Abstract
Deletion analysis of the promoter for the Staphylococcus aureus enterotoxin D determinant indicated that a 52-bp sequence, from −34 to +18, was sufficient for sed promoter function and agr regulation. A consensus −10 Pribnow box sequence, a less conserved −35 sequence, and a TG dinucleotide motif were present. Transcribed sequences (+1 to +18) are essential for promoter activity.
Staphylococcal food poisoning is an intoxication resulting from ingestion of foods contaminated with enterotoxin producing Staphylococcus aureus strains (16). The symptoms include emesis, diarrhea, abdominal cramping, and in severe cases, fever and shock (6, 30, 31). The staphylococcal enterotoxins are a group of secreted proteins that cause emesis when orally administered to primates (6). To date, a number of enterotoxins have been characterized based on their serological reactivities and designated SEA to SEJ, including subtypes SEC1 to SEC3 (5, 8, 24, 29, 32–34).
Though staphylococcal enterotoxins are similar in structure and biological properties (22), they differ with respect to genetic localization, amount of toxin produced, and mechanism of gene regulation. The genes for SEA and SEE are carried on prophage, some of which are defective prophage. SED and SEJ gene determinants are carried on the same penicillinase-type plasmid. The genes for SEB and SEC are chromosomal, but the nature of the genetic elements on which they reside has not been elucidated (7, 18, 19). SEB and SEC are expressed in greater quantities than the other enterotoxins, often on the order of 100 μg/ml of culture supernatant, whereas maximal production of SEA, SED, and SEE is usually less than 10 μg/ml (3). Furthermore, SEA is produced throughout the log phase of growth, while SEB, SEC, and SED are produced in greater quantities during the transition from the exponential to the stationary phases of growth (5). The latter expression pattern is characteristic of many staphylococcal exoprotein virulence factors which are under the control of the accessory gene regulator (agr) two-component regulatory system (25). The transcription of seb, sec, and sed is subject to regulation by the agr system. In agr mutant strains, mRNA steady-state levels were reduced 4-fold for seb, 5.5-fold for sed, and 2 to 3-fold for sec (2, 28).
Information regarding the promoter elements of the staphylococcal enterotoxin genes is very limited. The sea promoter has been identified by means of primer extension analysis in conjunction with deletion mutagenesis. However, detailed characterization of the promoters for the agr-regulated enterotoxin genes has not been reported. Promoter sequences for seb and sed were defined by mapping the transcription start site and by comparison to Escherichia coli promoter consensus −10 and −35 sequences. Mahmood and Khan found that a region upstream of the −35 element, at positions −93 to −58, is required for seb expression (21). Although primer extension studies have been carried out on sed, only a Pribnow box could be identified in the sequence (2). No convincing −35 element was obvious in the sequence analysis. As part of our effort to define the mechanism by which RNAIII regulates sed transcription, this study was undertaken to characterize the promoter and potential regulatory sequences for sed. In this study, the transcription start site was mapped by primer extension. The transcription start site and bases which are critical to promoter function were identified through characterization of site-specific sequence changes in the sed promoter.
Construction of the deletion mutants and promoter analysis.
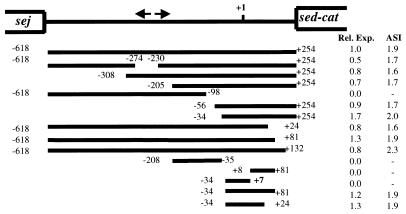
To characterize the promoter sequences for sed, we cloned 1.7 kb of DNA immediately 5′ to the ribosome binding site of sed and introduced a series of deletions into the sequence at either the 5′ or 3′ end by exonuclease III digestion (Erase-A-Base; Promega). All deletions were verified by sequencing, and nucleotide positions were numbered relative to the start site of transcription (+1). The deleted sed promoter region sequences were then fused with a promoterless chloramphenicol acetyltransferase (cat) gene carried by pMH109 (17). The sed promoter activity was then determined by measuring the expression level of cat (Fig. 1). The upstream boundary of the promoter element was defined by promoter active deletions which extended to position −34. The downstream boundary of the promoter element was identified as +24. Deletions up to this position did not appreciably affect sed promoter activity and the agr-regulatory effect, but deletions which extended beyond these endpoint sequences resulted in a loss of promoter function. The insert of plasmid pZS2688, containing sed sequences from −34 to +7, failed to drive measurable cat expression. This suggested that sequences beyond the start site of transcription were required for transcription from the sed promoter. Therefore, the essential promoter sequences appeared to be contained within the sequence between −34 to +24. To confirm this, plasmid pZS2074 containing sed promoter sequences from −34 to +24 was created. When CAT activity was measured from lysates of cells bearing this plasmid, the insert in pZS2704 retained promoter activity and the agr-stimulatory effect comparable to that of the 1.7-kb insert in pZS2605. Therefore, the 58-bp fragment contained in pZS2074 possessed all required promoter elements and the cis sequences responsible for the stimulation of transcription which occurs in agr wild-type hosts.
FIG. 1.
Deletion analysis of the sed promoter. Arrows indicate the inverted repeat sequence located within the intergenic region of sed and sej. The transcription start site of sed is indicated as +1. Lines represent the remaining part of the sed promoter which were fused with a promoterless cat gene in pMH109. Deletions are numbered according to distance from the transcription start site. Promoter activities were analyzed by measuring CAT expression levels in both KSI2054 (Agr+) and ISP546 (Agr−) host strains (33). CAT values were converted to relative expression (Rel. Exp.), which is the value obtained with the deletion mutant divided by the value obtained with the wild-type sed promoter in strain KSI2054. The agr stimulation index (ASI) is the CAT value obtained with KSI2054 divided by the value obtained from ISP546 cells bearing the same plasmid.
Mapping the sed transcription start site.
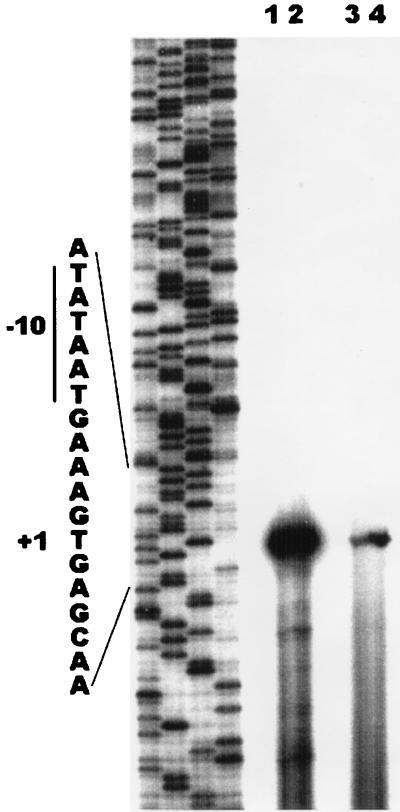
To confirm that the 58-bp promoter fragment accurately reflected the sed promoter activity, the transcription start site was mapped. First, we mapped the transcription start site when the intact upstream sequences were present. RNA was isolated (14) from S. aureus strains KSI2054 (agr+) and ISP546 (agr) harboring plasmid pZS2607 (carrying 1.7 kb of DNA immediately 5′ to the sed ribosome binding site [34]). Primer extension assays (9) revealed a reverse-transcribed DNA fragment at a position corresponding to the T residue located 265 bp upstream of the translation start site (S. Zhang and G. C. Stewart, unpublished data). This is in agreement with the results of Bayles and Iandolo (2). To map the transcription start site from the 58-bp promoter, RNA was isolated from KSI2054 and IPS546 harboring pZS2704. The same start site of transcription was identified with the 58-bp promoter fragment as was seen with the intact upstream sequence (Fig. 2). The signal is stronger with the RNA isolated from the agr+ host (compare lane 4 to lane 3), consistent with the results obtained with the CAT assays. The primer extension assay was also carried out with RNA isolated from cells bearing the lac-sed hybrid promoter. This promoter has the −35 and spacer sequences from the lac promoter and the Pribnow box and downstream sequences from the sed promoter. This promoter is stronger than the wild-type sed promoter (Fig. 3, pZS2743), but the start site of transcription is unchanged (Fig. 2, lanes 1 and 2).
FIG. 2.
Primer extension mapping of the transcription start site. RNA was isolated from S. aureus ISP546 (lanes 1 and 3) and KSI2054 (lanes 2 and 4) harboring plasmid pZS2704 (lanes 3 and 4) or pZS2743 (lanes 1 and 2) at an optical density at 540 nm of 1.2. Reverse transcription was carried out using the primer cat3, which anneals to a site near the 5′ end of the cat gene. The samples were loaded into adjacent lanes of a sequencing gel, and the lanes between the two sets were left blank. The sequence ladder was generated using the same primer and pZS2704 as the template. The transcription start site is indicated as +1.
FIG. 3.
(A) The promoter sequences of sed and lac are presented in the top and bottom lines, respectively. The −35, −10, and +1 sequences of the sed promoter are underlined. For the hybrid promoters, dashed lines indicate sed sequences and the indicated bases signify replacement by lac promoter sequences. (B) Activity of mutant forms of the sed promoter. Dashed lines indicate sed sequences, bases indicate base substitutions, and blank spaces indicate deleted bases. CAT assays were conducted with the wild-type (wt) strain KSI2054 harboring the corresponding plasmids. CAT values are expressed as nanomoles of chloramphenicol acetylated per minute per milligram (dry weight) of cells. Average values from three independent determinations and standard deviations are given.
Five bases upstream of the +1 position in the sed promoter there is a sequence, TATAAT, that matches the consensus Pribnow box of bacterial promoters. A less conserved −35 (ATGAAA) is located 17-bp upstream from the −10 element. The sed promoter also contains a TG dinucleotide at position −14/−13. This dinucleotide is an important element of procaryotic promoters with poor −35 elements (1, 27).
Promoter distal sequences are required for sed promoter function.
The 58-bp fragment, encompassing nucleotide positions −34 to +24, displayed comparable promoter activity to that of the undeleted sed promoter. However, the fragment containing sequences from −34 to +7 failed to drive cat gene expression. Sequences downstream of +1 appear to be required for sed promoter function. To test this possibility, we created a series of sed-lac hybrid promoters by fusing the 5′ portion of the sed promoter with the 3′ portion of the S. aureus lactose operon (lac) promoter sequences (26). This approach permitted a study of the role of the downstream sequence without interrupting the distance between the transcription and the translation start sites. Then the cat expression levels of these hybrid constructs were analyzed (Fig. 3A). The results showed that when the sed sequences from −12 to +24 were replaced with lac sequences (pZS2746, sed/lac), the promoter function was abolished. This hybrid promoter still had the −35 of sed, the −10 sequences are the same in the two promoters, and the spacer region was very similar to that of the sed (16 identical bases out of 17). Thus, the lack of promoter activity was most likely the result of the substitution of lac for sed sequences distal to the +1 position. When the sed sequence was extended to +7 and +14 to generate pZS2771 (M11) and pZS2781 (M12), these hybrid promoters had only barely detectable activity. To determine the 3′ boundary of the sed promoter, additional hybrid promoters (pZS2832, pZS2833, pZS2834, and pZS2835) were generated by replacing the last eight, six, four, and two bases of the 58-bp fragment with the corresponding lac promoter sequences. The CAT assay results indicated that the eight-base replacement (+17 to +24) abolished the sed promoter function, but a six-base replacement (+19 to +24) resulted in a 2.5-fold increase in promoter activity, relative to that obtained with the wild-type 58-bp sequence. The CAT values for pZS2834 (four-base replacement, +21 to +24) and pZS2835 (two-base replacement, +23 to +24) had essentially no effect on sed promoter activity. To further define the 3′ terminus of the sed promoter, plasmid pZS2852 was created by replacing the last seven bases of the 58-bp sed promoter (T+18 to C+24) with lac sequences. This hybrid promoter was active, although it resulted in a CAT expression level lower than obtained with the 58-bp sed promoter. Thus, the downstream boundary of the sed promoter is nucleotide position +17.
To confirm the activity of the −34 to +18 fragment, plasmid pZS2853 was created by PCR amplification of the −34 to +18 DNA fragment, which was then cloned into pMH109 to create a promoter which differed from that of pZS2852 in that it lacked the +19 to +24 lac sequences. The CAT assay results showed that the promoter strength of this 52-bp fragment was comparable to that of pZS2833 (six-base replacement) and was about 2.3-fold higher than that of the 58-bp sed promoter (Fig. 3B).
Contribution of the −35 sequences to sed promoter function.
To determine the role of the putative −35 sequence in sed promoter function, the 5′ end of the 58-bp fragment was either deleted or mutated. The promoter activity of the mutants were measured (Fig. 3B). Deletion of the first 5, 10, and 15 bases (pZS2782, pZS2783, and pZS2784) destroyed the promoter function completely. Two mutant promoters (pZS2836 and pZS2837) with the putative −35 element altered to give a consensus sequence (TTGACA) were created by oligonucleotide-mediated site-directed mutagenesis (34). The promoter of pZS2836 (M25) consists of bases −34 to +24, whereas pZS2837 (M25′) includes only bases −34 to +7. The promoter strengths of both pZS2836 and pZS2837 were dramatically increased. The cat expression level was increased almost fivefold for pZS2836 relative to that exhibited by the 58-bp wild-type sequence. The level of expression was 4.7-fold higher for pZS2837. More significantly, the activity exhibited by the pZS2837 insert indicated that the requirement for the +8 to +18 sequence to produce an active promoter was eliminated with the introduction of the improved −35 element sequence. However, introduction of the consensus −35 sequences caused instability of the plasmids. When pZS2836 and pZS2837 were transduced into S. aureus KSI2054, the majority of the transductants did not produce chloramphenicol resistance. Subsequent sequence analysis indicated that deletions occurred either within the promoter region or the cat structure gene. In addition, a strain bearing a point mutation (A−29 to T; pZS2719) within the −35 sequence was isolated. This mutation resulted in a 1.7-fold reduction in promoter activity.
Effect of point mutations on sed promoter function.
To determine the role of bases outside of the −35 and −10 elements, single or double mutations were introduced into the sed promoter region (Fig. 3B). Mutations within the spacer region, A−22 to C and A−21 to G (pZS2720 and pZS2721), led to moderate reductions in cat expression levels, whereas a G−27-to-T transversion mutation (pZS2773) had no effect on sed promoter activity. Base substitutions near the −10 sequence had profound effects on sed promoter function. When G−13 was changed to T (pZS2772), the cat expression level was reduced to barely detectable level. Mutations G−5 to T and C (pZS2874 and pZS2875) and A−4 to T (pZS2876) led to a three- to fourfold reduction in cat expression level. Double mutations A−4A−3 to TT (pZS2877) abolished sed promoter activity. Base changes adjacent to the transcriptional start site also influenced the promoter strength. When G−1 or G+2 was changed to T (pZS2774 or pZS2775), an increase in cat expression resulted. Base substitutions within the downstream region, G+8 to T and C (pZS2750 and pZS2751), increased expression about twofold.
The rate of transcription from constitutive promoters depends on the sequences of the −35 and −10 promoter elements (12). However, bases outside of these core promoter elements are also important for promoter function. Footprint analyses have revealed protected regions as far upstream as −70 and downstream to +20 in E. coli promoters (10, 11, 20). An AT-rich upstream region can enhance promoter activity by facilitating promoter bending and wrapping around the RNA polymerase. Mahmood and Khan demonstrated that the upstream region between −93 and −58 was required for seb expression but that sequences distal to +1 were not required (21). Bases upstream from the −35 element are not required for sed expression, but sequences downstream of +1 are essential for sed promoter function. Thus, the promoters for these two staphylococcal enterotoxin genes are fundamentally different. Hybrid promoter analysis revealed that the region downstream from +1 to +18 was required for sed promoter activity, and a base substitution (G+8 to T or C) within this region also affected the promoter strength. Studies have shown that the downstream regions of certain E. coli promoters can influence the transcription rate by affecting promoter clearance (13). During this process, sigma factor is released from RNA polymerase holoenzyme which allows the elongation steps to proceed. Expression of sed does not occur when the +1 distal sequences are derived from the staphylococcal lac promoter but is restored when sequences from the seb promoter are used (Zhang and Stewart, unpublished data). This is interesting because these sequences are not required for expression from the seb promoter.
When base substitutions were introduced into the sed promoter to create a consensus −35 element, the cat expression level was increased approximately fivefold and the downstream region requirement was eliminated. Based on these data, the role of the downstream region may be to compensate for the poor −35 sequence. This could be accomplished by DNA bending to enable a better interaction with RNA polymerase, rather than just playing a rate-limiting role in the late steps of transcription initiation.
A required T−14G−13 dinucleotide was found one base upstream of the Pribnow box sequence. In studies of E. coli promoters, it has been proposed that RNA polymerase makes an additional contact with the TG dinucleotide during transcription initiation, which lowers the thermal energy required for the strand separation (1, 27). Compilation analysis of Bacillus subtilis promoter sequences also indicated that this dinucleotide pair is conserved in extended −10 element promoter sequences from this organism (15). The TG motif was also shown to be a conserved feature in 26% of Lactobacillus promoters and is an important determinant of promoter strength (23).
In addition to the TG dinucleotide, bases immediately downstream of the Pribnow box also play a vital role in sed promoter function. Base substitutions within this region (G−5 to T or C, A−4 to T, A−3A−4 to TT) either abolished the promoter activity or reduced it dramatically. These findings suggest that not only the T−14G−13 dinucleotide but those bases downstream from the −10 element are involved in transcription initiation, possibly by facilitating strand separation and open complex formation.
Acknowledgments
This work was supported by Public Health Service grant AI45778 from the National Institutes of Health.
REFERENCES
- 1.Barne K A, Bown J A, Busby S J W, Minchin S D. Region 2.5 of the Escherichia coli RNA polymerase ς70 subunit is responsible for the recognition of the “extended −10” motif at promoters. EMBO J. 1997;16:4034–4040. doi: 10.1093/emboj/16.13.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayles K W, Iandolo J J. Genetic and molecular analysis of the gene encoding staphylococcal enterotoxin D. J Bacteriol. 1989;171:4799–4806. doi: 10.1128/jb.171.9.4799-4806.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergdoll M S. Staphylococcal intoxications. In: Riemann H, Bryan F L, editors. Foodborne infections and intoxications. New York, N.Y: Academic Press; 1979. pp. 443–493. [Google Scholar]
- 4.Bergdoll M S. Enterotoxins. In: Easmon C S F, Adlam C, editors. Staphylococcal and streptococcal infections. New York, N.Y: Academic Press; 1983. pp. 559–598. [Google Scholar]
- 5.Bergdoll M S, Czop J K, Gould S S. Enterotoxin synthesis by the staphylococci. Ann N Y Acad Sci. 1974;236:307–316. doi: 10.1111/j.1749-6632.1974.tb41500.x. [DOI] [PubMed] [Google Scholar]
- 6.Betley M J, Borst D W, Regassa L B. Staphylococcal enterotoxins, toxic shock syndrome toxin, and streptococcal pyrogenic exotoxins: a comparative study of their molecular biology. Chem Immunol. 1992;55:1–35. [PubMed] [Google Scholar]
- 7.Betley M J, Mekalanos J J. Staphylococcal enterotoxin A is encoded by a phage. Science. 1989;229:185–187. doi: 10.1126/science.3160112. [DOI] [PubMed] [Google Scholar]
- 8.Bohach, G. A., C. V. Stauffacher, D. H. Ohlendorf, Y. I. Chi, G. M. Vath, and P. M. Schlievert. The staphylococcal and streptococcal pyrogenic toxin family. Adv. Exp. Med. Biol. 391:131–154. [DOI] [PubMed]
- 9.Borst D W, Betley M J. Promoter analysis of the staphylococcal enterotoxin A gene. J Biol Chem. 1994;269:1883–1888. [PubMed] [Google Scholar]
- 10.Busby S D, Spassky A, Chan B. RNA polymerase makes important contacts upstream from base pair −49 at the Escherichia coli galactose operon p1 promoter. Gene. 1987;53:145–152. doi: 10.1016/0378-1119(87)90002-3. [DOI] [PubMed] [Google Scholar]
- 11.Coulombe B, Burton Z F. DNA bending and wrapping around RNA polymerase: a “revolutionary” model describing transcriptional mechanisms. Microbiol Mol Biol Rev. 1999;63:457–478. doi: 10.1128/mmbr.63.2.457-478.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craig M L, Suh W C, Record M T., Jr HO and DNase I probing of E ς70 RNA polymerase-λ PR promoter open complex: Mg2+ binding and its structural consequences at the transcription start site. Biochemistry. 1995;34:15624–15632. doi: 10.1021/bi00048a004. [DOI] [PubMed] [Google Scholar]
- 13.Ellinger T, Behnke D, Bujard H, Gralla J D. Stalling of Escherichia coli RNA polymerase in the +6 to +12 region in vivo is associated with tight binding to consensus promoter elements. J Mol Biol. 1994;239:455–465. doi: 10.1006/jmbi.1994.1388. [DOI] [PubMed] [Google Scholar]
- 14.Hart M E, Smeltzer M S, Iandolo J J. The extracellular protein regulator (xpr) affects exoprotein and agr mRNA levels in Staphylococcus aureus. J Bacteriol. 1993;24:7875–7879. doi: 10.1128/jb.175.24.7875-7879.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helmann J D. Compilation and analysis of Bacillus subtilis ςA-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 1995;23:2351–2360. doi: 10.1093/nar/23.13.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmberg S, Blake P A. Staphylococcal food poisoning in the United States. JAMA. 1984;251:487–489. [PubMed] [Google Scholar]
- 17.Hudson M C, Stewart G C. Differential utilization of Staphylococcus aureus promoter sequences by Escherichia coli and Bacillus subtilis. Gene. 1986;48:93–100. doi: 10.1016/0378-1119(86)90355-0. [DOI] [PubMed] [Google Scholar]
- 18.Iandolo J J. Genetic analysis of extracellular toxins of Staphylococcus aureus. Annu Rev Microbiol. 1989;43:375–402. doi: 10.1146/annurev.mi.43.100189.002111. [DOI] [PubMed] [Google Scholar]
- 19.Johns M B, Jr, Khan S A. Staphylococcal enterotoxin B gene is associated with a discrete genetic element. J Bacteriol. 1998;170:4033–4039. doi: 10.1128/jb.170.9.4033-4039.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kammerer W, Deuschle U, Gentz R, Bujard H. Functional dissection of Escherichia coli promoters: information in the transcribed region is involved in late steps of the overall process. EMBO J. 1986;5:2995–3000. doi: 10.1002/j.1460-2075.1986.tb04597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahmood R, Khan S A. Role of the upstream region in the expression of the staphylococcal enterotoxin B gene. J Biol Chem. 1990;265:4652–4656. [PubMed] [Google Scholar]
- 22.Marrack P, Kappler J. The staphylococcal enterotoxins and their relatives. Science. 1990;248:705–711. doi: 10.1126/science.2185544. [DOI] [PubMed] [Google Scholar]
- 23.McCracken A, Timms P. Efficiency of transcription from promoter sequence variants in Lactobacillus is both strain and context dependent. J Bacteriol. 1999;181:6569–6572. doi: 10.1128/jb.181.20.6569-6572.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munson S H, Tremaine M T, Betley M J, Welch R A. Identification and characterization of staphylococcal enterotoxin type G and I from Staphylococcus aureus. Infect Immun. 1998;66:3337–3348. doi: 10.1128/iai.66.7.3337-3348.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novick R P, Ross H F, Projan S J, Kornblum J, Kreiswirth B, Moghazeh S L. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oskouian B, Stewart G C. Repression and catabolite repression of the lactose operon of Staphylococcus aureus. J Bacteriol. 1990;172:3804–3812. doi: 10.1128/jb.172.7.3804-3812.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ponnambalam S, Webster C, Bingham A, Busby S. Transcription initiation at the Escherichia coli galactose operon promoters in the absence of the normal −35 region sequences. J Biol Chem. 1986;261:16043–16048. [PubMed] [Google Scholar]
- 28.Regassa L B, Couch J L, Betley M J. Steady-state staphylococcal enterotoxin type C mRNA is affected by a product of the accessory gene regulator (agr) and by glucose. Infect Immun. 1991;59:955–962. doi: 10.1128/iai.59.3.955-962.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren K, Bannan J D, Pancholi V V, Cheung A L, Robbins J C, Fischetti V A, Zabriskie J B. Characterization and biological properties of a new staphylococcal enterotoxin. J Exp Med. 1994;180:1675–1683. doi: 10.1084/jem.180.5.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rizkallah M F, Tolaymat A, Martinez J S, Schlievert P M, Ayoub E M. Toxic shock syndrome caused by a strain of Staphylococcus aureus that produces enterotoxin C but not toxic shock syndrome toxin-1. Am J Dis Child. 1989;143:848–849. doi: 10.1001/archpedi.1989.02150190098031. [DOI] [PubMed] [Google Scholar]
- 31.Schlievert P M. Staphylococcal enterotoxin B and toxic shock syndrome toxin-1 are significantly associated with nonmenstrual TSS. Lancet. 1986;i:1149–1150. doi: 10.1016/s0140-6736(86)91859-3. [DOI] [PubMed] [Google Scholar]
- 32.Su Y-C, Wang A C L. Identification and purification of a new staphylococcal enterotoxin, H. Appl Environ Microbiol. 1995;61:1438–1443. doi: 10.1128/aem.61.4.1438-1443.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van den Bussche R A, Lyon J D, Bohach G A. Molecular evolution of the staphylococcal pyrogenic toxin gene family. Mol Phylogenet Evol. 1993;2:281–292. doi: 10.1006/mpev.1993.1027. [DOI] [PubMed] [Google Scholar]
- 34.Zhang S, Iandolo J J, Stewart G C. The enterotoxin D plasmid of Staphylococcus aureus encodes a second enterotoxin determinant (sej) FEMS Microbiol Lett. 1998;168:227–233. doi: 10.1111/j.1574-6968.1998.tb13278.x. [DOI] [PubMed] [Google Scholar]