3, which shows less sequence identity (36%) to PhdC. It is not known whether the Tyr residues can act as ligands to the [4Fe-4S] cluster. The two Pro residues (Pro21 and Pro55 in PhdC) are also conserved in many ferredoxins, suggesting that these residues should be relevant to the function and/or protein conformation of the members in this ferredoxin family.
By a phylogenetic analysis of ferredoxin reductase components of the ring-hydroxylating dioxygenases, PhdD was shown to be distantly related to other reductases: PhdD constituted a cluster together with BphG (46), CarAd (41), CmtAa (12), and RedA2 (2) (data not shown), but all of these reductases branch near the root of this cluster and are equally distant from each other, showing a radial pattern of evolution. The ferredoxin reductase component of DitA dioxygenase (33), of which ferredoxin is of the [3Fe-4S] or [4Fe-4S] type, has not yet been identified. Furthermore, the ferredoxin and ferredoxin reductase components of NarA (AF082663) and Nid (50) dioxygenases from Rhodococcus sp. also remain to be clarified. The relationships between these unknown electron transport proteins and the PhdCD components are of interest.
It has been shown that all four proteins are required for the full activity of the PhdABCD dioxygenase. PhdA and PhdB were essential for the enzyme activity. On the other hand, the ferredoxin component, PhdC, and the ferredoxin reductase component, PhdD, were dispensable in E. coli (Fig. 5). This may have been due to replacement of the function of the electron transport proteins of the ring-hydroxylating dioxygenase by that of E. coli. This result has been observed for other ring-hydroxylating dioxygenases (5, 31, 33), suggesting the relatively low specificity of the electron transport systems toward the oxygenase components. This tolerance between the oxygenase components and the electron transport systems has been advocated to play a role in the evolutionary process of ring-hydroxylating dioxygenases (21).
ACKNOWLEDGMENTS
We thank Y. Inomata-Yamauchi and Y. Sasaki for technical assistance.
This work was performed as part of the Industrial Science and Technology Frontier Program supported by the New Energy and Industrial Technology Development Organization.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armengaud J, Happe B, Timmis K N. Genetic analysis of dioxin dioxygenase of Sphingomonas sp. strain RW1: catabolic genes dispersed on the genome. J Bacteriol. 1998;180:3954–3966. doi: 10.1128/jb.180.15.3954-3966.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology, Suppl. 11. unit 16.2. New York, N.Y: John Wiley & Sons; 1990. [Google Scholar]
- 4.Batie C J, Ballou D P, Correll C C. Phthalate dioxygenase reductase and related flavin-iron-sulfur containing electron transferases. In: Muller F, editor. Chemistry and biochemistry of flavoenzymes. Vol. 3. Boca Raton, Fla: CRC Press; 1992. pp. 543–556. [Google Scholar]
- 5.Bergeron J, Ahmad D, Barriault D, Larose A, Sylvestre M, Powlowski J. Identification and mapping of the gene translation products involved in the first steps of the Comamonas testosteroni B-356 biphenyl/chlorobiphenyl biodegradation pathway. Can J Microbiol. 1994;40:743–753. doi: 10.1139/m94-118. [DOI] [PubMed] [Google Scholar]
- 6.Bosch R, Garcia-Valdes E, Moore E R B. Genetic characterization and evolutionary implications of a chromosomally encoded naphthalene-degradation upper pathway from Pseudomonas stutzeri AN10. Gene. 1999;236:149–157. doi: 10.1016/s0378-1119(99)00241-3. [DOI] [PubMed] [Google Scholar]
- 7.Brosius J. Superpolylinkers in cloning and expression vectors. DNA. 1989;8:759–777. doi: 10.1089/dna.1989.8.759. [DOI] [PubMed] [Google Scholar]
- 8.Cerniglia C E, Heitkamp M A. Microbial degradation of polycyclic aromatic hydrocarbons (PAH) in the aquatic environment. In: Varanasi U, editor. Metabolism of polycyclic aromatic hydrocarbons in the aquatic environment. Boca Raton, Fla: CRC Press, Inc.; 1989. pp. 41–68. [Google Scholar]
- 9.Crespi M, Vereecke D, Temmerman W, Van Montagu M, Desomer J. The fas operon of Rhodococcus fascians encodes new genes required for efficient fasciation of host plants. J Bacteriol. 1994;176:2492–2501. doi: 10.1128/jb.176.9.2492-2501.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denome S A, Stanley D C, Olson E S, Young K D. Metabolism of dibenzothiophene and naphthalene in Pseudomonas strains: complete DNA sequence of an upper naphthalene catabolic pathway. J Bacteriol. 1993;175:6890–6901. doi: 10.1128/jb.175.21.6890-6901.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eaton R W. Organization and evolution of naphthalene catabolic pathways: sequence of the DNA encoding 2-hydroxychromene-2-carboxylate isomerase and trans-o-hydroxybenzylidenepyruvate hydratase-aldolase from the NAH7 plasmid. J Bacteriol. 1994;176:7757–7762. doi: 10.1128/jb.176.24.7757-7762.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eaton R W. p-Cumate catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA carrying the cmt operon. J Bacteriol. 1996;178:1351–1362. doi: 10.1128/jb.178.5.1351-1362.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eaton R W, Chapman P J. Bacterial metabolism of naphthalene: construction and use of recombinant bacteria to study ring cleavage of 1,2-dihydroxynaphthalene and subsequent reactions. J Bacteriol. 1992;174:7542–7554. doi: 10.1128/jb.174.23.7542-7554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott J I, Yang S S, Ljungdahl L G, Travis J, Reilly C F. Complete amino acid sequence of the 4Fe-4S, thermostable ferredoxin from Clostridium thermoaceticum. Biochemistry. 1982;21:3294–3298. doi: 10.1021/bi00257a007. [DOI] [PubMed] [Google Scholar]
- 15.Erickson B D, Mondello F J. Nucleotide sequencing and transcriptional mapping of genes encoding biphenyl dioxygenase, a multicomponent PCB-degrading enzyme in Pseudomonas strain LB400. J Bacteriol. 1992;174:2903–2912. doi: 10.1128/jb.174.9.2903-2912.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felsenstein J. Phylogeny Inference Package, version 3.5c. Seattle, Wash: Department of Genetics, University of Washington; 1993. . (Distributed by the author.) [Google Scholar]
- 17.Freiberg C, Fellay R, Bairoch A, Broughton W J, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 18.Fuenmayor S L, Wild M, Boyes A L, Williams P A. A gene cluster encoding steps in conversion of naphthalene to gentisate in Pseudomonas sp. strain U2. J Bacteriol. 1998;180:2522–2530. doi: 10.1128/jb.180.9.2522-2530.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson D T, Subramanian V. Microbial degradation of aromatic hydrocarbons. In: Gibson D T, editor. Microbial degradation of organic compounds. New York, N.Y: Marcel Dekker, Inc.; 1984. pp. 181–252. [Google Scholar]
- 20.Harayama S. Polycyclic aromatic hydrocarbon bioremediation design. Curr Opin Biotechnol. 1997;8:268–273. doi: 10.1016/s0958-1669(97)80002-x. [DOI] [PubMed] [Google Scholar]
- 21.Harayama S, Kok M, Neidle E L. Functional and evolutionary relationships among diverse oxygenases. Annu Rev Microbiol. 1992;46:565–601. doi: 10.1146/annurev.mi.46.100192.003025. [DOI] [PubMed] [Google Scholar]
- 22.Heltzel A, Smith E T, Zhou Z H, Blamey J M, Adams M W. Cloning, expression, and molecular characterization of the gene encoding an extremely thermostable [4Fe-4S] ferredoxin from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 1994;176:4790–4793. doi: 10.1128/jb.176.15.4790-4793.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irie S, Doi S, Yorifuji T, Takagi M, Yano K. Nucleotide sequencing and characterization of the genes encoding benzene oxidation enzymes of Pseudomonas putida. J Bacteriol. 1987;169:5174–5179. doi: 10.1128/jb.169.11.5174-5179.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwabuchi T, Harayama S. Biochemical and genetic characterization of 2-carboxybenzaldehyde dehydrogenase, an enzyme involved in phenanthrene degradation by Nocardioides sp. strain KP7. J Bacteriol. 1997;179:6488–6494. doi: 10.1128/jb.179.20.6488-6494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwabuchi T, Harayama S. Biochemical and genetic characterization of trans-2′-carboxybenzalpyruvate hydratase-aldolase from a phenanthrene-degrading Nocardioides strain. J Bacteriol. 1998;180:945–949. doi: 10.1128/jb.180.4.945-949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwabuchi T, Harayama S. Biochemical and molecular characterization of 1-hydroxy-2-naphthoate dioxygenase from Nocardioides sp. KP7. J Biol Chem. 1998;273:8332–8336. doi: 10.1074/jbc.273.14.8332. [DOI] [PubMed] [Google Scholar]
- 27.Iwabuchi T, Inomata-Yamauchi Y, Katsuta A, Harayama S. Isolation and characterization of marine Nocardioides capable of growing and degrading phenanthrene at 42°C. J Mar Biotechnol. 1998;6:86–90. [Google Scholar]
- 28.Kauppi B, Lee K, Carredano E, Parales R E, Gibson D T, Eklund H, Ramaswamy S. Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure. 1998;6:571–586. doi: 10.1016/s0969-2126(98)00059-8. [DOI] [PubMed] [Google Scholar]
- 29.Kiyohara H, Nagao K, Kouno K, Yano K. Phenanthrene-degrading phenotype of Alcaligenes faecalis AFK2. Appl Environ Microbiol. 1982;43:458–461. doi: 10.1128/aem.43.2.458-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiyohara H, Torigoe S, Kaida N, Asaki T, Iida T, Hayashi H, Takizawa N. Cloning and characterization of a chromosomal gene cluster, pah, that encodes the upper pathway for phenanthrene and naphthalene utilization by Pseudomonas putida OUS82. J Bacteriol. 1994;176:2439–2443. doi: 10.1128/jb.176.8.2439-2443.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurkela S, Lehvaslaiho H, Palva E T, Teeri T H. Cloning, nucleotide sequence and characterization of genes encoding naphthalene dioxygenase of Pseudomonas putida strain NCIB9816. Gene. 1988;73:355–362. doi: 10.1016/0378-1119(88)90500-8. [DOI] [PubMed] [Google Scholar]
- 32.Laurie A D, Lloyd-Jones G. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism. J Bacteriol. 1999;181:531–540. doi: 10.1128/jb.181.2.531-540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin V J J, Mohn W W. A novel aromatic-ring-hydroxylating dioxygenase from the diterpenoid-degrading bacterium Pseudomonas abietaniphila BKME-9. J Bacteriol. 1999;181:2675–2682. [Google Scholar]
- 34.Masai E, Yamada A, Healy J M, Hatta T, Kimbara K, Fukuda M, Yano K. Characterization of biphenyl catabolic genes of gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. Appl Environ Microbiol. 1995;61:2079–2085. doi: 10.1128/aem.61.6.2079-2085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mason J R, Cammack R. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu Rev Microbiol. 1992;46:277–305. doi: 10.1146/annurev.mi.46.100192.001425. [DOI] [PubMed] [Google Scholar]
- 36.Nelson K E, Clayton R A, Gill S R, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Nelson W C, Ketchum K A, McDonald L, Utterback T R, Malek J A, Linher K D, Garrett M M, Stewart A M, Cotton M D, Pratt M S, Phillips C A, Richardson D, Heidelberg J, Sutton G G, Fleischmann R D, White O, Salzberg S L, Smith H O, Venter J C, Fraser C M. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 37.O'Keefe D P, Gibson K J, Emptage M H, Lenstra R, Romesser J A, Litle P J, Omer C A. Ferredoxins from two sulfonylurea herbicide monooxygenase systems in Streptomyces griseolus. Biochemistry. 1991;30:447–455. doi: 10.1021/bi00216a021. [DOI] [PubMed] [Google Scholar]
- 38.Page R D. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 39.Saito A, Iwabuchi T, Harayama S. Characterization of genes for enzymes involved in the phenanthrene degradation in Nocardioides sp. KP7. Chemosphere. 1999;38:1331–1337. doi: 10.1016/s0045-6535(98)00534-7. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 41.Shepherd J M, Lloyd-Jones G. Novel carbazole degradation genes of Sphingomonas CB3: sequence analysis, transcription, and molecular ecology. Biochem Biophys Res Commun. 1998;247:129–135. doi: 10.1006/bbrc.1998.8750. [DOI] [PubMed] [Google Scholar]
- 42.Simon M J, Osslund T D, Saunders R, Ensley B D, Suggs S, Harcourt A, Suen W C, Cruden D L, Gibson D T, Zylstra G J. Sequences of genes encoding naphthalene dioxygenase in Pseudomonas putida strains G7 and NCIB 9816-4. Gene. 1993;127:31–37. doi: 10.1016/0378-1119(93)90613-8. [DOI] [PubMed] [Google Scholar]
- 43.Simons R, Priefer U, Puhler A. A broad host range mobilization system of in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–790. [Google Scholar]
- 44.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 46.Sylvestre M, Sirois M, Hurtubise Y, Bergeron J, Ahmad D, Shareck F, Barriault D, Guillemette I, Juteau J M. Sequencing of Comamonas testosteroni strain B-356-biphenyl/chlorobiphenyl dioxygenase genes: evolutionary relationships among Gram-negative bacterial biphenyl dioxygenases. Gene. 1996;174:195–202. doi: 10.1016/0378-1119(96)00039-x. [DOI] [PubMed] [Google Scholar]
- 47.Taira K, Hirose J, Hayashida S, Furukawa K. Analysis of bph operon from the polychlorinated biphenyl-degrading strain of Pseudomonas pseudoalcaligenes KF707. J Biol Chem. 1992;267:4844–4853. [PubMed] [Google Scholar]
- 48.Takizawa N, Kaida N, Torigoe S, Moritani T, Sawada T, Satoh S, Kiyohara H. Identification and characterization of genes encoding polycyclic aromatic hydrocarbon dioxygenase and polycyclic aromatic hydrocarbon dihydrodiol dehydrogenase in Pseudomonas putida OUS82. J Bacteriol. 1994;176:2444–2449. doi: 10.1128/jb.176.8.2444-2449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;11:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Treadway S L, Yanagimachi K S, Lankenau E, Lessard P A, Stephanopoulos G, Sinskey A J. Isolation and characterization of indene bioconversion genes from Rhodococcus strain I24. Appl Microbiol Biotechnol. 1999;51:786–793. doi: 10.1007/s002530051463. [DOI] [PubMed] [Google Scholar]
- 51.Trower M K, Lenstra R, Omer C, Buchholz S E, Sariaslani F S. Cloning, nucleotide sequence determination and expression of the genes encoding cytochrome P-450soy (soyC) and ferredoxinsoy (soyB) from Streptomyces griseus. Mol Microbiol. 1992;6:2125–2134. doi: 10.1111/j.1365-2958.1992.tb01386.x. [DOI] [PubMed] [Google Scholar]
- 52.Wang P L, Donaire A, Zhou Z H, Adams M W, La Mar G N. Molecular model of the solution structure for the paramagnetic four-iron ferredoxin from the hyperthermophilic archaeon Thermococcus litoralis. Biochemistry. 1996;35:11319–11328. doi: 10.1021/bi960783u. [DOI] [PubMed] [Google Scholar]
- 53.Wierenga R K, Terpstra P, Hol W G J. Prediction of the occurrence of the ADP-binding bab-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986;187:101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- 54.Yang Y, Chen R F, Shiaris M P. Metabolism of naphthalene, fluorene, and phenanthrene: preliminary characterization of a cloned gene cluster from Pseudomonas putida NCIB 9816. J Bacteriol. 1994;176:2158–2164. doi: 10.1128/jb.176.8.2158-2164.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 56.Zhou Z H, Adams M W W. Proton NMR investigation of the oxidized three-iron clusters in the ferredoxins from the hyperthermophilic archae Pyrococcus furiosus and Thermococcus litoralis. Biochemistry. 1992;31:11952–11962. doi: 10.1021/bi00162a038. [DOI] [PubMed] [Google Scholar]
- 57.Zhou Z H, Adams M W W. Site-directed mutations of the 4Fe-ferredoxin from the hyperthermophilic archaeon Pyrococcus furiosus: role of the cluster-coordinating aspartate in physiological electron transfer reactions. Biochemistry. 1997;36:10892–10900. doi: 10.1021/bi9708141. [DOI] [PubMed] [Google Scholar]
- 58.Zylstra G J, Gibson D T. Toluene degradation by Pseudomonas putida F1. Nucleotide sequence of the todC1C2BADE genes and their expression in Escherichia coli. J Biol Chem. 1989;264:14940–14946. [PubMed] [Google Scholar]