Abstract
The transcription factor PpsR from the facultative photoheterotroph Rhodobacter sphaeroides is involved in repression of photosystem gene expression under aerobic growth conditions. We have isolated a number of spontaneous mutations as well as constructed directed mutations and deletions in ppsR. Repressor activities and the oligomeric state of the wild-type and mutant proteins were assayed. Our results suggest that the wild-type PpsR exists in cell extracts as a tetramer. Analysis of the PpsR mutants confirmed that the carboxy-terminal region of PpsR (residues 400 to 464) is involved in DNA binding. The central region of the protein (residues 150 to 400) was found to contain two PAS domains (residues 161 to 259 and 279 to 367). PAS domains are ubiquitous protein modules involved in sensory transduction as well as in protein-protein interactions. All spontaneously isolated mutations, which significantly impaired repressor activity and which mapped outside the DNA binding region, were positioned in the PAS domains. None of these, however, affected the overall oligomeric state. This implies that the conformation of the PAS domains within the tetramer is critical for repressor activity. Upstream of the first PAS domain resides a putative glutamine-rich hinge (residues 127 to 136) that connects the first PAS domain to the amino-terminal region (residues 1 to 135). The role of the amino terminus of PpsR is not obvious; however, extended deletions within this region abolish repressor activity, thus suggesting that the amino terminus is essential for structural integrity of the protein. We present a model of the domain architecture of the PpsR protein according to which PpsR is comprised of three regions: the carboxy terminus responsible for DNA binding, the central region primarily involved in protein oligomerization and possibly signal sensing, and the amino terminus of unknown function. This model may prove useful for determining the mode of PpsR action.
The transition of the facultative photoheterotroph Rhodobacter sphaeroides from an aerobic to an anaerobic environment requires, among other things, a significant increase in expression of the photosystem genes. These genes encode structural and assembly proteins of the light harvesting and reaction center complexes as well as proteins involved in biosynthesis of the photopigments, bacteriochlorophyll (bch genes) and carotenoids (crt genes). When oxygen tension decreases below certain threshold level, several major regulatory circuits are recruited to increase photosystem gene expression and hence photosystem production (19, 30). One of these circuits involves derepression of the bch and crt genes, as well as the puc operon, encoding light harvesting II complex proteins, by the inactivation of the repressor PpsR.
The PpsR repressor blocks transcription by binding to the sequences TGTN12ACA (where N is nucleotide), which are positioned either downstream of or overlapping with the promoters of a number of bch and crt genes as well as the puc operon (9, 20, 22). Under aerobic conditions, expression of these genes is low, while under semiaerobic or anaerobic conditions, repression is greatly decreased or abolished. The extent of PpsR-mediated repression is controlled at the level of repressor activity rather than PpsR protein abundance (10). Very little is known about the mechanism(s) of such control, although we have presented evidence that PpsR most likely does not interact with oxygen directly. Furthermore, PpsR retains partial repressor activity under fully anaerobic, photosynthetic conditions, and this activity diminishes with decreased light intensity (10). Therefore, we were led to suggest that instead of sensing primary environmental signals, i.e., oxygen tension and light intensity, PpsR may sense cellular redox poise or the redox state of a specific redox carrier (12). In accord with this hypothesis, in vitro DNA binding by CrtJ, a PpsR homolog from a related bacterium, Rhodobacter capsulatus, was shown to depend on the redox compounds present in the buffer (21).
Initial analysis of PpsR/CrtJ revealed no significant similarities to other proteins in the databases, except for the helix-turn-helix (HTH) motif at the carboxy terminus, which was proposed to be involved in DNA binding (20). More recently, a PAS domain was identified in these proteins (24). PAS domains are universal modules for signal sensing and transduction in proteins from Bacteria, Archaea, and Eucarya. Many such proteins are involved in cellular responses to light, oxygen, or changes in redox poise (24, 28).
We undertook a mutational analysis of ppsR to gain greater insight into the structural organization and possible mode of action of the PpsR repressor. To isolate mutations in ppsR, we took advantage of an earlier observation that all spontaneously appearing suppressors of the R. sphaeroides appA null mutation map to the ppsR gene (10). AppA is a redox regulator of photosystem gene expression that antagonizes PpsR repressor activity in vivo by an as yet unknown mechanism (7, 8, 10). The spontaneous mutations were cloned, and their repressor activities were assayed. We also constructed a number of directed mutations. In addition to PpsR activity, we determined the oligomeric state of the wild-type and mutant proteins. Based on analyses of mutants and protein sequence motifs within PpsR, we propose a model domain architecture of this protein which could help in elucidating its mechanism of action.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strains and plasmids used in this work are listed in Table 1.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| R. sphaeroides | ||
| APP11 | ΔappA::Tpr | 7 |
| APS1 | APP11 ppsR::ΩKmr | 10 |
| PS3 | APP11 ppsR3 (L107-P) | 10 |
| PS8 | APP11 ppsR8 (L345-P) | 10 |
| PS18 | APP11 ppsR18 (C251-R) | 10 |
| PS23 | APP11 ppsR23 (H90-R) | 10 |
| PS33 | APP11 ppsR33 (L248-P) | 10 |
| PS36 | APP11 ppsR36 (K422-E) | 10 |
| PS40 | APP11 ppsR40 (C424-R)a | 10 |
| PS41 | APP11 ppsR41 (L403-P) | 10 |
| PS48 | APP11 ppsR48 (L28-P) | This work |
| PS50 | APP11 ppsR50 (L249-P) | This work |
| PS52 | APP11 ppsR52 (L27-P) | This work |
| PS58 | APP11 ppsR58 (R242-C) | This work |
| PS66 | APP11 ppsR66 (N176-I) | This work |
| PS69 | APP11 ppsR69 (G321-V) | This work |
| PS72 | APP11 ppsR72 (G117-E) | This work |
| PS77 | APP11 ppsR77 (G288-E) | This work |
| PS80 | APP11 ppsR80 (R339-Q) | This work |
| P. denitrificans ATCC 17741 | Wild type | American Type Culture Collection |
| E. coli | ||
| S17-1 | Tra+ donor for conjugations | 26 |
| DH5α phe | Plasmid maintenance | 5 |
| JM109 | Overexpression of the GST-PpsR protein | Lab collection |
| Plasmids | ||
| pCF400Δ | Smr/Spr Kmr IncQ, puc::lacZ | 9 |
| pLX200 | Smr/Spr IncQ, bchF::lacZ | 9 |
| pLX14b | Smr/Spr IncQ, bchF::lacZ | 10 |
| pLX14P | Smr/Spr IncQ, bchF::lacZ, ppsR | 10 |
| pRK415 | Tcr IncP, vector | 14 |
| pPNs | pRK415::ppsR (wild type) | 9 |
| pPS#c | pRK415::ppsR# | This work |
| pPSΔ10 | pRK415::ppsRΔ10 (deletion of residues 2–10 of PpsR) | This work |
| pPSΔ40 | pRK415::ppsRΔ40 (deletion of residues 2–39 of PpsR) | This work |
| pPSΔ59-117 | pRK415::ppsRΔ59-117 (deletion of residues 60–116 of PpsR) | This work |
| pP[R] | pRK415::ppsRΔ406-464 (deletion of residues 406–464 of PpsR) | 9 |
| pBg[Ns] | pRK415::ppsRΔ1-356 (deletion of residues 1–356 of PpsR) | 9 |
| pGEX-2TK | Vector for overexpression GST fusions | Amersham Pharmacia Biotech |
| pGpps | pGEX-2TK::ppsR | This work |
| pRK2013 | Helper plasmid for conjugations | 6 |
This mutation was erroneously described as C424-A in reference 10, although proper nucleotide substitution was presented.
Plasmid pLX14, similar to pLX200, contains a bchF::lacZ transcriptional fusion. The difference between these two plasmids is in the point of fusion, which does not affect bchF expression (9, 10).
Symbol # refers to the number of the ppsR mutation.
Microbiological and genetic techniques.
Escherichia coli strains were grown at 37°C on Luria-Bertani medium (17) supplemented, where required, with the following antibiotics at the indicated final concentrations: tetracycline, 10 μg/ml; ampicillin, 100 μg/ml; streptomycin and spectinomycin, 25 μg/ml each.
R. sphaeroides strains were grown anaerobically at 30°C on Sistrom's medium A (2) containing succinate as the carbon source in fully filled screw-cap tubes which were illuminated with light at 10 W/m2. Paracoccus denitrificans liquid cultures (10 ml) were grown on the same medium as R. sphaeroides in 125-ml flasks under vigorous shaking. Antibiotics for R. sphaeroides and P. denitrificans were used, where appropriate, at the indicated final concentrations: tetracycline, 1 μg/ml; streptomycin plus spectinomycin, 50 μg/ml each.
Conjugation was performed essentially as described previously (3). To introduce plasmids of interest into various R. sphaeroides and P. denitrificans strains, biparental matings (with E. coli S17-1 as a donor) or triparental [with E. coli DH5α phe(pRK2013) as a helper strain] were used (Table 1).
Spectrophotometric assays.
R. sphaeroides cell extracts were obtained by sonication of photosynthetically grown cells and assayed as described previously (16), using samples containing equal amounts of protein. Photosynthetic growth of R. sphaeroides strains was monitored using a Klett-Summerson photometer with filter no. 66 and is expressed in Klett units, where 1 Klett unit equals approximately 107 cells ml−1.
β-Galactosidase assays.
In aerobically grown P. denitrificans, β-galactosidase was assayed in whole cells treated with chloroform and sodium dodecyl sulfate as described earlier (9). β-Galactosidase activity is expressed in Miller units, where 1 Miller unit corresponds to 1 μmol of o-nitrophenyl-β-galactoside hydrolyzed per min per unit of optical density of culture at 600 nm (OD600). All assays were performed at least twice with standard deviations not exceeding 15%.
DNA manipulations and sequence analysis.
Standard recombinant DNA techniques (17) and molecular biological enzymes and reagents were used according to the specifications of the manufacturers. DNA sequencing of the ppsR mutant alleles was performed on an ABI 377 automatic DNA sequencer (Applied Biosystems) at the DNA Core Facility of the Department of Microbiology and Molecular Genetics.
Isolation of the ppsR mutations.
Mutations were isolated as described earlier (10). Briefly, the R. sphaeroides AppA null mutant, APP11, was plated under photosynthetic conditions, and photosynthesis-competent pseudorevertants were isolated and streak purified. ppsR mutant alleles were either cloned from minilibraries made from chromosomal DNA of the isolated clones (10) or amplified by high-fidelity PCR using Pfu polymerase and primers flanking the ppsR coding sequence. All ppsR alleles were sequenced using ppsR-specific primers at the DNA Core Facility of the Department of Microbiology and Molecular Genetics. The 1.5-kb NsiI-NcoI fragments containing mutant ppsR alleles were cloned into vector pRK415 in the identical fashion to create plasmids designated pPS# (# represents the number of a given mutation).
Purification of a GST-PpsR fusion protein.
The 1.6-kb BspEI-XmaI fragment containing the ppsR gene was cloned into the XmaI site of vector pGEX-2TK downstream of the gene encoding glutathione S-transferase (GST) to create plasmid pGpps. The GST-PpsR fusion protein was expressed in E. coli JM109(pGpps). Optimal production of a soluble GST-PpsR fusion occurred when mid-log cells (OD600 = 0.6), grown at 25°C, were induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside for 8 h. Two liters of E. coli was grown for production of GST-PpsR. Harvested cells were disrupted in a French cell pressure unit. The soluble extract was then fractionated using a DEAE ion-exchange column (80-ml bed volume) equilibrated with 50 mM Tris-HCl (pH 7.5) (elution buffer). GST-PpsR was eluted using a 0 to 500 mM NaCl elution buffer, and peak fractions of GST-PpsR were obtained at about 270 mM NaCl. Pooled fractions containing GST-PpsR were charged onto a glutathione-Sepharose column (bed volume, 15 ml). After washing with 80 mM phosphate (pH 7.7)–0.4% n-octylglucoside, the GST-PpsR fusion was eluted with 5 mM glutathione in 80 mM phosphate (pH 7.7)–0.4% n-octylglucoside. The protein was judged to be pure as indicated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis; we obtained a single polypeptide of 76 kDa, close to the expected size of a GST-PpsR fusion.
Production of PpsR-specific antibodies.
The purified GST-PpsR protein was used to immunize chickens. Antibodies were obtained from egg yolks by ammonium sulfate precipitation (27). PpsR-specific antibodies were purified by immunoaffinity chromatography; 7.5 mg of cell extract of E. coli JM109(pRK415) was coupled to a 1-ml HiTrap N-hydroxysuccinimide column (Amersham Pharmacia Biotech). The crude antibody mixture was then run through the column, and protein fractions that did not bind to the column were collected and tested for anti-PpsR reactivity.
Western blotting and Ferguson plot analysis.
Western blot analyses were performed by standard protocols. PpsR-specific antibodies were used in a 1:100 ratio as a primary antibody; a mouse anti-chicken secondary antibody coupled to alkaline phosphatase was used for development of the immunoblots.
The native molecular mass of PpsR was determined by Ferguson plot analysis of the relative mobility of the native protein and a series of protein standards (13). Electrophoretic analysis was performed by Western blotting at six different acrylamide concentrations between 7.5 and 10% (30:0.8 acrylamide/bisacrylamide). After the running of each gel, the proteins were transferred to nitrocellulose. The protein standards were stained with Ponceau S and washed with water. PpsR was detected in the cell extracts contained on the nitrocellulose membrane by Western blotting. The protein standards used were lactalbumin (14 kDa), bovine serum albumin (66 kDa), dimethyl sulfoxide reductase (82 kDa), amylase (180 kDa), and urease (262 kDa). The log10 Rf for each protein was plotted as a function of polyacrylamide gel concentration. The slope of each plot gave the retardation coefficient for each protein (25). This slope was then plotted as a function of native Mr, and from this calibration curve the native Mr of PpsR was determined.
RESULTS
Isolation and characterization of R. sphaeroides PpsR mutants.
We have previously described (10) the procedure for isolating mutations in the R. sphaeroides ppsR gene and have mapped some of these mutations. The procedure is based on selection of photosynthesis-competent pseudorevertants of an AppA null mutant, which itself is impaired in photosynthetic growth. The ppsR gene is the only site found to date for suppressor mutations, i.e., overcoming the initial AppA null phenotype. Using this procedure, we isolated ppsR mutations in addition to those previously described (Table 1). Here, we present a detailed characterization of the isolated ppsR mutations as a means to better understanding of PpsR structure and function.
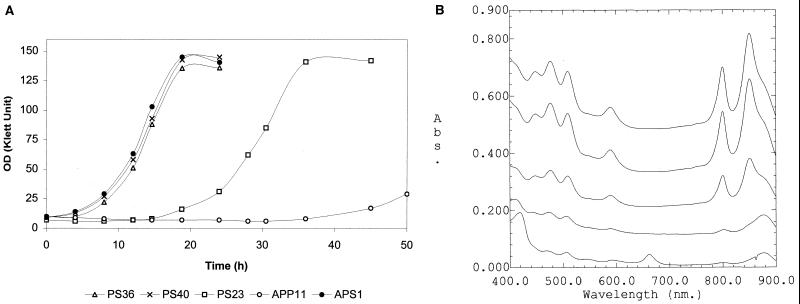
We tested for all of the mutants both photosynthetic growth and the abundance of photosynthetic complexes, traits which appear to correlate with the residual activity of the mutant PpsR repressor. The mutants of one class of suppressors (Table 2, class 1), were similar to the AppA PpsR double null mutant, APS1, in their rapid transition from aerobic to anaerobic photosynthetic growth conditions (Fig. 1A, strain PS40). These mutants accumulated photosynthetic complexes to high abundance (Fig. 1B, strain PS40) and, like mutant APS1, were genetically unstable when grown under aerobic conditions, apparently because of accumulation of photosynthetic complexes in the presence of oxygen (data not shown). The mutants of class 3 (Table 2) showed only slight improvement in the transition from aerobic to anaerobic photosynthetic growth compared to the parent AppA null mutant, APP11 (Fig. 1A, strain PS23). These contained only low levels of photosynthetic complexes (Fig. 1B, strain PS23). The phenotypes of the remaining mutants, placed in class 2 (Table 2), were intermediate (Fig. 1A and B, strain PS36). In contrast to mutants of class 1, class 2 mutants were genetically stable under aerobic growth conditions, and the abundance of photosynthetic complexes produced under anaerobic photosynthetic conditions was lower than in mutants of class 1 yet significantly higher than in mutants of class 3. Thus, the rather broad range of mutant phenotypes suggested that residual activities of the mutant PpsR varied widely, i.e., from minor deviations from the wild-type repressor activity (class 3) to the significant decrease or apparent absence thereof (class 1).
TABLE 2.
Analysis of PpsR mutations
| Mutation designation and/or description | Mutant classa | Expression levelb (unrepressed/repressed ratioc)
|
|
|---|---|---|---|
| puc::lacZ | bchF::lacZ | ||
| No PpsR presentd | NAe | 2,537 (1) | 636 (1) |
| None (wild-type PpsR) | NA | 9 (282) | 12 (53) |
| 40 | 1 | 2,278 (1) | 708 (1) |
| 18 | 1 | 1,202 (2) | 185 (3) |
| 50 | 1 | 1,227 (2) | 128 (5) |
| 66 | 1 | 1,214 (2) | 144 (4) |
| 41 | 1 | 977 (3) | 322 (2) |
| 69 | 1 | 517 (5) | 24 (27) |
| 48 | 1 | NDf | ND |
| 72 | 1 | ND | ND |
| 33 | 1 | ND | ND |
| 77 | 1 | ND | ND |
| 52 | 2 | 122 (21) | 46 (14) |
| 36 | 2 | 90 (28) | 71 (9) |
| 8 | 2 | ND | ND |
| 23 | 3 | 26 (98) | 27 (24) |
| 3 | 3 | 16 (158) | 20 (32) |
| 58 | 3 | 15 (169) | 21 (30) |
| 80 | 3 | ND | ND |
| Δ10 | NA | 9 (282) | 10 (64) |
| Δ40 | NA | 1,157 (2) | 230 (3) |
| Δ59-117 | NA | 2,165 (1) | 585 (1) |
Based on phenotypes of the R. sphaeroides PpsR mutants (Fig. 1) from which the ppsR alleles were cloned.
β-Galactosidase (Miller units) in aerobically grown P. denitrificans carrying two plasmids: (i) either pCF400Δ (contains puc::lacZ) or pLX14 (contains bchF::lacZ) and (ii) pPS# (contains the ppsR mutant allele).
Ratio of expression levels in the absence of PpsR and in the presence of the ppsR mutant allele. Higher ratios correspond to higher PpsR repressor activities.
P. denitrificans carrying vector pRK415 in place of plasmid pPSn.
NA, not applicable.
ND, not determined.
FIG. 1.
(A) Transition of the R. sphaeroides AppA PpsR mutant strains from aerobic to anaerobic photosynthetic (light intensity 10 W/m2) conditions. The strains were grown aerobically to approximately the same OD (25 to 30 Klett units), placed at 4°C for 10 h, diluted with fresh growth medium, and transferred to anaerobic light conditions (time zero). Representative growth curves from one experiment are shown. (B) Spectra of the AppA PpsR mutant strains grown under anaerobic photosynthetic conditions (from top to bottom, APS1, PS40, PS36, PS23, and APP11). Spectra were taken from the cultures at approximately the same OD i.e., 80 to 90 Klett units, except for APP11 (25 Klett units). Abs., absorbance.
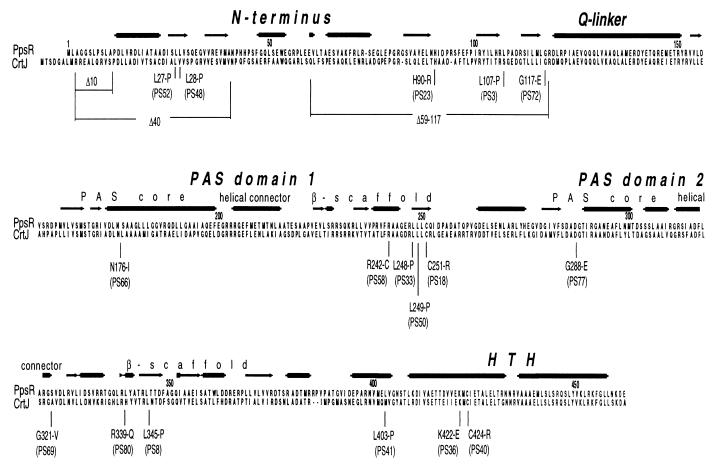
To understand the nature of the individual PpsR mutants, the ppsR alleles from representative mutants were cloned and sequenced. The mutations, most of which were single-base pair substitutions, were found to be positioned throughout the length of the ppsR gene (Fig. 2). Several identical amino acid changes were found among mutants isolated in independent rounds of selection; e.g., mutations identical to those present in mutants PS3 and PS18 were isolated three times, and mutations identical to those in mutants PS24 and PS33 were isolated twice. This indicated the presence of mutational hot spots in ppsR and imposed limits on the number of the spontaneous ppsR mutant alleles that we analyzed. Mutations which resulted in the presence of a premature stop codon or a frameshift will not be considered further. In total, 17 different single amino acid substitutions were obtained by this approach.
FIG. 2.
Domain structure of the PpsR protein and localization of the amino acid substitutions. Analysis of protein secondary structures was performed using the secondary structure prediction available as a part of the Jpred package (http://circinus.ebi.ac.uk:8081/). Consensus derived from several prediction algorithms is shown.
Repressor activities of the PpsR mutant forms in P. denitrificans.
To assess the extent of impairment to PpsR repressor function, we cloned representative ppsR alleles from each mutant class into a broad-host-range vector, pRK415, and introduced these into P. denitrificans. We have previously shown that this nonphotosynthetic species, which is closely related to R. sphaeroides, is useful for studying the expression of isolated photosystem genes in the absence of components of the photosystem and the numerous regulatory factors controlling photosystem gene expression (9–11).
To monitor PpsR repressor activity, two sets of plasmids were introduced in parallel into P. denitrificans. One, pCF400Δ, carries the puc::lacZ transcriptional fusion with two adjacently positioned PpsR binding sites in the puc operon upstream region. The other plasmid, pLX200, carries the bchF::lacZ transcriptional fusion which contains two PpsR binding sites separated by 125 bp.
The unrepressed level of puc::lacZ expression in P. denitrificans, i.e., the level of expression in the presence of the vector pRK415, was 282 times higher than the repressed level, i.e., in the presence of the wild-type PpsR protein expressed from plasmid pPNs (Table 2). For bchF::lacZ expression, the unrepressed level was 53 times higher than the repressed level (Table 2). The mutant PpsR proteins, produced from pRK415-derived plasmids (Table 1, pPS# series), varied widely in the ability to repress puc::lacZ and bchF::lacZ expression in P. denitrificans. In general, the PpsR mutant repressor activities in P. denitrificans (Table 2) correlated well with the abundance of photosynthetic complexes and the time required for the aerobic-to-anaerobic transition of R. sphaeroides strains from which these ppsR alleles were isolated (Fig. 1). For example, the repressor activity of the ppsR40 mutation, cloned from a mutant of class 1, was undetectable in P. denitrificans; i.e., the ratios of the unrepressed (ppsR absent) to repressed (mutant ppsR allele) levels of β-galactosidase were ∼1 for either puc::lacZ or bchF::lacZ expression (Table 2). The residual repressor activity in P. denitrificans of the ppsR36 mutation, which was cloned from a mutant of class 2, was higher than that of ppsR40, resulting in ratios of 28 and 9 for puc::lacZ and bchF::lacZ, respectively. The residual repressor activity of the ppsR23 mutation, which was cloned from a mutant of class 3, was higher still, resulting in ratios of 98 and 24 for puc::lacZ and bchF::lacZ, respectively (Table 2). The mutant proteins repressed puc::lacZ and bchF::lacZ in a similar, parallel fashion; however, the extent of repression for each fusion was not necessarily the same. The differences in the ability of a particular PpsR mutant protein to repress puc and bchF could be attributed to different arrangements of the PpsR binding sites; i.e., two neighboring TGTN12ACA sites upstream of puc versus two sites separated by ∼125 bp in the case of bchF.
As noted in a previous report (10), amino acid substitutions K422-E (PpsR36) and C424-R (PpsR40) are located in the HTH motif of PpsR (20) that is responsible for DNA binding (Fig. 2). Therefore, as a first approximation one can assume that these mutations impaired DNA binding capabilities of the altered repressor. Substitution L403-P (PpsR41) is located immediately upstream of the HTH motif (Fig. 2), and because of the nature of a proline residue, there is likely to be a perturbation in the secondary structure, thus changing the context of the downstream HTH compared to the rest of the protein. This could be the reason for low repressor activity of PpsR41 (Table 2). However, amino acid substitutions not localized to the DNA binding domain are likely to impair the oligomeric state of the PpsR repressor or/and its conformation. We set out to test these possibilities.
Genetic evidence for PpsR oligomerization and localization of the region of PpsR involved in oligomerization.
The DNA binding site for PpsR, TGTN12ACA, consists of an inverted repeat, implying that PpsR binds as a dimer. Further, the PpsR homolog from R. capsulatus, CrtJ, has been shown to bind DNA as a dimer and possibly form a tetramer (21).
We devised a genetic test to determine whether PpsR forms oligomeric structures. We introduced both the mutant form of PpsR (expressed from a plasmid of the pPS# series) and the wild-type PpsR (expressed from plasmid pLX14P) into P. denitrificans, Pd(pLX14P), and monitored bchF::lacZ expression. We assumed that a mutant PpsR protein, which is wholly defective in oligomerization, would not interfere with functioning of the wild-type PpsR; hence, the repression level would be similar to that of the wild-type PpsR alone. However, a subset of mutant PpsR proteins which are proficient in oligomerization yet defective in repression could have a dominant negative phenotype because of the formation of mixed oligomers with the wild-type PpsR.
We first tested PpsR40 (C424-R), containing a mutation in the HTH domain. Pd(pLX14P) carrying plasmid pPS40 showed an 1.9-fold-increased bchF::lacZ expression compared to Pd(pLX14P) containing vector pRK415 alone (Table 3). Hence, PpsR40 interfered with wild-type repressor presumably by forming mixed oligomers. We then tested the truncated derivative of PpsR (expressed from plasmid p[R]) that lacks the entire HTH domain. Pd(pLX14P) containing pP[R] showed an ∼2.7-fold increased bchF::lacZ expression compared to Pd(pLX14P) containing vector pRK415 alone (Table 3). Hence, this truncated protein is presumed to contain the regions involved in oligomerization. The truncated PpsR protein, which consisted of the 166 carboxy-terminal residues, did not affect repression levels in Pd(pLX14P) (Table 3, plasmid p[Bg]Ns). Hence, the carboxy terminus is likely to be incapable of interacting with the wild-type PpsR. We conclude that PpsR forms dimers or higher oligomers and that the regions involved in oligomerization are localized upstream of the carboxy-terminal DNA binding region.
TABLE 3.
Genetic evidence for PpsR oligomerization
| Plasmid carrying ppsR mutant allele | Mutation designation and/or description | bchF::lacZ expressiona | Derepression ratiob |
|---|---|---|---|
| pRK415 (vector) | No PpsR present | 22 | 1.0 |
| pPNs | None (wild-type PpsR) | 18 | 0.8 |
| pP[R] | Deleted HTH domain | 52 | 2.7 |
| pBg[Ns] | Present HTH domain only | 19 | 0.9 |
| pPS40 | 40 | 35 | 1.9 |
| pPS18 | 18 | 20 | 1.0 |
| pPS50 | 50 | 29 | 1.6 |
| pPS66 | 66 | 25 | 1.1 |
| pPS41 | 41 | 28 | 1.3 |
| pPSΔ40 | Δ40 | 23 | 1.0 |
| pPSΔ59-117 | Δ59-117 | 31 | 1.4 |
β-Galactosidase (Miller units) was measured in the aerobically grown P. denitrificans containing two plasmids, pLX14P (carries bchF::lacZ and the wild-type ppsR gene) and a plasmid carrying the ppsR mutant allele.
Ratio of bchF::lacZ expression level in P. denitrificans carrying pLX14P and the ppsR mutant allele and expression level in P. denitrificans, carrying pLX14P and pRK415 (vector).
We proceeded to test those ppsR point mutations which showed low-level repression in P. denitrificans for the ability to interfere with the wild-type repressor as an indication of their ability to oligomerize. Among the analyzed mutants, two, PpsR66 (N176-I) and PpsR18 (C251-R), showed no interference with the wild-type repressor (Table 3), while PpsR50 (L249-P) showed moderate interference. There are several possible explanations for this observation.
Residues N176 and C251 could be of critical importance for oligomerization; hence, PpsR66 and PpsR18 would not form oligomers and therefore would not interfere with the wild-type repressor. Alternatively, the P. denitrificans assay could lack sufficient sensitivity to detect interference derived from mixed oligomers employing either PpsR66 or PpsR18. The limited sensitivity of the P. denitrificans assay could arise from the significantly higher ratio of repressor to its DNA binding sites compared to that ratio in R. sphaeroides. The fact that the maximally observed increase in bchF::lacZ expression due to the formation of mixed oligomers was only 2.7-fold relative to the fully repressed value supports this prediction (Table 3, pPNs versus pP[R]).
As an independent approach to assess the oligomeric state of PpsR, we developed an in vitro assay.
The oligomeric state of native and mutant PpsR proteins.
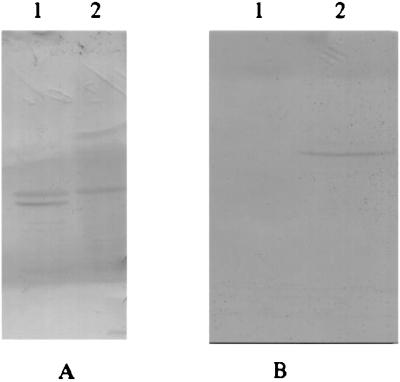
ppsR was overexpressed in E. coli as a fusion to GST and purified to apparent homogeneity (data not shown). Antibodies were raised against purified PpsR protein to examine its oligomeric state in cell extracts of R. sphaeroides. Figure 3A shows a Western blot from a nondenaturing gel probed with anti-PpsR antibody. Two cross-reacting proteins were present in extracts from wild-type R. sphaeroides. Since one of these cross-reacting bands disappeared in a ppsR null mutant, PPS1, we concluded that this protein corresponded to PpsR. The nature of the second band remains unknown. When PpsR was expressed in E. coli only, one PpsR-specific band was observed (Fig. 3B).
FIG. 3.
Western blot from a nondenaturing gel using PpsR-specific antibodies. (A) Cell extracts of R. sphaeroides. Lane 1, strain 2.4.1 (wild type); lane 2, strain PPS1 (ppsR null mutant). (B) Cell extracts of E. coli. Lane 1, JM109(pGEX-2TK); lane 2, JM109(pGpps).
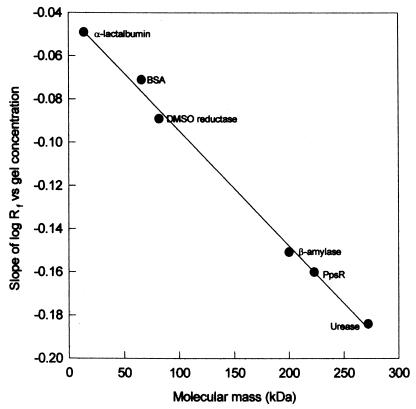
The native molecular mass of PpsR in cell extracts of E. coli JM109(pPNs) was determined by Ferguson plot analysis of the relative mobility of PpsR and a series of protein standards. Figure 4 shows the position of PpsR on a plot of protein retardation coefficient against molecular mass. The native molecular mass of PpsR was determined to be approximately 217 kDa. Since the subunit molecular mass of PpsR is 54 kDa, these data indicate that native PpsR in cell extracts is a tetramer.
FIG. 4.
Determination of molecular mass of the PpsR protein in cell extracts of JM109(pPNs) (Ferguson plot). BSA, bovine serum albumin; DMSO, dimethyl sulfoxide.
We tested the oligomeric state of the PpsR mutants. All mutant proteins tested were stable; i.e., their amounts in cell extracts of E. coli did not differ significantly from the amount of the wild-type protein. The mutant proteins were also stable in P. denitrificans. However, no differences in the native molecular masses were observed for any of the mutant proteins (not shown). This suggests that the main defect imposed by the mutations analyzed here (and localized outside the DNA binding region of PpsR [see below]) resulted from alteration in the conformation of the PpsR tetramer, not from a change in PpsR oligomeric state.
It is worth mentioning that substitutions in either of the two cysteine residues of PpsR, C251-R (PpsR18) and C424-R (PpsR40), had only a minor effect on the oligomeric state of PpsR. This result suggested that possible intermolecular disulfide bridges involving these cysteine residues, should they exist, may not be essential for oligomer formation.
Identification of a second PAS domain in PpsR.
To identify possible structural elements of PpsR involved in protein-protein interactions and conformational changes, we took a closer look at PpsR primary and secondary structures.
Previously, little sequence similarity between PpsR and other proteins in the databases could be identified, except for the carboxy-terminal HTH motif (20). Recent analysis of the PpsR protein primary structure (24) revealed that PpsR contains a PAS domain which is characteristic of a large group of prokaryotic and eukaryotic signal transduction proteins. Only limited primary sequence homology is found within the PAS domains, with highest amino acid conservation being within the PAS core and β scaffold. In many instances these domains are associated with binding-specific cofactors that lend them signal-sensing ability. Another role of PAS domains is involvement in protein-protein interactions. It has been shown that conformational changes within PAS domains play a critical role in signal transduction (reviewed in reference 28).
Closer analysis of the PpsR primary and secondary structures allowed us to identify a second PAS domain, in addition to the one previously suggested (24). In Figure 5 we present an alignment of the PpsR protein, as well as its homologue from R. capsulatus, CrtJ, with a subset of the bacterial signal transduction proteins with identifiable PAS domains. The putative first PAS domain of PpsR extends from P161 to T259; the second domain extends from G280 to R367. The two PAS domains show a noticeable, albeit not very high, degree of similarity to each other, i.e., 23% identity between core boxes of the first and second PAS domains of PpsR. The predicted secondary structure of the second PAS domain is similar to that of the first PAS domain (Fig. 2). Both predictions fit reasonably well with the secondary structure of PAS domains deduced from the crystal structures of PAS domain-containing proteins, i.e., primarily α-helical PAS core, helical connector, β scaffold (28).
FIG. 5.
Alignment of the two PAS domains from PpsR, CrtJ, and several representative signal transduction proteins from the proteobacteria. A portion of the alignment was taken from the World Wide Web (http://www.llu.edu/llu/medicine/micro/PAS/paslist.htm) (28) and modified. Consensus derived from the alignment of several hundred PAS domains (28) is shown below the alignment. Identical amino acids, i.e., those conserved in at least 50% of sequences, are shown in reverse contrast. Similar residues present in at least 75% of sequences are shaded. Designation of residues: c, charged (DEHKR), u, bulky hydrophobic (FILMVWY), h, hydrophobic (ACFGILMTVWY); o, hydroxy (ST); p, polar (CDEHKNQRST); t, turn-like (ACDEGHNQRST); s, small (ACDGNPSTV).
All analyzed mutations which drastically impaired PpsR repressor activity and which reside outside the DNA binding region of PpsR are positioned within the first or second PAS domain, e.g., N176-I, L248-P, L249-P, C251-R, or G288-E (Table 2, Fig. 2). Some of these mutations (e.g., N176-I and G288-E) represent substitutions of the most conserved residues of the PAS domains, while others (e.g., L248-P and L249-P) are likely to perturb local secondary structure because of the nature of the substitutions (see Discussion). This observation strengthens the importance of the PAS domains for maintaining proper PpsR conformation.
Construction and analysis of PpsR mutants containing deletions in the amino terminus of PpsR.
Given that the carboxy-terminal region of PpsR, encompassing approximately residues 400 to 464, is presumably involved in DNA binding, and the adjacent region containing two PAS domains, from residue 161 to 367, is presumably involved in protein oligomerization and/or signal sensing, the amino-terminal region of the protein remains to be studied.
Primary structure analysis revealed that a stretch of residues from 124 to 136 is a region of low complexity, highly enriched in glutamines. This resembles the so-called Q linkers, i.e., flexible hinges connecting neighboring protein domains (29). Identification of the putative Q linker immediately upstream of the first PAS domain enabled us to suggest that the amino terminus of PpsR represents a separate protein domain. The protein sequences of PpsR and CrtJ are most divergent in the amino-terminal region, i.e., only 38% identity, compared to 47% identity in the first PAS domain and 59% identity in the second PAS domain.
We have found in the databases no proteins bearing significant similarity to the amino terminus of PpsR (and CrtJ). We decided to test the significance of the amino-terminal region by constructing several deletions and testing for their effects on repressor function. Deletion of residues 2 to 10, which have no analogs in CrtJ, did not affect either PpsR repressor activity in P. denitrificans (Table 2, plasmid pPSΔ10) or oligomeric state. However, extended amino-terminal deletions, i.e., residues 2 to 39, or internal deletion of residues 60 to 116 resulted in marginally active and inactive repressor protein (Table 2, plasmids pPSΔ40 and pPSΔ59-117, respectively). According to our genetic test, PpsRΔ40 did not interfere with the wild-type repressor, suggesting a possible defect in oligomerization (Table 3), whereas PpsRΔ59-117 did interfere with the wild-type repressor (Table 3). However, the oligomeric state of neither of these proteins in vitro differed from that of the wild-type PpsR. Hence, conformational changes in the mutant PpsR proteins could be invoked to explain the deleterious effect of the removal of portions of the amino terminus on PpsR repressor activity. Taken together, these results suggest that the amino-terminal region is essential for proper tertiary structure of PpsR, albeit its precise role is as yet unclear.
DISCUSSION
R. sphaeroides PpsR protein functions as a key transcription factor that coordinately represses expression of photosystem genes, e.g., bch, crt, and puc, under aerobic conditions. PpsR repressor activity greatly decreases in response to anaerobiosis by as yet an unknown mechanism. Until recently, limited structural information pertaining to this protein was available. Here we attempted to gain further insight into PpsR structure and mode of action, based on analysis of the wild-type and numerous mutant proteins.
We presented genetic evidence that PpsR is likely to form oligomers. In agreement with this prediction, Ferguson plot analysis indicated that PpsR exists as a tetramer in cell extracts of E. coli. The homolog of PpsR from R. capsulatus, CrtJ, was suggested to bind its recognition site, TGTN12ACA, as a dimer. However, the presence of two recognition sites is required for efficient DNA binding by CrtJ; therefore, it was concluded that two CrtJ dimers must interact to form a tetramer (4, 21, 23). Although dimeric transcription factors are more common, a number of tetrameric DNA binding proteins have been described (1, 15, 18).
PpsR and CrtJ contain two conserved cysteine residues, C251 and C424. We considered the possibility that these cysteines could play a role in tetramer formation, e.g., by formation of intermolecular disulfide bridges. However, mutants PpsR18 (C251-R) and PpsR40 (C424-R), which contain substitutions in these conserved cysteine residues, still form tetramers. Therefore, disulfide bridges, if formed, may not be necessary for protein oligomerization.
None of the numerous ppsR mutations which led to a drastic impairment of repressor activity significantly affected the oligomeric state of the protein in the cell extracts of E. coli. This observation allows us to suggest that protein conformation within the tetramer must be critical for repressor activity. Further, it is possible that conformational changes rather than changes in the oligomeric state of PpsR represent a mechanism controlling repressor activity. The observation that PpsR contains two PAS domains strengthens this hypothesis. PAS domains are universal modules for signal sensing and transduction. In many instances these domains bind specific cofactors that respond to changes in environmental conditions. It is believed that either binding of a specific cofactor or changes in the state of a particular cofactor (e.g., changes in redox state) can significantly perturb protein conformation within a PAS domain, thus resulting in switching from the active to inactive protein conformation. Examples of redox cofactors include flavin adenine dinucleotide in the aerotaxis transducer Aer from E. coli and in the redox- and fixed nitrogen sensor NifL from Azotobacter vinelandii, heme in the oxygen-sensing histidine kinase FixL from Sinorhizobium meliloti, and 4-hydroxycinnamyl chromophore in the blue light receptor PYP from Ectothiorhodospira halophila (28).
Whether either or both of the PAS domains of PpsR is involved in cofactor binding is unknown. Neither PpsR (this work) nor CrtJ (21), purified using the E. coli overexpression systems, was found to contain cofactors. However, CrtJ showed DNA binding in vitro that was higher under oxidized compared to reduced conditions (21). One of the possibilities to explain this observation is that response to changes in redox poise could be an intrinsic property of the CrtJ and PpsR proteins. This could imply that direct oxidation-reduction of the conserved cysteine residues functions as a mechanism controlling PpsR repressor activity. One of these cysteines, C251, is present in the first PAS domain; the other, C424, is in the DNA binding region. Intriguingly, substitutions of either cysteine with arginine abolished repressor activity of PpsR (Table 2). The role of the cysteine residues requires further exploration.
Several mutations that either abolished or significantly impaired PpsR repressor activity were localized to the PAS domains of PpsR (Table 2; Fig. 5). N176-I (PpsR66) represents a substitution of one of the most conserved residues in the PAS core of the first PAS domain. Residues L248 (PpsR81) and L249 (PpsR50), located in the β scaffold of the first PAS domain, are not conserved within the PAS domains. However, their substitutions by prolines, which are known to bring about significant changes in protein secondary structure, could have had major effects on the conformation of the first PAS domain resulting in decreased repressor activity (Table 2). G288-E (PpsR77) represents a substitution of a well-conserved residue in the PAS core of the second PAS domain, hence the deleterious effect of the G288-E substitution on PpsR repressor activity (Table 2). Residue C251 (PpsR18) is not well conserved except in CrtJ; however, this position is usually occupied by a hydrophobic residue (Fig. 5). Therefore, substitution with a charged hydrophilic residue, C251-R, can have a dramatic effect on local conformation. It is also possible that this residue is specifically involved in signal sensing (see above).
Another recognized function of the PAS domains is their involvement in protein-protein interactions. These interactions may include homologous and/or heterologous partners. We suggest that each of the two PAS domains of PpsR is involved in interactions with other PpsR monomers. Surprisingly, none of the isolated mutations within the PAS domains resulted in a significant change in the oligomeric state of PpsR in E. coli cell extracts. This is even more striking when one considers that the changes are located in each module of the PAS domains, i.e., PAS core and β scaffold.
The amino termini of PpsR and CrtJ are the regions least conserved between these homologs. They show no significant similarity to known protein sequences. The fact that extended deletions in the amino terminus (e.g., in PpsRΔ40 and PpsRΔ58-117) impaired repressor activity of the mutant proteins suggests that the amino terminus plays an important role in overall protein folding. We found that the first PAS domains in both PpsR and CrtJ are preceded by a low-complexity region enriched in glutamines (residues 127 to 134 and possibly 127 to 144). We suggest that the location of this region, i.e., only several residues upstream of the first PAS domain, and its amino acid composition make it a good candidate for a flexible hinge. This hinge would link the PAS domains of PpsR with the amino-terminal domain. Despite uncertainty about the role of the amino terminus, we believe that it represents a separate protein domain with specific function.
We believe that the emerging structural organization of the PpsR and CrtJ proteins will help elucide their mode of action.
ACKNOWLEDGMENTS
This work was supported by the NIH grant GM15590 to S.K., Australian Research Council grant to J.M.P. and A.G.M., and Australian Postgraduate Award to I.M.H.
We are grateful to Joyce Marshall for her contributions in the early stages of the project.
REFERENCES
- 1.Chang M, Crawford I P. In vitro determination of the effect of indoleglycerol phosphate on the interaction of purified TrpI protein with its DNA-binding sites. J Bacteriol. 1991;173:1590–1597. doi: 10.1128/jb.173.5.1590-1597.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen-Bazire G, Sistrom W R, Stanier R Y. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J Cell Comp Physiol. 1957;49:25–68. doi: 10.1002/jcp.1030490104. [DOI] [PubMed] [Google Scholar]
- 3.Davis J, Donohue T J, Kaplan S. Construction, characterization, and complementation of a Puf− mutant of Rhodobacter sphaeroides. J Bacteriol. 1988;170:320–329. doi: 10.1128/jb.170.1.320-329.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elsen S, Ponnampalam S N, Bauer C E. CrtJ bound to distant binding sites interacts cooperatively to aerobically repress photopigment biosynthesis and light harvesting II gene expression in Rhodobacter capsulatus. J Biol Chem. 1998;273:30762–30769. doi: 10.1074/jbc.273.46.30762. [DOI] [PubMed] [Google Scholar]
- 5.Eraso J M, Kaplan S. prrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J Bacteriol. 1994;176:32–43. doi: 10.1128/jb.176.1.32-43.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Figurski D H, Helinski D R. Replication of an origin containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomelsky M, Kaplan S. appA, a novel gene encoding a trans-acting factor involved in the regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1995;177:4609–4618. doi: 10.1128/jb.177.16.4609-4618.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomelsky M, Kaplan S. AppA, a redox regulator of photosystem formation in Rhodobacter sphaeroides 2.4.1, is a flavoprotein. Identification of a novel FAD binding domain. J Biol Chem. 1998;273:35319–35325. doi: 10.1074/jbc.273.52.35319. [DOI] [PubMed] [Google Scholar]
- 9.Gomelsky M, Kaplan S. Genetic evidence that PpsR from Rhodobacter sphaeroides 2.4.1 functions as a repressor of puc and bchF expression. J Bacteriol. 1995;177:1634–1637. doi: 10.1128/jb.177.6.1634-1637.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomelsky M, Kaplan S. Molecular genetic analysis suggesting interactions between AppA and PpsR in regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1997;179:128–134. doi: 10.1128/jb.179.1.128-134.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomelsky M, Kaplan S. The Rhodobacter sphaeroides 2.4.1 rho gene: expression and genetic analysis of structure and function. J Bacteriol. 1996;178:1946–1954. doi: 10.1128/jb.178.7.1946-1954.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomelsky M, Lee H-J, Kaplan S. Mutation analysis and regulation of PpsR, the Rhodobacter sphaeroides repressor of photopigment and light harvesting complex-II expression. In: Peschek G A, Loffelhardt W, Schmetterer G, editors. The phototrophic prokaryotes. New York, N.Y: Kluwer Academic/Plenum Publishers; 1998. pp. 131–138. [Google Scholar]
- 13.Hedrick J L, Smith A J. Size and charge isomer separation and estimation of molecular masses of proteins by disc electrophoresis. Arch Biochem Biophys. 1968;126:155–164. doi: 10.1016/0003-9861(68)90569-9. [DOI] [PubMed] [Google Scholar]
- 14.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 15.Kullik I, Stevens J, Toledano M B, Storz G. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for DNA binding and multimerization. J Bacteriol. 1995;177:1285–1291. doi: 10.1128/jb.177.5.1285-1291.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J K, Kaplan S. cis-acting regulatory elements involved in oxygen and light control of puc operon transcription in Rhodobacter sphaeroides. J Bacteriol. 1992;174:1146–1157. doi: 10.1128/jb.174.4.1146-1157.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 18.Miller B E, Kredich N M. Purification of the cysB protein from Salmonella typhimurium. J Biol Chem. 1987;262:6006–6009. [PubMed] [Google Scholar]
- 19.Pemberton J M, Horne I, McEwan A G. Regulation of photosynthetic gene expression in purple bacteria. Microbiology. 1998;144:267–278. doi: 10.1099/00221287-144-2-267. [DOI] [PubMed] [Google Scholar]
- 20.Penfold R J, Pemberton J M. Sequencing, chromosomal inactivation, and functional expression of ppsR, a gene which represses carotenoid and bacteriochlorophyll synthesis in Rhodobacter sphaeroides. J Bacteriol. 1994;176:2869–2876. doi: 10.1128/jb.176.10.2869-2876.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ponnampalam S N, Bauer C E. DNA binding characteristics of CrtJ, a redox-responding repressor of bacteriochlorophyll, carotenoid, and light harvesting-II gene expression in Rhodobacter capsulatus. J Biol Chem. 1997;272:18391–18396. doi: 10.1074/jbc.272.29.18391. [DOI] [PubMed] [Google Scholar]
- 22.Ponnampalam S N, Buggy J J, Bauer C E. Characterization of an aerobic repressor that coordinately regulates bacteriochlorophyll, carotenoid, and light harvesting-II expression in Rhodobacter capsulatus. J Bacteriol. 1995;177:2990–2997. doi: 10.1128/jb.177.11.2990-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ponnampalam S N, Elsen S, Bauer C E. Aerobic repression of the Rhodobacter capsulatus bchC promoter involves cooperative interactions between CrtJ bound to neighboring palindromes. J Biol Chem. 1998;273:30757–30761. doi: 10.1074/jbc.273.46.30757. [DOI] [PubMed] [Google Scholar]
- 24.Pontig C P, Aravind L. PAS: a multifunctional domain family comes to light. Curr Biol. 1997;7:R674–R677. doi: 10.1016/s0960-9822(06)00352-6. [DOI] [PubMed] [Google Scholar]
- 25.Rodbard D, Chrambach A. Estimation of molecular radius, free mobility and valence using polyacrylamide gel electrophoresis. Anal Biochem. 1971;40:95–134. doi: 10.1016/0003-2697(71)90086-8. [DOI] [PubMed] [Google Scholar]
- 26.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:37–45. [Google Scholar]
- 27.Svendsen L, Crowley A, Ostergaard L H, Stoduski G, Hau J. Development and comparison of purification strategies for chicken antibodies from egg yolk. Lab Anim Sci. 1995;45:89–93. [PubMed] [Google Scholar]
- 28.Taylor B L, Zhulin I B. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wootton J C, Drummond M H. The Q-linker: a class of interdomain sequences found in bacterial multidomain regulatory proteins. Protein Eng. 1989;2:535–543. doi: 10.1093/protein/2.7.535. [DOI] [PubMed] [Google Scholar]
- 30.Zeilstra-Ryalls J, Gomelsky M, Eraso J, Yeliseev A, O'Gara J, Kaplan S. Control of photosystem formation in Rhodobacter sphaeroides. J Bacteriol. 1998;180:2801–2809. doi: 10.1128/jb.180.11.2801-2809.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]