Abstract
BACKGROUND
Intrinsic molecular subtypes define distinct biological breast cancers and can be used to further improve diagnosis and risk allocation.
METHODS
The Copenhagen Breast Cancer Genomics Study (CBCGS) prospectively included women diagnosed with breast cancer at Rigshospitalet from 2014 to 2021. Eligible patients were females with a primary invasive breast cancer (T1c, if N0M0; otherwise, any T, any N, or any M stage) and no prior malignancy. All patients underwent molecular profiling with the CIT256 and PAM50 molecular profile.
RESULTS
In the study period, 2,816 patients were included in the CBCGS. Molecular subtyping showed an increase in nonluminal (molecular-apocrine, luminal C, and Basal-like) as compared with luminal (luminal A, luminal B, and Normal-like) subtypes with increasing stage from I to IV. Across all stages, we found a significant difference in survival among subtypes; 91% of patients with LumA were alive at 5 years compared with 91% for LumB, 84% for LumC, 82% for mApo, and 80% for Basal-like. We identified 442 tumors (16%) that were discordant in subtype between CIT256 and IHC. Discordant subtype proved to be a risk factor of death among patients with IHC luminal breast cancer (hazard ratio [HR], 2.08; 95% CI, 1.51–2.86) in a multivariable Cox regression analysis. Discordance occurred more often among patients with N3, stage IV, or grade III disease.
CONCLUSION
Our findings indicate that molecular subtypes are a predominant classification for survival. Assessment is particularly crucial for patients with IHC luminal breast cancer with known high-risk factors, since they are at an increased risk of harboring an aggressive molecular subtype.
Keywords: Oncology
Keywords: Breast cancer, Clinical practice, Molecular pathology
Genomic alterations in breast cancer tumors better predict survival than traditional pathological assessments.
Introduction
Intrinsic molecular subtypes of breast cancer are biologic entities associated with specific prognostic and therapeutic features and provide further prognostic information than traditional clinical assessment with staging and receptor expression (1–3). Of the original 5 intrinsic subtypes, (Luminal A, Luminal B, HER2-enriched, Basal-like, and Normal-like) commercially available platforms have been made available based on 50- or 21-gene molecular classifiers (4, 5).
Within the traditional 3 IHC subgroups and with addition of other clinical and histologic factors, such as tumor size and grade, lymph node involvement, patients’ ages, and menopausal status, prognosis, and response to specific therapies is estimated. However, there remains a substantial difference in the behavior of breast cancers even within each one of such classifications. This discordance in breast cancer biology suggests that the true heterogeneity of epithelial breast cancers is much more vast than initially suspected.
A broad introduction of molecular classifiers into clinical practice is ongoing; however, issues have been raised due to the heterogeneity seen in ER+ tumors as well as the synonymous approach of Basal-like and triple-negative tumors (6, 7). The CIT256 intrinsic subtype has attempted to tackle these issues by integrative analysis and reclassification of intrinsic subtypes into 6 stable molecular subgroups that largely correspond to the original 5 subtypes, but without an ERBB2 subgroup and the separation of the large luminal ER+ group into 4 subgroups (8).
Metastatic development in breast cancer can be seen as either occurring due to diagnostic delay (either by patient or system) or due to clinicomolecular aggressive factors (9–11). Previous studies utilizing IHC subgroups as a surrogacy for intrinsic molecular subtypes have indicated a rise in aggressive nonluminal subtypes with increasing stage (12–14). Similarly, in primary metastatic tumors, IHC subgroups have been shown to be a risk factor for distant metastases in T1 tumors but not in T3/T4 tumors; this reflects the hypothesis that small tumors may harbor a potentially unavoidable systemic dissemination dependent on subtype. However, if that does not occur, continuous extension will lead to metastatic disease, regardless of subtype (15).
The Copenhagen Breast Cancer Genomics Study (CBCGS) was initiated in 2014 and provides unique insight in how molecular subtypes may be part of a standard of care diagnostic pipeline (16). In this study, we examine if there is a shift in intrinsic molecular subtypes across stages and tumor size in a consecutive breast cancer cohort. Furthermore, we wish to examine the clinically relevant information provided by intrinsic molecular subtypes, how it differs from IHC subgroups, and whether it may guide future treatment decisions.
Results
Patient demographics and flowchart.
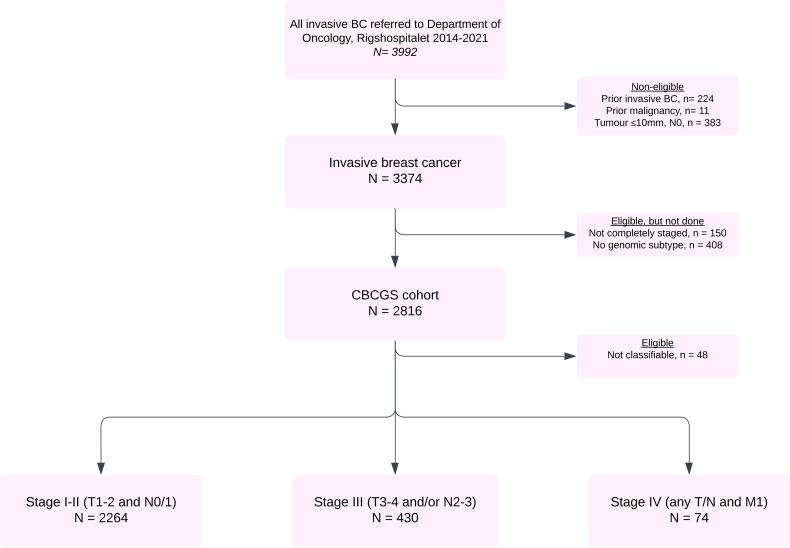
Between April 2014 and December 2021, 3,992 women were diagnosed with an invasive breast cancer at Copenhagen University Hospital, Rigshospitalet, and were referred to the Department of Oncology. Among the 3,768 women with a first invasive breast cancer, 383 had a tumor smaller than 10 mm and 11 had prior malignancy. Of the 3,374 eligible patients, 408 (12.1%) were not subtyped and 150 (4.4%) were biopsied with no further diagnostic workup. Thus, 2,816 women (83.5%) were included in the CBCGS cohort (Figure 1): 2,300 in stage I/II (81.7%), 442 in stage III (15.6%), and 74 in stage IV (2.7%). In total, 48 patients could not be classified by CIT256 and/or PAM50 (outliers) and were excluded from further analyses.
Figure 1. Flow diagram of patients diagnosed at Rigshospitalet from 2014 to 2021.
BC, breast cancer; CBCGS, Copenhagen Breast Cancer Genomics Study.
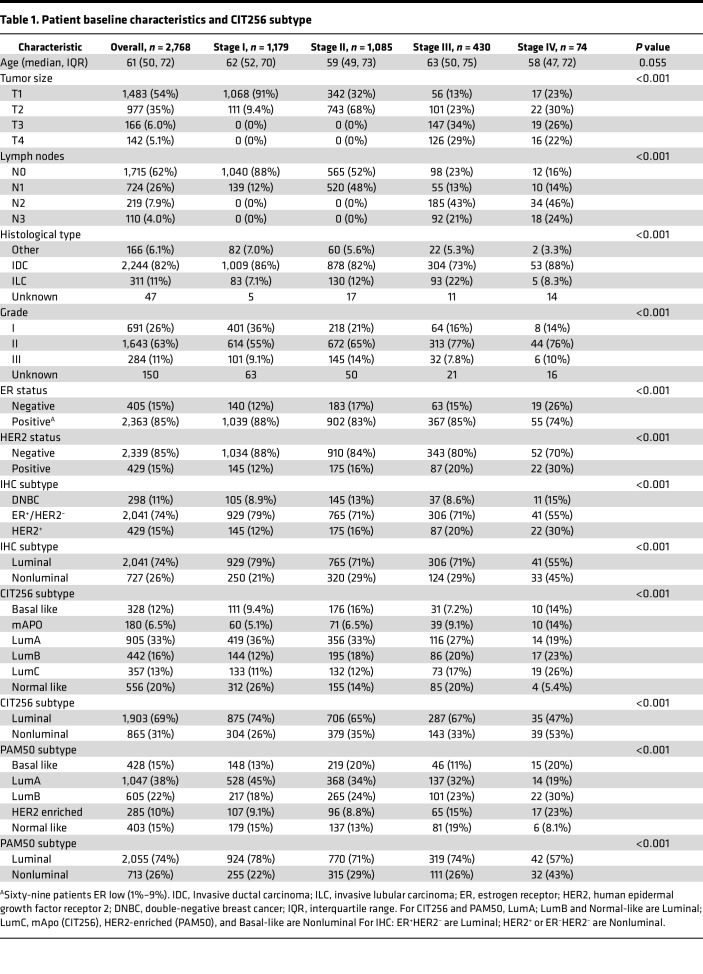
Table 1 shows patient and tumor characteristics. A stage-by-stage increase in HER2-positivity is seen (12%–30%) and an increase in ER– disease from stage I–III versus IV (12%, 17%, and 15% versus 26%, respectively), was identified. The CIT256 subtype differed significantly across stages. mApo increased from 5.1% in stage I to 14% in stage IV and Lumincal C (LumC) from 11% in stage I to 26% in stage IV; however, the same trend was not evident for Basal-like. A similar pattern was seen for PAM50.
Table 1. Patient baseline characteristics and CIT256 subtype.
Addition of molecular subtype at diagnosis.
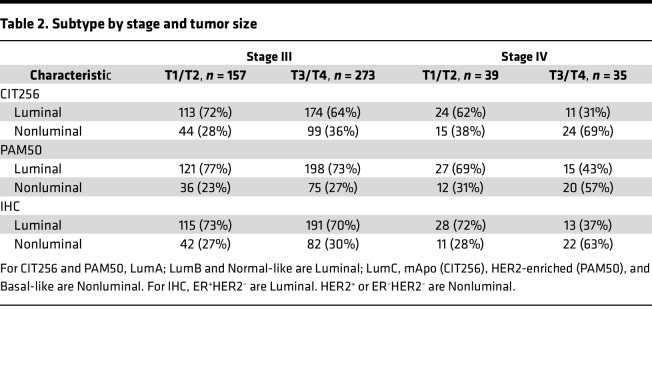
To investigate trends in molecular subtypes, stage, and tumor size, Table 2 demonstrates the distribution of CIT256 subtype, PAM50 subtype, and IHC subtype by stage (III and IV) and tumor size. No apparent difference was evident as to the classification of luminal versus nonluminal, whether it is examined by molecular subtyping scheme (CIT256; PAM50) or IHC. We did see a higher representation of nonluminal tumors in stage IV as compared with stage III and a shift toward more nonluminal tumors with increasing tumor stage — especially seen in stage IV with 69% nonluminal tumors in T3/T4. LumA was highly represented in T1 tumors in both CIT256 and PAM50, with an intermediate representation in T2 falling to almost half in T3/T4. We saw a higher representation of LumC, mApo, and HER2-enriched tumors in T3/T4 parallel to the increase of HER2+ tumors by stage (Supplemental Tables 1 and 2; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.178114DS1). The same was not evident for Basal-like tumors.
Table 2. Subtype by stage and tumor size.
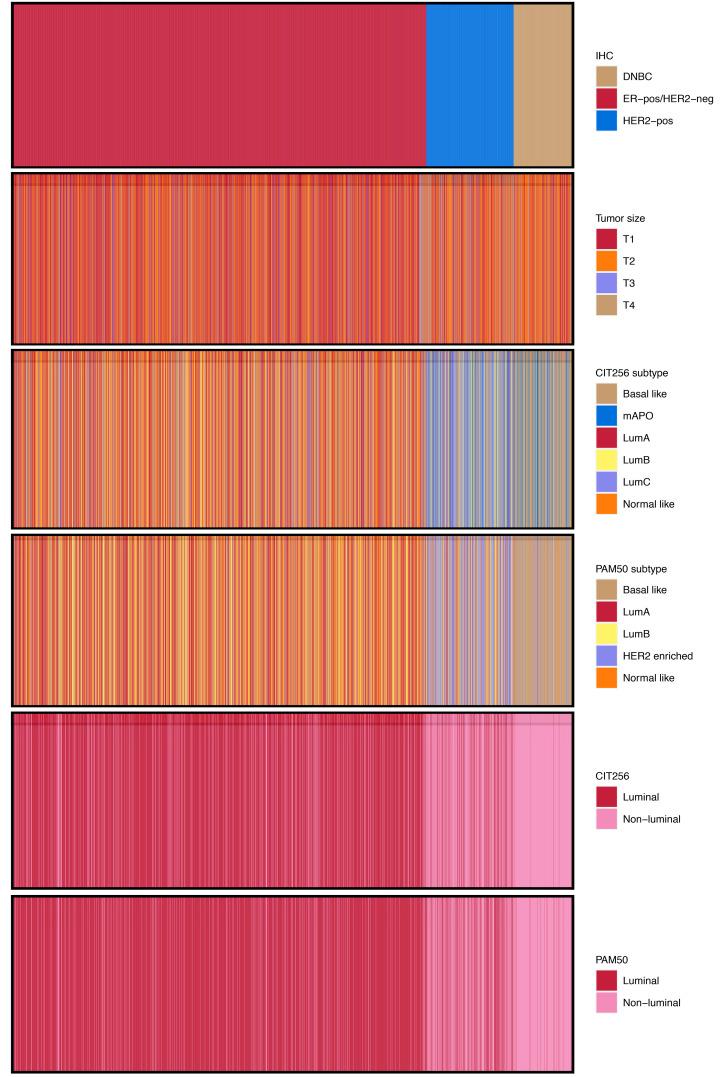
The associations between IHC, tumor size, and intrinsic subtypes are depicted in (Figure 2). No obvious associations are seen between IHC subtype and tumor size, nor in intrinsic subtypes. Overall trends indicate that most patients are classified correctly by crude IHC classification in luminal versus nonluminal subtypes. However, we found that 442 (16%) tumors were discordant, comparing IHC Lum versus nonluminal and CIT256 Lum versus nonluminal; discordance was especially evident among the 429 IHC HER2+ tumors, where 147 (34%) were assigned a luminal CIT256 subtype (185 for PAM50). Supplemental Figure 1 shows the correlation between PAM50 and CIT256 subtypes. In total, 91% of luminal PAM50 tumors are also luminal on CIT256; 94% of nonluminal PAM50 tumors are also nonluminal on CIT256.
Figure 2. Visual correlation of tumor characteristics.
Stacked Visual correlation of tumor characteristics of IHC, tumor size, and molecular subtypes (n = 2,768). Each line from top to bottom represents a patient.
Survival.
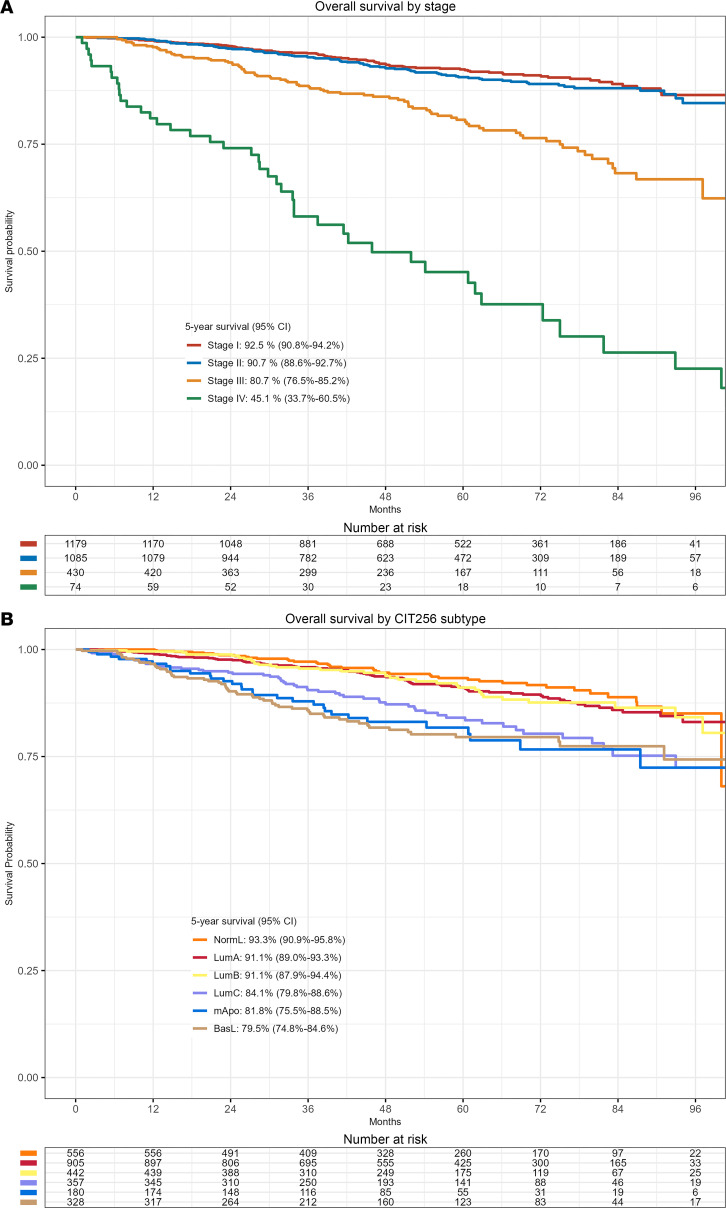
Estimated median potential follow-up was 57.9 months, with 308 deaths registered. Overall survival by stage shows a deteriorating survival probability with increased stage (P < 0.0001). Five-year survival was 92.5% (95% CI, 90.8%–94.2%), 90.7% (95% CI, 88.6%–92.7%), 80.7% (95% CI, 76.5%–85.2%), and 45.1% (95% CI, 33.7%–60.5%) for stage I, II, III, and IV, respectively (Figure 3A). Overall survival was significantly different among CIT256 subtypes (P < 0.0001); 91.1% (95% CI, 89.0%–93.3%) of patients with LumA were alive at 5 years compared with 91.1% (95% CI, 87.9%–94.4%) for LumB, 84.1% (95% CI, 79.8%–88.6%) for LumC, 81.8% (95% CI, 75.5%–88.5%) for mApo, and 79.5% (95% CI, 74.8%–84.6%) for Basal-like (Figure 3B). Supplemental Figure 2 shows 5-year survival estimates by stage and intrinsic molecular subtype.
Figure 3. Survival curves.
(A) Kaplan-Meier survival curve of overall survival by stage at diagnosis (n = 2,768). (B) Kaplan-Meier survival curve of overall survival by CIT256 subtype (n = 2,768).
Importance of IHC and CIT256 subtype.
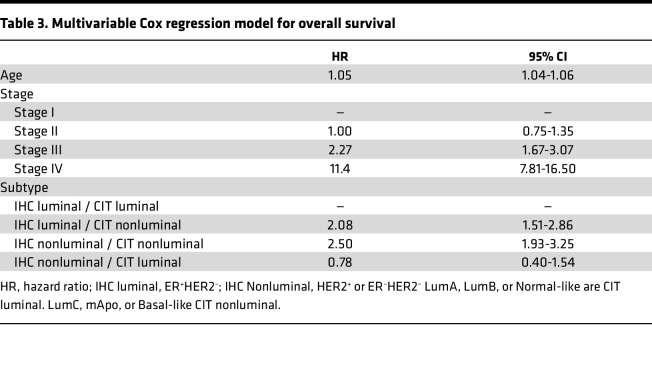
Among the 2,041 patients presenting with an IHC luminal tumor, 1,751 (85.8%) were, by CIT256, assigned a luminal subtype and 290 (14.2%) were assigned a nonluminal CIT256 subtype. Of the 727 patients with a nonluminal tumor by IHC, 575 (79.1%) were, by CIT256, assigned a nonluminal subtype, while 152 (20.9%) patients were assigned a luminal CIT256 subtype. The association between overall survival and subtype assignment by IHC and CIT256 was investigated in a multivariable Cox regression analysis including stage, age, IHC subtype, and CIT256 subtype (Table 3). Malignancy grade and histological subtype did not reach significance in univariable models and were not included. Compared with patients with concurring luminal IHC and CIT256 subtype (reference), a significantly higher mortality was detected in patients with dual nonluminal (IHC and CIT256) subtype (hazard ratio [HR], 2.50; 95% CI, 1.93–3.25) and in patients with a nonluminal CIT256 but with a luminal IHC subtype (HR, 2.08; 95% CI, 1.51–2.86). In contrast, overall survival was not significantly decreased in patients with luminal CIT256 but with a nonluminal IHC subtype (HR, 0.78; 95% CI, 0.40–1.54). This establishes CIT256 subtypes as being predominant for overall survival compared with IHC. This is further confirmed comparing a model with age, stage, and IHC subtype (P ≤ 0.001) with the model including age, stage, CIT256 subtype (P ≤ 0.001), and IHC subtype (P = 0.55). No interaction was detected between stage and IHC subtype.
Table 3. Multivariable Cox regression model for overall survival.
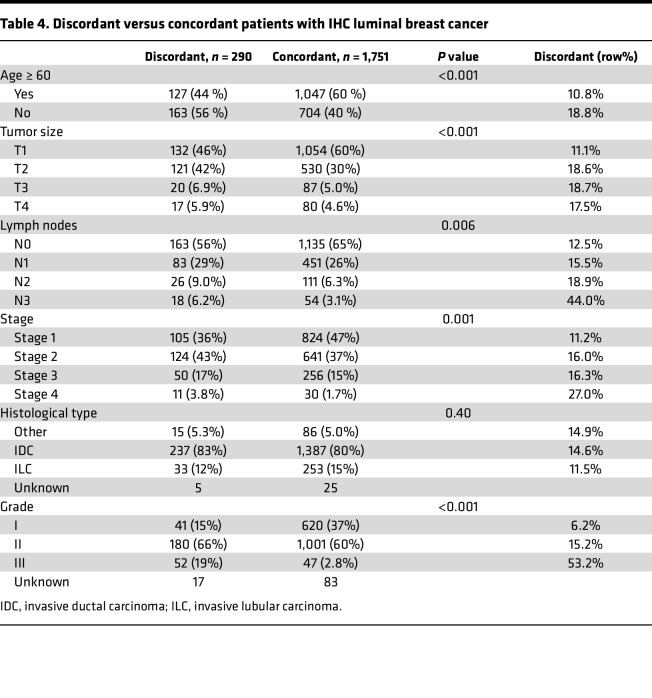
Table 4 displays differences in baseline characteristics for discordant and concordant IHC luminal tumors. Discordance was most pronounced in grade III, N3, and stage IV disease with 53.2%, 44.0%, and 27.0% of cases, respectively. To investigate this further, a logistic regression model was developed (Supplemental Table 3). This model incorporated age, stage of disease, grade, and lymph node metastases and shows an increased risk of discordance for patients with grade II, III, stage IV, N3 disease, or patients younger than 60. By applying our logistic model, we identified 18 individual risk groups in our cohort, and with a cut point of 14.2%, corresponding to the proportion of tumors with discordance among IHC luminal tumors in total, the model correctly classifies 68% of all samples corresponding to a sensitivity of 59% and a specificity of 70% (Supplemental Table 4) .
Table 4. Discordant versus concordant patients with IHC luminal breast cancer.
Discussion
The prospective CBCGS confirms the prognostic importance of stage at diagnosis of breast cancer with 5-year survival rates decreasing from 92.5% in stage I to 45.1% in stage IV. Across stages, we found a significant variation in the assignment of both IHC-based and intrinsic subtype with a decrease of luminal subtype with increasing stage and a particular clear distinction from stage III to IV.
We identified a large group of patients with luminal — i.e., ER+HER2– — breast cancer, who by CIT256 were assigned a nonluminal intrinsic subtype; these patients had a significantly impaired overall survival (HR, 2.08; 95% CI, 1.51–2.86) compared with patients with a dual (IHC and CIT256) luminal subtype. However, as compared with a dual luminal subtype, we found no significant difference in overall survival among patients assigned a luminal subtype by CIT256 and a nonluminal subtype by IHC (HR, 0.78; 95% CI, 0.40–1.54). Likewise, CIT256 remained statistically significant for overall survival in the model that included both CIT256 and IHC subtypes.
To better clarify which patients might be luminal by IHC but assigned a nonluminal intrinsic subtype, we developed a logistic model to help identify patients with potential discordant tumors. We found that age younger than 60 years (OR, 1.60; 95% CI, 1.22–2.10), stage IV (OR, 2.91; 95% CI, 1.30–6.14), grade II (OR, 2.56; 95% CI, 1.81–3.70), grade III (OR, 15.5; 95% CI, 9.36–26.0), and N3-disease (OR, 1.96; 95% CI, 1.07–3.46) were all risk factors for discordance. This accuracy of the risk model is 69% using a cut-point of 14.2% risk of discordance.
The current performance of stratification for early breast cancer treatment is based on stage; histopathological factors, including receptor status; and, for some patients, biomarker assays (4, 5, 17, 18). Our results could point toward incorporating intrinsic molecular subtypes as an element in risk stratification for all patients with breast cancer, as our results indicate that CIT256 better explains survival across all stages than does IHC, as IHC alone does not capture the genomic profile in tumors.
Progression of disease from stage I to IV can essentially be attributed to aggressive clinicomolecular risk factors or diagnostic delay. Our results indicate that diagnostic delays most likely are the main contributing factor for breast cancers being diagnosed in stage I–III, as a slight, nonlinear change in nonluminal subtypes is observed from stage I to III (26%, 35%, 33%, respectively) and then clinicomolecular factors in stage IV with 53% nonluminal tumors.
Our study has several strengths. The prospective inclusion of patients and unrestricted inclusion criteria limit the risk of selection bias in our cohort. The total number of included patients also gives certain strength to the study, especially regarding patients with stage I and II disease. The inclusion of patients across all stages also allowed us to examine differences in subtypes at time of diagnosis.
A limitation in our study is the number of patients diagnosed at Rigshospitalet who appear to have fulfilled our inclusion criteria but on whom we do not have a CIT256 intrinsic subtype. This is particularly evident for stage IV disease, which only accounts for 2.6% of our cohort. We are also limited by the patients undergoing neoadjuvant therapy, on whom we do not have a complete diagnostic workup regarding their lymph node statuses. We are also limited since these patients were exclusively treated according to their IHC subtype and not molecular subtype; this may influence any conclusions on outcomes. Furthermore, our models could be further enhanced by incorporation of the individual intrinsic subtype if more patients with nonluminal subtypes were available. Inclusion of Ki-67 was not done, as it was not readily available for all patients, and we have previously shown that Ki-67 is not optimal for identification of low- and high-risk patients compared with mRNA (16).
To our knowledge, cross-stage comparison of molecular subtypes has not been published on a scale as large as ours. The primary focus of molecular subtyping has primarily been on identifying patient subgroups who would benefit from a specific treatment or strategy. Among patients with early breast cancer, most studies have examined intrinsic subtypes in stage I–II disease (19–28), some of which have restricted their samples to ER+ tumors, resulting in assignment of more than 90% of tumors to a luminal intrinsic subtype (19–21, 23, 25–27). However, in cohorts unselected by ER, the proportion with an intrinsic luminal subtype has been around 80%, corresponding to our results (22, 24, 28). A few studies have included stage III tumors with proportions of stage III varying from 8% to 25% and nonluminal subtypes varying from 32% to 50% overall (29–35). Some of these studies also included PAM50 subtype by stage, with stage III differing from 19% to 57% of tumors being nonluminal (31, 32, 34, 35). A Swedish study on stage IV (mainly recurrent metastatic disease) identified 57% to be nonluminal, which is comparable to our results (36). Other cohorts have recognized an increasing trend toward nonluminal A subtypes with increasing tumor size and lymph node metastases; this differs from our results but, likewise, shows that intrinsic subtypes are a stronger prognostic indicator for outcomes than IHC subtypes (34, 37). Discordance between IHC and molecular subtyping by PAM50 has previously been reported as a risk factor for an event with discordance rates varying from 10% to 38% (21, 30, 34). As a surrogate for molecular subtypes, some de novo stage IV cohorts have reported comparable rates of ER negativity and HER2 positivity, akin to our findings (38–40). However, others have identified rates of more than a third of patients presenting as either ER– or HER2+ (41, 42). A recent study found a small improvement in predictive modeling by combining PREDICT with intrinsic subtypes, especially for patients with ER+ tumors, but it questions the economic burden to justify a broad implementation (43).
Intrinsic molecular subtyping provides clinically meaningful information for diagnostic workup and treatment considerations. Our results indicate that, without intrinsic molecular subtyping, more than 15% of patients are insufficiently classified by IHC alone, and molecular subtypes are a predominant classification for survival. This should serve as compelling evidence for the inclusion of molecular subtyping in the assessment of patients with breast cancer. Further trials are needed to establish how to optimize the evaluation and treatment of patients with either discordance between IHC and molecular subtype or with a low-stage, high-risk subtype.
Methods
Sex as a biological variable.
Only women were included, as female breast cancer accounts for more than 99% of cases of breast cancer (44).
Study population.
The CBCGS prospectively enrolled women aged 18 or older diagnosed with primary invasive breast cancer (T1c, if N0M0; otherwise, any T, N, and M stage) at the Department of Oncology, Copenhagen University Hospital, Rigshospitalet, between April 2014 and December 2021. The diagnostic workup has previously been described, but in short, all tumor tissue from biopsies and surgical specimens was collected and analyzed prospectively (16). Detailed information on diagnosis, genomic profiling, treatment, and follow-up was registered in the clinical Danish Breast Cancer Group (DBCG) database (www.dbcg.dk). Patients were recommended treatment according to national guidelines respecting stage at diagnosis.
Pathology.
Standard histopathological evaluation included tumor size, histological type according to WHO, and grade as defined by Elston and Ellis (45, 46). Resection margins, invasion into skin or deep fascia, lymphovascular invasion, number of axillary lymph nodes identified, and number of metastatic nodes (macro- and micrometastatic and isolated tumor cells) was likewise evaluated. ER was assessed by IHC using a cut-off point of > 1% for ER+ tumors, and scoring of HER2 was performed according to national guidelines (47–49).
Assessment of CIT256 and PAM50 subtype.
Fresh pretreatment or postsurgical breast biopsies (pretreatment in case of neoadjuvant treatment) were collected in RNAlater stabilization solution (Thermo Fisher Scientific), and total RNA was isolated as previously described (50). For the majority of samples, gene expression was measured using RNA microarrays (using Human Genome U133 Plus 2.0 Array; Affymetrix). For a subset of samples, expression was quantified using next-generation sequencing (RNA-Seq) — specifically, paired-end read sequencing (2 × 125 bp) on the Illumina HiSeq2500 platform. The library preparation, data preprocessing, and molecular subtype allocation has previously been described in detail (16, 51, 52). For the CIT256 scheme, 1 of 6 subtypes (Basal-like, mApo, LumA, LumB, LumC, Normal-like) was assigned to each sample by the CIT256 tool using a distance-to-centroid approach relying on expression of 375 probe sets (8, 51). For the PAM50 molecular subtyping scheme, for RNA-Seq samples, log2-transformed normalized expression values were used as input for the original predictor developed by Parker et al. (4). The classifier calculated Spearman’s rank correlation between each sample and each subtype centroid for the 50 genes of interest and assigns the class (LumA, LumB, HER2-enriched, Basal-like, Normal-like) of the most highly correlated centroid to each sample. For microarray normalized expression values, the genefu R package was used for assigning a PAM50 subtype based on the Pearson correlation to the PAM50 centroids.
For CIT256 and PAM50, LumA, LumB, and Normal-like are referred to as “luminal,” and LumC, mApo (CIT256), HER2-enriched (PAM50), and Basal-like are referred as “nonluminal.” For IHC, ER+ and HER2– are luminal and any HER2+ or double-negative breast cancer are “nonluminal.” This was chosen to reflect the clinical application of “luminal breast cancer” and due to lack of patients especially in stage III and IV.
Staging.
Anatomic staging was based on the eighth edition of the AJCC (53). In short, patients with pT1-2, pN0-1, cM0 were stage I–II; patients with pT3-4, pN2-3, cM0 were stage III; and patients with any-T, any-N, cM1 were stage IV (pMBC). Staging on patients allocated to neoadjuvant therapy was based on ultrasound or, if not present, MRI. Thus patients with cT1-2, cN0-1, cM0 were stage I–II and those with cT3-4, cN2-3, cM0 were stage III.
Statistics.
Patient demographics and disease characteristics were described with numbers and percentages for categorical variables and median ± IQR for age. Any difference was examined with a 2-tailed unpaired t test for age and χ2 or Fisher’s exact test for categorical variables, excluding unknowns. Overall survival was defined as time from diagnosis until death of any cause and was estimated using the Kaplan-Meier method. Groups were compared by log-rank test. Patients were censored March 1, 2023. Potential median follow-up was calculated by Schemper and Smiths’ method of reverse Kaplan-Meier (54). A multivariable Cox proportional hazards regression model was applied to assess hazard ratio of death for stage (individual stages), age (continuous), IHC subtype, and CIT subtype combined. Grade and histological subtype were, in a univariable model, found not significant (P > 0.1) and were not included in the final multivariable model. The proportional hazard assumption was tested by Schoenfeld residuals. Interaction between stage and IHC subtype was examined with a likelihood ratio test. A logistic regression model was applied to assess risk of discordance among patients with IHC luminal breast cancer adjusting for age (<60 versus ≥60 years), stage (stage I–III versus IV), malignancy grade (excluding unknowns), and nodal status (N0–N2 versus N3). The logistic regression model was performed including unknowns with similar results. Groups were combined using similar odds ratios (i.e., N0, N1, and N2 combined) to derive the most clinically applicable model. All tests were 2-sided, and a P value of < 0.05 was considered statically significant. All statistical analysis were performed using RStudio.
Study approval.
All participants provided written, informed consent before clinical and biomarker study data were entered in the CBCGS database hosted by DBCG. Since the study did not include any contact with patients nor did it include additional use of biological material, the need to obtain a reconsent from participants for this subanalysis was waived by the Ethical Committee of the Capital Region of Denmark. In compliance with Danish regulations, the CBCGS database was authorized by the Danish Data Protection Agency (2012-58-0004, 30-1504 I-Suite 03845), and the study was approved by the Danish Breast Cancer Group (jr.no. DBCG-2015-14). Furthermore, this register-based study was reported to the Capital Regions Research Overview (P-2020-861), approved by the Capital Regions Chart Data Unit (R-22036280).
Data availability.
All clinical data and molecular subtypes used in this study were obtained from the CBCGS data repository hosted by DBCG (www.dbcg.dk). Raw data have previously been made publicly available: microarray data reposited in GEO (GSE231629 and GSE196723) and RNA-Seq data in Zenodo (10.5281/zenodo.7898803). Data used for generation of tables and supplemental tables are not publicly available, due to institutional restrictions. The data set can be made available to qualified researchers through application to the Danish Breast Cancer Group. Please contact dbcg.rigshospitalet@regionh.dk. Values for all figures and supplemental figures can be found in the Supporting Data Values file.
Author contribution
TB contributed with designing the study, acquiring data, analyzing data, formal statistical analysis, and writing and approving the manuscript. MBJ contributed with analyzing the data, formal statistical analysis, and writing and approving the manuscript. AC contributed with acquiring data, analyzing data, and writing and approving the manuscript. MLT contributed with acquiring data, providing reagents, and writing and approving the manuscript. MAM contributed with acquiring data, and writing and approving the manuscript. ASK contributed with designing the study, analyzing the data, and writing and approving the manuscript. FCN contributed with acquiring the data, and writing and approving the manuscript. BE contributed with designing the study, analyzing the data, and writing and approving the manuscript. MR contributed with designing the study, acquiring data, analyzing the data, providing reagent, and writing and approving the manuscript.
Supplementary Material
Version 1. 04/08/2024
Electronic publication
Footnotes
Conflict of interest: TB has institutional grants from Pfizer, Astra Zeneca, Novartis, Samsung Bioepis, Seattle Genetics, Merck, Eli Lilly, and Daiichi Sankyo. TB was on the advisory board for Novartis and has traveled for Daiichi Sankyo. MJ was on the advisory board for Novartis. ASK has institutional grants from Pfizer, AstraZeneca, Merck, Eli Lilly, Seattle Genetics, Roche, and Novartis and has personal grants from Astra Zeneca (travel and advisory board), MSD (travel), Daiichi Sankyo (advisory board), Novartis (advisory board), Seagen (advisory board), and Gilead (advisory board). BE has institutional grants from:Astra Zeneca, Daiichi Sankyo, Eli Lilly, Merck, Novartis, and Pfizer; was on the advisory board for Eli Lilly and Medac; and has traveled for Daiichi Sankyo and Merck. MR was on the advisory board for Merck; has conducted talks for Merck and GSK; and has an institutional grant from Astra Zeneca.
Copyright: © 2024, Berg et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: JCI Insight. 2024;9(7):e178114.https://doi.org/10.1172/jci.insight.178114.
Contributor Information
Tobias Berg, Email: tobias.berg.01@regionh.dk.
Maj-Britt Jensen, Email: maj-britt.raaby.jensen@regionh.dk.
Alan Celik, Email: acel0013@regionh.dk.
Maj-Lis Talman, Email: mtal0011@regionh.dk.
Maria Anna Misiakou, Email: maria.anna.misaikou@regionh.dk.
Ann Søegaard Knoop, Email: ann.soeegaard.knop@regionh.dk.
Finn Cilius Nielsen, Email: fcn@rh.dk.
Bent Ejlertsen, Email: Bent.Ejlertsen@regionh.dk.
Maria Rossing, Email: caroline.maria.rossing@regionh.dk.
References
- 1.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Sørlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van’t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 4.Parker JS, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paik S, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 6.Gusterson B. Do ‘basal-like’ breast cancers really exist? Nat Rev Cancer. 2008;9(2):128–134. doi: 10.1038/nrc2571. [DOI] [PubMed] [Google Scholar]
- 7.Foulkes WD, et al. Triple-negative breast cancer. N Engl J Med. 2010;363(20):1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 8.Guedj M, et al. A refined molecular taxonomy of breast cancer. Oncogene. 2012;31(9):1196–1206. doi: 10.1038/onc.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caplan L. Delay in breast cancer: implications for stage at diagnosis and survival. Front Public Heal. 2014;2:87. doi: 10.3389/fpubh.2014.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heller DR, et al. Why has breast cancer screening failed to decrease the incidence of de novo stage IV disease? Cancers (Basel) 2019;11(4):500. doi: 10.3390/cancers11040500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seltzer S, et al. The clinicomolecular landscape of de novo versus relapsed stage IV metastatic breast cancer. Exp Mol Pathol. 2020;114:104404. doi: 10.1016/j.yexmp.2020.104404. [DOI] [PubMed] [Google Scholar]
- 12.Haque R, et al. Impact of breast cancer subtypes and treatment on survival: an analysis spanning two decades. Cancer Epidemiol Biomarkers Prev. 2012;21(10):1848–1855. doi: 10.1158/1055-9965.EPI-12-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howlader N, et al. Differences in breast cancer survival by molecular subtypes in the United States. Cancer Epidemiol Biomarkers Prev. 2018;27(6):619–626. doi: 10.1158/1055-9965.EPI-17-0627. [DOI] [PubMed] [Google Scholar]
- 14.Gong Y, et al. Impact of molecular subtypes on metastatic breast cancer patients: a SEER population-based study. Sci Rep. 2017;7:45411. doi: 10.1038/srep45411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barinoff J, et al. Clinicopathological differences between breast cancer in patients with primary metastatic disease and those without: a multicentre study. Eur J Cancer. 2013;49(2):305–311. doi: 10.1016/j.ejca.2012.07.027. [DOI] [PubMed] [Google Scholar]
- 16.Rossing M, et al. Molecular subtyping of breast cancer improves identification of both high and low risk patients. Acta Oncol. 2018;57(1):58–66. doi: 10.1080/0284186X.2017.1398416. [DOI] [PubMed] [Google Scholar]
- 17.Sestak I, et al. Comparison of the performance of 6 prognostic signatures for estrogen receptor-positive breast cancer: a secondary analysis of a randomized clinical trial. JAMA Oncol. 2018;4(4):545–553. doi: 10.1001/jamaoncol.2017.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cardoso F, et al. 70-gene signature as an aid to treatment decisions in early-stage breast cancer. N Engl J Med. 2016;375(8):717–729. doi: 10.1056/NEJMoa1602253. [DOI] [PubMed] [Google Scholar]
- 19.Wuerstlein R, et al. The West German study group breast cancer intrinsic subtype study: a prospective multicenter decision impact study utilizing the Prosigna assay for adjuvant treatment decision-making in estrogen-receptor-positive, HER2-negative early-stage breast cancer. Curr Med Res Opin. 2016;32(7):1217–1224. doi: 10.1185/03007995.2016.1166102. [DOI] [PubMed] [Google Scholar]
- 20.Martín M, et al. Prospective study of the impact of the Prosigna assay on adjuvant clinical decision-making in unselected patients with estrogen receptor positive, human epidermal growth factor receptor negative, node negative early-stage breast cancer. Curr Med Res Opin. 2015;31(6):1129–1137. doi: 10.1185/03007995.2015.1037730. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen TO, et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res. 2010;16(21):5222–5232. doi: 10.1158/1078-0432.CCR-10-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vallon-Christersson J, et al. Cross comparison and prognostic assessment of breast cancer multigene signatures in a large population-based contemporary clinical series. Sci Rep. 2019;9(1):12184. doi: 10.1038/s41598-019-48570-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terán S, et al. Analysis of the association of HER-2 low carcinomas and PAM50 assay in hormone receptor positive early-stage breast cancer. Breast. 2023;71:42–46. doi: 10.1016/j.breast.2023.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Viale G, et al. Immunohistochemical versus molecular (BluePrint and MammaPrint) subtyping of breast carcinoma. Outcome results from the EORTC 10041/BIG 3-04 MINDACT trial. Breast Cancer Res Treat. 2018;167(1):123–131. doi: 10.1007/s10549-017-4509-9. [DOI] [PubMed] [Google Scholar]
- 25.Dowsett M, et al. Comparison of PAM50 risk of recurrence score with oncotype DX and IHC4 for predicting risk of distant recurrence after endocrine therapy. J Clin Oncol. 2013;31(22):2783–2790. doi: 10.1200/JCO.2012.46.1558. [DOI] [PubMed] [Google Scholar]
- 26.Lænkholm AV, et al. PAM50 risk of recurrence score predicts 10-year distant recurrence in a comprehensive danish cohort of postmenopausal women allocated to 5 years of endocrine therapy for hormone receptor-positive early breast cancer. J Clin Oncol. 2018;36(8):735–740. doi: 10.1200/JCO.2017.74.6586. [DOI] [PubMed] [Google Scholar]
- 27.Gnant M, et al. Identifying clinically relevant prognostic subgroups of postmenopausal women with node-positive hormone receptor-positive early-stage breast cancer treated with endocrine therapy: a combined analysis of ABCSG-8 and ATAC using the PAM50 risk of recurrence score and intrinsic subtype. Ann Oncol. 2015;26(8):1685–1691. doi: 10.1093/annonc/mdv215. [DOI] [PubMed] [Google Scholar]
- 28.Ohnstad HO, et al. Prognostic value of PAM50 and risk of recurrence score in patients with early-stage breast cancer with long-term follow-up. Breast Cancer Res. 2017;19(1):120. doi: 10.1186/s13058-017-0911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohara AM, et al. PAM50 for prediction of response to neoadjuvant chemotherapy for ER-positive breast cancer. Breast Cancer Res Treat. 2019;173(3):533–543. doi: 10.1007/s10549-018-5020-7. [DOI] [PubMed] [Google Scholar]
- 30.Kim HK, et al. Discordance of the PAM50 intrinsic subtypes compared with immunohistochemistry-based surrogate in breast cancer patients: potential implication of genomic alterations of discordance. Cancer Res Treat. 2019;51(2):737–747. doi: 10.4143/crt.2018.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reid S, et al. Impact of molecular subtype and race on HR+, HER2- breast cancer survival. Breast Cancer Res Treat. 2021;189(3):845–852. doi: 10.1007/s10549-021-06342-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pu M, et al. Research-based PAM50 signature and long-term breast cancer survival. Breast Cancer Res Treat. 2020;179(1):197–206. doi: 10.1007/s10549-019-05446-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laurberg T, et al. Intrinsic subtypes and benefit from postmastectomy radiotherapy in node-positive premenopausal breast cancer patients who received adjuvant chemotherapy - results from two independent randomized trials. Acta Oncol. 2018;57(1):38–43. doi: 10.1080/0284186X.2017.1401735. [DOI] [PubMed] [Google Scholar]
- 34.Sweeney C, et al. Intrinsic subtypes from PAM50 gene expression assay in a population-based breast cancer cohort: differences by age, race, and tumor characteristics. Cancer Epidemiol Biomarkers Prev. 2014;23(5):714–724. doi: 10.1158/1055-9965.EPI-13-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kensler KH, et al. PAM50 molecular intrinsic subtypes in the nurses’ health study cohorts. Cancer Epidemiol Biomarkers Prev. 2019;28(4):798–806. doi: 10.1158/1055-9965.EPI-18-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tobin NP, et al. Molecular subtype and tumor characteristics of breast cancer metastases as assessed by gene expression significantly influence patient post-relapse survival. Ann Oncol. 2015;26(1):81–88. doi: 10.1093/annonc/mdu498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caan BJ, et al. Intrinsic subtypes from the PAM50 gene expression assay in a population-based breast cancer survivor cohort: prognostication of short- and long-term outcomes. Cancer Epidemiol Biomarkers Prev. 2014;23(5):725–734. doi: 10.1158/1055-9965.EPI-13-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plichta JK, et al. Novel prognostic staging system for patients with de novo metastatic breast cancer. J Clin Oncol. 2023;41(14):2546–2560. doi: 10.1200/JCO.22.02222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deluche E, et al. Contemporary outcomes of metastatic breast cancer among 22,000 women from the multicentre ESME cohort 2008-2016. Eur J Cancer. 2020;129:60–70. doi: 10.1016/j.ejca.2020.01.016. [DOI] [PubMed] [Google Scholar]
- 40.Hölzel D, et al. Survival of de novo stage IV breast cancer patients over three decades. J Cancer Res Clin Oncol. 2017;143(3):509–519. doi: 10.1007/s00432-016-2306-1. [DOI] [PubMed] [Google Scholar]
- 41.Choi SH, et al. Locoregional treatment of the primary tumor in patients with de novo stage IV breast cancer: a radiation oncologist’s perspective. Clin Breast Cancer. 2018;18(2):e167–e178. doi: 10.1016/j.clbc.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 42.Van Uden DJP, et al. Metastatic behavior and overall survival according to breast cancer subtypes in stage IV inflammatory breast cancer. Breast Cancer Res. 2019;21(1):113. doi: 10.1186/s13058-019-1201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chowdhury A, et al. Evaluation and comparison of different breast cancer prognosis scores based on gene expression data. Breast Cancer Res. 2023;25(1):17. doi: 10.1186/s13058-023-01612-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Larønningen S, Arvidsson G, Bray F, Engholm G, Ervik M, Guðmundsdóttir EM, Gulbrandsen J, Hansen HL, Hansen HM, Johannesen TB, Kristensen S, Kristiansen MF, Kønig SM, Lam F, Laversanne M, Miettinen J, Mørch LS, Ólafsdóttir E, Pejicic S, Petterson D, Skog A, Steig BÁ, Tian H, Aagnes B, Storm HH(2023). NORDCAN: Cancer Incidence, Mortality, Prevalence and Survival in the Nordic Countries, Version 9.3 (02.10.2023). Association of the Nordic Cancer Registries. Cancer Registry of Norway. Available from: https://nordcan.iarc.fr/ accessed on 11/03/2024.
- 45. Lakhani SR, et al, eds. WHO Classification of Tumours of the Breast. World Health Organization; 2012. [Google Scholar]
- 46.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19(5):403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 47.Hansen TO, et al. High-density SNP arrays improve detection of HER2 amplification and polyploidy in breast tumors. BMC Cancer. 2015;15(1):35. doi: 10.1186/s12885-015-1035-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolff AC, et al. Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline focused update. J Clin Oncol. 2018;36(20):2105–2122. doi: 10.1200/JCO.2018.77.8738. [DOI] [PubMed] [Google Scholar]
- 49.Allison KH, et al. Estrogen and progesterone receptor testing in breast cancer: ASCO/CAP guideline update. J Clin Oncol. 2020;38(12):1346–1366. doi: 10.1200/JCO.19.02309. [DOI] [PubMed] [Google Scholar]
- 50.Rossing M, et al. Clinical implications of intrinsic molecular subtypes of breast cancer for sentinel node status. Sci Rep. 2021;11(1):2259. doi: 10.1038/s41598-021-81538-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pedersen CB, et al. Using microarray-based subtyping methods for breast cancer in the era of high-throughput RNA sequencing. Mol Oncol. 2018;12(12):2136–2146. doi: 10.1002/1878-0261.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kinalis S, et al. Characterization of basal-like subtype in a Danish consecutive primary breast cancer cohort. Acta Oncol. 2018;57(1):51–57. doi: 10.1080/0284186X.2017.1398837. [DOI] [PubMed] [Google Scholar]
- 53.Amin MB, et al. The eighth edition AJCC Cancer Staging Manual: continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J Clin. 2017;67(2):93–99. doi: 10.3322/caac.21388. [DOI] [PubMed] [Google Scholar]
- 54.Schemper M, Smith TL. A note on quantifying follow-up in studies of failure time. Control Clin Trials. 1996;17(4):343–346. doi: 10.1016/0197-2456(96)00075-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All clinical data and molecular subtypes used in this study were obtained from the CBCGS data repository hosted by DBCG (www.dbcg.dk). Raw data have previously been made publicly available: microarray data reposited in GEO (GSE231629 and GSE196723) and RNA-Seq data in Zenodo (10.5281/zenodo.7898803). Data used for generation of tables and supplemental tables are not publicly available, due to institutional restrictions. The data set can be made available to qualified researchers through application to the Danish Breast Cancer Group. Please contact dbcg.rigshospitalet@regionh.dk. Values for all figures and supplemental figures can be found in the Supporting Data Values file.