The genomic contribution to inflammatory bowel diseases (IBDs) is complex and polygenic,1 with more than 240 susceptibility loci identified by genome-wide association studies.2 Although genome-wide association studies are powerful, those still carry limitations, and commonly are not powered toward rare subtype analysis within a disease group.3 Therefore, in this study, we set out to compare the genetics (exomes) of patients with proximal small-bowel–predominant Crohn’s disease (SB-CD) (L4) or colon-predominant Crohn’s disease (C-CD) (L2 and/or colon-predominant L3). We also examined a differentiating candidate gene in a mouse model of colitis to address the translational relevance of our findings. Susceptibility subject–gene bipartite networks in SB-CD and C-CD also were generated to study the polygenic background of the disease subtypes.
Eight SB-CD and 11 C-CD cases met inclusion criteria. The clinical characteristics of these patients are included in Table 1. With methods described in the Supplementary Materials, we identified 115 single-nucleotide polymorphisms (SNPs) with a combined annotation-dependent depletion (CADD) (a tool for scoring the deleteriousness of single-nucleotide variants) Phred score >10 associated with 97 genes, which had significantly (P < .01) different allele variation between C-CD and SB-CD. An SNP in the EFNA3 gene was among the top 28 candidates with a CADD score >20 to differentiate between the 2 phenotypically distinct CD groups (Supplementary Table 1). EFNA3 rs17723260 (predicted to be deleterious) was found to have a significantly lower allele frequency (4.5%) in C-CD, compared with its allele frequency of 37.5% in SB-CD (chi square P = .0097). This finding indicated that EFNA3 might play a role in modulating colonic inflammation, in which a deleterious genetic defect might provide protection against colitis (and direct autoimmunity against the proximal small bowel) in the polygenic background of CD. Importantly, EFNA3 has been linked to ulcerative colitis4 and CD5 as well. The other 4 genes associated with the top 5 SNP candidates (Supplementary Table 1) already have been connected with IBD or mammalian intestinal inflammation. ACACB up-regulated in small-bowel strictures of the marmoset,6 STEAP1B linked to adult IBD,7 DSG1as a serologic marker of complicated CD,8 and CYP4F2 in which the specific polymorphism identified in this study (rs2108622) also has been observed to associate significantly with CD by Costea et al.9
Table 1.
Demographic and Baseline Characteristics of Patients With Either C-CD or SB-CD
| C-CD (n = 11) | SB-CD (n = 8) | P value | |
|---|---|---|---|
| Mean age at diagnosis, y | 10.9 | 11 | .95 |
| Sex, % female | 36.3 | 25 | 1 |
| Ethnicity, % Caucasian | 72.7 | 87.5 | .6 |
| Paris location, % | |||
| L1 | 0 | 50 | .018 |
| L2 | 54.5 | 0 | .018 |
| L3 | 45.5 | 25 | .63 |
| L4a | 18.2 | 37.5 | .6 |
| L4b | 0 | 100 | .0001 |
| Paris behavior, % | |||
| B1 | 45.5 | 12.5 | .177 |
| B2 | 27.2 | 62.5 | .18 |
| B3 | 18.2 | 0 | .485 |
| B2/B3 | 9.1 | 25 | .546 |
| Perianal disease, % yes | 36.4 | 12.5 | .338 |
| Presence of granulomas, % yes | 33.3 (2/6) | 33.3 (3/9) | 1 |
| Surgical intervention required, % yes | 54.5 | 75 | .633 |
| Surgery in first 2 years, % yes | 33.3 (2/6) | 50 (3/6) | 1 |
| Type of surgery, % | n = 6 | n = 6 | |
| Partial colectomy | 50 | 50 | 1 |
| Total colectomy | 33.3 | 0 | .455 |
| Ileocecotomy | 0 | 16.7 | 1 |
| Enterectomy | 16.7 | 66.7 | .242 |
| Small-bowel diversion only | 16.7 | 0 | 1 |
| Biologic agents used before surgery, % | n = 6 | n = 6 | |
| Infliximab | 100 | 66.7 | .455 |
| Adalimumab | 83.3 | 33.3 | .242 |
| Ustekinumab | 16.7 | 16.7 | 1 |
| Vedolizumab | 16.7 | 0 | 1 |
| Biologic agents used in nonsurgical patients, % | n = 5 | n = 2 | |
| Infliximab | 80 | 100 | .49 |
| Adalimumab | 60 | 0 | .429 |
C-CD, colon-predominant Crohn’s disease; SB-CD, small-bowel–predominant Crohn’s disease.
A dextran sodium sulfate (DSS) model experiment with 5 wild-type and 5 Efna3 null-allele female mice found Efna3 null mice to be protected significantly (P < .0001) against colitis (Supplementary Figure 1). We validated these findings in a subsequent experiment (Supplementary Figure 2). Male Efna3 null-allele mice did not demonstrate a significant difference in DSS colitis severity compared with wild-type littermates (not shown). Based on the literature and our murine model findings, we propose that normal or increased EFNA3 expression in patients with CD might shift susceptibility toward complicated colonic and terminal ileal (ie, L1, L2, and L3) disease. On the contrary, decreased EFNA3 expression might shift the disease toward the more proximal L4b phenotype by protecting against colonic injury in the background of CD.
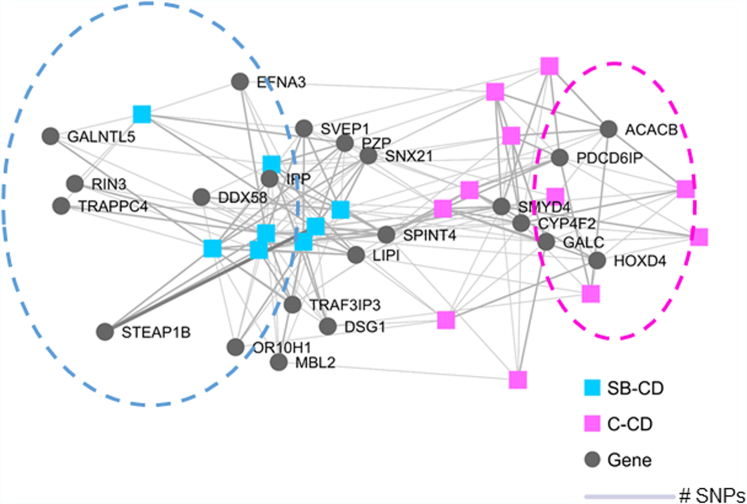
Recognizing the polygenic nature of most IBD cases, we studied the disease subtype differentiating subject–gene networks within our patients considering genes that had CADD >20 prediction for deleteriousness (ie, high susceptibility genes) (Supplementary Table 1). Differential gene networks separated SB-CD from C-CD (Figure 1), in which EFNA was associated with SB-CD, among others. Although there was no clear clustering of the differentiating genes in SB-CD when analyzed separately (Supplementary Figure 3A), we observed a commonly shared network between ACACB, CYP4F2, GALC, HOXD4, PDCD6IP, SPINT4, and SMYD4 in C-CD patients (Supplementary Figure 3B).
Figure 1.
Bipartite subject–gene networks between combined annotation-dependent depletion (CADD) >20, missense single-nucleotide polymorphism (SNP)-associated genes that differentiated between small-bowel–predominant Crohn’s disease (SB-CD) and colon-predominant Crohn’s disease (C-CD). The nodes represent either a subject or a gene and the edges represent the presence of polymorphism(s). The thickness of the edges is proportional to the number of SNPs in the corresponding subject–gene pair. Differential cluster of genes separated SB-CD (highlighted by blue circle) from C-CD (highlighted by pink circle).
This was a genetic study to address exome (coding) variation between C-CD (L2 or colon-predominant L3) and SB-CD (L4b). We describe a candidate gene compendium (Supplementary Table 1) in which SNPs predicted to be deleterious varied significantly in abundance between the 2 patient groups. Although our cohort sizes were small, they were sufficient to yield significant results, indicating that genetic predisposition may direct intestinal disease location in the background of pediatric CD. As a comparison, identical methodology (PLINK) (see Supplementary Materials) examining exome variation between granulomatous and nongranulomatous CD10 did not find significant separation, although the cohorts in that study were larger than within this work. The existing literature supports the significance of our results (Supplementary Materials). Even beyond the top candidates, there are numerous other genes within our compendium that have been implicated in IBD.
In summary, the biomedical literature and our mouse model findings implicate the translational relevance of our candidate gene compendium for directing colon- vs small-bowel–predominant CD development. We trust that our findings will be replicated in larger CD cohorts differentiated by disease location. Our work may set the nidus for CD subtype–based precision medicine by guiding individualized treatment strategies.
Acknowledgments
Author contributions
Halee Patel, Stanley Cho, Wenly Ruan, R. Alan Harris, Justin H. Qian, Savini Britto, Ashleigh Watson, Reka G. Szigeti, Antone Opekun, Numan Oezguen, Geoffrey Preidis, and Richard Kellermayer made substantial contributions to the conception and design of the study, or acquisition of data, or analysis and interpretation of data; Halee Patel, R. Alan Harris, and Richard Kellermayer drafted the article or revised it critically for important intellectual content; and Halee Patel, Stanley Cho, Wenly Ruan, R. Alan Harris, Justin H. Qian, Savini Britto, Ashleigh Watson, Reka G. Szigeti, Antone Opekun, Numan Oezguen, Geoffrey Preidis, and Richard Kellermayer gave final approval of the version to be submitted.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by the ProKIIDS Network of the Crohn’s and Colitis Foundation grant 585708 (R.K.), and Public Health Service grant DK56338, which funds the Texas Medical Center Digestive Diseases Center at Baylor College of Medicine and the Texas Medical Center Inflammatory Bowel Disease Tissue Bank. Supported by philanthropic funds from the Wagner, Frugoni, and Klaasmeyer families–led Gutsy Kids Fund and other generous donors contributing to the Gutsy Kids Fund. This work also was supported in part by the DR and GL Laws Fund.
Data Availability Data, analytic methods, and study materials will be made available to other researchers upon reasonable request to the corresponding author.
Note: To access the supplementary material accompanying this article, go to the full text version at http://doi.org/10.1016/j.jcmgh.2024.02.010
Supplementary Material
References
- 1.Serra E.G., et al. Nat Commun. 2020;11:995. doi: 10.1038/s41467-019-14275-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cordero R.Y., et al. Hum Mol Genet. 2023;32:873–882. doi: 10.1093/hmg/ddac269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tam V., et al. Nat Rev Genet. 2019;20:467–484. doi: 10.1038/s41576-019-0127-1. [DOI] [PubMed] [Google Scholar]
- 4.Fenton C.G., et al. Inflamm Bowel Dis. 2021;27:94–105. doi: 10.1093/ibd/izaa075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ventham N.T., et al. Cell Mol Gastroenterol Hepatol. 2023;16:431–450. doi: 10.1016/j.jcmgh.2023.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheh A., et al. Sci Rep. 2022;12:4430. doi: 10.1038/s41598-022-08255-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Q., et al. Gastroenterology. 2016;150:1196–1207. doi: 10.1053/j.gastro.2016.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yau Y.Y., et al. Mol Cell Proteomics. 2017;16:1244–1257. doi: 10.1074/mcp.M116.066506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costea I., et al. PLoS One. 2010;5 doi: 10.1371/journal.pone.0015672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris R.A., et al. J Pediatr Gastroenterol Nutr. 2023;77:354–357. doi: 10.1097/MPG.0000000000003873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.