Abstract
The SARS-CoV-2 pandemic has highlighted the need for broad-spectrum antiviral drugs to respond promptly to viral emergence. We conducted a preclinical study of molnupiravir (MOV) against SARS-CoV-2 to fully characterise its antiviral properties and mode of action. The antiviral activity of different concentrations of MOV was evaluated ex vivo on human airway epithelium (HAE) and in vivo in a hamster model at three escalating doses (150, 300 and 400 mg/kg/day) according to three different regimens (preventive, pre-emptive and curative). We assessed viral loads and infectious titres at the apical pole of HAE and in hamster lungs, and MOV trough concentration in plasma and lungs. To explore the mode of action of the MOV, the entire genomes of the collected viruses were deep-sequenced. MOV effectively reduced viral titres in HAE and in the lungs of treated animals. Early treatment after infection was a key factor in efficacy, probably associated with high lung concentrations of MOV, suggesting good accumulation in the lung. MOV induced genomic alteration in viral genomes with an increase in the number of minority variants, and predominant G to A transitions. The observed reduction in viral replication and its mechanism of action leading to lethal mutagenesis, supported by clinical trials showing antiviral action in humans, provide a convincing basis for further research as an additional means in the fight against COVID-19 and other RNA viruses.
Keywords: COVID-19, Antiviral drug, Nucleoside analog, Mutagenesis, Animal model, HAE
Highlights
-
•
We confirmed the antiviral activity of molnupiravir in human airway epithelium model.
-
•
The delay of administration is a key factor of antiviral activity in the hamster model.
-
•
Pulmonary rather than plasma concentrations are the determinants of antiviral activity.
-
•
Post-treatment mutation profile is found ex-vivo and in-vivo.
1. Introduction
The pandemic caused by SARS-CoV-2, since its emergence in December 2019, has highlighted the lack of rapidly available antiviral treatment in case of viral emergence. Several strategies have been employed to discover antiviral molecules against SARS-CoV-2, including drug repurposing, which has the theoretical advantage of saving time in the context of a health crisis, since pre-clinical and clinical research for these molecules has already been partially or totally conducted[1]. The off-target based drug repurposing, consisting in screening molecules marketed or under clinical evaluation for pharmacological properties other than those of antivirals, has been widely used, but to date has failed to identify any molecules showing efficacy in well-conducted clinical trials [2]. On the other hand, the on-target based drug repurposing strategy, involving known antiviral drugs, has also been exploited, in particular to investigate the efficacy of molecules that possess broad-spectrum antiviral properties [3]. Remdesivir is the most obvious example: promising in vivo and in vitro studies on Ebola and other coronaviruses have prompted the scientific community to investigate its activity against SARS-CoV-2[4].
Among broad-spectrum antivirals, molnupiravir (MOV) was among the first to prove in vitro and in vivo efficacy against SARS-CoV-2. Discovered in the early 70s, it is the prodrug of the β-D N4-hydroxycytidine (NHC; EIDD-1931), and has been described as having the ability to induce mutagenesis in bacteria [5]. In the 2000s, studies highlighted the antiviral activity of NHC, first against members of the Flaviviridae family [6], then on several members of the Coronaviridae family (HCoV-NL63 [7], SARS-CoV-1 [8]. Finally against many other RNA viruses (influenza virus, respiratory syncytial virus [9], noroviruses [10], Venezuelan equine encephalitis virus [11], enteroviruses [12], Chikungunya virus [13] and Ebolavirus [14]. Due to an unfavorable pharmacokinetic profile, resulting mainly from poor oral bioavailability, NHC was subsequently engineered by the addition of an isopropyl group to obtain a prodrug with improved bioavailability: MOV (EIDD-2801). The anti-influenza activity of MOV was therefore assessed in vitro and in vivo [15] shortly before the SARS-COV-2 outbreak.
Towards SARS-CoV-2, MOV displayed 50 % effective concentrations ranging from 0.08 to 3.4 μM in different cell lines (VeroE6 [[16], [17], [18], [19]], Calu-3 [19,20] and Huh 7 [16]), and was active in more complex ex vivo models such as human upper airway epithelium [19,21]. Studies in animal models have also demonstrated its potency in preventing transmission in ferret models [17,22] and limiting viral replication in multiple rodent models (mice [23,24], Roborowski [22] and golden hamster [16,20]).
Biochemical tests with MOV showed that its active tri-phosphorylated form (NHC-TP) can be incorporated by SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) instead of cytidine or uridine, causing the mis-incorporation of bases such as guanosine or adenosine and inducing lethal mutagenesis [25,26]. Few studies have investigated this mechanism in more complex systems. Using MERS-CoV, Sheahan et al. have shown increased mutation frequency ex vivo in an infected respiratory epithelium model and in vivo using a mouse model [19]. In a hamster model of SARS-CoV-2 infection, Abdelnabi et al. showed an increase in the mutation frequency of viral RNA recovered from the lungs following treatment with MOV [16].
Given the potential of MOV as a treatment for COVID-19 and its current use in specific patient populations [27], we further evaluated its preclinical efficacy both ex vivo using reconstituted human airway epithelial model and ex in vivo using a hamster model SARS-CoV2 infection. In addition, to better understand its mechanism of action, we characterized its mutagenic effect in both models and offers our knowledge the first detailed mutagenesis study in the HAE model.
2. Material and methods
2.1. Virus
All experiments with infectious virus were conducted in a biosafety level (BSL) 3 laboratory. The SARS-CoV-2 ancestral strain BavPat1, containing the spike D614G mutation, was used as previously described [28].
2.2. Human airway epithelia (HAE)
Mucilair™ HAE reconstituted from human primary cells of bronchial biopsies were purchased from Epithelix SARL (Geneva, Switzerland). The bronchial epithelium was maintained in air-liquid interface with specific media purchased from Epithelix.
2.2.1. MOV evaluation in HAE
2.2.1.1. Culture and infection
After a gentle wash with pre-warmed OPTI-MEM medium (Life technologies), epithelia were infected with SARS-COV-2 on the apical side using 4000 TCID50 per well as previously described [[29], [30], [31]]. Cells were cultivated in Epithelix media that contained different MOV concentrations (0.3, 1 or 3 μM) or without the drug (Virus control). Remdesivir (BLD pharm) was used at 10 μM as a positive drug control. At day 1, the apical side of the epithelia was washed with pre-warmed OPTI-MEM to eliminate the viral inoculum. Media were changed daily during the experiment (media composition remained identical throughout the experiment). For each condition, experiments were carried out in triplicates, using inserts from the same donor. Samples were collected at the apical side by washing with 200 μL of pre-warmed OptiMEM medium, then divided into two aliquots, with one used for TCID50 assay immediately, while the other was stored at −80° before viral extraction.
2.3. In vivo experiments
2.3.1. Approval and authorization
In vivo experiments were approved by the local ethical committee (C2EA—14) and the French “Ministère de l’Enseignement Supérieur, de la Recherche et de l’Innovation” (APAFIS#23975). All experiments were conducted in BSL 3 laboratory.
2.3.2. Animal handling, hamster infection and organ collection
Three-week-old female Syrian hamsters were provided by Janvier Labs. Animals handling, anesthesia, infection, nasal washes, euthanasia and organ collection were performed as previously described [28,29], briefly, animals received 50 μL of a solution containing either 104 TCID50 of virus diluted in 0.9 % sodium chloride (infected group) or 0.9 % sodium chloride solution (sham group) by intranasal injection. Nasal washes were conducted using 150 μL of 0.9 % sodium chloride solution. The samples were centrifuged and subsequently stored at −80 °C. Animals were euthanized 15 h following the final treatment intake, and lung and blood samples were promptly collected. Left lung lobes were washed with 10 mL of 0.9 % sodium chloride solution, weighed, and crushed with glass beads. The resulting supernatants were stored at −80 °C. Right apical lobes were ground with Qiazol (Qiagen) lysis reagent and stored at −80 °C. Blood samples were collected in tubes with EDTA, centrifuged, and the plasma was stored at −80 °C.
2.3.3. Antiviral administration
MOV solution used was obtained by diluting MOV powder (DNDI0003948820/Batch PCDRUG02-0051) in a vehicle composed of ultra-pure distilled water with 10 % polyethylene glycol 400 (PEG400) and 2.5 % polyethoxylated castor oil (Kolliphore EL). The MOV solution or vehicle was administrated to anesthetized (isoflurane) hamsters by oral gavage. Favipiravir (FAV) (Courtesy of Toyama-Chemical) was injected intraperitoneally to vigil animals as previously described [28].
2.3.4. Experimental timeline
For pre-emptive treatment (D0), the first dose (MOV, FAV, or vehicle) was administered at the time of infection, followed by subsequent doses every 12 h until the day of euthanasia. An additional dose was administered 12 h before infection for preventive treatment (D-1), while for curative treatment (D1), the first dose was delayed until 12 h after infection. In all experiments, except for the histological study where euthanasia was performed on D5, animals were euthanized on D3. The number of animals in each group was 6, except in the dose assessment experiment where groups of 10 (150 and 400 mg/kg/d) or 12 (FAV and VEH) animals were used.
2.4. Viral RNA yield
Before PCR amplification, RNA extraction was conducted in according to the manufacturer's guidelines using the QIAamp 96 DNA Kit and the Qiacube HT Kit, both from Qiagen as described before [29] with a final extract volume of 70 μL. Briefly, 100 μL of tissue clarified homogenates supplemented with 10 μL of internal control (bacteriophage MS2) or HAE supernatant were transferred to an S-block containing the appropriate amounts of VXL, proteinase K and RNA carrier. RT-PCR were performed with the GoTaq 1-step qRtPCR kit (Promega) using 3.8 μL of extracted RNA and 6.2 μL of RT-qPCR mix that contains 250 nM of each primer and 75 nM of probe which target for HAE SARS-CoV-2 N gene Fw: GGCCGCAAATTGCACAAT; Rev: CCAATGCGCGACATTCC; Probe: FAM-CCCCCAGCGCTTCAGCGTTCT-BHQ1 and SARS-CoV-2 RdRp gene for in vivo experiment Fw: GTGARATGGTCATGTGTGGCGG-; Rev: CARATGTTAAASACACTATTAGCATA; Probe: CAGGTGGAACCTCATCAGGAGATGC-TAMRA. Quantification was provided by four 2 log serial dilutions of an appropriate T7-generated synthetic RNA standard of known quantities (102 to 108 copies/reaction).
2.5. Viral infectious titers
To determine infectious titers confluent VeroE6 cells were inoculated with serial dilutions of each sample (tenfold or four-fold dilutions for cell supernatant media or lung clarified homogenates respectively) in 96-well culture plates. Six dilutions were replicated for each sample. Following a 5-day incubation period, observation for the presence or absence of cytopathic effect in each well was conducted. The estimation of infectious titers was carried out utilizing the methodology described by Reed & Muench[32], with a limit of quantification (L.O.Q) equal to 0,5 Log10 TCID50/mL.
2.6. Molnupiravir pharmacokinetics
Quantification of MOV in plasma and lung tissues was performed by a sensitive and selective validated high-performance liquid chromatography coupled with tandem mass spectrometry method (UPLC-TQD, Waters, USA) with a lower limit of quantification of 5 ng/mL (precision: 12.6 %; accuracy: 0.13 %). Precision and accuracy of the 3 quality control samples (QCs) were within 15 % over the calibration range (5 μg/mL to 2500 μg/mL). MOV was extracted by a simple protein precipitation method, using acetonitrile for plasma and ice-cold acetonitrile for clarified lung homogenates. To assess the selectivity and specificity of the method and matrix effect, blank plasma and tissues homogenates from 2 control animals (uninfected and untreated) were processed at each run. Moreover, the same control samples spiked with MOV concentration equivalent to the QCs (10, 250 and 2000 μg/mL) were also processed and compared to the QCs samples. As the animals were sacrifices 15 h after the last drug intake, and the drug was given twice daily, the observed concentration corresponds to a trough concentration. Drug accumulation in lung was assessed by calculating a tissue to plasma concentration ratio.
2.7. Expression level of seven cytokines in lungs
Expression level of seven cytokines in lungs was performed as described elsewhere [33]. In essence, the upper lobe shreds were processed with QUIAZOL-chloroform extraction protocol according to manufacturer's instructions. Quantitative RT-qPCR was performed using the primers described by Dias de Melo et al. [34] and the QuantiNova SYBR® Green RT-PCR Kit (Qiagen) according to the manufacturer's guidelines. Cytokine mRNA levels were quantified using serial dilutions of synthetic standards and normalised to the reference gene γ-actin (Fw: ACAGAGAGAAGATGACGCAGATAATG; Rev: GCCTGAATGGCCACGTACA; Probe:FAM-TTGAAACCTTCAACACCCCAGCC-TAMRA).
2.8. Histology
Animal handling, hamster infections and treatment administrations were performed as described above. Lungs were collected after intratracheal instillation of 4 % (w/v) formaldehyde solution, fixed 72 h at room temperature with a 4 % (w/v) formaldehyde solution and embedded in paraffin. Tissue sections of 3.5 μm, obtained following guidelines from the ‘global open RENI’ (The standard reference for nomenclature and diagnostic criteria in toxicologic pathology; https://www.goreni.org/), were stained with hematoxylin-eosin (H&E) and blindly analyzed by a certified veterinary pathologist. Microscopic examination was done using a Nikon Eclipse E400 microscope. Different anatomic compartments were examined (1) for bronchial and alveolar walls, a score of 0–4 was assigned based on severity of inflammation; (2) regarding alveoli, a score of 0–2 was assigned based on the presence and severity of hemorrhagic necrosis; (3) regarding vessel lesions (endothelitis/vasculitis), absence or presence was scored 0 or 1 respectively. A cumulative score was then calculated and assigned to a grade of severity (Supplemental Table 1).
2.9. Sequence analysis of the full-length genome
200 μL of lung clarified homogenate or infectious cell supernatant (virus stock and HAE) was inactivated with an equal volume of VXL lysis buffer (Qiagen) and viral RNA was extracted using an EZ1 Advanced XL robot with the EZ1 mini virus 2.0 kit (both from Qiagen) and linear acrylamide (ThermoFisher Scientific) in place of carrier RNA. The extracts were then subjected to quantitative real-time RT-PCR in order to standardize the amounts of viral RNA used during complete genome amplification (see below).
A specific set of primers (Supplemental Table 2) was used to generate thirteen amplicons covering the entire genome with the Superscript IV one step RT-PCR System (ThermoFisher Scientific). PCR mixes (final volume 25 μL) contained 2.5 μL containing a standard quantity of viral RNA from the nucleic acid extract, 0.75 μL of each primer (10 μM), 12.5 μL of 2X Platinum SuperFi RT-PCR Master Mix, 8.25 μL of RNA free water and 0.25 μL SuperScript IV RT Mix. Amplifications were performed with the following conditions: 15 s at 55 °C, 2 min at 98 °C, then 40 cycles of 10 s at 98 °C, 10 s à 56 °C and 1.5 min at 72 °C. Size of PCR products was verified by gel electrophoresis. For each sample, an equimolar pool of all amplicons was prepared and purified using Monarch PCR & DNA Cleanup Kit (New England Biolabs). After DNA quantification using Qubit dsDNA HS Assay Kit and Qubit 2.0 fluorometer (ThermoFisher Scientific), amplicons were fragmented by sonication into fragments of around 200bp long. Libraries were built by adding barcodes, for sample identification, and primers using AB Library Builder System (ThermoFisher Scientific). To pool equimolarly the barcoded samples a quantification step by real time PCR using Ion Library TaqMan Quantitation Kit (ThermoFisher Scientific) was performed. Then, emulsion PCR from pools and loading on 530 chip was performed using the automated Ion Chef instrument (ThermoFisher Scientific). Sequencing was performed using the S5 Ion torrent technology v5.12 (ThermoFisher Scientific) following manufacturer's instructions. Consensus sequence was obtained after trimming of reads (reads with quality score <0.99, and length <100 pb were removed and the 30 first and 30 last nucleotides were removed from the reads). Mapping of the reads on the reference was done using CLC genomics workbench software v.20 (Qiagen). Mutation frequency for each position was calculated as the number of reads with a mutation compared to the reference divided by the total number of reads at that site. Only substitutions with a frequency of at least 1 % were considered for the analysis (Supplementary Table 3).
2.10. Graphical representations and statistical analysis
Graphical representations and statistical analyses were performed with Graphpad Prism 7 (Graphpad software). Statistical details for each experiment are described in the figure legends and in corresponding supplemental tables (Supplemental Table 4.). When a value fell below the limit of quantification, it was treated as being equal to the limit for statistical analysis. When relevant, two-sided statistical tests were always used. For simple comparisons, depending on the data distribution assessed with the Shapiro-Wilk normality test, either the Mann-Whitney test or an unpaired t-test was used. Variance equality was evaluated using a Fisher test, and Welch's correction was applied if deemed necessary. A two-way Holm-Šídák ANOVA was used for Fig. 1, and a multiple Mann-Whitney test with the Holm-Šídák method was used for Fig. 5-K and Fig. 5-L. P-values lower than 0.05 were considered statistically significant. The statistical tests conducted are provided in Supplementary Table 4. Experimental timelines were created on biorender.com.
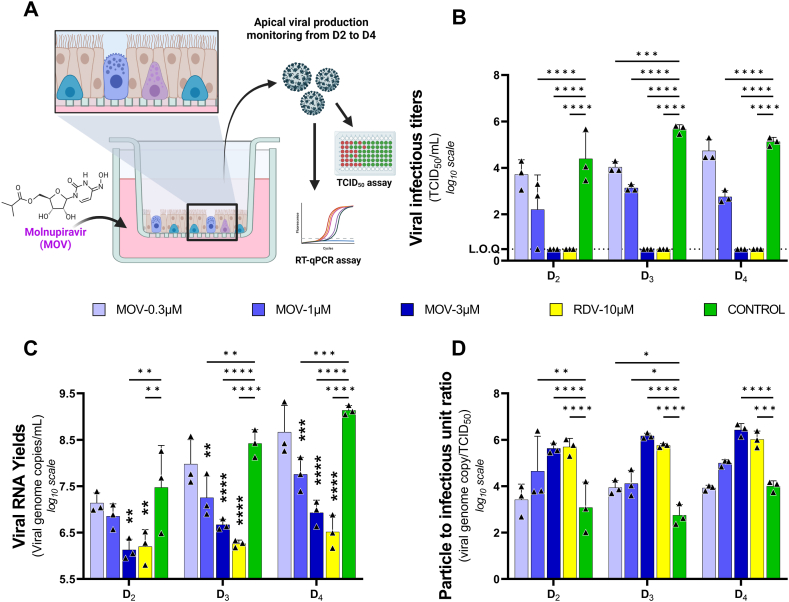
Fig. 1.
Antiviral activity of molnupiravir in reconstituted human airway epithelium. Experimental procedure (A). Kinetics of virus excretion at the apical side of the epithelium measured using a TCID50 (B) or RT-qPCR assay (C). Particle to infectious unit ratio calculated as follows: ratio of viral RNA yields over viral infectious titer (D). The dotted line represent limit of quantification (L.O.Q) Data represent mean ± SD of three independent inserts. Remdesivir at 10 μM was used as a positive drug control. The significance was tested two-way ANOVA test with holm-Šídák adjustment. *, **, ***, and **** symbolize statistically significant reductions compared to the control group, with associated p-values falling within the ranges of 0.01–0.05, 0.001–0.01, 0.0001–0.001, and <0.0001, respectively.
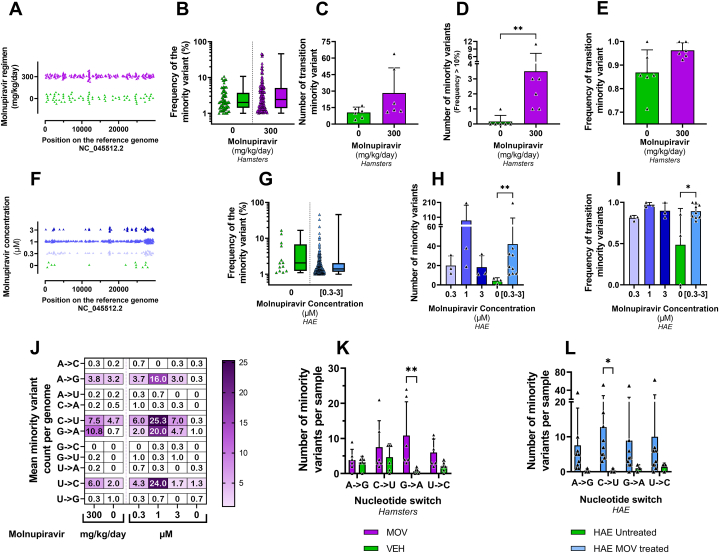
Fig. 5.
In vitro and in vivo Mutagenic effect of molnupiravir.Graphical representation of the distribution of minority variants (MVs) along the SARS-CoV-2 genome in clarified hamster lung homogenates (A) and in HAE supernatants (F), each triangle representing an individual MV. Frequency of MVs presented individually or as a box-and-whisker plot in purified hamster lung clarified homogenates (B) and in HAE supernatants pooled from the three exposed groups (G). Mean number of MVs in vivo (C) and ex-vivo (H). Mean number of mutations with a frequency greater than 10 % in purified hamster lung clarified homogenates (D). Average frequency of transition-type MVs in each group in vivo (E) or ex vivo (I). For each specimen, the frequency was calculated by dividing the number of transition-type MV identified within the sample by the total number of MV found in that sample. Heat map representing the mean number of each nucleotide exchange in each group (J). Mean number of the 4 types of transition in each group in vivo (K) and ex vivo (L). Two-sided statistical analysis was performed using Shapiro-Wilk normality test, Fisher's exact test, Student t-test, Mann-Whitney test for panel B to E and G to I, for panel K and L, multiple Mann-Whitney test with Holm-Šídák correction was used (detail in Supplemental Table 4). ** and * symbols indicate that the average value for the group is significantly higher than that of the untreated group with a p-value ranging between 0.001 and 0.01, and 0.01–0.05, respectively.
3. Results
3.1. Ex-vivo efficacy of molnupiravir
As an initial step, we assessed the antiviral activity of MOV in a HAE model reconstituted with primary bronchial cells. Epithelia were infected apically (D0) at a MOI (multiplicity of infection) of 0.1 with an ancestral G614 SARS CoV-2 strain (B.1 lineage) and exposed basolaterally to MOV at 0.3, 1 or 3 μM during three consecutive days. Remdesivir (10 μM) was used as positive control. Viral excretion was assessed from 2 to 4 days post infection (D2 to D4), by measuring viral RNA yields and infectious titers at the apical side of the epithelium (Fig. 1-A).
Significant decrease in viral infectious titres (Fig. 1-B) were observed from D2 to D4 for both 3 and 1 μM exposure (p < 0.0001 at each time point). With the concentration of 0.3 μM a significant decrease in infectious titres was observed only at D3 (p = 0.0008).
When viral excretion was assessed on viral RNA yields (Fig. 1-C), a significant decrease was observed from D2 to D4 with the concentration of 3 μM (p = 0.0011; p < 0.0001; p = 0.0001 respectively). With the 1 μM concentration, a significant reduction was observed at D3 and D4 (p = 0.0023; p = 0.0004 respectively). In contrast, no significant reduction in viral RNA yields was observed with 0.3 μM.
We then estimated the particle to infectious unit ratio (i.e., the ratio between the number of viral RNA molecules and the number of infectious particles) (Fig. 1-D). At 3 μM, the increase in the ratio, indicating a reduction in the relative infectivity of the virus particles, was significant from D2 to D4 (p < 0.001). At 1 μM, this increase was significant at D2 and D3 (p = 0.0047 and 0.014 respectively), but not at D4 (p = 0.0845). Finally, the 0.3 μM concentration produced a significant increase only at D3 (p = 0.0172).
3.2. In vivo efficacy of molnupiravir
We further investigated the potential antiviral activity of MOV in vivo using a previously described hamster model of SARS-CoV-2 infection[28,29,31,33,35].
In a first set of experiments, treatment was initiated at D0 (pre-emptive antiviral therapy) and ended at D2. Hamsters were intranasally infected with 104 TCID50 of SARS-CoV-2 (ancestral B.1 (614G) strain) and received MOV orally at doses of 150, 300 or 400 mg/kg/day BID (Fig. 2-A). Untreated group of hamsters received the vehicle (VEH) solution BID and a positive control group was treated with Favipiravir intraperitoneally (926 mg/kg/day BID). At D3, nasal washes were performed and lungs and plasma collected following the sacrifice of the animals to assess viral replication and determine NHC concentrations (hydrolysis product of MOV) in plasma and lung tissue.
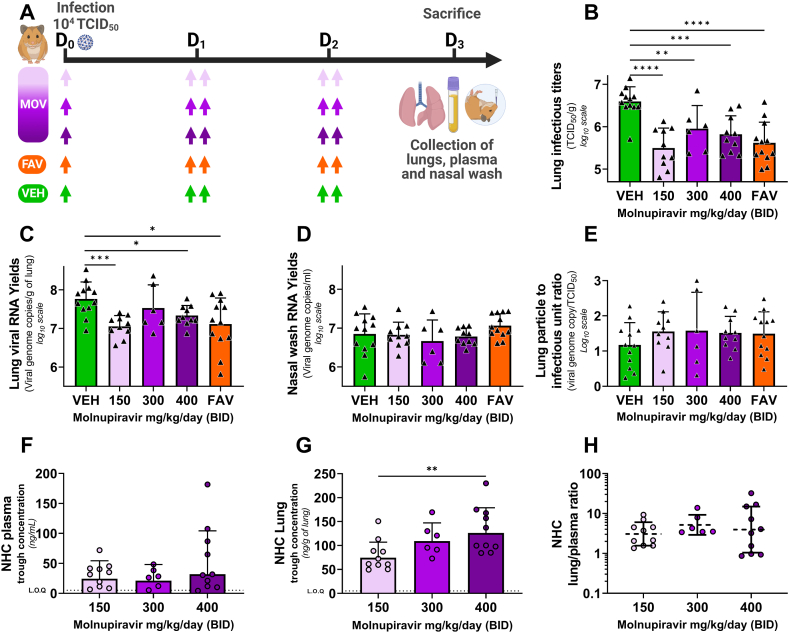
Fig. 2.
Antiviral activity and pharmacokinetics of oral molnupiravir treatment in a hamster model: Dose assessment experiment.
Groups of 6 to12 hamsters were intranasally infected with 104 TCID50 of virus. Animals received Monlupiravir (MOV) or the vehicle (VEH, untreated group) orally, and Favipiravir (FAV) intraperitoneally. Experimental timeline (A). Viral replication in lung based on infectious titers expressed in TCID50/g of lung (B) or on viral RNA yields (measured using an RT-qPCR assay) expressed in viral genome copies/g of lung (C). Viral replication in nasal wash based on viral RNA yields (measured using an RT-qPCR assay) expressed in viral genome copies/mL of nasal wash (D). Lung particle to infectious unit ratio, (E). NHC concentration in plasma (F) and in lung (G) 15 h after the last intake expressed in ng/mL of plasma and ng/g of lung tissue respectively. Lung/plasma ratio, calculated as the ratio of lung over plasma concentrations (H). Data represent mean ± SD for panel B to E. From panel F to H bars and dash line represent geometric mean ± SD, and the dotted line represent the limit of quantification (LOQ). Two-sided statistical analysis was performed using Shapiro-Wilk normality test, Fisher's exact test, Student t-test, Mann-Whitney test (detail in Supplemental Table 4). *, **, ***, and **** symbolize statistically significant reductions compared to the control group (or augmentation compared to the 150 mg/kg/d group for panel G), with associated p-values falling within the ranges of 0.01–0.05, 0.001–0.01, 0.0001–0.001, and <0.0001, respectively.
The infectious titers measured in the clarified lung homogenates of MOV-treated animals were significantly lower than those of untreated animals (p < 0.0001, p = 0.0074 and p = 0.0002 for MOV 150, 300 and 400 mg/kg/day BID respectively) : reduction of infectious titers ranged between 0.78 and 1.1 log10 (Fig. 2-B). Hamsters treated with 150 or 400 mg/kg/day BID of MOV expressed significant lower viral RNA yields in lungs than untreated animals (p = 0.003 and p = 0.0127 respectively). (Fig. 2-C). In contrast, no significant differences for RNA viral yields in nasal washes were observed for all treated animals compared to untreated animals (Fig. 2-D). For Fig. 2-B and Fig. 2-C, comparisons between the three treated groups showed no significant difference. (See Supplementary Table 4). No significant differences were observed when analysing the particle-to-infectious unit ratio (Fig. 2-E).
NHC plasma concentrations were lower than pulmonary ones and comparable between doses (Fig. 2-F). In contrast Analysis of trough NHC concentrations in lungs showed a dose-exposure relationship (r2 = 0.29) (Supplemental Fig. S6) (Fig. 2-G). In particular, we noticed that NHC concentration was significantly higher with the dose of 400 mg/kg/day BID compared to the dose of 150 mg/kg/day BID (p = 0.0087). Lung partition coefficients, calculated as the ratio of lung over plasma concentrations, were above 1 and constant at all doses (Fig. 2-H).
In a second set of experiments, treatments were initiated either one day before infection (D-1), at the day of infection (D0) or one day after the infection (D1) (preventive, pre-emptive or curative antiviral therapy respectively) resulting in a cumulative number of doses of MOV of 7, 5 and 3 doses respectively (Fig. 3-A). Untreated animals received the vehicle (VEH) solution BID starting on D-1 and the positive control group received FAV starting on D0 (925 mg/kg/day BID by intraperitoneal injection). Hamsters were intranasally infected with 104 TCID50 of SARS-CoV-2 (ancestral B.1 (614G) strain) and received MOV orally at the same regimen of 300 mg/kg/day BID. Lungs and plasma were collected at D3, to perform the same analyses as in the first experiment.
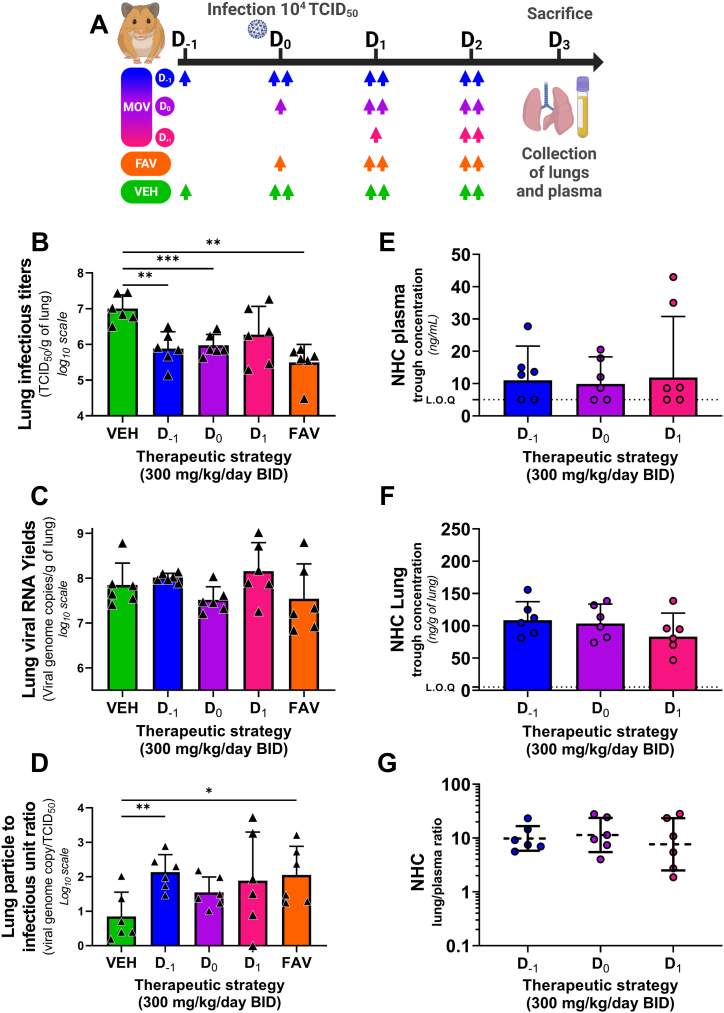
Fig. 3.
Antiviral activity and pharmacokinetics of oral molnupiravir treatment in a hamster model: Therapeutic strategies assessment experiment.Groups of 6 hamsters were intranasally infected with 104 TCID50 of virus. Animals received Monlupiravir (MOV) or the vehicle (VEH, untreated group) orally, and Favipiravir (FAV) intraperitoneally. Experimental timeline (A). Viral replication in lung based on infectious titers (measured using a TCID50 assay) expressed in TCID50/g of lung (B). Viral replication in lung based on viral RNA yields (measured using an RT-qPCR assay) expressed in viral genome copies/g of lung (C). Lung particle to infectious unit ratio (D). NHC trough concentration in plasma (E) and in lung (F) expressed in ng/mL of plasma and ng/g of lung tissue. Lung/plasma ratio (H). Data represent mean ± SD for panel B to D. From panel E to G bars and dash line represent geometric mean ± SD, and the dotted line represent the limit of quantification (LOQ). Two-sided statistical analysis was performed using Shapiro-Wilk normality test, Fisher's exact test, Student t-test, Mann-Whitney test (detail in Supplemental Table 4). *, **, ***, and **** symbolize statistically significant reductions (or augmentation for panel D) compared to the control group, with associated p-values falling within the ranges of 0.01–0.05, 0.001–0.01, 0.0001–0.001, and <0.0001, respectively.
The infectious viral titers found in the clarified lung homogenates of MOV-treated animals preventively (D-1) and pre-emptively (D0) were significantly lower than those of the untreated animals (p = 0.0087 and p = 0.0139 respectively), corresponding to a reduction of 1.12 and 1.03 log10 respectively (Fig. 3-B). In contrast, the viral RNA yields in lungs of the three treated groups did not differ significantly from that of the control group (Fig. 3-C). When examining the particles to infectious unit ratio (Fig. 3-D), a significant elevation was only found for preventively MOV-treated animals group (D-1) (p = 0.0048).
Analysis of lung and plasma NHC trough concentrations showed no difference between the different treatment strategies, (Fig. 3-E/Fig. 3-F). Once again, the pulmonary partition coefficient was above 1 and constant between administration regimens (Fig. 3-G).
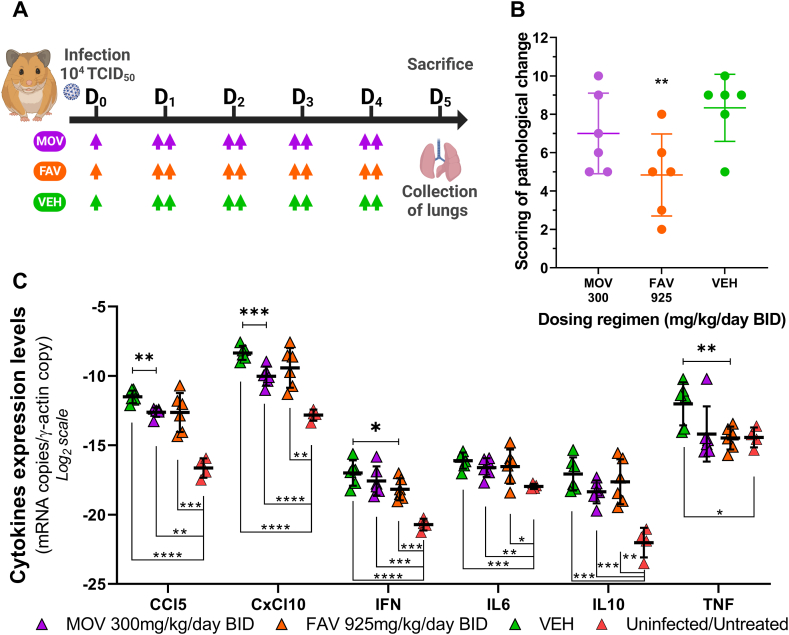
In a final set of experiments, we assessed the impact of MOV treatment on lung pathological changes and pulmonary cytokine expression. Treatment with MOV was initiated at D0. Animals were intranasally infected with 104 TCID50 of SARS-CoV-2 (same ancestral B.1 G 614 strain) and treated with MOV at 300 mg/kg/day BID. Control groups identical to the previous ones (untreated and positive) were also included. A group of untreated, uninfected animals was used as a control for the measure of lung cytokine expression levels. At D5, we harvested the lungs for histological study and measurement of lung cytokine expression levels (Fig. 4-A). Based on the severity of inflammation, alveolar hemorrhagic necrosis and vessel lesions, a cumulative score from 0 to 10 was calculated and assigned to a grade of severity (0 = normal; 1 = mild; 2 = moderate; 3 = marked and 4 = severe; details in Supplementary Table 1).
Fig. 4.
Bio-pathological effect of oral molnupiravir treatment in a hamster model: lung histology and expression of pulmonary cytokines.Two Groups of 6 hamsters were intranasally infected with 104 TCID50 of virus and a group of 4 hamsters uninfected and untreated was used as control. Animals received Monlupiravir (MOV) or the vehicle (VEH, untreated group) orally, and Favipiravir (FAV) intraperitoneally. Experimental timeline (A). Scoring of pathological changes (B). Expression level of cytokines genes in the lungs at D5 post infection (C). A panel of six cytokines is presented (IL1β not shown). Two-sided statistical analysis was performed using Shapiro-Wilk normality test, Fisher's exact test, Student t-test, Mann-Whitney test (detail in Supplemental Table 4). *, **, ***, and **** represent a significant decrease relative to the vehicle (VEH) group (panel B and Panel C when placed above the symbols) or an increase relative to the untreated/uninfected group (panel C when placed below the symbols), with associated p-values falling within the ranges of 0.01–0.05, 0.001–0.01, 0.0001–0.001, and <0.0001, respectively.
When comparing the cumulative pathological scores of the untreated and treated groups, no significant difference could be demonstrated for the MOV-treated animals, whereas treatment with FAV resulted in a significantly different score (p = 0.0112) (Fig. 4-B).
Regarding the measurement of pulmonary cytokine expression levels (Fig. 4-C), infection with SARS-CoV-2 in hamsters (green triangle) caused an increase in 6 of the 7 cytokines analyzed compared to uninfected and untreated animals (red triangle): CCL5, CxCL10, IFN, IL6, IL10 and TNF (p value range from 0.0207 to less than 0.0001). IL1β expression did not differ between groups (data not shown). MOV was able to significantly decrease the expression of cytokines CCl5 and CxCl10 compared to the vehicle receiving group (p = 0.0022 and p = 0.0007 respectively). For IL10 and TNF, although the reduction was not significant, it did appear to reduce the expression of these cytokines. Overall, the MOV treatment notably improved the cytokine profile compared to the untreated infected group, however, it failed to fully restore the baseline profiles observed in untreated and uninfected animals.
3.3. Mutagenic effect of molnupiravir
To investigate the genomic changes induced by MOV treatment, whole viral genome sequencing was performed on 12 clarified lung homogenates: 6 from hamsters treated with 300 mg/kg/day BID of MOV from D0 and 6 from untreated ones, all selected from the second experiment (Fig. 3-A). Whole viral genome sequencing was also performed on HAE culture supernatants: 3 replicates were analyzed for each group (0.3 μM, 1 μM, 3 μM or untreated).
The average sequencing coverage for each sample ranged from 2725 to 16,076 reads per genomic position and we further analyzed substitutions with frequency ≥1 %. Overall, no majority mutations (mutation frequency >50 %) were detected (Fig. 5-B/G), in both hamster lungs and HAE samples. Minority variants (MVs; mutation frequency <50 %) were homogeneously distributed along the viral genome (Fig. 5-A/F).
As previously described [28] and in order to only study the mutagenic effect induced during HAE and animal infections, all minority mutations previously present in the virus stocks used for infections were not taken into consideration (19 and 6 MVs were found in virus stocks used in the HAE and in vivo experiments respectively; See Supplementary Table 3).
3.3.1. Genomic analysis of SARS-COV-2 in hamster lungs
The mean number of MVs per genome was 12.3 (ranging from 4 to 20) in the untreated group and 29.5 (ranging from 11 to 68) in the group treated with MOV. There was no significant difference (p = 0.0621) between the two groups studied (Fig. 5-C). However, a significant higher number of MVs with a frequency greater than 10 % occurred in the MOV group (p = 0.0065; Fig. 5-D). We then explored which mutational bias was induced by MOV treatment. The mean frequency of transitions-type MVs was higher in the treated group compared to the untreated group, but not significantly (Fig. 5-E, p = 0.0621). We then looked at the individual trends for each of the 4 types of transition. The enhanced number of transitions is associated with an individual increase in each type of transition, with G to A MVs overrepresented and showing a significant increase over the untreated group. (Fig. 5-J/K, p = 0.0086).
3.3.2. Genomic analysis of SARS-COV-2 in HAE
The average number of MVs per genome was 20, 88 and 18.3 for the HAE exposed at 0.3, 1 and 3 μM, respectively, and 4.3 (ranging from 1 to 7) for the non-exposed groups. Exposure of the virus to MOV in HAE leads to an increase in the number of mutations compared with the untreated group, although this is not significant when each group is compared separately with the untreated group (Fig. 5-H, p = 0.0995; p = 0.2964; p = 0.1185 for 0.3, 1 and 3 μM respectively). When all samples exposed to MOV were pooled for analysis (mean number of MVs was 44.1 ranging from 11 to 207), the difference between exposed and non-exposed HAE was significant (Fig. 5-H, p = 0.0091). Analysis of the frequency of transition-type MVs seems to show that transitions were more frequent in the MOV-exposed groups, with mean frequencies of 0.81, 0.97 and 0.89 for the 0.3, 1 and 3 μM-exposed groups respectively, whereas the non-exposed group had a transition frequency of 0.48. The difference was significant only when exposed group were pooled (Fig. 5-I, p = 0.0161). Looking at the individual trends for the 4 transitions, the pooled analysis of exposed and non-exposed HAE showed an individual increase for each type of transition, with significance found for type C to U-MVs (Fig. 5-K/M, p = 0.036).
4. Discussion
In this study, we further evaluated the antiviral activity of MOV against SARS-CoV-2 in various pre-clinical models to better characterise its antiviral properties and its mechanism of action. Two different models were used. First, we used an ex-vivo model of human airway epithelium reconstituted from primary bronchial cells representing the target cells of SARS-CoV-2 infection and consequently of the treatment. This model had been developed to evaluate the antiviral treatment of respiratory viruses, in particular coronaviruses [19], and its use had been particularly useful during the pandemic [[29], [30], [31],36]. Then, an in vivo evaluation was carried out using the Syrian golden hamster model, which was established for the evaluation of antivirals and used extensively during the SARS-CoV-2 pandemic [16,28,29,31]. During this evaluation, we mainly focused on assessing its antiviral activity by measuring viral replication in the different models used, but given its mechanism of action we also considered genomic data to characterise potential alterations in the viral genome that arise during treatment. Finally, we also carried out a pharmacokinetic study to assess MOV exposure in order to better understand the concentration-response relationship and the determinants of antiviral activity.
We were able to show that MOV is active in HAE, which closely mimics the functional, structural and immune characteristics of the human airway epithelium [30]. Activity was observed at concentrations as low as 1 μM, reducing viral excretion at the apical pole in terms of both infectious titres and viral RNA yields. At 3 μM, infectious titres were significantly reduced, akin to the positive control RDV at 10 μM. The more pronounced reduction in infectious titres compared to viral RNA yields, leading to an increase in the particle to infectious unit ratio, aligns with observations for MOV and other mutagenic nucleotide analogues like FAV [28,37]. Our data are consistent and contribute with to sparse literature evaluating MOV in this model [19,21,22,38,39].
In the golden Syrian hamster model a reduction in pulmonary replication of SARS-CoV-2 was present for the three doses studied. Despite substantial within-group variability in lung infectious titres, this reduction was significant. However, as in HAE experiments, this reduction was less pronounced in terms of viral RNA load, resulting in an elevation of the particle to infectious unit ratio. The reduction of viral replication efficacy was not observed with nasal washes probably due to the low diffusion of MOV in the upper respiratory tract, the primary site of viral replication. It should be noted that the infectious titres were not measured in the nasal washes due to the poor results after D2 in this model [33]. These data are consistent with the findings of Abdelnabi et al. and Rosenke et al. who also observed decreases in infectious titres and viral loads in hamster lungs at comparable doses from 150 to 500 mg/kg/day[16,20,40]. In contrast, we were unable to reproduce dose-response relationship or histological improvement. The fact that Rosenke et al., although using groups of comparable size, used higher doses than ours (500 vs 300 mg/kg/d) and that the groups used by Abdelnabi et al. were larger (6–15 animals per group) could be factors that may help to explain these differences.
Our results showed that the antiviral effect of MOV in vivo depends on its prompt administration after infection. Experiments clearly showed that there is a loss of efficacy if treatment initiation is delayed by one day after the infection. This highlights the importance of rapid initiation of treatment in humans after infection, as seen with other antiviral/virus pairs [41]. This aspect was considered in the phase 2 study of the phase 2/3 MOVE OUT trial[42], where the inclusion criteria were up to 7 days after symptom onset, then reduced to 5 days for the phase 3 trial[43], in order to optimise the clinical benefit of antiviral therapy. Despite this, Results of the phase 3 MOVE OUT trial show modest efficacy in reducing the risk of hospitalisation or death, well below what was expected from the interim results [44]. In addition, there was no reduction in the risk of hospitalisation or death in vaccinated patients in the multicentre PANORAMIC trial, which involved more than 26,000 patients, but MOV was associated with reduced time to recovery overall and reduced viral load [45]. In these two studies, it should be noted that most of the patients had a delay of more than 3 days between symptom onset and treatment initiation, suggesting that one of the major determinants of MOV activity is early treatment initiation. Although Rosenke et al. [20] also assessed the effect of delay in treatment relative to infection, they combined for the analysis the groups that received treatment 12 h or 2 h before infection (pre-treatment group). Our evaluation provides further insight into the differences observed between the preventive and pre-emptive administration.
MOV is rapidly and extensively converted by host esterases to NHC, then to an active ribonucleotide analog, NHC-triphosphate, within cells. This pathway is more active in rodents, so the doses used are proportionally higher than in ferrets [17], rhesus macaques [46] and humans [47]. We determined trough plasma and lung concentrations to ensure that animals were properly exposed to MOV. We found a relationship between dose and lung trough concentration, while plasma trough concentrations were stable similar for the different doses. Pulmonary to plasma concentration ratios between 5 and 10 suggest substantial pulmonary accumulation, an important consideration when dealing with viruses having a respiratory tropism. Assuming a lung density of approximately 1 g/mL and a MOV IC50 of 1 μM, pulmonary inhibitory quotients (calculated as the ratio of tissue concentration to IC50) ranged from 0.28 to 0.48, 15 h after the last dose (Supplementary Fig. S7). These data, combined with the apparent pulmonary accumulation, tend to show that pulmonary rather than plasma concentrations are the determinants of antiviral activity.
Since NHC-triphosphate has the ability to bind to cytidine and uridine, thereby increasing the error rate during viral replication, we subsequently conducted a study assessing the genomic evolution of viruses exposed to MOV both ex vivo and in vitro. Our whole-genome sequencing approach, while more time and resource consuming than methods based on partial genome sequencing, provides a complete view of mutagenesis across the entire genome. As expected, we found an increase in the number of minority variants in cells and animals treated with MOV. This increase seems to be more significant in the HAE model than in the hamster model. We can assume that the cell exposure in the HAE model is greater and more stable over time than in the animal model, so that mutations are more likely to occur. A difference in the dynamic of viral replication between these two models could also be considered. It is also important to note the high variability observed within groups. This may be due to the randomness of mutation events, but also to differences in exposure between replicates, especially in the animal model. An important finding is the high predominance of transition-type mutations, particularly G to A and C to U mutations, which is consistent with the mechanism of action of MOV [25,26]. These data suggest that under the conditions of our experiments, MOV exerts its antiviral activity by inducing an error catastrophe in the replicating SARS-CoV-2 genome. This mechanism of action leverages the innate susceptibility of RNA viruses to high mutation rates and explains why MOV is also active against other RNA viruses with similar genetics finding, like influenza virus [15] or alphaviruses [48]. The analysis of the characteristics of the viral genomes of patients treated with MOV in the AGILE Phase IIa trial [49] showed similar patterns to those seen here (overrepresentation of G to A and C to U transitions). And on a larger scale, Sanderson and his colleagues have published the results of their analysis of SARS-CoV-2 genomic databases, which revealed the mutational motifs characteristic of MOV (notably G to A mutation) in post-marketing sequences [50]. These data may raise concerns that the use of MOV may be the cause of the emergence of new variants of SARS-CoV-2 or MOV-resistant strains. Given the random nature of mutations and the fact that their accumulation in the genome leads to a reduction in infectivity, the probability of the emergence of new SARS-COV-2 variants following the use of MOV seems not very different from the natural probability of occurrence. Following submission of our manuscript, an article with similar study in a Syrian hamster model was released [51]. In this study, they carried out a genomic analysis of the data obtained in previous research of Abdelnabi et al. [16,40]. Similar to our study, they found an accumulation of low frequency mutations, promoting the transition from C to U and G to A variants, without significant gains in higher frequency variants. They conclude that there is no conclusive evidence of either the emergence of antiviral resistance or the systematic emergence of more suitable variants that would argue against the clinical use of mutagenic drugs. Finally, to our knowledge, no MOV-resistant strain has yet been described. However, approvals are mainly limited to immunocompromised patients, and this population, characterized by impaired viral clearance, may not be the most appropriate for this treatment.
It is important to recognize several limitations of this study. The sample sizes used, although adapted to the main objective of the study (ie assessing antiviral activity), may limit the statistical power to detect certain effects. This is particularly true for the genomic study, where the random occurrence of minor variants combined with the small number of samples results in significant variability in the results. Research showing the reduced lower respiratory effect of the omicron variant [52] highlights another limitation: our use of the ancestral D614G strain. Our study confirmed the efficacy of MOV in reducing infectious titres in the lung. However, we did not observe a significant reduction in viral RNA levels in nasal washings. This change in tropism could alter the efficacy of MOV in this model, but other studies have shown that MOV remains effective against the more recent variants of SARS-CoV-2 [53,54]. In the pharmacokinetic study, we measured NHC, the intracellular concentration of NHC-TP, the ultimate active form, could provide valuable information. However, this determination is very difficult to perform, especially in small animals, because it requires large sample volumes and thus a higher number of animals.
5. Conclusion
MOV demonstrate antiviral activity against SARS-CoV-2 ex-vivo and in vivo, leading to a decrease in the infectious titres. Its prompt administration and accumulation in the lungs are factors that may influence its activity in vivo. This activity is paralleled by the generation of minority variants in the viral genome, which are related to the ability of NHC triphosphate to be incorporated into nascent RNA instead of cytidine triphosphate or uridine triphosphate. This study provides additional data to support the existing evidence for the efficacy of MOV both ex-vivo and in vivo and the determinants of its efficacy against SARS-CoV-2 raising questions about its use for the current indication and its place in the COVID-19 therapeutic strategy. To our knowledge, this is the first evaluation of the impact of SARS-CoV-2 exposure to MOV on genomic evolution in an HAE model. Besides, his simultaneous comparison of HAE and in vivo genomic data, demonstrating the strong relevance of the HAE model for future investigations in this area.
Data availability statement
All of the data generated or analyzed during this study are included in this published article or available on demand.
CRediT authorship contribution statement
Paul-Rémi Petit: Writing – review & editing, Writing – original draft, Visualization, Methodology, Investigation, Formal analysis, Conceptualization. Franck Touret: Writing – review & editing, Writing – original draft, Visualization, Methodology, Investigation, Formal analysis, Conceptualization. Jean-Sélim Driouich: Writing – review & editing, Writing – original draft, Visualization, Methodology, Investigation, Formal analysis, Conceptualization. Maxime Cochin: Investigation, Formal analysis. Léa Luciani: Investigation, Formal analysis. Ornéllie Bernadin: Investigation, Formal analysis. Caroline Laprie: Investigation, Formal analysis. Géraldine Piorkowski: Formal analysis. Laurent Fraisse: Writing – review & editing, Funding acquisition. Peter Sjö: Writing – review & editing, Funding acquisition. Charles E. Mowbray: Writing – review & editing, Funding acquisition. Fanny Escudié: Writing – review & editing, Funding acquisition, Conceptualization. Ivan Scandale: Writing – review & editing, Funding acquisition, Conceptualization. Eric Chatelain: Writing – review & editing, Funding acquisition, Conceptualization. Xavier de Lamballerie: Writing – review & editing, Resources, Funding acquisition. Caroline Solas: Writing – review & editing, Writing – original draft, Resources, Methodology, Funding acquisition, Conceptualization. Antoine Nougairède: Writing – review & editing, Writing – original draft, Visualization, Resources, Methodology, Investigation, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Rayane Amaral (UVE; Marseille), Hawa Sophia Bouzidi (UVE; Marseille), Magali Gilles (UVE; Marseille), Gregory Moureau (UVE; Marseille), Camille Placidi (UVE; Marseille) and Madeleine Giocanti (Laboratoire de Pharmacocinétique et Toxicologie, Hôpital de la Timone, Marseille) for their valuable technical contribution. We thank Pr. Drosten and Pr. Drexler for providing the SARS-CoV-2 strain through the European Research infrastructure EVA GLOBAL.
This work was supported by the ANRS-MIE (PRI projects of the EMERGEN research program), by European Virus Archive Global (EVA GLOBAL) funded by the European Union's Horizon 2020 research and innovation program under grant agreement No. 871029 and by DNDi under support by the Wellcome Trust Grant ref: 222489/Z/21/Z through the COVID-19 Therapeutics Accelerator. A part of the work was done on the Aix Marseille University antivirals platform “AD2P”.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e30862.
Funding acquisition, LF, CEM, EC, XdL, CS and AN.
Graphical representation of the linear regression between dosing regimen and trough pulmonary concentrations. For each dose, the coloured point represents the geometric mean of the concentration associated with the geometric SD.
Data represent mean ± SD for each different group of treatment.
Contributor Information
Paul-Rémi Petit, Email: paul.petit@ap-hm.fr.
Antoine Nougairède, Email: antoine.nougairede@univ-amu.fr.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Tobinick E.L. The value of drug repositioning in the current pharmaceutical market. Drug News Perspect. 2009;22 doi: 10.1358/dnp.2009.22.2.1303818. 119. [DOI] [PubMed] [Google Scholar]
- 2.Li G., Hilgenfeld R., Whitley R., De Clercq E. Therapeutic strategies for COVID-19: progress and lessons learned. Nat. Rev. Drug Discov. 2023;22:449. doi: 10.1038/s41573-023-00672-y. 475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simonis A., Theobald S.J., Fätkenheuer G., Rybniker J., Malin J.J. A comparative analysis of remdesivir and other repurposed antivirals against SARS‐CoV‐2. EMBO Mol. Med. 2021;13 doi: 10.15252/emmm.202013105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nili A., Farbod A., Neishabouri A., Mozafarihashjin M., Tavakolpour S., Mahmoudi H. Remdesivir: a beacon of hope from Ebola virus disease to COVID ‐19. Rev. Med. Virol. 2020;30:1–13. doi: 10.1002/rmv.2133. [DOI] [PubMed] [Google Scholar]
- 5.Popowska E., Janion C. N4-hydroxycytidine — a new mutagen of a base analogue type. Biochem. Biophys. Res. Commun. 1974;56:459. doi: 10.1016/0006-291X(74)90864-X. 466. [DOI] [PubMed] [Google Scholar]
- 6.Stuyver L.J., Whitaker T., McBrayer T.R., Hernandez-Santiago B.I., Lostia S., Tharnish P.M., Ramesh M., Chu C.K., Jordan R., Shi J., Rachakonda S., Watanabe K.A., Otto M.J., Schinazi R.F. Ribonucleoside analogue that blocks replication of bovine viral diarrhea and hepatitis C viruses in culture. Antimicrob. Agents Chemother. 2003;47:244–254. doi: 10.1128/AAC.47.1.244-254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pyrc K., Bosch B.J., Berkhout B., Jebbink M.F., Dijkman R., Rottier P., van der Hoek L. Inhibition of human coronavirus NL63 infection at early stages of the replication cycle. Antimicrob. Agents Chemother. 2006;50:2000–2008. doi: 10.1128/AAC.01598-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnard D.L., Hubbard V.D., Burton J., Smee D.F., Morrey J.D., Otto M.J., Sidwell R.W. Inhibition of severe acute respiratory syndrome-associated coronavirus (SARSCoV) by calpain inhibitors and β-D-N 4 -hydroxycytidine. Antivir. Chem. Chemother. 2004;15:15–22. doi: 10.1177/095632020401500102. [DOI] [PubMed] [Google Scholar]
- 9.Yoon J.-J., Toots M., Lee S., Lee M.-E., Ludeke B., Luczo J.M., Ganti K., Cox R.M., Sticher Z.M., Edpuganti V., Mitchell D.G., Lockwood M.A., Kolykhalov A.A., Greninger A.L., Moore M.L., Painter G.R., Lowen A.C., Tompkins S.M., Fearns R., Natchus M.G., Plemper R.K. Orally efficacious broad-spectrum ribonucleoside analog inhibitor of influenza and respiratory syncytial viruses. Antimicrob. Agents Chemother. 2018;62 doi: 10.1128/AAC.00766-18. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costantini V.P., Whitaker T., Barclay L., Lee D., McBrayer T.R., Schinazi R.F., Vinjé J. Antiviral activity of nucleoside analogues against norovirus. Antivir. Ther. 2012;17:981–991. doi: 10.3851/IMP2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Painter G.R., Bowen R.A., Bluemling G.R., DeBergh J., Edpuganti V., Gruddanti P.R., Guthrie D.B., Hager M., Kuiper D.L., Lockwood M.A., Mitchell D.G., Natchus M.G., Sticher Z.M., Kolykhalov A.A. The prophylactic and therapeutic activity of a broadly active ribonucleoside analog in a murine model of intranasal venezuelan equine encephalitis virus infection. Antivir. Res. 2019;171 doi: 10.1016/j.antiviral.2019.104597. [DOI] [PubMed] [Google Scholar]
- 12.Kozlovskaya L.I., Golinets A.D., Eletskaya A.A., Orlov A.A., Palyulin V.A., Kochetkov S.N., Alexandrova L.A., Osolodkin D.I. Selective inhibition of enterovirus A species members' reproduction by furano[2, 3- d ]pyrimidine nucleosides revealed by antiviral activity profiling against (+)ssRNA viruses. ChemistrySelect. 2018;3:2321–2325. doi: 10.1002/slct.201703052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehteshami M., Tao S., Zandi K., Hsiao H.-M., Jiang Y., Hammond E., Amblard F., Russell O.O., Merits A., Schinazi R.F. Characterization of β-d-N4-hydroxycytidine as a novel inhibitor of Chikungunya virus. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.02395-16. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reynard O., Nguyen X.-N., Alazard-Dany N., Barateau V., Cimarelli A., Volchkov V. Identification of a new ribonucleoside inhibitor of Ebola virus replication. Viruses. 2015;7:6233–6240. doi: 10.3390/v7122934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toots M., Yoon J.-J., Cox R.M., Hart M., Sticher Z.M., Makhsous N., Plesker R., Barrena A.H., Reddy P.G., Mitchell D.G., Shean R.C., Bluemling G.R., Kolykhalov A.A., Greninger A.L., Natchus M.G., Painter G.R., Plemper R.K. Characterization of orally efficacious influenza drug with high resistance barrier in ferrets and human airway epithelia. Sci. Transl. Med. 2019;11:eaax5866. doi: 10.1126/scitranslmed.aax5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdelnabi R., Foo C.S., Kaptein S.J.F., Zhang X., Do T.N.D., Langendries L., Vangeel L., Breuer J., Pang J., Williams R., Vergote V., Heylen E., Leyssen P., Dallmeier K., Coelmont L., Chatterjee A.K., Mols R., Augustijns P., De Jonghe S., Jochmans D., Weynand B., Neyts J. The combined treatment of Molnupiravir and Favipiravir results in a potentiation of antiviral efficacy in a SARS-CoV-2 hamster infection model. EBioMedicine. 2021;72 doi: 10.1016/j.ebiom.2021.103595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox R.M., Wolf J.D., Plemper R.K. Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets. Nat Microbiol. 2021;6:11–18. doi: 10.1038/s41564-020-00835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gidari A., Sabbatini S., Schiaroli E., Bastianelli S., Pierucci S., Busti C., Comez L., Libera V., Macchiarulo A., Paciaroni A., Vicenti I., Zazzi M., Francisci D. The combination of molnupiravir with nirmatrelvir or GC376 has a synergic role in the inhibition of SARS-CoV-2 replication in vitro. Microorganisms. 2022;10:1475. doi: 10.3390/microorganisms10071475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schäfer A., Dinnon K.H., Stevens L.J., Chappell J.D., Lu X., Hughes T.M., George A.S., Hill C.S., Montgomery S.A., Brown A.J., Bluemling G.R., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G., Harcourt J., Tamin A., Thornburg N.J., Swanstrom R., Denison M.R., Baric R.S. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenke K., Hansen F., Schwarz B., Feldmann F., Haddock E., Rosenke R., Barbian K., Meade-White K., Okumura A., Leventhal S., Hawman D.W., Ricotta E., Bosio C.M., Martens C., Saturday G., Feldmann H., Jarvis M.A. Orally delivered MK-4482 inhibits SARS-CoV-2 replication in the Syrian hamster model. Nat. Commun. 2021;12:2295. doi: 10.1038/s41467-021-22580-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Do T.N.D., Donckers K., Vangeel L., Chatterjee A.K., Gallay P.A., Bobardt M.D., Bilello J.P., Cihlar T., De Jonghe S., Neyts J., Jochmans D. A robust SARS-CoV-2 replication model in primary human epithelial cells at the air liquid interface to assess antiviral agents. Antivir. Res. 2021;192 doi: 10.1016/j.antiviral.2021.105122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lieber C.M., Cox R.M., Sourimant J., Wolf J.D., Juergens K., Phung Q., Saindane M.T., Smith M.K., Sticher Z.M., Kalykhalov A.A., Natchus M.G., Painter G.R., Sakamoto K., Greninger A.L., Plemper R.K. SARS-CoV-2 VOC type and biological sex affect molnupiravir efficacy in severe COVID-19 dwarf hamster model. Nat. Commun. 2022;13:4416. doi: 10.1038/s41467-022-32045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeong J.H., Chokkakula S., Min S.C., Kim B.K., Choi W.-S., Oh S., Yun Y.S., Kang D.H., Lee O.-J., Kim E.-G., Choi J.-H., Lee J.-Y., Choi Y.K., Baek Y.H., Song M.-S. Combination therapy with nirmatrelvir and molnupiravir improves the survival of SARS-CoV-2 infected mice. Antivir. Res. 2022;208 doi: 10.1016/j.antiviral.2022.105430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wahl A., Gralinski L.E., Johnson C.E., Yao W., Kovarova M., Dinnon K.H., Liu H., Madden V.J., Krzystek H.M., De C., White K.K., Gully K., Schäfer A., Zaman T., Leist S.R., Grant P.O., Bluemling G.R., Kolykhalov A.A., Natchus M.G., Askin F.B., Painter G., Browne E.P., Jones C.D., Pickles R.J., Baric R.S., Garcia J.V. SARS-CoV-2 infection is effectively treated and prevented by EIDD-2801. Nature. 2021;591:451–457. doi: 10.1038/s41586-021-03312-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon C.J., Tchesnokov E.P., Schinazi R.F., Götte M. Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template. J. Biol. Chem. 2021;297 doi: 10.1016/j.jbc.2021.100770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kabinger F., Stiller C., Schmitzová J., Dienemann C., Kokic G., Hillen H.S., Höbartner C., Cramer P. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat. Struct. Mol. Biol. 2021;28:740–746. doi: 10.1038/s41594-021-00651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dyer O. Covid-19: FDA expert panel recommends authorising molnupiravir but also voices concerns. BMJ. 2021;n2984 doi: 10.1136/bmj.n2984. [DOI] [PubMed] [Google Scholar]
- 28.Driouich J.-S., Cochin M., Lingas G., Moureau G., Touret F., Petit P.-R., Piorkowski G., Barthélémy K., Laprie C., Coutard B., Guedj J., de Lamballerie X., Solas C., Nougairède A. Favipiravir antiviral efficacy against SARS-CoV-2 in a hamster model. Nat. Commun. 2021;12:1735. doi: 10.1038/s41467-021-21992-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Driouich J.-S., Cochin M., Touret F., Petit P.-R., Gilles M., Moureau G., Barthélémy K., Laprie C., Wattanakul T., Chotsiri P., Hoglund R.M., Tarning J., Fraisse L., Sjö P., Mowbray C.E., Escudié F., Scandale I., Chatelain E., de Lamballerie X., Solas C., Nougairède A. Pre-clinical evaluation of antiviral activity of nitazoxanide against SARS-CoV-2. EBioMedicine. 2022;82 doi: 10.1016/j.ebiom.2022.104148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pizzorno A., Padey B., Julien T., Trouillet-Assant S., Traversier A., Errazuriz-Cerda E., Fouret J., Dubois J., Gaymard A., Lescure F.-X., Dulière V., Brun P., Constant S., Poissy J., Lina B., Yazdanpanah Y., Terrier O., Rosa-Calatrava M. Characterization and treatment of SARS-CoV-2 in nasal and bronchial human airway epithelia. Cell Reports Medicine. 2020;1 doi: 10.1016/j.xcrm.2020.100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Touret F., Driouich J.-S., Cochin M., Petit P.R., Gilles M., Barthélémy K., Moureau G., Mahon F.-X., Malvy D., Solas C., de Lamballerie X., Nougairède A. Preclinical evaluation of Imatinib does not support its use as an antiviral drug against SARS-CoV-2. Antivir. Res. 2021;193 doi: 10.1016/j.antiviral.2021.105137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reed L.J., Muench H. A simple method of estimating fifty per cent ENDPOINTS12. Am. J. Epidemiol. 1938;27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 33.Cochin M., Luciani L., Touret F., Driouich J.-S., Petit P.-R., Moureau G., Baronti C., Laprie C., Thirion L., Maes P., Boudewijns R., Neyts J., De Lamballerie X., Nougairède A. The SARS-CoV-2 Alpha variant exhibits comparable fitness to the D614G strain in a Syrian hamster model. Commun. Biol. 2022;5:225. doi: 10.1038/s42003-022-03171-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Melo G.D., Lazarini F., Larrous F., Feige L., Kornobis E., Levallois S., Marchio A., Kergoat L., Hardy D., Cokelaer T., Pineau P., Lecuit M., Lledo P., Changeux J., Bourhy H. Attenuation of clinical and immunological outcomes during SARS‐CoV‐2 infection by ivermectin. EMBO Mol. Med. 2021;13 doi: 10.15252/emmm.202114122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cochin M., Touret F., Driouich J.-S., Moureau G., Petit P.-R., Laprie C., Solas C., de Lamballerie X., Nougairède A. Hydroxychloroquine and azithromycin used alone or combined are not effective against SARS-CoV-2 ex vivo and in a hamster model. Antivir. Res. 2022;197 doi: 10.1016/j.antiviral.2021.105212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maisonnasse P., Guedj J., Contreras V., Behillil S., Solas C., Marlin R., Naninck T., Pizzorno A., Lemaitre J., Gonçalves A., Kahlaoui N., Terrier O., Fang R.H.T., Enouf V., Dereuddre-Bosquet N., Brisebarre A., Touret F., Chapon C., Hoen B., Lina B., Calatrava M.R., van der Werf S., de Lamballerie X., Le Grand R. Hydroxychloroquine use against SARS-CoV-2 infection in non-human primates. Nature. 2020;585:584–587. doi: 10.1038/s41586-020-2558-4. [DOI] [PubMed] [Google Scholar]
- 37.Kaptein S.J.F., Jacobs S., Langendries L., Seldeslachts L., ter Horst S., Liesenborghs L., Hens B., Vergote V., Heylen E., Barthelemy K., Maas E., De Keyzer C., Bervoets L., Rymenants J., Van Buyten T., Zhang X., Abdelnabi R., Pang J., Williams R., Thibaut H.J., Dallmeier K., Boudewijns R., Wouters J., Augustijns P., Verougstraete N., Cawthorne C., Breuer J., Solas C., Weynand B., Annaert P., Spriet I., Vande Velde G., Neyts J., Rocha-Pereira J., Delang L. Favipiravir at high doses has potent antiviral activity in SARS-CoV-2−infected hamsters, whereas hydroxychloroquine lacks activity. Proc. Natl. Acad. Sci. U.S.A. 2020;117:26955–26965. doi: 10.1073/pnas.2014441117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jonsdottir H.R., Siegrist D., Julien T., Padey B., Bouveret M., Terrier O., Pizzorno A., Huang S., Samby K., Wells T.N.C., Boda B., Rosa-Calatrava M., Engler O.B., Constant S. Molnupiravir combined with different repurposed drugs further inhibits SARS-CoV-2 infection in human nasal epithelium in vitro. Biomed. Pharmacother. 2022;150 doi: 10.1016/j.biopha.2022.113058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dichtl S., Diem G., Jäger M., Zaderer V., Lupoli G., Dächert C., Muenchhoff M., Graf A., Blum H., Keppler O.T., Lass-Flörl C., Weiss G., Wilflingseder D., Posch W. Antiviral drugs block replication of highly immune-evasive Omicron subvariants ex vivo, but fail to reduce tissue inflammation. Antivir. Res. 2023;213 doi: 10.1016/j.antiviral.2023.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abdelnabi R., Foo C.S., De Jonghe S., Maes P., Weynand B., Neyts J. Molnupiravir inhibits replication of the emerging SARS-CoV-2 variants of concern in a hamster infection model. J. Infect. Dis. 2021;224:749–753. doi: 10.1093/infdis/jiab361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muthuri S.G., Venkatesan S., Myles P.R., Leonardi-Bee J., Al Khuwaitir T.S.A., Al Mamun A., Anovadiya A.P., Azziz-Baumgartner E., Báez C., Bassetti M., Beovic B., Bertisch B., Bonmarin I., Booy R., Borja-Aburto V.H., Burgmann H., Cao B., Carratala J., Denholm J.T., Dominguez S.R., Duarte P.A.D., Dubnov-Raz G., Echavarria M., Fanella S., Gao Z., Gérardin P., Giannella M., Gubbels S., Herberg J., Iglesias A.L.H., Hoger P.H., Hu X., Islam Q.T., Jiménez M.F., Kandeel A., Keijzers G., Khalili H., Knight M., Kudo K., Kusznierz G., Kuzman I., Kwan A.M.C., Amine I.L., Langenegger E., Lankarani K.B., Leo Y.-S., Linko R., Liu P., Madanat F., Mayo-Montero E., McGeer A., Memish Z., Metan G., Mickiene A., Mikić D., Mohn K.G.I., Moradi A., Nymadawa P., Oliva M.E., Ozkan M., Parekh D., Paul M., Polack F.P., Rath B.A., Rodríguez A.H., Sarrouf E.B., Seale A.C., Sertogullarindan B., Siqueira M.M., Skręt-Magierło J., Stephan F., Talarek E., Tang J.W., To K.K.W., Torres A., Törün S.H., Tran D., Uyeki T.M., Van Zwol A., Vaudry W., Vidmar T., Yokota R.T.C., Zarogoulidis P., Nguyen-Van-Tam J.S. Effectiveness of neuraminidase inhibitors in reducing mortality in patients admitted to hospital with influenza A H1N1pdm09 virus infection: a meta-analysis of individual participant data. Lancet Respir. Med. 2014;2:395–404. doi: 10.1016/S2213-2600(14)70041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caraco Y., Crofoot G.E., Moncada P.A., Galustyan A.N., Musungaie D.B., Payne B., Kovalchuk E., Gonzalez A., Brown M.L., Williams-Diaz A., Gao W., Strizki J.M., Grobler J., Du J., Assaid C.A., Paschke A., Butterton J.R., Johnson M.G., De Anda C. Phase 2/3 trial of molnupiravir for treatment of covid-19 in nonhospitalized adults. NEJM Evidence. 2022;1 doi: 10.1056/EVIDoa2100043. [DOI] [PubMed] [Google Scholar]
- 43.Jayk Bernal A., Gomes da Silva M.M., Musungaie D.B., Kovalchuk E., Gonzalez A., Delos Reyes V., Martín-Quirós A., Caraco Y., Williams-Diaz A., Brown M.L., Du J., Pedley A., Assaid C., Strizki J., Grobler J.A., Shamsuddin H.H., Tipping R., Wan H., Paschke A., Butterton J.R., Johnson M.G., De Anda C. Molnupiravir for oral treatment of covid-19 in nonhospitalized patients. N. Engl. J. Med. 2022;386:509–520. doi: 10.1056/NEJMoa2116044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merck and Ridgeback Biotherapeutics . Press release; 2021. https://www.merck.com/news/merck-and-ridgebacks-investigational-oral-antiviral-molnupiravir-reduced-the-risk-of-hospitalization-or-death-by-approximately-50-percent-compared-to-placebo-for-patients-with-mild-or-moderat/ (Merck and Ridgeback’s Investigational Oral Antiviral Molnupiravir Reduced the Risk of Hospitalisation or Death by Approximately 50 Percent Compared to Placebo for Patients with Mild or Moderate COVID-19 in Positive Interim Analysis of Phase 3 Study.). [Google Scholar]
- 45.Butler C.C., Hobbs F.D.R., Gbinigie O.A., Rahman N.M., Hayward G., Richards D.B., Dorward J., Lowe D.M., Standing J.F., Breuer J., Khoo S., Petrou S., Hood K., Nguyen-Van-Tam J.S., Patel M.G., Saville B.R., Marion J., Ogburn E., Allen J., Rutter H., Francis N., Thomas N.P.B., Evans P., Dobson M., Madden T.-A., Holmes J., Harris V., Png M.E., Lown M., Van Hecke O., Detry M.A., Saunders C.T., Fitzgerald M., Berry N.S., Mwandigha L., Galal U., Mort S., Jani B.D., Hart N.D., Ahmed H., Butler D., McKenna M., Chalk J., Lavallee L., Hadley E., Cureton L., Benysek M., Andersson M., Coates M., Barrett S., Bateman C., Davies J.C., Raymundo-Wood I., Ustianowski A., Carson-Stevens A., Yu L.-M., Little P., Agyeman A.A., Ahmed T., Allcock D., Beltran-Martinez A., Benedict O.E., Bird N., Brennan L., Brown J., Burns G., Butler M., Cheng Z., Danson R., De Kare-Silver N., Dhasmana D., Dickson J., Engamba S., Fisher S., Fox R., Frost E., Gaunt R., Ghosh S., Gilkar I., Goodman A., Granier S., Howell A., Hussain I., Hutchinson S., Imlach M., Irving G., Jacobsen N., Kennard J., Khan U., Knox K., Krasucki C., Law T., Lee R., Lester N., Lewis D., Lunn J., Mackintosh C.I., Mathukia M., Moore P., Morton S., Murphy D., Nally R., Ndukauba C., Ogundapo O., Okeke H., Patel A., Patel K., Penfold R., Poonian S., Popoola O., Pora A., Prasad V., Prasad R., Razzaq O., Richardson S., Royal S., Safa A., Sehdev S., Sevenoaks T., Shah D., Sheikh A., Short V., Sidhu B.S., Singh I., Soni Y., Thalasselis C., Wilson P., Wingfield D., Wong M., Woodall M.N.J., Wooding N., Woods S., Yong J., Yongblah F., Zafar A. Molnupiravir plus usual care versus usual care alone as early treatment for adults with COVID-19 at increased risk of adverse outcomes (PANORAMIC): an open-label, platform-adaptive randomised controlled trial. Lancet. 2023;401:281–293. doi: 10.1016/S0140-6736(22)02597-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson D.M., Brasel T., Massey S., Garron T., Grimes M., Smith J., Torres M., Wallace S., Villasante-Tezanos A., Beasley D.W., Comer J.E. Evaluation of molnupiravir (EIDD-2801) efficacy against SARS-CoV-2 in the rhesus macaque model. Antivir. Res. 2023;209 doi: 10.1016/j.antiviral.2022.105492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khoo S.H., Fitzgerald R., Fletcher T., Ewings S., Jaki T., Lyon R., Downs N., Walker L., Tansley-Hancock O., Greenhalf W., Woods C., Reynolds H., Marwood E., Mozgunov P., Adams E., Bullock K., Holman W., Bula M.D., Gibney J.L., Saunders G., Corkhill A., Hale C., Thorne K., Chiong J., Condie S., Pertinez H., Painter W., Wrixon E., Johnson L., Yeats S., Mallard K., Radford M., Fines K., Shaw V., Owen A., Lalloo D.G., Jacobs M., Griffiths G. Optimal dose and safety of molnupiravir in patients with early SARS-CoV-2: a Phase I, open-label, dose-escalating, randomized controlled study. J. Antimicrob. Chemother. 2021;76:3286–3295. doi: 10.1093/jac/dkab318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Urakova N., Kuznetsova V., Crossman D.K., Sokratian A., Guthrie D.B., Kolykhalov A.A., Lockwood M.A., Natchus M.G., Crowley M.R., Painter G.R., Frolova E.I., Frolov I. β-d-N4-Hydroxycytidine is a potent anti-alphavirus compound that induces a high level of mutations in the viral genome. J. Virol. 2018;92 doi: 10.1128/JVI.01965-17. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donovan-Banfield I., Penrice-Randal R., Goldswain H., Rzeszutek A.M., Pilgrim J., Bullock K., Saunders G., Northey J., Dong X., Ryan Y., Reynolds H., Tetlow M., Walker L.E., FitzGerald R., Hale C., Lyon R., Woods C., Ahmad S., Hadjiyiannakis D., Periselneris J., Knox E., Middleton C., Lavelle-Langham L., Shaw V., Greenhalf W., Edwards T., Lalloo D.G., Edwards C.J., Darby A.C., Carroll M.W., Griffiths G., Khoo S.H., Hiscox J.A., Fletcher T. Characterisation of SARS-CoV-2 genomic variation in response to molnupiravir treatment in the AGILE Phase IIa clinical trial. Nat. Commun. 2022;13:7284. doi: 10.1038/s41467-022-34839-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanderson T., Hisner R., Donovan-Banfield I., Hartman H., Løchen A., Peacock T.P., Ruis C. A molnupiravir-associated mutational signature in global SARS-CoV-2 genomes. Nature. 2023 doi: 10.1038/s41586-023-06649-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Illingworth C.J.R., Guerra-Assuncao J.A., Gregg S., Charles O., Pang J., Roy S., Abdelnabi R., Neyts J., Breuer J. Genetic consequences of effective and suboptimal dosing with mutagenic drugs in a hamster model of SARS-CoV-2 infection. Virus Evolution. 2024;10:veae001. doi: 10.1093/ve/veae001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Halfmann, P.J. Iida S., Iwatsuki-Horimoto K., Maemura T., Kiso M., Scheaffer S.M., Darling T.L., Joshi A., Loeber S., Singh G., Foster S.L., Ying B., Case J.B., Chong Z., Whitener B., Moliva J., Floyd K., Ujie M., Nakajima N., Ito M., Wright R., Uraki R., Warang P., Gagne M., Li R., Sakai-Tagawa Y., Liu Y., Larson D., Osorio J.E., Hernandez-Ortiz J.P., Henry A.R., Ciuoderis K., Florek K.R., Patel M., Odle A., Wong L.-Y.R., Bateman A.C., Wang Z., Edara V.-V., Chong Z., Franks J., Jeevan T., Fabrizio T., DeBeauchamp J., Kercher L., Seiler P., Gonzalez-Reiche A.S., Sordillo E.M., Chang L.A., van Bakel H., Simon V., Consortium Mount Sinai Pathogen Surveillance (PSP) study group, Douek D.C., Sullivan N.J., Thackray L.B., Ueki H., Yamayoshi S., Imai M., Perlman S., Webby R.J., Seder R.A., Suthar M.S., García-Sastre A., Schotsaert M., Suzuki T., Boon A.C.M., Diamond M.S., Kawaoka Y. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603:687–692. doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosenke K., Okumura A., Lewis M.C., Feldmann F., Meade-White K., Bohler W.F., Griffin A., Rosenke R., Shaia C., Jarvis M.A., Feldmann H. Molnupiravir inhibits SARS-CoV-2 variants including Omicron in the hamster model. JCI Insight. 2022;7 doi: 10.1172/jci.insight.160108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vangeel L., Chiu W., De Jonghe S., Maes P., Slechten B., Raymenants J., André E., Leyssen P., Neyts J., Jochmans D. Remdesivir, Molnupiravir and Nirmatrelvir remain active against SARS-CoV-2 Omicron and other variants of concern. Antivir. Res. 2022;198 doi: 10.1016/j.antiviral.2022.105252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data generated or analyzed during this study are included in this published article or available on demand.