Significance
The basal forebrain (BF) is involved in control of sleep, cortical oscillations, attention, feeding, reward, and olfaction. Here, we characterize a major subset of BF GABAergic neurons which express neuronal PAS domain 1 (Npas1), a transcription factor linked to neuropsychiatric disorders. BF Npas1+ neurons are distinct from and more numerous than cholinergic, parvalbumin, or glutamate neurons. They project to brain areas involved in arousal, motivation, and olfaction. Activation of BF Npas1+ neurons in the light period increased wakefulness due to increased long wake bouts. Use of a developmental marker to identify this major subpopulation of BF GABAergic wake-promoting neurons allows cross-species studies of these neurons and their role in insomnia, dementia, neurodevelopmental disorders, and other conditions involving BF.
Keywords: nucleus basalis, ventral pallidum, sleep, parvalbumin, GABA
Abstract
Here, we describe a group of basal forebrain (BF) neurons expressing neuronal Per-Arnt-Sim (PAS) domain 1 (Npas1), a developmental transcription factor linked to neuropsychiatric disorders. Immunohistochemical staining in Npas1-cre-2A-TdTomato mice revealed BF Npas1+ neurons are distinct from well-studied parvalbumin or cholinergic neurons. Npas1 staining in GAD67-GFP knock-in mice confirmed that the vast majority of Npas1+ neurons are GABAergic, with minimal colocalization with glutamatergic neurons in vGlut1-cre-tdTomato or vGlut2-cre-tdTomato mice. The density of Npas1+ neurons was high, five to six times that of neighboring cholinergic, parvalbumin, or glutamatergic neurons. Anterograde tracing identified prominent projections of BF Npas1+ neurons to brain regions involved in sleep–wake control, motivated behaviors, and olfaction such as the lateral hypothalamus, lateral habenula, nucleus accumbens shell, ventral tegmental area, and olfactory bulb. Chemogenetic activation of BF Npas1+ neurons in the light period increased the amount of wakefulness and the latency to sleep for 2 to 3 h, due to an increase in long wake bouts and short NREM sleep bouts. NREM slow-wave and sigma power, as well as sleep spindle density, amplitude, and duration, were reduced, reminiscent of findings in several neuropsychiatric disorders. Together with previous findings implicating BF Npas1+ neurons in stress responsiveness, the anatomical projections of BF Npas1+ neurons and the effect of activating them suggest a possible role for BF Npas1+ neurons in motivationally driven wakefulness and stress-induced insomnia. Identification of this major subpopulation of BF GABAergic neurons will facilitate studies of their role in sleep disorders, dementia, and other neuropsychiatric conditions involving BF.
Sleep is one of the three pillars of health alongside diet and exercise, and disrupted sleep has been linked to a wide variety of physical and neuropsychiatric consequences (1). Sleep disturbances are common among the general public, with higher prevalence among current and former military personnel, the elderly, and those suffering from psychiatric disorders, neurodegenerative disease, or traumatic brain injury (1). Precise delineation of the brain circuits which control sleep and wakefulness is essential for the development of new treatments but is hampered by the lack of markers to identify the key neuronal subpopulations in evolutionarily conserved brain regions.
Different subgroups of neurons releasing the main inhibitory neurotransmitter in the brain, gamma-aminobutyric acid (GABA), are involved in a wide variety of different vigilance state regulatory roles, including the promotion of NREM and REM sleep, wakefulness, and the cortical oscillations which are typical of these states (2). The wide distribution of GABAergic neurons makes it challenging to find markers which identify key subpopulations, thus hindering targeted experiments to determine their functionality. An approach that has been productive in defining subpopulations of cerebral cortex GABAergic interneurons (3) and subpopulations of GABAergic projection neurons in the basal ganglia (4, 5) is the use of developmental transcription factors to serve as markers for specific neuronal populations. This approach has also been fruitful in identifying sleep–wake populations in the hypothalamus (6) and brainstem (7), but has so far not been applied to the basal forebrain (BF), a key brain region involved in the control of sleep–wake behavior, cortical activation, attention, and motivation (2, 8–10) which degenerates in dementia patients (11, 12) and is implicated in behavioral dysfunction in various psychiatric disorders (10, 13, 14).
Previous chemogenetic experiments showed that activation of all GABAergic neurons in the BF leads to a large increase in the amount of wakefulness (15, 16), whereas chemogenetic activation of cholinergic or glutamatergic BF neurons has less pronounced effects on sleep–wake behavior and cortical EEG (15). However, BF GABAergic neurons are heterogeneous in their neurochemical phenotype and properties (2, 17, 18). To date, only those GABAergic neurons which contain the calcium-binding protein parvalbumin (PV) or the neuropeptide somatostatin (SST) have been analyzed in detail. Optogenetic activation of BF PV+ neurons or their projections to the thalamic reticular nucleus (TRN) enhances cortical gamma oscillations and leads to rapid arousals from sleep (13, 19–21). However, arousals elicited by optogenetic activation of BF PV+ neurons are brief and only slightly increase the amount of wakefulness (20–22). In contrast, chemogenetic activation of BF SST neurons has little effect on sleep–wake states or cortical EEG (23), and strong optogenetic activation of BF SST neurons is sleep-promoting (22). Thus, the strong increase in wakefulness elicited by activation of all BF GABAergic neurons cannot be explained by activation of BF PV+ or SOM+ neurons, and markers for the wake-promoting GABAergic BF population(s) remain to be uncovered.
Neuronal Per-Arnt-Sim domain 1 (Npas1) is a basic helix–loop–helix class transcription factor (24) which has been linked by genetic and molecular studies to neuropsychiatric disorders (25, 26). Npas1 is expressed in immature and mature cortical interneurons and in their progenitors in the medial and caudal ganglionic eminences (27). Npas1 is also expressed in basal ganglia GABAergic projection neurons, also probably generated from the ganglionic eminences (27–30). In the globus pallidus pars externa (GPe), Npas1+ GABAergic projection neurons represent a distinct functional class of neurons from PV+ neurons (4), suggesting that Npas1 might represent a marker for non-PV+ GABAergic neurons in the BF. Npas1+ neurons have previously been reported within the BF region (4, 10, 25), but their neurochemical phenotype is unclear and their role in sleep–wake control has not been studied.
Here, we show that Npas1 identifies a major subpopulation of BF GABAergic neurons, distinct from and considerably more dense than previously characterized PV+, cholinergic, or glutamatergic neurons. BF Npas1+ neurons project to brain regions involved in sleep–wake control, motivated behavior, and olfaction. When activated chemogenetically, BF Npas1+ neurons promote wakefulness and disrupt NREM sleep and cortical oscillations.
Results
Many Small or Medium-Sized Npas1-tdTomato+ Neurons Are Present in the BF.
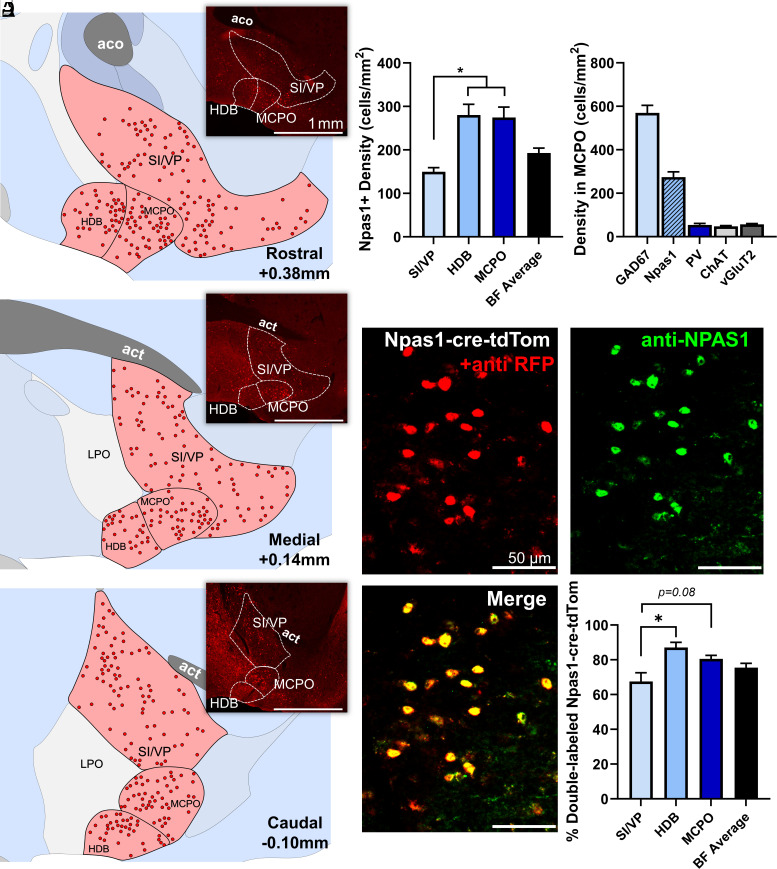
To study the distribution and neurochemical phenotype of BF Npas1+ neurons, we used previously validated Npas1-cre-2A-tdTomato mice (4) together with immunohistochemical staining. The fluorescent marker tdTomato is expressed at low levels in the brains of Npas1-cre-2A-tdTomato mice (4) (SI Appendix, Fig. S1A). Thus, to comprehensively visualize Npas1-tdTomato+ neurons, tdTomato fluorescence was amplified using a red fluorescent protein (RFP) immunohistochemical stain. Following staining, dense clusters of tdTomato+ neurons were observed in all three main regions of the intermediate BF, the substantia innominata/ventral pallidum (SI/VP), horizontal limb of the diagonal band (HDB), and magnocellular preoptic nucleus (MCPO) (Fig. 1 A–C), as well as in the globus pallidus pars externa and striatum, as previously described (4, 27, 30). Quantification of the densities of Npas1-tdTomato+ neurons in SI/VP, HDB, and MCPO, across rostral, medial, and caudal BF coronal sections revealed that the density was significantly higher in HDB (279.5 ± 24.8 neurons/mm2/section, n = 3 mice) and MCPO subregions (274.4 ± 24.6 neurons/mm2/section) compared to SI/VP (149.7 ± 9.3 neurons/mm2/section; P < 0.05) (Fig. 1D). Npas1+ neurons were also found in regions surrounding the BF. The densities of Npas1+ neurons in these areas are provided in SI Appendix, Table S1. Notably, only a few Npas1-tdTomato+ neurons (typically <5/section) were observed in the small sleep-promoting ventrolateral preoptic nucleus or median preoptic nucleus regions. Comparing the density of Npas1-tdTomato+ neurons with our previously described density measures for other neuronal phenotypes in the BF, it was clear that Npas1-tdTomato+ neurons are a major subpopulation (Fig. 1E). For instance, in the MCPO region, the density of Npas1-tdTomato+ neurons was 48.2 % of the density for all GABAergic neurons we calculated in a previous study (17). In this same subregion, the density of Npas1-tdTomato+ neurons was ~5 times that of PV+ neurons (17), 5.8 times that of cholinergic neurons (ChAT+) (31) and 4.8 times the density of vesicular glutamate transporter subtype 2 (vGlut2+) neurons (32).
Fig. 1.
Neurons expressing the transcription factor neuronal PAS domain 1 (Npas1) comprise a major cell population of the basal forebrain (BF). (A–C) Npas1+ neurons are abundant within the BF and its major subregions, including the substantia innominata and ventral pallidum (SI/VP), horizontal diagonal band (HDB), and magnocellular preoptic nucleus (MCPO), depicted across (A) rostral (+0.38 mm from bregma), (B) medial (+0.14 mm), and (C) caudal (−0.1 mm) coronal sections. Insets show low-power images of Npas1-tdTomato+ neurons stained with an anti-red fluorescent protein antibody to enhance endogenous tdTomato fluorescence in the same sections used for mapping the distribution of Npas1+ neurons. Other abbreviations: aco, anterior commissure, olfactory limb; act, anterior commissure, temporal limb; LPO, lateral preoptic area. (D) Npas1+ neurons are denser in ventral (HDB, MCPO) BF subregions compared to SI/VP (n = 3 animals; P < 0.05, one-way ANOVA, P = 0.0069). (E) Within the MCPO where GABAergic cell density is highest, Npas1-tdTomato+ cell density is 48.2% of that of all GABAergic cells (GAD67-GFP+ density from ref. 17). Npas1-tdTomato+ cell density is five to six times higher than other major cell subpopulations, such as parvalbumin (PV), choline acetyltransferase (ChAT), and vesicular glutamate transporter 2 (vGlut2) expressing neurons (Densities previously published in refs. 17, 31, and 32). (F) Although Npas1 is a transcription factor active early in development, the majority (75.5% ± 2.47%, n = 3 animals) of BF Npas1-tdTomato neurons (anti-RFP, red) in adult mice continue to express Npas1 as quantified through colocalization with an anti-NPAS1 nuclear stain (green). Colocalization was significantly higher in ventral subregions (HDB, MCPO) compared to the SI/VP (one-way ANOVA, P = 0.021). *P < 0.05.
Measurements of the long-axis diameter revealed that BF Npas1-tdTomato+ neurons were generally small-(10 to 15 μm) or medium-sized neurons (15 to 20 μm) (SI Appendix, Fig. S1B). Only 3 of 270 Npas1-tdTomato+ neurons evaluated had a long-axis diameter >20 μm (SI Appendix, Fig. S1C). Overall, BF Npas1-tdTomato+ neurons had a mean long-axis diameter of 13.2 ± 0.4 µm (270 neurons in three mice). Npas1-tdTomato+ neurons were slightly larger in the SI/VP (14.17 ± 0.35 µm) when compared to their counterparts in the HDB (12.63 ± 0.55 µm; P = 0.0010) and MCPO (12.70 ± 0.39 µm; P = 0.0012, ordinary one-way ANOVA, SI Appendix, Fig. S1D). BF Npas1-tdTomato+ neurons were smaller than BF PV+ neurons (mean long-axis diameter 20.2 µm) (17) or cholinergic neurons (24.4 µm) (17) but similar in size to all GAD67+ neurons (15.9 µm) (17), including large PV+ neurons, and vGlut2+ neurons (13.4 µm) (17).
Npas1 is a transcription factor which is active early in development in the ganglionic eminence regions which generate forebrain GABAergic neurons (27–30). Many developmental transcription factors are also active in adulthood and are involved in maintaining the postmitotic identity of the neurons in which they are expressed (33). Npas1 is expressed in adult cortical interneurons and adult basal ganglia projection neurons (4, 27). To determine the percentage of Npas1-tdTomato+ neurons which express NPAS1 protein in the BF in adult animals, we performed immunohistochemical staining for NPAS1 protein in Npas1-cre-2A-tdTomato mice (P90 or older; n = 3). As in our other experiments using these mice, low levels of tdTomato were amplified using an anti-RFP stain. As expected for a transcription factor, staining for NPAS1 protein was observed in the nucleus but not the cytoplasm. Overall, the distribution of NPAS1 protein was similar to the distribution of tdTomato+ neurons and most tdTomato+ neurons in the BF of adult Npas1-cre-2A-tdTomato mice also expressed NPAS1 protein (75.49 ± 2.47%, Fig. 1F). Expression of NPAS1 was not significantly different across Npas1-tdTomato+ cells from rostral, medial, and caudal BF sections. Highest levels of colocalization were found in the HDB (87.07 ± 3.12%) and MCPO (80.53 ± 2.38%) with slightly lower levels found in the fiber-dense SI/VP (67.51 ± 5.13%), possibly due to poor antibody penetration in the SI/VP region. Npas1-tdTomato+ neurons in regions neighboring the BF also expressed NPAS1 protein, as previously demonstrated in the globus pallidus pars externa (4). We note that the size and density measurements we report in the previous sections for Npas1-tdTomato+ neurons reflect values for all neurons derived from the Npas1 lineage, i.e., those that still express detectable NPAS1 protein (~75% in all BF), as well as those that expressed Npas1 during development but no longer have NPAS1 protein which is detectable by immunohistochemistry (~25%).
The Vast Majority of BF Npas1+ Neurons Are GABAergic but Do Not Express Parvalbumin, Choline Acetyltransferase, or Vesicular Glutamate Transporters.
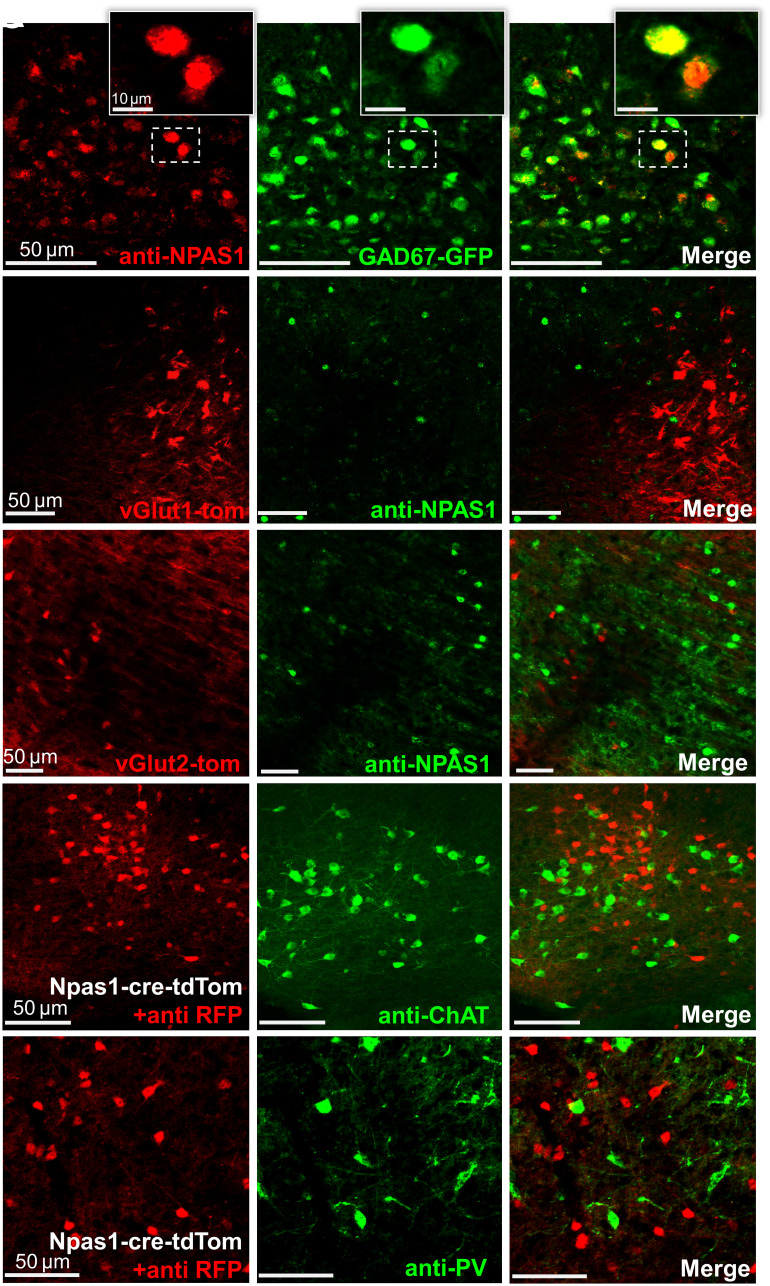
In other forebrain areas such as the cortex, hippocampus, and GPe, Npas1 is expressed in GABAergic neurons (4, 27). To test whether this is also the case in the BF, we performed immunohistochemistry for NPAS1 protein in neurons expressing fluorescent GABAergic (GAD67-GFP knock-in mice) (17) and glutamatergic markers (vGlut1-cre-tdTomato and vGlut2-cre-tdTomato mice). In GAD67-GFP knock-in mice (n = 3), the vast majority of NPAS1+ nuclei colocalized with GFP fluorescence (Fig. 2A). Overall: 98.24 ± 0.33% of NPAS1+ nuclei colocalized with GFP (SI/VP: 96.58 ± 0.43%; HDB: 96.84 ± 1.90%; MCPO: 99.05 ± 0.67%). No significant differences were found between rostral, medial, and caudal sections. Similarly high colocalization was also observed in one GAD67-GFP/Npas1-cre-2A-tdTomato crossed mouse (SI Appendix, Fig. S1E).
Fig. 2.
Neuronal PAS domain 1 (Npas1)-expressing neurons of the basal forebrain (BF) are GABAergic neurons distinct from parvalbumin (PV) and cholinergic (choline acetyltransferase, ChAT) neurons. (A) The vast majority of immunohistochemically stained NPAS1+ nuclei (red) colocalize with green fluorescent protein (GFP) in GAD67-GFP knock-in mice. NPAS1+ nuclei (green) do not colocalize with tdTomato (red) in (B) vesicular glutamate transporter 1 (vGlut1)-cre-tdTomato or (C) vesicular glutamate transporter 2 (vGlut2)-cre-tdTomato mice. (D and E) Npas1-tdTomato+ neurons (red) do not colocalize with neurons stained against ChAT (green, D) or PV (green, E).
In vGlut1-cre-tdTomato (n = 3) and vGlut2-cre-tdTomato mice (n = 5), anti-NPAS1 immunohistochemistry revealed minimal (<0.5%) colocalization of NPAS1 protein (green) with red fluorescent glutamatergic neurons (Fig. 2 B and C). To determine whether Npas1+ neurons express the cholinergic marker choline acetyltransferase (ChAT) or the calcium-binding protein PV, we performed immunohistochemistry in Npas1-cre-2A-tdTomato mice (n = 3 animals). Looking throughout the BF, there was similarly low colocalization between PV+ or ChAT+ neurons with Npas1-tdTomato+ neurons (Fig. 2 D and E). As described above, Npas1-tdTomato+ neurons also tended to be smaller than most PV+ or cholinergic neurons. Thus, together, our immunohistochemical data suggest that BF Npas1+ neurons represent a subset of noncholinergic, GABAergic neurons distinct from BF PV+ neurons.
Anterograde Tracing Revealed Projections of BF Npas1+ Neurons to Brain Areas Involved in Sleep–Wake Control, Motivated Behavior, and Olfaction.
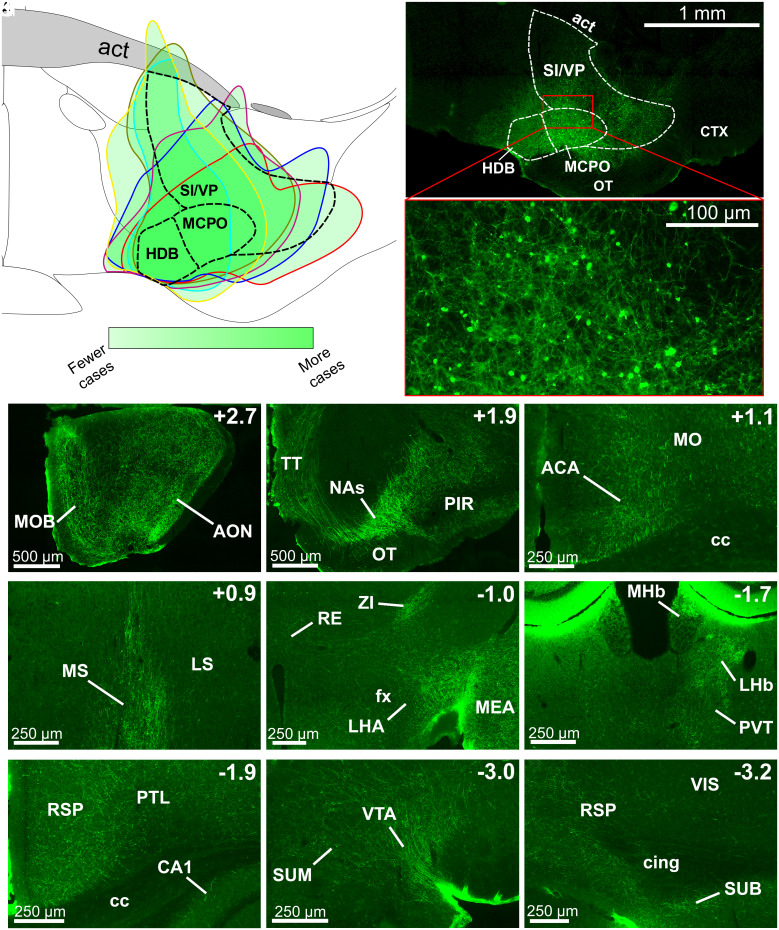
To determine the targets of BF Npas1+ neurons, we performed anterograde tracing experiments using unilateral injections of AAV5-DIO-ChR2-EYFP into the BF of adult Npas1-cre-tdTomato mice to specifically express ChR2-EYFP in BF Npas1+ neurons and their efferent projections (Fig. 3). The EYFP fluorescence was amplified with an anti-GFP immunohistochemical stain to more easily identify transduced axons of BF Npas1+ neurons. Post hoc analysis revealed good targeting of AAV5-DIO-ChR2-EYFP infusions to the ventral BF (HDB and MCPO) and dorsal SI/VP regions (n = 6 mice; Fig. 3 A and B). There was some limited transduction in a region medial to the BF which, in our previous studies and in the mouse brain atlas of Franklin and Paxinos (34), is labeled substantia innominata, but is included within the lateral preoptic area in the Allen Brain Atlas. Since this region is an area which contains cholinergic projection neurons (17), it can be considered as part of the BF, as conventionally described.
Fig. 3.
BF Npas1+ neurons project to brain areas involved in sleep–wake control, motivated behavior, and olfaction. (A) Composite schematic showing the boundaries of unilateral AAV5-DIO-ChR2-EYFP infusions in the medial BF in six mice, with darker green indicating more frequent transduction of the region. Transduced neurons were largely restricted to the BF. (B) Low (Top) and high (Bottom) magnification images of an individual injection. Abbreviations: act, anterior commissure temporal limb; CTX, cortex; OT, olfactory tubercle. (C–K) Rostral to caudal images of regions with prominent fiber projections. Top Right indicates position relative to bregma. (C and D) BF Npas1+ cells strongly project to olfactory regions, including the main olfactory bulb (MOB) and anterior olfactory nucleus (AON), taenia tecta (TT), and piriform cortex (PIR). (D) BF Npas1+ fibers densely innervate the nucleus accumbens shell (NAs). (E) In the neocortex, BF Npas1+ fibers innervated the anterior cingulate area (ACA) and motor cortex (MO) dorsal to the corpus callosum (cc) and (I and K) the retrosplenial cortex (RS). (F) BF Npas1+ fibers are denser in the medial septum (MS) compared to the lateral septum (LS). (G) Hypothalamic projections were strongest in the lateral hypothalamic area (LHA) close to the fornix (fx) with moderate projections to the zona incerta (ZI). Dense projections were also observed in the amygdala, including the medial amygdalar nucleus (MEA). RE, nucleus reuniens of the thalamus. (H) Projections to the thalamus were densest in the lateral habenula (LHb), dorsal part of the medial habenula (MHb) and paraventricular nucleus (PVT). (H, I, and K) Fibers were present throughout most of the hippocampal formation, including the cornu ammonis 1 (CA1) region, the retrosplenial cortex (RSP), and the subiculum (SUB). Additional abbreviations: Cing, cingulum bundle; PTL, parietal cortex; VIS, visual cortex. (J) Densest midbrain projections were to the ventral tegmental area (VTA) neighboring the supramammillary nucleus (SUM).
Anterograde tracing revealed prominent forebrain and midbrain projections of BF Npas1+ neurons (Fig. 3 and SI Appendix, Table S2). Projections were predominantly ipsilateral (SI Appendix, Table S2). In the neocortex, labeled fibers were observable in almost all regions but were generally sparse, with the exception of moderate fiber densities in the frontal cortex (prelimbic, infralimbic, anterior cingulate, and endopiriform cortices; Fig. 3E), and in the retrosplenial cortex (Fig. 3 I and K). Fibers were also evident in most subregions of the hippocampal formation (Fig. 3 I and K), most prominently in the entorhinal cortex and dorsal subiculum (Fig. 3K). The strongest projections to the thalamus targeted the lateral habenula (Fig. 3H) and the paraventricular thalamic nucleus (Fig. 3H), a stress-related region which also regulates wakefulness (35), although scattered fibers were also present in other associative and sensory/motor relay nuclei (SI Appendix, Table S2). There were only weak projections to the thalamic reticular nucleus, in marked contrast to the strong projections of GPe Npas1+ neurons (5), as well as BF PV+ and cholinergic neurons (15, 20, 36, 37). There were high fiber densities in rostral BF regions such as the medial septum, septofimbrial nucleus, and triangular nucleus of the septum, and a moderate density of fibers in the vertical limb of the diagonal band, but only low levels in the lateral septum (Fig. 3F and SI Appendix, Table S2). Amygdala regions also tended to have high or moderate fiber densities (Fig. 3G and SI Appendix, Table S2). A prominent finding was very high fiber densities in brain areas involved in olfaction, including the main olfactory bulb (Fig. 3C), anterior olfactory nucleus (Fig. 3C), and taenia tecta (Fig. 3D). In the basal ganglia, a strong projection was observed in the shell of the nucleus accumbens (Fig. 3D) as well as to the midbrain ventral tegmental area (Fig. 3J). BF Npas1+ fibers largely avoided the core of the nucleus accumbens (Fig. 3D) and the dorsal striatum (SI Appendix, Table S2). In the hypothalamus, the strongest projection was to the lateral hypothalamic area (Fig. 3G) which contains orexin/hypocretin and other wake-promoting neuronal phenotypes (38). The lateral supramammillary nucleus (Fig. 3J), posterior hypothalamus, and zona incerta (Fig. 3G) exhibited a moderate density of fibers, as did the anterior hypothalamic nucleus, dorsomedial hypothalamus, and medial preoptic area (SI Appendix, Table S2). Few fibers were observed in sleep-promoting areas such as the ventrolateral preoptic area and median preoptic area. The fiber density was generally low in the pons, but fibers could be observed in sleep–wake regulatory regions such as the raphe nuclei, pedunculopontine nucleus, and periaqueductal gray (SI Appendix, Table S2).
In summary, these cell-specific anterograde experiments revealed that BF Npas1+ cells are projection neurons that strongly innervate subcortical regions and have a dual role in control of sleep–wake regulation and motivated behavior. Additionally, there are strong projections of BF Npas1+ neurons to brain areas involved in olfaction. Given previous studies of BF GABAergic neurons and the projections of BF Npas1+ neurons to sleep–wake control regions, we next determined the effect of activating BF Npas1+ neurons on sleep–wake behavior and cortical oscillations.
Chemogenetic Stimulation of BF Npas1+ Neurons Promotes Wakefulness and Disrupts NREM Sleep Slow and Sigma Oscillations.
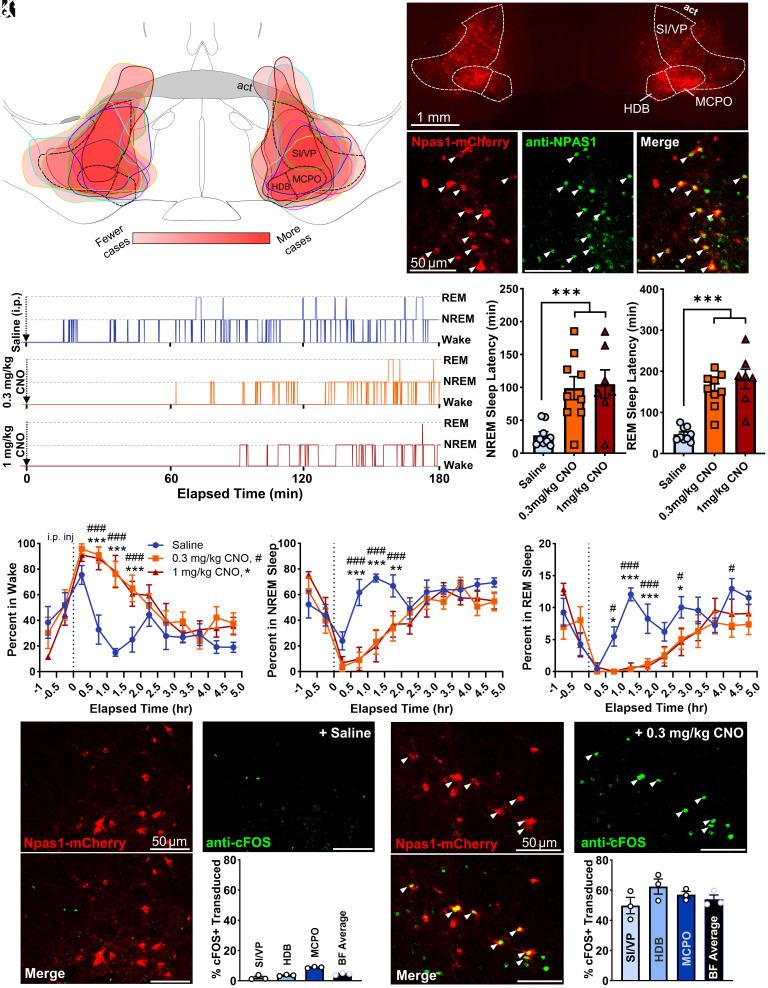
To determine the functional effect of activating BF Npas1+ neurons on sleep–wake behavior and cortical EEG, we performed chemogenetic experiments in Npas1-cre-2A-tdTomato mice. Viral vectors for Cre-dependent expression of the excitatory receptor hM3d(Gq) and a fluorescent marker of transduction (mCherry) were injected bilaterally into the BF (Fig. 4 A and B). Four or more weeks later, mice received intraperitoneal (i.p.) injections of saline, 0.3 mg/kg, or 1 mg/kg clozapine-N-oxide (CNO) in a within-subject design to activate BF Npas1+ neurons at the beginning of the light (inactive) period at zeitgeber time 2 (ZT2). Post hoc histological examination confirmed successful targeting of the BF (Fig. 4 A and B) and transduction of Npas1+ neurons (Fig. 4C). Npas1+ neurons transduced with mCherry were bright and easy to distinguish from Npas1+ neurons expressing low levels of unamplified tdTomato (SI Appendix, Fig. S1A). In total, 43.9% ± 6.6% of all neurons expressing NPAS1 within the BF (SI/VP: 48.1 ± 6.7%; HDB: 38.0 ± 6.3%; MCPO: 45.3 ± 8.8%) were transduced by hM3d(Gq)-mCherry, likely an undercount of the viral transduction efficiency, as not every viral infusion entirely filled all subregions of the BF (Fig. 4A). Among transduced Npas1-cre animals, 65.0 ± 2.7% of mCherry neurons expressed detectable NPAS1 protein, with a slightly lower percentage in the fiber dense SI/VP region (SI/VP: 59.0 ± 4.0%; HDB: 71.8 ± 4.0%; MCPO: 71.2 ± 2.7%). These numbers are comparable to those we reported for the percentage of Npas1-tdTomato+ neurons which expressed detectable NPAS1 protein in adults. Limited transduction was observed in preoptic areas medial to the BF. In a subset of cases (2/9 cases bilaterally and 2/9 unilaterally), there was spread dorsal to the anterior commissure. However, results were comparable to those cases without spillover across the anterior commissure and have been combined for the analysis.
Fig. 4.
Activation of basal forebrain (BF) neuronal PAS domain 1 (Npas1+) neurons promotes wakefulness and suppresses non-rapid-eye-movement (NREM) sleep and rapid-eye-movement (REM) sleep. (A) Injection composite indicating the spread of individual bilateral AAV8-hSyn-DIO-hM3D(Gq)-mCherry infusions in the medial BF across nine animals. Darker red indicates more frequent targeting of the region. Abbreviations: act, anterior commissure temporal limb; HDB, horizontal limb of the diagonal band; MCPO, magnocellular preoptic nucleus; SI/VP, substantia innominata/ventral pallidum. (B) Example bilateral infusion of AAV-hSyn-DIO-hM3D(Gq)-mCherry in a medial BF section. (C) The majority of transduced Npas1-cre cells expressing mCherry (red) also colocalize with an NPAS1 nuclear stain (green), indicating most transduced Npas1-cre-tdTomato+ neurons had active Npas1 expression in adults. (D) Representative hypnogram from one mouse following i.p. injections of 0.3 mg/kg clozapine-N-oxide (CNO, orange), 1 mg/kg CNO (red), or saline (blue). The onset of NREM sleep is <30 min following saline administration, but >60 min after CNO administration. Latency to REM sleep following saline injection is <90 min, but >150 min after CNO. (E) Compared to saline (n = 9), NREM sleep latency was significantly higher (P < 0.005) following administration of 0.3 mg/kg CNO (n = 9) or 1 mg/kg CNO (n = 7). (F) Additionally, REM sleep latency was significantly longer following injections of 0.3 mg/kg CNO and 1 mg/kg CNO compared to saline. (G) Administration of 0.3 or 1 mg/kg CNO promotes significantly more wakefulness vs. saline beginning 0.5 h after injection and persisting for approximately 2 h. (H) Concomitantly, 0.3 and 1 mg/kg CNO injections promote significantly less time in NREM sleep vs. saline. (I) REM sleep is also significantly suppressed after CNO administration vs. saline, starting 0.5 h postinjection and persisting approximately 3 h. Saline vs. 0.3 mg/kg CNO, P < 0.05 #, P < 0.01 ##, P < 0.005 ###. Saline vs. 1 mg/kg CNO: P < 0.05 *, P < 0.01 **, P < 0.005 ***. (J) Few BF NPAS1-mCherry cells (red) expressed cFOS (green) following saline administration 2 h before the mice were killed (4.5% ± 0.4%, n = 3). (K) Colocalization with immunohistochemical nuclear stains for cFOS (green) indicates the majority of transduced BF NPAS1-mCherry cells (red) were activated by 0.3 mg/kg CNO administered 2 h before the mice were killed (54.0% ± 2.8%, n = 3), with no significant difference between subregions. Colocalization is marked with an arrowhead.
Following saline injections (n = 9 mice), average sleep latency was <30 min and REM latency was <50 min. In contrast, injections of 0.3 mg/kg (n = 9) or 1 mg/kg CNO (n = 7) resulted in significantly longer latencies to both non-REM sleep (approximately 100 min) and REM sleep (>150 min) (Fig. 4D). Overall, compared to saline (27.2 ± 5.7 min), NREM sleep latency was significantly higher following administration of 0.3 mg/kg (98.8 ± 17.5 min; P = 0.0045) or 1 mg/kg CNO (105.0 ± 21.5 min; P = 0.0045, Fig. 4E). A one-way ANOVA finds a significant effect of treatment on NREM sleep latency (P = 0.0024). Similarly, in comparison to saline administration (47.0 ± 5.7 min), latency to REM sleep was also significantly higher following injections of 0.3 mg/kg CNO (155.3 ± 14.2 min; P < 0.0001) and 1 mg/kg CNO (181.6 ± 23.7 min; P < 0.0001, one-way ANOVA, P < 0.0001, Fig. 4F).
I.P. administration of 0.3 mg/kg and 1 mg/kg CNO significantly increased the amount of wakefulness and suppressed NREM and REM sleep vs. saline in the first 2 to 3 h postinjection (Fig. 4 G–I). Two-way ANOVA indicated a significant interaction of time and treatment on the proportion of time spent in wake (P = 0.0001), NREM sleep (P = 0.0002), and REM sleep (P = 0.0001). Latencies and amounts of wakefulness, NREM, or REM sleep were not significantly different between 0.3 and 1 mg/kg doses of CNO. 0.3 mg/kg CNO alone had no significant effect on sleep–wake behavior or cortical EEG in a subset of mice of the same background strain where Npas1+ neurons were not transduced with hM3d(Gq) (n = 4, SI Appendix, Fig. S2).
A subset of mice was given saline, 0.3 or 1 mg/kg CNO at ZT2, 2 hours before they were killed to confirm activation of BF Npas1-mCherry+ neurons by CNO using immunohistochemistry for the immediate early gene product cFOS. In saline-treated mice (n = 3; Fig. 4K), the percentage of mCherry+ neurons expressing cFOS was <5% (4.5 ± 0.4%, n = 3), whereas following 0.3 mg/kg CNO (Fig. 4J) it was >50% (54.0 ± 2.8%, n = 3; P < 0.0001, two-tailed unpaired t test), confirming strong activation of the Npas1+ neurons by CNO (Fig. 4 J and K). Similarly high percentages were seen following 1 mg/kg CNO (57.2 ± 3.9%; n = 2). The percentage of neighboring PV+ neurons which expressed cFOS also increased following chemogenetic activation of Npas1+ neurons when compared to saline injections (n = 3, P = 0.038; SI Appendix, Fig. S3), or in comparison to 0.3 mg/kg CNO administration in mice lacking hM3d(Gq) (n = 3, P = 0.045), consistent with their wake-active profile (15, 17, 22). The percentage of neighboring ChAT+ neurons which expressed cFOS was not significantly different following chemogenetic activation of Npas1+ neurons when compared to values in saline-treated mice or CNO injections in mice lacking hM3D(Gq) (SI Appendix, Fig. S3).
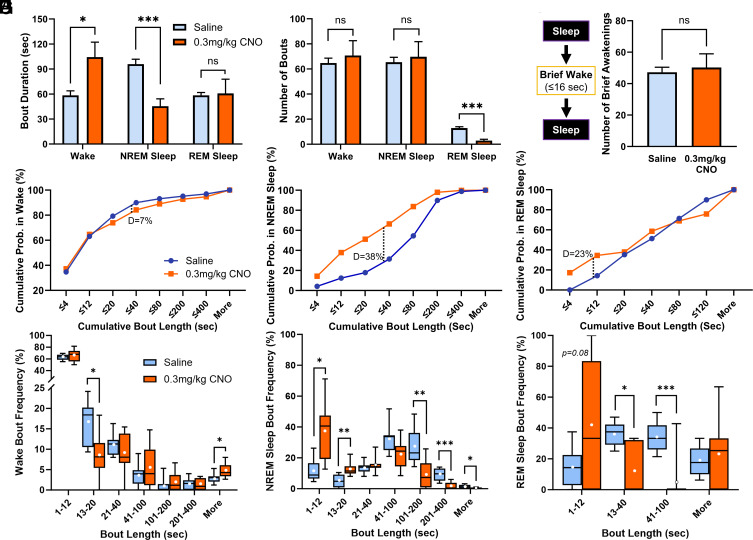
To further explore the increase in wakefulness following chemogenetic activation of BF Npas1+ neurons, we performed sleep–wake bout analysis. One mouse treated with 0.3 mg/kg and its saline control data was excluded since it exhibited continuous wakefulness for >3 h postinjection of CNO and was identified as an outlier by a Grubbs’ outlier test (P < 0.0001). In the first 3 h postinjection, the duration of wake bouts was increased by 0.3 mg/kg CNO (Saline 58.4 ± 5.4 s; 0.3 mg/kg CNO 104.3 ± 18.0 s; P = 0.021, two-tailed paired t test, Fig. 5A) and by 1 mg/kg CNO (213.2 ± 70.5 s, n = 7; P = 0.0039, SI Appendix, Fig. S4A). NREM sleep bout duration was reduced by 0.3 mg/kg CNO (saline 95.9 ± 5.9 s; 0.3 mg/kg CNO 40.3 ± 9.3 s; P = 0.002, two-tailed paired t test, Fig. 5A) and tended to be reduced by 1 mg/kg CNO (58.5 ± 22.2 s; P = 0.14, SI Appendix, Fig. S4A). The number of wake and NREM sleep bouts was unaltered (Fig. 5B). REM sleep bout duration was not significantly altered by 0.3 mg/kg (Fig. 5A) or 1 mg/kg CNO (SI Appendix, Fig. S4A), but the number of REM sleep bouts was strongly and significantly reduced (P = 0.0001, two-tailed paired t test, Fig. 5B and SI Appendix, Fig. S4B). Neither dose of CNO significantly changed the number of brief awakenings, defined as four or fewer epochs (<16 s) of wakefulness interrupting two sleep bouts (Fig. 5C and SI Appendix, Fig. S4C). Further analysis of individual bouts during the 3 h postinjection period revealed that following 0.3 mg/kg CNO administration, shorter wake bouts tended to be less frequent (P = 0.093, two-sample K-S test; Fig. 5D) and the frequency of very long wake bouts were significantly increased (Fig. 5G) whereas there was a significant increase in short NREM sleep bouts (Fig. 5 E and H). Across treatments, the cumulative distribution of REM sleep bout lengths was not significantly different (P = 0.16; Fig. 5F), although these data have high variability due to the low overall amount of REM sleep, and significant reductions of intermediate duration REM bouts were observed (Fig. 5I). Trends in cumulative wake, NREM sleep, and REM sleep bout length distributions and bout frequency comparisons were comparable following administration of 1 mg/kg CNO (SI Appendix, Fig. S4 D–I). Overall, these data suggest that increases in the percentage of time in wakefulness and corresponding decreases in NREM sleep time caused by chemogenetic activation of BF Npas1+ neurons are due primarily to an increase in long-duration bouts of wakefulness and an increase in short-duration NREM sleep bouts. In contrast, suppression of percentage time in REM sleep is due to a suppression of entries into REM sleep.
Fig. 5.
Activation of basal forebrain (BF) neuronal PAS domain 1 (Npas1+) neurons increases the duration of wake bouts while reducing the duration of non-rapid-eye-movement (NREM) sleep bouts and the number of rapid-eye-movement (REM) sleep bouts. (A) Compared to saline (n = 8), administration of 0.3 mg/kg clozapine-N-oxide (CNO, n = 8) produced significantly longer wake bouts (P = 0.021) and significantly shorter NREM sleep bouts (P = 0.0020) over the 3-h period postinjection period. REM sleep bout length was unaffected (P = 0.89). Comparisons were made via two-tailed paired t tests. (B) Administration of 0.3 mg/kg CNO did not change the number of wake or NREM sleep bouts. However, there were significantly fewer REM sleep bouts after 0.3 mg/kg CNO administration (P = 0.00013). (C) CNO administration did not change the number of brief awakenings, defined as ≤4 epochs (≤16 s) of wakefulness interspersing epochs of NREM sleep (P = 0.71). (D) Cumulative probability of wakefulness during the 3-h period following 0.3 mg/kg CNO administration trends toward less frequent short wake bouts (P = 0.093, two-sample Kolmogorov–Smirnov (K-S) test D = 0.070). (E) Shorter NREM sleep bouts are more frequent following 0.3 mg/kg CNO administration, producing a significantly different cumulative frequency distribution (P < 0.001, K-S D = 0.38). (F) Overall cumulative distributions of REM sleep bouts were not significantly different across treatments (P = 0.16, K-S D = 0.23). (G) Bout analysis indicates significantly fewer wake bouts between 13 and 20 s long (P = 0.015) and more frequent bouts over 400 s (P = 0.020). (H) CNO produces significantly more NREM sleep bouts between 1 to 12 and 13 to 20 s (P = 0.015 and P = 0.0098, respectively), and significantly fewer longer bouts between 101 to 200, 201 to 400, and >400 s (P = 0.0078, P = 0.0006, and 0.018, respectively). (I) CNO produced significantly fewer moderate-length REM sleep bouts between 13 to 40 and 41 to 100 s (P = 0.016 and P = 0.0007, respectively). Individual binned bout comparisons were made via two-tailed paired t tests. Whiskers on box and whisker plots range from min to max, the midline represents the median, and the mean is indicated with a white circle. Up to one animal per group bin may be excluded if it is alone in spending 0% or 100% of its time in that bin. P < 0.05 *, P < 0.01 **, P < 0.005 ***, ns: not significant.
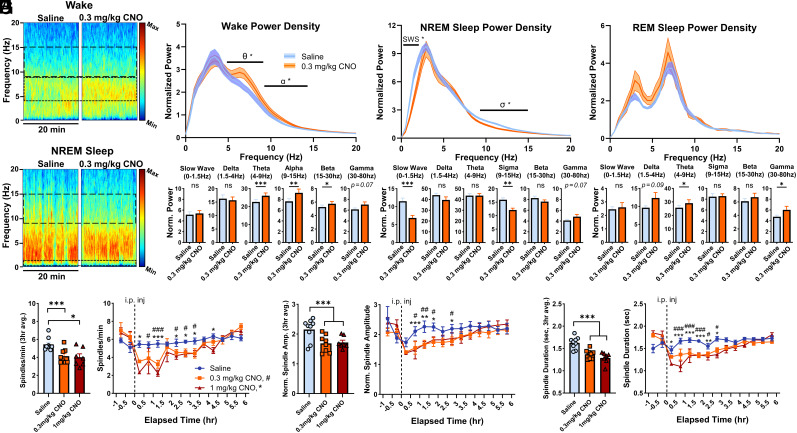
Next, we analyzed whether activation of BF Npas1+ neurons changed the quality of sleep–wake states, in terms of their spectral profile in the first 3 h after injection (Fig. 6 and SI Appendix, Fig. S5). During wakefulness, 0.3 mg/kg CNO in mice expressing hM3d(Gq) in BF Npas1+ neurons produced a significant increase in power in the theta (4 to 9 Hz, 115% of saline control, P = 0.0043), alpha (9 to 15 Hz bands, to 121% of control, P = 0.0051), and beta bands (15 to 30 Hz, 109% of control, P = 0.039; two-tailed paired t tests, Fig. 6C). Maximal differences compared to saline were observed at 6.8 to 7.3 Hz in the theta band (127% of control), 11.2 to 11.7 Hz in the alpha band (126% of control) and 25.4 to 25.9 Hz in the beta band (114% of control). Effects were similar but not as pronounced with 1 mg/kg CNO (115% of control in theta power, P = 0.22; 114% of control in alpha power, P = 0.090; 104% of control in beta power, P = 0.14, two-tailed paired t test, SI Appendix, Fig. S5C). In NREM sleep, 0.3 mg/kg CNO led to significant decreases in slow-wave activity (0 to 1.5 Hz, to 60% of control values, P < 0.001) and power in the sigma band (9 to 15 Hz, to 70% of control values; P = 0.0098, two-tailed paired t test, Fig. 6D). Maximal spectral differences during NREM sleep were seen between 0.49 and 0.98 Hz (56% of control) in the slow-wave band and 11.2 to 11.7 Hz (70% of control in the sigma band). Effects on slow-wave and sigma power were also observed following 1 mg/kg CNO (52.9% of control in the slow band, P = 0.004; 75.9% of control for the sigma band; P = 0.014, two-tailed paired t test, SI Appendix, Fig. S5D). Power spectral changes during REM sleep were less pronounced (Fig. 6E) with a smaller increase in the theta and gamma band power (30 to 80 Hz) with 0.3 mg/kg CNO (P < 0.05, two-tailed paired t test) which was not replicated with 1 mg/kg CNO (SI Appendix, Fig. S5E). Changes in power spectral measures were largely unchanged if we excluded the first 20 min postinjection from our analysis, when increased wakefulness is induced by handling and the i.p. injection. Further analysis of sleep and NREM sleep delta (1.5 to 4 Hz) power in the remainder of the light period did not identify any homeostatic sleep rebound in either measure (SI Appendix, Fig. S5 A and B).
Fig. 6.
Chemogenetic activation of basal forebrain (BF) neuronal PAS domain 1 (Npas1+) neurons alters cortical spectral components of sleep–wake states and suppresses NREM sleep spindles. (A) Representative EEG spectrogram during a continuous 20-min sample of predominantly wakefulness occurring during the first hour following intraperitoneal injection. More activity at a given frequency range is indicated by warmer colors, and less by cooler colors. Wake theta (4 to 9 Hz) and alpha (9 to 15 Hz) are outlined (dashed lines), and more relative activity in both bands was observed following 0.3 mg/kg CNO administration. (B) A continuous 20-min excerpt of predominantly NREM sleep EEG activity from the second posttreatment hour shows a decrease in NREM sleep slow wave (0 to 1.5 Hz) and sigma (9 to 15 Hz) activity. (C) A normalized power spectral density plot between 0 and 20 Hz during wakefulness (n = 9 animals) indicates 0.3 mg/kg CNO produces a significant increase within theta and alpha bands during the 3-h postinjection period, maximized at 6.8 to 7.3 Hz and 11.2 to 11.7 Hz, respectively. Individual power spectra normalized to total power and time in sleep–wake state finds significantly more activity in theta, alpha, and beta bands but not slow wave, delta, or gamma. (D) Power spectra density during NREM sleep indicates less power within the slow wave (0 to 1.5 Hz) and sigma (10 to 15 Hz) bands following 0.3 mg/kg CNO administration, with the greatest differences lying between 0.49 to 0.98 Hz and 11.2 to 11.7 Hz, respectively. Normalized power analysis concurrently finds significantly less slow wave and sigma activity, but no changes in delta, theta, beta, or gamma bands. (E) REM sleep power spectra densities between 0 and 20 Hz were not significantly different between saline or 0.3 mg/kg CNO groups, with no difference observed in slow wave, delta, sigma, or beta bands, but with a significant increase observed in the theta and gamma bands. (F) Spindle density (NREM sleep spindles/min) was significantly lower in mice administered 0.3 mg/kg (P = 0.0004) or 1 mg/kg CNO (P = 0.032) vs. saline. Compared to saline, CNO produced significantly lower spindle densities within the first 0.5 h after injection and persisting up to 3.5 h. (G) Normalized spindle amplitudes were significantly lower during the 3-h postinjection period in mice administered 0.3 mg/kg (P = 0.0048) or 1 mg/kg CNO (P = 0.0048) vs. saline. Compared to saline, CNO produced significantly shorter spindle amplitudes beginning 1 h after injection and persisting up to 3 h. (H) NREM Sleep Spindles were significantly shorter following administration of 0.3 mg/kg (P = 0.0029) or 1 mg/kg CNO (P = 0.0005) vs. saline. Spindles were significantly shorter within the first 0.5 h postinjection, persisting up to 3 h. Saline vs. 0.3 mg/kg CNO, P <0.05 #, P < 0.01 ##, P < 0.005 ###. Saline vs. 1 mg/kg CNO: P < 0.05 *, P < 0.01 **, P < 0.005 ***, ns: not significant.
Changes in sigma power during NREM sleep are often, but not always, indicative of changes in sleep spindles (39, 40), a feature of light NREM sleep associated with sleep stability and memory consolidation which is often impaired in many neuropsychiatric disorders (41). Thus, we examined changes in sleep spindles after i.p. injections of saline or CNO using a validated algorithm (39). Sleep spindle density, duration, and amplitude were stable following saline injections, but large reductions in all three measures were observed during the 3-h period following administration of 0.3 mg/kg or 1 mg/kg CNO (Fig. 6 F, G, and H). Spindle density (NREM sleep spindles/min) was significantly lower in mice administered 0.3 mg/kg CNO (4.14 ± 0.29 spindles/min; P = 0.0004) or 1 mg/kg CNO (4.01 ± 0.38 spindles/min; P = 0.032) vs. saline (5.44 ± 0.23 spindles/min; one-way ANOVA, P = 0.027; Fig. 6F). Additionally, normalized spindle amplitudes were significantly smaller in mice administered 0.3 mg/kg (1.69 ± 0.10, P = 0.0048) or 1 mg/kg CNO (1.72 ± 0.07; P = 0.0048) vs. saline (2.15 ± 0.12; one-way ANOVA, P = 0.0005; Fig. 6G), as were spindle durations (Fig. 6H) after administration of 0.3 mg/kg (1.37 ± 0.037 s, P = 0.0029) or 1 mg/kg CNO (1.29 ± 0.049 s; P = 0.0005) vs. saline (1.62 ± 0.045 s; one-way ANOVA, P = 0.0002). One 0.3 mg/kg CNO datapoint was excluded from 3 h averaged analyses due to exhibiting sustained wakefulness >3 h postinjection. These reductions were maximal within 90 min postinjection, typically lasted three or more hours, and did not significantly differ between doses of CNO. Two-way mixed-model ANOVAs find significant interactions of time and treatment for each measure (P ≤ 0.005). No effects of 0.3 mg/kg CNO were found on spectral measures or sleep spindles in control mice without expression of hM3d(Gq) (n = 4, SI Appendix, Fig. S6).
In summary, chemogenetic activation of BF Npas1+ neurons strongly increased wakefulness, delayed the onset of sleep and disrupted NREM sleep oscillations, features reminiscent of sleep disruption in a variety of psychiatric and neurodegenerative disorders (1, 41).
Discussion
Here, we characterized a major subpopulation of BF projection neurons which express Npas1, a developmental transcription factor linked to neuropsychiatric disorders (25, 26). Our results suggest that BF Npas1+ neurons are GABAergic, distinct from PV+ and cholinergic neurons, and project to brain regions involved in sleep–wake control, motivated behavior, and olfaction. When activated chemogenetically, BF Npas1+ neurons strongly enhance wakefulness, suppress NREM and REM sleep, and disrupt NREM sleep oscillations, reminiscent of sleep disruption in a variety of disorders.
Early studies reported expression of Npas1 in the globus pallidus and cortical/striatal interneurons (4, 25, 27, 28, 30). Previous studies also reported Npas1 expression in the BF (4, 25). Here, we comprehensively examine the location, density, size, and neurochemical phenotype of Npas1+ neurons in the three main BF subregions. In addition to Npas1+ neurons in the VP (10), substantial numbers of small or medium-sized Npas1 neurons were located in the HDB and MCPO regions, as well as in neighboring lateral and medial preoptic areas. The density of Npas1+ neurons was substantially (5 to 6×) higher than that of previously characterized BF PV+, ChAT+, and vGlut2+ neurons (17, 31, 32). Immunohistochemical staining for NPAS1 revealed that the majority (~75%) of BF Npas1-tdTomato+ neurons continue to express detectable levels of Npas1 into adulthood, likely an underestimate due to the fiber-dense nature of the BF which hinders antibody penetration. These results suggest a possible function for Npas1 in regulating the activity or phenotype of these neurons in adults, similar to other developmental transcription factors (33). Immunohistochemical staining in Npas1-cre-2A-tdTomato+ tissue against PV and ChAT found negligible overlap, comparable to findings in the GPe and cerebral cortex (4, 27, 30) and consistent with the expression of Npas1 in embryonic precursors which generate forebrain GABAergic neurons (25, 28–30). In the BF, NPAS1+ nuclei colocalized with GFP in GAD67-GFP knock-in mice, whereas virtually no overlap was found between NPAS1 and tdTomato in vGlut1-cre-tdTomato or vGlut2-cre-tdTomato mice, indicating that Npas1+ neurons are not glutamatergic. Surprisingly, Morais-Silva et al. (10) found that GABAergic markers were not enriched in VP Npas1+ neurons in a study which analyzed ribosome-associated mRNA in Npas1-cre-Ribotag mice, despite high expression of another transcription factor, Sox6, which is expressed in developing and adult forebrain GABAergic neurons (5, 42). The origin of this discrepancy is unclear, but could reflect insufficient sensitivity of the Ribotag method. In contrast, our immunohistochemical staining experiments and genetic cross strongly suggest that BF Npas1+ neurons are GABAergic, like their counterparts in GPe and cortex.
To determine the projections of BF Npas1+ neurons, we expressed ChR2-EYFP fusion proteins in Npas1+ neurons and examined the location and relative density of green (EYFP+) fibers. In general, the results of this experiment were similar to those of a study (10) which focused on Npas1+ neurons in the VP, using the same Npas1-cre-2A-tdTomato mice. Both our study and that of Morais-Silva et al. (10) revealed major projections to the lateral hypothalamus, medial septum, nucleus accumbens, lateral habenula, VTA, and anterior olfactory nucleus. However, our injections also transduced Npas1+ neurons in the HDB and MCPO BF subregions. We further identified additional projections to the neocortex, hippocampal formation, amygdala, supramammillary and posterior hypothalamus, and to the olfactory bulb. Overall, these results are consistent with BF Npas1+ neurons being involved in limbic basal ganglia circuits related to motivation, reward, stress, and sleep–wake control (10, 14, 16, 43, 44).
There are parallels between the previously described projections of Npas1+ neurons in the GPe involved in motor control and BF Npas1+ neurons involved in motivational circuitry. GPe Npas1+ neurons project to the dorsal striatum (45), whereas BF Npas1+ neurons project strongly to the ventral striatum (shell of the nucleus accumbens). GPe Npas1+ neurons project to the substantia nigra, whereas BF Npas1+ neurons project heavily to VTA. We also observed projections to the neocortex, especially frontal regions, similar to previously described neocortical projections of GPe Npas1+ neurons (5). One notable difference between the projections of GPe Npas1+ neurons and BF Npas1+ neurons was that we did not observe a strong projection of BF Npas1+ neurons to the TRN, whereas there is a strong output from GPe Npas1+ neurons (5). This finding also distinguishes BF Npas1+ neurons from BF PV+ and ChAT+ neurons which project heavily to the TRN (19, 20, 36, 37).
The prominence of BF Npas1+ projections to olfactory regions was a surprising finding of this study. A major projection from the BF to the olfactory bulb (OB) has been known for some time and comprises both cholinergic and GABAergic components (46, 47). The cholinergic BF projection to the olfactory bulb originates in the medial part of the HDB whereas the GABAergic component originates in lateral HDB and MCPO regions (46, 47). However, the identity of BF GABAergic neurons mediating this projection was unknown. Previous tracing studies found that BF PV+ and SST+ projections to OB are weak (37). In contrast, our results suggest that BF GABAergic neurons of the Npas1+ lineage are a major component of this projection, with strong projections to not only OB but also olfactory cortex regions such as the anterior olfactory cortex, piriform cortex, and taenia tecta. Whether olfactory region-projecting BF Npas1+ neurons also mediate arousal and motivational functions is an open question but conceivably, odors with strong survival/reproductive relevance could activate BF Npas1+ neurons to enhance olfactory discrimination, arousal, and appropriate motivational responses.
Previous chemogenetic studies had shown that activation of all GABAergic neurons in the BF (15) or in the VP subregion of BF (16) strongly enhances wakefulness, whereas chemogenetic activation of vGlut2+ or cholinergic neurons at the same dose of CNO did not (15). However, these previous studies did not identify which of the many subtypes of BF GABAergic neurons (18) are responsible for this wake-promoting action. Although optogenetic excitation of the BF PV-expressing GABAergic subpopulation promotes arousals from sleep (20, 21) and cortical gamma band activity (13, 19), this effect was transient and only slightly increased total wakefulness, making it unlikely that activation of BF PV+ neurons is responsible for the strong wake-promoting effect of activating BF GABAergic neurons. Thus, our findings here represent a major advance in identifying a marker (Npas1) for strongly wake-promoting BF GABAergic neurons. However, we note that compared to the results following chemogenetic activation of all BF GABAergic neurons (15) or those in the VP region (16), the wake-promoting effect was less profound and shorter-lasting. Thus, additional, non-Npas1+ BF GABAergic neuronal subpopulations may contribute to promotion of long-lasting wakefulness. Further analysis of our data revealed that the increase in wakefulness was due to an increase in long bouts of wakefulness and not due to more frequent awakenings. Concomitantly, the number of short NREM sleep bouts was increased at the expense of long NREM sleep bouts. Npas1+ neuron-mediated suppression of REM sleep was due to a reduced number of entries into REM sleep. Control experiments in a separate group of mice confirmed that 0.3 mg/kg CNO administered without hM3d(Gq) transduction did not affect the amount of sleep or sleep architecture, consistent with other published findings that low doses of CNO do not have marked effects on sleep or sleep physiology (15, 48). A previous study confirmed that CNO depolarizes and increases action potential discharge of BF Npas1+ neurons in the VP subregion transduced with hM3d(Gq)-mCherry (10), promoting activation of the immediate early gene product, cFOS. Here, we confirmed that injections of the low dose of CNO (0.3 mg/kg) strongly increased the percentage of Npas1+ neurons expressing cFOS using immunohistochemical staining. Collectively these results strongly suggest that the behavioral results are due to activation of BF Npas1+ neurons. Furthermore, chemogenetic activation of Npas1+ neurons is associated with small but significant increases in the percentage of BF PV+ expressing c-Fos following CNO injections, likely via long-loop circuits and consistent with their wake-active profile (22).
Several potential targets of BF Npas1+ neurons may be involved in promotion of wakefulness. Optogenetic activation of VP GABAergic terminals in the VTA or lateral hypothalamus promotes wakefulness, whereas activation of terminals in the lateral habenula or mediodorsal thalamus does not (16). Thus, the projections to the VTA and LH are strong candidate pathways to mediate the wake-promoting action. Additional projection sites which might mediate the wake-promoting actions of BF Npas1+ neurons are those to the supramammillary nucleus (49) and the shell of the nucleus accumbens (43). Overall, BF Npas1+ neurons appear to be part of subcortical circuits involved in motivationally driven wakefulness (16). Consistent with that interpretation, chemogenetic activation of Npas1+ neurons increased EEG power in the high theta/alpha EEG band which has been linked to goal-directed behavior during wakefulness (44, 50).
The functions of NREM sleep are linked to the specific cortical oscillations which define this state. Sleep spindles are linked to protection of sleep from the arousing effects of sensory stimuli and to sleep-dependent memory consolidation, especially when coupled to slow oscillations (41). Slow oscillations in NREM sleep are linked to glymphatic clearance of toxic proteins, synaptic homeostasis, and cellular metabolism. In our experiments, activation of Npas1+ neurons not only delayed the onset and amount of NREM sleep, but also reduced cortical slow wave and sigma band activity during NREM sleep. Notably, the density, amplitude, and duration of sleep spindles were significantly reduced, reminiscent of findings in various conditions such as schizophrenia, Alzheimer’s disease, and neurodevelopmental disorders (41). Thus, the insomnia-like phenotype and disrupted NREM sleep oscillations produced by increased activity of BF Npas1+ neurons have face validity to human sleep and neuropsychiatric disorders.
Several studies have linked the Npas1 transcription factor and BF Npas1+ neurons to aspects of neuropsychiatric disorders and stress responsivity. Erbel-Sieler et al. (25) found that Npas1 and Npas3 transcription factors are primarily expressed in inhibitory interneurons in cortical regions and, when knocked out, elicit behavioral and neurochemical abnormalities reminiscent of rodent models of neuropsychiatric disease such as schizophrenia and anxiety disorders. While abnormalities in cortical Npas1+ interneurons could mediate these behavioral abnormalities, a contribution of GPe or BF Npas1+ neurons cannot be ruled out. Michaelson et al. (26) found that Npas1 is a master regulator of neuropsychiatric risk genes in hippocampal interneurons which are likely derived from the same ganglionic precursors which generate BF and GP Npas1+ GABAergic projection neurons. More recently, a role for VP Npas1+ neurons in stress responsivity has been demonstrated (10), which may be mediated by their projections to a range of stress-mediating regions. Our study suggests a role for BF GABAergic neurons derived from the Npas1 lineage in sleep–wake control. Furthermore, as far as we are aware, this is the first study to examine the sleep–wake function of a subgroup of BF neurons based on a developmental lineage marker, emphasizing the usefulness of developmental transcription factors in identifying subpopulations of sleep–wake control neurons (6, 7). Collectively, these findings suggest that overactivity of BF Npas1+ neurons could be important in stress-induced insomnia or sleep disruptions in neuropsychiatric disorders. Identification of this major population of wakefulness-promoting BF GABAergic neurons will allow studies of their role in sleep disorders, dementia, and other neuropsychiatric conditions affecting the BF.
Materials and Methods
Detailed descriptions of immunohistochemistry, microscopy analysis, stereotaxic surgery, anterograde tracing, chemogenetics, and sleep–wake recordings/analysis can be found in SI Appendix, Supplementary Methods.
Animals.
Npas1-cre-2A-tdTomato mice (4) (Jackson Laboratory strain 027718) were used to identify neurons derived from the Npas1 lineage. To identify vGlut1+ or vGlut2+ glutamatergic neurons, we crossed vGlut1-cre (Strain 023527; Jackson Laboratory) or vGlut2-cre mice (Congenic strain 028863; Jackson Laboratory) with a Cre-reporter strain expressing a red fluorescent marker (tdTomato; Strain 007905; Jackson Laboratory). To identify BF GABAergic neurons, we used heterozygous GAD67-GFP knock-in mice (17) from our in-house colony. Both male and female animals were used for anatomical and sleep–wake experiments. We did not observe any obvious sex differences.
Target Area within the BF.
We focused on intermediate areas of BF approximately centered at AP +0.14 mm; ML 1.6 mm; and DV −5.3 mm. Mapping used semiautomated analyses based on the Allen Brain Atlas.
Immunohistochemistry and Microscopy Analysis.
Details of the primary and secondary antibodies are provided in SI Appendix, Tables S1 and S2. Npas1-cre-2A-tdTomato mice express tdTomato constitutively but at low levels (4). Thus, anti-RFP staining was used to enhance the endogenous red signal. For each fluorophore, representative BF sections [three 40 µm coronal sections per animal, spanning +0.38 mm (rostral), +0.14 mm (medial), and −0.10 mm (caudal) from bregma)] were imaged using StereoInvestigator and quantified using NeuroInfo software packages (Version 2022; Microbrightfield) and a Zeiss Imager.M2 fluorescent microscope.
Anterograde Tracing.
Viral vectors encoding Channelrhodopsin-2 (ChR2) and enhanced yellow fluorescent fusion proteins (AAV5-DIO-ChR2-EYFP) were used for anterograde tracing of the projections of BF Npas1 neurons. This vector was originally generated by the laboratory of Karl Deisseroth (Stanford University, CA) and was purchased from the University of North Carolina Vector Core facility. Three hundred nanoliters of viral vector was injected unilaterally at a flow rate of 50 nL/min targeting BF (AP +0.14 mm; ML 1.6 mm; DV −5.3 mm).
Chemogenetics and Stereotaxic Surgery.
To test the effect of activation of BF Npas1+ neurons on sleep–wake behavior, adeno-associated viral vectors expressing excitatory receptors and a fluorescent marker of expression (AAV8-hSyn-DIO-hM3D(Gq)-mCherry, plasmid #44361, Addgene, Watertown, MA) were injected bilaterally into BF (AP: +0.4, ML: ±1.6, DV: −5.3) of adult (3 to 6 mo old) mice. To record cortical electrical activity, bilateral frontal neocortical EEG screw electrodes (Pinnacle Technology Inc.; Part # 8403) were placed at AP: +1.9 mm, ML: ±1.5 with a ground electrode at AP: −3 mm, ML: −2.7 and a reference electrode at AP: −5.2 mm ML: 0. EMG electrodes were placed in the nuchal muscle. Intraperitoneal injections of saline (vehicle control) or 0.3 or 1 mg/kg CNO were given early in the light cycle at 9AM (ZT2) when mice normally sleep. Control CNO injection experiments were conducted in a different strain of transgenic mice (Lhx6-cre) on the same C57/BL6J background strain of mice using a control virus expressing a fluorescent marker without hM3d(Gq) (AAV-DIO-EYFP). Following completion of sleep–wake recordings and 120 min before being killed, mice received an additional injection of saline or CNO (0.3 or 1 mg/kg) at ZT2 to verify activation of Npas1+ neurons using immunohistochemical staining for the immediate early gene cFOS. Sleep–wake states were manually scored in 4 s epochs. Power spectral densities of wake, NREM sleep, and REM sleep were computed using the MATLAB pwelch function using a 4-s Hanning window with 50% overlap. To normalize EEG power, power in each frequency band per state was divided by the total power across all frequencies (0 to 500 Hz) from the entire 24 h record. Power spectra were binned in 3-h intervals, except for the NREM sleep delta power time course throughout the light cycle (SI Appendix, Fig. S5B), which used 1-h bins. We analyzed sleep spindles (≥0.5 s in length) using our validated automated spindle detection algorithm (39), where normalized amplitude is calculated by dividing against a threshold value based on putative spindle peaks in each EEG record. Representative time-frequency spectrograms were created using the mtspectrogramc function (Chronux Toolbox; https://Chronux.org). Latency to NREM sleep was defined as the time postinjection until the start of 10 continuous NREM sleep epochs (40 s). Latency to REM sleep was defined as the time postinjection until the start of five continuous REM sleep epochs (20 s). Brief awakenings were defined as a ≤4 epoch (≤16 s) long period of wake interrupting two NREM sleep epochs.
Ethics Approval.
All experiments were reviewed and approved by the institutional animal care and use, safety, biosafety, and research and development committees of Veterans Affairs Boston Healthcare System.
Consent for Publication.
All authors approved this submission.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by United States Veterans Administration Biomedical Laboratory Research and Development Service Merit Awards I01 BX004673 (including a mentored Research Supplement to Promote Diversity) and I01 BX004500 United States Veterans Administration Biomedical Laboratory Research and Development Service Career Development Award 2 IK2 BX004905. United States NIH support was from NIH awards R01 NS069777, R01 NS119227, R21 MH125242, and K01 AG068366. T.A.T., D.S.U., R.B., J.M.M., J.T.M., and R.E.B are Research Health Scientists at VA Boston Healthcare System, West Roxbury, MA. The contents of this work do not represent the views of the U.S. Department of Veterans Affairs or the United States Government.
Author contributions
T.A.T., J.T.M., and R.E.B. designed research; T.A.T., C.Y., F.K., I.L., D.D.A., T.S., and J.T.M. performed research; D.S.U. and C.S.C. contributed new reagents/analytic tools; T.A.T., C.Y., D.S.U., J.T.M., and R.E.B. analyzed data; R.B. and J.M.M. provided technical guidance and comments on the manuscript; R.E.B. obtained funding; and T.A.T., J.T.M., and R.E.B. wrote the paper.
Competing interests
J.T.M. received partial salary compensation and funding from Merck MISP (Merck Investigator Sponsored Programs) but has no conflict of interest with this work.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
Original study data (https://osf.io/p8ufj/) and the MATLAB script to identify sleep spindles in mouse EEG/EMG data (39) (https://osf.io/2ne8j/) are available at the Open Science Framework repository. Npas1-cre-2A-tdTomato mice are available at Jackson Laboratories (Bar Harbor, ME, Strain 027718).
Supporting Information
References
- 1.Brown R. E., Spratt T. J., Kaplan G. B., Translational approaches to influence sleep and arousal. Brain Res. Bull. 185, 140–161 (2022), 10.1016/j.brainresbull.2022.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown R. E., McKenna J. T., Turning a negative into a positive: Ascending GABAergic control of cortical activation and arousal. Front. Neurol. 6, 135 (2015), 10.3389/fneur.2015.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim L., Mi D., Llorca A., Marin O., Development and functional diversification of cortical interneurons. Neuron 100, 294–313 (2018), 10.1016/j.neuron.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hernandez V. M., et al. , Parvalbumin+ neurons and NPAS1+ neurons are distinct neuronal classes in the mouse external globus pallidus. J. Neurosci. 35, 11830–11847 (2015), 10.1523/JNEUROSCI.4672-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abecassis Z. A., et al. , Npas1+-Nkx2.1+ neurons are an integral part of the cortico-pallidal-cortical loop. J. Neurosci. 40, 743–768 (2020), 10.1523/JNEUROSCI.1199-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu K., et al. , Lhx6-positive GABA-releasing neurons of the zona incerta promote sleep. Nature 548, 582–587 (2017), 10.1038/nature23663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirjavainen A., et al. , Gata2, Nkx2-2 and Skor2 form a transcription factor network regulating development of a midbrain GABAergic neuron subtype with characteristics of REM-sleep regulatory neurons. Development 149, dev200937 (2022), 10.1242/dev.200937. [DOI] [PubMed] [Google Scholar]
- 8.Detari L., Rasmusson D. D., Semba K., The role of basal forebrain neurons in tonic and phasic activation of the cerebral cortex. Prog. Neurobiol. 58, 249–277 (1999), 10.1016/s0301-0082(98)00084-7. [DOI] [PubMed] [Google Scholar]
- 9.Lin S. C., Brown R. E., Hussain Shuler M. G., Petersen C. C., Kepecs A., Optogenetic dissection of the basal forebrain neuromodulatory control of cortical activation, plasticity, and cognition. J. Neurosci. 35, 13896–13903 (2015), 10.1523/JNEUROSCI.2590-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morais-Silva G., et al. , Molecular, circuit, and stress response characterization of ventral pallidum Npas1-neurons. J. Neurosci. 43, 405–418 (2023), 10.1523/JNEUROSCI.0971-22.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grothe M. J., et al. , Atrophy of the cholinergic basal forebrain in dementia with Lewy bodies and Alzheimer’s disease dementia. J. Neurol. 261, 1939–1948 (2014), 10.1007/s00415-014-7439-z. [DOI] [PubMed] [Google Scholar]
- 12.Pereira J. B., et al. , Longitudinal degeneration of the basal forebrain predicts subsequent dementia in Parkinson’s disease. Neurobiol. Dis. 139, 104831 (2020), 10.1016/j.nbd.2020.104831. [DOI] [PubMed] [Google Scholar]
- 13.McNally J. M., et al. , Optogenetic manipulation of an ascending arousal system tunes cortical broadband gamma power and reveals functional deficits relevant to schizophrenia. Mol. Psychiatry 26, 3461–3475 (2021), 10.1038/s41380-020-0840-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soares-Cunha C., Heinsbroek J. A., Ventral pallidal regulation of motivated behaviors and reinforcement. Front. Neural Circuits 17, 1086053 (2023), 10.3389/fncir.2023.1086053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anaclet C., et al. , Basal forebrain control of wakefulness and cortical rhythms. Nat. Commun. 6, 8744 (2015), 10.1038/ncomms9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y.-D., et al. , Ventral pallidal GABAergic neurons control wakefulness associated with motivation through the ventral tegmental pathway. Mol. Psychiatry 26, 2912–2928 (2021), 10.1038/s41380-020-00906-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenna J. T., et al. , Distribution and intrinsic membrane properties of basal forebrain GABAergic and parvalbumin neurons in the mouse. J. Comp. Neurol. 521, 1225–1250 (2013), 10.1002/cne.23290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang C., Thankachan S., McCarley R. W., Brown R. E., The menagerie of the basal forebrain: How many (neural) species are there, what do they look like, how do they behave and who talks to whom? Curr. Opin. Neurobiol. 44, 159–166 (2017), 10.1016/j.conb.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim T., et al. , Cortically projecting basal forebrain parvalbumin neurons regulate cortical gamma band oscillations. Proc. Natl. Acad. Sci. U.S.A. 112, 3535–3540 (2015), 10.1073/pnas.1413625112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thankachan S., et al. , Thalamic reticular nucleus parvalbumin neurons regulate sleep spindles and electrophysiological aspects of schizophrenia in mice. Sci. Rep. 9, 3607 (2019), 10.1038/s41598-019-40398-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKenna J. T., et al. , Basal forebrain parvalbumin neurons mediate arousals from sleep induced by hypercarbia or auditory stimuli. Curr. Biol. 30, 2379–2385 (2020), 10.1016/j.cub.2020.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu M., et al. , Basal forebrain circuit for sleep-wake control. Nat. Neurosci. 18, 1641–1647 (2015), 10.1038/nn.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anaclet C., et al. , Genetic activation, inactivation, and deletion reveal a limited and nuanced role for somatostatin-containing basal forebrain neurons in behavioral state control. J. Neurosci. 38, 5168–5181 (2018), 10.1523/JNEUROSCI.2955-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y.-D., et al. , Molecular characterization of two mammalian bHLH-PAS domain proteins selectively expressed in the central nervous system. Proc. Natl. Acad. Sci. U.S.A. 94, 713–718 (1997), 10.1073/pnas.94.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erbel-Sieler C., et al. , Behavioral and regulatory abnormalities in mice deficient in the NPAS1 and NPAS3 transcription factors. Proc. Natl. Acad. Sci. U.S.A. 101, 13648–13653 (2004), 10.1073/pnas.0405310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michaelson J. J., et al. , Neuronal PAS domain proteins 1 and 3 are master regulators of neuropsychiatric risk genes. Biol. Psychiatry 82, 213–223 (2017), 10.1016/j.biopsych.2017.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stanco A., et al. , NPAS1 represses the generation of specific subtypes of cortical interneurons. Neuron 84, 940–953 (2014), 10.1016/j.neuron.2014.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cobos I., Long J. E., Thwin M. T., Rubenstein J. L., Cellular patterns of transcription factor expression in developing cortical interneurons. Cereb. Cortex 16, 82–88 (2006), 10.1093/cercor/bhk003. [DOI] [PubMed] [Google Scholar]
- 29.Flandin P., Kimura S., Rubinstein J. L., The progenitor zone of the ventral medial ganglionic eminence requires Nkx2-1 to generate most of the globus pallidus by few neocortical interneurons. J. Neurosci. 30, 2812–2823 (2010), 10.1523/JNEUROSCI.4228-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nobrega-Pereira S., et al. , Origin and molecular specification of globus pallidus neurons. J. Neurosci. 30, 2824–2834 (2010), 10.1523/JNEUROSCI.4023-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang C., et al. , Cholinergic neurons excite cortically projecting basal forebrain GABAergic neurons. J. Neurosci. 34, 2832–2844 (2014), 10.1523/JNEUROSCI.3235-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKenna J. T., et al. , Characterization of basal forebrain glutamate neurons suggests a role in control of arousal and avoidance behavior. Brain Struct. Funct. 226, 1755–1778 (2021), 10.1007/s00429-021-02288-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deneris E.S., Hobert O., Maintenance of postmitotic neuronal cell identity. Nat. Neurosci. 17, 899–907 (2014), 10.1038/nn.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franklin K. B. J., Paxinos G., The Mouse Brain in Stereotaxic Coordinates (Academic Press, New York, 2008). [Google Scholar]
- 35.Ren S., et al. , The paraventricular thalamus is a critical thalamic area for wakefulness. Science 362, 429–434 (2018), 10.1126/science.aat2512. [DOI] [PubMed] [Google Scholar]
- 36.Hallanger A. E., Levey A. I., Lee H. J., Rye D. B., Wainer B. H., The origins of cholinergic and other subcortical afferents to the thalamus in the rat. J. Comp. Neurol. 262, 105–124 (1987), 10.1002/cne.902620109. [DOI] [PubMed] [Google Scholar]
- 37.Do J. P., et al. , Cell type-specific long-range connections of basal forebrain circuit. Elife 5, e13214 (2016), 10.7554/eLife.13214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arrigoni E., Chee M. J. S., Fuller P. M., To eat or to sleep: That is a lateral hypothalamic question. Neuropharmacology 154, 34–49 (2019), 10.1016/j.neuropharm.2018.11.017. [DOI] [PubMed] [Google Scholar]
- 39.Uygun D. S., et al. , Validation of an automated sleep spindle detection method for mouse EEG. Sleep 42, zsy218 (2019), 10.1093/sleep/zsy218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Astori S., et al. , The Cav3.3 calcium channel is the major sleep spindle pacemaker in thalamus. Proc. Natl. Acad. Sci. U.S.A. 108, 13823–13828 (2011), 10.1073/pnas.1105115108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fernandez L. M. J., Luthi A., Sleep spindles: Mechanisms and functions. Physiol. Rev. 100, 805–868 (2020), 10.1152/physrev.00042.2018. [DOI] [PubMed] [Google Scholar]
- 42.Azim E., Jabaudon D., Fame R. M., Macklis J. D., SOX6 controls dorsal progenitor identity and interneuron diversity during neocortical development. Nat. Neurosci. 12, 1238–1247 (2009), 10.1038/nn.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo Y. J., et al. , Nucleus accumbens controls wakefulness by a subpopulation of neurons expressing dopamine D1 receptors. Nat. Commun. 9, 1576 (2018), 10.1038/s41467-018-03889-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fifel K., El Farissi A., Cherasse Y., Yanagisawa M., Motivational and valence-related modulation of sleep/wake behavior are mediated by midbrain dopamine and uncoupled from the homeostatic and circadian processes. Adv. Sci. (Weinh.) 9, e2200640 (2022), 10.1002/advs.202200640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Glajch K. E., et al. , Npas1+ pallidal neurons target striatal projection neurons. J. Neurosci. 36, 5472–5488 (2016), 10.1523/JNEUROSCI.1720-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Záborszky L., Carlsen J., Brashear H. R., Heimer L., Cholinergic and GABAergic afferents to the olfactory bulb in the rat with special emphasis on the projection neurons in the nucleus of the horizontal limb of the diagonal band. J. Comp. Neurol. 243, 488–509 (1986), 10.1002/cne.902430405. [DOI] [PubMed] [Google Scholar]
- 47.Gracia-Llanes F. J., et al. , GABAergic basal forebrain afferents innervate selectively GABAergic targets in the main olfactory bulb. Neuroscience 170, 913–922 (2010), 10.1016/j.neuroscience.2010.07.046. [DOI] [PubMed] [Google Scholar]
- 48.Traut J., et al. , Effects of clozapine-N-oxide and compound 21 on sleep in laboratory mice. eLife 12, e84740 (2023), 10.7554/eLife.84740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pedersen N., et al. , Supramammillary glutamate neurons are a key node of the arousal system. Nat. Commun. 8, 1405 (2017), 10.1038/s41467-017-01004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vassalli A., Franken P., Hypocretin (orexin) is critical in sustaining theta/gamma-rich waking behaviors that drive sleep need. Proc. Natl. Acad. Sci. U.S.A. 114, E5464–E5473 (2017), 10.1073/pnas.1700983114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Original study data (https://osf.io/p8ufj/) and the MATLAB script to identify sleep spindles in mouse EEG/EMG data (39) (https://osf.io/2ne8j/) are available at the Open Science Framework repository. Npas1-cre-2A-tdTomato mice are available at Jackson Laboratories (Bar Harbor, ME, Strain 027718).