Abstract
Plasmids that carry one of several type II restriction modification gene complexes are known to show increased stability. The underlying mechanism was proposed to be the lethal attack by restriction enzyme at chromosomal recognition sites in cells that had lost the restriction modification gene complex. In order to examine bacterial responses to this postsegregational cell killing, we analyzed the cellular processes following loss of the EcoRI restriction modification gene complex carried by a temperature-sensitive plasmid in an Escherichia coli strain that is wild type with respect to DNA repair. A shift to the nonpermissive temperature blocked plasmid replication, reduced the increase in viable cell counts and resulted in loss of cell viability. Many cells formed long filaments, some of which were multinucleated and others anucleated. In a mutant defective in RecBCD exonuclease/recombinase, these cell death symptoms were more severe and cleaved chromosomes accumulated. Growth inhibition was also more severe in recA, ruvAB, ruvC, recG, and recN mutants. The cells induced the SOS response in a RecBC-dependent manner. These observations strongly suggest that bacterial cells die as a result of chromosome cleavage after loss of a restriction modification gene complex and that the bacterial RecBCD/RecA machinery helps the cells to survive, at least to some extent, by repairing the cleaved chromosomes. These and previous results have led us to hypothesize that the RecBCD/Chi/RecA system serves to destroy restricted “nonself” DNA and repair restricted “self” DNA.
A type II restriction enzyme, such as R.EcoRI, will make a double-stranded break at a specific sequence on DNA (49). A cognate modification enzyme (M.EcoRI) can methylate the same sequence and protect it from restriction cleavage. The genes involved are tightly linked and form a type II restriction- modification (RM) gene complex. Type II RM gene complexes will attack unmodified foreign DNA such as bacteriophage DNA but not the modified DNA of the cells where they reside. They have been considered to function as bacterial tools against invasion by foreign DNA.
We found that elimination of type II RM gene complexes from bacterial cells by a competing genetic element inhibits cell growth (43, 44). Our experiments suggested the following course of events after a cell has lost a type II RM gene complex. In the descendants of the cell that have lost the type II RM gene complex, the number of molecules of the modification enzyme will decrease with each cell division. Eventually, the capacity of the enzyme to modify the many sites needed to protect the newly replicated chromosomes from the remaining pool of restriction enzyme will become inadequate. Chromosomal DNA will then be cleaved at the unmodified sites, and the cells will be killed.
This is reminiscent of postsegregational cell killing mechanisms, which have been shown to contribute to the stable maintenance of plasmids (11, 12, 21). Indeed, linkage of several type II RM gene complexes stabilizes plasmids (29, 32, 43, 44, 45). In other postsegregational killing systems, the differential stabilities of the killer and the antikiller are considered to be essential. What is essential for RM systems may be the effective concentration of the two enzymes. The modification enzyme will need to modify all (or almost all) of the many chromosomal recognition sites along the chromosome in order to be effective, i.e., to prevent cell death. The restriction enzyme would need to cut only one (or a few) of these in order to be effective, i.e., to kill the cell. Even dilution of the two enzymes to the same extent might lead to cell death. Therefore, the RM systems may employ a novel strategy of postsegregational host killing.
Microscopic observation of individual cells demonstrated that cell death can happen after loss of an RM plasmid (15a). However, this early work employed a bacterial strain that is defective in its own major recombination repair pathway and, instead, expresses recombination repair and restriction alleviation functions of a bacteriophage (13, 24). The relevance of those data to bacterial biology is, therefore, not straightforward.
In the present work, we addressed the question of how bacterial cells respond to loss of an RM gene complex. We used Escherichia coli strains either wild type or mutant with respect to various recombination and repair functions for that purpose. The cleaved huge chromosomes were detected by pulsed-field gel electrophoresis. Our results strongly suggest that, after loss of an RM plasmid, the bacterial cells die as a result of chromosome cleavage and that the bacterial RecBCD/Chi/RecA machinery helps the cells to survive by repairing the cleaved chromosomes.
MATERIALS AND METHODS
Bacteria, bacteriophage, and plasmids.
All the bacterial strains used are derivatives of E. coli K-12 and are listed in Table 1. The recC1002 mutation was isolated as a suppressor mutation of the recC73 mutation, which produces a null phenotype (54). The resulting mutant allele with two mutations, recC73 and recC1002, is often referred to as recC1002. The recBCD mutant alleles were verified by UV sensitivity and/or plaque size of bacteriophage lambda (with or without Chi) (54). The recC73 mutant was as sensitive to postsegregational killing as the recB21 recC22 mutant used here (N. Handa, A. Ichige, and I. Kobayashi, unpublished data). Bacteriophage P1 vir from our laboratory collection was used in transduction. Plasmid pIK172 carries the temperature-sensitive replication initiator of pSC101 (16) and is EcoRI r+ m+ and ampicillin resistant (Apr) (43). pIK173 and pIK174 are its r− m+ (43) and r− m− versions (32), respectively. Details of plasmid construction was published elsewhere (15a).
TABLE 1.
Bacterial strains
| Strain | Genotype | Comments | Source/reference |
|---|---|---|---|
| AB1157 | supE44 thr-1 ara-14 leuB6 Δ(gpt-proA)62 lacY1 tsx-33 galK2 hisG4 rfbD1 mgl-51 rpsL31 kdgK51 xyl-5 mtl-1 argE3 thi-1 λ− F− | Same as BIK788 | Laboratory collection/4 |
| BIK733 | AB1157 ΔrecA306::Tn10 | K. Yamamoto/67 | |
| JC5519 | AB1157 recB21 recC22 | Same as BIK751 | T. Kato/66 |
| JC8679 | AB1157 recB21 recC22 sbcA23 | Same as BIK813 | A. J. Clark/13 |
| BIK1044 | recB21 recC22 sbcA23 recN1502::Tn5 | 63 | |
| BIK2565 | AB1157 recN1502::Tn5 | P1 from BIK1044 to BIK788 | This work |
| N2796 | AB1157 recB21 recC22 sbcA recG258::Tn10 mini-Kan | Same as BIK1400 | R. Lloyd/35 |
| BIK1538 | AB1157 recG258::Tn10 mini-Kan | P1 from BIK1400 to BIK788 | This work |
| HRS2302 | AB1157 ruvAB::Cm | Same as BIK1620 | H. Iwasaki/20 |
| HRS1100 | AB1157 ΔruvC100::Cm | Same as BIK1618 | H. Iwasaki/52 |
| V66 | recF143 argA his-4 met rpsL31 gal(?) xyl(?) ara(?) λ− F− | Same as BIK796 | G. Smith/54 |
| V69 | V66 recC1002 recC73 | Same as BIK1272 | G. Smith/54 |
| NK5992 | IN(rrnD-rrnE)1 F− λ−argA81::Tn10 | Same as BIK800 | A. Taylor |
| BIK3682 | V69 argA81::Tn10 | P1 from BIK800 to V69 | This work |
| BIK3686 | AB1157 recC73 recC1002 argA81::Tn10 | P1 from BIK3682 to AB1157 | This work |
| SG13171 | F− SA500 his leu sulA malF::Tn10 lexA3 strA | Same as BIK1206 | S. Mizusawa |
| BIK2574 | AB1157 malF::Tn10 | This work | |
| BIK2571 | BIK2574 lexA3 (Ind−) | This work | |
| GC3403 | sfiA::Mud (Ap lac) c62 trp::MuC Δlac thr leu pyrD thi malB (λs) rpsL (Strr) gal sfiC | Same as BIK1715 | NIGa (A. Nishimura)/19 |
| BIK3919 | JC5519 argA81::Tn10 | P1 from BIK800 to JC5519 | This work |
| BIK3920 | GC3403 recBC argA81::Tn10 | P1 from BIK3919 to GC3403; UVs | This work |
| BIK3921 | GC3403 rec+ argA81::Tn10 | P1 from BIK3919 to GC3403; UVr | This work |
NIG, National Institute of Genetics.
Media, DNA preparation, and transformation.
E. coli cells were grown in L broth and were supplemented, if necessary, with antibiotics at the following concentrations: ampicillin, 50 μg/ml; methicillin, 200 μg/ml; chloramphenicol, 25 μg/ml; kanamycin, 10 μg/ml; tetracycline, 10 μg/ml. Plasmids were introduced into E. coli cells by electroporation using a Bio-Rad Gene Pulser.
Other methods are described in the figure legends.
RESULTS
Growth inhibition following loss of the EcoRI RM gene complex.
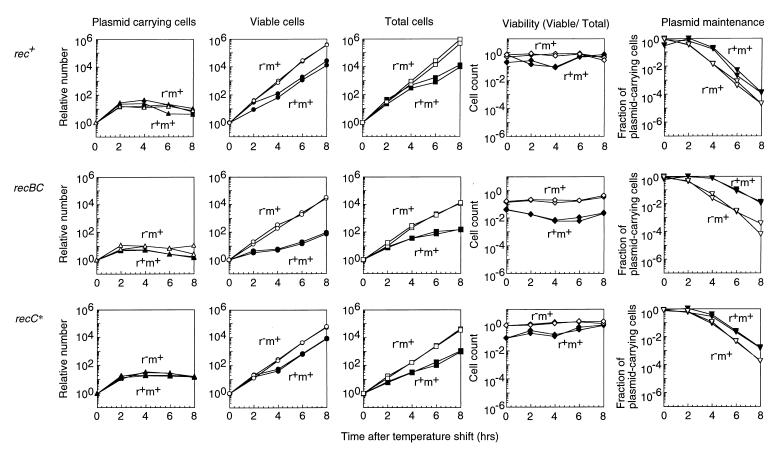
The EcoRI RM gene complex was inserted into a plasmid possessing a thermosensitive replication initiator in order to analyze the effect of loss of the gene complex on cell growth. With a rec+ bacterial strain (AB1157), the inhibition of plasmid replication following the shift of a r+ culture or r− culture to the nonpermissive temperature for plasmid replication stopped the increase in the number of plasmid-carrying viable cells (Fig. 1, first row, first column). The shift attenuated the increase in viable cell counts for the r+ culture but not for the r− control culture (second column), as reported previously for a different genetic background (15a, 43). Later, the increase in the total cell number as estimated under a microscope was also inhibited (third column). The cell viability (viable cell count/total cell count) dropped after the shift but recovered later (fourth column). As a result of cell growth inhibition, the fraction of viable cells carrying plasmids remained higher for the r+ plasmid than for the r− plasmid (fifth column). These features are qualitatively similar to those observed with classical postsegregational killing systems for plasmids (11, 12, 21).
FIG. 1.
Cell growth inhibition following loss of the EcoRI RM gene complex. Three E. coli strains, AB1157 (rec+), JC5519 (recB21 recC22), and BIK3686 (recC1002) carrying pIK172 (pSC101Ts, EcoRI r+ m+, Apr) or pIK173 (its r− version) were grown with aeration at 30°C in L broth with antibiotics to an optical density at 660 nm (OD660) of 0.3. Then the antibiotics were removed, and the culture was transferred to 42°C with aeration. The culture was diluted every time its OD660 reached 0.3. Total cells (first column from left) were counted under a microscope. The number of viable cells (second column) was estimated by counting the colonies on L agar without selective antibiotics at 30°C. The number of plasmid-carrying cells (third column) was estimated by counting the colonies on L agar with antibiotics at 30°C. The viable-cell number was divided by the total cell number for each point to calculate viability (fourth column). The number of plasmid-carrying viable cells was divided by the number of viable cells to calculate the fraction of plasmid-carrying cells (fifth column). The numbers at time zero were set to unity in the first to third columns. Solid symbols, cells losing the r+ m+ plasmid; open symbols, cells losing the r− m+ plasmid.
Cell filamentation and loss of nuclei.
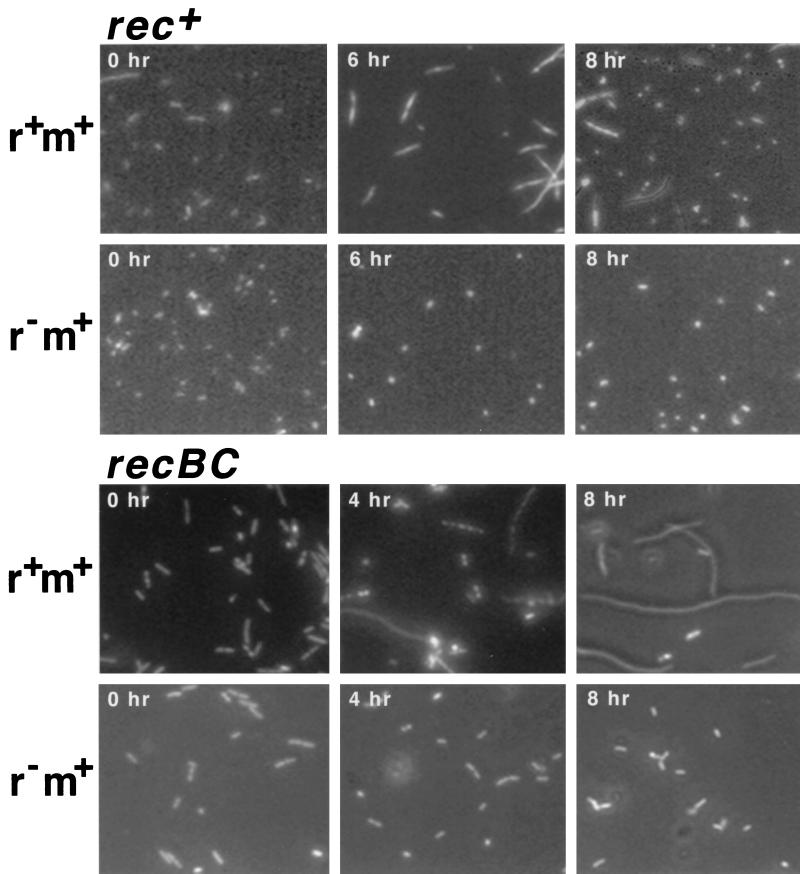
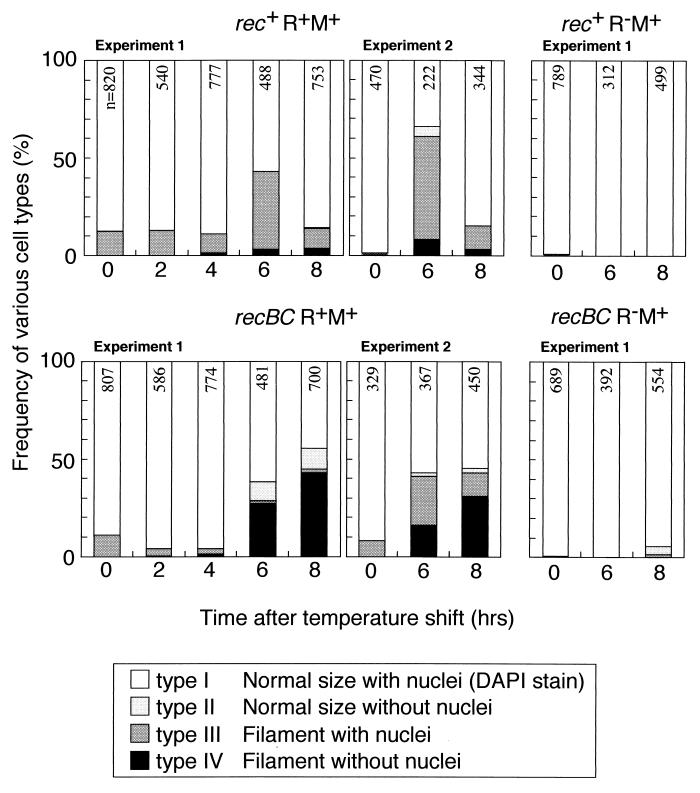
In a search for evidence of the death of individual cells, we observed cells after DAPI (4′,6′-diamidino-2-phenylindole) staining following loss of the RM plasmid. Some of the rec+ cells appeared elongated (Fig. 2, top). Most of these filaments contained multiple nuclei (DAPI-stained areas), while some lacked detectable nuclei. The cells were classified into four groups based on two criteria, cell length and nuclear content (Fig. 3, top). Many filaments appeared 6 h after the temperature shift. The filamentation was dependent on the r+ genotype (Fig. 3, top right). A small but significant fraction of the cells appeared anucleated. The r+ dependent formation of cell filaments was observed even at 30°C (time zero). These results demonstrate that cell death occurred after the loss of the type II restriction modification gene complex.
FIG. 2.
Cell morphology following loss of the EcoRI RM gene complex. E. coli strains AB1157 (rec+) and JC5519 (recB21 recC22) carrying pIK172 (pSC101Ts, EcoRI r+ m+, Apr) or pIK173 (its r− version) were aerated at 30°C in L broth with selective antibiotics to an optical density of 0.3 at 660 nm. Then the antibiotics were removed, and the culture was transferred to 42°C with aeration as described for Fig. 1. Cells were harvested at the indicated time intervals after the temperature shift and mixed with the same volume of methanol-HCO2H (2:1). After incubation on ice for 10 min, cells were collected by centrifugation, resuspended in 10 mM Tris-HCl (pH 7.5)–10 mM MgSO4, stained with DAPI, and observed under a microscope.
FIG. 3.
Morphological classification of cells following loss of the EcoRI RM gene complex. E. coli strains AB1157 (rec+) and JC5519 (recB21 recC22) carrying pIK172 (pSC101Ts, EcoRI r+ m+, Apr) or pIK173 (its r− version) were treated as described in the legend to Fig. 2. The cells were classified into four types by visual inspection. A cell was judged to be a filament when it was larger than twice the unit size of the r− cell before the temperature shift. The presence or absence of nuclei means that DAPI staining was positive or negative, respectively. The number at the top of each bar is the number of cells examined. Data for cell types I to IV are shown from top to bottom for each bar.
Increased cell death in a recBCD mutant.
We next looked for the E. coli mutants that enhance these death symptoms. The recBC mutants, which are defective in recombination repair of DNA double-stranded breaks, turned out to belong to this class.
The inhibition of cell growth, as seen in the reduction of the slope of the viable-cell count curve, was stronger in the recBC mutant culture (Fig. 1, second row, second column) than in the rec+ culture. Also, the inhibition lasted longer in the recBC mutant culture than in the rec+ culture. Total cell counts showed the same tendency (third column). Even at a lower temperature, the viability of the RM-carrying culture was low and decreased further after the temperature shift (fourth column). Because of this extensive cell death, the difference in the fraction of plasmid-carrying viable cells between the r+ m+ culture and r− m+ culture was larger in the recBC strain than in the rec+ strain (fifth column).
Extensive cell filamentation was observed, with the recBC strain undergoing loss of the r+ plasmid (Fig. 2, bottom; Fig. 3, bottom). The fraction of cells lacking nuclei was larger than in the rec+ strain (P < 0.0005 in experiment 1).
Chromosome cleavage and degradation.
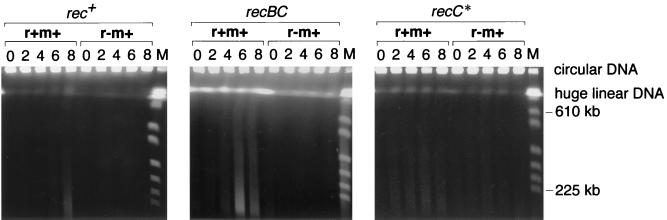
We then directly analyzed the chromosomal DNA in these cells by pulsed-field gel electrophoresis (Fig. 4). As in our earlier work, smears composed of relatively small DNA fragments in the central and lower parts of our pulsed-field gels were taken as evidence of chromosome degradation (32, 43). Under our gel assay conditions in the present study, large circular DNAs such as intact bacterial chromosomes are trapped in the well, whereas huge (>700-kbp) linear DNAs form a band which migrates just below the well in the upper part of the gel (10, 33). Chromosomes that had been cleaved at few sites and that had undergone only weak degradation (and also chromosomes partially restored from the broken pieces) were detected as bands in this area.
FIG. 4.
Chromosome cleavage following loss of the EcoRI RM gene complex. Cultures of three E. coli strains, AB1157 (rec+), JC5519 (recB21 recC22), and BIK3686 (recC1002), carrying pIK172 (pSC101Ts, EcoRI r+ m+, Apr) or pIK173 (its r− version) were transferred from 30 to 42°C as described in the legend to Fig. 1. The cells were mixed with 2,4-dinitrophenol to block energy metabolism at the indicated time intervals (in hours) after the temperature shift and were treated as described previously (33). DNA was electrophoresed through 1.0% agarose gel at 15°C in 45 mM Tris-borate–1.25 mM EDTA at 165 V with a pulse time of 50 s for 24 h using hexagonal electrodes in a Pharmacia LKB apparatus. Lane M contains Saccharomyces cerevisiae chromosomes.
The following are reproducible observations about the huge linear DNAs (Fig. 4, upper part of the gel) and the smear DNAs (central and lower parts of the gel) in our pulsed-field gel electrophoresis analysis.
In the rec+ strain (Fig. 4, left), (i) more huge linear DNAs are detected in r+ cells than in r− cells; (ii) r+, but not in r− cells, there are smear DNAs detected; and (iii) the smear DNAs increase over the incubation period after the temperature shift.
In the recBC mutant strains (Fig. 4, middle panel), (i) there are many huge linear DNAs in r+ cells; (ii) there are more of these in r+ cells than in r− (recBC) cells; (iii) there are more of these in recBC r+ cells than in rec+ r+ cells; and (iv) there are hugh linear DNAs even at the time of temperature shift (time zero) (the huge linear band at 8 h for r+ was shifted to the right lane (0 h for r−; this is an artifact inherent to the machine used). Also, (i) there are many smear DNAs in r+ cells; (ii) there are more of these in r+ cells than in r− (recBC) cells; and (iii) there are more of these in recBC r+ cells than in rec+ r+ cells. Finally, the smear DNAs increase with time until 6 h after the shift.
These results strongly suggest that insufficient ability to repair broken chromosomes after the temperature shift in the recBC mutant cells leads to their extensive degradation. We concluded that the RecBCD enzyme repairs chromosomal breakage caused by the restriction enzyme. The recBC mutant with the r− m+ plasmid showed lower, but still significant, levels of large linear DNAs. This restriction-independent chromosomal cleavage was again reduced in the rec+ strain, in agreement with another study (40).
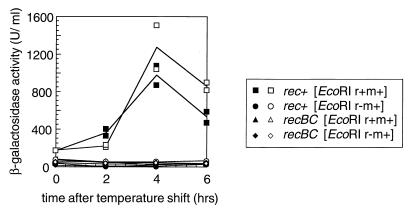
SOS induction.
The chromosomal DNA degradation and cell filamentation suggested that the SOS response had been induced. We used a chromosomal sfiA::lacZ (note that sfiA is the same as sulA) gene fusion as a reporter for SOS induction. In this system beta-galactosidase is induced in response to DNA breakage. The cells carrying the EcoRI r+ m+ plasmid were found to be slightly induced for the SOS response at 30°C (Fig. 5). The inhibition of r+ m+ plasmid replication by a temperature shift of a liquid culture increased SOS induction (Fig. 5). The SOS induction before and after the temperature shift was dependent on the presence of the r+ gene on the plasmid and on the recBC+ genotype of the host bacteria (Fig. 5).
FIG. 5.
SOS induction following loss of the EcoRI RM gene complex. Isogenic rec+ and recBC strains with a sfiA::lacZ promoter fusion (BIK3920 and BIK3921) carrying pIK172 (pSC101Ts, EcoRI r+ m+, Apr) or pIK173 (its r− version) were aerated at 30°C in L broth with antibiotics and grown to an optical density at 660 nm (OD660) of 0.3. Then the antibiotics were removed, and the culture was transferred to 42°C with aeration. The culture was diluted every time its OD660 reached 0.3. The beta-galactosidase activity was measured as described previously (41). Enzyme concentrations (in units per milliliter) were calculated from the following formula: 1,000 × (OD420 − 1.75 × OD550)/t× v× OD600, where t is the duration of the reaction in minutes and v is the volume of culture in milliliters. The results plotted were from two independent experiments.
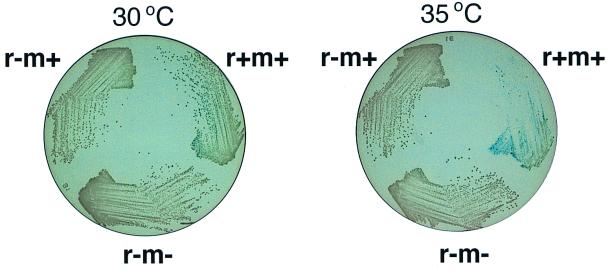
Plate assay for postsegregational killing.
In the next series of experiments, postsegregational killing was estimated from growth inhibition on solid media. An E. coli strain carrying a temperature-sensitive plasmid with the EcoRI r+ m+ gene complex was found to form smaller and fewer colonies at the semipermisssive temperature (35°C) than the control strain with an r− m+ or r− m− plasmid (Fig. 6). The inhibition appeared stronger at 37 (see Fig. 7) and 42°C (data not shown) than at 35°C. Because of the sfiA::lacZ reporter construct in these strains, SOS induction should result in a blue color on agar containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). The bacterial cells carrying this r+ m+ temperature-sensitive plasmid were found to be slightly induced for the SOS response at 30°C and to be more induced at 35°C (Fig. 6). These results are qualitatively similar to those obtained in liquid media (Fig. 5) and sugggest that the plate assay provides a reasonable measurement of postsegregational killing by RM systems, as shown previously for other classical postsegregational killing systems on plasmids (12).
FIG. 6.
Growth inhibition and SOS induction following loss of the EcoRI RM gene complex. An E. coli strain with the sfiA::lacZ promoter fusion (GC3403) carrying pIK172 (pSC101Ts, EcoRI r+ m+, Apr), pIK173 (its r− m+ version), or pIK174 (its r− m− version) was aerated in L broth containing selective antibiotics at 30°C and then streaked on L agar plates lacking selective antibiotics but containing 20 ng of X-Gal/ml and 0.25 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The plates were incubated at 30 or 35°C. Essentially the same results were obtained without IPTG.
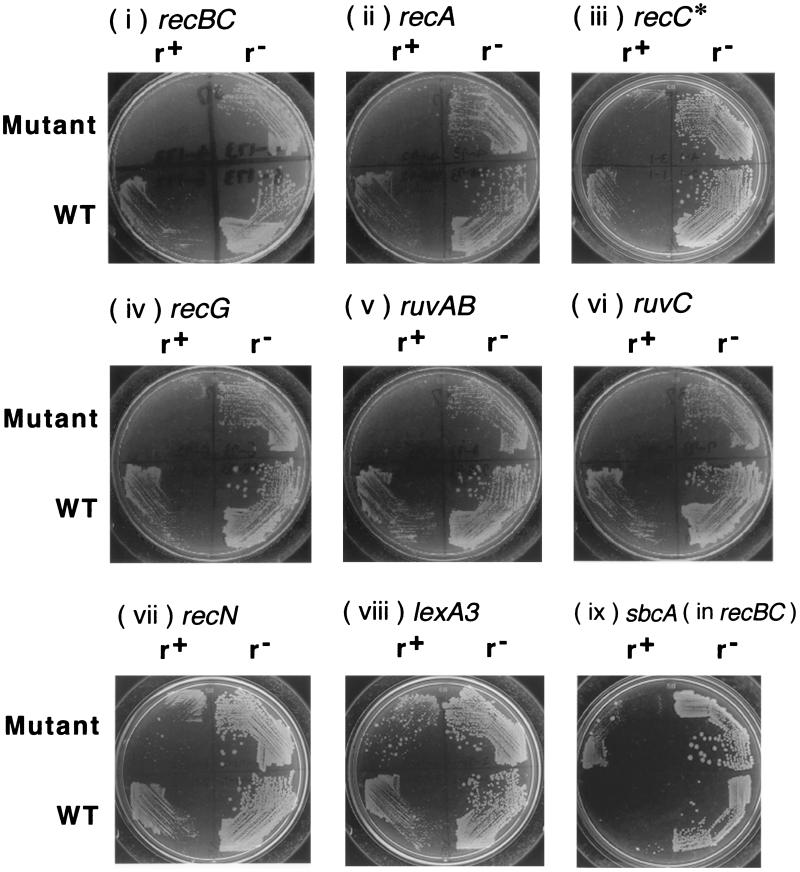
FIG. 7.
Effect of bacterial mutations on growth inhibition following loss of the EcoRI RM gene complex. Various E. coli strains carrying pIK172 (pSC101Ts, EcoRI r+ m+, Apr) or pIK173 (its r− version) were grown with selective antibiotics to log phase at 30°C and were then streaked on L agar for incubation at 37°C for 18 to 23 h. (i) JC5519 (recB21 recC22) and AB1157 (rec+). (ii) BIK733 (ΔrecA306::Tn10) and AB1157 (rec+). (iii) BIK3686 (recC73 recC1002 argA81::Tn10) and AB1157 (rec+). (iv) BIK1538 (recG258::Tn10 mini-Kan) and AB1157 (rec+). (v) HRS2302 (ruvAB::Cm) and AB1157 (rec+). (vi) HRS1100 (ruvC100::Cm) and AB1157 (rec+). (vii) BIK2565 (recN1502::Tn5) and AB1157 (rec+). (viii) BIK2571 (lexA3) and BIK2574 (lexA+). (ix) JC8679 (recB21 recC22 sbcA23) and JC5519 (recB21 recC22). r+, bacteria losing r+ m+ plasmid; r−, bacteria losing r− m+ plasmid; WT, wild type with respect to the gene in question.
Postsegregational killing in other bacterial mutants.
Using the plate assay, we examined the effects of several bacterial mutations on postsegregational killing. The extent of cell killing was inferred from the differences in colony growth between cells losing the EcoRI r+ m+ plasmid and those losing the EcoRI r− m+ control plasmid (Fig. 7; compare the right part and the left part of each plate). At 30°C, the cells losing the r+ and r− plasmid showed indistinguishable levels of growth (data not shown). The recBCD mutant showed strong growth retardation (Fig. 7i) as in the liquid media (Fig. 1, second row, second column). The RecBCD enzyme degrades DNA as an exonuclease from a double-stranded break. When the enzyme encounters a specific sequence called Chi (5-GCTGGTGG), it promotes recombination of this DNA with a homologous DNA molecule with the help of the RecA protein (9, 28, 34, 60). The recC1002 recC* mutant was found to be as proficient in recombination as the recBCD+ strain in an assay with a substrate DNA lacking Chi, but it is defective in recombination enhancement by Chi (54). The recC* mutation was found to slightly increase killing in the plate assay (Fig. 7iii).
In liquid assay, the growth inhibition of the recC* strain was not as severe as that of the recBC strain (Fig. 1, third row). The recC* mutant carrying the r+ plasmid showed low viability even at 30°C (Fig. 1, third row, fourth column), probably as a result of postsegregational killing. The cell population of the recC* strain that survived postsegregational killing showed increased viability in this (Fig. 1, third row, fourth column) and other independent experiments, presumbably because the cells did not carry the restriction gene any more. The recC* mutant culture with the EcoRI r+ m+ gene complex had a level of chromosome cleavage that was intermediate between that of the recBC mutant and that of the rec+ strain (Fig. 4, right). The recC* culture with the r− plasmid contained a level of chromosome cleavage that was intermediate between that of the recBC mutant and that of the rec+ strain.
The growth inhibition turned out to be severe in other mutants defective in the RecBCD pathway of recombination, including mutants of recA, ruvAB, ruvC, and recG (Fig. 7ii, v, vi, and iv, respectively). A less severe defect was observed in a mutant of the recN gene which is defective in DNA double-strand break repair (Fig. 7vii).
A lower level of inhibition in the lexA3 mutant (Fig. 7viii), defective in SOS induction, than in the recBC mutant was observed. This indicates that the RecBCD function contributes in other ways to cell survival than just the generation of SOS signals.
The sbcA mutations on the Rac prophage activate RecET-mediated homologous recombination (27), and this can repair type II restriction breaks at least on plasmids (31). As expected, an sbcA mutation partially suppressed the sensitivity of the recBC mutant to postsegregational killing (Fig. 7ix).
The extent of postsegregational killing in these mutants was measured quantitatively by comparing their colony-forming efficiencies at a lower temperature and at a higher, nonpermissive temperature (Table 2). The results roughly paralleled those from the above streak assay. The only and interesting exception is the recA mutant, which will be a subject of a future study of ours. Taken together, these results suggest that RecBCD-mediated recombination repair of restriction breaks contributes to cell survival after postsegregational killing by the RM systems.
TABLE 2.
Viability of rec mutants in postsegregational killing by EcoRI RM systema
| Relevant strain genotype | Expt 1 viabilities
|
Expt 2 viabilities
|
||||
|---|---|---|---|---|---|---|
| r+#1, r+#2 | r−#1, r−#2 | Avg (<[r+#1] + [r+#2]>/<[r−#1] + [r−#2]>) | r+#1, r+#2 | r−#1, r−#2 | Avg (<[r+#1] + [r+#2]>/<[r−#1] + [r−#2]>) | |
| rec+ | 0.46, 0.25 | 1.2, 1.7 | 0.30 | 0.12, 0.14 | 1.1, 1.2 | 0.11 |
| recA | 0.040, 0.034 | 0.54, 0.57 | 0.067 | 0.044, 0.028 | 1.0, 0.67 | 0.042 |
| recBC | 0.033, 0.044 | 0.98, 1.6 | 0.030 | 0.012, 0.010 | 1.1, 0.79 | 0.012 |
| recC* | 0.024, 0.012 | 1.3, 1.0 | 0.016 | 0.013, 0.015 | 1.2, 1.3 | 0.011 |
| recG | 0.0025, 0.0013 | 1.4, 1.4 | 0.0014 | 0.00088, 0.0011 | 1.2, 0.98 | 0.00094 |
| ruvAB | 0.00082, 0.00054 | 1.6, 1.3 | 0.00047 | 0.0030, 0.0039 | 1.3, 1.8 | 0.0023 |
| ruvC | 0.0059, 0.0039 | 1.2, 1.6 | 0.0035 | 0.0014, 0.0069 | 0.99, 1.1 | 0.0041 |
| recN | 0.038, 0.029 | 1.2, 1.0 | 0.031 | 0.037, 0.094 | 1.2, 1.3 | 0.054 |
| lexA3 | 0.11, 0.086 | 1.1, 0.87 | 0.097 | 0.093, 0.23 | 1.0, 1.5 | 0.13 |
| recBC sbcA | 0.12, 0.18 | 1.5, 1.7 | 0.093 | 0.27, 0.33 | 1.6, 1.6 | 0.19 |
Various E. coli strains carrying pIK172 or pIK173 were grown with selective antibiotics to early log phase at 30°C, were diluted with 10 mM Tris-HCl (pH 7.5)–10 mM MgSO4, and were then streaked on an L agar plate or an ampicillin plate for incubation at 37 and 30°C, respectively. Colonies were counted after 20 h of incubation. Viability was calculated as colony number on L plate at 37°C/colony number on ampicillin plate at 30°C. The viability for the r+ m+ plasmid divided by that for the r− m+ plasmid is indicated. #1 and #2 indicate duplicate measurements within one experiment.
DISCUSSION
Our results here demonstrate that (i) the cells of bacterium E. coli die after loss of an RM plasmid; (ii) this death is accompanied by cleavage and degradation of their chromosomes; (iii) a recBC mutant shows more extensive cell death and accumulates more huge cleaved chromosomes; (iv) recC*, recA, ruvAB, ruvC, recG, recN, and lexA mutants also show extensive cell death; and (v) SOS function is induced in a recBC-dependent manner.
Postsegregational cell killing by RM systems.
The observation of anucleated cells in an E. coli rec+ strain in our experiments clearly demonstrates that cell death can occur following the loss of an RM gene complex. This and other observations in the present work and previous works (15a, 26, 26a) strongly support the postsegregational cell killing concept for RM systems. The formation of anucleated cells was observed in postsegregational killing by ccd genes on F plasmids (21).
Type II RM gene complexes are prevalent on bacterial chromosomes (26a, 49). A chromosomally located type II RM gene complex also resists replacement by a homologous stretch of DNA (Y. Nakayama, N. Handa, and I. Kobayashi, unpublished data). Therefore, postsegregational killing appears to be an intrinsic property of RM gene complexes rather than of the plasmids that carry them. Postsegregational killing would assure maintenance of RM gene complexes in their competition against other genetic elements. This “selfish-gene hypothesis” can explain several properties of type II RM systems (1, 25, 26, 32, 45, 50). Their behavior as mobile genetic elements shaping the bacterial genome supports this hypothesis (26a, 45a).
We have been able to detect plasmid stabilization by all type II RM systems we have examined so far, including PaeR7I (43), EcoRI (43), and EcoRV (45). However, postsegregational killing has not been detected for type I RM systems (30, 46; Y. Naito, N. Handa, and I. Kobayashi, unpublished data).
Resistance to killing through recombination repair of cleaved chromosomes.
Mutations in recBC, recA, recG, recN, ruvAB, and ruvC increased postsegregational killing by the EcoRI gene complex. These genes are involved in homologous recombination and recombinational repair of DNA damage (28, 36). A simple explanation would be that the RecBCD machinery defends the host against attack by type II RM systems by repairing the restricted chromosomes. In support of this, it was found that many anucleated cells and cleaved chromosomes accumulated in the recBC mutant (Fig. 2 to 4). However, protection by the RecBCD system is incomplete, as shown in Fig. 1 to 4.
Many experiments in vivo and in vitro have suggested that the Chi sequence plays an important role in RecBCD-mediated recombinational repair of DNA (58). The strain with the recC1002 mutation promotes recombination as efficiently as the recBCD+ strain in a lambda recombination assay, but the recombination is not stimulated by the presence of Chi on the substrate DNA (54). The postsegregational killing in the plate assay was stronger (Fig. 7), and the level of cleaved chromosomes was higher (Fig. 4), in the recC1002 mutant than in the recBCD+ strain. This observation is consistent with the idea that Chi recognition is important for the recombinational repair of restricted chromosomes in postsegregational killing as in other forms of recombinational repair. Three recBCD mutants (recC2145, recB2154, and recB2155) showed essentially the same phenotype as this strain (N. Handa, A. Ichige, and I. Kobayashi, unpublished observation). A recD null mutant, which is insensitive to Chi but hyperrecombinogenic, is as resistant to postsegregational killing as the wild-type strain (N. Handa, A. Ichige, and I. Kobayashi, unpublished observation). Determination of these mutant sequences would help to elucidate the underlying molecular events.
The presence of the r+ m+ plasmid appears to result in damage to DNA even at a temperature permissive for plasmid replication, as suggested by the low level of SOS induction in the plate and liquid assays, the low level of chromosome cleavage, and the presence of a few filamentous cells. We do not know whether this reflects low-frequency plasmid loss at this temperature in the genetic backgrounds used or insufficient modification activity to protect chromosomal recognition sites.
Loss of an RM gene complex will generate unmethylated and hemimethylated chromosome sites at the same locus after several rounds of DNA replication. For recombinational repair of the restriction break at the former site, the latter site would be the most appropriate partner.
Some correlation between the amount of cleaved chromosomes and the level of cell death was observed, although we cannot exclude the possibility that, instead of double-stranded breaks, single-stranded breaks introduced at hemimethylated sites (22) represent the real source of lethal damage. Such a single-stranded break, if present, might be converted to a double-stranded break after passage of the replication fork, which would then be recognized and repaired by the RecBCD enzyme as proposed earlier (58).
In an earlier study, an E. coli strain bearing a temperature-sensitive mutant of the EcoRI restriction enzyme displayed induction of the SOS response and chromosome cleavage (17). Strains defective in SOS induction and recombination (recA and recB mutants) were no more sensitive to this in vivo DNA scission than were wild-type bacteria (17). A number of EcoRI restriction enzyme mutants are apparently temperature sensitive and create lesions that are much more toxic in recA and recB mutant cells (18).
Mutations in the recN gene are known to confer sensitivity to ionizing radiation but not to UV light. The sensitivity to ionizing radiation in these mutants correlates with a deficiency in their capacity to repair DNA double-stranded breaks. RecN and this repair capacity are SOS inducible (47, 51). The necessity for RecN as a defense against an RM gene system (Fig. 7vii) is consistent with our hypothesis that double-stranded breaks are the true source of lethal damage.
A mutation in the ruvAB, ruvC, or recG gene confers sensitivity to DNA-damaging agents and results in a defect in homologous recombination. RuvA and RuvB proteins together and the RecG protein by itself promote branch migration of Holliday structures (37, 65), and the RuvC protein resolves Holliday structures (65). These enzymes may be involved in the later steps of recombination repair of restriction breaks in vivo, although other possibilities (55) cannot be ruled out.
The severe postsegregational killing in the recBC mutant was partially suppressed by an sbcA mutation (Fig. 7ix), which activates RecET-mediated homologous recombination coded by the Rac prophage. The suppressive effect is likely to be due to repair of chromosomal double-stranded breaks by this homologous recombination machinery because RecET- and Red-mediated recombination can repair double-stranded breaks by type II restriction enzymes (56, 61, 62). However, the possible involvement of other Rac prophage genes (24) cannot be excluded.
The bacterial cells losing RM genes induced the SOS response in a restriction-dependent and RecBC-dependent manner. Presumably, the helicase activity of RecBCD generated single-stranded DNA from a restriction break and this in turn induced the SOS response (6, 53). This SOS induction is unlikely to be the only role of the RecBCD enzyme in cell defense because the lexA3 mutation, which tightly suppresses the SOS response (42), leads to less-severe postsegregational killing than recBC mutations (Fig. 7viii). The role of SOS induction in defense is unlikely to be restriction alleviation, which is not effective for EcoRI restriction (64). Its role might be the induction of RuvAB and/or RecN (51, 65) or induction of stable chromosomal replication (3).
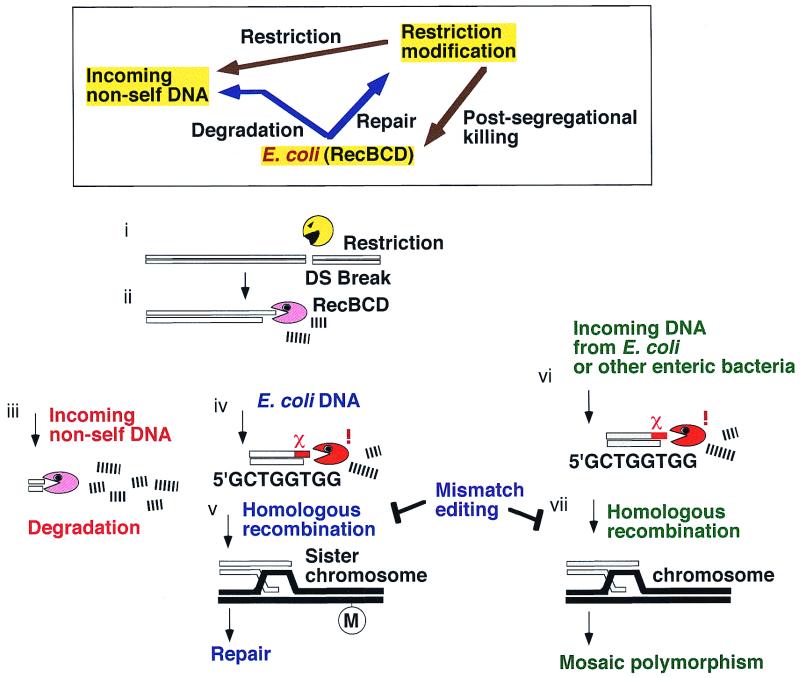
A self-recognition hypothesis for the destruction/repair behavior of the RecBCD system.
Acting at a DNA double-stranded break, the RecBCD enzyme shows two contrasting activities, exonucleolytic degradation and recombination. The exonucleolytic activity serves to destroy invading foreign DNAs after restriction breakage (5, 57). When the RecBCD enzyme encounters Chi, it stops degrading DNA and promotes recombination of this DNA with a homologous DNA molecule (9, 28, 34, 60). Regeneration of a collapsed replication fork was hypothesized to be the role for this unusual destruction/recombination activity (2, 7).
The ambivalent destruction and recombination action of the RecBCD/Chi/RecA machinery may be understood in terms of the interactions between three genetic elements within a cell, the RM system, the invading DNA, and the RecBCD/Chi system as illustrated in Fig. 8. An RM gene complex will attack any unmodified recognition site, whether it be on an invading DNA or on the chromosome. From the restriction break, the RecBCD enzyme will start exonucleolytic degradation of the DNA. This would destroy alien DNA. If the enzyme encounters a Chi sequence, which would serve as an identification marker for the genome, degradation would stop, and recombinational repair would start. In brief, we suggest that the bacterial RecBCD system destroys invading DNAs in collaboration with the RM gene complexes but protects its own chromosome from attack by RM gene complexes.
FIG. 8.
A self-recognition hypothesis for the destruction/repair behavior of the Rec/Chi system. (Box) Three genetic elements in the cell and their relationships. The restriction modification systems will attack invading nonself (unmethylated at recognition sites) DNA as well as the chromosomal DNA of their ex-host (in postsegregational killing). The RecBCD exonuclease/recombinase system will destroy invading nonself DNA (without a Chi sequence) but will repair self DNA (with a Chi sequence and a homologous DNA). After double-stranded cleavage by a restriction enzyme (i) (or by some other factor), the RecBCD enzyme enters a duplex DNA and initiates exonucleolytic degradation (ii). This would destroy incoming foreign DNAs (iii). For chromosomal DNA (iv), the enzyme encounters a Chi sequence, which serves as an identification marker for the chromosome. This results in the attenuation of degradation and promotes recombinational repair with the sister chromosome (v). For an incoming DNA with a Chi sequence in the proper configuration (such as those from E. coli and other closely related enteric bacteria) (vi), degradation would stop, and homologous recombination with the chromosome would follow, if not inhibited by the mismatch recognition system (vii).
Therefore, RecBCD exonuclease and Chi may be regarded as one self-recognizing system analogous to RM systems. This self-recognition concept was proposed (15, 26) on the basis of the observed postsegregational host killing by type II RM systems (43) and on the mutational alteration of the sequence specificity of the RecBCD enzyme (14). Earlier, homologous recombination and RM systems were proposed to have evolved under similar pressures associated with cell invasion by foreign DNAs (48).
Several other bacterial groups seem to have their own unique pairs of RecBCD-like enzymes and recognition sequences (23). If a piece of chromosomal DNA from one member were to enter another member of the same group sharing the same chromosomal identification sequence, Chi or a Chi equivalent in the proper configuration, a part of the invading DNA would be incorporated by the Rec machinery into the chromosome (Fig. 8) (8). The final discrimination between “self” and “nonself” will take place during the process of homologous pairing by the mismatch repair system (38). Therefore, the interactions between the RecBCD/Chi system, RM systems, and invading DNAs that we identified might have contributed to the observed genetic structure of chromosomes in bacterial populations, i.e., mosaics resulting from homologous replacement within a group of bacteria (39, 59).
ACKNOWLEDGMENTS
We thank the people listed in Table 1 for gifts of materials and Steve Kowalczykowski, Tom Bickle, Michael Yarmolinsky, Maurice Fox, Sota Hiraga, and Kenn Gerdes for discussion.
This work was supported by the Ministry of ESSC of the Japanese government (class B, DNA repair, genome), Nagase Science Foundation, Takeda Science Foundation, Japan Science Society, and NEDO. N.H. was supported by a JSPS Research Fellowship for Young Scientists.
Footnotes
Dedicated to Tokio Kogoma.
REFERENCES
- 1.Alm R A, Ling L S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 2.Asai T, Bates D B, Kogoma T. DNA replication triggered by double-stranded breaks in E. coli: dependence on homologous recombination functions. Cell. 1994;78:1051–1061. doi: 10.1016/0092-8674(94)90279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asai T, Sommer S, Bailone A, Kogoma T. Homologous recombination-dependent initiation of DNA replication from DNA damage-inducible origins in Escherichia coli. EMBO J. 1993;12:3287–3295. doi: 10.1002/j.1460-2075.1993.tb05998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachmann B J. Derivation and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 1190–1219. [Google Scholar]
- 5.Brammar W J, Murray N E, Winton S. Restriction of lambda trp bacteriophages by Escherichia coli K. J Mol Biol. 1974;90:633–647. doi: 10.1016/0022-2836(74)90529-4. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhury A M, Smith G R. Role of Escherichia coli RecBC enzyme in SOS induction. Mol Gen Genet. 1985;201:525–528. doi: 10.1007/BF00331350. [DOI] [PubMed] [Google Scholar]
- 7.Cox M M. A broadening view of recombinational DNA repair in bacteria. Genes Cells. 1998;3:65–78. doi: 10.1046/j.1365-2443.1998.00175.x. [DOI] [PubMed] [Google Scholar]
- 8.Dabert P, Smith G R. Gene replacement with linear DNA fragments in wild-type Escherichia coli enhancement by Chi sites. Genetics. 1997;145:877–889. doi: 10.1093/genetics/145.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon D A, Churchill J J, Kowalczykowski S C. Reversible inactivation of the Escherichia coli RecBCD enzyme by the recombination hotspot Chi in vitro: evidence for functional inactivation or loss of the RecD subunit. Proc Natl Acad Sci USA. 1994;91:2980–2984. doi: 10.1073/pnas.91.8.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Game J C, Sitney K C, Cook V E, Mortimer R K. Use of a ring chromosome and pulsed-field gels to study interhomolog recombination, double-strand DNA breaks and sister-chromatid exchange in yeast. Genetics. 1989;123:695–713. doi: 10.1093/genetics/123.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerdes K, Rasmussen P B, Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc Natl Acad Sci USA. 1986;83:3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerdes, K., S. Ayora, I. Canosa, P. Ceglowski, R. Diaz, T. Franch, A. P. Gultyaev, R. B. Jensen, I. Kobayashi, C. MacPherson, D. Summers, C. Thomas, and U. Zielenkiewicz. Plasmid maintenance systems. In C. M. Thomas (ed.), The horizontal gene pool: bacterial plasmids and gene spread, in press. Harwood Academic Publishers GmbH, Amsterdam, The Netherlands.
- 13.Gillen J R, Willis D K, Clark A J. Genetic analysis of the RecE pathway of genetic recombination in Escherichia coli K-12. J Bacteriol. 1981;145:521–532. doi: 10.1128/jb.145.1.521-532.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handa N, Ohashi S, Kusano K, Kobayashi I. χ*, aχ-related 11-mer partially active in an E. coli recC* strain. Genes Cells. 1997;2:525–536. doi: 10.1046/j.1365-2443.1997.1410339.x. [DOI] [PubMed] [Google Scholar]
- 15.Handa N, Ohashi S, Kobayashi I. Clustering of χ sequence in Escherichia coli genome. Microb Comp Genomics. 1997;2:287–298. doi: 10.1089/omi.1.1997.2.287. [DOI] [PubMed] [Google Scholar]
- 15a.Handa N, Kobayashi I. Post-segregational killing by restriction modification gene complexes: observations of individual cell death. Biochimie. 1999;81:931–938. doi: 10.1016/s0300-9084(99)00201-1. [DOI] [PubMed] [Google Scholar]
- 16.Hashimoto-Gotoh T, Franklin F C H, Nordheim A, Timmis K N. Specific-purpose plasmid cloning vectors. I. Low copy number, temperature-sensitive, mobilization-defective pSC101-derived containment vectors. Gene. 1981;16:227–235. doi: 10.1016/0378-1119(81)90079-2. [DOI] [PubMed] [Google Scholar]
- 17.Heitman J, Zinder N D, Model P. Repair of the Escherichia coli chromosome after in vivo scission by the EcoRI endonuclease. Proc Natl Acad Sci USA. 1989;86:2281–2285. doi: 10.1073/pnas.86.7.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heitman J, Ivanenko T, Kiss A. DNA nicks inflicted by restriction endonucleases are repaired by a RecA and RecB dependent pathway in Escherichia coli. Mol Microbiol. 1999;33:1141–1151. doi: 10.1046/j.1365-2958.1999.01556.x. [DOI] [PubMed] [Google Scholar]
- 19.Huisman O, D'Ari R. Effect of suppressors of SOS-mediated filamentation on sfiA operon expression in Escherichia coli. J Bacteriol. 1983;153:169–175. doi: 10.1128/jb.153.1.169-175.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishioka K, Iwasaki H, Shinagawa H. Roles of the recG gene product of Escherichia coli in recombination repair: effects of the ΔrecG mutation on cell division and chromosome partition. Genes Genet Syst. 1997;72:91–99. doi: 10.1266/ggs.72.91. [DOI] [PubMed] [Google Scholar]
- 21.Jaffe A, Ogura T, Hiraga S. Effects of the ccd function of the F plasmid on bacterial growth. J Bacteriol. 1985;163:841–849. doi: 10.1128/jb.163.3.841-849.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jen-Jacobson L, Engler L E, Lesser D R, Kurpiewski M R, Yee C, McVerry B. Structural adaptations in the interaction of EcoRI endonuclease with methylated GAATTC sites. EMBO J. 1996;15:2870–2882. [PMC free article] [PubMed] [Google Scholar]
- 23.Karoui M E, Ehrlich D, Gruss A. Identification of the lactococcal exonuclease/recombinase and its modulation by the putative Chi sequence. Proc Natl Acad Sci USA. 1998;95:626–631. doi: 10.1073/pnas.95.2.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King G, Murray N E. Restriction alleviation and modification enhancement by the Rac prophage of Escherichia coli K-12. Mol Microbiol. 1995;16:769–777. doi: 10.1111/j.1365-2958.1995.tb02438.x. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi I. DNA modification and restriction: selfish behavior of an epigenetic system. In: Russo V, Martienssen R, Riggs A, editors. Epigenetic mechanisms of gene regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 155–172. [Google Scholar]
- 26.Kobayashi I. Selfishness and death: raison d'être of restriction, recombination and mitochondria. Trends Genet. 1998;14:368–374. doi: 10.1016/s0168-9525(98)01532-7. [DOI] [PubMed] [Google Scholar]
- 26a.Kobayashi I, Nobusato A, Kobayashi-Takahashi N, Uchiyama I. Shaping the genome—restriction-modification systems as mobile genetic elements. Curr Opin Genet Dev. 1999;9:649–656. doi: 10.1016/s0959-437x(99)00026-x. [DOI] [PubMed] [Google Scholar]
- 27.Kolodner R, Hall S, Luisi-DeLuca C. Homologous pairing proteins encoded by the Escherichia coli recE and recT genes. Mol Microbiol. 1994;11:23–30. doi: 10.1111/j.1365-2958.1994.tb00286.x. [DOI] [PubMed] [Google Scholar]
- 28.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulakauskas S, Lubys A, Ehrlich S D. DNA restriction-modification systems mediate plasmid maintenance. J Bacteriol. 1995;177:3451–3453. doi: 10.1128/jb.177.12.3451-3454.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulik E M, Bickle T A. Regulation of the activity of the type IC EcoR124I restriction enzyme. J Mol Biol. 1996;264:891–906. doi: 10.1006/jmbi.1996.0685. [DOI] [PubMed] [Google Scholar]
- 31.Kusano K, Takahashi N, Yoshikura H, Kobayashi I. Involvement of RecE exonuclease and RecT annealing protein in DNA double-strand break repair by homologous recombination. Gene. 1994;138:17–25. doi: 10.1016/0378-1119(94)90778-1. [DOI] [PubMed] [Google Scholar]
- 32.Kusano K, Naito T, Handa N, Kobayashi I. Restriction-modification systems as genomic parasites in competition for specific sequences. Proc Natl Acad Sci USA. 1995;92:11095–11099. doi: 10.1073/pnas.92.24.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kusano K, Nakayama K, Nakayama H. Plasmid-mediated lethality and plasmid multimer formation in an Escherichia coli recBC sbcBC mutant. J Mol Biol. 1989;209:623–634. doi: 10.1016/0022-2836(89)90000-4. [DOI] [PubMed] [Google Scholar]
- 34.Kuzminov A, Schabtach E, Stahl F W. χ sites in combination with RecA protein increase the survival of linear DNA in Escherichia coli by inactivating exoV activity of RecBCD nuclease. EMBO J. 1994;13:2764–2776. doi: 10.1002/j.1460-2075.1994.tb06570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lloyd R G, Buckman C. Genetic analysis of the recG locus of Escherichia coli K-12 and of its role in recombination and DNA repair. J Bacteriol. 1991;173:1004–1011. doi: 10.1128/jb.173.3.1004-1011.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lloyd R G, Low K B. Homologous recombination. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2236–2255. [Google Scholar]
- 37.Lloyd R G, Sharples G J. Dissociation of synthetic Holliday junctions by E. coli RecG protein. EMBO J. 1993;12:17–22. doi: 10.1002/j.1460-2075.1993.tb05627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matic I, Taddei F, Radman M. Genetic barriers among bacteria. Trends Microbiol. 1996;4:69–72. doi: 10.1016/0966-842X(96)81514-9. [DOI] [PubMed] [Google Scholar]
- 39.McKane M, Milkman R. Transduction, restriction and recombination patterns in Escherichia coli. Genetics. 1995;139:35–43. doi: 10.1093/genetics/139.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michel B, Ehrlich S D, Uzest M. DNA double-strand breaks caused by replication arrest. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 42.Mount D W, Low K B, Edmiston S J. Dominant mutations (lex) in Escherichia coli K-12 which affect radiation sensitivity and frequency of ultraviolet light-induced mutations. J Bacteriol. 1972;112:886–893. doi: 10.1128/jb.112.2.886-893.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naito T, Kusano K, Kobayashi I. Selfish behavior of restriction-modification systems. Science. 1995;267:897–899. doi: 10.1126/science.7846533. [DOI] [PubMed] [Google Scholar]
- 44.Naito Y, Naito T, Kobayashi I. Selfish restriction modification genes: resistance of a resident R/M plasmid to displacement by an incompatible plasmid mediated by host killing. Biol Chem. 1998;379:429–436. doi: 10.1515/bchm.1998.379.4-5.429. [DOI] [PubMed] [Google Scholar]
- 45.Nakayama Y, Kobayashi I. Restriction-modification gene complexes as selfish gene entities: roles of a regulatory system in their establishment, maintenance, and apoptotic mutual exclusion. Proc Natl Acad Sci USA. 1998;95:6442–6447. doi: 10.1073/pnas.95.11.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45a.Nobusato A, Uchiyama I, Ohashi S, Kobayashi I. Insertion with long target duplication: a novel mechanism for bacterial gene mobility suggested from genome comparison. In: Asai K, Miyano S, Takagi T, editors. Genome informatics 1999. Tokyo, Japan: Universal Academy Press; 1999. pp. 346–347. [Google Scholar]
- 46.O'Neill M, Chen A, Murray N E. The restriction-modification genes of Escherichia coli K-12 may not be selfish: they do not resist loss and are readily replaced by alleles conferring different specificities. Proc Natl Acad Sci USA. 1997;94:14596–14601. doi: 10.1073/pnas.94.26.14596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Picksley S M, Attfield P V, Lloyd R G. Repair of DNA double-strand breaks in Escherichia coli K12 requires a functional recN product. Mol Gen Genet. 1984;195:267–274. doi: 10.1007/BF00332758. [DOI] [PubMed] [Google Scholar]
- 48.Price C, Bickle T A. A possible role for DNA restriction in bacterial evolution. Microbiol Sci. 1986;3:296–299. [PubMed] [Google Scholar]
- 49.Roberts R J, Macelis D. REBASE-restriction enzymes and methylases. Nucleic Acids Res. 1999;27:312–313. doi: 10.1093/nar/27.1.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rocha E P C, Viari A, Danchin A. Oligonucleotide bias in Bacillus subtilis: general trends and taxonomic comparisons. Nucleic Acids Res. 1998;26:2971–2980. doi: 10.1093/nar/26.12.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rostas K, Morton S J, Picksley S M, Lloyd R G. Nucleotide sequence and LexA regulation of the Escherichia coli recN gene. Nucleic Acids Res. 1987;15:5041–5049. doi: 10.1093/nar/15.13.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saito A, Iwasaki H, Ariyoshi M, Morikawa K, Shinagawa H. Identification of four acidic amino acids that constitute the catalytic center of the RuvC Holliday junction resolvase. Proc Natl Acad Sci USA. 1995;92:7470–7474. doi: 10.1073/pnas.92.16.7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sassanfar M, Roberts J W. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J Mol Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- 54.Schultz D W, Taylor A F, Smith G R. Escherichia coli recBC pseudorevertants lacking Chi recombinational hotspot activity. J Bacteriol. 1983;155:664–680. doi: 10.1128/jb.155.2.664-680.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seigneur M, Bidnenko V, Ehrlich S D, Michel B. RuvAB acts at arrested replication forks. Cell. 1998;95:419–430. doi: 10.1016/s0092-8674(00)81772-9. [DOI] [PubMed] [Google Scholar]
- 56.Silberstein Z, Tzfati Y, Cohen A. Primary products of break-induced recombination by Escherichia coli RecE pathway. J Bacteriol. 1995;177:1692–1698. doi: 10.1128/jb.177.7.1692-1698.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simmon V F, Lederberg S. Degradation of bacteriophage lambda deoxyribonucleic acid after restriction by Escherichia coli K-12. J Bacteriol. 1972;112:161–166. doi: 10.1128/jb.112.1.161-169.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith G R. DNA double-strand break repair and recombination in Escherichia coli. In: Nickoloff J A, Hoekstra M F, editors. DNA damage and repair. Totowa, N.J: Humana Press; 1998. pp. 135–162. [Google Scholar]
- 59.Smith J M, Dowson C G, Spratt B G. Localized sex in bacteria. Nature. 1991;349:29–31. doi: 10.1038/349029a0. [DOI] [PubMed] [Google Scholar]
- 60.Stahl M M, Kobayashi I, Stahl F W, Huntington S K. Activation of Chi, a recombinator, by the action of an endonuclease at a distant site. Proc Natl Acad Sci USA. 1983;80:2310–2313. doi: 10.1073/pnas.80.8.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takahashi N, Kobayashi I. Evidence for the double-strand break repair model of bacteriophage λ recombination. Proc Natl Acad Sci USA. 1990;87:2790–2794. doi: 10.1073/pnas.87.7.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takahashi N K, Sakagami K, Kusano K, Yamamoto K, Yoshikura H, Kobayashi I. Genetic recombination through double-strand break repair: shift from two-progeny mode to one-progeny mode by heterologous inserts. Genetics. 1997;146:9–26. doi: 10.1093/genetics/146.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takahashi N K, Kusano K, Yokochi T, Kitamura Y, Yoshikura H, Kobayashi I. Genetic analysis of double-strand break repair in Escherichia coli. J Bacteriol. 1993;175:5176–5185. doi: 10.1128/jb.175.16.5176-5185.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thoms B, Wackernagel W. UV-induced allevation of lambda restriction in Escherichia coli K-12: kinetics of induction and specificity of this SOS function. Mol Gen Genet. 1982;186:111–117. doi: 10.1007/BF00422921. [DOI] [PubMed] [Google Scholar]
- 65.West S C. Processing of recombination intermediates by the RuvABC proteins. Annu Rev Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- 66.Willetts N S, Clark A J. Characteristics of some multiply recombination-deficient strains of Escherichia coli. J Bacteriol. 1969;100:231–239. doi: 10.1128/jb.100.1.231-239.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamamoto K, Yoshikura H, Takahashi N, Kobayashi I. Apparent gene conversion in Escherichia coli rec+ strain is explained by multiple rounds of reciprocal crossing-over. Mol Gen Genet. 1988;212:393–404. doi: 10.1007/BF00330842. [DOI] [PubMed] [Google Scholar]