Abstract
Rhoptry neck protein 2 (RON2) binds to the hydrophobic groove of apical membrane antigen 1 (AMA1), an interaction essential for invasion of red blood cells (RBCs) by Plasmodium falciparum (Pf ) parasites. Vaccination with AMA1 alone has been shown to be immunogenic, but unprotective even against homologous challenge in human trials. However, the AMA1-RON2L (L is referred to as the loop region of RON2 peptide) complex is a promising candidate, as preclinical studies with Freund’s adjuvant have indicated complete protection against lethal challenge in mice and superior protection against virulent infection in Aotus monkeys. To prepare for clinical trials of the AMA1-RON2L complex, identity and integrity of the candidate vaccine must be assessed, and characterization methods must be carefully designed to not dissociate the delicate complex during evaluation. In this study, we developed a native Tris-glycine gel method to separate and identify the AMA1-RON2L complex, which was further identified and confirmed by Western blotting using anti-AMA1 monoclonal antibodies (mAbs 4G2 and 2C2) and anti-RON2L polyclonal Ab coupled with mass spectrometry. The formation of complex was also confirmed by Capillary Isoelectric Focusing (cIEF). A short-term (48 h and 72 h at 4°C) stability study of AMA1-RON2L complex was also performed. The results indicate that the complex was stable for 72 h at 4°C. Our research demonstrates that the native Tris-glycine gel separation/Western blotting coupled with mass spectrometry and cIEF can fully characterize the identity and integrity of the AMA1–RON2L complex and provide useful quality control data for the subsequent clinical trials.
Keywords: Apical membrane antigen 1, Capillary isoelectric focusing, Malaria vaccine, Native gel electrophoresis, Rhoptry neck protein 2
1. Introduction
Malaria remains a major global health problem, most recently causing an estimated 229 million cases and more than 4 00 000 deaths worldwide in 2019 [https://www.who.int/publications-detail/world-malaria-report-2020], with Plasmodium falciparum(Pf) infection accounting for most cases [https://www.who.int/news-room/fact-sheets/detail/malaria]. An effective vaccine is urgently needed, especially as existing disease control methods are waning in efficacy. An effective blood stage vaccine would prevent clinical disease and also potentially reduce transmission of the parasite within a community [1].
One promising blood stage vaccine target is Plasmodium falciparum apical membrane antigen 1 (PfAMA1), which has been characterized and demonstrated to play an essential role in infection. It was further shown that binding of AMA1 to a 47 amino acid region of rhoptry neck protein 2 (RON2L) [2,3] comprises an essential step in the invasion of red blood cells (RBCs) by Pf merozoites [4]. Recent study has shown that immunization with a preformed AMA1–RON2L complex, but not AMA1 alone, formulated with Freund’s adjuvant provided complete protection against a lethal Plasmodium yoelii challenge in mice, and superior protection against virulent Pf infection in Aotus monkeys [5,6]. To prepare clinical trials using AMA1–RON2L complex as a candidate vaccine, quality control (QC) studies confirming the AMA1–RON2L complex identity and integrity must be conducted. Since the complex is formed by noncovalent interactions, the characterization methods must be carefully designed to prevent dissociation of the complex during evaluation.
SDS-PAGE is the most widely used technique to analyze mixtures of proteins [7–9]. However, SDS denatures secondary and nondisulfide-linked tertiary structures [10,11], thus, this method is unfeasible to characterize proteins which require intact higher order structures and ability for protein–protein interactions. A native PAGE, nevertheless, may be able to circumvent the problem of dissociation and is worth further investigation.
Capillary isoelectric focusing (cIEF) is a high-resolution technique that separates protein/peptide mixtures of various glycoforms and charges, based on their pI [12–14]. This technique has been increasingly applied in recent years [15,16], but analysis of a protein–protein complex by cIEF in the field of vaccine research and development has not been reported. We hypothesize the AMA1–RON2L complex can be analyzed by charge using the exceptionally high-resolution power of cIEF.
A high-binding affinity (apparent equilibrium dissociation constant (KD) ∼20 nM) between PfAMA1–3D7 and RON2L has been reported [17], which suggests the complex may be analyzed by a native PAGE and cIEF. In this study, the AMA1–RON2L complex was first separated by simplified native electrophoresis using commercially pre-cast Tris-glycine PAGE without addition of anionic reagents. Following electrophoresis, the protein complex integrity was further identified and confirmed by Western blotting using anti-AMA1 and anti-RON2L antibodies and by MS. The formation of complex was also confirmed by cIEF.
Our research demonstrates that this methodology can successfully characterize the identity and integrity of the AMA1–RON2L complex, and enables QC analysis for subsequent clinical trials. Using these techniques, a short-term (48 and 72 h at 4°C) stability study of AMA1–RON2L complex was performed and results indicate this complex was stable for 72 h at 4°C.
2. Materials and methods
2.1. Animal study ethics statement
Rabbits were used to generate anti-RON2L antibodies. All experiments adhered to animal study protocol Laboratory of Malaria Immunology and Vaccinology (LMIV) 1E that was approved by the Institutional Animal Care and Use Committees of the National Institute of Allergy and Infectious Diseases, NIH. LMIV, as part of the Public Health Service, Department of Health and Human Services, NIH Intramural Research Program, is accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care, (AAALAC file # 777; last accredited 2017), and holds a PHS Assurance on file with the National Institute of Health, Office of Laboratory Animal Welfare as required by the US Public Health Service Policy on Humane Care and Use of Laboratory Animals. The PHS Animal Welfare Assurance (File Number # D16–00602) was last approved on June 10, 2019. The USDA certification file number is # 51-F0016.
2.2. Reagents
Recombinant P. falciparum AMA1–3D7, which has the sequences YVQNYWEHPYQNSDVYRPIN…TYDKMKTSHHHHHH and molecular weight of 61 973.1 Daltons, was manufactured and characterized by the LMIV, National Institute of Allergy and Infectious Disease, National Institutes of Health [18].
Rhoptry neck protein 2 peptide (RON2L), which has the sequences of DITQQAKDIGAGPVASCFTTRMSPPQQICLNSVVNTALSTSTQSAMK with acetylated N-terminus and amidated at the C-terminus, and molecular weight of 4898.5 Daltons, was synthesized by Lifetein LLC (Somerset, NJ, USA). The synthesized RON2L was analyzed by MS for mass accuracy, and by HPLC for purity. The synthetic RON2L peptide used was more than 95% in purity.
Rabbit anti-RON2L antibodies were generated in LMIV. Briefly, 2 mg of RON2L peptide was conjugated to 2 mg of Keyhole Limpet Hemocyanin (KLH) by using the Readilink KLH conjugation kit (AAT Bioquest, Sunnyvale, CA, USA). Two New Zealand female white rabbits (∼11 weeks old, body weight 2.2–2.6 kg) were used in this study. Each rabbit was immunized with 27.5 μg peptide in 0.5 mL formulation containing 70% of Montanide ISA 720 VG adjuvant (SEPPIC Inc., Fairfield, NJ, USA). Following three immunizations at 3 week intervals, animals were bled 2 weeks after the third immunization. Rabbit anti-sera were aliquoted and stored at −80°C until used.
2.3. Preparation of AMA1–RON2L complex
2.3.1. Complex preparation for Native PAGE gel and LC-MS/MS
Ninety microliters of 16 μM AMA1–3D7 in saline and 30 μL of 612 μM of RON2L in PBS were mixed in a 1.5 mL autoclaved Eppendorf microcentrifuge tube (Eppendorf North America, Enfield, CT, USA) and incubated at room temperature for 30 min.
2.3.2. Complex preparation for Western blot
Ninety microliters of 16 μM AMA1–3D7 in saline and 15 μL of 612 μM of RON2L in PBS were mixed in a 1.5 mL autoclaved Eppendorf microcentrifuge tube and incubated at room temperature for 30 min.
2.3.3. Complex preparation for cIEF
AMA1–3D7 (16 μM) in saline was buffer-exchanged with 20 mM Tris buffer at pH 8.0 by loading 0.5 mL of AMA1–3D7 into Centrifugal Filter tubes (Millipore UFV5BCC25, 5k NMWL) and centrifuging in an Eppendorf 5415D Digital Centrifuge (Eppendorf North America, Framingham, MA, USA) for 8 min at 12 000 g, adding 500 μL of 20 mM Tris buffer to the retentate, centrifuging for 8 min at 12 000 g. Centrifugation and buffer replacement cycles were repeated three times to concentrate AMA1–3D7 to 81 μM. The concentrated AMA1 was further mixed with 612 μM RON2L at a desired mass ratio according to Table 1 in autoclaved 1.5 mL Eppendorf microcentrifuge tube and incubated at room temperature for 30 min to form the AMA1–RON2L complex.
Table 1.
Preparation of AMA1–RON2L complex at various molar ratios for capillary isoelectric focusing
| Molar ratio | Volume used (μL) |
Final concentration (μM) |
||
|---|---|---|---|---|
| RON2L:AMA1 | AMA1a | RON2Lb | AMA1 | RON2L |
| 0 | 10 | 0 | 81 | 0 |
| 1 | 10 | 1.35 | 71 | 73 |
| 2 | 10 | 2.7 | 64 | 130 |
| 4.1 | 10 | 5.4 | 52 | 215 |
| 12.7 | 10 | 16.67 | 30 | 383 |
AMA1 staring concentration was 81 μM.
ROM2L starting concentration was 612 μM.
2.3.4. Complex preparation for stability study
Three vials of complex were prepared in 1.5 mL autoclaved Eppendorf microcentrifuge tubes by mixing 50 μL of 16 μM AMA1–3D7 in saline and 50 μL of 612 μM of RON2L in PBS and incubated at room temperature for 30 min. The vials were prepared fresh (0 h) and stored at 4°C for 48 and 72 h, respectively.
2.4. Native PAGE gel
All reagents used in this section were from Thermo Fisher Scientific (Waltham, MA, USA) unless specified.
Twelve-well Novex® 4–20% Tris-Glycine Mini Protein Gels (Thermo Fisher Scientific) were run at 150 V for 90 min with Novex® Native Tris-Glycine Running Buffer System at room temperature. Novex® Unstained Protein Standard and Novex® Native 2X Sample Buffer were used in the electrophoresis. The gels were stained by Coomassie Blue Staining (GE Healthcare Bio-Science AB, Uppsala, Sweden) for visualization or Imperial™ Protein Stain for visualization and further mass spectrometric analysis. The gel image was captured using a Bio-Rad GS-800 calibrated densitometer (Bio- Rad Laboratories, Hercules, CA, USA).
2.5. Western blot Removed
For Western blotting, proteins separated by the Novex® 4–20% Tris-Glycine Mini Protein Gels were transferred to nitrocellulose membranes (Invitrogen Corp. Carlsbad, CA, USA) by iBlot® Dry Blotting System (Thermo Fisher Scientific) according to manufacturer’s instructions. The membranes were blocked with 3% skim milk (BD Difco, Franklin Lakes, NJ, USA) in 1x TBS (Quality Biological, Gaithersburg, MD, USA), and then probed with 2.5 μg per milliliters of monoclonal antibodies 4G2, whose binding site is near the PfAMA1 hydrophobic pocket in the flexible domain II loop [2,19]; 0.82 μg per milliliters of 2C2 which recognizes an unknown epitope of AMA1; and 1:500 dilution of rabbit anti-RON2L sera. The secondary reagent used was anti-mouse or anti-rabbit IgG labeled with alkaline phosphatase, and the substrate 5-bromo-4-chloro-3-indolyl-phosphate/nitroblue tetrazolium (BCIP/NBT) (KPL Labs, Gaithersburg, MD, USA). The membranes were scanned with an Epson Perfection scanner (Epson America, Los Alamitos, CA, USA).
2.6. LC-MS/MS
2.6.1. Sample preparation
The AMA1 lane and the AMA1-RON2L lane were cut at three locations of the native PAGE gel: slightly below and similarly to the 146 kDa marker (Bands 1a and 3a); slightly below and similarly to 66 kDa marker (Bands 1b and 3b); and below Bands 1b/3b (Band 1c and 3c) (Fig. 1B). Each gel strip was diced into small pieces (1 mm3) and placed into labeled Protein LoBind tubes (Thermo Fisher Scientific). The gel pieces were destained twice by sequentially adding 100 μL of 50 mM ammonium bicarbonate (NH4HCO3)/50% ACN for 10 min and 100 μL of 50 mM NH4HCO3. The samples were reduced in a solution of 20 mM DTT and 50 mM NH4HCO3 for 20 min at 60°C, alkylated in a solution containing 50 mM iodoacetamide and 50 mM NH4HCO3 for 20 min in the dark at room temperature. The gel pieces were washed with 50 mM NH4CO3 and dehydrated in a solution containing 50 mM NH4HCO3/50% ACN. The samples were dried using the SpeedVac. The samples were rehydrated and digested with trypsin in solution with a ratio of 1:50 (enzyme: total protein) then incubated overnight at 37°C. Digestion was stopped by extraction with 50% ACN/0.1% formic acid (FA) and the extracted peptide samples were dried using the SpeedVac to evaporate ACN, then reconstituted with 0.1% FA. All samples were desalted using C18 zip tips (Millipore, Burlington, MA, USA).
Figure 1. Native polyacrylamide gel electrophoresis with imperial protein stain.
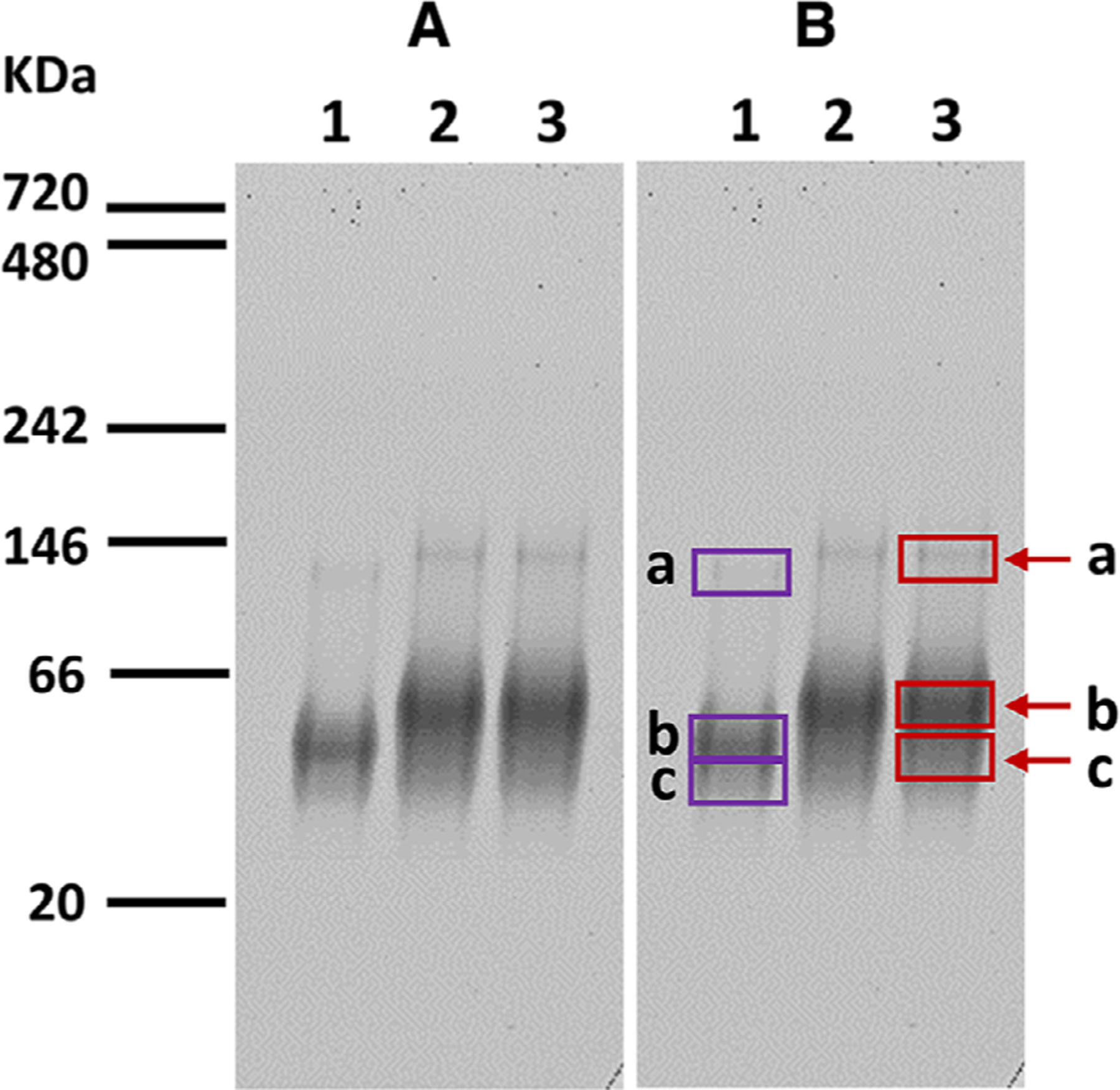
(A) For visualization; (B) for mass spectrometric analysis; Lane 1. 5 μL of 16 μM AMA1–3D7 loaded; Lanes 2 and 3. 12 μL of AMA1-RON2L complex loaded. Bands 1a, 1b, 1c, 3a, 3b, and 3c in Panel B were cut for mass spectrometry analysis. Molecular weight markers are indicated in kDa.
2.6.2. LC-MS/MS
One micrograms of tryptic-digested peptide mixtures was loaded on to a reverse-phase C-18 precolumn in line with an analytical column (Acclaim PepMap, 75 μm × 15 cm, 2 μm, 100Å) connected to the EASY-Nano LC II (Thermo Fisher Scientific). The peptides were separated using a gradient of 5 to 30% of solvent B (0.1% FA, acetonitrile) for 75 min, and then to 95% solvent B for an additional 50 min at a flow rate of 300 μL/min followed by solvent A (0.1% FA, water) for 5 min at a temperature of 25°C. The peptides were analyzed in datadependent mode and positive ionization in the LTQ Orbitrap Velos, and the top 20 precursors were fragmented using CID in an ion trap with a collision energy of 35 eV. The mass window for precursor ion selection was 2 Da and a minimum of 5000 counts were needed to trigger the MS/MS. The MS1 and MS2 were acquired in the Orbitrap, at a resolution of 60 000 and 15 000, respectively.
2.6.3. Bioinformatic analysis
Acquired spectra were analyzed using PEAKS® Studio 7.5 (Bioinformatics Solutions, Inc, Waterloo, ON, Canada) with a precursor tolerance of 10 ppm and 0.02 for Orbitrap for MS/MS for CID against a combined database composed of AMA1 protein sequence and RON2L peptide sequence. Sequences of known contaminants from the cRAP database were added (115 sequences). Carbamidomethyl (C), deamidation (N,Q), and oxidation (M) were selected as variable modifications, and one nonspecific cleavage at the peptide terminus and two missed cleave sites were allowed. The false discovery rate for peptides was set to 1% by applying the target-decoy strategy in PEAKS® Studio. The output of PEAKS® Studio contains the database peptides and proteins.
2.7. Capillary isoelectric focusing (cIEF)
cIEF gel polymer solution and cIEF Peptide Marker Kit (pI 4.1, 5.5, 7.0, 9.5, and 10.0) were purchased from SCIEX (Framingham, MA, USA). Pharmalyte pH 3–10 carrier ampholytes were obtained from GE Healthcare (Chicago, IL, USA). Anolyte (200 mM phosphoric acid), Catholyte (300 mM sodium hydroxide), chemical mobilizer (350 mM acetic acid), cathodic stabilizer (500 mM arginine), and anodic stabilizer (200 mM iminodiacetic acid) were prepared with deionized water (Neu-Ion Inc, Baltimore, MD, USA).
The cIEF sample was prepared by mixing the following reagents in a 0.5 mL Eppendorf tube: 200 μL of cIEF Gel; 12.0 μL of Pharmalyte 3–10 carrier ampholytes; 20.0 μL of cathodic stabilizer; 2.0 μL of anodic stabilizer; 2.0 μL each of pI markers 10, 9.5 and 4.1; and 10 μL of AMA1 alone or AMA1–RON2L complex. The cIEF sample was centrifuged for 20 s at 1000 × g to remove air bubbles.
The cIEF was performed using the PA 800 Plus Pharmaceutical Analysis System (SCIEX) equipped with a UV detector and a 280 nm filter. All separations were carried out using a Neutral Capillary (SCIEX), which was 30.2 cm long and 20 cm from inlet to detector. An aperture of 200 μm was used in the capillary cartridge. The capillary temperature was maintained at 20°C in all separations, unless otherwise specified. Chemical mobilization was used in all cIEF experiments and voltage was applied in normal polarity. The cIEF separation method started by performing two 50 psi rinses in the forward direction: capillary cleaning solution of 0.1 M NaOH and 0.1 M HCL for 3 min and then filtered water for 2 min. The mixture of sample, ampholytes, and pI markers were introduced into the capillary by performing a 99.9 s injection at 25 psi. Immediately, both capillary ends were cleaned by submerging both capillary ends in filtered water for a few seconds. Focusing was performed at 25 kV for 5 min under normal polarity with the inlet side of the capillary submerged in anolyte and the outlet side submerged in catholyte. Chemical mobilization was carried out at 30 kV for 30 min under normal polarity, with the inlet side of the capillary submerged in anolyte and the outlet side in chemical mobilizer. The voltage ramp of 0.17 min was used for both focusing and mobilization steps. At the end of the mobilization step, the data collection was stopped, the capillary was rinsed with filtered water for 2 min at 50 psi, and then both capillary ends were submerged in filtered water.
cIEF Data were analyzed using 32 Karat Software. A standard curve of standard pI markers (pI 10, 9.5, 7, 5.5, 4.1, SCIEX) versus migration time was used to interpolate the unknown pI of AMA1-RON2L, AMA1, or RON2L. During data analysis, the integration parameters were optimized by adjusting width, shoulder sensitivity, threshold, and integration for each individual run.
3. Results
3.1. Native PAGE gel analysis of AMA1-RON2L complex
When AMA1 or AMA1-RON2L complex was separated by native Tris-Glycine gels and visualized by Imperial Protein Stain, bands that migrated slower than AMA 1 alone (Fig. 1,Bands 3a and 3b) were demonstrated in the AMA1–RON2L complex (Fig. 1, Lane 2 and 3). The majority of the band that migrated slower than AMA1 alone (Fig. 1, Lane 1 or Band 3c) was at a molecular mass of approximately 66 kDa, and this band appeared to have a calculated molecular mass of the AMA1–RON2L complex. In addition, a protein band (Fig. 1, Band 3a) that migrated similarly to the 146 kDa marker appeared to be a dimer for AMA1–RON2L complex by calculation (Fig. 1, Lanes 2 and 3).
3.2. Western Blot analysis of AMA1-RON2L complex
Anti-AMA1 mAb 4G2 detected AMA1 (Fig. 2, Panel A, Lane 1, AMA1 alone) and the complex band that migrated slower than AMA1 alone (Panel A, Lanes 2 and 3, AMA1-RON2L complex). Further, anti-AMA1 mAb 2C2 recognized both AMA1 monomers and dimers in AMA1 alone sample (Fig. 2, Panel C, Lane 1); this antibody also recognized complex bands which migrated slower than AMA1 monomers and dimers in the AMA1–RON2L complex lane (Panel C, Lanes 2 and 3). In addition, anti-RON2L antibody detected the complex band which migrated slower than AMA1 alone (Fig. 2, Panel B, Lanes 2 and 3) at a migration position similar to those detected by anti-AMA1 mAbs 4G2 and 2C2 (Panels A and C, Lanes 2 and 3). Anti-RON2L antibody did not detect the AMA1 alone sample (Panel B, Lane 1).
Figure 2. Western blot results.
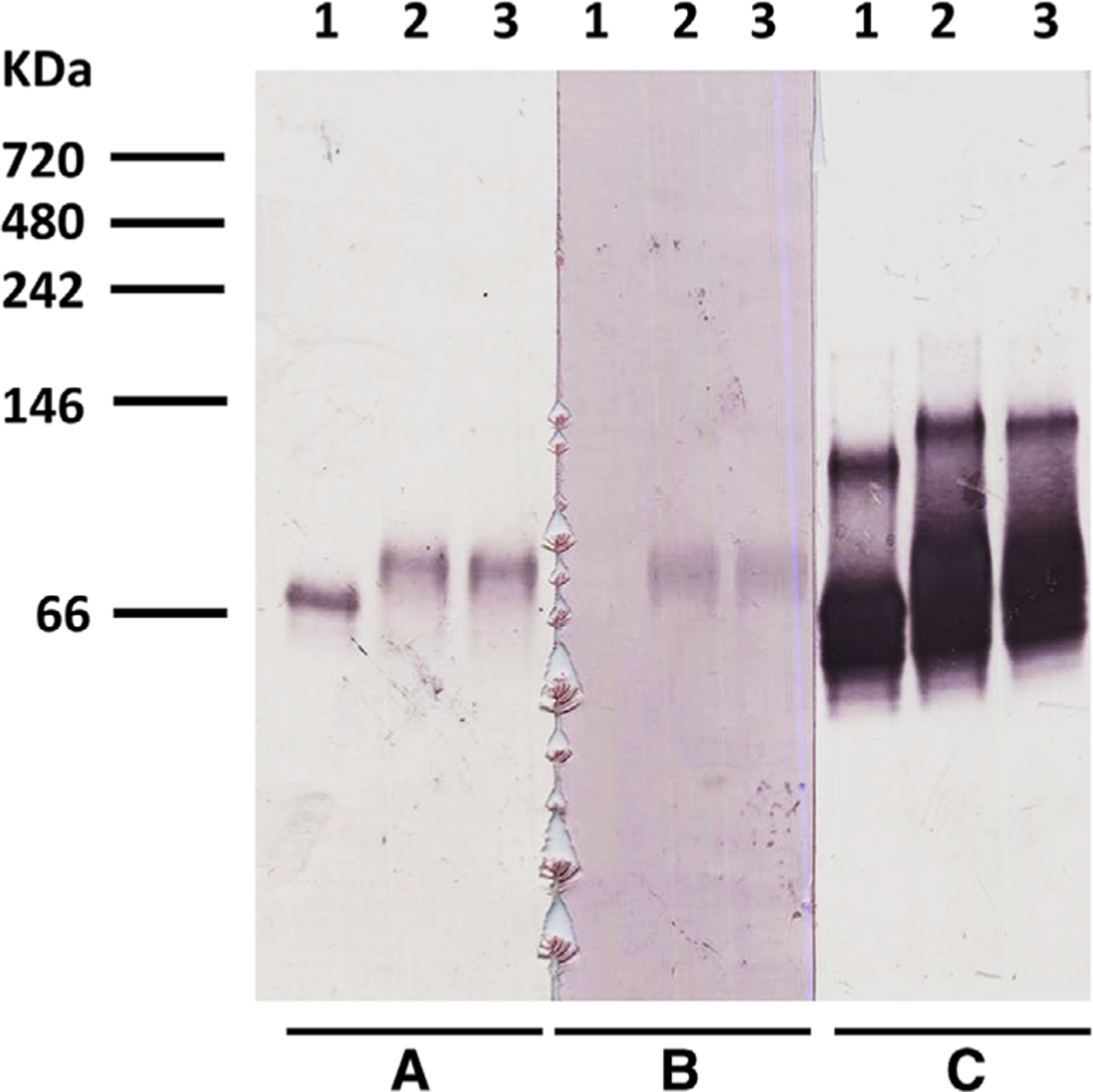
After Native polyacrylamide gel electrophoresis, the membranes were probed with (A): anti-AMA1 mAb 4G2; (B): rabbit anti-RON2L sera; (C): anti-AMA1mAb 2C2. Lane 1, 2.6 μL of 16 μM AMA1–3D7 loaded; Lanes 2 and 3, 3 μL of AMA1-RON2L complex loaded.Molecular weight markers are indicated in kDa.
3.3. LC-MS/MS results
For the AMA1 alone lane (Fig. 1B, Lane 1), the gel band at below 66 kDa (Lane 1b) had the highest number of peptides identified as AMA1 with 47 peptides identified, representing a protein sequence coverage of 63% of AMA1. The gel band below the 146 kDa marker (Lane 1a) contained 20 peptides representing an AMA1 protein sequence coverage of 33%; the other gel band below the 66 kDa marker (Lane 1c) contained 21 peptides representing an AMA1 protein sequence coverage of 34%.
For the AMA1–RON2L complex lane (Fig. 1B, Lane 3), the gel band similar to the 146 kDa marker (Lane 3a) had the highest number of peptides identified as RON2L with two peptides identified, representing a protein coverage sequence of 45%. In this band, 27 peptides were identified as AMA1, representing a protein sequence coverage of 42%. In the gel band at the 66kDa marker (Lane 3b), 44 peptides of AMA1 were identified representing an AMA1 protein sequence coverage of 62%, and 1 peptide of RON2L was identified, representing a RON2L protein sequence coverage of 30%. Finally, in the gel band below 66kDa (Lane 3c), 48 peptides were identified as AMA1, representing a protein sequence coverage of 65%.
3.4. Capillary isoelectric focusing (cIEF)
AMA1–3D7 alone had a cIEF profile of multiple peaks between peak migration time of approximately 34.56 and 35.54 min, representing a pI range of 5.50 to 5.88 (Fig. 3, Table 2). At RON2L to AMA1–3D7 ratios of 1:1, 2:1, 4.1:1, and 12.7:1, peaks were detected between peak migration times of 34.68 to 35.26 min (pI range of 5.61 to 5.83), 34.52 to 35.16 min (pI range of 5.65 to 5.89), 34.46 to 35.10 min (pI range of 5.67 to 5.92), and 34.55 to 35.06 min (pI range of 5.69 to 5.88), respectively. The peak migration time decreased with a concomitant increase in pI range when RON2L to AMA1 molar ratio increased from 1:1 to 2:1, 4.1:1 or 12.7:1 (Fig. 3, Table 2). RON2L peptide alone was not detectable due to lack of absorbance at 280 nm.
Figure 3. Capillary Isoelectric Focusing profiles of RON2L and AMA1 at various molar ratios:
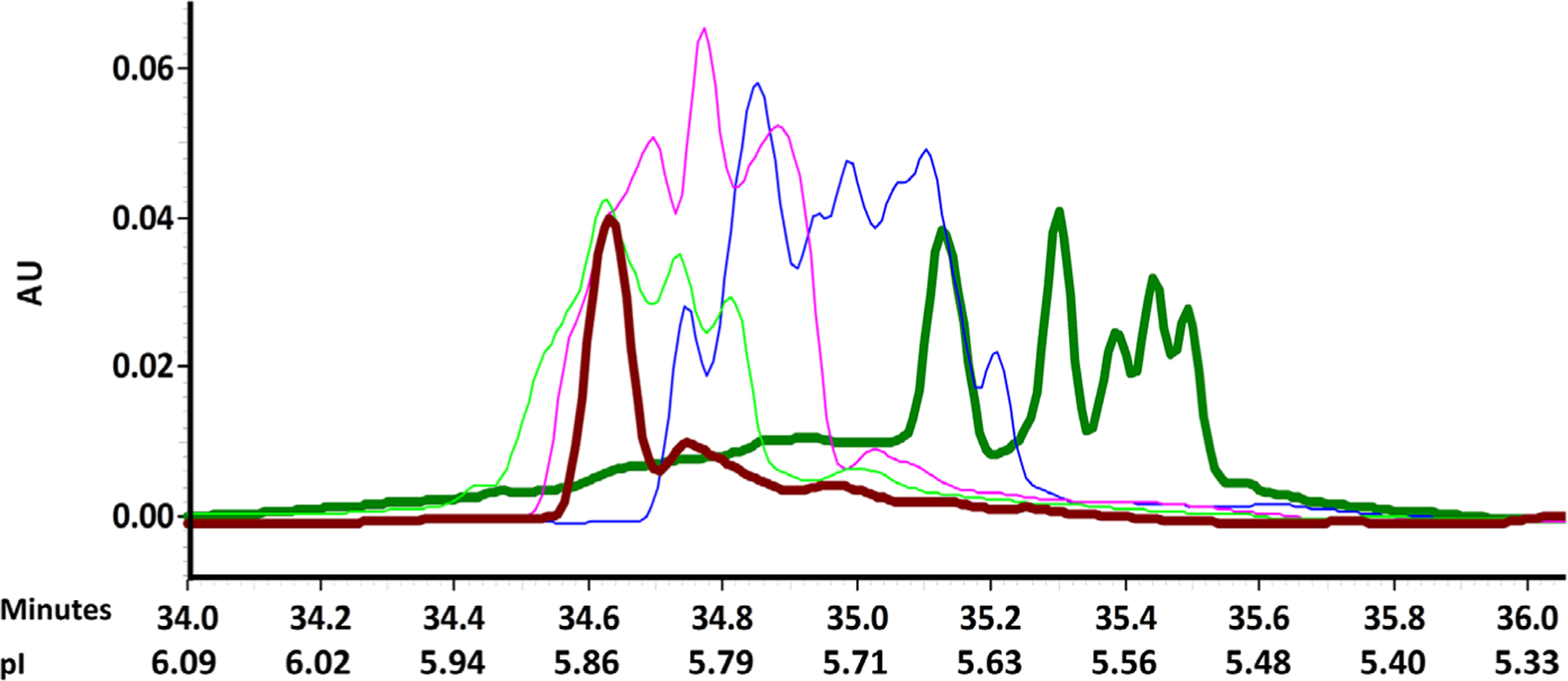
Dark green line: AMA1–3D7 alone; Blue line: Amixture of RON2L to AMA1–3D7 at 1:1 molar ratio; Magenta line: A mixture of RON2L to AMA1–3D7 at 2:1 molar ratio; Green line: A mixture of RON2L to AMA1–3D7 at 4.1:1molar ratio; Burgundy line: A mixture of RON2L to AMA1–3D7 at 12.7:1 molar ratio.
Table 2.
Capillary isoelectric focusing profiles of RON2L and AMA1 at various molar ratios
| Molar Ratio RON2L: AMA1 | Time range (min) | pI rangea |
|---|---|---|
| 0 | 34.56–35.54 | 5.50–5.88 |
| 1 | 34.68–35.26 | 5.61–5.83 |
| 2 | 34.52–35.16 | 5.65–5.89 |
| 4.1 | 34.46–35.10 | 5.67–5.92 |
| 12.7 | 34.55–35.06 | 5.69–5.88 |
pI is calculated using the standard curve generated by the migration time of pI marker versus the value of pI marker (y = –0.3833x + 19.125. R2 = 1).
3.5. AMA1–RON2L stability
Following storage at 4°C for 48 and 72 h, the AMA1–RON2L complex appeared to remain undissociated when analyzed by native PAGE gel (Fig. 4).
Figure 4. AMA1–RON2L stability by native polyacrylamide gel electrophoresis with Coomassie Blue Stain.
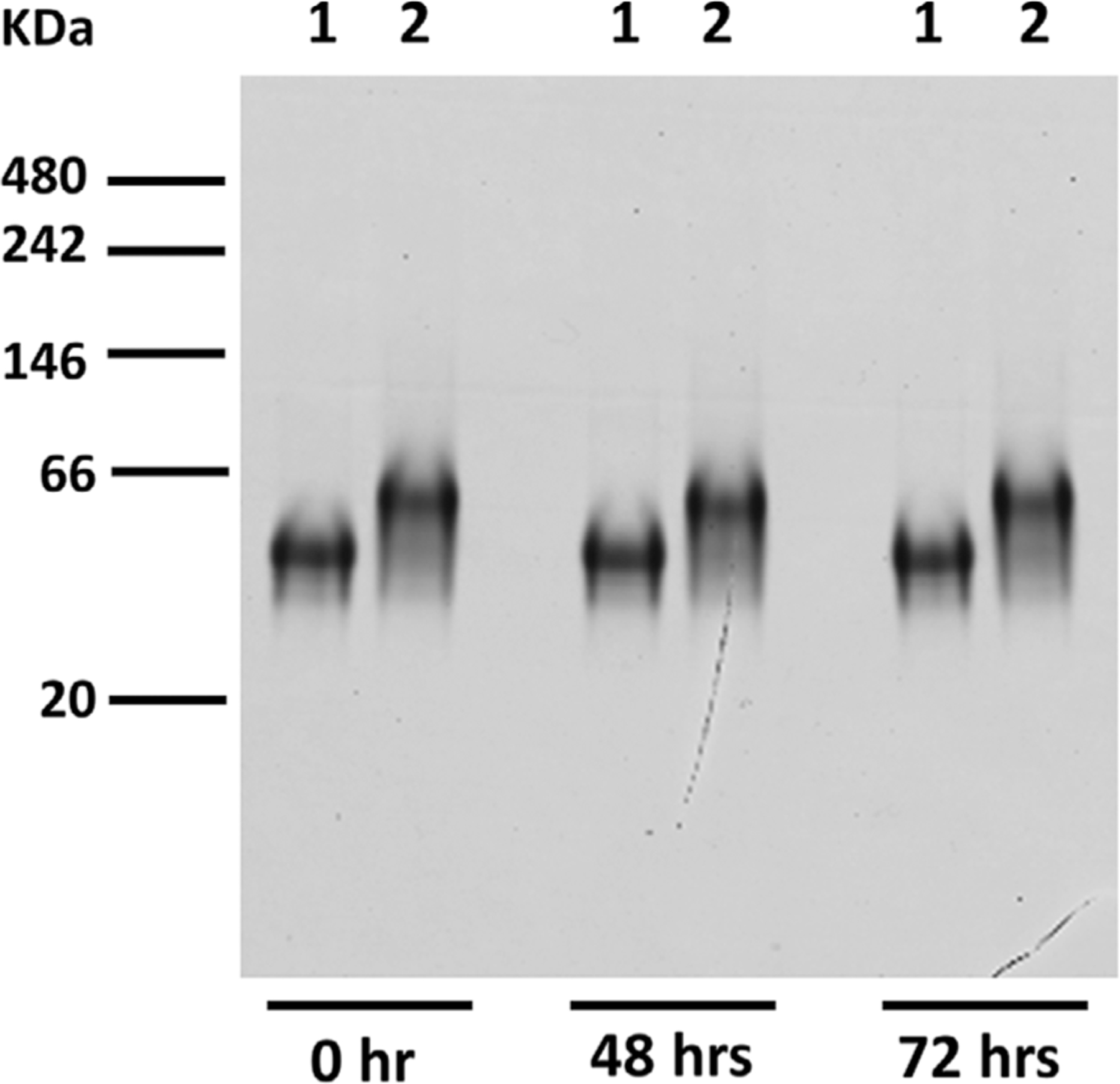
Lane 1. 5 μL of 16μM AMA1–3D7 loaded; Lane 2. 10 μL of AMA1–RON2L complex loaded; the complexes were kept at 4°C as the time indicated. Molecular weight markers are indicated in kDa.
4. Discussion
The theoretical pIs of AMA1–3D7 and RON2L are 5.55 and 8.05, respectively. However, with N-acetyl and C-amide modifications, the pI of RON2L will be substantially higher. Nevertheless, under the electrophoresis running conditions of pH 8.3–8.6 using commercial reagents [20], AMA1–3D7 and AMA1–RON2L complex are negatively charged and able to move toward the anode, and thus, anionic/neutral detergents are not required to induce a negative charge shift on these proteins. As for RON2L alone, it may not migrate into the native PAGE due to high pI. This is the first report to perform electrophoresis of the AMA1–RON2L complex without the use of a charge shift reagent, which not only simplifies the procedure, but also eliminates the risk of protein complex dissociation.
The aggregation (dimer formation) of AMA1–3D7 was observed in the native gel, as was the formation of AMA1–RON2L complex in aggregated status, indicating that aggregation of AMA1 did not affect the binding between AMA1 and RON2L.
The cIEF data demonstrated increases in pI when RON2L and AMA1 were mixed at increasing molar ratios, indicating that RON2L and AMA1 were complexed and RON2L in AMA1 may have caused an increase in pI of the complex. However, it is interesting that AMA1 alone showed a broad cIEF profile of multiple peaks, and we speculate that AMA1, an apparently homogenous preparation, may slightly differ in surface charges; binding of RON2L, a small peptide with a pI above 8.05, increased the overall pI of AMA1, and thus, caused the shift of migration patterns. We will continue to improve techniques for protein complex analysis by investigating the dynamics of protein–protein interaction and complex stability during electrophoresis.
In summary, a native precast Tris-glycine gel without the need for additional anionic reagents to impose a charge shift on proteins has been developed to characterize the AMA1– RON2L complex. The operating procedure using the commercially precast Tris-glycine gel is simple and effective. In addition, cIEF is also a sensitive and practical method to characterize the AMA1–RON2L complex. All these techniques supported the notion that AMA1–RON2L was formed in solution and can be analyzed for protein complex analysis, vaccine development and QC purposes.
Acknowledgments
The authors would like to thank Dr. David L. Narum for providing AMA1–3D7, Ms. Beth Chen for making RON2L-KLH conjugate, Ms. Lynn Lambert, Ms. Kelly M. Rausch, and Ms. Emma Barnafo for generating rabbit anti-RON2L polyclonal antibody, and Mr. J. Patrick Gorres for assisting in writing and editing the manuscript. This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Glossary
- AMA1
apical membrane antigen 1
- cIEF
capillary isoelectric focusing
- LMIV
Laboratory of Malaria Immunology and Vaccinology
- Pf
Plasmodium falciparum
- RON2
Rhoptry neck protein 2
Footnotes
The authors have declared no conflict of interest.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
5 References
- [1].Ellis DR, Sagara I, Doumbo O, Wu Y, Hum. Vaccines 2010, 6, 627–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Srinivasan P, Beatty WL, Diouf A, Herrera R, Ambroggio X, Moch JK, Tyler JS, Narum DL, Pierce SK, Boothroyd JC, Haynes JD, Miller LH, Proc. Natl. Acad. Sci. U. S. A 2011, 108, 13275–13280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Normand BV, Tonkin ML, Lamarque MH, Langer S, Hoos S, Roques M, Saul FA, Faber BW, Bentley GA, Boulanger MJ, Lebrun M, PLoS Pathog 2012, 8, e1002755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Srinivasan P, Yasgar A, Luci DK, Beatty WL, Hu X, Andersen J, Narum DL, Moch JK, Sun H, Haynes JD, Maloney DJ, Jadhav A, Simeonov A, Miller LH, Nat. Commun 2013, 4, 2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Srinivasan P, Ekanem E, Diouf A, Tonkin ML, Miura K, Boulanger MJ, Long CA, Narum DL, Miller LH, Proc. Natl. Acad. Sci. U. S. A 2014, 111, 10311–10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Srinivasan P, Baldeviano GC, Miura K, Diouf A, Ventocilla JA, Leiva KP, Lugo-Roman L, Lucas C, Orr-Gonzalez S, Zhu D, Villasante E, Soisson L, Narum DL, Pierce SK, Long CA, Diggs C, Duffy PE, Lescano AG, Miller LH, NPJ Vaccines 2017, 2, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chrambach A, Rodbard D, Science 1971, 172, 440–451. [DOI] [PubMed] [Google Scholar]
- [8].Smith BJ, Methods Mol Biol 1994, 32, 23–34. [DOI] [PubMed] [Google Scholar]
- [9].Laemmli UK, Nature 1970, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- [10].Brunelle JL, Green R, Methods Enzymol 2014, 541, 151–159. [DOI] [PubMed] [Google Scholar]
- [11].Smith BJ, Methods Mol Biol 1984, 1, 41–55. [DOI] [PubMed] [Google Scholar]
- [12].Hjerten S, Zhu MD, J. Chromatogr 1985, 346, 265–270. [Google Scholar]
- [13].Rodriguez-Diaz R, Wehr T, Zhu MD, Electrophoresis 1997, 18, 2134–2144. [DOI] [PubMed] [Google Scholar]
- [14].Pergande MR, Cologna SM, Proteomes 2017, 5, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kilár F, Electrophoresis 2003, 24, 3908–3916. [DOI] [PubMed] [Google Scholar]
- [16].Silvertand LHH, Toraño JS, van Bennekom WP, de Jong GJ, J. Chromatogr. A 2008, 1204, 157–170. [DOI] [PubMed] [Google Scholar]
- [17].Vulliez-Le Normand B, Tonkin ML, Lamarque MH, Langer S, Hoos S, Roques M, Saul FA, Faber BW, Bentley GA, Boulanger MJ, Lebrun M, PLoS Pathog 2012, 8, e1002755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kennedy MC, Wang J, Zhang Y, Miles AP, Chitsaz F, Saul A, Long CA, Miller LH, Stowers AW, Infect. Immun 2002, 70, 6948–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kocken CH, van der Wel AM, Dubbeld MA, Narum DL, van de Rijke FM, van Gemert GJ, van der Linde X, Bannister LH, Janse C, Waters AP, Thomas AW, J. Biol. Chem 1998, 273,15119–15124. [DOI] [PubMed] [Google Scholar]
- [20].Invitrogen, Novex® Pre-Cast Gel Electrophoresis Guide: General information and protocols for using Novex® pre-cast gels, Pub No. MAN0003187 Part no. IM-1002, Invitrogen, Carlsbad, CA 2010. https://tools.thermofisher.com/content/sfs/manuals/electrophoresisguide_man.pdf. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


