Abstract
STUDY QUESTION
What is the effect of the chemical in vitro activation (cIVA) protocol compared with fragmentation only (Frag, also known as mechanical IVA) on gene expression, follicle activation and growth in human ovarian tissue in vitro?
SUMMARY ANSWER
Although histological assessment shows that cIVA significantly increases follicle survival and growth compared to Frag, both protocols stimulate extensive and nearly identical transcriptomic changes in cultured tissue compared to freshly collected ovarian tissue, including marked changes in energy metabolism and inflammatory responses.
WHAT IS KNOWN ALREADY
Treatments based on cIVA of the phosphatase and tensin homolog (PTEN)-phosphatidylinositol 3-kinase (PI3K) pathway in ovarian tissue followed by auto-transplantation have been administered to patients with refractory premature ovarian insufficiency (POI) and resulted in live births. However, comparable effects with mere tissue fragmentation have been shown, questioning the added value of chemical stimulation that could potentially activate oncogenic responses.
STUDY DESIGN, SIZE, DURATION
Fifty-nine ovarian cortical biopsies were obtained from consenting women undergoing elective caesarean section (C-section). The samples were fragmented for culture studies. Half of the fragments were exposed to bpV (HOpic)+740Y-P (Frag+cIVA group) during the first 24 h of culture, while the other half were cultured with medium only (Frag group). Subsequently, both groups were cultured with medium only for an additional 6 days. Tissue and media samples were collected for histological, transcriptomic, steroid hormone, and cytokine/chemokine analyses at various time points.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Effects on follicles were evaluated by counting and scoring serial sections stained with hematoxylin and eosin before and after the 7-day culture. Follicle function was assessed by quantification of steroids by ultra-performance liquid chromatography tandem-mass spectrometry at different time points. Cytokines and chemokines were measured by multiplex assay. Transcriptomic effects were measured by RNA-sequencing (RNA-seq) of the tissue after the initial 24-h culture. Selected differentially expressed genes (DEGs) were validated by quantitative PCR and immunofluorescence in cultured ovarian tissue as well as in KGN cell (human ovarian granulosa-like tumor cell line) culture experiments.
MAIN RESULTS AND THE ROLE OF CHANCE
Compared to the Frag group, the Frag+cIVA group exhibited a significantly higher follicle survival rate, increased numbers of secondary follicles, and larger follicle sizes. Additionally, the tissue in the Frag+cIVA group produced less dehydroepiandrosterone compared to Frag. Cytokine measurement showed a strong inflammatory response at the start of the culture in both groups. The RNA-seq data revealed modest differences between the Frag+cIVA and Frag groups, with only 164 DEGs identified using a relaxed cut-off of false discovery rate (FDR) <0.1. Apart from the expected PI3K–protein kinase B (Akt) pathway, cIVA also regulated pathways related to hypoxia, cytokines, and inflammation. In comparison to freshly collected ovarian tissue, gene expression in general was markedly affected in both the Frag+cIVA and Frag groups, with a total of 3119 and 2900 DEGs identified (FDR < 0.001), respectively. The top enriched gene sets in both groups included several pathways known to modulate follicle growth such as mammalian target of rapamycin (mTOR)C1 signaling. Significant changes compared to fresh tissue were also observed in the expression of genes encoding for steroidogenesis enzymes and classical granulosa cell markers in both groups. Intriguingly, we discovered a profound upregulation of genes related to glycolysis and its upstream regulator in both Frag and Frag+cIVA groups, and these changes were further boosted by the cIVA treatment. Cell culture experiments confirmed glycolysis-related genes as direct targets of the cIVA drugs. In conclusion, cIVA enhances follicle growth, as expected, but the mechanisms may be more complex than PI3K–Akt–mTOR alone, and the impact on function and quality of the follicles after the culture period remains an open question.
LARGE SCALE DATA
Data were deposited in the GEO data base, accession number GSE234765. The code for sequencing analysis can be found in https://github.com/tialiv/IVA_project.
LIMITATIONS, REASONS FOR CAUTION
Similar to the published IVA protocols, the first steps in our study were performed in an in vitro culture model where the ovarian tissue was isolated from the regulation of hypothalamic–pituitary–ovarian axis. Further in vivo experiments will be needed, for example in xeno-transplantation models, to explore the long-term impacts of the discovered effects. The tissue collected from patients undergoing C-section may not be comparable to tissue of patients with POI.
WIDER IMPLICATIONS OF THE FINDINGS
The general impact of fragmentation and short (24 h) in vitro culture on gene expression in ovarian tissue far exceeded the effects of cIVA. Yet, follicle growth was stimulated by cIVA, which may suggest effects on specific cell populations that may be diluted in bulk RNA-seq. Nevertheless, we confirmed the impact of cIVA on glycolysis using a cell culture model, suggesting impacts on cellular signaling beyond the PI3K pathway. The profound changes in inflammation and glycolysis following fragmentation and culture could contribute to follicle activation and loss in ovarian tissue culture, as well as in clinical applications, such as fertility preservation by ovarian tissue auto-transplantation.
STUDY FUNDING/COMPETING INTEREST(S)
This study was funded by research grants from European Union’s Horizon 2020 Research and Innovation Programme (Project ERIN No. 952516, FREIA No. 825100), Swedish Research Council VR (2020-02132), StratRegen funding from Karolinska Institutet, KI-China Scholarship Council (CSC) Programme and the Natural Science Foundation of Hunan (2022JJ40782). International Iberian Nanotechnology Laboratory Research was funded by the European Union’s H2020 Project Sinfonia (857253) and SbDToolBox (NORTE-01-0145-FEDER-000047), supported by Norte Portugal Regional Operational Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund. No competing interests are declared.
Keywords: fertility preservation, primary ovarian insufficiency, follicle development, gene expression, ovary
WHAT DOES THIS MEAN FOR PATIENTS?
Premature ovarian insufficiency leads to fertility loss before 40 years of age. Scientists are exploring ‘in vitro activation’ protocols to help these women: this involves using mechanical or mechanical and chemical methods to ‘wake up’ immature follicles within excised ovarian tissue in a culture dish in the laboratory. However, the effectiveness and safety of these methods remain uncertain, and our study was designed to investigate the impact and underlying mechanisms of in vitro activation of the follicles. Our study on donated human ovarian tissue found that chemical treatment of the tissue promoted follicle growth (compared to untreated tissue), yet changes in gene expression between the treated and non-treated groups, which could indicate important responses to the treatment, were minimal. Importantly, we found that gene expression in both treated and untreated tissue differed significantly from healthy intact ovaries, particularly in terms of energy metabolism and inflammation: this suggests that ovary fragmentation itself can trigger extensive damage responses in the tissue, which would also contribute to follicle loss. This finding has implications for the quality of ovarian tissue that is often handled in laboratories for fertility preservation in cancer patients, as well as for attempts to grow mature eggs from excised human ovarian tissue in the laboratory. Our results lead us to believe that extensive changes in metabolism and inflammation take place in ovarian tissue as soon as it is removed from the body and cut into pieces for such medical treatments and laboratory experiments. We therefore suggest that minimizing tissue processing could improve these methods in the future, benefiting women who face fertility challenges.
Introduction
Premature ovarian insufficiency (POI) is a heterogeneous condition characterized by the loss of ovarian function before the age of 40 years (Nelson, 2009). The prevalence of POI is between 1% and 3.5% in the general population worldwide (Lagergren et al., 2018; Li et al., 2023a). Hormonal replacement therapy (HRT) can effectively mitigate the symptoms of estrogen deficiency and decrease the cardiovascular risks associated with early menopause (Sullivan et al., 2016). Nevertheless, for those patients of childbearing age who are still seeking further fertility, options are very limited. Even with the aid of ART, the chance of obtaining biological offspring is usually no higher than 5–10% (Ishizuka et al., 2021).
Residual primordial follicles that persist in the ovaries of patients with POI who have entered menopause remain dormant (van Kasteren and Schoemaker, 1999; Donnez and Dolmans, 2014). The reason why these follicles are not able to activate and grow in situ has not been fully elucidated. However, a few studies suggested that these follicles might not be responsive to regulation by gonadotrophins, and they might exhibit inherent dormancy (Aittomäki et al., 1996; Liu et al., 2017). In general, the regulation of primordial follicle activation involves several sophisticated mechanisms including the phosphatidylinositol 3-kinase (PI3K)–protein kinase B (Akt) pathway. In a mouse model, oocyte-specific deletion of phosphatase and tensin homolog (PTEN), a critical negative regulator of PI3K–Akt signaling, results in global activation of the entire primordial follicle pool (Reddy et al., 2008; Adhikari and Liu, 2009). This demonstrates the key role of the PI3K–Akt pathway in maintaining the dormancy of primordial follicles. The loss of PTEN leads to increased levels of phosphatidylinositol (3,4,5)-trisphosphate (PIP3) and phosphorylated Akt (p-Akt) (John et al., 2008). Subsequently, it promotes the phosphorylation and translocation of downstream target forkhead transcription factor 3 (Foxo3) from the nucleus to the cytoplasm, ultimately activating the follicles (Zhang et al., 2020). Consistent with this mechanism, in multiple studies, short-term treatment of mouse ovary in vitro with a PTEN inhibitor has been proven to be effective in activating primordial follicles after allografting (Li et al., 2010; Adhikari et al., 2012). Activation of follicles based on PI3K–Akt pathway modification has also been validated in other in vitro or in vivo experiments in swine and humans (Novella-Maestre et al., 2015; Sun et al., 2015; Raffel et al., 2019).
Based on above findings, a clinical trial involving the use of a PTEN inhibitor combined with a PI3K stimulator, thereby activating the PI3K–Akt pathway, was carried out in patients with POI. The protocol, known as in vitro activation (IVA), centered on the stimulation of excised human ovarian cortical fragments with bpV (HOpic), a PTEN inhibitor, and 740Y-P, a PI3K stimulator, in tissue culture before autologous transplantation. This chemical IVA (cIVA) protocol led to successful live births following gonadotrophin stimulation, oocyte retrieval, and ICSI–embryo transfer (ICSI–ET) (Kawamura et al., 2013). Two subsequent cohort studies also reported full-term delivery in POI patients following this protocol (Suzuki et al., 2015; Zhai et al., 2016). However, concerns have been raised as the PI3K–Akt pathway plays crucial roles in various physiological processes, including cell survival, proliferation, and metabolism (Katso et al., 2001; Engelman et al., 2006), which are also associated with cancer (Russo et al., 2018; Shi et al., 2019). In addition to potentially increasing the risk for tumorigenesis, PTEN inhibition may lead to the accumulation of DNA damage (Puc and Parsons, 2005). For example, Maidarti et al. (2019) reported that high levels of intracellular Akt might reduce the homologous recombination repair capacity of DNA double-strand breaks in oocytes.
Owing to the aforementioned concerns, a modified protocol, focusing on Hippo signaling modification, also known as ‘drug-free’ or mechanical IVA, has been developed (Zhang et al., 2019; Ferreri et al., 2020; Tanaka et al., 2020). Mechanical IVA is based on observations suggesting that fragmentation of ovarian cortex disrupts Hippo signaling, leading to a reduced degradation and an increased nuclear accumulation of the downstream transcription factor Yes-associated protein (YAP). Increased nuclear YAP levels promotes follicle activation (Kawamura et al., 2013; Hsueh and Kawamura, 2020). This protocol circumvents the chemical PTEN-PI3K modulation of ovarian tissue. However, independent replications of these different protocols have yielded conflicting results (Dolmans et al., 2019; Lunding et al., 2019; Ferreri et al., 2021; Díaz-García et al., 2022; Méndez et al., 2022). Therefore, the effectiveness and safety of cIVA and mechanical IVA protocols, as well as their molecular mechanisms and overall impacts on ovarian tissue, remain controversial and unexplored. Elucidating the mechanism of these protocols on early follicle development at a molecular level would not only benefit patients with POI but also potentially other disorders leading to early loss of ovarian function, such as Turner syndrome. By obtaining mature oocytes from residual follicles, IVA protocols offer the possibility of restoring fertility in these patients, providing them with the opportunity to obtain biological offspring. Moreover, on a broader scale, different IVA approaches may offer alternatives to traditional ovarian stimulation for ART, potentially reducing the administration of exogenous hormone and associated risks. Finally, a holistic assessment of transcriptomic changes in ovarian tissue upon fragmentation is also relevant for fertility preservation, where the activation and loss of follicles in fragmented and transplanted tissue is a known issue with no solutions.
In this study, our aim was to investigate the impacts and mechanisms of mechanical IVA (fragmentation only, ‘Frag’) and cIVA (fragmentation and cIVA, ‘Frag+cIVA’) on human ovarian tissue using transcriptomic profiling alongside traditional histological tissue assessment and steroid, cytokine, and chemokines profiling.
Materials and methods
Study participants and ethical approval
Ovarian tissue samples were collected from women undergoing elective caesarean section (C-section) at Karolinska University Hospital Huddinge, Sweden, during years 2015–2017 and 2022–2023. Patients received both oral and written information about the research project and signed a consent form following the Declaration of Helsinki, agreeing to donate a small superficial piece of ovarian cortical tissue (5 mm × 5 mm × 2 mm) during C-section. Samples collected in 2015–2017 were anonymized, while samples collected in 2022–2023 were pseudonymized and biobanked following the Swedish Biobanking in Health Care Act. Data analysis and sample handling were performed in accordance with European General Data Protection Regulation. The study was approved by the Swedish Ethical Review Authority (license numbers 2010/549-31/2 and 2015/798-31/2).
Human ovarian tissue culture and in vitro activation
The effects of Frag+cIVA versus Frag on ovarian tissue and follicles were investigated in a tissue culture system similar to the original protocol (Suzuki et al., 2015). In total, we collected ovarian cortex biopsies from 59 C-section patients (mean age ± SD: 33.25 ± 5.31 years). Tissues from 33 patients were utilized for morphological assessment of follicle health and growth after a 7-day culture. For transcriptomic profiling by RNA-sequencing (RNA-seq), tissues from 11 patients were used after 24-h culture. For further validation of the RNA-seq findings, 15 tissue samples were cultured and sampled at various time points (Days 1, 3, 5, and 7) for quantitative PCR (qPCR), immunofluorescence staining, and analysis of steroids (n = 7) or cytokines/chemokines (n = 3) in the culture media (Supplementary Fig. S1). The general characteristics of the validation sample donors are shown in Supplementary Table S1. Owing to the restrictions in the old ethical permit, characteristics of the other samples are not available.
The tissue culture system has been previously described (Hao et al., 2020). In brief, tissue samples used for histological assessment and RNA-seq were cultured in a 24-well plate (Sarstedt, Numbrecht, Germany) with Millicell cell culture plate inserts (Merck Millipore, Darmstadt, Germany) that were coated either with 10 mg/ml human recombinant laminin 221 (LN221, Biolamina, Sundbyberg, Sweden) or with Matrigel (hESC qualified, Corning Inc., Corning, NY, USA). Samples used for validation experiments (n = 15) were cultured only on LN221. On the day of tissue collection, the laminin-coated plates were pre-warmed and washed before use. The ovarian biopsy retrieved from the operating room was transferred to the laboratory within 10 min. Tissues were cut into 1 mm × 1 mm × 1 mm pieces and placed on the insert for subsequent culture, one per insert. Fragments of each patient were cultured using two substrates, Matrigel and LN221. The basal culture media consisted of DMEM/high glucose (Life Technologies, Paisley, UK), 1% insulin–transferrin–selenium (Life Technologies, Grand Island, NY, USA), 1% antibiotic–antimycotic (Life Technologies, Grand Island, NY, USA), 1% 100× GlutaMAX™ (Life Technologies, Grand Island, NY, USA), 10% human serum albumin (Vitrolife, Frolunda, Sweden), and 0.5 IU/ml human recombination FSH (Fostimon, IBSA Farmaceutici, Lodi, Italy). The samples were cultured under 37°C and 5% CO2 for 7 days. For the first 24 h, tissue in the Frag+cIVA group was treated with 30 mM of bpV (HOpic) (Merck Millipore), a PTEN inhibitor, and 150 mg/ml of 740Y-P (Tocris, Bristol, UK), a PI3K stimulator, following the published IVA protocol (Suzuki et al., 2015). Thereafter, both groups were cultured in basal media. A full medium change was performed first after the initial 24-h culture step, and then every 48 h. Spent media were stored in microtubes at −80°C for steroid and cytokine/chemokine analyses. From every sample, a fresh piece was collected, without culture (fresh control). Tissues used for histological analyses were fixed in Bouin’s solution (Sigma Aldrich, St Louis, MO, USA). For immunofluorescence staining, tissues were fixed in 4% formaldehyde (Thermo Fisher Scientific, Rockford, IL, USA) and embedded in paraffin for sectioning. For transcriptomic analyses, one piece of tissue per patient and group was collected in RNALater (Invitrogen, Vilnius, Lithuania), and stored in a −80°C freezer for later RNA extraction.
Follicle quantification and measurement of oocytes
Paraffin-embedded sections from 33 patients were prepared by the FENO morphological phenotype analysis core facility (Karolinska Institutet, Sweden), where tissues were sectioned into 4 µm sections through paraffin blocks and every seventh and eighth sections were stained with hematoxylin and eosin (H&E). H&E-stained sections were digitalized using a Pannoramic scanner (3DHistech, Budapest, Hungary). Follicles with a visible nucleus in the oocytes were counted and the diameters of oocytes and follicles were measured using Panoramic Viewer software (3DHistech). Follicles were classified into four categories, namely primordial, primary, secondary, and atretic, following the criteria in our previous work (Li et al., 2023b). Atretic follicles were classified by the presence of >10% of atretic granulosa cells, condensed nucleus, and/or eosinophilic cytoplasm. Follicles devoid of these features were considered healthy. Oocyte and secondary follicle diameters were measured in two perpendicular directions of maximum diameter. All follicles were quantified by one observer and 20% of the tissue sections were double quantified by a second observer as a quality control.
RNA extraction, library preparation, RNA-seq, and data analysis
Ovarian cortical tissues from 11 patients were homogenized with FastPrep-24 dissociator (MP Biomedicals, Solon, OH, USA). Tissues were homogenized in 150 µl of RLT buffer (Qiagen, Hilden, Germany) in a 2-ml tube with a speed of 4 m/s for 40 s, followed by 5 min cooling on ice. The step was repeated three times to fully homogenize the tissue. Total RNA of tissue was extracted with the RNeasy Micro Kit (Qiagen) following the manufacturer’s instructions. Briefly, proteinase K solution (600 mAU/ml, Qiagen) was diluted 1:60 and added to the lysate, followed by 10-min incubation at 55°C. After centrifuging for 3 min at 10 000 g, the supernatant was transferred to a new tube and washed with 0.5 volumes of absolute ethanol (Histolab, Askim, Sweden). The lysate was then transferred to a RNeasy MiniElute spin column and washed with RW1 buffer before a 15-min DNA digestion using DNase I (Qiagen). Subsequently, the membrane was washed with buffer RW1, RPE, and 80% ethanol before RNA elution.
The concentration of the bulk RNA was measured by both Nanodrop (IMPLEN, Nordic Biolabs, Taby, Sweden) and a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA). After that, the RNA integrity (RIN) was analyzed by RNA 6000 Nano assay (>15 ng/µl) using an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). RIN values were equal to or higher than 8.5 for all samples. After dilutions, 10 ng RNA of each sample was processed using the original single-cell tagged reverse transcription (STRT) RNA-seq protocol (Islam et al., 2014) with bulk RNA setup. Briefly, RNA samples were placed in a 48-well plate in which a universal primer, template-switching oligos with a well-specific 6-bp barcode sequence, and a 6-bp random unique molecular identifier sequences (Kivioja et al., 2011; Krjutškov et al., 2016) were added to each well. Reverse transcription reagents and ERCC RNA Spike-In Mix I (Thermo Fisher Scientific Baltic UAB, Vilnius, Lithuania) were added to generate first-strand cDNA. The synthesized cDNA from the samples were then pooled into one library, fragmented, end-polished, ligated, and amplified by single-primer PCR with the universal primer sequence. The resulting amplified library was then quantified and sequenced in three lanes using the Illumina HiSeq2000 instrument (Illumina, San Diego, CA, USA) at the Bioinformatics and Expression Analysis (BEA) core facility at Karolinska Institutet, Sweden.
Fastq files were mapped to the human genome GRCh38 using Hisat2 aligner (version 2.2.1) and assigned using featureCount function in subread (version 2.0.1). Subsequently, genes were annotated using an Ensembl gtf file (Homo_sapiens.GRCh38.105.chr.gtf). Downstream analysis was performed in R/Bioconductor through RStudio (R Studio Team, 2020; R Core Team, 2021). Differential expression analysis was performed using the DESeq2 package where comparisons between Frag+cIVA and fresh, Frag and fresh, and Frag+cIVA and Frag were performed separately. Differentially expressed genes (DEGs) were identified as genes with a false discovery rate (FDR) <0.1 for Frag+cIVA and Frag comparison, and FDR <0.001 for the rest. The stricter FDR criteria were applied for the Frag+cIVA and Frag versus fresh group comparisons because of the large effect sizes. Gene set enrichment analysis (GSEA) was performed using the molecular signatures database MSigDB hallmark gene sets (Liberzon et al., 2011) with clusterProfiler (Yu et al., 2012), pathview (Luo and Brouwer, 2013), DOSE (Yu et al., 2015), and apeglm package (Zhu et al., 2019). Disease enrichment analysis was performed on DEGs ranked by log2 fold change using clusterProfiler against the human Disease Gene Network (DisGeNET) database (Piñero et al., 2015). Plotting was performed using the ggplot2 package in R (version 4.1.2).
Validation of DEGs by qPCR in tissue and KGN cells
DEGs were chosen for validation based on the transcriptomic profiling and GSEA analysis. Primers were designed and validated as described (Li et al., 2023b), and they are listed in Supplementary Table S2. Time-course tissue culture samples were used for RNA extraction. Tissues were homogenized with a GentleMACS™ dissociator (Miltenyi Biotec, Bergisch Gladbach, Germany). Total tissue RNA was extracted with the RNeasy Micro Kit (Qiagen) as described above. Total RNA of KGN cells (human ovarian granulosa-like tumor cell line: culture described below) was extracted using a RNeasy Mini kit (Qiagen) following the manufacturer’s protocol. DNase I (Qiagen) was applied to remove DNA from the total RNA.
Conversion of total RNA to cDNA was performed with an iScript cDNA Synthesis Kit (BIO-RAD, Hercules, CA, USA). RT-qPCR was performed using a SsoAdvanced Universal SYBR Green Supermix Kit (BIO-RAD) to study the expression of selected targets. Each target was normalized to a housekeeping gene, and because gene expression changes dramatically in tissue culture, we carefully identified suitable housekeeping genes. We downloaded a list of 2833 potential housekeeping genes in human tissue from the Housekeeping and Reference Transcript Atlas (Hounkpe et al., 2021) and checked their expression in our RNA-seq data. Three targets: the 60S ribosomal protein L22 (RPL22), ribosomal protein lateral stalk subunit P2 (RPLP2), and ubiquitin C (UBC) were found to have high and relatively stable expression in both fresh and cultured tissues. These three genes, together with two of our earlier used housekeeping genes, namely hypoxanthine phosphoribosyltransferase 1 (HPRT1) and ribosomal protein lateral stalk subunit P0 (RPLP0) (Hao et al., 2020), were validated using the time-course samples. The comparison of the selected housekeeping genes was performed by Genorm (Vandesompele et al., 2002), Normfinder (Andersen et al., 2004), and Bestkeeper (Pfaffl et al., 2004), independently. The most stable housekeeping gene in tissue was selected by combining the results of the three methods, leading to the selection of RPL22. For the KGN cells, HPRT1 was used as a housekeeper. Technical duplicates and three biological replicates were run on 40 cycles of qPCR. Relative fold change was calculated with the Livak method (ddCT) for each gene compared to fresh control group (Livak and Schmittgen, 2001).
Additional validation of cIVA treatment targets was carried out in cell culture. The KGN cell line, obtained from RIKEN Cell Bank (RBRC-RCB1154, Tokyo, Japan) and authenticated by the Eurofins Genomics Europe Applied Genomics GmbH, was cultured in 12-well plates (Sarstedt). Culture media consisted of DMEM/F12 (Life Technologies, Grand Island, NY, USA), 10% heat-inactivated fetal bovine serum (HI-FBS, Life Technologies, Grand Island, NY, USA), 1% penicillin–streptomycin (100×) (Life Technologies, Grand Island, NY, USA), and 1% 100× GlutaMAX™ (Life Technologies, Grand Island, NY, USA). KGN cells were cultured under 5% CO2 at 37°C and passaged with 0.05% trypsin–EDTA (Life Technologies, Grand Island, NY, USA) when reaching 80% confluency. To study the effects of cIVA, KGN were seeded for 24 h and exposed to 15 mg/ml 740Y-P and 3 µM bpV (HOpic) for 24 h. A 10-fold lower concentration of the cIVA drugs was used compared to tissue culture to avoid cytotoxicity. We also included control and cIVA groups treated with 0.05, 0.5, and 1 IU/ml FSH. Cells were then washed with Dulbecco’s PBS without calcium or magnesium (Life Technologies, Grand Island, NY, USA), harvested in 350 µl of RLT buffer, and stored at −80°C until further RNA extraction and qPCR analysis, as described above.
Immuno- and TUNEL staining
To validate selected DEGs at the protein level and to identify the affected cell types, immunostainings were carried out. Tissues were processed to formaldehyde-fixed and paraffin-embedded tissue sections by the FENO core facility. The sections were rehydrated using xylene and serial ethanol solutions, followed by antigen retrieval with Tris/EDTA solution (Sigma Aldrich) at 96°C for 30 min, unless stated otherwise. After cooling down, blocking solution, consisting of 5% w/v bovine serum albumin (Sigma Aldrich), 20% v/v normal donkey serum (Life Technologies, Grand Island, NY, USA), and Tris buffered saline, was added to the sections for 1 h at room temperature and then the sections were incubated with primary antibodies against DEAD-box helicase 4 (DDX4), alpha-enolase (ENO1), lactate dehydrogenase A (LDHA), and hypoxia inducible factor 1 subunit alpha (HIF1α) (Supplementary Table S3) overnight at 4°C. Afterwards, diluted secondary antibody was added to sections for a 2-h incubation in the dark at room temperature. DAPI stain (Thermo Fisher Scientific, Rockford, IL, USA, 1:1000 dilution) was applied to the sections after washing, followed by mounting using DAKO mounting medium (Agilent Technologies). Isotype antibodies matched with the primary antibody’s species were used as negative controls.
Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) was performed using a TUNEL Assay Kit (Fluorescence, 594 nm, Cell Signaling Technology, Danvers, MA, USA) to assess the apoptosis in fresh and cultured tissues, following the manufacturer’s protocol. Briefly, deparaffinization and antigen retrieval were performed following the same protocol as immunofluorescence, except that citrate solution (pH = 6, Sigma Aldrich) was used instead of Tris/EDTA. Subsequently, sections were washed and incubated with TUNEL equilibration buffer followed by TUNEL reaction mix. A negative control was incubated with reaction buffer without terminal transferase instead of TUNEL reaction mixture. A positive control was prepared by treating tissue samples with DNase I (Roche Diagnostics, Mannheim, Germany) for 10 min after equilibration. The labelled sections were then proceeded for simultaneous immunofluorescence staining of γH2AX (a marker of typical DNA damage) and Ki67 (a cell proliferation marker) (Supplementary Table S3), as described above.
Imaging of the sections was performed using a widefield microscope at the Live Cell Imaging (LCI) facility at Karolinska Institutet. An air objective (20×/0.75) together with a 1.5x lens were used for imaging in brightfield, blue, green, red, and far-red channels. OMERO.figure was used for imaging assembling. All images were brightness- and contrast-adjusted for clarity and are presented under the same scale for better visualization.
Quantification of immunofluorescence signals
CellProfiler was used for the quantification of immunofluorescence signals (Stirling et al., 2021). All Images were corrected for illumination differences between the center and the edge before being used for quantification. For Ki67 and γH2AX quantification, the percentage of cells positive for these two markers was calculated. For ENO1, LDHA, and HIF1α quantification, masks for follicles and oocytes were created and employed for the identification of granulosa cells and stroma. Mean intensities were calculated by dividing the total fluorescence intensities by the area of the corresponding region. HIF1α signal was included in the measurement only when it was within the nucleus.
Steroid, cytokine, and chemokine measurements
Culture medium was collected for steroid hormone and cytokine/chemokine measurement at Day 1, Day 3, Day 5, and Day 7 of culture. Pregnenolone, progesterone, dehydroepiandrosterone (DHEA), androstenedione, testosterone, and estradiol were measured using a liquid chromatography tandem-mass spectrometry-based method, as described in our previous study (Li et al., 2023b). The concentrations of the cytokines and chemokines were measured according to the manufacturer’s instructions using the bead-based multiplex immunoassay system (Multiplex kit Cytokine & Chemokine 34-Plex Human ProcartaPlex™ Panel 1A, Thermo Fisher Scientific, Vienna, Austria). All samples were analyzed at the same time to prevent batch effects and run in duplicate. Briefly, supernatants were thawed on ice, mixed by vortexing, and centrifuged to remove cellular debris. Two hundred microliters of magnetic bead mix were washed using a hand-held magnetic plate washer to retain the beads and subsequently incubated in the dark with 50 µl of undiluted samples at room temperature for 90 min, shaking at 500 rpm. The beads were then washed twice and incubated in the dark with the detection antibody mix, at room temperature for 30 min, shaking at 500 rpm. After two washes, 50 μl of streptavidin–phycoerythrin solution was added and incubated in the dark, for 30 min at room temperature shaking at 500 rpm. Finally, the beads were washed twice and resuspended in 120 µl of reading buffer. After 5 min of shaking at 500 rpm, the plate was read on a Luminex FLEXMAP 3D instrument in combination with xPONENT 4.3 (Diasorin, Saluggia, Italy), and mean fluorescence intensity was converted to pg/ml by interpolation from a five-parameter logistic standard curve.
Statistical analysis
Statistical analyses were performed using R (R Core Team, 2021) and RStudio (R Studio Team, 2020). A Chi-square test was employed to compare the proportion of follicles in different categories and P-values were adjusted for multiple comparisons using the Bonferroni method. Student’s t-test was used for the comparison of follicle and oocyte diameters. For tissue qPCR validation, immunofluorescence signals, steroid hormone, and cytokine measurement, statistical analysis was performed using two-way ANOVA with interaction and Tukey’s post hoc correction on untransformed (cytokine), log2- (qPCR, immunofluorescence), or log10- (steroids) transformed data. For qPCR results of KGN exposure, the statistics were performed using one-way ANOVA with Tukey’s post hoc correction on log2-transformed data. A P-value <0.05 was considered as statistical significance.
Results
Frag+cIVA boosted follicle growth in human ovarian cortical tissue culture
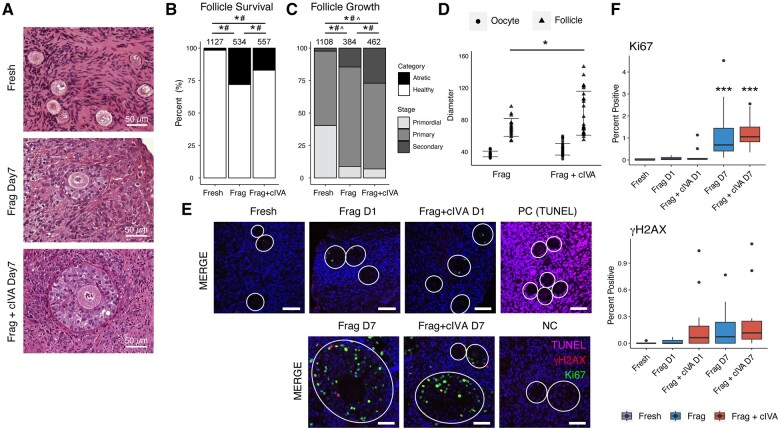
We utilized ovarian biopsies collected from 33 C-section patients to study the effects of Frag and Frag+cIVA on ovarian follicle growth in vitro. We did not include an additional ‘non-fragmented’ control group because entire ovaries cannot be collected for research for ethical reasons and, in contrast to mouse ovaries, human ovaries are too large to be cultured in vitro without cutting into fragments. In addition, the cIVA and mechanical IVA protocols are both based on short-term culture of ovarian tissue fragments, as originally described (Adhikari et al., 2012; Kawamura et al., 2013; Zhai et al., 2016; Zhang et al., 2019; Ferreri et al., 2020; Tanaka et al., 2020). This histological material encompassed 2218 visible follicles in total, and the subset of samples that was double counted by two observers showed high concordance (96.6% in follicle number, 99.3% in follicle category), suggesting that the follicle assessment was reliable. Similar to our previous work (Hao et al., 2020), we cultured half of the tissue on Matrigel and the other half on human recombinant LN221 to assess the suitability of laminin as a defined and xeno-free growth substrate for ovarian tissue culture. In agreement with our previous study, there were no major differences in follicle survival and growth between the two growth substrates (Supplementary Table S4), and therefore the data were pooled for downstream analysis. The pooled data showed decreased primordial and primary follicles as well as an increased proportion of secondary and atretic follicles after a 7-day culture in both the Frag and Frag+cIVA groups compared to fresh tissues (Fig. 1A–C). Moreover, cIVA treatment led to a significantly higher proportion of secondary follicles when compared to both fresh and Frag groups (Fig. 1C). Meanwhile, a lower proportion of primary follicles was also recorded in the Frag+cIVA group compared to the Frag group (Fig. 1C). We further assessed the size of the secondary follicles, and a significant increase in follicle diameters was detected in the Frag+cIVA group compared to the Frag group (Fig. 1D).
Figure 1.
Follicle survival and growth in human ovarian cortex cultured with and without chemical in vitro activation at different time points. Ovarian cortical tissue fragments from 33 women were cultured for a total of 7 days with half of the samples treated with cIVA protocol during the first 24 h. (A) Representative histological images of follicles in fresh, Frag, and Frag+cIVA group after 7-day culture. Scale bars 50 µm. (B) Follicle survival and (C) growth in fresh, Frag, and Frag+cIVA groups at the end of the 7-day culture (*P < 0.05, # P < 0.05, ^ P < 0.05 fresh versus Frag versus Frag+cIVA, chi-square test with Bonferroni’s correction). The total number of follicles is indicated above the bars. In Figure 1B, the asterisk (*) represents statistics for atretic follicles while the hashtag (#) indicates statistics for healthy follicles. In Figure 1C, the asterisk (*) represents statistics for secondary follicles, hashtag (#) for primary, and circumflex (^) for primordial. (D) Comparison of the diameter of the oocyte from the secondary follicle and follicle diameters (*P < 0.05, Student’s t-test) in the Frag and Frag+cIVA groups after 7-day culture. (E) Detection of DNA damage and proliferation by TUNEL and immunofluorescence staining of γH2AX and Ki67 in fresh tissue and in Frag and Frag+cIVA groups on Days 1 and 7 of culture. Dark blue indicates DAPI; green Ki67; red γH2AX; and magenta TUNEL. Scale bars 50 µm. Follicles are indicated with white circles. Images were processed using OMERO.figure and are presented under the same scale of brightness and contrast. (F) Quantification of the percentage of cells positive for Ki67 and γH2AX staining in fresh, Frag, and Frag+cIVA group on fresh, Day 1, and Day 7 samples. Statistics were performed using two-way ANOVA with interaction and Tukey’s correction after log2 transformation in RStudio. Asterisks represent the comparison between groups. *P < 0.05, ***P < 0.001. cIVA, chemical in vitro activation; Frag, fragmentation; NC, negative control; PC, positive control; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling.
To further investigate the health of the tissues after culture, DNA damage and proliferation were assessed by TUNEL, γH2AX, and Ki67 staining in fresh samples and tissues cultured for 1 and 7 days. Our TUNEL staining results suggested that no profound apoptosis was detected in culture (Fig. 1E). The γH2AX staining indicated scattered positive signals in granulosa cells of secondary follicles in both the Frag+cIVA and Frag groups on Day 7 (Fig. 1E). Quantification of these signals revealed a higher proportion of γH2AX-positive cells in the Frag+cIVA group already on Day 1, while the level of DNA damage was similar on Day 7 between the two groups (Fig. 1F). Finally, Ki67 staining suggested granulosa cell proliferation starting at Day 1 of culture in both groups, with significantly higher percentages of positive cells present in follicles and stroma by Day 7 (Fig. 1E and F). However, no significant difference in proliferation and DNA damage was recorded between the Frag and Frag+cIVA groups (Fig. 1F).
Transcriptomics data overview
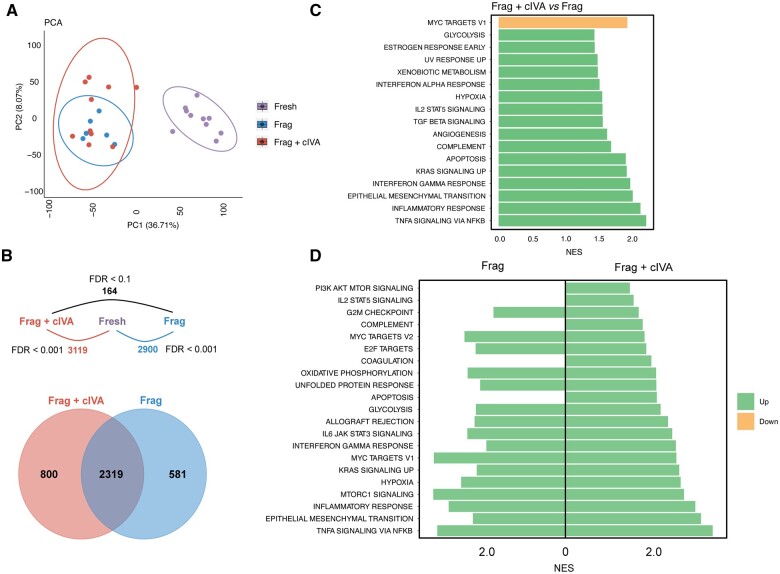
To study the overall gene expression changes in response to Frag and Frag+cIVA, we conducted transcriptomic profiling of the tissue fragments after 24-h culture using RNA-seq. Ovarian cortical tissue from 11 women was fragmented and the fragments were divided into three groups: fresh (uncultured, n = 11), 24-h culture (Frag, n = 6), and 24-h culture with cIVA (Frag+cIVA, n = 10). After quality control, 26 samples remained in the dataset. One sample in the fresh group was removed due to a low library size. Principal component analysis (PCA) showed that the dataset was mainly driven by a strong PC1 that divided the samples into ‘fresh’ and ‘cultured’ and explained 36.71% of the variation in the data (Fig. 2A). PCA did not separate the two cultured groups, Frag and Frag+cIVA, from each other (Fig. 2A). In agreement with the results from histological analysis and our earlier studies (Hao et al., 2020), we did not observe a separation between samples cultured on Matrigel or LN221 (Supplementary Fig. S2). When performing differential expression analysis on samples cultured on the two substrates in the Frag+cIVA group and Frag group separately, no DEGs were identified using an FDR < 0.05 as the cut-off. Thus, the samples were pooled for downstream analyses. A direct comparison of the Frag+cIVA and Frag groups with each other showed modest transcriptomic differences, with only a low number of DEGs (n = 164) identified when the significance cut-off was relaxed to an FDR < 0.1. To investigate affected signaling pathways and processes connected to the gene expression changes, GSEA using all expressed genes ranked by log2 fold change was carried out, using the hallmark gene set as a reference. The results suggested that compared to Frag, cIVA treatment enriched gene sets (FDR < 0.1) related to inflammation (e.g. tumor necrosis factor (TNF)α signaling via nuclear factor (NF)-κB, interferon-gamma response, interferon alpha (IFN-α) response), hypoxia, and glycolysis, while it downregulated MYC targets (Fig. 2C).
Figure 2.
Overview of transcriptomic changes in human ovarian cortex cultured with or without chemical in vitro activation. Ovarian cortical tissue fragments from 11 women were cultured for a total of 24 h with half of the samples treated with the cIVA protocol. The fragments were collected for transcriptomic profiling by RNA-seq. (A) PCA of the RNA-seq data using all expressed genes. (B) Number of DEGs between the different comparisons, and a Venn diagram showing the overlap in genes altered in culture in the Frag+cIVA and Frag groups compared to the fresh samples. The used FDR cut-off to determine DEGs is indicated. (C) Barplot displaying selected significantly enriched hallmark gene sets from GSEA analysis using all expressed genes in Frag versus Frag+cIVA comparison (FDR < 0.1). (D) Barplot showing selected significantly enriched hallmark gene sets from GSEA analysis using significant DEGs in Frag versus fresh and Frag+cIVA versus fresh comparisons (FDR<0.1). The normalized enrichment score (NES) is presented on the x-axis. cIVA, chemical in vitro activation; DEGs, differentially expressed genes; FDR, false discovery rate; Frag, fragmentation; GSEA, gene set enrichment analysis; NES, normalized enrichment score; PCA, principal component analysis.
The modest effects of Frag+cIVA compared to the Frag group were in striking contrast to the changes that took place in both cultured groups when they were compared to the fresh uncultured control samples: 3119 DEGs were found in the Frag+cIVA group and 2900 DEGs in the Frag group at FDR < 0.001. Among these, 2319 DEGs were shared between these two comparisons, suggesting highly similar overall changes during culture in both groups (Fig. 2B). GSEA showed that the overall effects of Frag and Frag+cIVA on the tissue transcriptome were similar. In general, gene sets related to inflammation (e.g. TNFα signaling via NF-κB, interferon response), glycolysis, hypoxia, and mammalian target of rapamycin (mTOR)C1 signaling were significantly upregulated (FDR < 0.1) in both culture conditions compared to fresh control tissue. However, we also found gene sets that were enriched only in one of the two conditions. For example, Frag+cIVA treatment significantly upregulated gene sets related to PI3K–AKT–mTOR signaling and IL2–STAT5 signaling, but also apoptosis (Fig. 2D). To study treatment-specific changes further, we carried out GSEA analysis based on those DEGs that were only found in Frag (n = 581) or in Frag+cIVA (n = 800) samples (Fig. 2B). Our results showed enrichment of similar gene sets, as already found by analyzing the commonly altered DEGs. For example, Frag-specific DEGs were enriched for gene sets related to MYC targets, oxidative phosphorylation, and interferon responses, while Frag+cIVA-specific DEGs were related to TNFα signaling via NF-κB, hypoxia, and mTORC1 signaling (Supplementary Table S5). This confirms that the overall effects of both treatments on the tissue transcriptome are similar.
Expression of steroidogenesis and granulosa cell-related genes significantly changed during culture
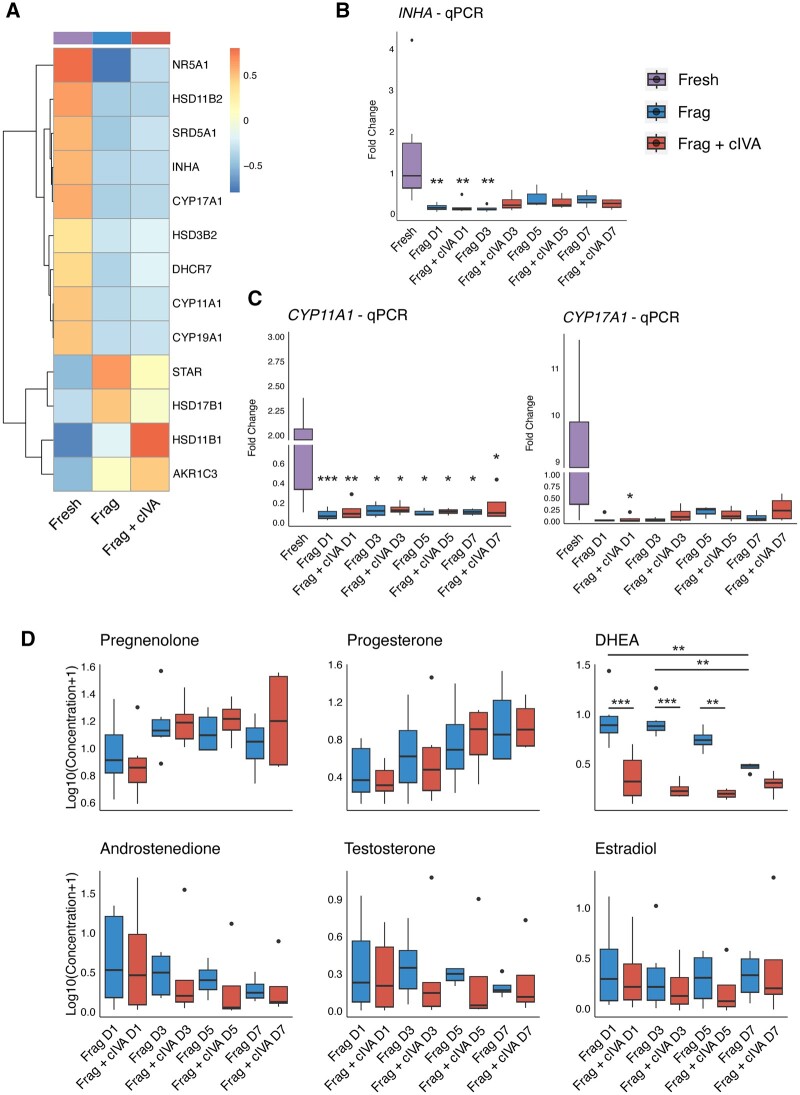
To further investigate the RNA-seq dataset, we focused specifically on genes relating to granulosa cell functions, as follicle growth in culture is characterized by granulosa cell proliferation. We noticed a significant downregulation of typical steroidogenesis-related genes CYP11A1, CYP17A1, NR5A1, HSD11B2, and INHA in the RNA-seq data in both Frag+cIVA and Frag groups (Fig. 3A), despite the histologically confirmed follicle growth (Fig. 1A–C). A similar trend of down-regulation was also observed in other related genes such as HSD3B2, CYP19A1, SRD5A1, and DHCR7 (Fig. 3A). However, some steroidogenesis genes, namely STAR, HSD17B1, HSD11B1, and AKR1C3, showed an opposite, upregulated trend (Fig. 3A). Additionally, lower expression of classical granulosa cell markers (i.e. FOXL2, AMHR2, CDH2) and nuclear steroid hormone receptors (i.e. PGR, ESR1, ESR2, AR) was found in both cultured groups compared to the fresh samples (Supplementary Fig. S3).
Figure 3.
Changes in steroidogenesis during human ovarian cortical tissue culture in fragmentation and fragmentation+chemical in vitro activation groups. Ovarian cortical tissue fragments from 15 women were cultured for a total of 7 days with half of the samples exposed to the cIVA protocol. Fragments and culture media were collected for analyses on Days 1, 3, 5, and 7. (A) Clustered heatmap of average expression of steroidogenesis-related genes at 24 h in the RNA-seq data. Data were normalized with DESeq2 normalization and scaled to obtain mean equal to 0 and SD equal to 1. Color bar above the heatmap represents the group. (B) Results of INHA and (C) CYP11A1 and CYP17A1 expression by quantitative PCR in fresh, Day 1, Day 3, Day 5, and Day 7-cultured tissue, normalized to the housekeeping gene RPL22. (D) Log10-transformed levels of pregnenolone, progesterone, DHEA, androstenedione, testosterone, and estradiol during the 7-day culture. Results are presented as boxplots where the box represents the interquartile range, the horizontal line denotes the median, whiskers mark the non-outlier range, and outliers are depicted as dots. Statistics were performed using two-way ANOVA with interaction and Tukey’s post hoc correction after data was log2- (qPCR) and log10-transformed (steroids) in RStudio. Asterisks represent the comparison between fresh and each culture group. *P < 0.05, **P < 0.01, ***P < 0.001. cIVA, chemical in vitro activation; CYP11A1, cytochrome P450 family 11 subfamily A member 1; CYP17A1, cytochrome P450 family 17 subfamily A member 1; DHEA, dehydroepiandrosterone; Frag, fragmentation; INHA, inhibin subunit alpha; qPCR, quantitative PCR; RPL22, ribosomal protein L22.
To further validate this downregulation, we carried out a new culture experiment to study the time-dependent changes in gene expression over the 1-week culture using qPCR. This method is highly dependent on robust housekeeping genes that are used for normalization of the data. While analyzing the RNA-seq data, we noticed that the expression of many commonly used housekeeping genes (e.g. ACTB, GAPDH, B2M) significantly changed during culture, which makes them unfit for normalization purposes. Therefore, we set out to identify more robust housekeeping genes, and identified RPL22 as the most stable candidate based on results from Normfinder, Bestkeeper, and GeNorm analyses (Supplementary Tables S6, S7, and S8). Therefore, RPL22 was used in subsequent qPCR validations.
We selected INHA, CYP11A1, and CYP17A1 for qPCR validation. The qPCR experiments confirmed a dramatic downregulation of all selected genes in both culture groups starting from Day 1 of culture (Fig. 3B and C). Notably, the basal expression levels of these genes varied widely in the fresh control samples. We then assessed if the downregulation of steroidogenic genes corresponded to changes in functional outcomes by measuring steroid concentrations in the culture media. Significant differences were found in the steroidogenic pattern between the Frag+cIVA and Frag groups starting from the first day of culture: the concentration of DHEA was significantly reduced in the media of tissue treated with cIVA (Fig. 3D, Supplementary Table S9). In addition, median concentrations of androstenedione, testosterone, and estradiol tended to be lower in the Frag+cIVA group compared to Frag; however, these did not reach statistical significance (Fig. 3D, Supplementary Table S9). The only steroids that increased during culture in both groups were pregnenolone and progesterone (Fig. 3D, Supplementary Table S9).
No profound differences in cytokine and chemokine profiles between Frag and Frag+cIVA
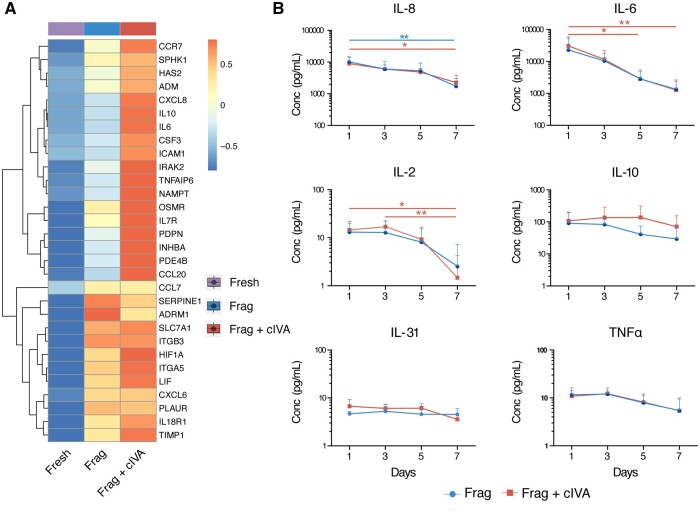
In the RNA-seq data, a significant upregulation of various gene sets related to inflammatory responses was found in both culture groups, including gene sets related to IFN-α and gamma (IFN-γ), IL-2, and TNFα signaling (Fig. 2C, Supplementary Table S5). On an individual gene level, multiple cytokines were found among the DEGs, and some of them also differed between the Frag+cIVA and Frag groups (i.e. CXCL8/IL-8 and IL-6) at 24 h (Fig. 4A). Therefore, we measured time-dependent changes in cytokine and chemokine secretion to further investigate the effects of Frag+cIVA and Frag on inflammatory responses. In agreement with the RNA-seq data, most of the selected cytokines and chemokines were detected in the samples. In general, there was a decreasing trend in most cytokines and chemokines measured across the culture period (Fig. 4B). There were no significant differences between levels of IL-8, IL-6, IL-31, IL-2, IL-10, and TNFα in Frag+cIVA versus Frag group at any time point (Fig. 4B). The levels of IL-2, IL-6, and IL-8 significantly declined during culture (Fig. 4B).
Figure 4.
Changes of cytokines and chemokines during human ovarian cortical tissue culture in fragmentation and fragmentation+chemical in vitro activation groups. (A) Clustered heatmap of average expression of inflammatory response-related genes at 24 h in the RNA-seq data. Data was normalized with DESeq2 normalization and scaled to obtain mean equal to 0 and SD equal to 1. Color bar above the heatmap represents the group (n = 11). (B) Ovarian cortical tissue fragments from three women were cultured for a total of 7 days with half of the samples exposed to the cIVA protocol. Concentrations of IL-8, IL-6, IL-31, IL-2, IL-10, and TNFα were measured in culture media collected on Days 1, 3, 5, and 7 of culture, shown as average (+SD). Statistics were performed using two-way ANOVA with interaction and Tukey’s post hoc correction in RStudio. No significant differences were observed between Frag+cIVA and Frag at any time point. Blue line represents statistical comparison within the Frag group while red represents the Frag+cIVA group. Blue dot represents the Frag group and red square represents the Frag+cIVA group. *P < 0.05, **P < 0.01. cIVA, chemical in vitro activation; Conc, concentration; Frag, fragmentation; TNFα, tumor necrosis factor alpha.
In vitro culture activated glycolysis-related genes in ovarian tissues
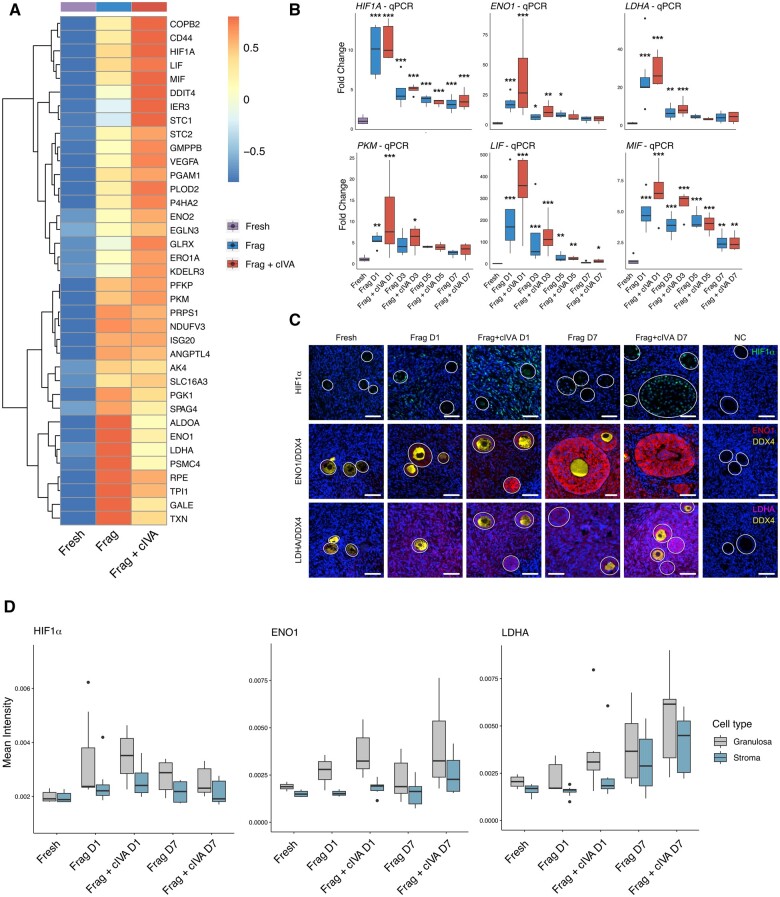
The activation and growth of follicles were confirmed by the histological follicle counting in both the Frag and Frag+cIVA groups (Fig. 1). To elucidate mechanisms underlying follicle activation and growth in the in vitro system, we analyzed the RNA-seq data more deeply. We specifically focused on the DEGs and gene set enrichment results found in the Frag versus fresh and Frag+cIVA versus fresh comparisons. Interestingly, glycolysis was found to be affected by Frag+cIVA treatment compared both to Frag and to the fresh group (Fig. 2C and D). As glycolysis is a central energy metabolic pathway in tissues, we focused on the further characterization of possible differences in energy metabolism between Frag and Frag+cIVA. We plotted the expression of representative genes related to glycolysis (e.g. ENO1, LDHA, PKM) and their upstream regulators HIF1A, LIF, and MIF (Benigni et al., 2000; Lum et al., 2007; Toso et al., 2008). Consistent with our gene set enrichment results where induction of glycolysis was observed (Fig. 2C), upregulation of these genes and the upstream regulators HIF1A, LIF, and MIF were shown in the two cultured groups in the RNA-seq data at 24 h (Fig. 5A), although few were significant in the differential expression analysis between the Frag and Frag+cIVA groups.
Figure 5.
Upregulation of glycolysis-related genes in human ovarian tissue during in vitro culture. Ovarian cortical tissue from 11 women was cultured for 24 h for RNA-seq, and from 15 women for 7 days for validation experiments by qPCR and immunostainings. (A) Clustered heatmap of average expression of glycolysis-related genes at 24 h in the RNA-seq data. Data was normalized with DESeq2 normalization and scaled to obtain mean equal to 0 and SD equal to 1. Color bar above the heatmap represents the group. (B) qPCR results of HIF1A, ENO1, LDHA, PKM, LIF, and MIF, normalized to RPL22, on Day 1, Day 3, Day 5, Day 7 of culture and in freshly collected tissues. (C) Immunostaining of DDX4, HIF1α, LDHA, and ENO1 in fresh, Day 1, and Day 7 of Frag and Frag+cIVA groups. Isotype control (IgG) was used as the negative control. Dark blue represents DAPI signal; green indicates HIF1α; red ENO1; yellow DDX4; and magenta LDHA. Scale bars 50 µm. All figures are presented under the same scale of brightness and contrast. Figures were processed using OMERO.figure. (D) Quantification of immunofluorescence intensity of HIF1α, ENO1, and LDHA in granulosa and stroma cells. Gray indicates signals in granulosa cells while blue represents signals in stroma. Results are presented as boxplots where the box represents the interquartile range, the line denotes the median, whiskers mark the non-outlier range, and outliers are depicted as dots. Statistics were performed using two-way ANOVA with interaction and Tukey’s post hoc correction after data was log2-transformed in RStudio. *P < 0.05, **P < 0.01, ***P < 0.001. cIVA, chemical in vitro activation; DDX4, DEAD-box helicase 4; ENO1, alpha-enolase; Frag, fragmentation; HIF1A/HIF1α, hypoxia inducible factor 1 subunit alpha; LDHA, lactate dehydrogenase A; LIF, leukemia inhibitory factor; MIF, macrophage migration inhibitory factor; NC, negative control; PKM, pyruvate kinase M1/2; qPCR, quantitative PCR; RPL22, ribosomal protein L22.
Next, we validated the changes in glycolysis-related pathways in an independent set of samples using qPCR. Consistent with our RNA-seq results (Fig. 5A), a highly significant upregulation of ENO1, LDHA, PKM, HIF1A, LIF, and MIF was observed already on Day 1 of culture (Fig. 5B). The expression slowly returned towards baseline during the culture (Fig. 5B). Although the median expression levels tended to be higher in the Frag+cIVA group compared to Frag at most time points, this difference did not reach statistical significance. To further validate the changes in glycolysis and assess which cell types were affected, we chose ENO1, LDHA, and HIF1A for immunofluorescence staining experiments. Our results suggested marked upregulation of ENO1 and LDHA proteins during culture: LDHA starting on Day 1 across stromal and follicular cells, and ENO1 being strongly expressed by granulosa cells of growing follicles as well as some stromal cells on Day 7 (Fig. 5C). The expression of ENO1 and LDHA was low and mainly localized in granulosa cells in the fresh samples. However, in the cultured tissues, their immunofluorescence signal was found in the cytoplasm of both granulosa cells and the surrounding stromal compartment (Fig. 5C). On the other hand, the upstream regulator HIF1α showed a nuclear localization pattern, and it was mainly localized in granulosa cells in the Day 7-cultured samples while stromal cells showed HIF1α staining on Day 1 (Fig. 5C). Samples treated with cIVA tended to show stronger HIF1α staining (Fig. 5C). When we quantified the fluorescence signals separately in stromal and granulosa cells, the results suggested that granulosa cells displayed a stronger expression of HIF1α, ENO1, and LDHA (Fig. 5D). Notably, protein levels of HIF1α peaked on Day 1 while ENO1 and LDHA exhibited a gradual increase trend during the culture (Fig. 5D). In agreement with the staining, increased trends of HIF1α, ENO1, and LDHA levels were found in the cultured group compared to the fresh control (Fig. 5D). Similar to the qPCR results, we observed a higher expression of quantified proteins (except for HIF1α on Day 7 culture) in the Frag+cIVA group compared to the Frag group, even though these trends did not reach statistical significance (Fig. 5D).
cIVA induced the expression of hypoxia- and glycolysis-related genes in KGN cells
Our RNA-seq results suggested that Frag+cIVA treatment might induce a higher expression of some glycolysis-related genes compared to the Frag group (Fig. 5), although statistical significance was not reached using clinical patient samples. Besides, immunofluorescence results suggested that this effect mainly took place in granulosa cells (Fig. 5C and D). Therefore, we wanted to further study if glycolysis might be a direct target of cIVA drugs in granulosa cells using a simpler model, the human granulosa cell-like cancer cell line KGN. Additionally, to ensure that the results were comparable to tissue culture, we included groups cultured with 0.05, 0.5, and 1 IU/ml FSH in the experiment. The results showed that the expression of HIF1A, LDHA, and MIF was significantly induced by 24 h bpV (HOpic) and 740Y-P exposure without FSH treatment (Supplementary Fig. S4). A clear upregulation trend was also found in ENO1, PKM, and LIF in KGN without FSH treatment although no statistical significances were observed (Supplementary Fig. S4). Interestingly, FSH alone did not affect the expression of these genes, but it reduced the effects of cIVA treatment, especially concerning the expression of LDHA (Supplementary Fig. S4).
Safety of IVA on the ovary
To assess the safety of the IVA protocols, we performed disease enrichment analysis using DEGs ranked by log2 fold change against the human Disease Gene Network (DisGeNET) database (Piñero et al., 2015). Comparing Frag+cIVA to Frag, the identified DEGs were associated with increased risk of PCOS (Supplementary Fig. S5). However, when compared to the fresh control, both Frag and Frag+cIVA induced transcriptomic changes related to ovarian cancer, with more ovarian-related diseases enriched in the Frag+cIVA group (Supplementary Fig. S5).
Discussion
In the current study, we employed histological assessment, steroid, cytokine/chemokine measurements, and transcriptomic profiling to evaluate the effects of mechanical (Frag) and chemical (Frag+cIVA) IVA on human ovarian tissues. Current opinions on the benefits of performing IVA using both protocols on ovarian tissue are contradictory (Dolmans et al., 2019; Lunding et al., 2019). Therefore, investigating the effects and underlying mechanisms allows us to assess the risks and benefits of this procedure.
Despite extensive efforts, ovarian tissue culture has shown limited success in promoting follicle survival and maturation, with no ground-breaking improvements observed across various tested conditions and substrates (Scott et al., 2004; Telfer et al., 2008; Hao et al., 2020; Bjarkadottir et al., 2021; Ghezelayagh et al., 2021; Pais et al., 2021). The few successful protocols report very low success rates and have not been widely adopted (Xiao et al., 2015; McLaughlin et al., 2018; Xu et al., 2021). The bottleneck that remains is efficient activation of primordial follicles and their survival to healthy secondary follicles that could be isolated for further IVM. Most follicles are lost in the primary-to-secondary growth step in culture in all reported systems. Historical studies from our department have shown that Matrigel improves the survival of follicles in tissue culture (Hovatta et al., 1997); however, this mouse-derived undefined matrix is not suitable for possible future clinical translations. Therefore, we tested an alternative growth substrate in the first part of our study: human laminin LN221. In agreement with our previous study (Hao et al., 2020), LN221 supported ovarian tissue in culture equally well as Matrigel. We therefore pooled the results from the two substrates for downstream analyses and carried out the validation part of the study using LN221 only. As a xeno-free and defined component of the extracellular matrix, LN221 is suitable for eventual clinical applications.
As extensively documented by us and others, the follicles within an ovarian tissue sample activate to grow in culture (Scott et al., 2004; Telfer et al., 2008; Hao et al., 2020). This can be seen as a significantly declined proportion of primordial follicles together with an increased number of secondary follicles, which was also recorded in the current study in both cultured groups. Ki67 staining indicated increased proliferation in the granulosa cells, which aligned with the increased number of secondary follicles observed in both the Frag and Frag+cIVA groups. Many studies have documented the effects of tissue fragmentation on follicle growth, with the hypothesis that the effects were possibly attributed to the disruption of the Hippo signaling pathway through actin polymerization (Hsueh and Kawamura, 2020). Intriguingly, we did not find transcriptomic evidence of Hippo signaling being activated at 24 h of culture. This could depend on the post-translational nature—phosphorylation and ubiquitination—of the Hippo-mechanism, which could be better captured by protein-level studies (Kawamura et al., 2013; He et al., 2016). Future studies utilizing spatial proteomics technology should be performed to thoroughly characterize the change of Hippo signaling induced by fragmentation. In line with previous studies (Li et al., 2010; Raffel et al., 2019), our results confirmed that cIVA treatment resulted in increased follicle survival and growth on Day 7 compared to the Frag group. Maidarti et al. (2019) reported increased DNA damage in cultured bovine ovaries after cIVA treatment. However, in our samples only a small number of cells were positive for typical DNA damage (γH2AX) and apoptosis markers (TUNEL). We did not identify clear apoptotic signatures in the RNA-seq data between fresh and cultured samples either.
To the best of our knowledge, this is the first study to analyze the transcriptomic changes by RNA-seq in ovarian tissue culture. Here, we had the unique opportunity to unravel tissue-wide changes in the transcriptome following short-term culture. Perhaps not surprisingly, the mere process of culture caused major changes in the tissue transcriptome. The many upregulated gene sets related to tissue damage and inflammation revealed the stress that fragmented ovarian tissue is exposed to. Tissue fragmentation is a necessity when the ovarian cortex is collected from patients and prepared for culture, but this may activate, for example, TNFα, IL-6, IL-8, and IL-10, as suggested by our RNA-seq results and cytokine measurements. These potent signaling molecules could, in turn, induce expression of factors that modulate follicle dormancy and growth. For example, LIF can be activated by TNFα in the ovary, where LIF is shown to be involved in primordial follicle activation in rats(Nilsson et al., 2002). TNFα has also been shown to induce the expression and secretion of the pro-inflammatory cytokine MIF (Rossi et al., 1998). As the downstream target of both TNFα and HIF1α, MIF subsequently stimulates macrophages to release cytokines IL-6 and IL-8 (Winner et al., 2007; Kasama et al., 2010). Apart from inducing inflammatory-related signaling, the increased expression of LIF and MIF might contribute to increased granulosa cell proliferation, leading to follicle growth observed in both the Frag and Frag+cIVA groups (Matsuura et al., 2002; Field et al., 2014). Interestingly, ovarian tissue is also fragmented in fertility preservation, where small tissue fragments are cryopreserved for later transplantation. Over-activation and loss of follicles are known problems following tissue transplantation, where ischemia-reperfusion damage likely plays a role (Rodrigues et al., 2023). However, our data suggest that the fragmentation itself can trigger extensive damage responses in the tissue, which would also contribute to the follicle loss.
We observed a dramatic decrease in the expression of the central steroidogenic genes CYP11A1 and CYP17A1 after 24 h culture. These findings do not necessarily correlate with follicle death or the poor quality of follicles, as histological assessment suggested a higher number of secondary follicles in cultured tissue. One of the key functions of granulosa cells is steroidogenesis, boosted by gonadotrophins from the secondary stage onwards. FSH in culture medium was shown to regulate the steroidogenesis in 3D mouse follicle culture (Kreeger et al., 2005). However, despite the presence of FSH in the culture medium, steroid output did not consistently increase during the culture period in our study, suggesting a potential lack of typical follicular functionality. On one hand, the follicles did not develop to advanced secondary or antral stages in the 1-week culture, and robust steroid output might not be expected either owing to the limited theca cells recruitment. On the other hand, the cellular stress caused by tissue fragmentation and culture could also lead to a temporary decline in the functionality of steroidogenesis-related cells and compromised hormone production. In accordance with this hypothesis, decreased functionality of cultured bovine granulosa cells was reported where the expression of FSHR, CHD1, CYP19A1, and ESR1 was reduced in short-term culture (Yenuganti and Vanselow, 2017). Furthermore, the overall low hormone synthesis level and high inter-individual variation made it challenging to detect statistically significant differences between the groups. The higher number of follicles present in the fresh samples compared to the cultured tissues could also explain the lower expression of granulosa cell markers in the RNA-seq data. Intriguingly, Frag+cIVA treatment resulted in a lower DHEA concentration in the culture medium, suggesting that cIVA might somehow affect the activity of steroidogenic enzymes. We observed a trend towards increased STAR and HSD17B1 expression as well as significant AKR1C3 upregulation in Frag+cIVA compared to the fresh group. Especially AKR1C3 is related to DHEA conversion to androstenediol. This might partially explain the decrease of DHEA in the Frag+cIVA group. CYP17A1 is also centrally involved in DHEA production, and although we did not find differences in mRNA expression levels between Frag+cIVA and Frag groups, future studies could determine the effect of cIVA on CYP17A1 enzymatic activity. Nevertheless, our current results do not clearly explain the impact of cIVA on steroidogenesis. Further studies could focus on outlining the transcriptomic profiles of isolated secondary follicles to form a reference. Such a reference would enable conclusions on appropriate levels of steroidogenesis and granulosa cell markers in culture-produced secondary follicles, which would be helpful in further optimization of culture systems.
Unexpectedly, our RNA-seq data revealed major changes in energy metabolism. We discovered that multiple central glycolysis-related genes and their upstream regulators were heavily upregulated during the first day of culture in both groups. Glycolysis is the preferable energy metabolic pathway in granulosa cells, and the mutual interaction between granulosa cells and oocyte in cellular metabolism plays an important role in folliculogenesis (Fontana et al., 2020). Interestingly, enhanced glycolysis has been shown to induce primordial follicles activation through mTOR signaling in mice and human granulosa cells (Zhang et al., 2022).
A clustered heatmap of glycolysis-related genes indicated that cIVA might have an additional boosting effect on this pathway. This hypothesis gained support from the qPCR validation of KGN cells exposed to the IVA drugs for 24 h. This indicates that although glycolysis genes were upregulated in tissue culture per se, their expression is likely further boosted by cIVA. Surprisingly, we found that FSH reduced the cIVA-stimulated glycolysis induction. This suggests that the effectiveness and precise molecular impacts of the cIVA protocol might differ in patients with different FSH levels.
To our knowledge, this is the first study that employed transcriptomic profiling to thoroughly characterize the effects of Frag and Frag+cIVA on human-cultured ovarian tissues. The use of a substantial number of patient samples enhances the robustness of the findings. Extensive validation and independent experiments on RNA and protein levels were also performed to test our hypothesis. The results shed light on the potential underlying molecular mechanisms of mechanical and chemical IVA on follicle activation and growth in culture. However, it is acknowledged that there is considerable inter-patient variation, which is one of the drawbacks in our study, but at the same time reflects the reality at reproductive medicine clinics. Moreover, the study was performed in an in vitro model where tissue was isolated from regulation by the hypothalamic–pituitary–ovarian axis and, thus, further experiments with xeno-transplantation may be needed to explore the effects of cIVA in vivo in the future. However, it should be noted that the different IVA protocols are based on the culture of fragmented tissue and hence our model and findings are relevant.
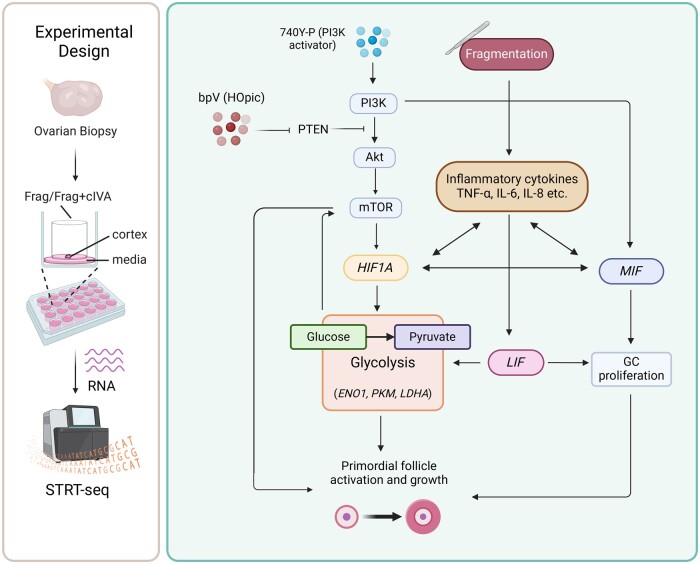
Taken together, we propose that tissue fragmentation alone triggers a large-scale inflammatory response, leading to the activation of downstream targets such as LIF and MIF (Rossi et al., 1998; Göllner et al., 1999; Malkov et al., 2021). These molecules, in turn, activate genes associated with glycolysis (ENO1, PKM, and LDHA), ultimately enhancing the activation and growth of primordial follicles in the culture (Matsuura et al., 2002; Winner et al., 2007; Field et al., 2014; Zhang et al., 2022). We further hypothesize that the inflammatory cytokines MIF and LIF can also directly contribute to primordial follicle activation by promoting the proliferation of granulosa cells (Matsuura et al., 2002; Field et al., 2014). Finally, the application of cIVA treatment further amplifies these mechanisms: the activation of PI3K–Akt–mTOR signaling leads to the activation of LIF and MIF, and the mTOR signaling also activates HIF1A and downstream glycolysis-related genes (Düvel et al., 2010). Altogether, this complex interplay of signaling networks could synergistically enhance the primordial follicle activation and growth characterizing both chemical and mechanical (drug-free) IVA (Zhang et al., 2022) (Fig. 6). Similar mechanisms could also be involved in the well-documented loss of follicles in ovarian tissue transplantation in fertility preservation.
Figure 6.
Working model of putative pathways involved in mediating effects of fragmentation and fragmentation+chemical in vitro activation on cultured human ovarian tissue. Fragmentation of tissue promotes the production of inflammatory cytokines, MIF, LIF, and glycolysis-related genes ENO1, PKM, and LDHA, contributing to primordial follicle activation. In the context of Frag+cIVA, activation of PI3K–Akt–mTOR signaling results in the upregulation of the downstream targets that include HIF1A, MIF, and LIF, which enhance the expression of glycolysis-related genes and granulosa cells proliferation. These mechanisms contribute to the growth activation of primordial follicles in culture. Akt, protein kinase B; bpV (HOpic), bisperoxovanadium (HOpic); cIVA, chemical in vitro activation; ENO1, alpha-enolase; Frag, fragmentation; HIF1A, hypoxia inducible factor 1 subunit alpha; GC, granulosa cells; LDHA, lactate dehydrogenase A; LIF, leukemia inhibitory factor; MIF, macrophage migration inhibitory factor; mTOR, mammalian target of rapamycin; PI3K, phosphoinositide 3 kinase; PTEN, phosphatase and tensin homolog; STRT-seq, single-cell tagged reverse transcription sequencing; TNF-α, tumor necrosis factor alpha.
The safety of the cIVA protocol has been a concern as it might initiate malignancy when activating the important PI3K–Akt–mTOR signaling. To identify disease related to the changes in the transcriptome after cIVA treatment, DEGs identified in the Frag+cIVA versus fresh comparisons were used for enrichment analysis. Our results showed that these DEGs were related to ovarian diseases, particularly malignant tumors, supporting concerns that cIVA may stimulate tumorigenic pathways. However, some tumor-related pathways were also found in the Frag groups, indicating that risks also exist with mechanical follicle activation. Therefore, not limited to IVA, all clinical interventions involving ovarian tissue manipulation, such as ovarian tissue cryopreservation, should minimize the duration of in vitro culture of ovarian cortex before transplantation, as well as reduce mechanical stimulation, such as tissue traction and compression, to mitigate associated risks. Additionally, the health status of the transplanted tissue should be carefully monitored, for example, by ultrasound scanning and measurement of tumor markers.
Conclusion
In conclusion, this study provides evidence that both tissue fragmentation (mechanical IVA) and a combination of tissue fragmentation with chemical in vitro activation (cIVA) enhances the activation of human ovarian follicles in culture, with Frag+cIVA demonstrating a more pronounced effect on follicle growth. However, the comprehensive changes in the ovarian tissue transcriptome triggered by dissection and short-term culture far exceed the effects of added IVA chemicals. The tremendous changes in the transcriptomic profile, particularly the significant inflammatory and metabolic alterations, emphasize the need to study the quality of any follicles derived from fragmented and/or cultured tissue in detail before use in clinical trials. Clinical applications of IVA protocols aimed at offspring production should proceed with caution until further evidence of safety is obtained. The risks of tumor development should be followed up in patients who have received ovarian tissue transplantation.
Supplementary Material
Acknowledgements
We would like to acknowledge the Morphological Phenotype Analysis facility (FENO) for the preparation and scan of the histological slides. Imaging of the fluorescent slides was performed at the Live Cell Imaging Core facility/Nikon Center of Excellence, at Karolinska Institutet, supported by the KI infrastructure council. We also would like to thank BEA, the Bioinformatics and Expression Analysis core facility, which is supported by the board of research at the Karolinska Institutet and the research committee at the Karolinska Hospital. We wish to thank the Biobank and Study Support at Karolinska University Hospital for their contribution including professional service and support. The authors thank Professor Matti Poutanen for discussions relating to steroidogenesis. Finally, we extend our sincerest gratitude to Professor Emerita Outi Hovatta for her invaluable initiation and support of the studies on ovarian tissue culture conducted at our department.
Contributor Information
Jie Hao, Department of Reproductive Medicine, Xiangya Hospital, Central South University, Changsha, P.R. China; Department of Gynecology and Reproductive Medicine, Karolinska University Hospital, Stockholm, Sweden; Division of Obstetrics and Gynecology, Department of Clinical Science, Intervention and Technology, Karolinska Institutet, Stockholm, Sweden.
Tianyi Li, Department of Gynecology and Reproductive Medicine, Karolinska University Hospital, Stockholm, Sweden; Division of Obstetrics and Gynecology, Department of Clinical Science, Intervention and Technology, Karolinska Institutet, Stockholm, Sweden.
Manuel Heinzelmann, Department of Environment and Health, Amsterdam Institute for Life and Environment, Amsterdam, The Netherlands.
Elisabeth Moussaud-Lamodière, Department of Gynecology and Reproductive Medicine, Karolinska University Hospital, Stockholm, Sweden; Division of Obstetrics and Gynecology, Department of Clinical Science, Intervention and Technology, Karolinska Institutet, Stockholm, Sweden.
Filipa Lebre, Nanosafety Group, International Iberian Nanotechnology Laboratory, Braga, Portugal.
Kaarel Krjutškov, Faculty of Medicine, Institute of Clinical Medicine, University of Tartu, Tartu, Estonia; Competence Centre on Health Technologies, Tartu, Estonia.
Anastasios Damdimopoulos, Bioinformatics and Expression Analysis Core Facility, Karolinska Institutet, Stockholm, Sweden.
Catarina Arnelo, Division of Obstetrics and Gynecology, Department of Clinical Science, Intervention and Technology, Karolinska Institutet, Stockholm, Sweden.
Karin Pettersson, Division of Obstetrics and Gynecology, Department of Clinical Science, Intervention and Technology, Karolinska Institutet, Stockholm, Sweden.
Ernesto Alfaro-Moreno, Nanosafety Group, International Iberian Nanotechnology Laboratory, Braga, Portugal.
Cecilia Lindskog, Department of Immunology, Genetics and Pathology, Cancer Precision Medicine Research Program, Uppsala University, Uppsala, Sweden.
Majorie van Duursen, Department of Environment and Health, Amsterdam Institute for Life and Environment, Amsterdam, The Netherlands.
Pauliina Damdimopoulou, Department of Gynecology and Reproductive Medicine, Karolinska University Hospital, Stockholm, Sweden; Division of Obstetrics and Gynecology, Department of Clinical Science, Intervention and Technology, Karolinska Institutet, Stockholm, Sweden.
Data availability
Data were deposited in GEO data base (accession number GSE234765). The code for sequencing analysis can be found in https://github.com/tialiv/IVA_project.
Authors’ roles
Conception and design of study: P.D., J.H. Experimental work and data acquisition: J.H., T.L., M.H., F.L., K.K., C.A., K.P., E.A.-M., C.L., M.v.D. Data analysis and interpretation: P.D., A.D., J.H., T.L., E.M.-L., F.L. Manuscript preparation: J.H., T.L., P.D. All authors participated in the critical discussion of the findings and revision of the manuscript. All authors approved the final version of the manuscript.
Funding
European Union’s Horizon 2020 Research and Innovation Programme (Project ERIN No. 952516, FREIA No. 825100); Swedish Research Council VR (2020-02132); StratRegen funding from Karolinska Institutet, KI-China Scholarship Council (CSC) Programme; Natural Science Foundation of Hunan (2022JJ40782). International Iberian Nanotechnology Laboratory Research was funded by the European Union’s H2020 Project Sinfonia (857253) and SbDToolBox (NORTE-01-0145-FEDER-000047), supported by Norte Portugal Regional Operational Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund.
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
References
- Adhikari D, Gorre N, Risal S, Zhao Z, Zhang H, Shen Y, Liu K. The safe use of a PTEN inhibitor for the activation of dormant mouse primordial follicles and generation of fertilizable eggs. PLoS One 2012;7:e39034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari D, Liu K. Molecular mechanisms underlying the activation of mammalian primordial follicles. Endocr Rev 2009;30:438–464. [DOI] [PubMed] [Google Scholar]
- Aittomäki K, Herva R, Stenman UH, Juntunen K, Ylöstalo P, Hovatta O, la CAd Clinical features of primary ovarian failure caused by a point mutation in the follicle-stimulating hormone receptor gene. J Clin Endocrinol Metab 1996;81:3722–3726. [DOI] [PubMed] [Google Scholar]
- Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 2004;64:5245–5250. [DOI] [PubMed] [Google Scholar]
- Benigni F, Atsumi T, Calandra T, Metz C, Echtenacher B, Peng T, Bucala R. The proinflammatory mediator macrophage migration inhibitory factor induces glucose catabolism in muscle. J Clin Invest 2000;106:1291–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarkadottir BD, Walker CA, Fatum M, Lane S, Williams SA. Analysing culture methods of frozen human ovarian tissue to improve follicle survival. Reprod Fertil 2021;2:59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-García C, Herraiz S, Pamplona L, Subirá J, Soriano MJ, Simon C, Seli E, Pellicer A. Follicular activation in women previously diagnosed with poor ovarian response: a randomized, controlled trial. Fertil Steril 2022;117:747–755. [DOI] [PubMed] [Google Scholar]
- Dolmans M-M, Cordier F, Amorim CA, Donnez J, Vander Linden C. In vitro activation prior to transplantation of human ovarian tissue: is it truly effective? Front Endocrinol (Lausanne) 2019;10:520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnez J, Dolmans M-M. Transplantation of ovarian tissue. Best Pract Res Clin Obstet Gynaecol 2014;28:1188–1197. [DOI] [PubMed] [Google Scholar]
- Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell 2010;39:171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet 2006;7:606–619. [DOI] [PubMed] [Google Scholar]
- Ferreri J, Fàbregues F, Calafell JM, Solernou R, Borrás A, Saco A, Manau D, Carmona F. Drug-free in-vitro activation of follicles and fresh tissue autotransplantation as a therapeutic option in patients with primary ovarian insufficiency. Reprod Biomed Online 2020;40:254–260. [DOI] [PubMed] [Google Scholar]
- Ferreri J, Méndez M, Calafell JM, Fábregues F. Long-term outcome of ovarian function after drug-free in vitro activation (IVA) in primary ovarian insufficiency patient. JBRA Assist Reprod 2021;25:318–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field SL, Dasgupta T, Cummings M, Orsi NM. Cytokines in ovarian folliculogenesis, oocyte maturation and luteinisation. Mol Reprod Dev 2014;81:284–314. [DOI] [PubMed] [Google Scholar]
- Fontana J, Martínková S, Petr J, Žalmanová T, Trnka J. Metabolic cooperation in the ovarian follicle. Physiol Res 2020;69:33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezelayagh Z, Abtahi NS, Khodaverdi S, Rezazadeh Valojerdi M, Mehdizadeh A, Ebrahimi B. The effect of agar substrate on growth and development of cryopreserved-thawed human ovarian cortical follicles in organ culture. Eur J Obstet Gynecol Reprod Biol 2021;258:139–145. [DOI] [PubMed] [Google Scholar]
- Göllner G, Bug G, Rupilius B, Peschel C, Huber C, Derigs HG. Regulatory elements of the leukaemia inhibitory factor (LIF) promoter in murine bone marrow stromal cells. Cytokine 1999;11:656–663. [DOI] [PubMed] [Google Scholar]
- Hao J, Tuck AR, Prakash CR, Damdimopoulos A, Sjödin MOD, Lindberg J, Niklasson B, Pettersson K, Hovatta O, Damdimopoulou P. Culture of human ovarian tissue in xeno-free conditions using laminin components of the human ovarian extracellular matrix. J Assist Reprod Genet 2020;37:2137–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Zhou Z, Shah AA, Hong Y, Chen Q, Wan Y. New insights into posttranslational modifications of Hippo pathway in carcinogenesis and therapeutics. Cell Div 2016;11:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hounkpe BW, Chenou F, de Lima F, De Paula EV. HRT Atlas v1.0 database: redefining human and mouse housekeeping genes and candidate reference transcripts by mining massive RNA-seq datasets. Nucleic Acids Res 2021;49:D947–D955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovatta O, Silye R, Abir R, Krausz T, Winston RM. Extracellular matrix improves survival of both stored and fresh human primordial and primary ovarian follicles in long-term culture. Hum Reprod 1997;12:1032–1036. [DOI] [PubMed] [Google Scholar]
- Hsueh AJW, Kawamura K. Hippo signaling disruption and ovarian follicle activation in infertile patients. Fertil Steril 2020;114:458–464. [DOI] [PubMed] [Google Scholar]
- Ishizuka B, Furuya M, Kimura M, Kamioka E, Kawamura K. Live birth rate in patients with premature ovarian insufficiency during long-term follow-up under hormone replacement with or without ovarian stimulation. Front Endocrinol (Lausanne) 2021;12:795724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, Lönnerberg P, Linnarsson S. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods 2014;11:163–166. [DOI] [PubMed] [Google Scholar]
- John GB, Gallardo TD, Shirley LJ, Castrillon DH. Foxo3 is a PI3K-dependent molecular switch controlling the initiation of oocyte growth. Dev Biol 2008;321:197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasama T, Ohtsuka K, Sato M, Takahashi R, Wakabayashi K, Kobayashi K. Macrophage migration inhibitory factor: a multifunctional cytokine in rheumatic diseases. Arthritis 2010;2010:106202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katso R, Okkenhaug K, Ahmadi K, White S, Timms J, Waterfield MD. Cellular function of phosphoinositide 3-kinases: implications for development, homeostasis, and cancer. Annu Rev Cell Dev Biol 2001;17:615–675. [DOI] [PubMed] [Google Scholar]
- Kawamura K, Cheng Y, Suzuki N, Deguchi M, Sato Y, Takae S, Ho C, Kawamura N, Tamura M, Hashimoto S et al. Hippo signaling disruption and Akt stimulation of ovarian follicles for infertility treatment. Proc Natl Acad Sci USA 2013;110:17474–17479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivioja T, Vähärautio A, Karlsson K, Bonke M, Enge M, Linnarsson S, Taipale J. Counting absolute numbers of molecules using unique molecular identifiers. Nat Methods 2011;9:72–74. [DOI] [PubMed] [Google Scholar]
- Kreeger PK, Fernandes NN, Woodruff TK, Shea LD. Regulation of mouse follicle development by follicle-stimulating hormone in a three-dimensional in vitro culture system is dependent on follicle stage and dose. Biol Reprod 2005;73:942–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krjutškov K, Katayama S, Saare M, Vera-Rodriguez M, Lubenets D, Samuel K, Laisk-Podar T, Teder H, Einarsdottir E, Salumets A et al. Single-cell transcriptome analysis of endometrial tissue. Hum Reprod 2016;31:844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagergren K, Hammar M, Nedstrand E, Bladh M, Sydsjö G. The prevalence of primary ovarian insufficiency in Sweden; a national register study. BMC Womens Health 2018;18:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kawamura K, Cheng Y, Liu S, Klein C, Liu S, Duan E-K, Hsueh AJW. Activation of dormant ovarian follicles to generate mature eggs. Proc Natl Acad Sci USA 2010;107:10280–10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhu Y, Wei J, Chen L, Chen S, Lai D. The global prevalence of premature ovarian insufficiency: a systematic review and meta-analysis. Climacteric 2023a;26:95–102. [DOI] [PubMed] [Google Scholar]
- Li T, Vazakidou P, Leonards PEG, Damdimopoulos A, Panagiotou EM, Arnelo C, Jansson K, Pettersson K, Papaikonomou K, van DM et al. Identification of biomarkers and outcomes of endocrine disruption in human ovarian cortex using In Vitro Models. Toxicology 2023b;485:153425. [DOI] [PubMed] [Google Scholar]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011;27:1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Xu X, Han T, Yan L, Cheng L, Qin Y, Liu W, Zhao S, Chen Z-J. A novel homozygous mutation in the FSHR gene is causative for primary ovarian insufficiency. Fertil Steril 2017;108:1050–1055.e2. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- Lum JJ, Bui T, Gruber M, Gordan JD, DeBerardinis RJ, Covello KL, Simon MC, Thompson CB. The transcription factor HIF-1α plays a critical role in the growth factor-dependent regulation of both aerobic and anaerobic glycolysis. Genes Dev 2007;21:1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunding SA, Pors SE, Kristensen SG, Landersoe SK, Jeppesen JV, Flachs EM, Pinborg A, Macklon KT, Pedersen AT, Andersen CY et al. Biopsying, fragmentation and autotransplantation of fresh ovarian cortical tissue in infertile women with diminished ovarian reserve. Hum Reprod 2019;34:1924–1936. [DOI] [PubMed] [Google Scholar]
- Luo W, Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013;29:1830–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maidarti M, Clarkson YL, McLaughlin M, Anderson RA, Telfer EE. Inhibition of PTEN activates bovine non-growing follicles in vitro but increases DNA damage and reduces DNA repair response. Hum Reprod 2019;34:297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkov MI, Lee CT, Taylor CT. Regulation of the hypoxia-inducible factor (HIF) by pro-inflammatory cytokines. Cells 2021;10:2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura T, Sugimura M, Iwaki T, Ohashi R, Kanayama N, Nishihira J. Anti-macrophage inhibitory factor antibody inhibits PMSG-hCG-induced follicular growth and ovulation in mice. J Assist Reprod Genet 2002;19:591–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin M, Albertini DF, Wallace WHB, Anderson RA, Telfer EE. Metaphase II oocytes from human unilaminar follicles grown in a multi-step culture system. Mol Hum Reprod 2018;24:135–142. [DOI] [PubMed] [Google Scholar]
- Méndez M, Fabregues F, Ferreri J, Calafell JM, Villarino A, Otero J, Farre R, Carmona F. Biomechanical characteristics of the ovarian cortex in POI patients and functional outcomes after drug-free IVA. J Assist Reprod Genet 2022;39:1759–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson LM. Clinical practice. Primary ovarian insufficiency. N Engl J Med 2009;360:606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson EE, Kezele P, Skinner MK. Leukemia inhibitory factor (LIF) promotes the primordial to primary follicle transition in rat ovaries. Mol Cell Endocrinol 2002;188:65–73. [DOI] [PubMed] [Google Scholar]
- Novella-Maestre E, Herraiz S, Rodríguez-Iglesias B, Díaz-García C, Pellicer A. Short-term PTEN inhibition improves in vitro activation of primordial follicles, preserves follicular viability, and restores AMH levels in cryopreserved ovarian tissue from cancer patients. PLoS One 2015;10:e0127786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pais AS, Reis S, Laranjo M, Caramelo F, Silva F, Botelho MF, Almeida-Santos T. The challenge of ovarian tissue culture: 2D versus 3D culture. J Ovarian Res 2021;14:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol Lett 2004;26:509–515. [DOI] [PubMed] [Google Scholar]
- Piñero J, Queralt-Rosinach N, Bravo À, Deu-Pons J, Bauer-Mehren A, Baron M, Sanz F, Furlong LI. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database (Oxford) 2015;2015:bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puc J, Parsons R. PTEN loss inhibits CHK1 to cause double stranded-DNA breaks in cells. Cell Cycle 2005;4:927–929. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2021. https://www.R-project.org/ [Google Scholar]
- R Studio Team. RStudio: Integrated Development for R. Boston, MA: RStudio, PBC, 2020. http://www.rstudio.com/ [Google Scholar]
- Raffel N, Klemm K, Dittrich R, Hoffmann I, Söder S, Beckmann MW, Lotz L. The effect of bpV(HOpic) on in vitro activation of primordial follicles in cultured swine ovarian cortical strips. Reprod Domest Anim 2019;54:1057–1063. [DOI] [PubMed] [Google Scholar]
- Reddy P, Liu L, Adhikari D, Jagarlamudi K, Rajareddy S, Shen Y, Du C, Tang W, Hämäläinen T, Peng SL et al. Oocyte-specific deletion of Pten causes premature activation of the primordial follicle pool. Science 2008;319:611–613. [DOI] [PubMed] [Google Scholar]
- Rodrigues AQ, Silva IM, Goulart JT, Araújo LO, Ribeiro RB, Aguiar BA, Ferreira YB, Silva JKO, Bezerra JLS, Lucci CM et al. Effects of erythropoietin on ischaemia-reperfusion when administered before and after ovarian tissue transplantation in mice. Reprod Biomed Online 2023;47:103234. [DOI] [PubMed] [Google Scholar]
- Rossi AG, Haslett C, Hirani N, Greening AP, Rahman I, Metz CN, Bucala R, Donnelly SC. Human circulating eosinophils secrete macrophage migration inhibitory factor (MIF). Potential role in asthma. J Clin Invest 1998;101:2869–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A, Czarnecki AA, Dean M, Modi DA, Lantvit DD, Hardy L, Baligod S, Davis DA, Wei J-J, Burdette JE. PTEN loss in the fallopian tube induces hyperplasia and ovarian tumor formation. Oncogene 2018;37:1976–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JE, Carlsson IB, Bavister BD, Hovatta O. Human ovarian tissue cultures: extracellular matrix composition, coating density and tissue dimensions. Reprod Biomed Online 2004;9:287–293. [DOI] [PubMed] [Google Scholar]
- Shi X, Wang J, Lei Y, Cong C, Tan D, Zhou X. Research progress on the PI3K/AKT signaling pathway in gynecological cancer (Review). Mol Med Rep 2019;19:4529–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling DR, Swain-Bowden MJ, Lucas AM, Carpenter AE, Cimini BA, Goodman A. CellProfiler 4: improvements in speed, utility and usability. BMC Bioinformatics 2021;22:433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan SD, Sarrel PM, Nelson LM. Hormone replacement therapy in young women with primary ovarian insufficiency and early menopause. Fertil Steril 2016;106:1588–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Su Y, He Y, Zhang J, Liu W, Zhang H, Hou Z, Liu J, Li J. New strategy for in vitro activation of primordial follicles with mTOR and PI3K stimulators. Cell Cycle 2015;14:721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Yoshioka N, Takae S, Sugishita Y, Tamura M, Hashimoto S, Morimoto Y, Kawamura K. Successful fertility preservation following ovarian tissue vitrification in patients with primary ovarian insufficiency. Hum Reprod 2015;30:608–615. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Hsueh AJ, Kawamura K. Surgical approaches of drug-free in vitro activation and laparoscopic ovarian incision to treat patients with ovarian infertility. Fertil Steril 2020;114:1355–1357. [DOI] [PubMed] [Google Scholar]
- Telfer EE, McLaughlin M, Ding C, Thong KJ. A two-step serum-free culture system supports development of human oocytes from primordial follicles in the presence of activin. Hum Reprod 2008;23:1151–1158. [DOI] [PubMed] [Google Scholar]
- Toso C, Emamaullee JA, Merani S, Shapiro AMJ. The role of macrophage migration inhibitory factor on glucose metabolism and diabetes. Diabetologia 2008;51:1937–1946. [DOI] [PubMed] [Google Scholar]
- van Kasteren YM, Schoemaker J. Premature ovarian failure: a systematic review on therapeutic interventions to restore ovarian function and achieve pregnancy. Hum Reprod Update 1999;5:483–492. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002;3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winner M, Koong AC, Rendon BE, Zundel W, Mitchell RA. Amplification of tumor hypoxic responses by macrophage migration inhibitory factor-dependent hypoxia-inducible factor stabilization. Cancer Res 2007;67:186–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Zhang J, Romero MM, Smith KN, Shea LD, Woodruff TK. In vitro follicle growth supports human oocyte meiotic maturation. Sci Rep 2015;5:17323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Lawson MS, Bean Y, Ting AY, Pejovic T, De Geest K, Moffitt M, Mitalipov SM, Xu J. Matrix-free 3D culture supports human follicular development from the unilaminar to the antral stage in vitro yielding morphologically normal metaphase II oocytes. Hum Reprod 2021;36:1326–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yenuganti VR, Vanselow J. Cultured bovine granulosa cells rapidly lose important features of their identity and functionality but partially recover under long-term culture conditions. Cell Tissue Res 2017;368:397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 2012;16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang L-G, Yan G-R, He Q-Y. DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics 2015;31:608–609. [DOI] [PubMed] [Google Scholar]
- Zhai J, Yao G, Dong F, Bu Z, Cheng Y, Sato Y, Hu L, Zhang Y, Wang J, Dai S et al. In vitro activation of follicles and fresh tissue auto-transplantation in primary ovarian insufficiency patients. J Clin Endocrinol Metab 2016;101:4405–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lin F, Zhao J, Wang Z. Expression regulation and physiological role of transcription factor FOXO3a during ovarian follicular development. Front Physiol 2020;11:595086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Han T, Yan L, Jiao X, Qin Y, Chen Z-J. Resumption of ovarian function after ovarian biopsy/scratch in patients with premature ovarian insufficiency. Reprod Sci 2019;26:207–213. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang W, Wang Z, Zheng N, Yuan F, Li B, Li X, Deng L, Lin M, Chen X et al. Enhanced glycolysis in granulosa cells promotes the activation of primordial follicles through mTOR signaling. Cell Death Dis 2022;13:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A, Ibrahim JG, Love MI. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics 2019;35:2084–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data were deposited in GEO data base (accession number GSE234765). The code for sequencing analysis can be found in https://github.com/tialiv/IVA_project.