Abstract
Computer analysis of the Bacillus subtilis genome sequence revealed a gene with no previously attributed function, yhaG, specifying a transcript containing a presumptive binding site for the tryptophan-activated regulatory protein, TRAP. The presumptive TRAP binding site overlaps the yhaG Shine-Dalgarno sequence and translation initiation region. TRAP was shown to regulate expression of yhaG translationally. Production of the yhaG transcript in vivo was found to compete for the binding of TRAP to other known TRAP binding sites. YhaG is likely to be a transmembrane protein involved in tryptophan transport.
Transcription of the trp operon of Bacillus subtilis and some related species is regulated by transcription attenuation. A tryptophan-activated RNA-binding regulatory protein, TRAP, binds to the leader segment of the trp operon transcript and promotes formation of an RNA terminator structure, causing transcription termination before RNA polymerase can reach the first structural gene. TRAP recognizes a series of (G/U)AG repeats in the antiterminator segment of the trp leader transcript; the antiterminator precedes and partially overlaps the terminator. TRAP binding to the antiterminator prevents its formation, thereby favoring terminator formation (3, 4, 12, 15). The structures of TRAP and the TRAP-RNA complex have been described previously (1, 2). These structures define how the TRAP protein wraps the leader transcript around its surface and prevents the formation of the antiterminator. TRAP also binds to a (G/U)AG repeat region overlapping the Shine-Dalgarno sequence of the trpG segment of the folate transcript; TRAP binding regulates translation of trpG (4, 6, 17).
In an effort to identify all the potential TRAP binding sites in the B. subtilis genome, we designed an appropriate computer program and searched for (G/U)AG repeats (14). Four (G/U)AG repeat regions were identified, two in addition to the (G/U)AG repeats mentioned above, in the trp operon and trpG. One of these TRAP binding sites overlaps the Shine-Dalgarno sequence of a gene of unknown function, ycbK, and presumably is used to regulate translation of the ycbK coding region (14). The second (G/U)AG repeat region overlaps the yhaG Shine-Dalgarno sequence and presumably regulates initiation of synthesis of its encoded polypeptide. The experiments described in this paper were performed to determine whether yhaG expression was regulated by the TRAP protein and to analyze the function of its encoded polypeptide.
Features of the yhaG gene.
To determine the location of the promoter responsible for yhaG expression, primer extension analysis was performed to identify the start point of transcription (data not shown). A site was identified which is preceded by sequences which exhibit high similarity to the consensus sequence of the −10 and −35 regions of vegetative B. subtilis promoters (Fig. 1A). There is a short leader sequence of 82 bases upstream of an open reading frame encoding a predicted product containing 172 amino acid residues. The open reading frame is preceded by a sequence closely resembling a Shine-Dalgarno sequence. A sequence encoding a putative transcription terminator is present downstream of the yhaG coding region.
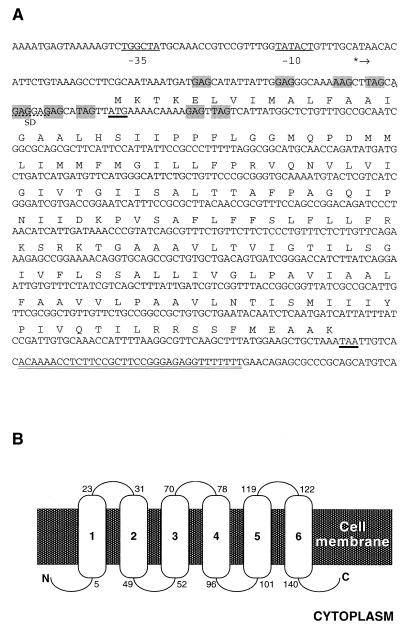
FIG. 1.
(A) Features of the yhaG region of the B. subtilis chromosome. Promoter regions and translation start and stop codons are underlined. The start point of transcription is marked by an asterisk and an arrow. The Shine-Dalgarno sequence (SD) is indicated by a dashed underline. A sequence corresponding to a Rho-independent transcription terminator is indicated by a double underline. Trinucleotide repeats constituting a putative TRAP binding site are shaded. Nucleotide coordinates shown are from 1074054 to 1074773 according to the B. subtilis genome database (8). (B) Predicted topological model of the YhaG protein, generated by the TopPred II computer program using a window size of 21 (5). Oval structures represent alpha-helical transmembrane spans. Predicted amino acid residues at the membrane interface are indicated.
Nine (G/U/A)AG trinucleotide repeats are positioned in the leader transcript encompassing the Shine-Dalgarno sequence and the beginning of the yhaG coding region (Fig. 1A). These consist of five GAG motifs, three UAG motifs, and a single AAG triplet. To achieve effective TRAP binding the optimal spacing between repeats is two nucleotides (3). The identified sequence contains five consecutive triplets separated by exactly two nucleotides. The greatest distance between any two trinucleotide motifs in the yhaG sequence is 14 nucleotides. In comparison, the TRAP binding sites of both the trp operon and trpG also have no more than a stretch of four consecutive repeats separated by only two nucleotides (4, 17). The largest interval between triplet sequences in the trp operon leader transcript is three nucleotides, and there are as many as eight nucleotides separating triplets in trpG. The position of the putative TRAP binding site overlapping the beginning of the yhaG coding region is in a position similar to that found in trpG. The five optimally spaced triplets in the yhaG sequence overlap the Shine-Dalgarno sequence. Previous results have demonstrated that TRAP can bind to transcripts containing only six GAG or UAG repeats (3).
Regulation of yhaG expression.
To determine whether expression of yhaG is regulated by TRAP in the presence of tryptophan, strains that contained a yhaG′-′lacZ translational fusion were constructed. A 352-bp fragment encompassing the yhaG promoter, leader region, putative TRAP binding site, and the beginning of the yhaG coding region was cloned into the integration vector ptrpBG1-PLK (10, 16), to produce plasmid pJPS648. The fusion was then integrated into the amyE locus of the chromosome. Strains containing this integrated fusion, in different genetic backgrounds that influence TRAP production, were grown in the presence and absence of tryptophan and assayed for β-galactosidase activity (11). In an mtrB+ strain, yhaG′-′lacZ expression was significantly reduced by the presence of tryptophan in the growth medium (Table 1; rows 1 and 2). Deleting mtrB, the structural gene for TRAP, resulted in elevated expression of the yhaG′-′lacZ translational fusion, regardless of the presence of added tryptophan. These findings establish that yhaG expression is regulated in response to tryptophan and that the TRAP protein is required for this regulation. Considering the location of the TRAP binding site in the yhaG transcript (Fig. 1A), it is likely that yhaG is translationally regulated by tryptophan-activated TRAP.
TABLE 1.
Regulation of yhaG expression and effect of multicopy expression of the yhaG leader region on trpE and trpG gene expression
| Fusion (integrated in amyE locus) | Genetic background | Plasmid | β-Galactosidase sp act of strain grown under conditiona
|
|
|---|---|---|---|---|
| −Tryptophan | + Tryptophan | |||
| yhaG′-′lacZ | mtrB+ | None | 20 | 1.1 |
| yhaG′-′lacZ | ΔmtrB | None | 2,270 | 2,200 |
| trpE′-′lacZ | mtrB+ | None | 13 | 0.1 |
| trpE′-′lacZ | mtrB+ | pHY300PLKb | 11 | 0.3 |
| trpE′-′lacZ | mtrB+ | pJPS647c | 248 | 246 |
| trpE′-′lacZ | mtrB264 | None | 215 | 187 |
| trpG′-′lacZ | mtrB+ | None | 2.3 | 0.4 |
| trpG′-′lacZ | mtrB+ | pHY300PLK | 1.9 | 0.4 |
| trpG′-′lacZ | mtrB+ | pJPS647 | 11 | 9.7 |
| trpG′-′lacZ | mtrB264 | None | 13 | 12 |
Strains were grown overnight in Vogel-Bonner medium, subcultured in the same medium with or without 50 μg of l-tryptophan per ml, and grown for three to four generations. Assays were performed in duplicate on three separate occasions. Values are presented in Miller units.
Parental multicopy shuttle vector.
pHY300PLK containing a 352-bp yhaG promoter-leader fragment.
In order to ascertain if tryptophan affected transcriptional regulation of the gene, dot blot analyses were carried out. RNA was extracted using the cold phenol method described by Putzer et al. (13), from a wild-type strain grown in Vogel-Bonner minimal medium in the presence or absence of 50 μg of tryptophan per ml. Serial twofold dilutions of RNA samples were applied to a nylon membrane followed by hybridization with a probe corresponding to the first 278 nucleotides of the transcript (positions 1074107 to 1074384). The intensity of each spot was quantitated using the public domain NIH Image program. For each condition tested, the average spot intensity (per microgram of RNA) was calculated using a subset of the serially diluted samples which exhibited proportionality. In arbitrary units produced by the program, it was found that, in the absence of tryptophan, the level of yhaG transcript produced is 12,300 U per μg of RNA. In the presence of tryptophan, there were 11,200 U of yhaG transcript per μg of RNA. The presence of tryptophan appears to make no discernible difference in the level of yhaG transcription. Therefore, tryptophan regulation of yhaG expression appears to be solely translational.
The effect of overexpression of yhaG on trp operon and trpG expression.
If the yhaG transcript contains a TRAP binding site, then overproduction of this transcript should result in increased expression of the trp operon and of trpG of the folate operon. It was shown previously that overexpression of the TRAP binding site of the trp operon leader region results in increased expression of the trp operon (9). The same 352-bp fragment used to construct the yhaG′-′lacZ fusion was cloned into the multicopy shuttle vector pHY300PLK (7). The resulting plasmid, pJPS647, was introduced into strains which contained integrated trpE′-′lacZ and trpG′-′lacZ reporters. It is evident that both trpE′-′lacZ expression and trpG′-′lacZ expression were increased appreciably by the presence of pJPS647 (Table 1, rows 3 to 10). Addition of tryptophan had no effect on expression of either reporter. The presence of the parental plasmid pHY300PLK alone had no effect. These findings demonstrate that the presence of a multicopy plasmid containing a fragment from yhaG results in considerable relief of TRAP regulation. This presumably occurs because the yhaG transcripts that are produced from the plasmid bind to and remove some of the available TRAP.
Analysis of the likely function of the YhaG protein.
On the basis of hydropathy profiles, it appears that the product of the yhaG gene is a polytopic protein with intervening hydrophobic and hydrophilic regions. We suggest that the YhaG protein may be a membrane-embedded protein containing six or possibly seven transmembrane spans. One possible topological model predicted by the TopPred II computer program (5) is presented in Fig. 1B. We therefore considered the possibility that YhaG is a tryptophan transport protein or an aromatic amino acid transporter. The YhaG protein is not homologous to known aromatic amino acid transporters of Escherichia coli or other organisms.
To examine the role of YhaG, a segment of the chromosome encompassing the yhaG promoter and part of the coding region was deleted and replaced by a spectinomycin resistance determinant, to produce a ΔyhaG strain. This was performed by deletion of a segment of the yhaG sequence present on a plasmid vector and insertion of a spectinomycin resistance determinant in its place. Linearized plasmid DNA was then used to transform B. subtilis to spectinomycin resistance. Integration into the yhaG locus on the chromosome occurs by a homologous recombination event taking place in each of the flanking regions surrounding the spectinomycin resistance determinant. Formation of the desired construct was confirmed by performing PCR analysis on chromosomal DNA.
Strains lacking yhaG are not auxotrophic, nor do they appear to have a metabolic defect; therefore, it is unlikely that yhaG encodes a protein performing an essential function. A possible role for YhaG in tryptophan or aromatic amino acid transport was examined by comparing the sensitivity of strains with and without yhaG to growth-inhibiting levels of analogs of the various aromatic amino acids.
Filter disks separately containing 5 or 25 μg of 5-fluorotryptophan, 4-methyltryptophan, 5-methyltryptophan, 7-azatryptophan, tryptamine, 5-methyltryptamine, tryptophanol, 5-fluorotyrosine, p-aminophenylalanine, β-thienylalanine, or p-fluorophenylalanine were applied to plates seeded with wild-type or ΔyhaG strains of B. subtilis. Zones of growth inhibition were measured after 24 h. A marked difference in the level of sensitivity between the two strains was produced by 5-fluorotryptophan. The zone of growth inhibition of the wild-type strain was much greater than that of the ΔyhaG strain in the presence of this analog. There was not a large difference in the growth of the two strains in response to the other analogs tested. Streaks of the two strains on plates containing various concentrations of 5-fluorotryptophan indicated that the ΔyhaG strain was able to grow on concentrations of the analog some 40-fold higher than the level that inhibited the wild-type strain. 5-Fluorotryptophan is a potent tryptophan analog as it is capable of being charged onto tRNATrp. This may account for the observation that 5-fluorotryptophan was the only analog to produce a difference in the level of sensitivity between the wild-type and ΔyhaG strains. These results imply that yhaG is concerned with tryptophan transport, consistent with the predicted transmembrane structure of its product, and with its regulation by tryptophan-activated TRAP. Preliminary assays measuring the transport of radiolabeled tryptophan also suggest that the ΔyhaG strain has an impaired ability to transport tryptophan (data not shown).
Conclusions.
A gene of B. subtilis of previously unknown function specifies a transcript that has a binding site for tryptophan-activated TRAP. Overproduction of the yhaG transcript increases expression of the trp operon and trpG, indicating that this transcript can sequester TRAP in vivo. We have assigned a stretch of nine (G/U/A)AG trinucleotide repeats as a putative TRAP binding site, but further experiments are necessary to ascertain the exact contacts made between TRAP and the yhaG transcript. The predicted TRAP binding site in the yhaG transcript overlaps the Shine-Dalgarno sequence of the yhaG coding region. TRAP was shown to regulate yhaG expression translationally, presumably by binding at this Shine-Dalgarno sequence. TRAP does not regulate transcription of yhaG. The predicted structure of the YhaG protein is that of a transmembrane protein, raising the possibility that YhaG is a transport protein. Amino acid analog resistance-sensitivity tests and labeled tryptophan uptake analyses suggest that the YhaG protein transports tryptophan. Additional experiments are required to unambiguously confirm the function of this protein.
Acknowledgments
We thank P. Gollnick and P. Babitzke for critical reading of the manuscript.
This study was supported by National Institutes of Health grant GM09738. J.P.S. was supported by a Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation Fellowship, DRG-1315.
REFERENCES
- 1.Antson A A, Dodson E J, Dodson G, Greaves R B, Chen X, Gollnick P. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature. 1999;401:235–242. doi: 10.1038/45730. [DOI] [PubMed] [Google Scholar]
- 2.Antson A A, Otridge J, Brzozowski A M, Dodson E J, Dodson G G, Wilson K S, Smith T M, Yang M, Kurecki T, Gollnick P. The structure of trp RNA-binding attenuation protein. Nature. 1995;374:693–700. doi: 10.1038/374693a0. [DOI] [PubMed] [Google Scholar]
- 3.Babitzke P, Bear D G, Yanofsky C. TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a toroid-shaped molecule that binds transcripts containing GAG or UAG repeats separated by two nucleotides. Proc Natl Acad Sci USA. 1995;92:7916–7920. doi: 10.1073/pnas.92.17.7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babitzke P, Stults J T, Shire S J, Yanofsky C. TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a multisubunit complex that appears to recognize G/UAG repeats in the trpEDCFBA and trpG transcripts. J Biol Chem. 1994;269:16597–16604. [PubMed] [Google Scholar]
- 5.Claros M G, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 6.Du H, Tarpey R, Babitzke P. The trp RNA-binding attenuation protein regulates TrpG synthesis by binding to the trpG ribosome binding site of Bacillus subtilis. J Bacteriol. 1997;179:2582–2586. doi: 10.1128/jb.179.8.2582-2586.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishiwa H, Shibahara H. New shuttle vectors for Escherichia coli and Bacillus subtilis. II. Plasmid pHY300PLK, a multipurpose cloning vector with a polylinker derived from pHY460. Jpn J Genet. 1985;60:235–243. [Google Scholar]
- 8.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 9.Kuroda M I, Henner D, Yanofsky C. cis-Acting sites in the transcript of the Bacillus subtilis trp operon regulate expression of the operon. J Bacteriol. 1988;170:3080–3088. doi: 10.1128/jb.170.7.3080-3088.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merino E, Babitzke P, Yanofsky C. trp RNA-binding attenuation protein (TRAP)-trp leader RNA interactions mediate translational as well as transcriptional regulation of the Bacillus subtilis trp operon. J Bacteriol. 1995;177:6362–6370. doi: 10.1128/jb.177.22.6362-6370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 12.Otridge J, Gollnick P. MtrB from Bacillus subtilis binds specifically to trp leader RNA in a tryptophan-dependent manner. Proc Natl Acad Sci USA. 1993;90:128–132. doi: 10.1073/pnas.90.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Putzer H, Gendron N, Grunberg-Manago M. Co-ordinate expression of the two threonyl-tRNA synthetase genes in Bacillus subtilis: control by transcriptional antitermination involving a conserved regulatory sequence. EMBO J. 1992;11:3117–3127. doi: 10.1002/j.1460-2075.1992.tb05384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarsero, J. P., E. Merino, and C. Yanofsky. A Bacillus subtilis operon containing genes of unknown function senses tRNATrp charging and regulates expression of the genes of tryptophan biosynthesis. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 15.Shimotsu H, Kuroda M I, Yanofsky C, Henner D J. Novel form of transcription attenuation regulates expression of the Bacillus subtilis tryptophan operon. J Bacteriol. 1986;166:461–471. doi: 10.1128/jb.166.2.461-471.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sudershana S, Du H, Mahalanabis M, Babitzke P. A 5′ RNA stem-loop participates in the transcription attenuation mechanism that controls expression of the Bacillus subtilis trpEDCFBA operon. J Bacteriol. 1999;181:5742–5749. doi: 10.1128/jb.181.18.5742-5749.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang M, de Saizieu A, Loon A P, Gollnick P. Translation of trpG in Bacillus subtilis is regulated by the trp RNA-binding attenuation protein (TRAP) J Bacteriol. 1995;177:4272–4278. doi: 10.1128/jb.177.15.4272-4278.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]