Abstract
Haemophilus ducreyi is the etiologic agent of chancroid, a genital ulcer disease. The lipooligosaccharide (LOS) is considered to be a major virulence determinant and has been implicated in the adherence of H. ducreyi to keratinocytes. Strain A77, an isolate from the Paris collection, is serum sensitive, poorly adherent to fibroblasts, and deficient in microcolony formation. Structural analysis indicates that the LOS of strain A77 lacks the galactose residue found in the N-acetyllactosamine portion of the strain 35000HP LOS as well as the sialic acid substitution. From an H. ducreyi 35000HP genomic DNA library, a clone complementing the defect in A77 was identified by immunologic screening with monoclonal antibody (MAb) 3F11, a MAb which recognizes the N-acetyllactosamine portion of strain 35000HP LOS. The clone contained a 4-kb insert that was sequenced. One open reading frame which encodes a protein with a molecular weight of 33,400 was identified. This protein has homology to glycosyltransferases of Haemophilus influenzae, Haemophilus somnus, Neisseria species, and Pasteurella haemolytica. The putative H. ducreyi glycosyltransferase gene was insertionally inactivated, and an isogenic mutant of strain 35000HP was constructed. The most complex LOS glycoform produced by the mutant has a mobility on sodium dodecyl sulfate-polyacrylamide gel identical to that of the LOS of strain A77 and lacks the 3F11-binding epitope. Structural studies confirm that the most complex glycoform of the LOS isolated from the mutant lacks the galactose residue found in the N-acetyllactosamine portion of the strain 35000HP LOS. Although previously published data suggested that the serum-sensitive phenotype of A77 was due to the LOS mutation, we observed that the complemented A77 strain retained its serum-sensitive phenotype and that the galactosyltransferase mutant retained its serum-resistant phenotype. Thus, the serum sensitivity of strain A77 cannot be attributed to the galactosyltransferase mutation in strain A77.
Chancroid is a genital ulcerative disease caused by Haemophilus ducreyi which is prevalent in several developing countries (30, 46). Although chancroid has been characterized histologically (16, 17), little is known about the molecular basis for ulcer formation. A number of putative virulence determinants of H. ducreyi have been described, including a hemolytic cytotoxin (3, 35, 36, 45), cytolethal distending toxin (11), pili (9), a hemoglobin binding protein (15, 44), and lipooligosaccharide (LOS) (10, 18, 25, 32). The structure of the oligosaccharide portion of the LOS resembles the structure of human paraglobosides. Thus, it has been proposed that the LOS may help the organism evade the human immune response by molecular mimicry (26, 27). Direct injection of H. ducreyi LOS causes ulcers in rabbits and mice (10, 25, 47). Injection of H. ducreyi LOS causes a stronger inflammatory response in rabbits than injection of rough lipopolysaccharide from Escherichia coli (10). Other studies have implicated LOS in adherence to fibroblasts and keratinocytes (1, 18).
Odumeru and coworkers demonstrated that strain A77 was killed by incubation in normal human serum (32, 33). Further, they observed that the bactericidal activity in normal human serum could be removed by incubation with heat-killed whole cells or LOS from serum-sensitive strains but not by incubation with heat-killed serum-resistant strains or LOS from serum-resistant strains. They concluded that LOS composition contributed to the susceptibility of H. ducreyi to the bactericidal activity of normal human serum. A serum-resistant derivative of A77 was also isolated, and the LOS composition of this derivative appeared to differ from that of A77.
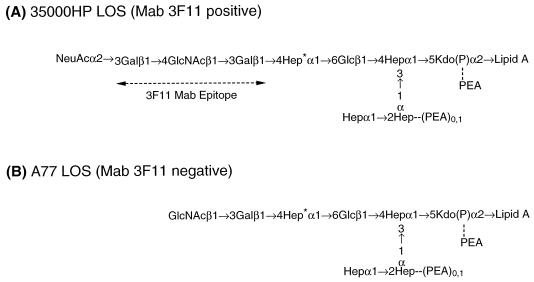
In order to increase our understanding of the role of LOS in the pathogenesis of chancroid, the structures of the major glycoforms of the LOS from H. ducreyi strains have been determined (8, 28, 29, 41, 42). The major glycoform from strain 35000HP has the following structure: Galβ1→4GlcNAcβ1→ 3Galβ1→4Hepα1→6Glcβ1→(Hepα1→2Hepα1→)3,4Hepα1→ 5Kdo. Approximately one-third of the terminal galactose residues are substituted with sialic acid (8, 28). The structure of the LOS of strain A77 was also determined (B. W. Gibson, W. Melaugh, N. J. Phillips, and A. A. Campagnari, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. B/D-51, p. 39, 1999). The most complex glycoform of the A77 LOS lacks the galactose found in the N-acetyllactosamine portion of strain 35000HP LOS. The monoclonal antibody (MAb) 3F11 binds to the terminal Galβ1→4GlcNAcβ1→3Gal epitope; thus A77 does not bind MAb 3F11 (40).
In this study, we demonstrate that the LOS biosynthetic defect in strain A77 is in the galactosyltransferase II gene, designated lgtB, and that this defect is not responsible for the serum-sensitive phenotype.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
H. ducreyi strains (Table 1) were grown at 35°C with 5% CO2 on chocolate agar (Becton Dickinson). Chocolate agar plates supplemented with streptomycin at 20 μg/ml, kanamycin at 20 μg/ml, and/or X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) at 40 μg/ml were prepared as previously described (18, 34). Brain heart infusion broth supplemented with 5% fetal calf serum, 0.0025% hemin chloride solution (Sigma; predissolved in 20 mM NaOH), and 1% IsoVitaleX (sBHI) was used for growth of H. ducreyi in liquid medium. E. coli strains were grown on Luria-Bertani (LB) plates or in LB broth supplemented with appropriate antibiotics. Kanamycin was used at 20 μg/ml, chloramphenicol was used at 30 μg/ml, and ampicillin was used at 50 μg/ml where appropriate.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | Host strain used for cloning | Gibco/BRL |
| H. ducreyi | ||
| 35000HP | Virulent wild-type strain; isolate from Winnipeg; passaged through a human volunteer | 37 |
| A77 | Serum sensitive; lacks terminal galactose residue on its LOS | 32, 33 |
| 35000HP-RSM210 | LOS mutant derived from strain 35000HP by insertion of the ΩKm-2 cassette into the galactosyltransferase II gene | This study |
| Plasmids | ||
| pUC19 | Cloning vector; Ampr | |
| pLS88 | Shuttle vector capable of replication in H. ducreyi and E. coli; Smr Sulr Kanr | 13 |
| pSuperCosI | Cloning vector used as source of cassette for modification of pLS88 | Stratagene |
| pUC4DEcat | Source of the chloramphenicol acetyltransferase gene (cat) | Bruce Green |
| pRSM1937 | pLS88 carrying pSuperCosI cloning region with cat gene in the BamHI site | This study |
| pLS99 | pLS88 with the cloning region of pSuperCosI inserted into EcoRI site | This study |
| pRSM1955 | Recombinant clone from H. ducreyi 35000HP plasmid library which complements the LOS defect in strain A77 | This study |
| pRSM1791 | Suicide vector for selection of H. ducreyi mutants | 7 |
| pRSM1956 | pUC19 containing the 3-kb EcoRI fragment from pRSM1955 | This study |
| pJRS102.0 | Source of the ΩKm-2 cassette | 38 |
| pRSM1975 | pRSM1956 with ΩKm-2 insertion in the putative glycosyltransferase II gene | This study |
| pRSM1977 | pRSM1791 carrying EcoRI fragment from pRSM1975 in the NotI site | This study |
Modification of shuttle vector pLS88.
The H. ducreyi-E. coli shuttle vector pLS88 (13) was modified by addition of the cloning site from pSuperCosI (Stratagene, La Jolla, Calif.) which contains the following restriction sites in the indicated sequence: EcoRI-NotI-BamHI-NotI-EcoRI. To facilitate construction, the chloramphenicol acetyltransferase gene (cat) from pUC4DEcat was cloned into the BamHI site of pSuperCosI. The EcoRI fragment from this construct was then ligated to EcoRI-digested, dephosphorylated pLS88. The ligation mixture was transformed into DH5α, and clones were selected on LB plates supplemented with kanamycin and chloramphenicol. A plasmid with the appropriate restriction map, designated pRSM1937, was digested with BamHI, religated, and transformed into DH5α, and clones were selected on kanamycin plates. A plasmid lacking the chloramphenicol resistance gene was saved as pLS99. This plasmid has a unique BamHI cloning site flanked by NotI sites.
Construction of an H. ducreyi 35000HP genomic DNA library.
Chromosomal DNA from H. ducreyi 35000HP was purified with the AGTC bacterial genomic DNA purification kit (Edge Biosystems, Gaithersburg, Md.) and was partially digested with Sau3AI. Fragments 3 to 5 kb in size were isolated on a 1% agarose gel, purified with the Geneclean kit (Bio 101, Inc., La Jolla, Calif.), and then ligated to BamHI and calf intestine alkaline phosphatase-treated pLS99. The ligation reaction mixture was electroporated into DH5α as previously described (34), yielding approximately 1.2 × 104 recombinant clones. These transformants were harvested from plates and suspended in 100 ml of LB broth containing kanamycin. Some cells were kept in 10% glycerol and stored at −70°C for future usage. Plasmid DNA was purified from the remainder of the preparation using the Wizard DNA purification system (Promega Corp., Madison, Wis.).
Immunologic screening of library.
The plasmid library was electroporated into H. ducreyi strain A77 as previously described (34) with a Bio-Rad Gene Pulser II at 200 Ω, 2.5 kV, and 25 μF. Transformants were transferred to nitrocellulose filters (Schleicher & Schuell, Keene, N.H.) and screened immunologically by colony blot assay as described previously using MAb 3F11 (18). An immunologically reactive clone was identified. The plasmid from this clone was designated pRSM1955.
Nucleotide sequence analysis.
The DNA sequence was determined in both directions using an ABI 377 automated DNA sequencer. Contig assembly and sequence analysis were performed with DNASTAR (Madison, Wis.) software. Homology was determined through the National Center for Biotechnology Information BLAST network server, the Megalign program (DNASTAR), and the GAP program from the Genetics Computer Group, Madison, Wis.
Construction of an H. ducreyi 35000HP isogenic mutant deficient in galactosyltransferase II.
The insert in pRSM1955 contains an internal EcoRI site as well as the two EcoRI sites at the vector insert junction. The large fragment in the insert was isolated as an EcoRI fragment and subcloned into pUC19 to form pRSM1956. Plasmid pRSM1956 was partially digested with MunI. Full-length linear DNA was isolated on an agarose gel, dephosphorylated, and ligated to the ΩKm-2 element, which had been isolated as an EcoRI fragment from pJRS102.0 (38). The ligation mixture was transformed into DH5α, and clones were isolated on LB plates supplemented with kanamycin and ampicillin. EcoRI and BamHI restriction enzyme analysis was performed to select a plasmid in which the kanamycin cassette was inserted in the putative galactosyltransferase II gene. A plasmid with the appropriate restriction map was saved as pRSM1975.
We recently reported a novel strategy to perform allele replacement in H. ducreyi (7). The ColEI-type suicide vector pRSM1791, which contains the lacZ gene, was constructed. We observed that the hydrolysis product of X-Gal is toxic to H. ducreyi, and thus β-galactosidase can be used as a counter-selectable marker. The EcoRI fragment of pRSM1975 was isolated, blunt ended, and cloned into the NotI site of pRSM1791 to form pRSM1977. pRSM1977 DNA was electroporated into H. ducreyi 35000HP, and kanamycin-resistant clones were selected. Kanamycin-resistant clones were then streaked for isolation on chocolate agar containing both kanamycin and X-Gal. Clones that were white and that grew normally were further characterized by Southern blotting to verify the allele exchange and loss of plasmid sequences. A representative mutant, designated 35000HP-RSM210, was chosen for LOS biochemical structure analysis.
Southern blot hybridization.
Chromosomal DNA was isolated from H. ducreyi strains and digested with EcoRI, subjected to electrophoresis on a 0.7% agarose gel, and transferred to a nylon membrane using the Turbo Blotter kit (Schleicher & Schuell). The plasmid DNA of pRSM1956 was labeled with 32P using the RadPrime DNA labeling system (Gibco/BRL, Gaithersburg, Md.). Hybridization and posthybridization were performed as described previously (34).
Complementation of the glycosyltransferase II mutation.
Competent cells of 35000HP-RSM210 were prepared and transformed with pRSM1955 as described above. Clones were selected on chocolate plates supplemented with streptomycin.
Preparation and analysis of LOS.
Crude LOS preparations from H. ducreyi strains were prepared by a modified microphenol method and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 14% acrylamide gel as previously reported (10). To obtain larger amounts of the mutant strain LOS (1 mg) for subsequent mass spectrometric characterization studies, this strain was grown in liquid media (1 liter) and LOS was prepared as described elsewhere (5, 24, 48). Crude LOS preparations isolated from both the mutant and wild-type strains were O-deacylated with hydrazine (37°C for 30 min) (21) to increase solubility and make the LOS more amenable to mass spectrometric analysis (19). In addition, a portion of the O-deacylated LOS (150 μg) was further treated with 48% aqueous HF (at 4°C for 16 h) to remove phosphoethanolamine (PEA) and phosphate groups (29). Finally, an oligosaccharide fraction was prepared by hydrolysis of the crude LOS (175 μg) in a solution of 1% acetic acid (350 μl) at 100°C for 2 h (29). The resulting water-soluble oligosaccharides were then separated from the lipid A fraction by centrifugation (5,000 × g, 4°C) and purified by size exclusion chromatography using two 300- by 7.8-mm BioSelect SEC 125-5 columns (Bio-Rad) connected in series. Samples were eluted in 50 mM pyridinium acetate (pH 5.2) at a flow rate of 1 ml min−1. Fractions were collected in 0.5- to 1-ml volumes. The O-deacylated LOS, the HF-treated and O-deacylated LOS, and the acid-released oligosaccharide fractions were all analyzed by mass spectrometry as described below.
Mass spectra were obtained for all samples by matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF) mass spectrometry on a Voyager DE or Voyager DESTR instrument (PE Biosystems, Framingham, Mass.). Both instruments were equipped with a nitrogen laser (337 nm) and were run under delayed-extraction conditions (19): the delay times were 100 (Voyager DE) and 100 to 200 ns (Voyager DESTR); the grid voltages were 92 to 94% of full-acceleration voltage (20 to 30 kV) under linear conditions and 72.5 to 75% of full-acceleration voltage (20 to 25 kV) under reflectron conditions. Mass spectra were run in both the positive- and negative-ion modes and under post-source-decay (PSD) conditions (43). The obtained mass spectra were externally calibrated with an equimolar mixture of angiotensin II, bradykinin, LHRH, bombesin, α-MSH (CZE mixture; Bio-Rad), and adrenocorticotropin 1-24 (Sigma). All samples were prepared using a 320 mM 2,5-dihydroxybenzoic acid solution in 4:1 (vol/vol) acetone-water containing 175 mM 1-hydroxyisoquinoline (19). In all cases, 1 μl of analyte (0.1 to 1 μg of material) was mixed with 1 μl of matrix solution, desalted with cation exchange resin beads (DOWEX 50X, NH4+) (31), and then air dried at room temperature on a stainless steel target. Typically, 20 to 50 laser shots were used to record each linear and reflectron spectrum, or spectral segment if PSD conditions were used.
Bactericidal assay.
H. ducreyi cells grown for 48 h on a chocolate agar plate were washed in phosphate-buffered saline, then subcultured into sBHI broth, and grown to early log phase (optical density at 600 nm [OD600] = 0.2). The cells were pelleted and resuspended to the same OD in Hanks' balanced salt solution–0.1% gelatin (HBSSg). The cells were diluted 1:100 in HBSSg, and 30 μl was used in each sample. The cells were assayed in HBSSg with 40 and 60% normal human serum (pooled from four subjects) in a total volume of 300 μl. Two controls were employed for each sample. A control without serum was used to determine the total cell count, and a heat-inactivated serum control was used to verify that killing of the bacteria was due to complement. Serum was inactivated by heating at 56°C for 30 min. Except for the control without serum, samples were incubated at 35°C with shaking (160 rpm). Dilutions of the control without serum were plated immediately. The other samples were plated after 60 and 120 min. All dilutions were plated in triplicate on chocolate agar plates. Bactericidal activity was measured as percent survival compared to that of the time zero control.
Nucleotide sequence accession number.
The sequence of the insert of pRSM1955 obtained in this study has been assigned GenBank accession no. AF224466.
RESULTS AND DISCUSSION
Structural characterization of strain A77 LOS.
H. ducreyi strain A77, a strain from the Paris collection, is serum sensitive (32, 33), poorly adherent to fibroblasts, and deficient in microcolony formation (2, 4). The structure of the LOS of strain A77 was determined by nuclear magnetic resonance, carbohydrate, and mass spectrometric analysis (Gibson et al., Abstr. 99th Gen. Meet. Am. Soc. Microbiol.). This LOS lacks the sialic acid substitution as well as the galactose residue found in the N-acetyllactosamine portion of strain 35000HP and therefore is defective in 3F11 binding (Fig. 1). Since galactose was present in the LOS of strain A77, we hypothesized that A77 could synthesize UDP-galactose, produced a functional galactosyltransferase I, and was likely deficient in the galactosyltransferase II gene.
FIG. 1.
Major LOS structures from H. ducreyi strains 35000HP (A) and A77 (B). All core heptoses are l-glycero-d-manno-heptose with the exception of the branch heptose (asterisk), which is d-glycero-d-manno-heptose.
Screening of H. ducreyi 35000HP genomic DNA library.
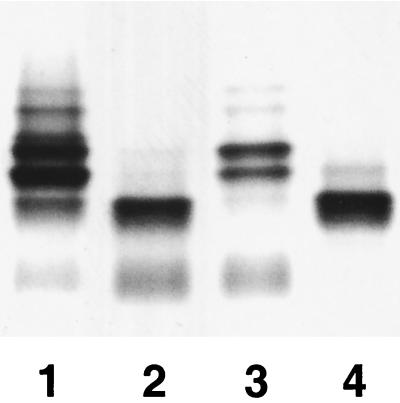
The A77 strain was used as the recipient for an H. ducreyi 35000HP genomic plasmid library. Clones which reacted with MAb 3F11 were identified. One plasmid, designated pRSM1955, conferred 3F11 reactivity to strain A77. The SDS-PAGE profile of the LOS isolated from strain A77/pRSM1955 was similar to the profile of the LOS from strain 35000HP (Fig. 2).
FIG. 2.
SDS-PAGE silver-stained gel of LOS preparations. LOS preparations were from strains 35000HP (lane 1), A77 (lane 2), A77/pRSM1955 (lane 3), and 35000HP-RSM210 (lane 4).
Nucleotide sequence analysis.
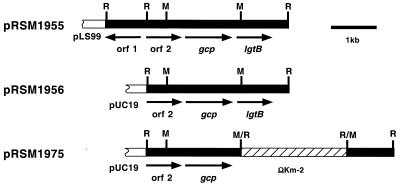
The sequence of the insert of pRSM1955 was determined. This insert contains three complete open reading frames (ORFs) (Fig. 3). The 5′ portion of the sequence contains a partial ORF, designated orf1, encoding the H. ducreyi homologue of pseudouridylate synthase (P44445). orf2 begins at nucleotide 905 and is 783 bp in length. The derived amino acid sequence has homology with those of hypothetical proteins from Haemophilus influenzae (HI0177) and other gram-negative organisms. orf3 begins at nucleotide 1772 and is 1,047 bp in length. The derived amino acid sequence is 87% identical to that of the product of the sialylglycoprotease (gcp) gene of P. haemolytica, and therefore orf3 has been designated gcp. orf4 starts at nucleotide 2833 and is 843 bp in length. The hypothetical protein encoded by orf4 has homology with the products of numerous glycosyltransferase genes including genes from H. influenzae (12, 22), Haemophilus somnus (Genbank accession no. AF096997) (unpublished data), Neisseria meningitidis (23), Neisseria gonorrhoeae (20), and P. haemolytica (39). As orf4 encodes a glycosyltransferase that catalyzes the transfer of galactose to GlcNAc (see below) and has homology to Neisseria galactosyltransferases with similar specificities (encoded by a gene previously designated lgtB), we have tentatively designated this gene lgtB pending a unified nomenclature for the glycosyltransferase genes of Haemophilus and Neisseria.
FIG. 3.
Partial restriction and ORF map of the plasmids characterized in this study. The plasmid pRSM1955 contains a 4-kb genomic fragment of H. ducreyi DNA cloned into the shuttle vector pLS99. Arrows indicate the position and direction of transcription of the ORFs. Plasmid pRSM1975 has the ΩKm-2 cassette inserted in the MunI site in the galactosyltransferase II (lgtB) gene in pRSM1956. The restriction site designations are as follows: R, EcoRI; M, MunI.
The gene arrangement we observed is reminiscent of that seen in P. hemolytica in that the P. hemolytica gcp gene is also 5′ to a glycosyltransferase gene (39). In contrast, the H. influenzae gcp gene is 5′ to the thymidine kinase gene. The function of the gcp genes in H. ducreyi, H. influenzae, and E. coli is unknown. We have failed in numerous attempts to construct a mutant deficient in the Haemophilus homologues of gcp, suggesting that the mutation is lethal, and a recent report indicates that the homologues of gcp in E. coli and Bacillus subtilis are essential for growth of these bacteria as well (6).
Insertional mutagenesis of the putative glycosyltransferase gene.
Given the observed homology between the H. ducreyi lgtB gene and other glycosyltransferase genes, it was extremely likely that the observed complementation in A77/pRSM1955 was due to the lgtB gene. In order to demonstrate directly that a mutation in lgtB would result in a strain producing a truncated LOS, we insertionally inactivated the lgtB gene in pRSM1956 and constructed an isogenic mutant in the 35000HP background. Presumptive mutants were identified as described in Materials and Methods, and Southern blot hybridization was performed to confirm proper allelic exchange. The plasmid pRSM1956 containing the glycosyltransferase gene was used to probe blots containing EcoRI-digested chromosomal DNA from six putative mutants. A hybridization band of 5.5 kb was observed with strain 35000HP. All mutants had a single hybridizing band of 8.5 kb (data not shown), and one mutant, designated 35000HP-RSM210, was saved for further analysis.
Analysis of the LOS from the isogenic galactosyltransferase mutant of H. ducreyi.
LOS isolated from strain 35000HP-RSM210 was separated by SDS-PAGE along with LOS obtained from the parental strain, 35000HP (Fig. 2). The most complex major glycoform produced by strain 35000HP-RSM210 ran with a mobility identical to that of the major glycoform produced by strain A77. The major glycoform produced by strains A77 and 35000HP-RSM210 ran considerably faster than the two major glycoforms present in strain 35000HP LOS. One of these two major glycoforms terminates in N-acetyllactosamine, and the other terminates in sialyl-N-acetyllactosamine (Fig. 1).
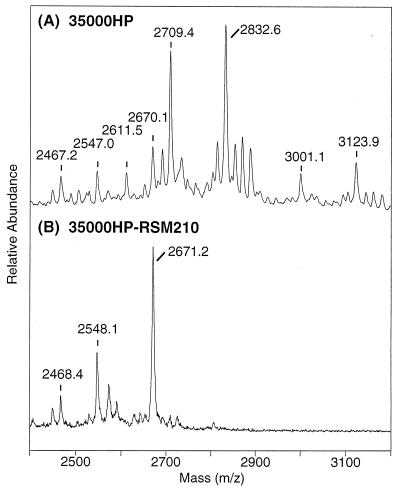
The structure of the strain 3500HP-RSM210 LOS glycoforms was determined by mass spectrometric experiments on several chemically degraded forms of the LOS in a fashion similar to that previously reported for structural studies on the parental strain 35000 (8, 18). To obtain the precise molecular masses of the LOS glycoforms, LOSs from both the mutant and parental wild type strains were converted into their O-deacylated forms and analyzed by negative-ion MALDI-TOF mass spectrometry. As shown in Fig. 4A, the wild-type O-deacylated LOS gave a series of singly deprotonated molecular ion peaks, with the four most abundant peaks corresponding to the LOS glycoforms terminated in N-acetyllactosamine (m/z 2709.4) and sialyl-N-acetyllactosamine (m/z 3001.1), both of which were partially replaced with a single PEA group (m/z 2832.6 and 3123.9, respectively). These masses and assignments were consistent with structures previously reported for the wild-type strain passaged in humans (8). In contrast, O-deacylated LOS from the mutant showed three abundant peaks at lower masses, (M − H)− at m/z 2671.2, 2548.1, and 2468.4 (Fig. 4B). The two higher peaks at m/z 2671.2 and 2548.1 yielded molecular masses consistent with the expected composition for this O-deacylated LOS, as shown in Fig. 1B, involving the loss of terminal Gal (−162 Da; m/z 2710→2548 and 2833→2671) or its sialylated counterpart, NeuAcα2→3Gal (−453 Da; m/z 3001→2548 and 3124→2671), from the four major LOS glycoforms present in the parental strain. Indeed, these same species are also present in the parental wild-type strain but at much lower abundances. The remaining lower-mass peak at m/z 2468 appears to arise from the additional loss of a HexNAc (−203-Da) moiety, and can be attributed to the loss of terminal GlcNAc from the major LOS glycoform at m/z 2671. In addition to the molecular ions, several “prompt fragments” (data not shown) resulting from gas phase fragmentation at the labile Kdo-lipid A were present. Fragmentation of this bond among the three molecular ions gives rise to the conserved diphosphoryl-di-N-acyl lipid A peak at m/z 952 and the three oligosaccharides that differ in PEA content at m/z 1718.8, 1595.7, and 1515.2. Fragmentation analysis of the major LOS glycoform at m/z 2671.4 by MALDI-PSD, a technique that allows for specific ion selection followed by detection of metastable decomposition fragments, confirmed this interpretation. Moreover, the PSD spectrum showed that the oligosaccharide fragment at m/z 1718.2 undergoes further decomposition with losses of PEA (m/z 1595.9) and/or phosphorylphosphoethanolamine (PPEA; m/z 1515.7), consistent with a structure containing a single PEA linked exclusively to the phosphate of phospho-Kdo, i.e., Kdo(PPEA), and not the heptose core. As the pattern of phosphorylation can vary in LOS, particularly when oligosaccharide truncations are introduced, the corresponding dephosphorylated O-deacylated LOS from this mutant was examined by MALDI and MALDI-PSD after HF treatment. The most abundant molecular ion species was (M + Na)+ at m/z 2331.60 (exact mass), consistent with the expected loss of two phosphates (−160 Da) from the lipid A moiety and a loss of PPEA (−203 Da) from the oligosaccharide moiety. This phosphate substitution pattern is therefore similar, if not identical, to that defined in the parental wild-type strain.
FIG. 4.
Molecular ion region of the negative-ion MALDI-TOF (linear mode) mass spectra of O-deacylated LOS from the parental 35000HP strain (A) and mutant 35000HP-RSM210 strain (B).
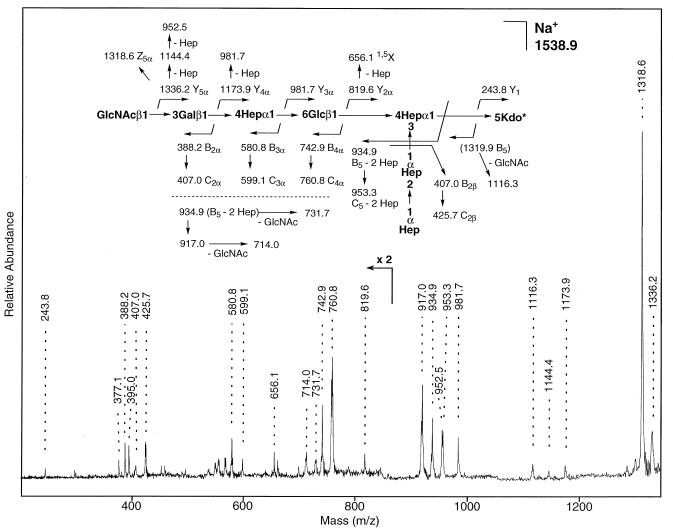
Since alternative biosynthesis has been observed in other Haemophilus mutants deficient in LOS biosynthesis, the oligosaccharide portion of the LOS was subjected to sequence analysis. A MALDI-reflectron spectrum of the resulting oligosaccharide fraction obtained after mild acid hydrolysis of LOS was first and revealed a major (M + Na)+ oligosaccharide peak at m/z 1538.49 (exact mass) and a second minor sodiated molecular ion at m/z 1335.36 (data not shown). These masses agree with those expected from the three previously identified major LOS glycoform species (Fig. 4B). These oligosaccharide masses are also consistent with the substitution of a PPEA group on Kdo, since it has been previously noted that phosphate groups substituted at the 4 position of Kdo undergo β-elimination under mild acid conditions, yielding terminal anhydro-Kdo. A MALDI-PSD spectrum of the most abundant oligosaccharide (Fig. 5) yielded a series of sequence ions that correspond to cleavage of the carbohydrate chain, including a complete Y-ion series (m/z 1336.2, 1173.9, 981.7, 819.6, and 243.8) and B-ion series (m/z 388.2, 580.8, 742.9, and 934.9). (Ion nomenclature is according to that proposed by Domon and Costello [14]). Together with previous mass spectrometry data, these studies support a structure in which the most abundant LOS glycoforms from 35000HP-RSM210 are identical to those from the parental strain LOS except that they now terminate prior to the second galactose residue. Furthermore, the LOS of this mutant strain directly corresponds to the LOS of the “atypical” wild-type strain A77.
FIG. 5.
Partial positive-ion MALDI-PSD mass spectrum of sodiated oligosaccharide (M + Na)+ (timed ion selection of m/z 1539; average masses) isolated after acetic acid hydrolysis of LOS from the mutant strain 35000HP-RSM210. Kdo*, anhydro-Kdo.
The mutation in strain 35000HP-RSM210 was complemented by transformation with pRSM1955, the plasmid initially shown to complement the defect in strain A77. Strain 35000HP-RSM210/pRSM1955 produces an LOS that contains the galactose- and sialic acid-containing glycoforms observed in H. ducreyi 35000HP (data not shown).
Sequence of the galactosyltransferase II gene from strain A77.
The galactosyltransferase II genes from strains 35000HP and A77 were amplified by PCR. Residual PCR primers were removed by spin column purification, and the PCR products were directly sequenced. The nucleotide sequence of the gene from 35000HP agreed with the sequence determined from the plasmid clone, while the sequence of the gene amplified from strain A77 differed by a single nucleotide. The derived amino acid sequence of the A77 protein differs from the sequence of the strain 35000HP protein by an alanine-to-threonine change at position 207 (data not shown).
Serum sensitivities of strains 35000HP and 35000HP-RSM210.
In order to determine whether the serum sensitivity of strain A77 was due to the LOS mutation, as previous data suggested, we determined the serum sensitivities of strains 35000HP, A77, 35000HP-RSM210, and A77/pRSM1955. As reported by other investigators, strain 35000HP is resistant to the bactericidal activity of 40 or 60% normal human serum. In our experiments, the survival of strain 35000HP in 60% normal human serum was 269% ± 26%, while the survival of strain A77 under the same conditions was 1% ± 1%. Surprisingly, complementation of the LOS defect in strain A77 (strain A77/pRSM1955) did not result in a serum-resistant phenotype (<1% survival in 60% serum). Similarly, introduction of a galactosyltransferase II mutation into strain 35000HP did not make the strain serum sensitive (222% ± 53% survival).
LOS (lipopolysaccharide) is a critical virulence factor in many gram-negative pathogens, and mutations in LOS biosynthesis frequently result in strains with a decreased resistance to the bactericidal activity of normal human serum. Characterization of the LOS from strain A77 and identification of the glycosyltransferase mutation in strain A77 were of particular interest as strain A77 was considered to be serum sensitive because of a LOS defect. However, our data fail to support this earlier hypothesis, as repair of the LOS defect in A77 failed to confer a serum-resistant phenotype to the strain and an isogenic mutant of strain 35000HP deficient in the lgtB gene retained its serum-resistant phenotype.
ACKNOWLEDGMENTS
We thank Huachun Zhong for excellent technical assistance.
This work was supported by National Institutes of Health grants R01 AI38444 (R.S.M.) and R01 AI31254 (B.W.G.) and by PE Biosystems, which kindly provided instrumentation to B.W.G. The DNA sequence was determined by the Core Facility at Children's Research Institute, which was supported in part by National Institutes of Health grant HD34615.
ADDENDUM IN PROOF
Elkins and coworkers recently identified a protein, designated DrsA, that is in part responsible for the serum resistance of Haemophilus ducreyi (C. Elkins, K. J. Morrow, Jr., and B. Olsen. Infect. Immun. 68:1608–1619, 2000).
REFERENCES
- 1.Alfa M J, DeGagne P. Attachment of Haemophilus ducreyi to human foreskin fibroblasts involves LOS and fibronectin. Microb Pathog. 1997;22:39–46. doi: 10.1006/mpat.1996.0089. [DOI] [PubMed] [Google Scholar]
- 2.Alfa M J, DeGagne P, Hollyer T. Haemophilus ducreyi adheres to but does not invade cultured human foreskin cells. Infect Immun. 1993;61:1735–1742. doi: 10.1128/iai.61.5.1735-1742.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alfa M J, DeGagne P, Totten P A. Haemophilus ducreyi hemolysin acts as a contact cytotoxin and damages human foreskin fibroblasts in cell culture. Infect Immun. 1996;64:2349–2352. doi: 10.1128/iai.64.6.2349-2352.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alfa M J, Stevens M K, DeGagne P, Klesney-Tait J, Radolf J D, Hansen E J. Use of tissue culture and animal models to identify virulence-associated traits of Haemophilus ducreyi. Infect Immun. 1995;63:1754–1761. doi: 10.1128/iai.63.5.1754-1761.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Apicella M A, Griffiss J M, Schneider H. Isolation and characterization of lipopolysaccharides, lipooligosaccharides, and lipid A. Methods Enzymol. 1994;235:242–252. doi: 10.1016/0076-6879(94)35145-7. [DOI] [PubMed] [Google Scholar]
- 6.Arigoni F, Talabot F, Peitsch M, Edgerton M D, Meldrum E, Allet E, Fish R, Jamotte T, Curchod M L, Loferer H. A genome-based approach for the identification of essential bacterial genes. Nat Biotechnol. 1998;16:851–856. doi: 10.1038/nbt0998-851. [DOI] [PubMed] [Google Scholar]
- 7.Bozue J A, Tarantino L, Munson R S., Jr Facile construction of mutations in Haemophilus ducreyi using lacZ as a counter-selectable marker. FEMS Microbiol Lett. 1998;164:269–273. doi: 10.1111/j.1574-6968.1998.tb13097.x. [DOI] [PubMed] [Google Scholar]
- 8.Bozue J A, Tullius M V, Wang J, Gibson B W, Munson R S., Jr Haemophilus ducreyi produces a novel sialyltransferase. Identification of the sialyltransferase gene and construction of mutants deficient in the production of the sialic acid-containing glycoform of the lipooligosaccharide. J Biol Chem. 1999;274:4106–4114. doi: 10.1074/jbc.274.7.4106. [DOI] [PubMed] [Google Scholar]
- 9.Brentjens R J, Ketterer M, Apicella M A, Spinola S M. Fine tangled pili expressed by Haemophilus ducreyi are a novel class of pili. J Bacteriol. 1996;178:808–816. doi: 10.1128/jb.178.3.808-816.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campagnari A A, Wild L M, Griffiths G E, Karalus R J, Wirth M A, Spinola S M. Role of lipooligosaccharides in experimental dermal lesions caused by Haemophilus ducreyi. Infect Immun. 1991;59:2601–2608. doi: 10.1128/iai.59.8.2601-2608.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cope L D, Lumbley S, Latimer J L, Klesney-Tait J, Stevens M K, Johnson L S, Purven M, Munson R S, Jr, Lagergard T, Radolf J D, Hansen E J. A diffusible cytotoxin of Haemophilus ducreyi. Proc Natl Acad Sci USA. 1997;94:4056–4061. doi: 10.1073/pnas.94.8.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cope L D, Yogev R, Mertsola J, Latimer J L, Hanson M S, McCracken G H, Jr, Hansen E J. Molecular cloning of a gene involved in lipooligosaccharide biosynthesis and virulence expression by Haemophilus influenzae type B. Mol Microbiol. 1991;5:1113–1124. doi: 10.1111/j.1365-2958.1991.tb01884.x. [DOI] [PubMed] [Google Scholar]
- 13.Dixon L G, Albritton W L, Willson P J. An analysis of the complete nucleotide sequence of the Haemophilus ducreyi broad-host-range plasmid pLS88. Plasmid. 1994;32:228–232. doi: 10.1006/plas.1994.1060. [DOI] [PubMed] [Google Scholar]
- 14.Domon B, Costello C E. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconjugate J. 1988;5:397–409. [Google Scholar]
- 15.Elkins C, Chen C J, Thomas C E. Characterization of the hgbA locus encoding a hemoglobin receptor from Haemophilus ducreyi. Infect Immun. 1995;63:2194–2200. doi: 10.1128/iai.63.6.2194-2200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freinkel A L. Histological aspects of sexually transmitted genital lesions. Histopathology. 1987;11:819–831. doi: 10.1111/j.1365-2559.1987.tb01885.x. [DOI] [PubMed] [Google Scholar]
- 17.Gaisin A, Heaton C L. Chancroid: alias the soft chancre. Int J Dermatol. 1975;14:188–197. doi: 10.1111/ijd.1975.14.3.188. [DOI] [PubMed] [Google Scholar]
- 18.Gibson B W, Campagnari A A, Melaugh W, Phillips N J, Apicella M A, Grass S, Wang J, Palmer K L, Munson R S., Jr Characterization of a transposon Tn916-generated mutant of Haemophilus ducreyi 35000 defective in lipooligosaccharide biosynthesis. J Bacteriol. 1997;179:5062–5071. doi: 10.1128/jb.179.16.5062-5071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson B W, Engstrom J J, John C M, Hines W, Falick A M. Characterization of bacterial lipooligosaccharides by delayed extraction matrix-assisted laser desorption ionization time-of-flight mass spectrometry. J Am Soc Mass Spectrom. 1997;8:645–658. [Google Scholar]
- 20.Gotschlich E C. Genetic locus for the biosynthesis of the variable portion of Neisseria gonorrhoeae lipooligosaccharide. J Exp Med. 1994;180:2181–2190. doi: 10.1084/jem.180.6.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helander I M, Nummila K, Kilpelainen I, Vaara M. Increased substitution of phosphate groups in lipopolysaccharides and lipid A of polymyxin-resistant mutants of Salmonella typhimurium and Escherichia coli. Prog Clin Biol Res. 1995;392:15–23. [PubMed] [Google Scholar]
- 22.High N J, Deadman M E, Moxon E R. The role of a repetitive DNA motif (5′-CAAT-3′) in the variable expression of the Haemophilus influenzae lipopolysaccharide epitope alpha Gal(β1-4)Gal. Mol Microbiol. 1993;9:1275–1282. doi: 10.1111/j.1365-2958.1993.tb01257.x. [DOI] [PubMed] [Google Scholar]
- 23.Jennings M P, Hood D W, Peak I R, Virji M, Moxon E R. Molecular analysis of a locus for the biosynthesis and phase-variable expression of the lacto-N-neotetraose terminal lipopolysaccharide structure in Neisseria meningitidis. Mol Microbiol. 1995;18:729–740. doi: 10.1111/j.1365-2958.1995.mmi_18040729.x. [DOI] [PubMed] [Google Scholar]
- 24.Johnson K G, Perry M B. Improved techniques for the preparation of bacterial lipopolysaccharides. Can J Microbiol. 1976;22:29–34. doi: 10.1139/m76-004. [DOI] [PubMed] [Google Scholar]
- 25.Lagergard T. The role of Haemophilus ducreyi bacteria, cytotoxin, endotoxin and antibodies in animal models for study of chancroid. Microb Pathog. 1992;13:203–217. doi: 10.1016/0882-4010(92)90021-f. [DOI] [PubMed] [Google Scholar]
- 26.Mandrell R E, Apicella M A. Lipo-oligosaccharides (LOS) of mucosal pathogens: molecular mimicry and host-modification of LOS. Immunobiology. 1993;187:382–402. doi: 10.1016/S0171-2985(11)80352-9. [DOI] [PubMed] [Google Scholar]
- 27.Mandrell R E, McLaughlin R, Aba Kwaik Y, Lesse A, Yamasaki R, Gibson B, Spinola S M, Apicella M A. Lipooligosaccharides (LOS) of some Haemophilus species mimic human glycosphingolipids, and some LOS are sialylated. Infect Immun. 1992;60:1322–1328. doi: 10.1128/iai.60.4.1322-1328.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melaugh W, Campagnari A A, Gibson B W. The lipooligosaccharides of Haemophilus ducreyi are highly sialylated. J Bacteriol. 1996;178:564–570. doi: 10.1128/jb.178.2.564-570.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melaugh W, Phillips N J, Campagnari A A, Tullius M V, Gibson B W. Structure of the major oligosaccharide from the lipooligosaccharide of Haemophilus ducreyi strain 35000 and evidence for additional glycoforms. Biochemistry. 1994;33:13070–13078. doi: 10.1021/bi00248a016. [DOI] [PubMed] [Google Scholar]
- 30.Morse S A. Chancroid and Haemophilus ducreyi. Clin Microbiol Rev. 1989;2:137–157. doi: 10.1128/cmr.2.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nordhoff E, Ingendoh A, Cramer R, Overberg A, Stahl B, Karas M, Hillenkamp F, Crain P F. Matrix-assisted laser desorption/ionization mass spectrometry of nucleic acids with wavelengths in the ultraviolet and infrared. Rapid Commun Mass Spectrom. 1992;6:771–776. doi: 10.1002/rcm.1290061212. [DOI] [PubMed] [Google Scholar]
- 32.Odumeru J A, Wiseman G M, Ronald A R. Role of lipopolysaccharide and complement in susceptibility of Haemophilus ducreyi to human serum. Infect Immun. 1985;50:495–499. doi: 10.1128/iai.50.2.495-499.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Odumeru J A, Wiseman G M, Ronald A R. Virulence factors of Haemophilus ducreyi. Infect Immun. 1984;43:607–611. doi: 10.1128/iai.43.2.607-611.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmer K L, Goldman W E, Munson R S., Jr An isogenic haemolysin-deficient mutant of Haemophilus ducreyi lacks the ability to produce cytopathic effects on human foreskin fibroblasts. Mol Microbiol. 1996;21:13–19. doi: 10.1046/j.1365-2958.1996.00615.x. [DOI] [PubMed] [Google Scholar]
- 35.Palmer K L, Grass S, Munson R S., Jr Identification of a hemolytic activity elaborated by Haemophilus ducreyi. Infect Immun. 1994;62:3041–3043. doi: 10.1128/iai.62.7.3041-3043.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmer K L, Munson R S., Jr Cloning and characterization of the genes encoding the hemolysin of Haemophilus ducreyi. Mol Microbiol. 1995;18:821–830. doi: 10.1111/j.1365-2958.1995.18050821.x. [DOI] [PubMed] [Google Scholar]
- 37.Palmer K L, Thornton A C, Fortney K R, Hood A F, Munson R S, Jr, Spinola S M. Evaluation of an isogenic hemolysin-deficient mutant in the human model of Haemophilus ducreyi infection. J Infect Dis. 1998;178:191–199. doi: 10.1086/515617. [DOI] [PubMed] [Google Scholar]
- 38.Perez-Casal J, Caparon M G, Scott J R. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J Bacteriol. 1991;173:2617–2624. doi: 10.1128/jb.173.8.2617-2624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Potter M D, Lo R Y. Cloning and characterization of a gene from Pasteurella haemolytica A1 involved in lipopolysaccharide biosynthesis. FEMS Microbiol Lett. 1995;129:75–81. doi: 10.1016/0378-1097(95)00140-Z. [DOI] [PubMed] [Google Scholar]
- 40.Preston A, Mandrell R E, Gibson B W, Apicella M A. The lipooligosaccharides of pathogenic gram-negative bacteria. Crit Rev Microbiol. 1996;22:139–180. doi: 10.3109/10408419609106458. [DOI] [PubMed] [Google Scholar]
- 41.Schweda E K, Jonasson J A, Jansson P E. Structural studies of lipooligosaccharides from Haemophilus ducreyi ITM 5535, ITM 3147, and a fresh clinical isolate, ACY1: evidence for intrastrain heterogeneity with the production of mutually exclusive sialylated or elongated glycoforms. J Bacteriol. 1995;177:5316–5321. doi: 10.1128/jb.177.18.5316-5321.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schweda E K, Sundstrom A C, Eriksson L M, Jonasson J A, Lindberg A A. Structural studies of the cell envelope lipopolysaccharides from Haemophilus ducreyi strains ITM 2665 and ITM 4747. J Biol Chem. 1994;269:12040–12048. [PubMed] [Google Scholar]
- 43.Spengler B, Kirsch D, Kaufmann R, Jaeger E. Peptide sequencing by matrix-assisted laser-desorption mass spectrometry. Rapid Commun Mass Spectrom. 1992;6:105–108. doi: 10.1002/rcm.1290060207. [DOI] [PubMed] [Google Scholar]
- 44.Stevens M K, Porcella S, Klesney-Tait J, Lumbley S, Thomas S E, Norgard M V, Radolf J D, Hansen E J. A hemoglobin-binding outer membrane protein is involved in virulence expression by Haemophilus ducreyi in an animal model. Infect Immun. 1996;64:1724–1735. doi: 10.1128/iai.64.5.1724-1735.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Totten P A, Norn D V, Stamm W E. Characterization of the hemolytic activity of Haemophilus ducreyi. Infect Immun. 1995;63:4409–4416. doi: 10.1128/iai.63.11.4409-4416.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trees D L, Morse S A. Chancroid and Haemophilus ducreyi: an update. Clin Microbiol Rev. 1995;8:357–375. doi: 10.1128/cmr.8.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tuffrey M, Alexander F, Ballard R C, Taylor-Robinson D. Characterization of skin lesions in mice following intradermal inoculation of Haemophilus ducreyi. J Exp Pathol (Oxford) 1990;71:233–244. [PMC free article] [PubMed] [Google Scholar]
- 48.Westphal O, Jann K. Bacterial lipopolysaccharides. Extraction with phenol-water and further applications of the procedure. Methods Carbohydr Chem. 1965;5:83–91. [Google Scholar]