Abstract
Two new tprD alleles have been identified in Treponema pallidum: tprD2 is found in 7 of 12 T. pallidum subsp. pallidum isolates and 7 of 8 non-pallidum isolates, and tprD3 is found in one T. pallidum subsp. pertenue isolate. Antibodies against TprD2 are found in persons with syphilis, demonstrating that tprD2 is expressed during infection.
Syphilis, caused by Treponema pallidum subsp. pallidum, is a chronic disease characterized by periods of activity and latency, with clearance of early lesions but persistence of infection. The mechanisms T. pallidum uses to persist in humans are still not known, but it is possible that alterations in surface proteins will be involved because of their interaction with the host and their visibility to the immune response.
A family of 12 tpr genes is contained in the T. pallidum subsp. pallidum Nichols strain genome (7), some of which code for candidate surface-exposed proteins (3, 7, 12, 13). The tpr genes of T. pallidum are of interest for a variety of reasons. Their gene products are homologous to the Msp proteins of Treponema denticola, which have been implicated in cell attachment and porin function (6, 11). Anti-TprK antibody has been shown to opsonize T. pallidum Nichols strain for phagocytosis (3); thus, TprK may be exposed at the cell surface. Immunization with TprK is partially protective against challenge with T. pallidum Nichols strain (3), suggesting some tpr gene products are a focus of the protective immune response. Finally, the variable nature of the tpr genes suggests a role in immune evasion and persistence if different tpr genes are sequentially expressed. The 12 tpr gene products can be categorized into three subfamilies (3). Subfamilies I (TprCDFI) and II (TprEGJ) have conserved amino- and carboxyl-terminal sequences, but variable central amino acid sequences (3). Subfamily III Tpr proteins (TprABHKL) have scattered variable and conserved sequences throughout their length (3).
A novel tpr gene was discovered.
Multiple tprK alleles have been found in recent isolates of T. pallidum (5), in contrast to the single tprK allele identified in the laboratory-adapted Nichols strain (7). Because of our interest in tpr heterogeneity, we compared tpr gene sequences from other T. pallidum isolates to those from the Nichols strain. For example, genomic DNA from the T. pallidum subsp. pallidum Mexico A isolate was used as a template for PCR with primers A and B (Table 1), which are complementary to conserved regions flanking the central variable domains of tpr subfamilies I and II. The rabbit propagation, sources of the treponeme isolates, extraction of genomic DNA, and PCR conditions have been described previously (2–5). One of the amplicons was homologous to subfamily I (tprCDFI) at the conserved 5′ and 3′ ends, yet distinct from all of the subfamily I genes in much of the variable region (GenBank accession no. AF187953). This novel tpr was also found in the T. pallidum subsp. pallidum Bal-3 isolate (GenBank accession no. AF187952).
TABLE 1.
Oligonucleotides used in this study
| Primer | Characteristic | Sequence |
|---|---|---|
| A | tpr conserved sense | CGACTCACCCTCGAACCA |
| B | tpr conserved antisense | GGTGAGCAGGTGGGTGTAG |
| C | tprD2-specific sense | CACTAGTCTTGGGGACACGC |
| D | tprD2-specific antisense | TACGTGAATTGCAACCAGGA |
| E | tprA-specific sense | TACCTACCGGGATACGAACAGT |
| F | tprA-specific antisense | TGCAAGGCATGGGTGTAATCAT |
| G | tprD2 antisense 2 | TGACTTCATGGACCCTCTGTG |
| H | Tp0130 antisense bp 45–66 | CATGGCATTGGTGAGAAAGACG |
| I | Tp0132 sense bp 137–157 | CGCGTACCGCTTTGCAGTTCA |
The novel tpr gene occupies the tprD locus, and thus was termed tprD2.
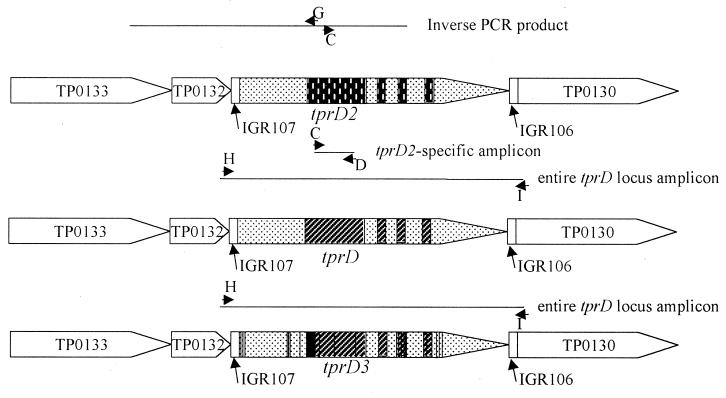
In order to localize the novel tpr gene in the genome, inverse PCR was used to amplify a fragment of genomic DNA containing the 5′ portion of the novel tpr gene and the 5′ flanking DNA. Genomic Bal-3 DNA (100 ng) was digested with Sau3A1 and ligated with T4 DNA ligase (New England Biolabs, Beverly, Mass.) in a 120-μl volume, such that circles were likely to be formed. This was used as a template in a PCR with primers C and G (Table 1 and Fig. 1) and yielded a 1.7-kb amplicon. Sequencing of this amplicon demonstrated that the novel tpr gene is flanked at the 5′ end by DNA with almost complete identity to the TP0132 and TP0133 genes, which are at the 5′ flanking end of tprD in the T. pallidum Nichols genome (Fig. 1).
FIG. 1.
Diagram of the tprD locus. Three different alleles of tprD (tprD, tprD2, and tprD3) are present in the tprD locus in the strains examined. The regions denoted as arrows and with TP or tpr are predicted coding regions with putative start codons at the beginning of the arrow and putative stop codons at the point of the arrow. IGR, intergenic region. PCR products are shown as thin lines, and the primers used to produce the products are shown as arrowheads. The different shading patterns and lines within the tprD, tprD2, and tprD3 alleles demonstrate the differences between the alleles.
The entire tprD locus and flanking regions (Fig. 1, primers H and I) were amplified from T. pallidum subsp. pallidum isolates Bal-3, Mexico A, Sea 81-3, Sea 81-4, and Bal-7 and from the Gauthier strain of T. pallidum subsp. pertenue. The sequence of these amplicons demonstrated that a novel sequence is found in the tprD locus of the Bal-3, Mexico A, Sea 81-4, and Sea 81-3 isolates (GenBank accession no. AF187952, AF217539, AF217540, and AF217541, respectively), and this allele was termed tprD2 (Fig. 1). The sequences of the tprD2 allele and flanking regions were identical in all four of these isolates. The amplicon from Bal-7 (GenBank accession no. AF217537) was identical to the tprD gene found in the Nichols strain, and the amplicon from the Gauthier strain (GenBank accession no. AF217538) was different from both tprD and tprD2 and was termed tprD3.
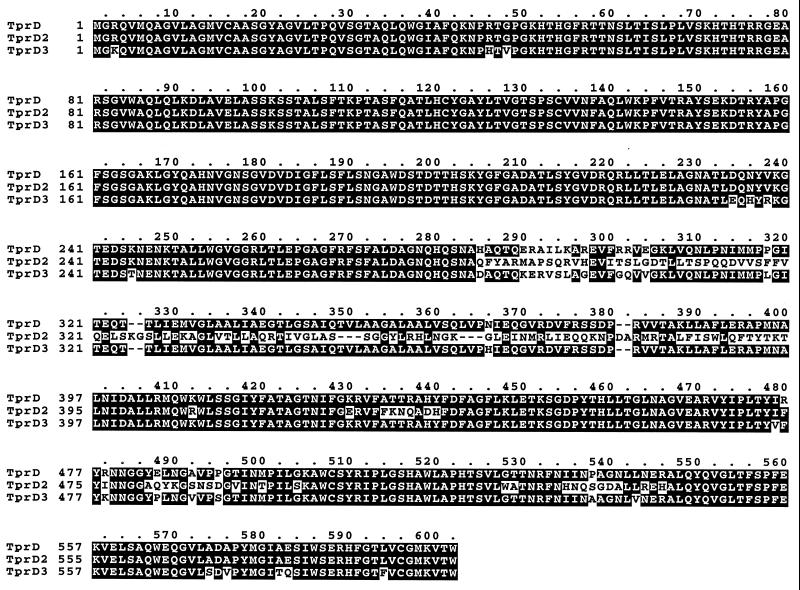
There are four regions of heterogeneity between tprD2 and tprD: a 330-bp central variable region and three smaller variable regions to the 3′ end of the open reading frames. No significant homology (>18-bp identity) to these four variable regions of tprD2 is present anywhere in the T. pallidum Nichols genome. These four variable regions are also reflected by differences in the predicted amino acid sequences of tprD2 and tprD (Fig. 2). Both TprD and TprD2 have predicted cleavable signal sequences (at amino acid 17), but TprD2 is highly predicted to be in the outer membrane, while TprD is predicted to localize in the inner membrane by PSORT analysis (http://psort.nibb.ac.jp/). Outer membrane expression of TprD2 could increase the repertoire of variable Tpr proteins on the surface for antigenic variation or to change the functionality of the protein. TprD3 is 95% identical to TprD, with the major region of heterogeneity located between amino acid residues 285 and 304, but with scattered amino acid differences found throughout the sequence. Like TprD, TprD3 is predicted by PSORT analysis to have a cleavable signal sequence, but to be located in the inner membrane. It is noteworthy that, while the 5′ conserved region (amino acids 1 to 284) of TprD2 is identical to the genome sequence for TprD, the same region in TprD3 has the signature at amino acids 234 to 245 (EQHYRKGTEDST) that characterizes TprF and TprI predicted from the genome sequence (7).
FIG. 2.
Amino acid sequence alignments of tprD, tprD2, and tprD3. The alignments of the predicted amino acid sequences of the tprD, tprD2, and tprD3 alleles are shown.
tprD2 was identified in about half of the Treponema isolates.
Primers C and D (Table 1) were designed to amplify a tprD2-specific 273-bp amplicon (Fig. 1) to test for the presence of tprD2 in a variety of Treponema isolates. We examined 12 isolates of T. pallidum subsp. pallidum, 4 isolates of T. pallidum subsp. pertenue (yaws spirochetes), 1 isolate of T. pallidum subsp. endemicum (endemic syphilis spirochete), the Simian isolate (primate spirochete [8]), and 2 isolates of Treponema paraluiscuniculi (rabbit venereal spirochetes) (Table 2). tprD2 was detected in 7 of 12 T. pallidum subsp. pallidum genomes and in 7 of 8 of the non-syphilis treponeme genomes (Table 2). Primers specific for tprA (primers E and F, Table 1) gave the predicted 315-bp amplicon with each of these isolates, demonstrating that the DNA from each treponeme isolate was intact and amplifiable (not shown). Rabbit DNA, a likely contaminant, did not give an amplicon with these sets of primers. Thus, it appears that the tprD2 allele is present in about half of the T. pallidum subsp. pallidum isolates tested, but not the Nichols strain that was used for the genome sequencing project (7).
TABLE 2.
tpr variable region amplification of genomic DNA from Treponema isolates
| Isolate or sample | Designation | tprD2 amplicon |
|---|---|---|
| T. pallidum subsp. pallidum | Nichols, Bal 73-1, Bal-7, Sea 83-1, Chicago | − |
| Bal-2, Bal-3, Bal-8, Sea 81-3, Sea 81-4, Sea 84-2, Mexico A | + | |
| T. pallidum subsp. pertenue | Gauthier | − |
| Haiti B,a Samoa D, Samoa F | + | |
| T. pallidum subsp. endemicum | Iraq B | + |
| Unnamed subspecies | Simian | + |
| T. paraluiscuniculi | Cuniculi A, Cuniculi M | + |
| Rabbit DNA | − |
Molecular analysis suggests that the Haiti B isolate may be a T. pallidum subsp. pallidum isolate (3a).
Humans with syphilis make antibodies to TprD2-specific peptide.
An amplicon encoding a 90-amino-acid peptide (amino acids 301 to 391 in Fig. 2) unique to the predicted TprD2 protein was amplified (primers C and D, Table 1 and Fig. 1), expressed as a six-histidine fusion protein, and purified (TprD2-specific peptide) (3). This TprD2-specific peptide had no homology to any of the other predicted tpr gene products or homology to any of the predicted open reading frames of the T. pallidum Nichols strain genome.
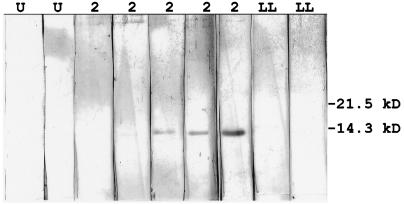
To test for immunoreactivity to the TprD2-specific peptide, human sera (obtained with informed consent as approved by the University of Washington Institutional Review Board for human subjects) were preabsorbed overnight with 5% Escherichia coli lysate containing an irrelevant recombinant Trypanosoma cruzi SA85-1.1-III protein in pRSET (9). Western blotting with 100 ng of recombinant TprD2-specific peptide, 1:100 diluted human sera, and 1:3,000 alkaline phosphatase-conjugated goat anti-human immunoglobulin G (Sigma, St. Louis, Mo.) was performed as previously described (1). Antibodies in sera from five of seven persons with secondary syphilis were reactive with the TprD2-specific peptide, while no anti-TprD2 activity was detected in sera from seven uninfected persons, two persons with primary syphilis, and three persons with late latent syphilis. Representative immunoblots are shown in Fig. 3. The antibodies to TprD2-specific peptide that were generated during syphilis infection demonstrate that TprD2 is expressed by T. pallidum subsp. pallidum in humans. The proportion of syphilitic sera reactive with TprD2-specific peptide (5 of 12 tested) is about the same proportion of T. pallidum subsp. pallidum isolates carrying the tprD2 allele (7 of 12 tested). This suggests that most isolates containing the tprD2 allele also express it.
FIG. 3.
Western blots demonstrating antibodies in sera from persons with secondary syphilis react with the TprD2-specific peptide. Shown are immunoblots with the 14-kDa TprD2-specific recombinant peptide from a representative experiment. The immunoblots were reacted with sera from two uninfected controls (U), five persons with secondary syphilis (noted by no. 2), and two persons with late latent syphilis (LL). Antibody reactivity to the tprD2-specific peptide is seen with the sera from three of five persons with secondary syphilis. Shown to the right are the positions of the molecular mass markers.
We have demonstrated that 7 of 12 of the T. pallidum subsp. pallidum isolates and most of the non-syphilis treponemes tested have a novel tprD2 allele. The one non-syphilis treponeme that did not contain tprD2, the Gauthier strain of T. pallidum subsp. pertenue, contains another variant of tprD, termed tprD3. In the four tprD2-containing isolates examined, the tprD2 gene occupies the position of the tprD gene, as defined in the Nichols strain; similarly, tprD3 occupies the same position in the Gauthier genome. Substitution of tprD2 and tprD3 for tprD generates additional diversity of the tpr genes in some strains. The differences in the predicted amino acid sequences of tprD and tprD2 are localized to a 110-amino-acid central variable region and three smaller variable regions toward the carboxyl terminus of TprD2. If these differences are in the immunodominant or exposed regions of these molecules, this could help to explain why immunity from heterologous challenge is not as complete as it is with homologous challenge (10). Alternatively, these variable domains may provide a functional capacity, like cell binding, and the sequence variation may extend the functional capacity of individual isolates. It is notable that TprD2, but not TprD or TprD3, is predicted to be in the outer membrane, suggesting a different functional role for TprD2.
Acknowledgments
We thank Barbara Molini for excellent technical assistance.
This work was supported by Public Health Service grants AI34616, AI42143, and AI31448 (Sexually Transmitted Diseases Cooperative Research Center New Investigator Award to A.C.-L.).
REFERENCES
- 1.Cameron C E, Castro C, Lukehart S A, Van Voorhis W C. Function and protective capacity of Treponema pallidum subsp. pallidum glycerophosphodiester phosphodiesterase. Infect Immun. 1998;66:5763–5770. doi: 10.1128/iai.66.12.5763-5770.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centurion-Lara A, Arroll T, Castillo R, Shaffer J M, Castro C, Van Voorhis W C, Lukehart S A. Conservation of the 15-kilodalton lipoprotein among Treponema pallidum subspecies and strains and other pathogenic treponemes: genetic and antigenic analyses. Infect Immun. 1997;65:1440–1444. doi: 10.1128/iai.65.4.1440-1444.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centurion-Lara A, Castro C, Barrett L, Cameron C, Mostowfi M, Van Voorhis W C, Lukehart S A. Treponema pallidum major sheath protein homologue Tpr K is a target of opsonic antibody and the protective immune response. J Exp Med. 1999;189:647–656. doi: 10.1084/jem.189.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Centurion-Lara A, Castro C, Castillo R, Shaffer J M, Van Voorhis W C, Lukehart S A. The flanking region sequences of the 15-kDa lipoprotein gene differentiate pathogenic treponemes. J Infect Dis. 1998;177:1036–1040. doi: 10.1086/515247. [DOI] [PubMed] [Google Scholar]
- 4.Centurion-Lara A, Castro C, Shaffer J M, Van Voorhis W C, Marra C M, Lukehart S A. Detection of Treponema pallidum by a sensitive reverse transcriptase PCR. J Clin Microbiol. 1997;35:1348–1352. doi: 10.1128/jcm.35.6.1348-1352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centurion-Lara A, Gordones C, Castro C, Van Voorhis W C, Lukehart S A. The tprK gene is heterogeneous among Treponema pallidum strains and has multiple alleles. Infect Immun. 2000;68:824–831. doi: 10.1128/iai.68.2.824-831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fenno J C, Müller K-H, McBride B C. Sequence analysis, expression, and binding activity of recombinant major outer sheath protein (Msp) of Treponema denticola. J Bacteriol. 1996;178:2489–2497. doi: 10.1128/jb.178.9.2489-2497.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraser C M, Norris S J, Weinstock G M, White O, Sutton G G, Dodson R, Gwinn M, Hickey E K, Clayton R, Ketchum K A, Sodergren E, Hardham J M, McLeod M P, Salzberg S, Peterson J, Khalak H, Richardson D, Howell J K, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton M D, Venter J C, et al. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 8.Fribourg-Blanc A, Niel G, Mollaret H H. Note sur quelques aspects immunologiques du cynocephale africain. Bull Soc Pathol Exot. 1963;56:474–485. [PubMed] [Google Scholar]
- 9.Kahn S J, Wleklinski M. The surface glycoproteins of Trypanosoma cruzi encode a superfamily of variant T cell epitopes. J Immunol. 1997;159:4444–4451. [PubMed] [Google Scholar]
- 10.Magnuson H J, Thompson F A., Jr Heterologous strain immunity in experimental syphilis. J Immunol. 1951;67:35–40. [PubMed] [Google Scholar]
- 11.Mathers D A, Leung W K, Fenno J C, Hong Y, McBride B C. The major surface protein complex in Treponema denticola depolarizes and induces ion channels in HeLa cell membranes. Infect Immun. 1996;64:2904–2910. doi: 10.1128/iai.64.8.2904-2910.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stamm L V, Greene S R, Bergen H L, Hardham J M, Barnes N Y. Identification and sequence analysis of Treponema pallidum tprJ, a member of a polymorphic multigene family. FEMS Microbiol Lett. 1998;169:155–163. doi: 10.1111/j.1574-6968.1998.tb13312.x. [DOI] [PubMed] [Google Scholar]
- 13.Stebeck C E, Shaffer J M, Arroll T W, Lukehart S A, Van Voorhis W C. Identification of the Treponema pallidum subsp. pallidum glycerophosphodiester phosphodiesterase homologue. FEMS Microbiol Lett. 1997;154:303–310. doi: 10.1111/j.1574-6968.1997.tb12660.x. [DOI] [PubMed] [Google Scholar]