Abstract
Thermus thermophilus HB8 can grow anaerobically by using a membrane-bound nitrate reductase to catalyze the reduction of nitrate as a final electron acceptor in respiration. In contrast to other denitrifiers, the nitrite produced does not continue the reduction pathway but accumulates in the growth medium after its active extrusion from the cell. We describe the presence of two genes, narK1 and narK2, downstream of the nitrate reductase-encoding gene cluster (nar) that code for two homologues to the major facilitator superfamily of transporters. The sequences of NarK1 and NarK2 are 30% identical to each other, but whereas NarK1 clusters in an average-distance tree with putative nitrate transporters, NarK2 does so with putative nitrite exporters. To analyze whether this differential clustering was actually related to functional differences, we isolated derivatives with mutations of one or both genes. Analysis revealed that single mutations had minor effects on growth by nitrate respiration, whereas a double narK1 narK2 mutation abolished this capability. Further analysis allowed us to confirm that the double mutant is completely unable to excrete nitrite, while single mutants have a limitation in the excretion rates compared with the wild type. These data allow us to propose that both proteins are implicated in the transport of nitrate and nitrite, probably acting as nitrate/nitrite antiporters. The possible differential roles of these proteins in vivo are discussed.
Nitrate can be used as an alternative to oxygen in respiration by many bacteria and by the mitochondria of certain fungi (1, 17). In most cases, nitrate reduction is the first step in a pathway catalyzed by a series of membrane-bound reductases whose final products are dinitrogen and ammonia (1, 17).
We have recently described the presence of a nitrate reductase operon in the extreme thermophile Thermus thermophilus HB8, a bacterium formerly considered a strict aerobe (11). Interestingly, the absence of a nitrite reductase in this bacterium results in the long-term accumulation of nitrite in the medium of anaerobically grown cultures. Accordingly, a very active nitrite export system should function in this bacterium to escape from its high toxicity.
Genes encoding polytopic membrane proteins belonging to the major facilitator superfamily of transporters (9) have been found close to the nar gene cluster for all such genes so far sequenced (1, 17). The role of such proteins is still controversial as nitrate/nitrite or H+/nitrite antiporters and nitrate/H+ symporters (17). There is experimental evidence that the NarK protein, encoded upstream of the narGHJI operon of Escherichia coli, is essentially implicated in nitrite export by using the electrochemical gradient as the energy source (an H+/nitrite antiporter) (14). On the other hand, and despite their similarity to the E. coli NarK, the NarT and NasA proteins from Staphylococcus carnosus and Bacillus subtilis, respectively, have been proposed to function as nitrate/H+ symporters (5, 10). Thus, the role of such membrane transporters in different bacteria is still unclear due to the intrinsic difficulties in measuring the transport of these anions and also because of the ability of these bacteria to overcome mutations in the corresponding genes through secondary transporters (17). Moreover, the ability of most denitrifiers to use alternative anaerobic pathways for growth increases the difficulties in analyzing the effects of mutations in the corresponding nitrate/nitrite transporters.
Since T. thermophilus HB8 does not have any alternative way for anaerobic growth than nitrate respiration, and keeping in mind that this bacterium lacks a nitrite reductase, a requirement for nitrite extrusion proteins seems crucial for its viability. Thus, any mutation in the putative nitrite transporters should have a strong phenotypic consequence in this bacterium. Here we describe two NarK homologues (named NarK1 and NarK2) encoded by genes downstream of the narGHJI operon of T. thermophilus HB8, one of which (narK2) overlaps the replicative origin of the nar-carrying conjugative plasmid (13). We demonstrate that mutations in each of the coding genes (narK1 and narK2) have minor effects on the ability to grow anaerobically, whereas double narK1 narK2 mutants are unable to grow under these conditions. This, and the analysis of nitrite excretion, led us to conclude that both proteins are implicated in nitrate import and nitrite export during anaerobic growth.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
T. thermophilus HB8 (ATCC 27634) was obtained from the American Type Culture Collection (Rockville, Md.). Its nitrate reductase (NR) deletion derivative (narGH::kat) was described previously (12). E. coli strains TG1 [supE Δ(nsdM mcrB) 5(rK−mK− mcrB) thi Δ(lac-proAB) F′ (traD36 proAB+ laqIqZΔM15)] and DH5αF′ [F′ supE44 Δ(lacZYA-argF)U169 Φ80lacZΔM15 hsdR17 recA1 endA1 gyrA96 thi-1 relA1] (Bethesda Research Laboratories, Gaithersburg, Md.) were used as hosts for plasmid construction. Plasmids pNIT5 and pNIT9 (12) were pUC119 derivatives containing the narK1 and narK2 genes, respectively. Plasmid pKT1 was the source of the kat gene, encoding thermostable resistance to kanamycin (7).
Aerobic growth conditions for T. thermophilus were attained by incubation at 70°C in a nitrate-free rich medium (12) with vigorous shaking. For NR induction, cells were grown in the same medium containing 40 mM KNO3 without stirring. For anaerobic growth, cells grown under aerobic conditions up to an optical density at 550 nm (OD550) of 0.05 were divided into 10-ml aliquots inside 15-ml tubes filled to the top with mineral oil (12). Plates were incubated at 70°C in a water-saturated atmosphere. For liquid and solid selection, kanamycin (30 μg/ml) was added. E. coli strains were grown at 37°C in Luria-Bertani (LB) medium (8) or in LB agar plates. Ampicillin (100 μg/ml) or kanamycin (30 μg/ml) was added when needed.
Mutant isolation.
Insertional mutagenesis was done through electroporation of T. thermophilus HB8 (3) with HindIII-linearized forms of defined plasmids, followed by selection on kanamycin-containing plates.
Plasmid pNIT9Kkat1 was used for the inactivation of narK1. It was obtained by insertion of the kat cassette (7) into the KpnI site of pNIT9 (12), interrupting the sequence of the narK1 gene at a position corresponding to amino acid 41 of the encoded protein. Isolation of the narK2::kat mutant was achieved by transformation with plasmid pNIT5kat1. In this plasmid, the kat gene replaced a SmaI fragment internal to the narK2 gene, resulting in a truncated protein of 200 amino acids. To obtain double narK1 narK2::kat mutants, plasmid pNIT9Kkat1 was digested with EcoRI. After treatment with the Klenow fragment of DNA polymerase I from E. coli, the linearized plasmid was partially digested with SmaI and religated. The new plasmid, pNIT9Kkat2, was used to produce a deletion of positions corresponding to encoded amino acids 41 of NarK1 to 304 of NarK2.
Genetic analysis.
General methods were used for DNA manipulation (15). Southern blots of total DNA from putative mutants digested with the appropriate enzyme(s) were hybridized with fluorescein-labeled oligonucleotides O-70 (5′CGGAGAGGAAGATGCCG3′), which hybridized to the sequence of narK2, and Okat-2 (5′GAAACTTCTGGAATCGC3′), directed against the 3′ region of the kat gene, and revealed with the ECL kit (Amersham Ibérica SA). DNA was sequenced by automatic methods (Applied Biosystems) with synthetic primers (Isogen Bioscience, Maarssen, The Netherlands). Partial sequences were assembled with the software of the University of Wisconsin Genetics Computer Group (4). DNA amplification from colonies of the putative mutants was developed in an MJ Research minicycler (MJ Research Inc., Watertown, Mass.). Oligonucleotides O-28 (5′CACCCTCATGTTCGCCG3′), O-54 (5′CCACCCTCCTCCTTCTC3′), O-65 (5′CGGGCCGATGAACTTGG3′) and O-70 (see above) were used for the amplification of specific fragments. Their approximate hybridization sites are labeled in Fig. 2A.
FIG. 2.
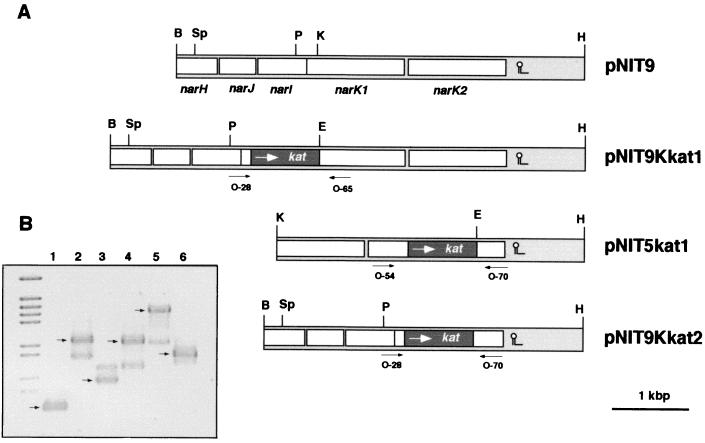
Isolation and analysis of narK::kat mutants. (A) Restriction maps of the plasmids used to get single and double narK::kat mutants. Enzymes: B, BamHI; E, EcoRI; H, HindIII; K, KpnI; P, PstI; Sp, SphI. (B) PCR analysis of narK::kat mutants. PCR amplification products from the DNA of single narK1 (lane 2) and narK2 (lane 4) and double narK1 narK2 (lane 6) mutants were compared with those obtained from the DNA of the parental strain (lanes 1, 3, and 5). Primers used for amplification: lanes 1 and 2, O-28 and O-65; lanes 3 and 4, O-54 and O-70; lanes 5 and 6, O-28 and O-70. Arrows indicate the product expected from the corresponding amplifications. The approximate positions at which these primers hybridize are shown in panel A.
NR activity and nitrite analysis.
For induction of the NR, cells of T. thermophilus HB8 were grown at 70°C in a shaker bath to an OD550 of 0.5. After addition of potassium nitrate (40 mM), the cultures were incubated at 70°C without stirring for an additional 2 h. Cells were then recovered by centrifugation, washed twice with phosphate buffer (50 mM, pH 7) by centrifugation (10,000 × g, 5 min, room temperature), and disrupted by sonication at a cell density of ∼24 OD550 units/ml. Intracellular concentrations of nitrite were measured in the above cell extracts by assuming a cell volume and a ratio of OD550 to cell mass similar to those of E. coli (10−12 ml/cell and 109 cells per OD550 unit per ml). The NR activity was measured as described before (12) after 5 min of incubation at 80°C with methyl viologen as the electron donor (16) and nitrate as the electron acceptor. The nitrite excreted was measured in cell-free samples of the growth medium.
RESULTS
Sequence of the region downstream of the narGHJI operon.
Analysis of the sequence downstream of the narGHJI operon from T. thermophilus HB8 (accession number AJ237974) revealed the presence of two open reading frames encoding 435 and 443 amino acids, both of them preceded by putative Shine-Dalgarno sequences. Due to their similarities to the sequences for nitrite transporters found close to nar operons from other bacteria (see below), the corresponding encoding genes were named narK1 and narK2, respectively.
The putative translation ATG start codon of narK1 overlaps the last codon of narI, the gene encoding the cytochrome b (γ subunit) component of the NR. Thus, a translational coupling from a common mRNA seems very likely. By contrast, 22 bp separate the last codon of narK1 and the ATG start codon of narK2, keeping the possibility of differential expression between the proteins open. However, the absence of putative transcription terminator sequences between the genes suggests that narK2 could be cotranscribed within the same mRNA.
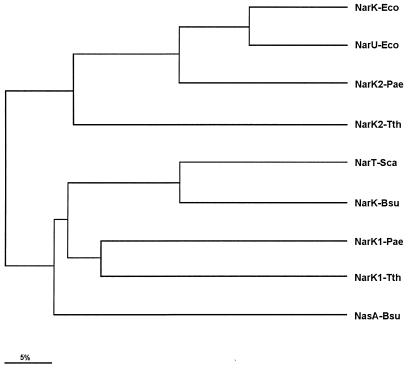
Secondary-structure predictions support a polytopic integral membrane nature for NarK1 and NarK2, with 12 putative α-helix domains spanning the cytoplasmic membrane. No typical general secretory pathway-dependent signal peptide was present in any of them. Comparison of their sequences with those in the data banks revealed similarities to membrane transporters belonging to the major facilitator superfamily of proteins (9). High scores were found when NarK1 and NarK2 were compared with proteins implicated in the extrusion of nitrite and/or nitrate transport from different bacteria. As shown in Fig. 1, the calculation of the average-distance tree from the alignment of the NarK1 and NarK2 sequences with those from proteins implicated in the transport of nitrate and/or nitrite in different bacteria revealed a differential grouping of both proteins. Whereas NarK1 groups with NarT and NasA, two proteins suggested to participate in nitrate transport in S. carnosus (5) and B. subtilis (10), respectively, NarK2 groups with the nitrite extrusion proteins NarK and NarU from E. coli.
FIG. 1.
Average-distance tree for NarK proteins. The sequences of the putative nitrate and nitrite transporters were aligned through the CLUSTAL program, and the average distance was represented through the JALview editor. In addition to the NarK1 and NarK2 proteins from T. thermophilus HB8 (Tth), the proteins used in the alignment included NarK (P10903) and NarU (P37958) from E. coli (Eco), NarK1 and NarK2 from P. aeruginosa (accession no. Y15252) (Pae), NarT from S. carnosus (accession no. U40014) (Sca), and NarK (P46907) and NasA (P42432) from B. subtilis (Bsu).
Consequently, a different role for these proteins is suggested from these comparisons, in which NarK1 could be responsible for nitrate transport and NarK2 for the extrusion of nitrite. This putative difference in roles is also supported by the low identity (30%) found between the two proteins.
The presence of two in-tandem genes encoding NarK homologues (narK1 and narK2) has been identified close to the nar operon within the genome sequence of Pseudomonas aeruginosa (accession number Y15252) and, more recently, in Pseudomonas stutzeri, where they have been named narK and narC (6). Interestingly, inclusion of the sequences of NarK1 and NarK2 from P. aeruginosa in the above comparison revealed a clustering similar to that of the NarK1 and NarK2 proteins from T. thermophilus HB8. Moreover, the NarK1 proteins from both organisms were 43% identical and 54.5% similar, whereas their corresponding NarK2 proteins were 41% identical and 51.3% similar. Thus, although in P. aeruginosa the narK1 and narK2 genes are located upstream of the corresponding narGHJI cluster, the high similarity found between these proteins supports similar roles for them in both organisms.
Insertional inactivation of narK1 and narK2.
The analysis described above suggested a role for NarK1 and NarK2 in nitrate transport and nitrite extrusion, respectively, suggesting that both proteins were required for nitrate respiration. To analyze this hypothesis, we isolated individual narK1::kat and narK2::kat mutants as well as a double narK1K2::kat mutant by using the plasmids shown in Fig. 2A (see Materials and Methods for construction details). After transformation and selection for kanamycin resistance, the presence of the expected mutations was confirmed by DNA amplification with primer pairs that hybridized at the approximate positions shown in Fig. 2A. As shown in Fig. 2B, the use of primers O-28 and O-65 allowed the amplification of a 0.4-kbp fragment in the wild-type strain (lane 1), whereas in the narK1::kat mutant, this fragment was replaced by a 1.3-kbp fragment as a result of insertion of the kat gene (lane 2). In the narK2::kat mutant, the use of primers O-54 and O-70 resulted in amplification of the expected 1.29-kbp fragment (Fig. 2B, lane 4) instead of the 0.69-kbp DNA obtained from the wild type (lane 3). Finally, the 2.2-kbp fragment amplified from the wild type with primers O-28 and O-70 (Fig. 2B, lane 5) was replaced by a 1-kbp fragment in the double mutant as a consequence of replacement with the kat cassette of most of the sequence from both genes (lane 6). These results were further confirmed by Southern blot analysis (not shown).
Phenotypic analysis of narK::kat mutants.
As could be expected, the growth of single and double mutants under aerobic conditions was indistinguishable from that of the wild-type strain (not shown), indicating that neither of these genes is required for the aerobic metabolism of the bacterium.
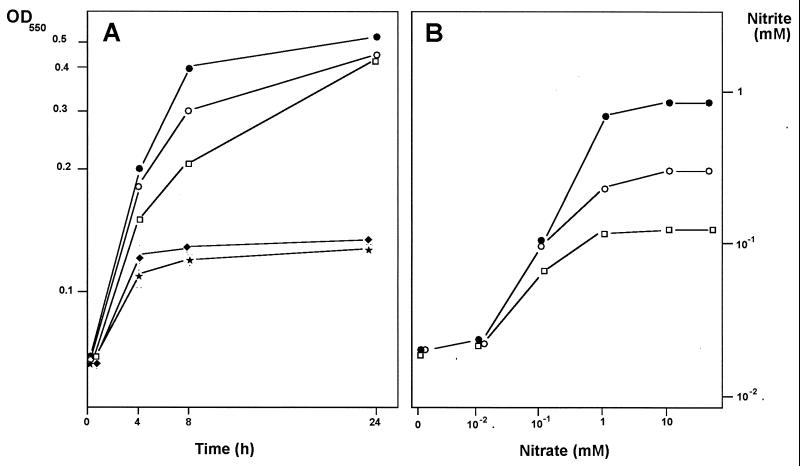
Under anaerobic conditions, however, single mutants grew at slightly lower rates than the wild type, reaching 5 to 10% lower cell mass densities than the wild type (Fig. 3A). By contrast, the double narK1K2::kat mutant was unable to grow anaerobically, a behavior indistinguishable from that of the narGH::kat mutant used as a negative growth control (Fig. 3A).
FIG. 3.
Anaerobic growth and nitrite export of narK::kat mutants. (A) Anaerobic growth. Growth curves under anaerobic conditions were performed as described in Materials and Methods. Tubes were taken at the indicated times to measure the OD550. (B) External concentration of nitrite at different nitrate concentrations. Samples from induced cultures of single and double narK mutants were incubated in preheated culture medium containing increasing concentrations of nitrate. After 10 min, the nitrite produced was measured and plotted against the external concentration of nitrate (see the text for details). Symbols: ●, wild type; ○, narK1::kat; □, narK2::kat; ⧫, narK1K2::kat; ★, narGH::kat.
In order to analyze whether the lack of anaerobic growth in the double mutant was due to a defect in nitrate transport or to the absence of an active NR, induction experiments of this enzyme were undertaken. The results showed that the NR activity in single and double mutants was similar to that in the wild type (from 300 to 600 nmol of nitrite produced per min per OD550 unit). Thus, the inability to grow anaerobically had nothing to do with a defect in NR induction, suggesting instead a defect in nitrate and/or nitrite transport.
To analyze this, cells were induced as above, washed twice by centrifugation, resuspended to an OD550 of 0.5 in nitrate-free medium, and separated into 2-ml aliquots. These aliquots were subsequently incubated at 70°C with increasing concentrations of nitrate, and the nitrite excreted was measured at different times. In Fig. 3B, the amount of nitrite excreted by each mutant after 10 min of incubation is plotted against the amount of nitrate added. As can be observed, the amount of nitrite excreted by the single mutants reached a plateau far below that of the wild type. By contrast, nitrite was not detected in the medium when the double mutant was assayed under these conditions.
DISCUSSION
The narK1 and narK2 genes described in this study are predicted to encode proteins belonging to the major facilitator superfamily of transporters. Proteins from this family have a broad substrate spectrum that includes H+, sugars, and antibiotics, and its members share 12 membrane-spanning α-helix and diverse sequence motifs (9). This fact, and the location of the genes downstream of the narGHJI cluster, suggested that NarK1 and NarK2 were involved in the transport of nitrate and/or nitrite. The average-distance tree shown in Fig. 1, in which each protein clusters in a different group, suggested a role in nitrate transport for NarK1 and a more specific role in nitrite extrusion for NarK2.
The results shown in Fig. 3A demonstrate that a double mutation in narK1 and narK2 abolishes the ability of T. thermophilus HB8 to grow anaerobically. Thus, either the substrate (nitrate) or the product (nitrite), or both, has no alternative way to cross the membrane than by using the NarK1 or NarK2 protein. On the other hand, as the presence of either of these proteins suppresses this defect in anaerobic growth, it follows that they share at least one of these functions. Whether this function is nitrate transport or nitrite excretion cannot be deduced from this experiment. However, a detailed analysis of our present results led us to support a nitrate/nitrite antiporter capability for both proteins.
The arguments in favor of this interpretation are various. First, if both proteins shared only the ability to extrude nitrite, the presence of an alternative way to bring nitrate into the cell should result in a dramatic intracellular accumulation of nitrite because of the presence of a normal level of NR. Most probably, such levels of intracellular nitrite would be lethal to the cell. In fact, in the double mutant, intracellular nitrite remains at concentrations similar to or even lower than that of the wild type (0.5 to 1 mM) in all the experiments in which it was checked. Thus, we concluded that there is not an alternative way to transport nitrate into T. thermophilus HB8 than the NarK1/NarK2 proteins. In consequence, both proteins should have the ability to act as nitrate transporters into the cell.
On the other hand, the inability of the double mutant to excrete a detectable amount of nitrite at the highest nitrate concentration used (40 mM) in the experiment shown in Fig. 3B could be expected from the absence of nitrate transporters. In addition, the plateau of nitrite secretion reached by each single mutant could reflect the existence of a limiting step only in nitrate transport. Alternatively, it could be related to a limitation in the excretion of nitrite that could be expected from the absence of nitrite transporters outside the nar cluster. In this sense, the fact that the nar cluster of T. thermophilus HB8 is located within a self-mobilizable element, along with the small size of its chromosome, strongly argues against the existence of nitrite extrusion transporters outside this genetic element. In fact, no homologous genes could be detected either with a labeled probe of narK1 and narK2 in Southern blot analysis or by comparison with the unfinished genome sequence of T. thermophilus HB27, a closely related aerobic strain to which the nar cluster can be transferred and expressed. Thus, although the inability of the double mutant to excrete a detectable amount of nitrite (Fig. 3B) does not exclude it, the putative existence of an alternative nitrite extruder other than the NarK1/NarK2 proteins seems unlikely.
As noted above, the T. thermophilus HB8 chromosome is quite small (1.8 Mbp) (2), and consequently, genetic redundancy is rare (e.g., there are only two copies of DNA encoding rRNA). Thus, a good but yet-unknown reason should justify the presence of two genes encoding functionally redundant proteins in the nar cluster for anaerobic respiration of T. thermophilus. Despite the above discussion, the low similarity between the two proteins suggests different roles for each protein in the natural environment in which these microorganisms live. For instance, the extrusion of nitrite and the transport of nitrate would require the proton motive force at low nitrate concentrations. In this scenario, one of the proteins could have the ability to function as an H+/nitrate symporter and the other as an H+/nitrite antiporter, and only at higher concentrations of nitrate could they function as nitrate/nitrite antiporters. Alternatively, the presence of two enzymes with redundant enzymatic activities could be related to putative differential expression between them. In this sense, the distance between the genes (20 bp) and the existence of a T-rich sequence overlapping the C-encoding region of narK1 could be related to differential expression of each protein.
Interestingly, the two narK genes identified upstream of the nitrate respiration cluster of P. aeruginosa encode proteins with high similarity to those described here from T. thermophilus (Fig. 1). Moreover, this similarity is conserved at the DNA sequence level, not only between the narK genes but also along many stretches of the whole nar cluster. Keeping in mind the plasmidic nature of the genetic element that encodes anaerobic respiration in T. thermophilus HB8 and the ubiquity, respiratory character, and similarity in codon usage of Thermus spp. and Pseudomonas spp., it is tempting to speculate about a common origin for both groups of genes. Meanwhile, future work with P. aeruginosa would confirm the functional relationship between the tandemly organized narK genes of these bacteria.
ACKNOWLEDGMENTS
This work has been supported by projects BIO98-0183, from the Comisión Interministerial de Ciencia y Tecnología, and 2FD97-0127-C02-01, cofunded by the European Union and the Spanish Ministerio de Educación y Cultura. An institutional grant from the Fundación Ramón Areces is also acknowledged. Sandra Ramirez-Arcos held a scholarship from the Instituto de Cooperación Iberoamericana during the development of this work.
REFERENCES
- 1.Berks B C, Ferguson S J, Moir J W, Richardson D J. Enzymes and associated electron transport systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim Biophys Acta. 1995;1232:97–173. doi: 10.1016/0005-2728(95)00092-5. [DOI] [PubMed] [Google Scholar]
- 2.Borges K M, Bergquist P L. Genome restriction map of the extremely thermophilic bacterium Thermus thermophilus HB8. J Bacteriol. 1993;175:103–110. doi: 10.1128/jb.175.1.103-110.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Grado M, Castán P, Berenguer J. A high-transformation-efficiency cloning vector for Thermus thermophilus. Plasmids. 1999;42:241–245. doi: 10.1006/plas.1999.1427. [DOI] [PubMed] [Google Scholar]
- 4.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fast B, Lindgren P-E, Götz F. Cloning, sequencing, and characterization of a gene (narT) enconding a transport protein involved in dissimilatory nitrate reduction in Staphylococcus carnosus. Arch Microbiol. 1997;166:361–367. doi: 10.1007/BF01682980. [DOI] [PubMed] [Google Scholar]
- 6.Hartig E, Schiek U, Vollack K U, Zumft W G. Nitrate and nitrite control of respiratory nitrate reduction in denitrifying Pseudomonas stutzeri by a two-component regulatory system homologous to NarXL of Escherichia coli. J Bacteriol. 1999;181:3658–3665. doi: 10.1128/jb.181.12.3658-3665.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lasa I, Castón J R, Fernandez-Herrero L A, Pedro M A, Berenguer J. Insertional mutagenesis in the extreme thermophilic eubacteria Thermus thermophilus. Mol Microbiol. 1992;11:1555–1564. doi: 10.1111/j.1365-2958.1992.tb00877.x. [DOI] [PubMed] [Google Scholar]
- 8.Lennox E X. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955;1:190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- 9.Marger M D, Saier M H. A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem Sci. 1993;18:13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- 10.Ogawa K-I, Akagawa E, Yamane K, Sun Z-W, LaCelle M, Zuber P, Nakano M M. The nasB operon and nasA gene are required for nitrate/nitrite assimilation in Bacillus subtilis. J Bacteriol. 1995;177:1409–1413. doi: 10.1128/jb.177.5.1409-1413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oshima M, Imahori K. Description of Thermus thermophilus (Yoshida and Oshima) comb. nov., a nonsporulating thermophilic bacterium from a Japanese hot spa. Int J Syst Bacteriol. 1974;24:102–112. [Google Scholar]
- 12.Ramírez-Arcos S, Fernández-Herrero L A, Berenguer J. A thermophilic nitrate reductase is responsible for the strain specific anaerobic growth of Thermus thermophilus HB8. Biochim Biophys Acta. 1998;1396:215–227. doi: 10.1016/s0167-4781(97)00183-8. [DOI] [PubMed] [Google Scholar]
- 13.Ramírez-Arcos S, Fernández-Herrero L A, Marín I, Berenguer J. Anaerobic growth, a property horizontally transferred by an Hfr-like mechanism among extreme thermophiles. J Bacteriol. 1998;180:3137–3143. doi: 10.1128/jb.180.12.3137-3143.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rowe J J, Ubbink-Kok T, Molenaar D, Konings W N, Driessen A J. NarK is a nitrite-extrusion system involved in anaerobic nitrate respiration by Escherichia coli. Mol Microbiol. 1994;12:579–586. doi: 10.1111/j.1365-2958.1994.tb01044.x. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 16.Snell F D, Snell C T. Colorimetric methods of analysis. New York, N.Y: Van Nostrand; 1949. [Google Scholar]
- 17.Zumft W G. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]