Abstract
The first step in the degradation of dibenzofuran and dibenzo-p-dioxin by Sphingomonas sp. strain RW1 is carried out by dioxin dioxygenase (DxnA1A2), a ring-dihydroxylating enzyme. An open reading frame (fdx3) that could potentially specify a new ferredoxin has been identified downstream of dxnA1A2, a two-cistron gene (J. Armengaud, B. Happe, and K. N. Timmis, J. Bacteriol. 180:3954–3966, 1998). In the present study, we report a biochemical analysis of Fdx3 produced in Escherichia coli. This third ferredoxin thus far identified in Sphingomonas sp. strain RW1 contained a putidaredoxin-type [2Fe-2S] cluster which was characterized by UV-visible absorption spectrophotometry and electron paramagnetic resonance spectroscopy. The midpoint redox potential of this ferredoxin (E′0 = −247 ± 10 mV versus normal hydrogen electrode at pH 8.0) is similar to that exhibited by Fdx1 (−245 mV), a homologous ferredoxin previously characterized in Sphingomonas sp. strain RW1. In in vitro assays, Fdx3 can be reduced by RedA2 (a reductase similar to class I cytochrome P-450 reductases), previously isolated from Sphingomonas sp. strain RW1. RedA2 exhibits a Km value of 3.2 ± 0.3 μM for Fdx3. In vivo coexpression of fdx3 and redA2 with dxnA1A2 confirmed that Fdx3 can serve as an electron donor for the dioxin dioxygenase.
Sphingomonas sp. strain RW1 was isolated from the River Elbe in Germany on the basis of its ability to grow aerobically on dibenzofuran and dibenzo-p-dioxin as the sole sources of carbon and energy (35). The degradative pathways of these two compounds in RW1 have been extensively studied, resulting in the biochemical and genetic characterization of the key enzymes. The initial reaction is an oxygenolytic attack on the carbon atoms adjacent to the ether bridge, and it is carried out by a three-component dioxin dioxygenase system. Unlike the well-known class IIB and class III dioxygenases, which involve electrons provided by a Rieske-type [2Fe-2S] ferredoxin, the dioxin dioxygenase has been shown to be associated with a putidaredoxin-type [2Fe-2S] ferredoxin (8). The gene of this ferredoxin, designated fdx1, has been cloned and characterized (4), as has the gene of its cognate reductase (5). In this type of ferredoxin, the prosthetic group is coordinated with the polypeptide by four cysteine residues arranged in the pattern Cys-X5-Cys-X2-Cys-Xn-Cys and thus differs significantly from the Rieske-type [2Fe-2S] ferredoxins containing a [2Fe-2S] cluster bound to the polypeptide by two cysteine and two histidine residues. The nature and the position of the ligands influence the properties of the Fe-S cluster. The ferredoxins containing a Rieske-type cluster exhibit a high redox potential (−150 mV for ferredoxins associated with class IIB dioxygenases [19]), whereas putidaredoxin-type [2Fe-2S] ferredoxins exhibit lower redox potentials (4, 12, 13), and ferredoxins containing a plant-type [2Fe-2S] cluster, also bound to the polypeptide by four cysteines, have even lower potential (9, 14, 33).
In a recent report (2), we described the cloning and characterization of the cistrons coding for the two subunits of the dioxin dioxygenase. Surprisingly, an open reading frame encoding a putative polypeptide exhibiting similarity to Fdx1 is located 1,923 nucleotides downstream of these two cistrons (6). The product of this gene, Fdx3, is thus an alternative candidate for the electron donor of the dioxin dioxygenase. In order to facilitate the isolation and biochemical characterization of the fdx3 product, an expression system was designed to allow purification from Escherichia coli of a recombinant holoferredoxin. The biochemical, spectroscopic, and redox properties of this third ferredoxin identified in Sphingomonas sp. strain RW1 are presented in this study and compared with those of Fdx1.
MATERIALS AND METHODS
Bacterial strains, plasmids, and reagents.
Restriction enzymes and reagents for genetic procedures were purchased from New England Biolabs, Boehringer Mannheim, Promega, United States Biochemicals, and Sigma. The medium used for bacterial growth was Luria-Bertani medium. The bacterial strains, plasmids, and oligonucleotides used in this study are listed in Table 1 and 2.
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or characteristics | Source or reference |
|---|---|---|
| Strains | ||
| BL21(DE3) | F−ompT [lon] hsdSB (rB− mB−; an E. coli B strain) with DE3 prophage | Novagen |
| DH5α | F−endA1 hsdR17(rK− mK+) supE44 thi-1 λ− recA1 gyrA96 relA1 Δ(argF-lacZYA) U169 φ80d lacZΔM15 | Gibco BRL |
| Plasmids | ||
| pBluescript II KS | Amprlac′IPOZ′, expression vector | Stratagene |
| pET9a | Kmr, T7 RNA polymerase promoter, expression vector | Novagen |
| pGEM-T | Ampr, T7 RNA polymerase promoter, T-cloning vector | Promega |
| pAJ104 | Kmr, expression vector derived from pET9a encompassing fdx1 | 4 |
| pAJ108 | Tcr, cosmid vector derived from pLARF3 encompassing redA2 | 5 |
| pAJ111 | Kmr, expression vector derived from pET9a encompassing redA2 | 5 |
| pAJ114 | Tcr, cosmid vector derived from pLARF3 encompassing fdx3 | 2 |
| pAJ127 | Kmr, expression vector derived from pBBR1-MCS encompassing dxnA1A2 | 2 |
| pAJ128 | 1.29-kb redA2 EcoRI-NotI PCR-amplified fragment into pGEM-T | 2 |
| pAJ129 | 0.33-kb fdx1 NotI-BamHI PCR-amplified fragment into pGEM-T | 2 |
| pAJ130 | Inserts from pAJ129 and pAJ128 cloned into pBluescript | 2 |
| pAJ146 | 0.34-kb fdx3 NdeI-BamHI PCR-amplified fragment into pGEM-T | This study |
| pAJ147 | 0.33-kb fdx3 NdeI-BamHI fragment from pAJ146 cloned into pET9a | This study |
| pAJ148 | 0.56-kb fdx3 NotI-BamHI PCR-amplified fragment into pGEM-T | This study |
| pAJ149 | Inserts from pAJ148 and pAJ128 cloned into pBluescript | This study |
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Primer sequencea | Hybridization region |
|---|---|---|
| AJ286 | cgggcatATGCCCAAAGTAATTTATG | 5′ end of fdx3 |
| NdeI | ||
| AJ287 | gcgcggatccGTTCAATCAAGATTGTTG | 3′ end of fdx3 |
| BamHI | ||
| AJ316 | ccccggatccatgatgcggccGCCAGATTCTAGACCGGGATCAG | 3′ end of redA2 |
| BamHINotI | ||
| AJ317 | cgcggaaTTCTGGAAGAGAAGGAAAG | 5′ end of redA2 |
| EcoRI | ||
| AJ320 | ccgggcggccGCTCAATCTGATGCACCA | 5′ end of fdx3 |
| NotI | ||
| AJ321 | cgccggaTCCCATTAGAGATTACATCT | 3′ end of fdx3 |
| BamHI |
Relevant recognition sequences for the restriction enzymes are underlined, and lowercase characters indicate the nonhomologous sequences to the corresponding DNA region from Sphingomonas sp. strain RW1.
Construction of the expression system pAJ147.
Standard DNA manipulations were carried out as described by Sambrook et al. (30). NdeI and BamHI restriction sites were introduced by PCR amplification at the 5′ and 3′ ends of the fdx3 gene, respectively. Oligonucleotides AJ286 and AJ287 were synthesized for this purpose on an Applied Biosystems DNA synthesizer model 381A and used as primers without further purification. The PCR was performed as previously described for the amplification of another ferredoxin gene (3). Cosmid pAJ114 (2), isolated and purified with the Qiawell-8 kit (Qiagen), was used as a PCR template. A fragment of the predicted size (340 bp) was purified from a 2% agarose gel by means of a QiaexII extraction kit (Qiagen) and cloned into pGEM-T plasmid (Promega) to produce plasmid pAJ146. The integrity of the insert was verified by restriction analysis with NotI-PstI double digestion, followed by DNA sequencing. Nucleotide sequencing was performed on a double-stranded template, prepared by means of a Qiawell-8 kit (Qiagen). Sequencing reactions were carried out with an Applied Biosystem DNA sequencing kit in the presence of 5% dimethyl sulfoxide, and processed on a 373 Stretch Applied Biosystems automated sequencer. The 326-bp fragment encompassing fdx3 was excised from pAJ146 by restriction with NdeI and BamHI, purified, and ligated into the expression plasmid pET9a, which had been digested with the same endonucleases. The plasmid generated, pAJ147, contains the ferredoxin gene inserted downstream of the Φ10 promoter.
Expression of fdx3 and purification of the recombinant ferredoxin.
Plasmid pAJ147 was introduced into E. coli BL21(DE3). The resulting strain was cultivated at 30°C in LB medium supplemented with 50 μM Fe2(SO4)3 and 50 μg of kanamycin per ml. Expression of fdx3 was induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) when cultures reached an optical density at 600 nm of 0.8. Cells from 6 liters of culture were harvested 3 h after induction and washed with 0.5 liters of 50 mM Tris-HCl–15 mM EDTA (pH 8.0). The cell paste obtained after centrifugation was frozen in liquid nitrogen and kept at −70°C. Purification of the ferredoxin was carried out according to a protocol based on that described for the purification of Fdx1 (4). Briefly, the cell pellet was thawed on ice and then resuspended in 110 ml of 100 mM Tris-HCl–15 mM EDTA (pH 8.0), containing 1 mM dithiothreitol. All steps were subsequently performed at 4°C. Cells were broken by means of a French pressure cell (Aminco Corp.) operated as recommended by the supplier at a pressure of 1,000 lb/in2. The crude extract was centrifuged at 18,000 × g for 30 min. The resulting supernatant fluid was diluted twofold in 25 mM Tris-HCl–1 mM EDTA (pH 8.0) containing 1 mM dithiothreitol (buffer A) and loaded onto a 20-ml Fast-flow DEAE-Sepharose mixture (Amersham Pharmacia Biotech) poured into a Φ 2.5-cm-diameter Econo-column (Bio-Rad) previously equilibrated in buffer A. After a 2-column-volume wash with buffer A containing 100 mM NaCl, a reddish protein fraction was eluted with buffer A containing 500 mM NaCl. The DEAE fraction was concentrated to 3 ml by ultrafiltration with an Amicon filtration unit, loaded onto a 120-ml Superdex 200 gel filtration Φ 1.6-cm-diameter column (Amersham Pharmacia Biotech), and eluted with buffer A containing 100 mM NaCl at a flow rate of 0.5 ml/min. The red fraction collected in a 6-ml volume was concentrated a second time with an Amicon filtration unit and desalted by repeated cycles of concentration and dilution. Finally, the protein was applied to a 5-ml Mono-Q (Amersham Pharmacia Biotech) Φ 1.0-cm-diameter Econo-column (Bio-Rad) which was developed with a 30-min linear gradient from 0 to 500 mM NaCl in buffer A by using an Amersham Pharmacia Biotech fast protein liquid chromatography system at a flow rate of 1 ml/min. Fdx3 eluted at approximately 250 mM NaCl and was desalted by dialysis against buffer A. The purified protein was concentrated on an Amicon filtration unit and stored in liquid N2.
Analytical methods.
Protein concentration determination, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), molecular weight determination, UV-visible absorption spectrophotometry, electron paramagnetic resonance (EPR) spectroscopy, and electrospray mass spectrometric measurements were carried out as previously described (1, 3). The redox potential of the [2Fe-2S] cluster of the recombinant Fdx3 was determined spectrophotometrically with 5-deazariboflavin as a photoreductant and phenosafranin as a redox indicator. By means of the Nernst equation, the intermediate redox potentials were determined by stepwise photoreduction of Fdx3 at 25°C (pH 8.0) inside an anaerobic glove box and measurement of the extent of phenosafranin reduction, as A250, and the ratio of the oxidized and reduced forms of Fdx3, calculated from the absorption at 410 nm. Cuvettes of a 0.2-cm light path contained 8 μM 5-deazariboflavin, 16 mM sodium oxalate, 52.6 μM phenosafranin, and 100 μM Fdx3 in 25 mM potassium phosphate buffer (pH 8.0).
In vitro electron transfer assays.
Recombinant reductase RedA2 and ferredoxin Fdx1, used as a control, were purified as previously described (5) with E. coli BL21(DE3)(pAJ111) and E. coli BL21(DE3)(pAJ104) cells, respectively. RedA2 activity was routinely assayed as reduction of 2,6-dichlorophenolindophenol (Cl2IND) at 25°C, monitored at 600 nm in a UV-2401 PC Shimadzu UV-visible recording spectrophotometer under the same conditions as previously reported (5) and assuming a molar absorption coefficient of ɛ600nm = 20.6 mM−1 · cm−1 (10). The assay system contained 100 μM Cl2IND, 150 μM NADH, and 6.8 nM RedA2 in 50 mM Tris-HCl (pH 8.0). In addition, ferredoxin-dependent cytochrome c reduction was measured by the decrease in A550 by using a molar absorption coefficient of ɛ550nm = 20 mM−1 · cm−1 (10). In this case, the reaction mixture (0.5 ml) contained 50 μM cytochrome c, 150 μM NADH, various amounts (from 2 to 20 μM) of ferredoxin Fdx3 (or Fdx1 as a control), and 22.7 nM RedA2 in 50 mM Tris-HCl (pH 8.0). The reductase was shown to not directly reduce cytochrome c under these conditions. The Km value for Fdx3 and corresponding standard deviation were determined from a linear regression algorithm of Lineweaver-Burk plots made in duplicate from independent kinetic data obtained at 25°C. RedA2 activity was measured as a function of NADH concentration in the presence of Cl2IND as an electron acceptor and as a function of Fdx3 concentration in the cytochrome c-Fdx3-dependent reduction.
Construction of a redA2/fdx3 expression vector and resting cell assays.
A 1,289-bp fragment containing the redA2 gene and 23 upstream nucleotides, which includes the putative ribosome binding site of redA2, was PCR amplified from cosmid pAJ108 with the oligonucleotides AJ317 and AJ316, which introduced EcoRI and NotI restriction sites at the 5′ and 3′ ends of this fragment, respectively. A fragment of the predicted size was purified from a 1% agarose gel and cloned into the pGEM-T plasmid to generate plasmid pAJ128. A 555-bp fragment containing the fdx3 gene was also PCR amplified from cosmid pAJ114 by means of oligonucleotides AJ320 and AJ321, which introduced NotI and BamHI sites at the 5′ and 3′ ends of the fragment, respectively. A fragment of the predicted size was purified from a 1% agarose gel and cloned into pGEM-T plasmid to generate plasmid pAJ148. The integrity of the two PCR-amplified fragments was checked by sequencing both strands. Plasmids pAJ128 and pAJ148 were digested with NotI and EcoRI and with NotI and BamHI, respectively, and the resulting 1,262- and 541-bp fragments were purified and ligated to EcoRI- and BamHI-cleaved plasmid pBluescript II KS to produce plasmid pAJ149 carrying the redA2 and fdx3 genes expressed from the Plac promoter.
Dioxin dioxygenase activity in resting cells of E. coli DH5α(pAJ127)(pAJ149), with E. coli DH5α(pAJ127)(pAJ130) as a control, cultured in LB medium containing 30 μg of kanamycin per ml and 100 μg of ampicillin per ml was measured under conditions identical to those previously described (2). Relative activities correspond to the average of activities measured in duplicate assays after 1, 2, and 3 h of incubation.
Secondary structural element prediction and three-dimensional structure modeling.
Prediction of secondary structural elements of Fdx1 and Fdx3 was carried out by using the Jpred2 resources (a consensus method for protein secondary structure prediction) of the EMBL-European BioInformatics Institute (http://jura.ebi.ac.uk:8888/). A three-dimensional structural model was calculated according to the method of Lund et al. (20) by using the CPHmodels resources from the Center for Biological Sequence analysis (http://genome.cbs.dtu.dk/services/CPHmodels) from the amino acid sequences of Fdx1 and Fdx3 with the putidaredoxin three-dimensional structure (28) as a template. The three-dimensional structures were viewed with WebLab ViewerPro 3.0 software (Molecular Simulations, Inc.).
RESULTS
Expression of the fdx3 gene and purification of the product.
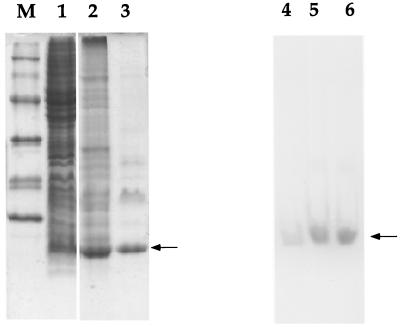
Cells of E. coli strain BL21(DE3) carrying plasmid pAJ147, which contains the fdx3 gene downstream of a T7 expression system and optimized translational signals, were induced with IPTG for 3 h. Analysis of cell extracts by SDS-PAGE revealed the accumulation of a polypeptide with an apparent molecular mass lower than 14 kDa (Fig. 1, lane 1). The recombinant ferredoxin was also seen as a brown band migrating close to the buffer front in a 10% native polyacrylamide gel (Fig. 1, lane 4), which suggested that this protein was produced in a holoform. Recombinant Fdx3 was purified by a simple three-step purification protocol, previously used for Fdx1, a protein exhibiting a similar theoretical isoelectric point and molecular weight (4), and consisting of DEAE-Sepharose chromatography followed by gel filtration on a Superdex 200 Prep Grade column and Mono-Q chromatography. One major contaminant protein, 18 kDa in size, was still present at this stage (Fig. 1, lane 3) and could not be removed without a dramatic decrease in the holo-Fdx3/apo-Fdx3 ratio. The purification yielded approximately 2.3 mg of recombinant Fdx3 per liter of culture.
FIG. 1.
SDS-PAGE and native PAGE analyses of purification fractions of recombinant Fdx3. Protein samples were separated by electrophoresis on a 15% Tricine–polyacrylamide gel and stained with Coomassie blue (lanes M, 1, 2, and 3) and on a 10% Tris–borate–polyacrylamide gel (lanes 4, 5, and 6). On the nondenaturing gel, holoferredoxins were observed without staining. Lanes: M, protein standards (lysozyme, 14.3 kDa; trypsin inhibitor, 21.5 kDa; carbonic anhydrase, 30 kDa; ovalbumin, 46 kDa; bovine serum albumin, 66 kDa; phosphorylase b, 97.4 kDa); 1, E. coli BL21(DE3)(pAJ147) crude extract; 2, Fast-Flow DEAE-cellulose fraction eluted with high-ionic buffer; 3, Fdx3 fraction eluted from Mono-Q; 4, E. coli BL21(DE3)(pAJ147) crude extract concentrated five times by ultrafiltration with an Amicon filtration unit; 5, purified recombinant Fdx1; 6, purified recombinant Fdx3.
Biochemical and spectroscopic characterization of recombinant Fdx3. (i) Biochemical properties.
The purified recombinant Fdx3 was subjected to electrospray mass spectrometry in the positive mode. The average mass of the apoferredoxin (11,259 ± 3 Da) determined by electrospray ionization mass spectrometry matches within 3 Da the expected mass calculated from the nucleotide-derived amino acid sequence with the initial methionine removed (11,262 Da). The removal of the initial methionine was expected (the second amino acid residue in the polypeptide chain is a proline, a small-side-chain-length residue [10]), despite the high levels of expression, because efficient processing of the N-terminal amino acid has been previously observed in E. coli in such cases (32). The molecular mass of the purified Fdx3 was determined to be 13 ± 2 kDa by gel filtration on a calibrated Superdex 200 Prep Grade chromatography column. Therefore, this ferredoxin behaves as a monomeric protein under the conditions employed. The recombinant Fdx3 exhibits behavior similar to that of Fdx1 (Fig. 1, lanes 5 and 6) during electrophoresis on a 10% polyacrylamide gel, under nondenaturing conditions. In that case, both proteins (which have close molecular masses), present very theoretical acidic isoelectric points (3.9 to 4.0) and migrate close to the buffer front. They thus have similar physicochemical properties.
(ii) Spectroscopic properties.
The UV-visible absorption spectrum of recombinant Fdx3 in the oxidized state shows the general features of a ferredoxin containing a [2Fe-2S] cluster. In the native state, broad charge transfer bands centered around 314, 412, and 460 nm were evident. These maxima are typical of ferredoxins containing a putidaredoxin-type [2Fe-2S] cluster and differ slightly from those of ferredoxins containing a plant-type [2Fe-2S] cluster characterized by a maximum at 420 nm rather than 414 nm. They strongly differ from those of ferredoxins containing a Rieske-type [2Fe-2S] cluster, which exhibit maxima at 325 and 460 nm with a broad shoulder around 575 nm (11, 31), or containing a [3Fe-4S] or [4Fe-4S] cluster, which are both characterized by broad charge transfer bands around 390 to 410 and 280 to 300 nm (15, 16). Upon reduction with excess dithionite, the recombinant ferredoxin changes color, and the UV-visible absorption spectrum shows only one maximum, centered at 538 nm, as is the case with other reduced ferredoxins containing a [2Fe-2S] cluster (3).
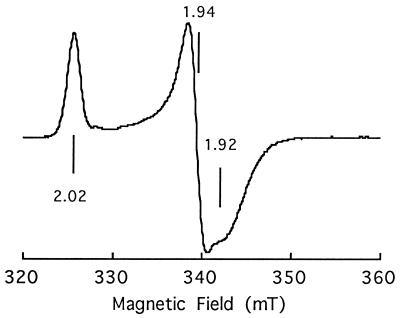
The [2Fe-2S] cluster in Fdx3 was further characterized by EPR spectroscopy. In the native state, the recombinant ferredoxin is EPR silent, whereas in the dithionite-reduced state, a nearly axial signal with apparent g values at 1.92, 1.94, and 2.02 was evident (Fig. 2). This EPR signal is typical of a ferredoxin containing a putidaredoxin-type [2Fe-2S] cluster. A comparison of the EPR spectra of reduced Fdx1 and Fdx3 shows a very minor difference at high field strength, the Fdx1 signal being slightly more axial, suggesting that the geometries of their Fe-S clusters are highly similar. This indicates that the environments of the Fe-S cluster are probably nearly the same in both proteins.
FIG. 2.
EPR analysis of recombinant Fdx3. EPR spectrum of reduced Fdx3 was recorded at 10 K, with a modulation frequency of 100 kHz and a modulation amplitude of 1 mT. The protein concentration was 170 μM in 20 mM Tris-HCl (pH 7.5) containing 50 mM NaCl. It was fully reduced with an excess (5 reducing equivalents) of sodium dithionite (1 mM) under anaerobic conditions (monitored by optical absorption). The spin concentration was estimated to be 250 μM by integration of the EPR signal and comparison with a reference.
Electron transfer properties of recombinant Fdx3. (i) Determination of the midpoint redox potential.
The redox potential of the [2Fe-2S] cluster of the recombinant Fdx3 was determined spectrophotometrically as described in Materials and Methods. The midpoint redox potential obtained was E0′ = 247 (± 10) mV versus that of the normal hydrogen electrode. This potential is in the range of values reported for homologous ferredoxins (4, 12, 13), except for the hydrogenosomal [2Fe-2S] ferredoxin from the protozoan Trichomonas vaginalis, which exhibits an atypical redox potential of −347 mV (34).
Cytochrome c-mediated reduction.
Purified recombinant RedA2 flavoprotein exhibited a specific activity of 95 μmol · min−1 · mg−1 in the Cl2IND reduction assay and 0.54 μmol · min−1 · mg−1 in the Fdx3-dependent cytochrome c reduction assay. The specific activity of RedA2 in the same assay carried out with Fdx1 is similar (0.55 μmol · min−1 · mg−1). The Km value for Fdx3 was determined to be 3.2 ± 0.3 μM, slightly lower than that recorded for Fdx1, 3.7 ± 0.4 μM, but statistically, the measured values were not significantly different.
The recombinant Fdx3 is able in vivo to transfer electrons to the dioxin dioxygenase.
The dioxin dioxygenase DxnA1A2 has been previously shown to function efficiently with Fdx1 and RedA2, when expressed from a two-plasmid expression system in E. coli DH5α cells (2). We designed a new two-expression system by using the pAJ127 plasmid containing the cistrons of the α and β subunits and the pAJ149 plasmid containing the genes of the Fdx3 ferredoxin and the RedA2 reductase from Sphingomonas sp. strain RW1. These two plasmids are derivatives of pBBR1-MCS2 and pBluescript, respectively, and are therefore compatible with one another. The levels of expression of dxnA1, like those of dxnA2, in both strains were estimated to be similar. Dioxin dioxygenase activity in the Fdx3 strain E. coli DH5α(pAJ127)(pAJ149) and in the Fdx1 strain E. coli DH5α(pAJ127)(pAJ130) was measured as conversion of dibenzofuran and dibenzo-p-dioxin to 2,2′,3-trihydroxybiphenyl (2,2′,3-THB) and 2,2′,3-trihydroxydiphenyl-ether (2,2′,3-THD-ether), respectively. Both bacteria exhibited similar activities toward both substrates (for the Fdx3 strain, 17 ± 4 nmol of 2,2′,3-THB/min/mg of protein formed from dibenzofuran and 16 ± 4 nmol of 2,2′,3-THD-ether/min/mg of protein formed from dibenzo-p-dioxin). In all cases, only one product was formed by the dioxygenase from each substrate tested: namely 2,2′,3-THB from dibenzofuran, with UV-visible maxima at 206, 244, and 283 nm; and 2,2′,3-THD-ether from dibenzo-p-dioxin, with maxima at 202 and 275 nm, both of which are produced via an angular attack at carbons involved in the bridges of the multi-ring aromatic molecules.
Predicted secondary structural elements and three-dimensional structure modeling.
At present some experimental three-dimensional data for [2Fe-2S] putidaredoxin-type ferredoxins are available: two models for the solution structure of putidaredoxin (27) and terpredoxin (21) were obtained from nuclear magnetic resonance data, and the structure of a truncated version of bovine adrenodoxin (Adx4-108) was obtained after crystallization and X-ray diffraction analysis (22). A multiple alignment of sequences homologous to Fdx1 and Fdx3 and a prediction of the secondary structure of these ferredoxins were obtained by a consensus method (http://jura.ebi.ac.uk:8888/). The predicted presence of three α-helices and several β-sheets in these two metalloproteins is in agreement with the secondary structural elements observed in putidaredoxin and adrenodoxin structures (23). This indicates that the global folds among all the putidaredoxin-type ferredoxins should be close, the [2Fe-2S] cluster environment being the most similar. In order to spatially locate the 25 amino acids differing in these two ferredoxins, three-dimensional structural models of Fdx1 and Fdx3 were designed by comparative modeling with putidaredoxin structure coordinates as a template, using a fully automated structure prediction web server (20). Fourteen out of these 25 residues are conservatively replaced and can be considered to not drastically modify the structure and therefore the function of the protein. The 11 nonconserved residues are indicated in the Fdx1/Fdx3 model (Fig. 3). The major differences between Fdx1 and Fdx3 are located on the molecular surface formed by juxtaposition of an N-terminal loop with the C terminus of the molecule, the position of which is probably influenced by the residue at position 102 (a proline in Fdx1 replaced by a serine in Fdx3). The global charge distributions in this region are similar in both proteins in accordance with the electric dipole vector configuration detected in [2Fe-2S] ferredoxins (23). However, local changes should modify the contact sites with possible interacting partners. (Two negatively charged residues, one positively charged residue, and one neutral residue in Fdx1 are modified for one negatively charged residue and three neutral residues in Fdx3.) The second molecular surface containing four nonconserved residues (residues 59, 63, 65, and 67) is located opposite the former, the accessible surface being less polar in Fdx1 than in Fdx3 (Fig. 3). These differences, except G59S, do not affect the two short α-helices (the bottom of the model presented in Fig. 3) known to be implemented in the interaction between putidaredoxin and putidaredoxin reductase (28). These differences should reflect the functional importance of these molecular surfaces. This structural model indicates that the two ferredoxins should function with very similar mechanisms, but may differ in at least one of their physiological partners.
FIG. 3.
Three-dimensional structure model of Fdx1 and Fdx3. The two models calculated with the CPHmodels resources from the Center for Biological Sequence analysis with the sequences of Fdx1 and Fdx3 were almost identical. The solid ribbon drawing was prepared with WebLab ViewerPro 3.0 software from Molecular Simulations, Inc. Residue side chains of Fdx1 were included in the drawing. Residues liganding the [2Fe-2S] cluster and key residues discussed in the text are indicated by peach and light green background colors, respectively. P102S indicates that a proline in Fdx1 is replaced by a serine in Fdx3.
DISCUSSION
The initial step in dibenzo-p-dioxin and dibenzofuran degradation by Sphingomonas sp. strain RW1 is carried out by a class IIA dioxygenase system. This is a multicomponent enzyme that functions with a reductase and a ferredoxin. From RW1 bacteria grown on salicylate, a monocyclic intermediate compound in the dibenzofuran degradation pathway, Bünz and Cook (8) have previously purified a ferredoxin (Fdx1) able to transfer electrons to the dioxin dioxygenase DxnA1A2 and two reductases (RedA1 and RedA2) which can transfer electrons from NADH to this ferredoxin. A new ferredoxin gene (fdx3) was recently identified downstream of the dxnA1A2 genes encoding the dioxygenase (6). The expression system based on the T7 promoter we developed and the three-step purification procedure described in this report allowed us to obtain milligram quantities of recombinant Fdx3 holoferredoxin. EPR analysis showed it to contain a putidaredoxin-type [2Fe-2S] cluster. Analysis of the redox properties of this new ferredoxin confirmed the strong similarity of Fdx3 and Fdx1 in the [2Fe-2S] cluster environment, even though their polypeptide chains differ in 25 of their 105 amino acids. A detailed comparison of Fdx1 and Fdx3 by using a three-dimensional model based on putidaredoxin structure confirms this point and pinpoints the major differences at two molecular surfaces on the proteins. This indicates that the two ferredoxins may differ in at least one of their physiological partners.
The Fdx1 ferredoxin was shown to represent up to 5% of total protein in exponentially growing bacteria, and Fdx1 was thus concluded to be the electron donor of the dioxin dioxygenase (8). However, we show here that Fdx3 has biochemical, spectroscopic, and redox properties strikingly similar to those of Fdx1. The two isoferredoxins are able to convey electrons from the reductase RedA2 to the dioxygenase DxnA1A2, but the exact nature of the partners of the two ferredoxins in Sphingomonas sp. strain RW1 remains to be clarified. The Fdx3 ferredoxin from Sphingomonas sp. strain RW1 shares 35 to 45% identity with a ferredoxin from Rhodococcus sp. strain NI86/21 (24), the putidaredoxin from Pseudomonas putida (17, 25), and the terpredoxin from a Pseudomonas strain (26). These three ferredoxins are involved in electron transfer to monooxygenases involved in the degradation of S-ethyl-dipropyl-carbamothioate, camphor, and α-terpineol, respectively. In these cases, the ferredoxin genes are located just upstream (rhodocoxin) or downstream (putidaredoxin and terpredoxin) of the gene of the cognate reductase and are probably cotranscribed. Tentative identification of the physiological partner of the reductase or the ferredoxin is therefore straightforward. In Sphingomonas sp. strain RW1, however, the genes encoding Fdx3 and Fdx1 are not located in the neighborhood of a reductase or oxidoreductase, and hence genetic evidence for partnership is lacking. The generation of knockout mutants will probably not clarify this point, since both the two ferredoxins and the two reductases have similar properties and thus constitute genetic redundancy. The existence in some bacteria of a multiple catabolic enzyme system, such as ring-cleavage dioxygenases—three in the polychlorobiphenyl-degrading bacterium Rhodococcus globerulus P6 (7) and up to seven in Rhodococcus erythropolis TA421 (18)— and of several ring-hydroxylating dioxygenases—at least four in Sphingomonas sp. strain RW1 (2) and five in Sphingomonas aromaticivorans strain F199 (29)—is now well established. Such a diversity of isoenzymes in the same organism is often interpreted to reflect a broader spectrum of substrates that the microorganism can handle under disparate environmental conditions than could be metabolized by a pathway containing unique enzymes. However, the presence in Sphingomonas sp. strain RW1 of two complete electron transport chains composed of two isoferredoxins (Fdx1 and Fdx3) and two isoreductases (RedA1 and RedA2) seems to be redundant, because they appear to be biochemically equivalent, at least as far as electron transfer to the dioxin dioxygenase is concerned. Whether or not these two systems are truly redundant, or each serves one function but lacks a distinct metabolic role, should be resolved by estimation of in vivo concentrations of Fdx1 and Fdx3 in Sphingomonas sp. strain RW1 and analysis of the regulation of their expression.
ACKNOWLEDGMENTS
We gratefully acknowledge Y. Jouanneau (CEA-Grenoble, Grenoble, France) for welcoming us to his team and allowing us to use his facilities for redox potential measurements.
This research was funded in part by the German Ministry for Education and Research (BMBF grant no. 0318896C). K. N. Timmis gratefully acknowledges the generous support of the Fonds der Chemischen Industrie.
REFERENCES
- 1.Armengaud J, Gaillard J, Forest E, Jouanneau Y. Characterization of a 2[4Fe-4S] ferredoxin obtained by chemical insertion of the Fe-S clusters into the apoferredoxin II from Rhodobacter capsulatus. Eur J Biochem. 1995;231:396–404. doi: 10.1111/j.1432-1033.1995.tb20712.x. [DOI] [PubMed] [Google Scholar]
- 2.Armengaud J, Happe B, Timmis K N. Genetic analysis of dioxin dioxygenase of Sphingomonas sp. strain RW1: catabolic genes dispersed on the genome. J Bacteriol. 1998;180:3954–3966. doi: 10.1128/jb.180.15.3954-3966.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armengaud J, Meyer C, Jouanneau Y. Recombinant expression of the fdxD gene of Rhodobacter capsulatus and characterization of its product, a [2Fe-2S] ferredoxin. Biochem J. 1994;300:413–418. doi: 10.1042/bj3000413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armengaud J, Timmis K N. Molecular characterization of Fdx1, a putidaredoxin-type [2Fe-2S] ferredoxin able to transfer electrons to the dioxin dioxygenase of Sphingomonas sp. RW1. Eur J Biochem. 1997;247:833–842. doi: 10.1111/j.1432-1033.1997.00833.x. [DOI] [PubMed] [Google Scholar]
- 5.Armengaud J, Timmis K N. The reductase RedA2 of the multicomponent dioxin dioxygenase system of Sphingomonas sp. RW1 is related to class-I cytochrome P450-type reductases. Eur J Biochem. 1998;253:437–444. doi: 10.1046/j.1432-1327.1998.2530437.x. [DOI] [PubMed] [Google Scholar]
- 6.Armengaud J, Timmis K N, Wittich R-M. A functional 4-hydroxysalicylate/hydroxyquinol degradative pathway gene cluster is linked to the initial dibenzo-p-dioxin pathway genes in Sphingomonas sp. strain RW1. J Bacteriol. 1999;181:3452–3461. doi: 10.1128/jb.181.11.3452-3461.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asturias J A, Timmis K N. Three different 2,3-dihydroxybiphenyl-1,2-dioxygenase genes in the gram-positive polychlorobiphenyl-degrading bacterium Rhodococcus globerulus P6. J Bacteriol. 1993;175:4631–4640. doi: 10.1128/jb.175.15.4631-4640.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bünz P V, Cook A M. Dibenzofuran 4,4a-dioxygenase from Sphingomonas sp. strain RW1: angular dioxygenation by a three-component enzyme system. J Bacteriol. 1993;175:6467–6475. doi: 10.1128/jb.175.20.6467-6475.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cammack R, Rao K K, Bargeron C P, Hutson K G, Andrew P W, Rogers L J. Midpoint redox potentials of plants and algal ferredoxins. Biochem J. 1977;168:205–209. doi: 10.1042/bj1680205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalboge H, Bayne S, Pedersen J. In vivo processing of N-terminal methionine in E. coli. FEBS Lett. 1990;266:1–3. doi: 10.1016/0014-5793(90)90001-b. [DOI] [PubMed] [Google Scholar]
- 11.Haddock D, Peletier D A, Gibson D T. Purification and properties of ferredoxinBPH, a component of biphenyl 2,3-dioxygenase of Pseudomonas sp. strain LB400. J Ind Microbiol Biotechnol. 1997;19:355–359. doi: 10.1038/sj.jim.2900429. [DOI] [PubMed] [Google Scholar]
- 12.Huang Y Y, Hsu D K, Kimura T. The effect of pH on the formal reduction potential of adrenodoxin in the presence and absence of adrenodoxin reductase: the implication in the electron transfer mechanism. Biochem Biophys Res Commun. 1983;115:116–122. doi: 10.1016/0006-291x(83)90977-4. [DOI] [PubMed] [Google Scholar]
- 13.Huang Y Y, Kimura T. Reduction potential and thermodynamic parameters of adrenodoxin by the use of an anaerobic thin-layer electrode. Anal Biochem. 1983;133:385–393. doi: 10.1016/0003-2697(83)90099-4. [DOI] [PubMed] [Google Scholar]
- 14.Hugo N, Armengaud J, Gaillard J, Timmis K N, Jouanneau Y. A novel [2Fe-2S] ferredoxin from Pseudomonas putida mt2 promotes the reductive reactivation of catechol 2,3-dioxygenase. J Biol Chem. 1998;273:9622–9629. doi: 10.1074/jbc.273.16.9622. [DOI] [PubMed] [Google Scholar]
- 15.Jouanneau Y, Meyer C, Gaillard J, Forest E, Gagnon J. Purification and characterization of a novel dimeric ferredoxin (FdIII) from Rhodobacter capsulatus. J Biol Chem. 1993;268:10636–10644. [PubMed] [Google Scholar]
- 16.Jouanneau Y, Meyer C, Gaillard J, Vignais P M. Purification and characterization of a 7Fe-ferredoxin from Rhodobacter capsulatus. Biochem Biophys Res Commun. 1990;171:273–279. doi: 10.1016/0006-291x(90)91388-9. [DOI] [PubMed] [Google Scholar]
- 17.Koga H, Yamaguchi E, Matsunaga K, Aramaki H, Horiuchi T. Cloning and nucleotide sequences of NADH-putidaredoxin reductase gene (camA) and putidaredoxin gene (camB) involved in cytochrome P-450cam hydroxylase of Pseudomonas putida. J Biochem (Tokyo) 1989;106:831–836. doi: 10.1093/oxfordjournals.jbchem.a122939. [DOI] [PubMed] [Google Scholar]
- 18.Kosono S, Maeda M, Fuji F, Arai H, Kudo T. Three of the seven bphC genes of Rhodococcus erythropolis TA421, isolated from a termite ecosystem, are located on an indigenous plasmid associated with biphenyl degradation. Appl Environ Microbiol. 1997;63:3282–3285. doi: 10.1128/aem.63.8.3282-3285.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Link T A, Hatzfeld O M, Unalkat P, Shergill J K, Cammack R, Mason J R. Comparison of the “Rieske” [2Fe-2S] center in the bc1 complex and in bacterial dioxygenases by circular dichroism spectroscopy and cyclic voltammetry. Biochemistry. 1996;35:7546–7552. doi: 10.1021/bi960004+. [DOI] [PubMed] [Google Scholar]
- 20.Lund O, Frimand K, Gorodkin J, Bohr H, Bohr J, Hansen J, Brunak S. Protein distance constraints predicted by neural networks and probability density functions. Protein Eng. 1997;10:1241–1248. doi: 10.1093/protein/10.11.1241. [DOI] [PubMed] [Google Scholar]
- 21.Mo H, Pochapsky S S, Pochapsky T C. A model for the solution structure of oxidized terpredoxin, a 2Fe-2S ferredoxin from Pseudomonas. Biochemistry. 1999;38:5666–5675. [Google Scholar]
- 22.Müller A, Müller J J, Müller Y A, Uhlmann H, Bernhardt R, Heinemann U. New aspects of electron transfer revealed by the crystal structure of a truncated bovine adrenodoxin, Adx(4-108) Structure. 1998;15:269–280. doi: 10.1016/s0969-2126(98)00031-8. [DOI] [PubMed] [Google Scholar]
- 23.Müller J J, Müller A, Rottmann M, Bernhardt R, Heinemann U. Vertebrate-type and plant-type ferredoxins: crystal structure comparison and electron transfer pathway modelling. J Mol Biol. 1999;294:501–513. doi: 10.1006/jmbi.1999.3253. [DOI] [PubMed] [Google Scholar]
- 24.Nagy I, Schoofs G, Compernolle F, Proost P, Vanderleyden J, de Mot R. Degradation of the thiocarbamate herbicide EPTC (S-ethyl dipropylcarbamothioate) and biosafening by Rhodococcus sp. strain NI86/21 involve an inducible cytochrome P-450 system and aldehyde dehydrogenase. J Bacteriol. 1995;177:676–687. doi: 10.1128/jb.177.3.676-687.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peterson J A, Lorence M C, Amarneh B. Putidaredoxin reductase and putidaredoxin. Cloning, sequence determination, and heterologous expression of the proteins. J Biol Chem. 1990;265:6066–6073. [PubMed] [Google Scholar]
- 26.Peterson J A, Lu J Y, Geisselsoder J, Graham-Lorence S, Carmona C, Witney F, Lorence M C. Cytochrome P-450terp. Isolation and purification of the protein and cloning and sequencing of its operon. J Biol Chem. 1992;267:14193–14203. [PubMed] [Google Scholar]
- 27.Pochapsky T C, Jain N U, Kuti M, Lyons T A, Heymont J. A refined model for the solution structure of oxidized putidaredoxin. Biochemistry. 1999;38:4681–4690. doi: 10.1021/bi983030b. [DOI] [PubMed] [Google Scholar]
- 28.Pochapsky T C, Mei Ye X, Ratnaswamy G, Lyons T A. An NMR-derived model for the solution structure of oxidized putidaredoxin, a 2Fe-2S ferredoxin from Pseudomonas. Biochemistry. 1994;33:6424–6432. doi: 10.1021/bi00187a006. [DOI] [PubMed] [Google Scholar]
- 29.Romine M F, Stillwell L C, Wong K-K, Thurston S J, Sisk E C, Sensen C W, Gaasterland T, Fredrickson J K, Saffer J D. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans strain F199. J Bacteriol. 1998;181:1585–1602. doi: 10.1128/jb.181.5.1585-1602.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Subramanian V, Liu T N, Yeh W K, Serdar C M, Wackett L P, Gibson D T. Purification and properties of ferredoxinTOL. A component of toluene dioxygenase from Pseudomonas putida F1. J Biol Chem. 1985;260:2355–2363. [PubMed] [Google Scholar]
- 32.Ta D T, Vickery L E. Cloning, sequencing, and overexpression of a [2Fe-2S] ferredoxin gene from Escherichia coli. J Biol Chem. 1992;267:11120–11125. [PubMed] [Google Scholar]
- 33.Verhagen M F, Link T A, Hagen W R. Electrochemical study of the redox properties of [2Fe-2S] ferredoxins. Evidence for superreduction of the Rieske [2Fe-2S] cluster. FEBS Lett. 1995;361:75–78. doi: 10.1016/0014-5793(95)00152-y. [DOI] [PubMed] [Google Scholar]
- 34.Vidakovic M S, Fraczkiewicz G, Germanas J P. Expression and spectroscopic characterization of the hydrogenosomal [2Fe-2S] ferredoxin from the protozoan Trichomonas vaginalis. J Biol Chem. 1996;271:14734–14739. doi: 10.1074/jbc.271.25.14734. [DOI] [PubMed] [Google Scholar]
- 35.Wittich R-M, Wilkes H, Sinnwell V, Francke W, Fortnagel P. Metabolism of dibenzo-p-dioxin by Sphingomonas sp. strain RW1. Appl Environ Microbiol. 1992;58:1005–1010. doi: 10.1128/aem.58.3.1005-1010.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]