Summary
Germline pathogenic variants in BRCA1 and BRCA2 (gpath(BRCA1/2)) represent genetic susceptibility for hereditary breast and ovarian cancer syndrome. Tumor-immune interactions are key contributors to breast cancer pathogenesis. Although earlier studies confirmed pro-tumorigenic immunological alterations in breast cancer patients, data are lacking in healthy carriers of gpath(BRCA1/2). Peripheral blood mononuclear cells of 66 women with or without germline predisposition or breast cancer were studied with a mass cytometry panel that identified 4 immune subpopulations of altered frequencies between healthy controls and healthy gpath(BRCA1) carriers, while no difference was observed in healthy gpath(BRCA2) carriers compared to controls. Moreover, 3 (one IgD-CD27+CD95+ B cell subpopulation and two CD45RA-CCR7+CD38+ CD4+ T cell subpopulations) out of these 4 subpopulations were also elevated in triple-negative breast cancer patients compared to controls. Our results reveal an activated peripheral immune phenotype in healthy carriers of gpath(BRCA1) that needs to be further elucidated to be leveraged in risk-reducing strategies.
Subject areas: Cancer, Cell biology, Immunology
Graphical abstract

Highlights
-
•
We report a stimulated peripheral immunophenotype in healthy gpath(BRCA1) carriers
-
•
1 IgD- B cell and 2 CD45RA-CD4+ T cell subsets were also elevated in TNBC patients
-
•
No alterations were found in healthy gpath(BRCA2) carriers
Cancer; Cell biology; Immunology
Introduction
Breast cancer is a common disease with a high degree of heterogeneity. Although the majority of these tumors expresses estrogen- and/or progesterone-receptors and are termed hormone-responsive (HRBC), cancers lacking these receptors as well as the HER-2 antigen are known as triple negative (TNBC). Historically considered immunologically neutral, recent advancements in molecular pathology revealed that more aggressive subtypes, including TNBCs, exhibit a considerable amount of immune cell infiltration.1 Higher frequencies of adaptive immune cells (CD8+ T cells, and B cells) are markers of better prognosis in multiple types of breast cancer, while the abundance of specific members of the myeloid lineage (myeloid-derived suppressor cells [MDSCs] and M2 macrophages) are considered to signal worse prognosis.1,2 Using single-cell mass cytometry, our group showed that low-dose cisplatin reduced the accumulation of MDSCs in a murine 4T1 TNBC model.3 Proliferating CD8+ T cells have been described as a characteristic of the TNBC microenvironment4 and their potentiation by the PD-1 immune checkpoint inhibitor (ICI) pembrolizumab has been shown to exert a clear benefit regarding an increased rate of patients achieving pathological complete response compared to chemotherapy alone in the neoadjuvant setting.5 Even without the occurrence of metastases, breast cancer significantly alters the peripheral immune phenotype.6,7 Several lines of evidence have confirmed the association of the elevated rate of immunosuppressive regulatory T cells (Tregs),8,9 of CD8+ T cells with stimulated/exhausted phenotype10,11 and of MDSCs8,12 in the peripheral blood of breast cancer patients, while other studies signaled phenotypic changes toward memory phenotypes in B cells13 and noncytotoxic CD56bright populations in the natural killer (NK) cell compartment.6,14
Hereditary breast and ovarian cancer syndrome (HBOC) is a common cancer predisposition syndrome, with approximately half of the cases related to germline pathogenic variants in BRCA1 (gpath(BRCA1)) and BRCA2 (gpath(BRCA2)).15,16 Women carrying gpath(BRCA1/2) have a ∼70% risk of developing breast cancer and ∼20–60% risk of developing ovarian cancer throughout their lifetime.16 gpath(BRCA1)-associated breast cancers usually occur at an earlier age with a predominance of the TNBC phenotype, while women with gpath(BRCA2) are typically diagnosed later in their life and a larger proportion of them develop HRBC.16 Tumors with pathogenic variants of BRCA1 (path(BRCA1)) and BRCA2 (path(BRCA2)) share distinct homologous recombination deficiency-related genomic scars,17,18 but also have differences in effecting the genomic architecture of breast cancers with the probably causative association of path(BRCA1) with the small tandem duplication mutator phenotype.17 Concerning the breast cancer tumor microenvironment, a recent study confirmed that BRCA1-deficient TNBCs were enriched in an immunoregulatory gene set as compared to BRCA1-proficient cancers.19 In another study, Samstein et al. aimed to analyze if somatic BRCA1/2 status correlated with the clinical outcome of ICI therapy.19 In an exploratory pan-cancer cohort of 95 patients, they have found that those with path(BRCA2) associated with improved overall survival, also suggesting an altered immune microenvironment based on the BRCA1/2 status.19
The high probability of breast cancer development can even justify risk-reducing surgical procedures to remove healthy breast tissue in women carrying gpath(BRCA1/2); therefore, large efforts are dedicated to alternative approaches to tailor personalized risk management. Recent research studies were directed to detect preneoplastic lesions and systemic biomarkers in healthy women with gpath(BRCA1/2). Several lines of evidence confirmed that luminal progenitor cells are the cells-of-origin in gpath(BRCA1)-related tumorigenesis20,21,22,23 and a recent study highlighted the pathogenetic contribution of the premalignant stromal compartment via the production of tumorigenic factors in preneoplastic breast tissues in healthy women with gpath(BRCA1).20 Moreover, it was shown that immune cell densities are elevated in normal breast tissues of women with gpath(BRCA1/2).24
Since the evaluation of the peripheral immune phenotype in healthy women with gpath(BRCA1/2) has not yet been implemented, in the present study we aimed to perform multiparametric single-cell mass cytometry experiments to investigate this issue. By comparing the peripheral immune phenotype of healthy women with and without gpath(BRCA1/2) and that of treatment-naive HRBC and TNBC patients we found several differences in the frequencies of naive and stimulated T and B cells. Our results show that even without cancer, carriers of gpath(BRCA1), but not of gpath(BRCA2) exhibit a stimulated peripheral immune phenotype that may mirror preneoplastic antitumor mechanisms.
Results
Analysis of main immune cell populations
Altogether 10,382,648 CD45+ live cells (average 157,312 ± 46,950 cells/sample) originating from 66 individuals (20 healthy women without hereditary cancer predisposition, 12 healthy women with gpath(BRCA1), 10 healthy women with gpath(BRCA2), and 12-12 women with HRBC and TNBC, Figure 1A and Table S1) were subjected to the analysis. Study cohorts were age-matched and the number of analyzed cells in each group did not differ (Figure S1). Following clustering and manual curation, 10 independent main immune clusters were identified including B cells, plasmablasts, CD4+, CD8+, γδ and double-negative T cells, monocytes, myeloid (mDC) and plasmacytoid (pDC) dendritic cells, and NK cells (Figures 1B and 1C). Monocytes were the only main immune population exhibiting altered frequency between study groups and were more frequent in cancer patients as compared to controls (Figure 1D); however, no individual marker showed a difference in expression between study groups within monocytes.
Figure 1.
Identification and characterization of major immune cell clusters
(A) Graphical representation of the study procedure. Abbreviations: LN2,liquid nitrogen; PBMC, peripheral blood mononuclear cells.
(B) t-SNE plot of the identified 10 immune cell population.
(C) Heatmap of median expression of each surface marker in each main immune cell cluster and frequencies of each main immune cell cluster in each study sample.
(D) Frequencies of monocytes in each study groups. Median frequencies (dashed lines) and interquartile range (between dotted lines) are included on violin plots. Asterisks correspond to statistical significance: p < 0.05∗ by Kruskal-Wallis test.
See also Figures S1, S3, and S4; and Table S1.
Phenotypic differences in B cells reveal a shift to an activated state in healthy gpath(BRCA1) carriers
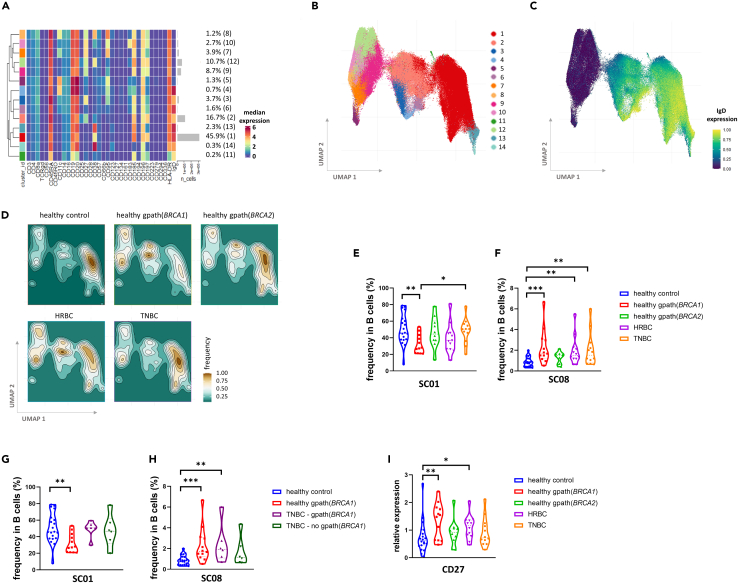
Subclustering of B cells revealed 14 subclusters (SCs) with IgD expression being one of the main differentiating factors between SCs (Figures 2A–2C). SCs 5, 7–10, and 12 are IgD− and SCs 1–4, 6, 11, and 13–14 are IgD+ (Figures 2A and 2C). We compared the relative abundance of each SC within the B cell compartment and found that SC01 and SC08 differed significantly between the study groups (Figures 2D–2F). SC01 is the most abundant B cell SC characterized by an IgD+/CD185(CXCR5)+/CD197(CCR7)+/CD11c−/CD95− phenotype reflecting a large naive B cell population being recruited toward lymphoid follicles. This SC is less frequent in the healthy gpath(BRCA1) group compared to healthy controls (Figure 2E). SC08 on the other hand is an IgD−/CD27+/CD95+/CD11c+/CD38+ B cell subset exhibiting the highest median CD95 expression within B cell SCs (Figures 2A and S2A). This SC corresponds to a class-switched memory B cell phenotype and was found to be more frequent in healthy women living with gpath(BRCA1) as well as in cancer cohorts compared to healthy controls (Figure 2F).
Figure 2.
Phenotyping of B cells and their distribution in the study cohorts
(A) Median expression of the investigated markers in each of the 14 subclusters and the frequency of each subcluster.
(B and C) UMAP representation of the B cell subclusters color coded by subclusters (B) and by IgD expression (C).
(D) Density heatmap of B cell subclusters in each study group.
(E and F) Frequencies of SC01 (E) and of SC08 (F) in each study groups.
(G and H) Subgroup analyses of TNBC compared to healthy control and healthy gpath(BRCA1) carriers demonstrating frequencies of SC01 (G) and SC08 (H). Frequencies of healthy individuals (also displayed on E and F) on these panels are included for optimal comparison.
(I) Median expression of CD27 on B cells in various study groups. Median values (dashed lines) and interquartile range (between dotted lines) are included on all violin plots. Asterisks correspond to statistical significance: p < 0.05∗; <0.01∗∗; <0.001∗∗∗ by one-way ANOVA test (E and G) or Kruskal-Wallis test (F, H, and I).
See also Figure S2.
To assess if these changes within healthy women based on germline BRCA1 status are maintained within the TNBC group, we performed subgroup analyses within the TNBC samples and compared those to the samples of healthy women with or without gpath(BRCA1). In this size-limited subgroup analysis, we have found no difference within the TNBC cohort based on germline BRCA1 status (Figures 2G and 2H). However, we confirmed the higher frequency of SC08 in gpath(BRCA1) carriers regardless of cancer status but not in TNBC patients without gpath(BRCA1), raising the possibility that germline BRCA1 status might strongly affect the frequency of SC08 (Figure 2H).
Next, we checked whether there are any marker expression changes between each study group. Interestingly, CD27 expression on B cells was elevated in healthy gpath(BRCA1) carriers, reflecting a more pronounced memory B cell phenotype in this study group (Figure 2I).
To investigate phenotypic differences between study groups regarding SC01 and SC08, we analyzed marker expressions restricted to these SCs. In SC08, elevated CD95 levels were confirmed in TNBC patients compared to healthy controls (Figure S2B), also reflecting a more activated SC08 phenotype in this disease group.
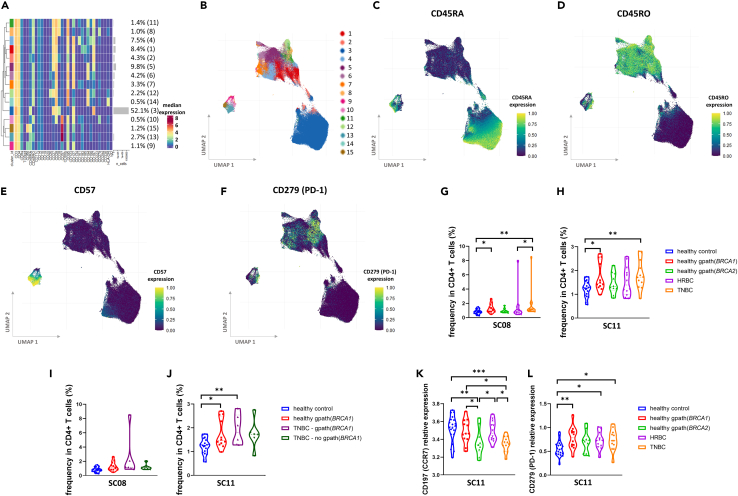
Two CD38-expressing CD4+ T cell subpopulations are more frequent in healthy gpath(BRCA1) carriers
Subclustering of CD4+ T cells yielded 15 SCs that formed three larger groups based on the similarity of the investigated marker expressions (Figures 3A and 3B). The most abundant SC, SC03, together with members of SC02 forms the CD45RA+/CD45RO− naive cell cluster, while the majority of the less frequent SCs represent a memory phenotype being CD45RO+/CD45RA− (Figures 3C and 3D). A smaller group including SCs 9, 10, 13, and 15 is distinguished mainly based on their pronounced CD57 expression (Figure 3E). Among the SCs with memory phenotype, SC08 and SC11 were more frequent in healthy gpath(BRCA1) carriers as well as in patients with TNBC (Figures 3G and 3H). These two SCs exhibit a similar, CD45RO+/CD38+/CD95+ phenotype, with SC08 also expressing the highest levels of PD-1 (Figures 3A and 3F). Subgroup analysis in the TNBC cohort revealed that the abundance of SC11 was predominantly elevated in TNBC patients with gpath(BRCA1) suggesting a significant germline genetic contribution on the frequency of this SC (Figures 3I and 3J). No difference was observed regarding median marker levels between study cohorts in the whole CD4+ T cell population; however, the levels of CD197(CCR7) on SC11 cells showed high variability with a striking decrease in the TNBC group compared to healthy controls (Figure 3K). Moreover, PD-1 expression on SC11 cells was higher in healthy gpath(BRCA1) carriers and in cancer patients compared to healthy controls (Figure 3L).
Figure 3.
CD4+ T cell phenotypes in the study cohorts
(A) Median expression of the investigated markers in each of the 15 subclusters and the frequency of each subcluster.
(B–F) UMAP representation of the CD4+ T cell subclusters color coded by subclusters (B) by CD45RA (C), CD45RO (D), CD57 (E), and PD-1 (F) expressions.
(G and H) Frequencies of SC08 (G) and of SC11 (H) in each study groups.
(I and J) Subgroup analyses of TNBC compared to healthy control and healthy gpath(BRCA1) carriers demonstrating frequencies of SC08 (I) and SC11 (J). Frequencies of healthy controls (also displayed on G and H) on these panels are included for optimal comparison.
(K and L) Median expression of CD197 (CCR7) and CD279 (PD-1) on SC11 cells in various study groups. Median values (dashed lines) and interquartile range (between dotted lines) are included on all violin plots. Asterisks correspond to statistical significance: p < 0.05∗; <0.01∗∗; <0.001∗∗∗ by Kruskal-Wallis test (G and I) or one-way ANOVA (H and J–L).
Phenotyping of additional immune subpopulations yielded no differences in subcluster frequencies between study groups.
In order to investigate if the observed differences were confounded by environmental and lifestyle factors, we performed further analyses to assess the distribution of these factors between study groups. We found no difference regarding which time of the year the samples were collected or where the study subjects lived within the country (Figures S3A and S3B). Moreover, in the cases of the HRBC and TNBC groups, data were available for additional comparisons. In these latter examinations, no differences were found between the highest level of education, smoking status, alcohol consumption, body mass index (BMI) and physical exercise, which would have confounded our results (Figures S3C–S3G).
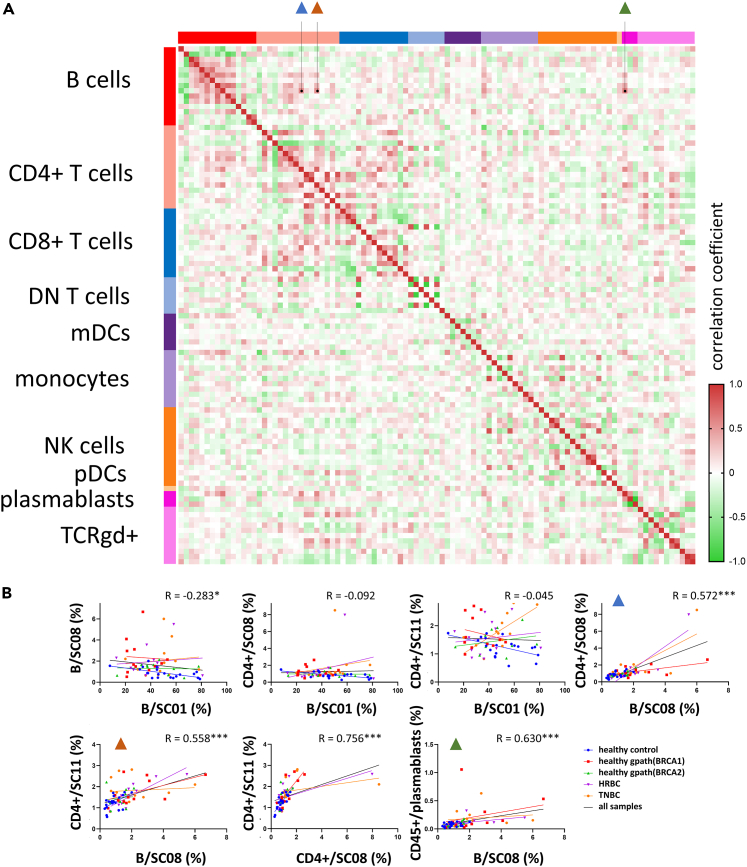
Correlation analysis of immune subpopulation frequencies proposes a functional link between activated T and B cells
To investigate the interdependence between immune subpopulations, we performed a correlation analysis between each of the 10 main immune populations and their SCs (Figure 4A). As expected, SCs of the same main immune populations correlated well with each other (e.g., lower frequencies of naive CD8+ populations with higher frequencies of stimulated CD8+ T cells), with additional correlative patterns between CD4+ and CD8+ SCs. Focusing on the 4 immune subpopulations with significant alterations between study groups, the B cell subpopulation SC08 displayed the highest levels of correlation with other significantly altered subpopulations (Figure 4B). SC08 and SC11 CD4+ subpopulations and plasmablasts were the three immune populations most strongly correlating with this SC, underlining the potential functional link between these CD4+ T cell and B cell populations.
Figure 4.
Correlation analysis of immune cell populations
(A) Heatmap of pairwise Spearman’s R correlation coefficient values between each investigated immune cell population. In each main immune cell population the first column/row corresponds to the main immune population frequency in the CD45+ cells, while following columns/rows correspond to each subcluster (SC01→SCNN).
(B) Correlation between frequencies of subclusters with significant alterations between study groups. R values displayed on each graph correspond to correlation analysis performed on all samples. Blue triangles correspond to B cell SC08 vs. CD4+ T cell SC08 analysis, while brown and green triangles correspond to B cell SC08 vs. CD4+ T cell SC11 and B cell SC08 vs. plasmablasts analyses, respectively. Asterisks correspond to statistical significance: p < 0.05∗; <0.001∗∗∗ by Spearman’s correlation.
Discussion
Germline pathogenic variants in BRCA1 and BRCA2 are the most frequent genetic defects resulting in HBOC with an estimated lifelong breast and ovarian cancer risk of 70% and 20–60%, respectively. Homologous recombination deficiency and genomic instability are hallmarks of HBOC-associated tumors priming these lesions as targets for antineoplastic immune response. ICI therapies potentiating this intrinsic immune activation have been successfully introduced in TNBC.5 However, cancer-mediated immunoediting influences the effectiveness of immune response explaining the disappointing clinical results of ICI in ovarian cancer.25 Although previous studies described luminal progenitor cells as cells-of-origin in gpath(BRCA1)-related breast tumorigenesis,20,21,22 no study has been conducted to investigate the systemic immunological characteristics of HBOC.
Therefore, we performed a mass cytometry study to compare the peripheral immune phenotype of healthy women carrying gpath(BRCA1/2) with that of healthy women without HBOC and breast cancer patients. We detected an elevated proportion of monocytes in cancer patients, which confirms earlier findings.8 By analyzing immune cell SCs in each study group we detected two B cell and two CD4+ T cell SCs with altered frequency, all of which demonstrated abundance changes in healthy gpath(BRCA1) carriers and TNBC patients, while neither of the SC frequencies differed in healthy gpath(BRCA2) carriers. This mirrors a significantly different immunogenic landscape of BRCA1- and BRCA2-related tumorigenesis that correspond to the clinical observation of gpath(BRCA1) predisposing to immunologically active TNBC phenotype while gpath(BRCA2) predisposes to immunologically more neutral HRBC.16
In particular, the largest, IgD+ B cell SC demonstrated decreased frequency in healthy gpath(BRCA1) carriers compared to controls. Deep phenotyping of this SC revealed elevated CXCR5 and CCR7 expression and decreased CD95 and CD11c expression reflecting a naive B cell phenotype being recruited to secondary lymphoid follicles for germinal center reaction.26 The lower frequency of the naive cell subpopulation correlates with a larger frequency of a CD95+/CD11c+ class-switched memory B cell population in healthy gpath(BRCA1) carriers.27,28,29 This, together with the observed higher median CD27 expression within B cells jointly confirms a more stimulated B cell phenotype in this study group. Additionally, it was interesting to find that the elevated SC08 frequency in gpath(BRCA1) carriers compared to controls was upheld in TNBC patients with this genetic predisposition, signaling a strong linkage of this subpopulation with gpath(BRCA1).
Within CD4+ T cells, two CD45RA−/CCR7+/CD38+ subpopulations (SC08 and SC11) demonstrated elevated frequencies in healthy gpath(BRCA1) carriers as well as in TNBC patients. These SCs correspond to an activated, central memory T helper phenotype. CD38 is an NAD+ glycohydrolase and is widely considered as an activation marker on T cells.30,31 CD38 expression on CD4+ T cells has been shown to correlate with reduced proliferation and increased proinflammatory cytokine-producing (TNFα, IFNγ, and IL-2) capabilities31,32 and T cell differentiation30 and was found to be elevated in chronic viral infections.33 Clinically, a CD4+CD38+ subpopulation in the peripheral blood as well as in the tumor tissue has been shown to exert poor prognosis in metastatic melanoma patients undergoing ICI therapy.34,35 These results point to an orchestrated crosstalk between CD4+ T cells and other immune populations involved in adaptive and innate immunity.
To investigate this possible crosstalk, we performed correlation analysis between SC frequencies and found that the CD95+/CD11c+ class-switched B cell SC correlated positively with the two CD4+CD38+ central memory SCs. This points to a potential mechanistic link that needs to be elucidated in further studies. A widely studied crosstalk between CD4+ T cells and B cells is that of follicular helper T cells (Tfh), helping B cell proliferation, survival, and differentiation in a variety of diseases including in cancer-associated tertiary lymphoid structures.36 However, the two CD38+ SCs in our study do not express canonical Tfh markers CXCR3 and CXCR5. Nevertheless, we hypothesize that altered cytokine production of these SCs might affect the activation of certain memory B cell populations.
A collaborative international initiative analyzing the contributors of interindividual variations in the peripheral immune phenotype concluded that the main factors driving these differences are age and household.37 Since all our study subjects lived in different households, similarly represented different regions in Hungary and study groups were age-matched, we can confirm that these factors did not confound the presented alterations. Moreover, additional analyses regarding environmental, socioeconomical, and lifestyle factors did not detect significant differences between HRBC and TNBC patients that would have confounded our results.
In conclusion, this is the first study investigating the peripheral immune phenotype of healthy women living with gpath(BRCA1/2). We report an altered CD4+ T cell and B cell phenotype in healthy gpath(BRCA1) carriers largely shared in TNBC patients and consisting of higher frequencies of stimulated phenotype. Correlations between these altered immune cell subpopulations link to a viable CD4+ T cell-B cell crosstalk that needs to be further investigated to tailor immune-mediated risk-reducing strategies in women with HBOC.
Limitations of the study
It is important to note the limitations of this study. As a single-center examination of limited sample size, the results of our study cannot be directly extrapolated to the general HBOC population. Confirmatory studies in independent cohorts as well as further mechanistic investigations into the proposed CD4+ T cell-B cell crosstalk in HBOC are needed to validate our results. Moreover, the cross-sectional nature of our study design prevented us to analyze how stable the detected alterations are through time. Since all cancer patients involved in this study underwent surgery and pharmacological treatments and a significant proportion of healthy gpath(BRCA1/2) carriers underwent risk-reducing surgical interventions, these questions should be analyzed in further studies specifically aiming to dissect longitudinal sample collections. However, analyzing the peripheral immune phenotype in longitudinal samples from 177 individuals, Carr et al. showed that only 1.4% of the total variation between samples are attributable to intraindividual variation between visits.37 This stability of the peripheral immune phenotype within individuals is consistent throughout additional landmark studies.37,38,39,40 An additional limitation of the study is the low sample size in the subgroup analyses in the TNBC cohort. Further studies should analyze the peripheral immune phenotype in gpath(BRCA1) carriers with and without TNBC.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-human-CD45 (HI30) - 89Y | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD196 (CCR6) (G034E3) - 141Pr | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD123 (6H6) - 143Nd | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD19 (HIB19) - 144Nd | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD4 (RPA-T4) - 145Nd | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD8a (RPA-T8) - 146Nd | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD11c (Bu15) - 147Sm | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD16 (3G8) - 148Nd | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD45RO (UCHL1) - 149Sm | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD45RA (HI100) - 150Nd | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD161 (HP-3G10) - 151Eu | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD194 (CCR4) (L291H4) - 152Sm | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD25 (BC96) - 153Eu | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD27 (O323) - 154Sm | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD57 (HNK-1) - 155Gd | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD183 (CXCR3) (G025H7) - 156Gd | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD185 (CXCR5) (J252D4) - 158Gd | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD28 (CD28.2) - 160Gd | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD38 (HB-7) - 161Dy | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD56 (NCAM) (NCAM16.2) - 163Dy | Standard BioTools | Cat. No.: 201325 |
| anti-human-TCRγδ (B1) - 164Dy | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD294 (BM16) - 166Er | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD197 (CCR7) (G043H7) - 167Er | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD14 (63D3) - 168Er | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD3 (UCHT1) - 170Er | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD20 (2H7) - 171Yb | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD66b (G10F5) - 172Yb | Standard BioTools | Cat. No.: 201325 |
| anti-HLA-DR (LN3) - 173Yb | Standard BioTools | Cat. No.: 201325 |
| anti-human-IgD (IA6-2) - 174Yb | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD127 (A019D5) - 176Yb | Standard BioTools | Cat. No.: 201325 |
| Cell-ID Intercalator - 103Rh | Standard BioTools | Cat. No.: 201325 |
| anti-human-CD45 (HI30) - 106Cd | Standard BioTools | Cat. No.: 3106001B |
| anti-human-CD45 (HI30) - 110Cd | Standard BioTools | Cat. No.: 3110001B |
| anti-human-CD45 (HI30) - 111Cd | Standard BioTools | Cat. No.: 3111001B |
| anti-human-CD45 (HI30) - 112Cd | Standard BioTools | Cat. No.: 3112001B |
| anti-human-CD45 (HI30) - 113Cd | Standard BioTools | Cat. No.: 3113001B |
| anti-human-CD45 (HI30) - 114Cd | Standard BioTools | Cat. No.: 3114001B |
| anti-human-CD45 (HI30) - 116Cd | Standard BioTools | Cat. No.: 3116001B |
| anti-human-CD366 (Tim-3) (F38-2E2) - 159Tb | Standard BioTools | Cat. No.: 3159037B |
| anti-human-CD95 (FAS) (DX2) - 162Dy | Standard BioTools | Cat. No.: 3162038B |
| anti-human-CD223 (LAG-3) (11C3C65) - 165Ho | Standard BioTools | Cat. No.: 3165037B |
| anti-human-CD279 (PD-1) (EH12.2H7) - 175Lu | Standard BioTools | Cat. No.: 3175008B |
| anti-human-CD134 (OX40) (ACT35) - 142Nd | Standard BioTools | Cat. No.: 3142018B |
| Biological samples | ||
| 66 PBMC samples | this paper | |
| Chemicals, peptides, and recombinant proteins | ||
| Gentra Puregene Blood DNA Isolation Kit | Qiagen | Cat. No.: 158389 |
| BRCA MASTR Plus Dx kit | Agilent Technologies | Cat. No.: PR7000-1420 |
| TruSight Hereditary Cancer Panel | Illumina Inc | Cat. No.: 20029551 |
| BigDye Terminator kit | Thermo Fisher Scientific | Cat. No.: 4337455 |
| Pancoll solution | PanBiotech GmbH | Cat. No.: P04-601000 |
| RPMI-1640 | Capricorn | Cat. No.: RPMIA |
| Penicillin/ streptomycin | Capricorn | Cat. No.: PS-B |
| Fetal Bovine Serum | Euroclone | Cat. No.: ECS0180L |
| glutamine | Capricorn | Cat. No.: GLN-B |
| Maxpar Cell Staining Buffer | Standard Biotools | Cat. No.: 201068 |
| TrueStain FcX™ Fc receptor blocking solution | BioLegend | Cat. No.: 422302 |
| formaldehyde solution | Pierce™, Thermo Scientific | Cat. No.: 28908 |
| Fix & Perm solution | Standard BioTools | Cat. No.: 201067 |
| Cell Acquisition Solution | Standard BioTools | Cat. No.: 201240 |
| Deposited data | ||
| Mass cytometry dataset | this paper, Mendeley Data | https://doi.org/10.17632/pkx5wn7cb3.1 |
| Software and algorithms | ||
| Graphpad Prism | Dotmatics | version 8.0.1. https://www.graphpad.com/ |
| MASTR Reporter software | Agilent Technologies | Cat. No.: PR7000-1420 |
| DRAGEN environment | Illumina Inc | Cat. No.: 20060397, https://www.illumina.com/products/by-type/informatics-products/dragen-secondary-analysis.html |
| CyTOF software v.07 | Standard Biotools | Cat. No.: FLDM-400338, version: 7.0.8493 |
| Cytobank Premium analysis platform | Beckman Coulter | Cat. No.: C47384 https://www.beckman.com/flow-cytometry/software/cytobank-premium |
| flowcore | Ellis B et al.41 | https://bioconductor.org/packages/release/bioc/html/flowCore.html |
| CATALYST | Crowell H et al.42 | https://bioconductor.org/packages/release/bioc/html/CATALYST.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Vince Kornél Grolmusz (grolmusz.vince@oncol.hu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Original mass cytometry data has been deposited on Mendeley Data (https://doi.org/10.17632/pkx5wn7cb3.1) and is publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
We designed and executed a cross-sectional study. All study subjects were White Hungarian females and study groups were matched for age (healthy controls nn = 20, healthy gpath(BRCA1) carrier n = 12, healthy gpath(BRCA2) carrier n = 10, HRBC patient n = 12, TNBC patient n = 12, Table S1 and Figure S1A). Only individuals without fever, respiratory or gastrointestinal infections at the time of sample collection and during the preceding two week time frame were involved. Moreover, subjects with prior diagnosis of any autoimmune diseases and those under the treatment of immune-modulating treatments (e.g., local or systemic corticosteroid treatments) were excluded. Individuals included in the study were informed of the study procedure and signed an informed consent form. The study was approved by the Scientific and Research Ethics Committee of the Medical Research Council of Hungary (ETT-TUKEB 53720-7/2019/EÜIG).
Method details
Patients and genetic analysis
Healthy women referred to genetic counseling and testing and women diagnosed with breast cancer were included at the Department of Molecular Genetics, National Institute of Oncology between April 1, 2021 and March 31, 2022. Following genetic counseling and informed consent, DNA was isolated from peripheral blood using Gentra Puregene Blood Kit (Qiagen, Hilden, Germany), according to the manufacturer’s recommendations. In breast cancer patients, the germline mutational analysis of BRCA1 and BRCA2 was analyzed by next-generation sequencing. Libraries were constructed by either BRCA MASTR Plus Dx kit (Agilent Technologies, Santa Clara, CA, USA) or TruSight Hereditary Cancer Panel (Illumina Inc, San Diego, CA, USA), sequencing was run on a MiSeq instrument (Illumina Inc, San Diego, CA, USA) and bioinformatic analyses have been performed either using the MASTR Reporter software (Agilent Technologies, Santa Clara, CA, USA) or the DRAGEN environment (Illumina Inc, San Diego, CA, USA). In healthy individuals analyzed through cascade testing for the familial pathogenic variants, PCR-amplified amplicons were subjected to gold standard Sanger dideoxy sequencing using BigDye Terminator kit (Thermo Fisher Scientific, Waltham, MA, USA) followed by capillary electrophoresis detection on an Applied Biosystems 3500 Genetic Analyzer (Thermo Fisher Scientific, Waltham, MA, USA) instrument. Variant interpretation was performed following the ACMG/AMP guideline43 and variants were cross-checked in the ENIGMA consortium database (https://brcaexchange.org). The performed genetic analyses confirmed gpath(BRCA1) in 6 out of 12 TNBC patients and gpath(BRCA2) in 3 out of 12 HRBC patients (Table S1).
Twenty healthy women who were found to be negative for the respective familial germline pathogenic variant constituted the healthy control group, while healthy women with germline pathogenic variants in BRCA1 and BRCA2 (n = 12 and n = 10, respectively) were also included. The lack of breast and ovarian cancer in healthy women carrying pathogenic germline variants in BRCA1 and BRCA2 was confirmed with mammography and/or ultrasound screening and transvaginal ultrasonography within 1 year. Additionally, we included 12-12 women diagnosed with hormone-responsive breast cancer (HRBC) and TNBC. All cancer patients were treatment-naive and did not undergo surgical resection at the time of inclusion and sample accrual. Baseline characteristics of the study subjects are detailed in Table S1. Individuals included in the study were informed of the study procedure and signed an informed consent form. The study was approved by the Scientific and Research Ethics Committee of the Medical Research Council of Hungary (ETT-TUKEB 53720-7/2019/EÜIG).
PBMC isolation
Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation.44 Briefly, 9 mL whole blood was layered on 5 mL of Pancoll solution (PanBiotech GmbH, Germany) within 4 h of venipuncture and was centrifuged at 800g for 20 min. PBMCs were washed twice with PBS (centrifugation at 300g for 10 min) and a high degree of viability (>90%) was confirmed with trypan blue staining. PBMCs were aliquoted and cryopreserved in a 90% FBS-10% DMSO solution in liquid nitrogen storage.
Cell preparation for mass cytometry
Cryotubes were thawed in a 37°C water bath for 2 min and cells were transferred into 9 mL 37°C warm complete RPMI-1640 cell culture media (cRPMI) containing 100 U/ml penicillin and 100 μg/mL streptomycin (Merck, USA), 10% FBS (Euroclone), 2mM glutamine (200mM 100x diluted Capricorn), and centrifugated at 370g for 6 min at room temperature (RT).45 PBMCs were washed one more time with 10 mL cRPMI and cells were counted using a standard counting chamber and viability was determined with trypan blue exclusion. 3 × 106 PBMCs/sample were plated on 96-well repellent plate (GreinerBio-One Ltd.) separately in 200 μL cRPMI and rested 2h in an incubator with 5% CO2 at 37°C. Rested cells were collected and washed twice with Maxpar Cell Staining Buffer (MCSB, Standard Biotools, San Francisco, CA, USA).
Cell barcoding and antibody staining
The Maxpar Direct Immune Profiling Assay46,47 was complemented with the addition of antibodies against cytotoxical marker CD95 and four immune checkpoint molecules (CD366/Tim-3, CD223/LAG-3, CD279/PD-1 and CD134/OX40) as ICI therapies are becoming widely used oncopharmacological agents especially in TNBC.5
Cells were resuspended in 200 μL MCSB and transferred into 1.5 mL centrifuge tube. Subsequently, 800 μL MCSB was added, and centrifuged at 370 g at RT for 6 min. Cells were suspended in 47.5 μL MCSB supplemented with 2.5 μL TrueStain FcX 1:20 (V/V%) Fc receptor blocking solution (BioLegend, San Diego, CA, USA) and incubated at RT for 10 min. After 10 min incubation in the cell culture incubator, 8 patient-derived samples were barcoded individually by adding 50-50 μL of different metal-tagged (89Y, 106Cd, 110Cd, 111Cd, 112Cd, 113Cd, 114Cd, 116Cd) CD45 antibodies (clone: HI30; Standard Biotools, San Francisco, CA, USA) at a final concentration of 1:100 (V/V%) per antibody and incubated at 4°C for 30 min. The CD45 live cell barcoding was described previously by Fish et al.48 Cells were washed two times with 1 mL MCSB, centrifuged at 370 g at RT for 6 min after which they were resuspended in 1 mL MCSB and counted. 5x105 cells from each patient were pooled into a 5 mL centrifuge tube, centrifuged at 370 g at RT for 6 min. Cells were resuspended in 275 μL MCSB and stained with 1:100 (V/V%) of 5 markers (Standard Biotools, San Francisco, CA, USA): anti-human CD366/Tim-3 (clone F38-2E2)-159Tb; anti-human CD95/Fas (clone DX2)-162Dy; anti-human CD223/LAG-3 (clone 11C3C65)-165Ho; anti-human CD279/PD-1 (clone EH12.2H7)-175Lu; anti-human CD134/OX40 (clone ACT35)-142Nd and transferred into a single tube of Maxpar Direct Immune Profiling Assay (Standard Biotools, San Francisco, CA, USA) and incubated at RT for 30 min (key resources table). Cells were washed with 2 mL MCSB two times, centrifuged at 370 g at RT for 6 min. Cells were fixed in 1.6% formaldehyde solution (Pierce, Thermo Scientific, Waltham, MA, USA) and incubated at RT for 10 min. Stained and fixed cells were centrifuged at 800 g at RT for 6 min and resuspended in 800 μL Fix & Perm solution supplemented with 1:1000 (V/V%) 191Ir-193Ir DNA intercalator (all from Standard Biotools, San Francisco, CA, USA) for overnight incubation.49,50
Mass cytometry data acquisition and preprocessing
Samples were washed three times with 4 mL MCSB, (centrifugation at 800g, RT for 6 min) before being filtered through a 30 μm CellTrics gravity filter (Sysmex, Görlitz, Germany). The cell concentrations were adjusted to 8x105/mL in Cell Acquisition Solution (CAS, Standard Biotools, San Francisco, CA, USA). Finally, EQ four-element calibration beads (Standard Biotools, San Francisco, CA, USA) were added at a 1:10 ratio (V/V%) and acquired on a properly tuned Helios mass cytometer with WB injector (Standard Biotools, San Francisco, CA, USA). We acquired min 1.2 × 106 events from the pooled samples (∼3 × 105 events/individual PBMC sample) to identify rare cell subsets. The generated flow cytometry standard (FCS) files were randomized and normalized with the default setting of the internal FCS-processing unit of the CyTOF Software (Standard Biotools, San Francisco, CA, USA, version: 7.0.8493).
Quantification and statistical analysis
Data processing
Randomized and normalized FCS files were uploaded to the Cytobank Premium analysis platform (Beckman Coulter). Exclusion of normalization beads, dead cells, debris, doublets and manual debarcoding were performed (Figure S4). There were no significant differences in cell counts between the examined groups (Figure S1B). The FCS files with the CD45 positive living singlets were exported, deposited on Mendeley Data and further analyzed in R.
Compensation methodology, FlowSOM clustering and dimensionality reduction were adapted from Crowell et al.51,52 FCS files were integrated, compensated and transformed utilizing the CATALYST and flowCore R packages.41,42 After signal spillover compensation, CyTOF marker intensities were inverse hyperbolic sine transformed (arcsinh) with cofactor 5. For the main population definition, we carried out self-organizing-map based method (FlowSOM) clustering on compensated and transformed files.53 We identified 10 different clusters which were separately subclustered with another round of FlowSOM. Cluster annotation by manual curation of the 10 main immune cell types were performed using the relative expression levels of CD3, CD4, CD8a, CD11c, CD14, CD19, CD20, CD56, TCRgd and HLA-DR. Dimensionality reduction and visualization were performed using the t-distributed stochastic neighbor embedding (t-SNE) algorithm. A total of 300,000 cells and 35 markers (key resources table) were used to create the peripheral human immune system t-SNE map. On every identified main population, we carried out subclustering with FlowSOM and visualized it using Uniform Manifold Approximation and Projection (UMAP). Each UMAP represented 3000 cells/sample.
Statistical analysis
Median signal intensities, cell frequencies and subpopulation frequencies were analyzed with GraphPad Prism 8.0.1. The normality of distributions were tested with D’Agostino & Pearson test and passed if all groups’ alpha value were under 0.05. Normally distributed datasets were compared with ordinary one-way ANOVA or Brown-Forsythe ANOVA when the standard deviations were not equal. For non-parametric analysis, Kruskal-Wallis test was applied. Significance tests were corrected for multiple comparisons by controlling for the False Discovery Rate (FDR) with a two-stage Benjamini, Krieger and Yekutieli approach with an FDR cutoff of 10%. Correlation analyses were performed using Spearman’s method. Comparisons of categorical values were performed using Fisher’s exact test. Differences are considered significant at p < 0.05∗; <0.01∗∗; <0.001∗∗∗.
Acknowledgments
The project was implemented with the support from the National Research, Development and Innovation Fund of the Ministry of Culture and Innovation under the National Laboratories Program (National Tumor Biology Laboratory (2022-2.1.1-NL-2022-00010)) and the Hungarian Thematic Excellence Program (under project TKP2021-EGA-44) Grant Agreements with the National Research, Development and Innovation Office. The authors would like to acknowledge the financial support of the National Research, Development and Innovation Office under grants Investment into Future (2020-1.1.6-JÖVŐ−2021-00003 and 2020-1.1.6-JÖVŐ-2021-00008), NKFIH-FK-142877, and NKFIH-FK-138377. G.J.S. is the recipient of the János Bolyai Postdoctoral Scholarship (BO/00582/22/8) and the New National Excellence Program Bolyai+ Scholarship (ÚNKP-23-5-SZTE-694). V.K.G. is the recipient of the János Bolyai Postdoctoral Scholarship (BO/00141/21) and the New National Excellence Program Bolyai+ Scholarship (ÚNKP-22-5-SE-25, ÚNKP-23-5-SE-16). V.K.G. is supported by COST Action CA20122 Harmonisation.
Author contributions
V.K.G. and G.J.S. designed the project. H.B., A.P., and V.K.G. performed genetic counseling and together with A.B. and J.P. were involved in the genetic diagnosis. V.K.G. and K.H.-O. performed sample collection. J.Á.B. and G.J.S. performed mass cytometry measurements and together with V.K.G. and D.K. were involved in data analysis and interpretation. V.K.G. and G.J.S. wrote the manuscript. All authors read and approved the final version.
Declaration of interests
The authors declare no competing interests.
Published: May 4, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109882.
Contributor Information
Gábor János Szebeni, Email: szebeni.gabor@brc.hu.
Vince Kornél Grolmusz, Email: grolmusz.vince@oncol.hu.
Supplemental information
References
- 1.Hammerl D., Smid M., Timmermans A.M., Sleijfer S., Martens J.W.M., Debets R. Breast cancer genomics and immuno-oncological markers to guide immune therapies. Semin. Cancer Biol. 2018;52:178–188. doi: 10.1016/j.semcancer.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Szebeni G.J., Vizler C., Kitajka K., Puskas L.G. Inflammation and Cancer: Extra- and Intracellular Determinants of Tumor-Associated Macrophages as Tumor Promoters. Mediators Inflamm. 2017;2017:9294018. doi: 10.1155/2017/9294018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balog J.A., Hackler L., Jr., Kovacs A.K., Neuperger P., Alfoldi R., Nagy L.I., Puskas L.G., Szebeni G.J. Single Cell Mass Cytometry Revealed the Immunomodulatory Effect of Cisplatin Via Downregulation of Splenic CD44+, IL-17A+ MDSCs and Promotion of Circulating IFN-gamma+ Myeloid Cells in the 4T1 Metastatic Breast Cancer Model. Int. J. Mol. Sci. 2019;21:170. doi: 10.3390/ijms21010170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pal B., Chen Y., Vaillant F., Capaldo B.D., Joyce R., Song X., Bryant V.L., Penington J.S., Di Stefano L., Tubau Ribera N., et al. A single-cell RNA expression atlas of normal, preneoplastic and tumorigenic states in the human breast. EMBO J. 2021;40:e107333. doi: 10.15252/embj.2020107333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmid P., Cortes J., Pusztai L., McArthur H., Kümmel S., Bergh J., Denkert C., Park Y.H., Hui R., Harbeck N., et al. Pembrolizumab for Early Triple-Negative Breast Cancer. N. Engl. J. Med. 2020;382:810–821. doi: 10.1056/NEJMoa1910549. [DOI] [PubMed] [Google Scholar]
- 6.Batalha S., Ferreira S., Brito C. The Peripheral Immune Landscape of Breast Cancer: Clinical Findings and In Vitro Models for Biomarker Discovery. Cancers. 2021;13:1305. doi: 10.3390/cancers13061305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verronese E., Delgado A., Valladeau-Guilemond J., Garin G., Guillemaut S., Tredan O., Ray-Coquard I., Bachelot T., N'Kodia A., Bardin-Dit-Courageot C., et al. Immune cell dysfunctions in breast cancer patients detected through whole blood multi-parametric flow cytometry assay. OncoImmunology. 2016;5:e1100791. doi: 10.1080/2162402X.2015.1100791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foulds G.A., Vadakekolathu J., Abdel-Fatah T.M.A., Nagarajan D., Reeder S., Johnson C., Hood S., Moseley P.M., Chan S.Y.T., Pockley A.G., et al. Immune-Phenotyping and Transcriptomic Profiling of Peripheral Blood Mononuclear Cells From Patients With Breast Cancer: Identification of a 3 Gene Signature Which Predicts Relapse of Triple Negative Breast Cancer. Front. Immunol. 2018;9:2028. doi: 10.3389/fimmu.2018.02028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jorgensen N., Laenkholm A.V., Saekmose S.G., Hansen L.B., Hviid T.V.F. Peripheral blood immune markers in breast cancer: Differences in regulatory T cell abundance are related to clinical parameters. Clin. Immunol. 2021;232:108847. doi: 10.1016/j.clim.2021.108847. [DOI] [PubMed] [Google Scholar]
- 10.Poschke I., De Boniface J., Mao Y., Kiessling R. Tumor-induced changes in the phenotype of blood-derived and tumor-associated T cells of early stage breast cancer patients. Int. J. Cancer. 2012;131:1611–1620. doi: 10.1002/ijc.27410. [DOI] [PubMed] [Google Scholar]
- 11.Song G., Wang X., Jia J., Yuan Y., Wan F., Zhou X., Yang H., Ren J., Gu J., Lyerly H.K. Elevated level of peripheral CD8(+)CD28(-) T lymphocytes are an independent predictor of progression-free survival in patients with metastatic breast cancer during the course of chemotherapy. Cancer Immunol. Immunother. 2013;62:1123–1130. doi: 10.1007/s00262-013-1424-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szebeni G.J., Vizler C., Nagy L.I., Kitajka K., Puskas L.G. Pro-Tumoral Inflammatory Myeloid Cells as Emerging Therapeutic Targets. Int. J. Mol. Sci. 2016;17:1958. doi: 10.3390/ijms17111958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuda B., Miyamoto A., Yokoyama K., Ogiya R., Oshitanai R., Terao M., Morioka T., Niikura N., Okamura T., Miyako H., et al. B-cell populations are expanded in breast cancer patients compared with healthy controls. Breast Cancer. 2018;25:284–291. doi: 10.1007/s12282-017-0824-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mamessier E., Pradel L.C., Thibult M.L., Drevet C., Zouine A., Jacquemier J., Houvenaeghel G., Bertucci F., Birnbaum D., Olive D. Peripheral blood NK cells from breast cancer patients are tumor-induced composite subsets. J. Immunol. 2013;190:2424–2436. doi: 10.4049/jimmunol.1200140. [DOI] [PubMed] [Google Scholar]
- 15.Buys S.S., Sandbach J.F., Gammon A., Patel G., Kidd J., Brown K.L., Sharma L., Saam J., Lancaster J., Daly M.B. A study of over 35,000 women with breast cancer tested with a 25-gene panel of hereditary cancer genes. Cancer. 2017;123:1721–1730. doi: 10.1002/cncr.30498. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida R. Hereditary breast and ovarian cancer (HBOC): review of its molecular characteristics, screening, treatment, and prognosis. Breast Cancer. 2021;28:1167–1180. doi: 10.1007/s12282-020-01148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nik-Zainal S., Davies H., Staaf J., Ramakrishna M., Glodzik D., Zou X., Martincorena I., Alexandrov L.B., Martin S., Wedge D.C., et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies H., Glodzik D., Morganella S., Yates L.R., Staaf J., Zou X., Ramakrishna M., Martin S., Boyault S., Sieuwerts A.M., et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat. Med. 2017;23:517–525. doi: 10.1038/nm.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samstein R.M., Krishna C., Ma X., Pei X., Lee K.W., Makarov V., Kuo F., Chung J., Srivastava R.M., Purohit T.A., et al. Mutations in BRCA1 and BRCA2 differentially affect the tumor microenvironment and response to checkpoint blockade immunotherapy. Nat. Cancer. 2021;1:1188–1203. doi: 10.1038/s43018-020-00139-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nee K., Ma D., Nguyen Q.H., Pein M., Pervolarakis N., Insua-Rodríguez J., Gong Y., Hernandez G., Alshetaiwi H., Williams J., et al. Preneoplastic stromal cells promote BRCA1-mediated breast tumorigenesis. Nat. Genet. 2023;55:595–606. doi: 10.1038/s41588-023-01298-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim E., Vaillant F., Wu D., Forrest N.C., Pal B., Hart A.H., Asselin-Labat M.L., Gyorki D.E., Ward T., Partanen A., et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat. Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 22.Molyneux G., Geyer F.C., Magnay F.A., McCarthy A., Kendrick H., Natrajan R., Mackay A., Grigoriadis A., Tutt A., Ashworth A., et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7:403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 23.Shalabi S.F., Miyano M., Sayaman R.W., Lopez J.C., Jokela T.A., Todhunter M.E., Hinz S., Garbe J.C., Stampfer M.R., Kessenbrock K., et al. Evidence for accelerated aging in mammary epithelia of women carrying germline BRCA1 or BRCA2 mutations. Nat. Aging. 2021;1:838–849. doi: 10.1038/s43587-021-00104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogony J., Hoskin T.L., Stallings-Mann M., Winham S., Brahmbhatt R., Arshad M.A., Kannan N., Peña A., Allers T., Brown A., et al. Immune cells are increased in normal breast tissues of BRCA1/2 mutation carriers. Breast Cancer Res. Treat. 2023;197:277–285. doi: 10.1007/s10549-022-06786-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kandalaft L.E., Dangaj Laniti D., Coukos G. Immunobiology of high-grade serous ovarian cancer: lessons for clinical translation. Nat. Rev. Cancer. 2022;22:640–656. doi: 10.1038/s41568-022-00503-z. [DOI] [PubMed] [Google Scholar]
- 26.Forster R., Mattis A.E., Kremmer E., Wolf E., Brem G., Lipp M. A putative chemokine receptor, BLR1, directs B cell migration to defined lymphoid organs and specific anatomic compartments of the spleen. Cell. 1996;87:1037–1047. doi: 10.1016/s0092-8674(00)81798-5. [DOI] [PubMed] [Google Scholar]
- 27.Rathmell J.C., Townsend S.E., Xu J.C., Flavell R.A., Goodnow C.C. Expansion or elimination of B cells in vivo: dual roles for CD40- and Fas (CD95)-ligands modulated by the B cell antigen receptor. Cell. 1996;87:319–329. doi: 10.1016/s0092-8674(00)81349-5. [DOI] [PubMed] [Google Scholar]
- 28.Golinski M.L., Demeules M., Derambure C., Riou G., Maho-Vaillant M., Boyer O., Joly P., Calbo S. CD11c(+) B Cells Are Mainly Memory Cells, Precursors of Antibody Secreting Cells in Healthy Donors. Front. Immunol. 2020;11:32. doi: 10.3389/fimmu.2020.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rincon-Arevalo H., Wiedemann A., Stefanski A.L., Lettau M., Szelinski F., Fuchs S., Frei A.P., Steinberg M., Kam-Thong T., Hatje K., et al. Deep Phenotyping of CD11c(+) B Cells in Systemic Autoimmunity and Controls. Front. Immunol. 2021;12:635615. doi: 10.3389/fimmu.2021.635615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kar A., Mehrotra S., Chatterjee S. CD38: T Cell Immuno-Metabolic Modulator. Cells. 2020;9:1716. doi: 10.3390/cells9071716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh A., Khanam A., Ray K., Mathur P., Subramanian A., Poonia B., Kottilil S. CD38: an ecto-enzyme with functional diversity in T cells. Front. Immunol. 2023;14:1146791. doi: 10.3389/fimmu.2023.1146791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandoval-Montes C., Santos-Argumedo L. CD38 is expressed selectively during the activation of a subset of mature T cells with reduced proliferation but improved potential to produce cytokines. J. Leukoc. Biol. 2005;77:513–521. doi: 10.1189/jlb.0404262. [DOI] [PubMed] [Google Scholar]
- 33.Ramzaoui S., Jouen-Beades F., Gilbert D., Borsa-Lebas F., Michel Y., Humbert G., Tron F. During HIV infection, CD4+ CD38+ T-cells are the predominant circulating CD4+ subset whose HLA-DR positivity increases with disease progression and whose V beta repertoire is similar to that of CD4+ CD38- T-cells. Clin. Immunol. Immunopathol. 1995;77:33–41. doi: 10.1016/0090-1229(95)90134-5. [DOI] [PubMed] [Google Scholar]
- 34.Woods D.M., Laino A.S., Winters A., Alexandre J., Freeman D., Rao V., Adavani S.S., Weber J.S., Chattopadhyay P.K. Nivolumab and ipilimumab are associated with distinct immune landscape changes and response-associated immunophenotypes. JCI Insight. 2020;5:e137066. doi: 10.1172/jci.insight.137066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitra A., Thompson B., Strange A., Amato C.M., Vassallo M., Dolgalev I., Hester-McCullough J., Muramatsu T., Kimono D., Puranik A.S., et al. A Population of Tumor-Infiltrating CD4+ T Cells Co-Expressing CD38 and CD39 Is Associated with Checkpoint Inhibitor Resistance. Clin. Cancer Res. 2023;29:4242–4255. doi: 10.1158/1078-0432.CCR-23-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui C., Craft J., Joshi N.S. T follicular helper cells in cancer, tertiary lymphoid structures, and beyond. Semin. Immunol. 2023;69:101797. doi: 10.1016/j.smim.2023.101797. [DOI] [PubMed] [Google Scholar]
- 37.Carr E.J., Dooley J., Garcia-Perez J.E., Lagou V., Lee J.C., Wouters C., Meyts I., Goris A., Boeckxstaens G., Linterman M.A., Liston A. The cellular composition of the human immune system is shaped by age and cohabitation. Nat. Immunol. 2016;17:461–468. doi: 10.1038/ni.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brodin P., Jojic V., Gao T., Bhattacharya S., Angel C.J.L., Furman D., Shen-Orr S., Dekker C.L., Swan G.E., Butte A.J., et al. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160:37–47. doi: 10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lakshmikanth T., Muhammad S.A., Olin A., Chen Y., Mikes J., Fagerberg L., Gummesson A., Bergström G., Uhlen M., Brodin P. Human Immune System Variation during 1 Year. Cell Rep. 2020;32:107923. doi: 10.1016/j.celrep.2020.107923. [DOI] [PubMed] [Google Scholar]
- 40.Tsang J.S., Schwartzberg P.L., Kotliarov Y., Biancotto A., Xie Z., Germain R.N., Wang E., Olnes M.J., Narayanan M., Golding H., et al. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014;157:499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ellis B., Haaland P., Hahne F., Le Meur N., Gopalakrishnan N., Spidlen J., Jiang M., Finak G. 2024. flowCore: flowCore: Basic Structures for Flow Cytometry Data. [DOI] [Google Scholar]
- 42.Crowell H., Zanotelli V., Chevrier S., Robinson M. 2023. CATALYST: Cytometry dATa anALYSis Tools. [DOI] [Google Scholar]
- 43.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brosseron F., May C., Schoenebeck B., Tippler B., Woitalla D., Kauth M., Brockmann K., Meyer H.E., Berg D., Bufe A., Marcus K. Stepwise isolation of human peripheral erythrocytes, T lymphocytes, and monocytes for blood cell proteomics. Proteomics Clin. Appl. 2012;6:497–501. doi: 10.1002/prca.201200032. [DOI] [PubMed] [Google Scholar]
- 45.Neuperger P., Szalontai K., Gémes N., Balog J.Á., Tiszlavicz L., Furák J., Lázár G., Puskás L.G., Szebeni G.J. Single-cell mass cytometric analysis of peripheral immunity and multiplex plasma marker profiling of non-small cell lung cancer patients receiving PD-1 targeting immune checkpoint inhibitors in comparison with platinum-based chemotherapy. Front. Immunol. 2023;14:1243233. doi: 10.3389/fimmu.2023.1243233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.IMPACC Manuscript Writing Team. IMPACC Network Steering Committee Immunophenotyping assessment in a COVID-19 cohort (IMPACC): A prospective longitudinal study. Sci. Immunol. 2021;6:eabf3733. doi: 10.1126/sciimmunol.abf3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bagwell C.B., Hunsberger B., Hill B., Herbert D., Bray C., Selvanantham T., Li S., Villasboas J.C., Pavelko K., Strausbauch M., et al. Multi-site reproducibility of a human immunophenotyping assay in whole blood and peripheral blood mononuclear cells preparations using CyTOF technology coupled with Maxpar Pathsetter, an automated data analysis system. Cytometry B Clin. Cytom. 2020;98:146–160. doi: 10.1002/cyto.b.21858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fish M., Ellis R., Bishop C., Todd K., Petrov N., Singer M., Swanson C.M., Shankar-Hari M. Utilising mass cytometry with CD45 barcoding and standardised leucocyte phenotyping for immune trajectory assessment in critically ill patients. Br. J. Anaesth. 2021;126:e149–e152. doi: 10.1016/j.bja.2021.01.006. [DOI] [PubMed] [Google Scholar]
- 49.Balog J.Á., Honti V., Kurucz É., Kari B., Puskás L.G., Andó I., Szebeni G.J. Immunoprofiling of Drosophila Hemocytes by Single-cell Mass Cytometry. Dev. Reprod. Biol. 2021;19:243–252. doi: 10.1016/j.gpb.2020.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neuperger P., Balog J.Á., Tiszlavicz L., Furák J., Gémes N., Kotogány E., Szalontai K., Puskás L.G., Szebeni G.J. Analysis of the Single-Cell Heterogeneity of Adenocarcinoma Cell Lines and the Investigation of Intratumor Heterogeneity Reveals the Expression of Transmembrane Protein 45A (TMEM45A) in Lung Adenocarcinoma Cancer Patients. Cancers. 2021;14:144. doi: 10.3390/cancers14010144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crowell H.L., Chevrier S., Jacobs A., Sivapatham S., Tumor Profiler Consortium. Bodenmiller B., Robinson M.D. An R-based reproducible and user-friendly preprocessing pipeline for CyTOF data. F1000Res. 2020;9:1263. doi: 10.12688/f1000research.26073.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nowicka M., Krieg C., Crowell H.L., Weber L.M., Hartmann F.J., Guglietta S., Becher B., Levesque M.P., Robinson M.D. CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. F1000Res. 2017;6:748. doi: 10.12688/f1000research.11622.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Gassen S., Callebaut B., Van Helden M.J., Lambrecht B.N., Demeester P., Dhaene T., Saeys Y. FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A. 2015;87:636–645. doi: 10.1002/cyto.a.22625. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Original mass cytometry data has been deposited on Mendeley Data (https://doi.org/10.17632/pkx5wn7cb3.1) and is publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.