Abstract
Nature possesses an inexhaustible reservoir of agents that could serve as alternatives to combat the growing threat of antimicrobial resistance (AMR). While some of the most effective drugs for treating bacterial infections originate from natural sources, they have predominantly been derived from fungal and bacterial species. However, a substantial body of literature is available on the promising antibacterial properties of plant-derived compounds. In this comprehensive review, we address the major challenges associated with the discovery and development of plant-derived antimicrobial compounds, which have acted as obstacles preventing their clinical use. These challenges encompass limited sourcing, the risk of agent rediscovery, suboptimal drug metabolism, and pharmacokinetics (DMPK) properties, as well as a lack of knowledge regarding molecular targets and mechanisms of action, among other pertinent issues. Our review underscores the significance of these challenges and their implications in the quest for the discovery and development of effective plant-derived antimicrobial agents. Through a critical examination of the current state of research, we give valuable insights that will advance our understanding of these classes of compounds, offering potential solutions to the global crisis of AMR.
© 2017 Elsevier Inc. All rights reserved.
Keywords: Plants, Antibacterials, Resistances, Gram-negative bacteria
Graphical abstract
This review focuses on the translational challenges of plant-derived antibacterials and offers suggestions that could potentially lead to more promising plant-derived antimicrobials.
Abbreviations
- ADMET
Absorption, Distribution, Metabolism, Elimination, Toxicity
- AMP
Antimicrobial peptides
- AMR
Antimicrobial resistance
- BCE
Before the common era
- BGC
Biosynthetic gene clusters
- CBC
Cannabichromene
- CBD
Cannabidiol
- CBG
Cannabigerol
- CBN
Cannabinol
- DMPK
Drug metabolism and pharmacokinetics
- GNPS
Global Natural Products Social Molecular Networking
- LPS
Lipopolysaccharide
- MADByTE
Metabolomics and Dereplication by Two-Dimensional Experiments
- MDR-TB
Multidrug-resistant tuberculosis
- MOA
Mechanism of action
- MRCNS
Methicillin-resistant coagulase-negative staphylococci
- MRSA
Methicillin-resistant Staphylococcus aureus
- MSCNS
Methicillin-susceptible coagulase-negative staphylococci
- MSSA
Methicillin-susceptible Staphylococcus aureus
- Mtb
Mycobacterium tuberculosis
- NAD
Nicotinamide adenine dinucleotide
- NP
Natural Products
- PBP2
Penicillin-binding protein 2a
- PSM
Plant's secondary metabolites
- RND
Resistance-nodulation-cell division (RND)
- SAR
Structure-activity relationship
- SMAR
Small Molecule Accurate Recognition Technology
- SNEDDS
Self-nanoemulsifying drug delivery systems
- SPI-1 S
Pathogenicity island-1
- SPR
Structure property relationship
- T3SS
Type III secretion system
- TCMs
Traditional Chinese Medicines
- THC
Δ9-Tetrahydrocannabinol
1. Introduction
From time immemorial, natural products (NPs) have been the veritable source of human remedies. One of the earliest writings on the use of NPs as medicine, Pen T'Sao, was written in 2500 BCE by Emperor Shen Nung. This work detailed 365 drugs of plant origins [1]. The work of Hippocrates in 459–370 BCE contained over 300 plants [2]. It is known that the ancient Egyptians used a poultice of mouldy bread to treat infected wounds. Similarly, the ancient Chinese treated boils and foot infections with mouldy soybean curd. In the Middle Ages, honey was used to treat arrow wounds [3]. Today, 80 % of the world population still rely on plant-derived medicine [4]. Moreover, about 50 % of approved drugs are either NPs or their derivatives [5]. Of the 258 small molecules approved for treating infectious diseases between 1981 and 2019, 136 (52.7 %) are NPs or their derivatives [6]. Nature is replete with inexhaustible chemical entities that are products of evolution [5]. As a result of the millions of years of natural selection, these compounds have been optimized to meet the physicochemical requirements for bacterial cells and bind to cellular targets with high efficiency [7].
There was tremendous progress in the discovery of antibiotics (antibacterials from fungi and bacteria), following the discovery of penicillin by Alexander Fleming in 1928 (Fig. 1). Around this period, sulfonamide, the first class of synthetic antimicrobials, was also developed [8]. Although sulfonamide was first synthesized around 1908 by German Chemists, it was not until 1931 before Bayer AG began screening sulfa-containing dyes for antibacterial activity in mice. By 1932, the first commercially available sulfonamide called Prontosil was identified and by 1935 its safe use as an oral antibacterial in humans was reported (Fig. 1) [8]. The 1950s, dubbed the “golden age” of antibiotic development, saw the discovery of most of the powerful antibiotic classes that are clinically available to date [9]. Advancements in the fields of natural product chemistry and synthetic organic chemistry led to the development of semisynthetic antibiotics or several other synthetic antimicrobial agents. Regrettably, since the end of the golden era, discovery and development progress has slowed. While we lowered our guard, there continued to be the development of resistance to the existing antimicrobial chemotherapy, thereby depleting our armamentarium. Bacterial antimicrobial resistance (bacterial AMR) is a natural phenomenon that allows bacteria to evade destruction from toxic chemicals through natural selection [10]. However, the rapid rise in AMR development has largely been fueled by the indiscriminate use and abuse of antibiotics. By the late 1940s, Mary Barber had begun raising awareness about ARM and had published about Penicillin-resistant Staphylococcus aureus infection [11]. And by the 1950s, resistant S. aureus was already implicated as a source of hospital outbreaks and deaths worldwide [12]. By 1962, S. aureus had developed methicillin resistance, just two years after the semisynthetic antibiotic was discovered [3].
Fig. 1.
Examples of early antibacterial agents. Early antibiotics (i.e., antibacterial agents from bacterial and fungal species) (top). Early synthetic antibacterials (bottom).
Today, bacterial AMR has emerged as a significant and urgent global health threat. It is directly responsible for the loss of an estimated 1.27 million lives and has been implicated in over 3 million additional deaths, highlighting the gravity of the situation [13]. Resistance to Gram-negative bacteria is of particular concern [14]. Four of the six “ESKAPE” pathogens (Enterococcus faecium, S. aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp), which are highly implicated in nosocomial infections and renowned for their multidrug resistance and virulence, are Gram-negative [[15], [16], [17], [18]]. The challenge of developing drugs active against Gram-negative bacteria is related to the difficulty of compounds accumulating within the cell [18]. Another cause of concern is the rapid rise in multidrug-resistant tuberculosis (MDR-TB) [[19], [20], [21], [22], [23], [24]]. This is caused by Mycobacterium tuberculosis (Mtb), which is resistant to rifampicin and isoniazid, the main components of the first-line combination of drugs used to treat tuberculosis [19]. There were 450,000 reported cases of MDR-TB or rifampicin-resistant (MDR/RR-TB) infections globally in 2022 [25,26] and approximately 20 % of patients with MDR-TB died during treatment [27]. It has been projected that the death toll from AMR will reach 10 million by 2050 if urgent measures to combat it are not escalated, highlighting the importance and significance of discovering and developing novel antibacterial agents [28,29].
Despite the extensive range of plant-derived antimicrobial agents that have been investigated [[2], [3], [4],6,[30], [31], [32], [33], [34]], clinical trials have been limited to a select few phytochemicals, including berberine, sanguinarine, and licochalcone [30,35]. Additionally, only a few botanicals, such as sinecatechins (Veregen) for treating genital and perianal warts, and crofelemer (Fulyzaq) for managing diarrhea in patients undergoing antiretroviral therapy, have received approval for the treatment of infections [4]. In this review, we comprehensively explore the challenges that have hindered the discovery, development, and clinical utilization of plant-derived antimicrobials. We emphasize strategies employed to address these obstacles, which include limited sources, the risk of agent rediscovery, suboptimal drug metabolism and pharmacokinetics (DMPK) properties, and a lack of knowledge regarding molecular targets and mechanisms of action. Our review underscores the significance of these challenges and their implications in the pursuit of effective plant-derived antimicrobial agents. We critically examine the current state of research and provide valuable insights that contribute to our understanding of these compound classes, offering potential solutions to the global crisis of antimicrobial resistance (AMR). While we provide a concise background on drug resistance and briefly touch upon selected plant secondary metabolites, our primary focus remains on the challenges and opportunities related to promising plant antibacterials.
2. Mechanism of antimicrobial resistance
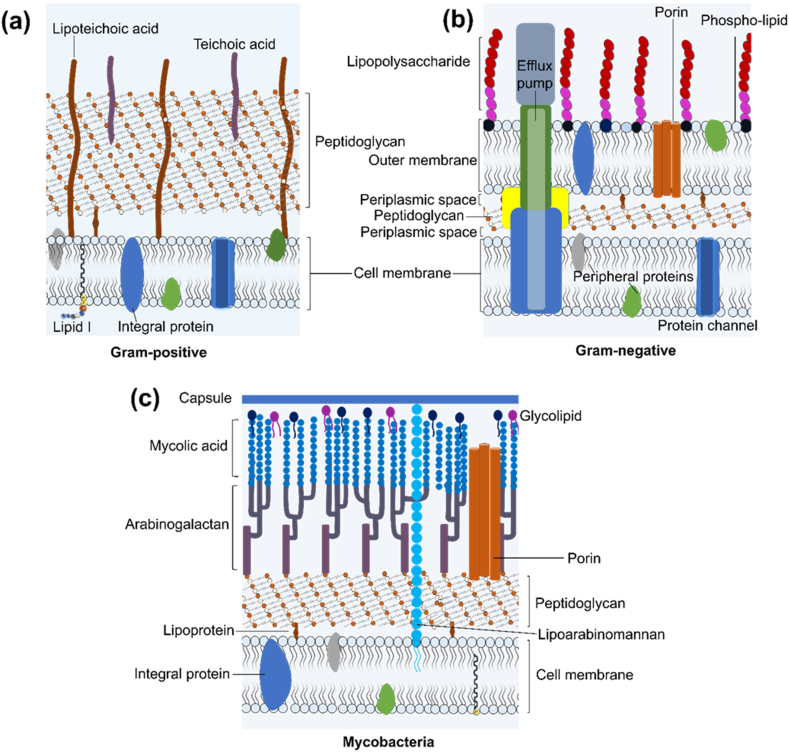
Antimicrobial resistance can arise from intrinsic mechanisms, irrespective of previous antibiotic exposure. Notable examples of inherent mechanisms are the presence of an outer membrane in Gram-negative bacteria that constitutes a major barrier to antimicrobial agents’ penetration [18,36]. While the phospholipid of the inner leaflet prevents the penetration of hydrophilic molecules, the polar glycolipids of the outer leaflet which is composed of lipopolysaccharide (LPS) in most Gram-negative species, limit the passage of hydrophobic molecules in the outer leaflet (Fig. 2) [37,38]. In Mtb, the presence of a structurally complex waxy cell envelope in Mtb plays a major role in hindering the penetration of antimicrobial agents and in the survival of Mtb within the macrophage, where it can persist and manipulate the human immune system to its advantage [19,39]. The expression of efflux pumps is another commonly observed inherent mechanism that impedes the accumulation of antimicrobial drugs, as these membrane proteins continually pump antibacterial agents out of the cell (Fig. 2) [14,40].
Fig. 2.
The cell wall of Gram-positive bacteria (a), Gram-negative bacteria (b) and mycobacteria (c).
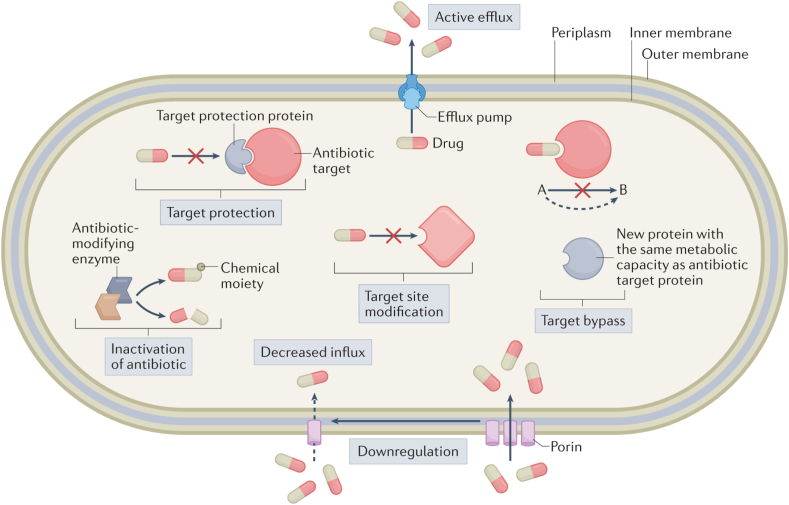
While the knowledge of intrinsic mechanisms is important in the development of new antimicrobial agents and adjuvants, when discussing antimicrobial resistance, the focus is on the development of acquired resistance in a bacterial population that was initially susceptible to antimicrobial agents in clinical settings [41,42]. Acquired resistance can emerge through chromosomal gene mutations or the acquisition of resistance-associated external genetic elements, which are likely from inherently resistant organisms, through horizontal gene transfer (Fig. 3) [42]. The genetic mutations that confer drug resistance are usually associated with genes coding for drug targets, enzymes that modify agents into active or inactive metabolites, and efflux pumps. For example, the most significant threat to β-lactam antibiotics is the enzymatic destruction caused by β-lactamases also called penicillinase [43,44]. These enzymes can break down the β-lactam ring structure present in penicillin and other beta-lactam antibiotics, rendering them inactive and unable to target and inhibit bacterial cell wall. Additionally, resistance to beta-lactams arises from the development of mutations that alter penicillin target proteins, referred to as penicillin-binding proteins (PBPs), and the overexpression of efflux pump systems. In methicillin-resistant S. aureus (MRSA), mutations in the mecA gene lead to the production of PBP2a (penicillin-binding protein 2a), a variant of PBP with reduced affinity for beta-lactam antibiotics, making them less effective in inhibiting bacterial cell wall synthesis [43,44]. Resistance to fluoroquinolones has been associated with mutation in DNA gyrase (gyrA, gyrB) and topoisomerase (parC and parE) genes that encode for the active agent [45]. Additionally, exposure of bacteria to fluoroquinolone can result in the selection of bacteria mutants that exhibit increased expression of efflux systems [46]. Resistance to the TB drug, isoniazid has been linked to mutation in katG and inhA genes of which 60–90 % is attributable to the former [[47], [48], [49]]. Isoniazid is a prodrug and requires activation by the endogenous enzyme KatG, a mycobacterial catalase-peroxidase, to generate an isonicotinic acyl radical that forms an adduct with nicotinamide adenine dinucleotide (NAD) [49]. An excellent overview of the molecular mechanism of drug resistance is illustrated by Darby et al. (2022) in Fig. 3 [50].
Fig. 3.
Overview of the molecular mechanisms underlying antibiotic resistance. Figure obtained from article “Molecular mechanisms of antibiotic resistance revisited” by Elizabeth M. Darby et al. and reused with permission from Springer Nature [50].
3. Plants as potential source of new antibacterial agents
NPs and their derivatives dominate clinically available antibiotics and pipelines [6]. Despite this, only a minuscule fraction of the world's biodiversity has been explored. In fact, only 300,000 of the millions of living species have been evaluated for bioactivity [51]. Therefore, nature remains a promising source of antibiotics with new structures. In the past few decades more and more isolation, characterization, chemical modification, and evaluation of fungi, bacteria, and plant's secondary metabolites (PSM) have taken place [2,4,30]. In contrast to the successes with the development of antibiotics (antibacterials from fungi and bacteria), the development of antibacterial from plants has rather been discouraging despite the many published articles showing the efforts made for the discovery and the development of plant antimicrobials. Many reviews referenced herein [[2], [3], [4],6,[30], [31], [32], [33], [34]] highlight the antibacterial activity of plant-derived extracts, fractionated extracts, and isolated metabolites and their potential to halt antimicrobial resistance. Since discussing hundreds of thousands of antimicrobial plant extracts might not be feasible and would be beyond the scope of this article, below, we highlight selected plant secondary metabolites that have been isolated, tested, and found to possess promising antimicrobial activity.
3.1. Plant secondary metabolites with antimicrobial activity
Plants owe their antimicrobial activities to their secondary metabolites. PSMs constitute more than 12,000 known alkaloids, over 8000 phenolic compounds, and over 25,000 different terpenoids with a good number of these PSMs showing strong antimicrobial properties [52]. Phenolic compounds are the most identified antimicrobial agents reported in the literature [30]. Phenolic compounds may bear a single substituted phenolic ring or multiple units of phenols, in which case they are referred to as polyphenols. Phenols can be further subdivided into phenolic acids, flavonoids, coumarins, tannins, stilbenes, and lignans. Terpenes and terpenoids (class of modified terpenes bearing different functional groups and oxidized methyl groups) are well known for their antibacterial activities. Terpenes and terpenoids are the primary components of essential oils and they are derived from the isoprenoid pathway [53]. Another class of plant-derived antimicrobials worth mentioning is alkaloids. This is a large class of nitrogen-bearing NPs known for their physiological activity [30,54]. Below we discuss the antimicrobial activity of selected plant secondary metabolites and present a summary of these compounds in Table 1.
Table 1.
Selected plant derived antimicrobial compounds.
| SN | Compound | Plant source | Activity (MIC) | Reference |
|---|---|---|---|---|
| 1 | Panduratin A | Kaempferia pandurata Roxb | ≤1 μg/mL against Methicillin-resistant Staphylococcus aureus (MRSA) strains. | [55,125] |
| 2 | Punicalagin | Punica granatum L. | 0.3–1.2 μg/mL against P. aeruginosa ATCC 9027, Staphylococcus epidermidis ATCC 12228, Staphylococcus xylosus LC 57, S. aureus ATCC 6538, Enterococcus mundtii LC E23, Enterococcus casseliflavus LC E1. | [68] |
| 3 | Scandenone | Derris scandens (Roxb.), from Maclura pomifera (Raf.) Schneid | 0.5–8 μg/mL against Gram-positive bacteria, 2 μg/mL E. coli, < 1 μg/mL against M. smegmatis | [69,70] |
| 4 | Mammea B/BA | Mammea americana L. | 0.5–1 μg/mL against methicillin-sensitive and methicillin-resistant S. aureus strains | [74,77] |
| 5 | Cannabinoids (e.g., cannabidiol, cannabigerol) | Cannabis sativa L. | 1–4 μg/mL against MRSA, multi-drug resistant Streptococcus pneumoniae, Enterococcus faecalis, and Clostridioides difficile | [85,86] |
| 6 | 27-epi-Scutianine N | Scutia buxifolia Reissek | 1.56 μg/mL against Enterobater aerogenes | [126] |
| 7 | Berberine | Hydrastis canadensis. L, Coptis chinensis Franch., Berberis integerrima Bonge. Berberis aristate DC, Berberis petiolaris Kunth, Berberis vulgaris L | 1.56–3.8 μg/mL against Brucella abortus | [127] |
| 8 | Kuwanon A | Morus alba L. | 1 μg/mL against S. aureus ATCC29213, 2 μg/mL against S. epidermidis ATCC14990, 8 μg/mL against E. faecalis ATCC29212, 4 μg/mL against B. subtilis ATCC6633 | [128] |
| 9 | Kuwanon B | Morus alba L. | 1 μg/mL against S. aureus ATCC29213, 1 μg/mL against S. epidermidis ATCC14990, 1 μg/mL against E. faecalis ATCC29212, 1 μg/mL against B. subtilis ATCC6633 | [128] |
| 10 | Sanguinarine | Sanguinaria canadensis L., Macleaya cordata (Willd.) R.Br., Eschscholzia californica Cham | 0.5–1 μg/mL against S. aureus ATCC 25923, S. epidermitis ATCC 14990, Bacillus subtilis ATCC 6051, MRSA MR 13165 and 13023. | [129] |
| 11 | Liridine | Artabotrys crassifolius Hook.f. & Thomson | 0.625 μg/mL against Bacillus cereus | [130] |
| 12 | Moronic acid | Schinus lentiscifolius Marchand | 1.5–3 μg/mL against B. subtilis, S. aureus, Streptococcus pyogenes, Staphylococcus saprophyticus, E. coli, Shigella sonnei | [131] |
| 13 | 8,9-Oxoisopropanyldshamirone | Ferula ferulioides (Steud.) Korovin | 0.5–128 mg/L against multi-drug resistant S. aureus and MRSA | [132] |
| 14 | Rhodomyrtosone B | Rhodomyrtus tomentosa (Aiton) Hassk | 0.62–2.5 μg/mL against MRSA, and vancomycin-resistant Enterococcus faecium (VRE) (2.5 μg/mL). | [133] |
| 15 | Artabotrine | Artabotrys crassifolius Hook.f. & Thomson | 1.25–5 μg/mL against B. cereus, Listeria monocytogenes, Proteus vulgaris, Rhodococcus equi, Micrococcus luteus, methicillin-sensitive S. aureus, Streptococcus agalactiae. | [130] |
| 16 | Licochalcone A | Glycyrrhiza inflata Batalin, Glycyrrhiza glabra L. | 1–8 μg/mL against S. aureus | [134] |
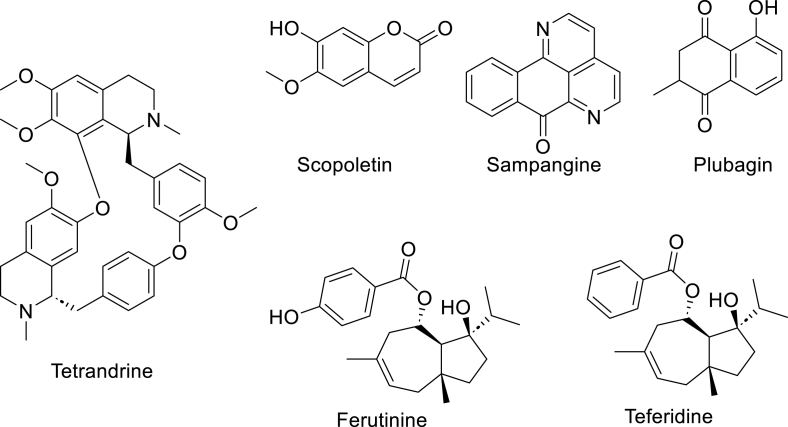
| 18 | Tetrandrine | Stephenia tetrandra S Moore | 0.25–1.25 μg/mL against isoniazid and ethambutol dual drug-resistant Mycobacterium tuberculosis clinical isolates | [135] |
| 19 | Scopoletin | Hymenodictyon floribundum (Hochst. & Steud.) B.L.Rob, Fatoua pilosa Gaud. , Morinda citrifolia L, Pelargonium sidoides DC. | 7.8 μg/mL against Mycobacterium smegmatis, 42.1 μg/mL against Mtb H37Rv | [116,136] |
| 20 | Sampangine | Cananga odorata (Lam.) Hook. f. & Thomson. | 0.78 μg/mL against M. intracellulare | [122,137] |
| 21 | Plubagin | P. indica L, P. rosea L, and Diospyros abyssinica Hiern | 21.1 μM against Mtb H37Rv and 13.3 μM against M. smegmatis mc2155 (0.02–2% glucose) | [120] |
| 22 | Ferutinine | Ferula hermonis Boiss. | 2 μg/mL against Mtb H37Rv, 1.56 μg/mL against M. bovis BCG Pasteur 1173P2 | [123] |
| 23 | Teferidine | Ferula hermonis Boiss. | 0.69 μg/mL against Mtb H37Rv, MIC 3.125 μg/mL against μg/mL | [123] |
3.1.1. Panduratin A
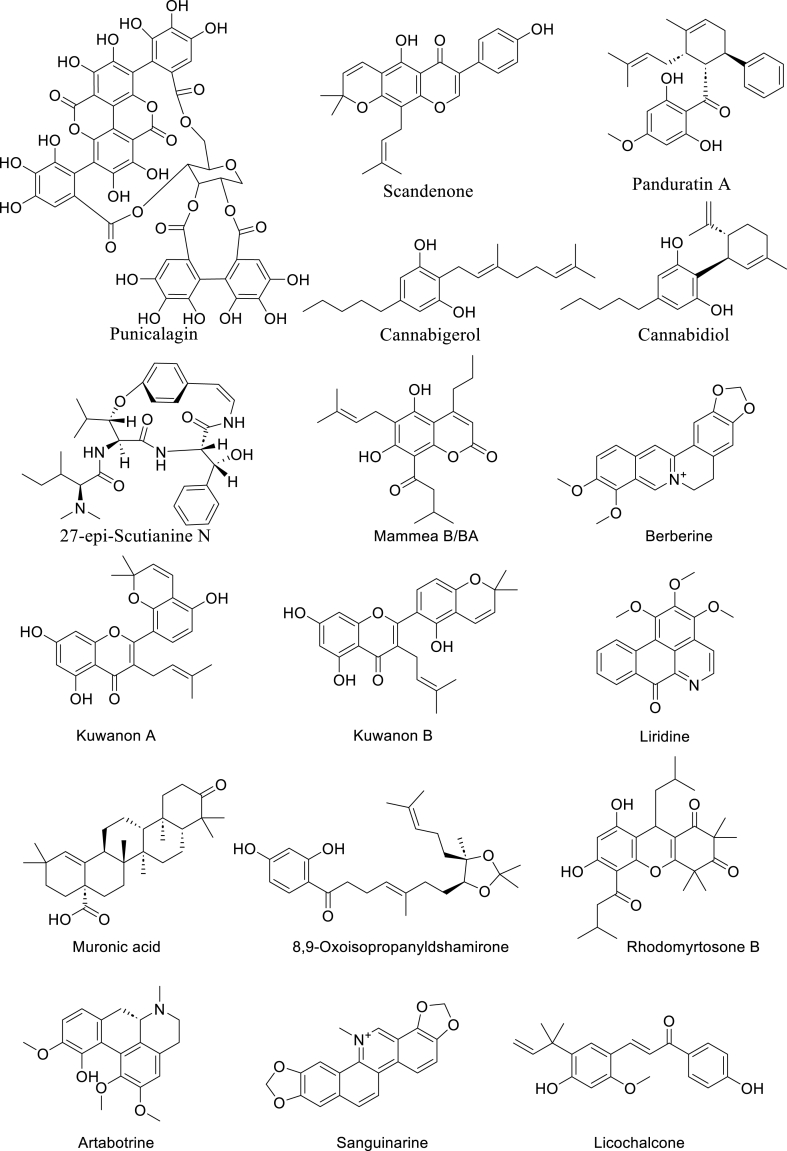
Panduratin A (Fig. 4) is a chalcone isolated from the rhizome of the perennial herb, Kaempferia pandurata Roxb. (Zingiberaceae) commonly known as Fingerroot [55]. The antibacterial activity of the chalcone against multiple organisms including those implicated in the pathogenesis of periodontitis has been reported by the Hwang group [55]. They first reported panduratin A as showing an MIC of 4 μg/mL against Gram-positive and cariogenic Streptococcus mutans and S. sobrinus. The compound also showed potent activity against Gram-negative periodontal bacteria including Prevotella intermedia (2 μg/mL), P. loescheii (4 μg/mL), and Porphyromonas gingivalis (8 μg/mL) [55]. In another study, the group reported the activity of panduratin A against 108 clinical isolates of Staphylococcus strains including MRSA, methicillin-susceptible S. aureus (MSSA), methicillin-resistant coagulase-negative Staphylococci (MRCNS), and methicillin-susceptible coagulase-negative Staphylococci (MSCNS) [56]. While MRSA was resistant to erythromycin, gentamicin, levofloxacin, oxacillin and tetracycline, the activity of Panduratin A with MIC of 1 ≤ μg/mL for all strains of MRSA was comparable to that of vancomycin (≤1 μg/mL) and better than Linezolid (≤2 μg/mL). For all clinical isolates of S. aureus Panduratin A maintained an impressive MIC of ≤2 μg/mL [56]. Similarly, 23 clinical isolates of clinical enterococci were susceptible to this compound with MIC of ≤2 μg/mL [57]. The activity of Panduratin A and its analogue isopanduratin A were determined against Cutibacterium acnes (formerly Propionibacterium acnes), a skin bacterium implicated in acnes vulgaris. Panduratin A and isopanduratin A showed MIC of 2 μg/mL and 4 μg/mL against P. acnes respectively [58]. Furthermore, Panduratin A has been shown to prevent and reduce multispecies oral biofilm of Actinomyces viscosus, S. mutans, and Streptococcus sanguis in vitro [59]. The compound also displays antiviral activity [60] and anticancer activity [61].
Fig. 4.
Selected plant-derived antibacterial compounds with impressive antibacterial activity.
3.1.2. Punicalagin
Punicalagin (Fig. 4) is a hydrolyzable tannin obtained from the fruit of the deciduous shrub, Punica granatum L. (Lythraceae), commonly known as pomegranate. The medicinal benefits of pomegranate have been known since ancient Egypt and appeared in several ancient medical texts such as the Eber papyrus, and the writings of Ayurveda and Unani. It is used in folk medicine for the management of asthma, bleeding disorders, bronchitis, fever, infection, and inflammation [62]. Today the popularity of pomegranate continues to rise globally due to its medicinal properties. It is consumed as fresh fruit, juice, jam, and used for numerous recipes. Pomegranate fruit is high in anthocyanin, tannins, and other polyphenol content, particularly in the peel which has 10-fold the amount of phenol in the arils [63]. The most abundant of these polyphenolic compounds are the hydrolysable tannins, mainly punicalagin, a tannin composed of multiple esters of gallic acid and glucose. The antimicrobial activity of pomegranate fruit extract has been reported in the literature and the higher antimicrobial activity of the peel extract compared to that of the juice and seed extract has been demonstrated [[64], [65], [66], [67]]. However, there have been conflicting reports on the specific agents responsible for this antibacterial activity [68]. Only recently punicalagin in its α and β anomeric forms was identified as the specific agent responsible for the antimicrobial activity of pomegranate peel using bioassay-guided purification techniques [68]. The α and β anomeric forms showed a range of MIC between 0.3 and 1.2 μg/mL and are reported to be interconvertible with an equilibrium ratio of 3/7 for α and β [68].
3.1.3. Scandenone
Scandenone (Fig. 4), an isoflavone with potent antibacterial activity, was first reported to be isolated from Derris scandens (Roxb.) (Fabaceae) [69]. Over the years, this compound has been isolated from other members of Fabaceae especially the Erythrina species [70]. The compound has also been isolated from Maclura pomifera (Raf.) Schneid (Moraceae) [71]. As with most plant antibacterial, scandenone is more potent against Gram-positive bacteria displaying a MIC value of 0.5–8 μg/mL against different Gram-positive strains. The highest potency reported for Gram-negative bacteria is that of Escherichia coli with MIC of 2 μg/mL. It is however not as potent against Pseudomonas aeruginosa as it only shows a poor MIC of 32 μg/mL. Nkengfack et al. (1997) reported MIC of 12.5 μg/mLof scandenone against Mycobacterium smegmatis, however, the MIC was recently reported to be < 1 μg/mL by Peleyeju et al. (2019) [70,72]. In addition to testing the activity of the compound against M. smegmatis, the compound was tested against several Gram-negative and Gram-positive bacteria strains and generally reported a higher MICs of 12–64 μg/mL compared to previous reports [70,73].
3.1.4. Mammea B/BA
Mammea B/BA (Fig. 4) is a naturally occurring coumarin first isolated from Mammea americana L. (Calophyllaceae) in 1952 by Moris and Pagan [74]. It has also been isolated from other Mammea plants such as Mammea africana where it was tested against Staphylococcus species [75]. Coumarins in the genus Mammea have been chemically categorized into 4-(phenyl)coumarins, 4-(alkyl)coumarins and 4-(1-acetoxypropyl)coumarins [76]; Mammea B/BA is a 4-(alkyl)coumarin. In a recent bio-guided fractionation study, Pájaro-González et al. (2022) reported the isolation of mamey coumarin-type Mammea B/BA from the ethanol seed extract of Mammea americana (mamey), and its anti-Stapylococcal activity [77].
Mammea B/BA showed significant activity against strains of methicillin-sensitive S. aureus ATCC 29213 (MIC, 1 μg/mL); and methicillin-resistant S. aureus ATCC 33591 (MIC, 0.5 μg/mL), ATCC 43300 (MIC, 0.5 μg/mL), and USA300-0114 (MIC, 1 μg/mL). The antibacterial activity of Mammea B/BA (MIC 0.5–1 μg/mL) was comparable to that of vancomycin (MIC, 1 μg/mL) and gentamicin (MIC, 0.25–>8 μg/mL) [77]. Pájaro-González et al. (2022) proposed that the antibacterial activity of Mammea B/BA is due to the presence of a prenyl group at its position C6 and acyl group at position C8. Prenyl group is a common feature of many antibacterial agents including three of the other compounds (cannabigerol, scandenone and Panduratin A) highlighted in this review (Fig. 4). Mammea B/BA had also been shown to have high activity (LD50, 5.3 μg/mL) against Artemia salina Leach [78]. It showed significant activities against SW-480, HT-29 and HCT-116 colon cancer cells (IC50, 13.7–17.5 μM) and antioxidant activity in the DPPH assay (IC50, 90 ± 0.7 μM) [79]. Related coumarins from the Mammea genus have been reported with high antibacterial activity. An example is Mammea A/AA, which is an isoprenylated mamey-type coumarin with activity against Campylobacter jejuni (MIC, 0.5 μg/mL), Streptococcus pneumoniae (MIC, 0.25 μg/mL) and Clostridium difficile (MIC, 0.5 μg/mL) [80].
3.1.5. Cannabigerol, cannabidiol and other cannabinoids
Cannabinoids are the major NPs of Cannabis sativus L. and related species. These compounds are of great importance to researchers and the public because of their medicinal properties and more importantly, because of the psychoactive properties of cannabinoids, the most important being Δ9-tetrahydrocannabinol (THC) [81]. Although the antimicrobial activity of cannabinoids has been known as far back as the 1960s [82,83], it was only recently that detailed studies of the antimicrobial properties of the cannabinoids and their potential to tackle antimicrobial resistance began to emerge [84,85]. Farha et al. (2020) investigated the antimicrobial activity of several commercially available cannabinoids against MRSA and identified seven potent cannabinoids with MIC of 2 μg/mL. This included cannabichromene (CBC), cannabidiol (CBD), cannabigerol (CBG), cannabinol (CBN), and THC and its regioisomers: Δ8-and exo olefin [85] (Fig. 4). They further reported that the cannabinoids inhibit biofilm formation in a manner that correlated with their antibacterial potency. CBG displayed superior activity against persister cells compared to other cannabinoids and existing antibiotics such as oxacillin and vancomycin. The profile of CBG as a non-psychotropic and non-sedative cannabinoid in addition to its antibiofilm and anti-persister activities, therefore, positioned it as the most promising antimicrobial cannabinoid. The researchers then determined the in vivo efficacy of CBG against MRSA using a mouse systemic infection model and determined 100 mg/kg as the most effective tolerable dose. They reported that in vivo efficacy of CBG was comparable to the control, vancomycin. Using multiple strategies to determine the mode of action, they concluded that CBG acted by disrupting the cytoplasmic membrane [85].
As with many antimicrobials from plants, CBG, and the other potent cannabinoids are generally inactive against the tested Gram-negative bacteria due to their inability to cross the outer membrane (OM). However, the cannabinoids regained potency in the presence of membrane destabilizers like polymyxin B or polymyxin B nonapeptide [85]. They reported that CBG in combination with a sublethal concentration of polymyxin B (0.062 μg/mL) was active against E. coli with a MIC of 1 μg/mL. This implies that CBG has the potential as a combination therapy for the treatment of Gram-negative infections [85].
Blaskovic et al. (2021) did a thorough study on the antimicrobial activity of CBD, another non-psychoactive cannabinoid [86] (Fig. 4). Like CBG, CBD acts by disrupting the cell membrane. CBD is potent against 20 Gram-positive pathogens including those among ESKAPE pathogens with MIC of 1–4 μg/mL. The compound was inactive against a panel of 20 Gram-negative pathogens including members of the ESKAPE pathogens. Interestingly, they reported that CGB had potent activity against a subset of Gram-negative including Legionella pneumophila (MIC, 1 μg/mL), Moraxella catarrhalis (MIC, 1 μg/mL), Neisseria gonorrhoeae (MIC, 1–2 μg/mL), and Neisseria meningitidis (MIC, 0.25 μg/mL). The compound displayed modest activity against M. smegmatis with a MIC of 16 μg/mL. On the other hand, it was poorly active against Mtb H37Rv with MIC greater than >64 μg/mL (70 % inhibition at 64 μg/mL). Furthermore, the efficacy of CBD as a topical agent was demonstrated in the ex vivo pig skin and mouse topical infection models. They report that the pig model was highly dependent on the composition of topical formulation [86].
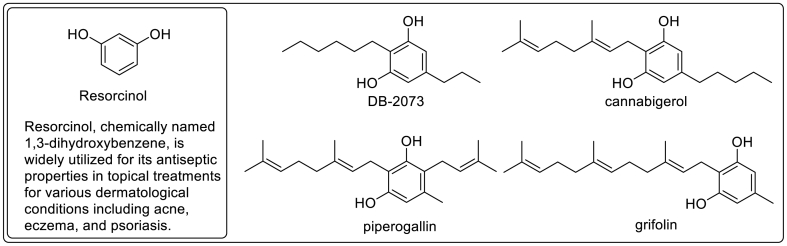
Structurally, CBD and CBG can be described as geranyl-resorcinol derivatives (Fig. 5). CBG bears great resemblance to piperogalin, a geranyl-resorcinol first isolated from Peperomia galioides [87] and grifolin, a farnesyl-resorcinol antibiotic isolated from basidiomycetes, Grifola confluens [88]. Similarly, structurally related alkylresorcinol antibiotic, DB-2073, isolated from Pseudomonas sp was reported by Kitahara et al. (1975) to be active against Gram-positive bacteria, mycobacteria, and fungi [89]. A major concern that hinders the clinical development of the resorcinol-containing antibacterial such as CBD and CBG is the inactivation in the serum and their irritant property [82,84]. Several cannabinoids have been approved for other clinical uses in humans other than bacterial infections [90]. Nabiximols (Sativex) is a standardized botanical drug which contains THC and CBD as principal active ingredients. It was approved in Europe in the year 2010 for neuropathic pain, overactive bladder, and spasticity. Epidiolex, an oral solution of CBD, was approved in 2018 for the treatment of a rare form of children epilepsy. Cannabinoids have poor oral bioavailability [91] and the dose that may be required to achieve antimicrobial effect in humans may aggravate side effects. However, cannabinoids remain promising sources of topical antimicrobial agents [84,86]. Indeed, synthetic CBD has been evaluated in phase-II trials for acne (BTX 1503, NCT03573518) and atopic dermatitis (BTX 1204, NCT03824405) by Botanix Pharmaceuticals (Perth, Australia). Additionally, CBD, BTX 1801 has been subjected to a phase-II trial (ACTRN126200004569540) for the eradication of nasally colonized S. aureus [92]. No rigorous clinical trials of CBD and other cannabinoids for systemic antimicrobial properties have been undertaken to date.
Fig. 5.
Naturally occurring resorcinols with antimicrobials property.
3.1.6. 27-Epi-scutianine N
27-epi-Scutianine N (Fig. 4) is an alkaloid found in Scutia buxifolia (Reissek) family Rhamnaceae [93]. A reinvestigation of the chemical constituents of the stem barks of Scutia buxifolia was done and concluded with the isolation of three undescribed diastereoisomeric alkaloids – scutianine N, 27-epi-scutianine N and 3, 4, 7-tri-epi-scutianine N in addition to the known alkaloids scutianine C and scutianene L [93]. The in vitro evaluation revealed some stereochemistry-activity relationships for the antibacterial activity of diastereoisomeric alkaloids against the Gram-negative bacteria Enterobacter aerogenes. The alkaloid 27-epi-scutianine N showed similar activity compared to the standard antibiotic chloramphenicol (MIC, 1.56 μg/mL). On the other hand, scutianine N and 3,4,27-tris-epi-Scutianine N had no activity (MIC, >100 μg/mL) [93].
3.1.7. Berberine
Berberine (Fig. 4) is a quaternary isoquinoline-based alkaloid that was first isolated from Hydrastis canadensis L. in 1917 [94]. The alkaloid has also been isolated from several Berberis species including B. aristate DC., B. darwinii Hook.f., B. petiolaris Kunth., and B. vulgaris L. Berberine has also been reported in Argemone Mexicana L., Coptis chinensis Franch., Coptis teeta Wall. ex Hook.f. & Thomson, Eschscholzia californica Cham., Mahonia aquifolium Nutt., Phellodendron amurense Rupr, Tinospora cordifolia (Willd.) Hook.f. & Thomson, Xanthorhiza simplicissima Marshall [95]. Berberine is known to possess multiple biological activities and has been studied for its use in the treatment of cardiovascular disease [96,97], cancer [98] immune system diseases, metabolic diseases [99], neurological disorders [100] and antimicrobial infection [[104], [103], [102], [101]]. Alone, berberine has been demonstrated to possess moderate to weak activity against Gram-negative and Gram-positive organisms. Wu et al. (2022) [104] reported a MIC of value 51 μg/mLfor berberine against S. aureus strain ATCC 25923, while Xia et al. (2022) [105] reported a MIC range of from 256 to 64 mg/L against MRSA clinical isolates [105]. Berberine acts synergistically against S. aureus when combined with clindamycin, rifamycin and vancomycin separately. When used alone, berberine displayed a weak antimicrobial activity (e.g., MIC ≥256 mg/L) against MDR A. baumannii. resensitize MDR Acinetobacter baumanni towards antibiotics ciprofloxacin, meropenem, sulbactam, and tigecycline [106]. Berberine is thought to elicit its antimicrobial activity by interfering with the shikimic acid pathway, inducing oxidative damage in bacteria and disrupting cell membrane integrity [102,104]. It is also thought to act as an efflux pump inhibitor [107,108]. A recent report, however, suggests that berberine is a good substrate for the efflux pump in A. baumannii. and could be competing for the efflux pump with antibiotics thereby leading to the sensitization of antibiotics [106]. The authors revealed that berberine binds to the AdeB transporter protein, an efflux pump belonging to the resistance-nodulation-cell division (RND) family and boosts the expression of the AdeB gene [106]. Berberine has also been reported to inhibit DNA synthesis [109], and to inhibit FtsZ A, cell division protein [110] and has been shown to inhibit biofilm formation in MRSA [111]. A search through the clinical trial database (https://www.clinicaltrials.gov/), reveals that the clinical safety and efficacy of berberine alone or in combination with known drugs have been tested for several disease conditions including polycystic ovary syndrome (POC), obesity, Schizophrenia and in the eradication of Helicobacter pylori infection in ulcer. It has been demonstrated in clinical trials that berberine, when used as an adjunct to standard triple therapy, increased in H. pylori eradication rates and clinical symptom remission rates [112].
3.1.8. Plant derived antimycobacterial agents
Plant-derived NPs have been investigated as potential sources of alternative compounds that are much needed to combat MDR-TB [113]. A major barrier to developing antitubercular agents is the thick waxy mycobacterial cell wall rich in mycolic acid which gives extra and intrinsic resistance (Fig. 2). Tetrandrine, a bis-benzylisoquinoline alkaloid first reported from a popular East Asian medicinal plant, Stephania tetrandra S. Moore is known for its antimicrobial and several other biological activities [114]. Zhang et al., 2015 reported that the administration of tetrandrine in combination with either isoniazid or ethambutol resulted in a reduction of the MICs from the drug resistance levels observed in clinical settings to the effective ranges (80 μg/mL to 1.25 μg/mL, and 8 μg/mL to 0.25 μg/mL, respectively) [115] Scopoletin (Fig. 6), a broad-spectrum antibacterial agent has been isolated from Hymenodictyon floribundum Roxb, Pelargonium sidoides DC., Morinda citrifolia L., Helichrysum italicum (Roth) G.Don, Convolvulus pluricaulis Choisy, Fatoua Pilosa L., Artemisia annua L. and Lasianthus lucidus Blume Morus alba L [116]. Scopoletin inhibits M. smegmatis with and Mtb H37Rv with MIC 7.8 μg/mL and 42.1 μg/mLrespectively [117,118]. Plumbagin (Fig. 4), is a plant-derived hydroxy naphthoquinone primarily found in roots, stems, and leaves Plumbaginaceae family, such as Plumbago indica L. and Plumbago rosea L., and in Diospyros abyssinica Hiern [119]. Plumbagin inhibits Mtb H37Rv with MIC 21.1 μM and M. smegmatis mc2155 (0.02–2% glucose) at an MIC of 13.3 μM [120]. Plumbagin activity has been linked to its inhibition of Mtb thymidylate synthase (ThyX), an enzyme responsible for the synthesis of dTMP from dUMP [121]. The azaoxoaporphine alkaloid, sampangine (Fig. 6), was first isolated from the stem bark of the tropical Indian plant, Cananga odorata (Lam.) Hook. f. & Thomson. Sampangine inhibits M. intracellulare at an MIC value, 0.78 μg/mL [122]. Sampangine is reported to elicits its antimicrobial activities by disruption of mitochondrial function through redox cycling of its quinone and semiquinone intermediates resulting in the excessive production of cellular ROS and consumption of cellular oxygen [122]. Ferutinin and Teferidin (Fig. 6) are broad-spectrum antimicrobial sesquiterpenoid esters isolated from the root of Ferula hermonis Boiss. Apiaceae. Teferidin and Ferutinin possess MIC values of 2 and 0.69 μg/mLagainst Mtb H37Rv respectively and MIC values of 1.56 and 3.125 μg/mL against M. bovis BCG Pasteur 1173P2 [123]. Ozturk et al. (2021) demonstrated that berberine is inactive against Mtb axenic cultures at a concentration ranging from 3.9 to 250 μM. Berberine was however shown to increase bacterial killing in human monocyte-derived macrophages and primary murine bone marrow-derived macrophages [124]. In vivo, berberine is beneficial in reducing lung pathology when used along with rifampicin and isoniazid but does not to result in the reduction of bacterial burden. It's in vivo benefit stems from its ability to mitigate lung inflammation by modulation of immune cell recruitment and the suppression of inflammatory cytokines [124].
Fig. 6.
Selected plant-derived antimycobacterial compounds.
4. Discovery and development challenges of plant-derived antimicrobials and possible solutions
4.1. Sourcing of natural products
Isolation of compounds from plants and microorganisms is at the heart of natural product chemistry but this is often a tedious, time-consuming, and expensive expedition that may lead to little or no yield of the desired product (see Fig. 7 for the example of berberine). Typically, isolation from plants begins with organic or aqueous extraction of several grams to several Kilograms of previously dried and ground plant material such as root, stem bark, leaves etc. The percentage yield of plant metabolite is about 1 % and could be less in some plant species like Taxus brevifolia Nutt. bark which contains only 0.01 % paclitaxel (dry weight) [138] and Morus alba L. root which contains 0.00035 % w/w of antibacterial agent, kuwanon A [139]. Efforts at increasing the yield will often not only impact cost and time but will lead to the destruction of plants and pollution of the environment. Hence, the traditional extraction of material sourced from the wild is not sustainable especially when a large quantity of compounds is required. Another challenge with the sourcing of plant-derived NPs is the variation of the chemical compositions or quantity of desired NPs that can be due to differences in seasonal variation, the geographical location of plants and extraction techniques. For instance, the discrepancy in the chemical compositions of pomegranate, P. granatum, is thought to be due to the difference in geographical source, method of collection, or chemical composition across varieties [68]. Similarly, the method of extraction of cannabis affects the chemical composition of the extract [140]. Bowen et al. (2021) compared the chemical profile of a single cultivar of cannabis extract obtained by two commonly used extraction methods: alcohol solvent extraction and supercritical CO2 extraction. They observed a marked difference in the profile of bioactive compounds and especially noted that alcohol (ethanol vs isopropyl alcohol) extraction generally yielded more CBD, Δ9THC, and other phyto-cannabinoids compared to supercritical CO2 extraction [140]. Variation in chemical composition presents the problem of uncertainty in the number of targeted NP yields to expect during the isolation process. In the case of botanical drugs or sourcing raw materials for their production, variation in chemical profiles can make the standardization of products elusive. Possible alternatives to obtaining this chemical entity from the wild include chemical synthesis [141], synthetic biology [142,143], plant cell or tissue culture [144] etc.
Fig. 7.
Sourcing of natural product, a berberine example.
4.1.1. The chemical synthesis
Numerous plant-derived antimicrobials have been accessed through total synthesis. For instance, the synthesis of berberine was initially achieved by Kametani and co-workers in 1969 [145]. Subsequently, several other research groups have made attempts to synthesize it through shorter and more environmentally friendly routes over the years [[146], [147], [148]]. Nevertheless, the total synthesis of complex NPs often involves numerous steps, resulting in increased costs and time investment, thereby limiting the feasibility of commercial production [141,149]. In fact, total synthesis of NPs has mainly found use in the structural elucidation, confirmation of chemical structure, development of new methodologies and anticipation of NPs and less for industrial scale production [149,150]. Yet, total synthesis can be a useful means of obtaining highly important compounds of extremely low-yielding, rare or endangered plants, thereby allowing early biological testing and structure-activity relationship (SAR) studies [141]. In one recent example, Dong et al. (2023) were motivated to undertake the total synthesis of kuwanon A and its regio-isomer, kuwanon B because of the extremely low yield of the compounds from the root of Morus alba [151]. Only 28 mg of kuwanon A was obtained from 8 kg of plant root leading to a yield of (0.00035 %) [151]. In contrast, the total synthesis of kuwanon A and B from commercially available intermediate was achieved at an overall yield of 6.6 % and 11.6 %, respectively [128].
Semi-synthesis can also be employed to modify NPs in other to modulate biological activity. This approach has been heavily exploited in the development of antibiotics and led to many successful semi-synthetic antibiotics such as ampicillin, methicillin, and ceftriaxone that are available for clinical use today [6].
4.1.2. Plant cell and tissue culture
Plant cell and tissue culture are techniques used to aseptically grow the cell, tissue, or organ of a plant under a controlled environment. Tissue culture can be used for the rapid production of plants at a commercial scale while avoiding limitations such as the spread of plant-pathogen and inconsistent chemical composition that are associated with conventional cultivation. This is made possible by the controlled conditions that are applied in the cultivation and the relative ease in the standardization of propagation [152]. As regulations around the growing of Cannabis dampens, the interest in the large-scale and cost-effective production of pathogen/disease-free plants with consistent chemical and morphological character by tissue culture/micropropagation is on the rise [152,153]. Ioannidis et al. (2020) developed an efficient protocol for the in vitro micropropagation of CBD and CBG-rich Cannabis sativa L. varieties [154]. They established that the chemical profiles of C. sativa varieties obtained by micropropagation were identical to those grown through conventional methods [154].
4.1.3. Genome mining and engineering
The genes responsible for the biosynthesis of secondary metabolites in microorganisms are organized into gene clusters on a chromosome, commonly known as biosynthetic gene clusters (BGC) [155]. Although similar gene clusters are found for some secondary metabolites in plants, they are reported to be less tightly clustered as some important genes in the pathway are often dispersed in different chromosomal locations [156,157].
The discovery of the BGC has greatly facilitated the identification and elucidation of the metabolic pathways, engineering of metabolic pathways, and transfer of pathways into heterologous systems using the combination of synthetic biology and bioengineering methods. Reconstruction of biosynthetic pathways in heterologous hosts has been used to carry out large-scale production of some plant-derived natural products such as artemisinin and taxol [158]. Recently, Han and Li demonstrated the biosynthesis of berberine, an antimicrobial alkaloid, in Saccharomyces cerevisiae providing a more cost-effective, rapid, efficient, and environmental-friendly alternative to isolation and synthesis (Fig. 5) [159].
Earlier synthetic biology efforts at growing metabolites relied on E. coli and Saccharomyces cerevisiae as host due to their biosynthetic capabilities, growth rate, inexpensive growth, and relative ease of genetic and metabolic manipulation. However, the use of these model organisms especially for complex plant-derived natural products has proven to be challenging due to limitations in the function and optimization of their pathways in bacterial or yeast hosts [143]. Hence, there is an increasing interest in the use of engineered plant hosts for plant-derived natural product biosynthesis due to their advantages over single-celled organisms [158]. Some of these advantages include the existence of plant-derived enzymes or partial or complete plant metabolic pathways suitable for biosynthesis [160]; the presence of intracellular membranes which are lacking in E. coli [143] and the tolerance toxicity of plant-derived natural products [142]. The proper elucidation of the plant biosynthetic gene cluster is critical for successful heterologous expression. In recent years several bioinformatics and computational tools have been developed to aid the discovery and elucidation of biosynthetic pathways such as AntiSmash [161], plantiSMASH [162], PlantClusterFinder [163], and MIBiG [164]. These tools can be employed in the discovery of plant-derived antimicrobials as well as increase production.
4.2. Rediscovery and the need for dereplication
The problem of rediscovery of known metabolites has plagued NP research for decades, resulting in waste of scarce resources and loss of man-hours [165]. Berberine, for example, was first isolated from the H. canadensis (goldenseal, family Berberidaceae) in 1917 but has been found in numerous other plant species from several other genera and families to date. As recently demonstrated by Hang group when they dereplicated berberine from the area part of Coptis chinensis, a lot of resources would be saved if the presence of berberine could be determined earlier in the isolation workflow and the isolation of newer compounds were prioritized (Fig. 5). Several dereplication strategies have been employed over the years to identify these known compounds early enough in the natural product isolation workflow thereby cutting costs and efforts [[165], [166], [167], [168], [169]]. These strategies rely on the fact that structurally similar compounds often share similar physical characteristics, such as chromatographic retention times, molecular masses, MS fragmentation patterns, NMR chemical shifts, or in some cases, biological properties [165,169]. The processed data of physical or biological properties obtained from one or a combination of techniques are compared to previously annotated datasets on a local database or publicly available databases like Global Natural Products Social Molecular Networking (GNPS) library (http://gnps.ucsd.edu), the Dictionary of Natural Products (http://dnp.chemnetbase.com), MarinLit (http://pubs.rsc.org/marinlit), Metlin (http://metlin.scripps.edu/index.php), RIKEN tandem mass spectral database (ReSpect) [170] to identify same or structurally related compounds.
4.2.1. MS-based dereplication strategies
With advances in bioinformatics and increased availability of LC-MS/MS, the MS-based Molecular Networks algorithm has become one of the most useful dereplication tools for the natural product chemist [171]. It works on the assumption that similar molecules have the same molecular masses and similar MS/MS fragmentation patterns. GNPS, developed by the Dorrestein group, is an interactive online curation and analysis platform that allows researchers to dereplicate NPs and annotate newly identified molecules for the benefit of other members of the community [172]. Molecular networks are constructed by matching an MS/MS spectrum against another or against the GNPS Spectral Libraries [172]. Metabolites, represented as nodes, are connected by edges to the same or structurally related metabolites in the network are connected. A cosine score ranging from 0 to 1 is ascribed to the edges based on the level of similarity of this metabolite [172]. Dorrestein and co-workers later developed a method that integrates molecular networking with a “bioactivity score”, a number that measures the likelihood that a molecule is biologically active [173]. Inspired by the work of Dorrestein, Ge et al. (2022) introduced the Bioactive Fractions Filtering Platform (BFFP), which also integrates LC-MS data and bioactivity scoring to assist in bioassay-guided fractionation [174]. The BFFP is particularly useful for molecules with structures prone to generating less MS/MS debris. Ge et al. (2022) uses the concept of statistically bioactive blocks, which quantifies biological activity indicators within specified retention time and molecular weight ranges in extracts [174]. Together these approaches help in the early identification of potentially bioactive compounds thereby allowing for a more focused, time and resource-saving isolation process.
Guided by a UHPLC-ESI-QTOF-MS/MS-based molecular networking strategies, Azizah et al. (2020), perform isolation and dereplication of antibacterial and antifungal compounds from the trunk and back of Ventilago denticulata Willd [175]. A total of 93 putative known and unknown compounds were annotated by comparing the MS/MS spectra from the crude extract to various databases including the Agilent MassHunter METLIN Metabolomics Database, online database METLIN, the Human Metabolome Database and the GNPS. Their effort led to the isolation of 8 antibacterial among which was a new naphthopyrone, ventilatone, and identification of several other antibacterial compounds through their dereplication strategy [175]. Cruz Pontes et al. (2022) used a combination of GC-MS and LC-MS/MS-based methods for the dereplication of the metabolite from Bromelia laciniosa. For non-polar compounds, they undertook a GC-MS analysis of petroleum ether fraction and then matched the spectral against Wiley 7 lib and NIST 08 lib databases. For polar compounds, spectral data obtained from HPLC-ESI-IT MS/MS were cross-referenced against GNPS. Overall, 39 chemical constituents were reported to be present in B. laciniosa for the first time [176]. They also isolated cirsilineol, a flavonoid with anti-proliferative and antibacterial properties [177]. GNPS has enjoyed widespread adoption among natural product chemists and its utility for the dereplication of plant-derived antimicrobials is well demonstrated in numerous pieces of literature [[178], [179], [180], [181], [182], [183]].
While MS-based dereplication strategies are the most common today, they face several challenges related to the inherent limitation of MS analysis [184,185]. Limitations, including challenges in accurately distinguishing isomers and instances where a single mass represents multiple metabolites, potentially leading to misidentification of compounds, alongside issues such as ion suppression and variable analyte ionization, emphasize the attractiveness of alternative methods like NMR [186].
4.2.2. NMR-based dereplication strategies
Since NMR uses direct structural information, it can avoid some of the challenges associated with MS/MS-based strategies and indeed complement the method [184]. In recent years, several platforms such as COLMAR [187], DEREP-NP [168], MetaboMiner [188] and SMART 2.0 (Small Molecule Accurate Recognition Technology) [189] have been developed for the dereplication of targeted metabolite [190]. Recently, the Roger group developed MADByTE (Metabolomics and Dereplication by Two-Dimensional Experiments) as a strategy particularly suited for complex mixtures of NPs [184,185]. MADByTE works by associating 1H–13C connectivity from HSQC spectra with a spin system from TOCSY spectra to decipher scaffold substructures in a complex mixture. Flores-Bocanegran et al. (2022) demonstrated the potential of MADByTE when they dereplicated seven fungal extracts against a database they created from the HSQC and TOCSY data of resorcylic acid lactones and 10 spirobisnaphthalenes [185].
4.3. Drug metabolism and pharmacokinetics (DMPK) properties
A major challenge hindering the translation of potent plant-derived antimicrobials is their suboptimal DMPK properties also referred to as ADMET (Absorption, Distribution, Metabolism, Elimination, Toxicity) properties (Table 2). DMPK properties are largely determined by physicochemical properties like aqueous solubility, chemical stability, hydrogen bonding capability, lipophilicity, size and structure, polar surface area, and enzymatic reaction susceptibility [191]. Hence an ideal antibacterial drug candidate would possess optimal physicochemical properties to proceed to in vivo preclinical and beyond. For example, a certain degree of lipophilicity is necessary for a drug to persist in the body for a sufficient duration before being eliminated through excretion. However, excessive hydrophobicity can result in poor aqueous solubility and poor bioavailability. Cannabinoids including cannabichromene (CBC), cannabidiol (CBD), cannabigerol (CBG), cannabinol (CBN), and THC and its regioisomers: Δ8-and exo olefin inhibits MRSA at MIC of 2 μg/mL but are limited by their suboptimal DMPK [85,86]. These cannabinoids are highly lipophilic and generally have poor aqueous solubility and poor oral bioavailability. The pharmacokinetic of cannabidiol (CBD) has been described as complex and variable [192]. The compound suffers significant first-pass metabolism in the liver and possesses low oral bioavailability (6 %) that can increase as much as fourfold with high-fat meal. This implies that a higher dose may be required to achieve the desired biological effect, thereby increasing the risk of developing side effects [91,192,193].
Table 2.
In silico physicochemical and DMPK properties of selected plant-derived antimicrobialsa.
| SN. | Compound | MW (g/mol) | # of H-bond acceptor | # of H-bond donor | ClogPd | # of rotatable bonds | LogS (Aq. Solubility)e | GI absorption | Fraction unbound in plasma (Fu) % |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Panduratin A | 406.51 | 4 | 2 | 4.77 | 6 | −7.14 (poor) | high | 4.645 |
| 2 | Punicalagin | 1084.72 | 30 | 17 | 0.07 | 0 | −12.26 (insoluble) | Low | 150.420 |
| 3 | Scandenone | 404.46 | 5 | 2 | 4.72 | 3 | −7.32 (poor) | High | 3.178 |
| 4 | Mammea B/BA | 372.45 | 5 | 2 | 4.53 | 7 | −7.11 (poor) | High | 4.856 |
| 5 | Cannabidiol | 314.46 | 2 | 2 | 5.20 | 6 | −7.17 (poor) | High | 1.151 |
| 6 | Cannabigerol | 316.48 | 2 | 2 | 5.74 | 9 | −8.10 (poor) | High | 2.510 |
| 7 | 27-epi-scutianine N | 550.69 | 6 | 4 | 2.7 | 9 | −6.94 (poor) | High | 3.845 |
| 8 | Berberine | 336.36 | 4 | 0 | 2.53 | 2 | −4.16 (moderate) | High | 2.335 |
| 9 | Kuwanon A | 420.45 | 6 | 3 | 4.29 | 3 | −7.38 (poor) | High | 3.283 |
| 10 | Kuwanon B | 420.45 | 6 | 3 | 4.32 | 3 | −7.38 (poor) | High | 3.851 |
| 11 | Liridine | 321.33 | 5 | 0 | 3.02 | 3 | −4.38 (moderate) | High | 5.292 |
| 12 | Moronic acid | 454.68 | 3 | 1 | 6.12 | 1 | −8.39 (poor) | Low | 4.817 |
| 13 | 8,9-Oxoisopropanyldshamirone | 430.58 | 5 | 2 | 5.45 | 10 | −7.81 (poor) | High | 2.195 |
| 14 | Rhodomyrtosone B | 442.54 | 6 | 2 | 4.39 | 5 | −7.65 (poor) | High | 1.724 |
| 15 | Artabotrine | 341.40 | 5 | 1 | 2.77 | 3 | −3.29 (soluble) | High | 15.441 |
| 16 | Sanguinarine | 332.33 | 4 | 0 | 2.88 | 0 | −5.03 (moderate) | High | 1.419 |
| 17 | Licochalcone A | 338.40 | 4 | 2 | 3.93 | 6 | −6.04 (poor) | High | 0.578 |
| 18 | Tetrandrine | 622.75 | 8 | 0 | 5.49 | 4 | −7.76 (poor) | High | 30.984 |
| 19 | Scopoletin | 192.17 | 4 | 1 | 1.52 | 1 | −2.39 (soluble) | High | 19.387 |
| 20 | Sampangine | 232.24 | 3 | 0 | 2.35 | 0 | −3.09 (soluble) | High | 3.132 |
| 21 | Plumbagin | 190.20 | 3 | 1 | 1.67 | 0 | −2.87 (soluble) | High | 9.127 |
| 22 | Ferutinine | 358.47 | 4 | 2 | 3.86 | 4 | −5.60 (moderate) | High | 2.523 |
| 23 | Teferidine | 342.47 | 3 | 1 | 4.38 | 4 | −5.55 (moderate) | High | 2.807 |
| 24 | Penicillin Gb | 334.39 | 4 | 2 | 1.37 | 5 | −3.80 (soluble) | High | 55.960 |
| 25 | Erythromycinb | 733.93 | 14 | 5 | 1.97 | 7 | −6.80 (poor)a | High | 29.158 |
| 26 | Ciprofloxacinc | 331.34 | 5 | 2 | 1.10 | 3 | 0.00 (high) | High | 78.856 |
fGastrointestinal absorption was determined according to the Brain Or IntestinaL EstimateD permeation method (BOILED-Egg). Fraction unbound (free) drug in plasma (Fu) typically constitutes the portion responsible for exerting its pharmacological effect
Physicochemical properties were calculated online using the SwissADME server (accessed on July 22, 2023), except for Fu (fraction unbound in plasma), which was determined using the ADMETLab 2.0 server (accessed on July 22, 2023).
Penicillin G, antibiotic derived from the fungi, Penicillium chrysogenum and Erythromycin, derived from the bacteria, Saccharopolyspora erythraea are included as references.
Ciprofloxacin is a synthetic antimicrobial included here as a reference.
cLogP represents an average of five different partition coefficients (Log Po/w) calculated using different methods.
Log S was determined using the Ali topological method implemented by Ali et al. (2012) [208].
4.3.1. Plant antimicrobial peptides and poor DMPK profiles
Antimicrobial peptides (AMPs) constitute a group of small peptides present across various organisms in nature, playing a crucial role in the innate immune responses of amphibians, mammals, microorganisms, and plants [194]. AMPs have long been explored for plant pest control and treating infections in humans and animals. They generally elicited their antimicrobial activity by disrupting the bacterial cell membrane or inhibiting the growth of bacteria. Important AMPs from microorganisms have undergone clinical trials, and several have received FDA approval for treating infectious diseases. Notable examples of microorganism-derived AMPs in clinical use include polymyxins derived from Gram-positive Paenibacillus polymyxa, gramicidin from Gram-positive Brevibacillus brevis, nisin (also known as nisin A) produced by lactic acid bacteria like Lactococcus lactis, and daptomycin, a cyclic lipopeptide naturally synthesized by Streptomyces roseosporus [195]. However, research on plant antimicrobial peptides (PAMP) as potential alternatives to conventional antibiotics in combating drug resistance over the past three decades has not achieved the same level of success as microbe-derived AMPs. PAMPs are crucial for plants' defense against pests and pathogens. They primarily exist in cysteine-rich linear peptide forms, with major classes including thionins, defensins, hevein-like peptides, knottin-type peptides, and cyclotides. Defensin, BcDef1, isolated from Brugmansia candida demonstrated antibacterial efficacy against both Gram-positive and Gram-negative pathogens, with Staphylococcus epidermidis exhibiting the lowest MIC at 15.70 μM [196]. Kalata B1, a cyclotide, demonstrated significant antibacterial activity against Gram-positive bacteria, particularly S. aureus (MIC value of 0.26 μM under low salt conditions). However, no activity was observed against Gram-negative E. coli under the same conditions (MIC value > 500 μM). Another cyclotide, Circulin A, displayed potent antibacterial activity against Gram-positive S. aureus (MIC value of 0.19 μM) under low salt conditions, while exhibiting lower activity against Gram-negative P. vulgaris (MIC value of 54.6 μM). Circulin B exhibited moderate activity against S. aureus (MIC value of 13.5 μM) and strong activity against various Gram-negative bacteria, notably E. coli (MIC value of 0.41 μM).
The clinical development of AMPs has generally been bogged down by challenges particularly those related to suboptimal DMPK properties [197]. These classes of plant-derived antimicrobials are inherently unstable metabolically, have short half-lives, and poor oral bioavailability [197,198]. Also, AMPs suffer from reduced antimicrobial efficacy in physiologically relevant conditions. For instance, they can lose their bactericidal potency in environments with physiological salt concentrations, attributed to the disruption of electrostatic interactions between AMPs and cell membranes [199]. Furthermore, toxicity is a major concern with the development of plant-derived AMP due to their potential hemolytic property and poor selectivity to microbial membranes [198]. Although these challenges are not peculiar to PAMPs, they appear to have encountered the greatest obstacles as none have successfully transitioned into clinical use. Moreover, all approved AMPs are derived from Gram-positive soil bacteria, underscoring the notable discrepancy in the clinical success between microorganism-derived and plant-derived AMPs.
4.3.2. Toxicity of plant-derived antimicrobials
It is frequently suggested that herbal medicine and PSMs are generally less toxic [9,200]. However, caution must be exercised as numerous toxic PSMs have been identified [200,201]. Additionally, toxicity can escalate when the dose required for physiological activity is reached, especially for compounds with low antimicrobial potency. For example, reserpine, a plant alkaloid that acts as an inhibitor of the NorA efflux system, enhancing the activity of orfloxacin and ciprofloxacin in S. aureus, exhibits neurotoxic effects at concentrations that inhibit the NorA efflux system [202]. Therefore, researchers should incorporate toxicological screening of antibacterial agents into their workflow and, if possible, counter-screening against mammalian cell lines such as Vero and human cell lines [200,203].
4.3.3. Strategies to improving DMPK
Formulation strategies have been used to improve the clinical potential of poorly bioavailable molecules like cannabinoids. A recent study by Kok et al. (2022) focused on the formulation and pharmacokinetic evaluation of CBD in self-nanoemulsifying drug delivery systems (SNEDDS) [204]. The group prepared CBD-SNEDDS formulations and compared the pharmacokinetic parameters of CBD to those obtained following the administration of CBD in two oil-based formulations. They report that the SNEDDS formulations led to more rapid absorption and more than two-fold systemic exposure to CBD compared to the oil-based formulations [204].
Optimization for DMPK property can be achieved through structure-property relationship (SPR) studies which allow for the medicinal chemist to optimize ADMET properties of compounds. SPR helps the medicinal chemist identify chemical liabilities and metabolic hotspots, enabling necessary chemical modifications to be made. Guidelines such as the Ghose rule, Lipinski's rule of five, Veber's rule of three, and 3Entry rule can be used to predict drug-likeness of plant-derived agents and guide in the selection of lead compounds for further development. Furthermore, these tools can also be useful in prioritizing compounds for synthetic modification during DMPK optimization. Lipinski's rule of five, the most popular of these rules, requires a molecule to have a Molecular mass of less than 500 Da, LogP of less than 5, less than 5 hydrogen bond donors, and less than 10 hydrogen bond acceptors (Table 2). These rules are however not sacrosanct as many approved compounds including antibiotics do not follow all the rules [205].
Online servers such as SwissADME (http://www.swissadme.ch/index.php) and ADMETLab 2.0 (https://admetmesh.scbdd.com/) are resources for in silico determination of drug-likeness of compounds using their molecular structures [206,207]. Using the above platforms, we determined the in silico physicochemical properties and DMPK of selected compounds presented in Table 2 to highlight some of the challenges associated with plant antimicrobials.
4.4. Relative low potency compared to antibiotics and synthetic antibacterials
Plant-derived antimicrobials are generally less potent compared to those derived from fungi and bacteria [9] Although the major focus of natural product research is isolation and determining the single molecule responsible for biological activity, it has been suggested that the antimicrobial activity of plants may result from the combination of molecules acting in an additive or synergistic manner [209]. Perhaps, this synergism or additive property explains why isolated compounds sometimes lose their biological activity or exhibit reduced activity compared to the crude extract or fractions during bioactivity-guided isolation. Antibacterial agents that synergize or possess additive properties act through unique mechanisms or in a way that allows complementary biological action with other NPs present in the crude mixture. One example of synergistic biological effects can be seen in Berberis fremontii Torr., a native American plant that produces both antibacterial berberine and a multidrug resistance pump inhibitor called 5′-methoxyhydnocarpin. This inhibitor prevents the efflux of berberine from pathogenic S. aureus expressing the NorA MDR pump [210]. It is noteworthy that herbal products, including a few examples approved for treating infections, are composed of numerous plant metabolites that collectively elicit their biological activity through synergism [4].
Optimization of antimicrobial activity can be achieved by chemical modification and expansive SAR understanding can yield a semisynthetic analogue with improved potency or broadened spectrum of activity. However, it is apparent that there is relatively limited chemical modification and SAR research focused on plant-derived antimicrobials, in contrast to the successful utilization of this approach in various classes of antibiotics such as penicillin, cephalosporins, tetracyclines, and macrolide antibiotics [[211], [212], [213]].
4.5. Unknown molecular targets or mechanism of action
To date, successful antibiotic projects have been based on phenotypic screening. In contrast, mechanism of action (MOA) or target-based drug discovery programs have been met with failure related to poor whole-cell potency or poor in vivo efficacy later in the program [214]. Yet, understanding MOA and molecular targets is instrumental in structure-based optimization of potency, selectivity, and pharmacokinetic profile, to improve its efficacy and safety [215]. Understanding MOA or ligand-target interaction is even more pertinent for plant-derived antimicrobials since they are generally less potent compared to antibiotics and are faced with suboptimal physicochemical property profiles. Unfortunately, most plant-derived agents’ molecular targets or MOAs are not clearly understood. Various strategies, including mutant selection, chemical genomics, hypersensitivity screening, activity-based profiling, affinity chromatography, and fluorescence labelling, can be employed to identify the molecular target or MOA of plant antibacterials [216]. Other methods such as X-ray crystallography, Cryo-electron microscopy (EM), and photo-proximity labelling offer additional opportunities for 3D elucidation of the binding mode of ligands to their protein target and thus making useful information available to lead optimization [217]. To comprehend the MOA of flavonoids obtained from Traditional Chinese Medicines (TCMs), Tsou et al. (2016) developed alkyne-flavonoid probes for activity-based protein profiling studies, utilizing click chemistry. Their research found that baicalein, a specific flavonoid derived from Scutellaria baicalensis Georgi, targets the bacterial virulence pathway in Salmonella enterica serovar Typhimurium. This pathway, known as S. pathogenicity island-1 (SPI-1), involves the type III secretion system (T3SS) effectors and translocases that are crucial for the invasion of epithelial cells by bacteria [218]. This approach, along with others, should be utilized to investigate isolated plant-derived antimicrobials to determine their drug targets and study their mechanisms of action.
However, this biological experimental determination of drug-target interactions may be tedious and time-consuming. Therefore, various computational methods such as molecular docking, molecular dynamic simulation, and machine learning have been developed to predict possible drug-target interactions [219]. In 2019, the Rastelli group employed in silico modelling to demonstrate that CBG inhibits enoyl acyl carrier protein reductase (InhA), a known target in mycobacteria [220]. While computational techniques, like in silico modelling, can provide a quicker and cost-effective starting point for target identification and elucidation, it is essential to emphasize that biophysical or biochemical methodologies are crucial to confirm molecular targets. In the case of the Rastelli group, they verified the in silico findings of CBG's activity through in vitro testing, revealing that CBG effectively inhibits enoyl acyl carrier protein with an IC50 value of 5.2 ± 0.1 μM [220].
4.5.1. In silico approaches to determining molecular targets or mechanism of action of plant-derived antibacterials
Given that biophysical determination of drug-target interactions may be tedious, costly, and time-consuming, various computer-aided drug design (CADD) strategies have been employed to predict possible drug-target interactions [219]. CADD has played important roles in drug discovery over some decades [221]. Models such as molecular docking, density functional theory (DTF), quantitative structure-activity relationship (QSAR), molecular dynamics simulations, virtual high throughput screening (vHTS), pharmacophore modelling, and drug-likeness studies, have been used to screen thousands of phyto-compounds for their activities against relevant protein targets [[222], [223], [224]]. CADD, which helps in finding biological targets and studying receptor-ligand interactions, has the advantage of reducing the time and cost of experimental work [224,225]. In 2019, the Rastelli group employed in silico modelling to demonstrate that CBG inhibits enoyl acyl carrier protein reductase (InhA), a known target in mycobacteria [220]. Lariciresinol, isolated from the ethyl acetate fraction of Zingiber officinale displays a good binding affinity with crystal structure of the multidrug resistance regulator RamA (PDB ID: 6IE8) (−7.4 kcal/mol) and RamR (PDB ID: 6IE9) (−8.2 kcal/mol) AcrAB-TolC efflux pumps of MDR Salmonella enterica serovar typhimurium. The result was comparable with that of the standard drug tetracycline (6IE8: −8.0 kcal/mol; 6IE9: −8.3 kcal/mol) [226] Geethalakshmi et al. (2018) has found β-hydroxyacyl-(acyl carrier protein) (ACP) dehydratase (FabZ) (PDB ID: 1U1Z) as an important target in the identification of effective anti-Pseudomonas aeruginosa flavonoids from Trianthema decandra L [227]. The ligand interactions with FabZ were attributed to four critical residues namely, GLY58, MSE56, GLU63, and ARG98.
Molecular docking analysis showed that betulinic acid and four other compounds isolated from Ocimum cufodontii (Lanza) A.J. Paton have strong docking efficiency with DNA gyrase, forming hydrophobic interactions and hydrogen bonding with key residues of the enzyme. This indicated that compounds from C. cufodontii could serve as potential DNA gyrase inhibitors [228]. Setzer et al. (2016) investigated 561 phytochemicals listed in the Dictionary of Natural Products for antibacterial properties using in silico approach. The phyto-compounds include alkaloids, terpenoids, flavonoids, chalcones, coumarins, stilbenes, tannins, and quinones, against six bacterial protein targets including DNA gyrase/topoisomerase IV, protein tyrosine phosphatase, peptide deformylase, and cytochrome P450 CYP121 [229]. Prenylated polyphenolics were found to have the best docking profiles, while peptide deformylases and NAD + -dependent DNA ligases were observed to be most susceptible to the compounds. Some of the best docked compounds include cochinchinenene B–D, mulberrofuran D, mulberrofuran Y, eryvarin Q, 5’-(1,1-dimethyl-2-propenyl)-2′,4′,5,7-tetrahydroxy-8-prenylflavanone, 5’-(1,1-dimethyl-2-propenyl)-4′,5,7-trihydroxy-2′-methoxy-8-prenylflavanone. Molecular docking analysis of 70 phyto-compounds from 35 medicinal plants from the Northwestern Himalayas with RamR of Salmonella typhimurium was carried out using AutoDock vina software. The results showed that asiaticoside, betasitosterol, bryophyllin A, mahanimbine, madecassoside, solasonine, solamargine, pennogenin, rutin, withanone, and withaferin A, exhibited the highest binding energy comparable to that of chenodeoxycholic acid [226] While computational techniques, like in silico modelling, can provide a quicker and cost-effective starting point for target identification and elucidation, it is essential to emphasize that biophysical or biochemical methodologies are crucial to confirm molecular targets. In the case of the Rastelli group, they verified the in silico findings of CBG's activity through in vitro testing, revealing that CBG effectively inhibits enoyl acyl carrier protein with an IC50 value of 5.2 ± 0.1 μM [220].
4.6. Lack of innovation and poor quality of research
The WHO's innovative research criteria for the development of antibacterials include the absence of known cross-resistance, a new target, a new MOA, and/or a new chemical class. With these criteria in mind, it is obvious that the lack of innovation poses a significant barrier to the research and development (R&D) of antibacterial agents from all sources and plant-derived antimicrobials in particular [230,231]. Many published articles only focus on testing crude extracts on laboratory bacterial isolates, and when compounds are isolated, extended biological testing to determine their MOA, target, and the presence or absence of cross-resistance is often missing. There is also concern over the lack of standard antimicrobial testing and reporting methods [4]. The agar diffusion method is inappropriate for quantitative analysis of plant extracts since non-polar compounds may fail to diffuse, leading to false negative results. Despite its drawbacks, some researchers still utilize the agar diffusion assay instead of more suitable options such as broth microdilution or agar dilution assays. Furthermore, the lack of proper botanical authentication and deposition of voucher specimens for the plants used in many studies raises concerns about the reliability of their results [4,232].
Most antimicrobial drug discovery projects based on plant-derived agents primarily originate from the Global South (India, Bangladesh, Nigeria, Brazil, Kenya etc), which possesses abundant and diverse forest resources. However, these regions often encounter known disparities and face research resource constraints that hinder the pursuit of advanced and innovative scientific research when compared to the Global North (France, United States, UK, Germany etc.) [233,234]. These challenges can be overcome through international and multidisciplinary research collaborations involving laboratories equipped with state-of-the-art instruments.
4.7. Lack of interest by large pharmaceutical companies
There is no gainsaying that taking a drug from bench to bedside is capital intensive and often beyond the reach of regular academic laboratories. It costs approximately $2 billion for research and development of a new drug for clinical use [235]. Hence, proprietary rights and profit are great incentives for the pharmaceutical industry to pursue a drug discovery program. Unfortunately, the process of obtaining patent protection for NPs has become increasingly challenging. In addition to various business, economic, societal, and regulatory issues, the antibiotics market has faced a decline in appeal, resulting in the withdrawal of large pharmaceutical companies from antibiotic drug discovery efforts [236]. As a result, the responsibility for such discoveries has largely shifted to academic laboratories and small to medium-scale companies [236]. Getting the pharmaceutical industries interested in plant-derived antibacterials will be a herculean task considering all the challenges raised above. Academic laboratories alone are simply not resourceful enough for translational science. We suggest a strategic alliance between academia, government organizations, the private sector, and non-profit organizations interested in solving the problem of AMR.
5. Future outlook and conclusions
Biomedical sciences have made significant advancements surpassing the “golden age” of antibiotic discovery, during which most of today's clinically available antibiotics were identified. Despite this scientific progress, combating AMR remains an unresolved and pressing health concern. Researchers have increasingly focused on studying and evaluating secondary metabolites derived from fungi, bacteria, and plants for antibacterial development. In our report, we examine the reasons behind the relatively limited success in discovering and developing plant-based antimicrobials, despite numerous research efforts in this field.
While challenges like “limited sources” and “dereplication” are common across NPs from various sources and for different biological activities, certain hurdles are more prevalent for plant-derived compounds. These include relatively low antimicrobial activity, a lack of innovation, and a dearth of high-quality research. To address these challenges, we propose fostering extensive international scientific collaborations. Collaboration is particularly encouraged between the Global South, with its vast tropical forest resources, and the Global North which possesses advanced scientific capabilities, to overcome the aforementioned obstacles. Regarding research funding, we recommend establishing a collaborative partnership among academic institutions, government organizations, private enterprises, and non-profit organizations. Only through this collective effort can we address the challenge of AMR by exploring innovative approaches and harnessing the potential of medicinal plants to discover new compounds that facilitate advancements in antibacterial drug development.
CRediT authorship contribution statement
Tomayo I. Berida: Writing – review & editing, Writing – original draft, Software, Methodology, Formal analysis, Data curation, Conceptualization. Yemi A. Adekunle: Writing – review & editing, Writing – original draft, Software, Methodology, Formal analysis, Data curation, Conceptualization. Hannah Dada-Adegbola: Writing – review & editing, Writing – original draft, Methodology, Formal analysis, Data curation, Conceptualization. Ayoub Kdimy: Writing – review & editing, Writing – original draft, Methodology, Formal analysis, Data curation, Conceptualization. Sudeshna Roy: Writing – review & editing, Writing – original draft, Supervision, Formal analysis, Conceptualization. Satyajit D. Sarker: Writing – review & editing, Writing – original draft, Supervision, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Funding
T.B. was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R21AI142210). S.R. was supported by the National Institute of General Medical Sciences of the National Institutes of Health (Award Number R35GM150768). Y.A.A. was supported by the Commonwealth Scholarships Commission, UK (NGCN-2021-184). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Contributor Information
Tomayo I. Berida, Email: tberida@go.olemiss.edu.
Yemi A. Adekunle, Email: y.a.adekunle@2022.ljmu.ac.uk.
Satyajit D. Sarker, Email: s.sarker@ljmu.ac.uk.
References
- 1.Wiart C. Springer Science \& Business Media; 2007. Ethnopharmacology of Medicinal Plants: Asia and the Pacific. [Google Scholar]
- 2.Cowan M.M. Plant products as antimicrobial agents. Clin. Microbiol. Rev. 1999 Oct 1;12(4):564 LP–582. doi: 10.1128/cmr.12.4.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rahman M., Sarker S.D. 2020. Antimicrobial Natural Products; pp. 77–113. [Google Scholar]
- 4.Chassagne F., Samarakoon T., Porras G., Lyles J.T., Dettweiler M., Marquez L., et al. A systematic review of plants with antibacterial activities: a Taxonomic and Phylogenetic Perspective. Front. Pharmacol. 2021 Jan 8;11 doi: 10.3389/fphar.2020.586548. https://www.frontiersin.org/articles/10.3389/fphar.2020.586548/full [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bérdy J. Thoughts and facts about antibiotics: where we are now and where we are heading. J. Antibiot. (Tokyo) 2012 Aug 18;65(8):385–395. doi: 10.1038/ja.2012.27. http://www.nature.com/articles/ja201227 [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 6.Newman D.J., Cragg G.M. Natural products as sources of new drugs over the Nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020 Mar 27;83(3):770–803. doi: 10.1021/acs.jnatprod.9b01285. [DOI] [PubMed] [Google Scholar]
- 7.Atanasov A.G., Zotchev S.B., Dirsch V.M., Supuran C.T. Natural products in drug discovery: advances and opportunities. Nat. Rev. Drug Discov. 2021 Mar 28;20(3):200–216. doi: 10.1038/s41573-020-00114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shambaugh G.E. History of sulfonamides. Arch. Otolaryngol. Head Neck Surg. 1966 Jan 1;83(1):1–2. http://archotol.jamanetwork.com/article.aspx?articleid=599738 [Internet] Available from: [Google Scholar]
- 9.Wright G.D. Opportunities for natural products in 21st century antibiotic discovery. Nat. Prod. Rep. 2017;34(7):694–701. doi: 10.1039/C7NP00019G. [Internet] [DOI] [PubMed] [Google Scholar]
- 10.Holmes A.H., Moore L.S.P., Sundsfjord A., Steinbakk M., Regmi S., Karkey A., et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet. 2016 Jan;387(10014):176–187. doi: 10.1016/S0140-6736(15)00473-0. https://linkinghub.elsevier.com/retrieve/pii/S0140673615004730 [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 11.Barber M. Staphylococcal infection due to penicillin-resistant strains. BMJ. 1947 Nov 29;2(4534):863–865. doi: 10.1136/bmj.2.4534.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Podolsky S.H. The evolving response to antibiotic resistance (1945–2018) Palgrave Commun. 2018 Oct 23;4(1):124. [Google Scholar]
- 13.Murray C.J., Ikuta K.S., Sharara F., Swetschinski L., Robles Aguilar G., Gray A., et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022 Feb;399(10325):629–655. doi: 10.1016/S0140-6736(21)02724-0. https://linkinghub.elsevier.com/retrieve/pii/S0140673621027240 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richter M.F., Drown B.S., Riley A.P., Garcia A., Shirai T., Svec R.L., et al. Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature. 2017 May 10;545(7654):299–304. doi: 10.1038/nature22308. http://www.nature.com/articles/nature22308 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rice L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008 Apr 15;197(8):1079–1081. doi: 10.1086/533452. https://academic.oup.com/jid/article-lookup/doi/10.1086/533452 [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 16.De Oliveira D.M.P., Forde B.M., Kidd T.J., Harris P.N.A., Schembri M.A., Beatson S.A., et al. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020 May 13;33(3) doi: 10.1128/CMR.00181-19. http://cmr.asm.org/lookup/doi/10.1128/CMR.00181-19 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parker E.N., Drown B.S., Geddes E.J., Lee H.Y., Ismail N., Lau G.W., et al. Implementation of permeation rules leads to a FabI inhibitor with activity against Gram-negative pathogens. Nat Microbiol. 2020 Jan 18;5(1):67–75. doi: 10.1038/s41564-019-0604-5. https://www.nature.com/articles/s41564-019-0604-5 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muñoz K.A., Hergenrother P.J. Facilitating compound Entry as a means to discover antibiotics for gram-negative bacteria. Acc. Chem. Res. 2021 Mar 16;54(6):1322–1333. doi: 10.1021/acs.accounts.0c00895. https://pubs.acs.org/doi/10.1021/acs.accounts.0c00895 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pai M., Behr M.A., Dowdy D., Dheda K., Divangahi M., Boehme C.C., et al. Tuberculosis. Nat Rev Dis Primers. 2016 Dec 22;2(1) doi: 10.1038/nrdp.2016.76. http://www.nature.com/articles/nrdp201676 [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 20.Fernandes G.F.S., Thompson A.M., Castagnolo D., Denny W.A., Dos Santos J.L. Tuberculosis drug discovery: challenges and new horizons. J. Med. Chem. 2022 Jun 9;65(11):7489–7531. doi: 10.1021/acs.jmedchem.2c00227. [DOI] [PubMed] [Google Scholar]
- 21.Dean A.S., Tosas Auguet O., Glaziou P., Zignol M., Ismail N., Kasaeva T., et al. 25 years of surveillance of drug-resistant tuberculosis: achievements, challenges, and way forward. Lancet Infect. Dis. 2022 Jul;22(7):e191–e196. doi: 10.1016/S1473-3099(21)00808-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma A., De Rosa M., Singla N., Singh G., Barnwal R.P., Pandey A. Tuberculosis: an overview of the immunogenic response, disease progression, and medicinal chemistry efforts in the last decade toward the development of potential drugs for extensively drug-resistant tuberculosis strains. J. Med. Chem. 2021 Apr 22;64(8):4359–4395. doi: 10.1021/acs.jmedchem.0c01833. [DOI] [PubMed] [Google Scholar]
- 23.Gupta V.K., Kumar M.M., Bisht D., Kaushik A. Plants in our combating strategies against Mycobacterium tuberculosis: progress made and obstacles met. Pharm. Biol. 2017 Dec;55(1):1536–1544. doi: 10.1080/13880209.2017.1309440. https://pubmed.ncbi.nlm.nih.gov/28385088 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seung K.J., Keshavjee S., Rich M.L. Multidrug-resistant tuberculosis and extensively drug-resistant tuberculosis. Cold Spring Harb Perspect Med. 2015 Sep;5(9):a017863. doi: 10.1101/cshperspect.a017863. http://perspectivesinmedicine.cshlp.org/lookup/doi/10.1101/cshperspect.a017863 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bagcchi S. WHO's global tuberculosis report 2022. Lancet Microbe. 2023 Jan;4(1) doi: 10.1016/S2666-5247(22)00359-7. [DOI] [PubMed] [Google Scholar]
- 26.Geneva: World Health Organization . 2023. Global Tuberculosis Report 2023.https://www.who.int/publications/i/item/9789240083851 [Internet]. Geneva. [cited 2024 Mar 26]. Available from: [Google Scholar]
- 27.Alemu A., Bitew Z.W., Worku T., Gamtesa D.F., Alebel A. Predictors of mortality in patients with drug-resistant tuberculosis: a systematic review and meta-analysis. PLoS One. 2021 Jun 28;16(6) doi: 10.1371/journal.pone.0253848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Kraker M.E.A., Stewardson A.J., Harbarth S. Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 2016 Nov 29;13(11) doi: 10.1371/journal.pmed.1002184. https://dx.plos.org/10.1371/journal.pmed.1002184 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Neill J. Antimicrobial Resistance: tackling a crisis for the health and wealth of nations. The Review on Antimicrobial Resistance. 2014 https://amr-review.org/ [Internet] Available from: [Google Scholar]
- 30.Porras G., Chassagne F., Lyles J.T., Marquez L., Dettweiler M., Salam A.M., et al. Ethnobotany and the role of plant natural products in antibiotic drug discovery. Chem Rev. 2021 Mar 24;121(6):3495–3560. doi: 10.1021/acs.chemrev.0c00922. https://pubs.acs.org/doi/10.1021/acs.chemrev.0c00922 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salam A.M., Quave C.L. Opportunities for plant natural products in infection control. Curr. Opin. Microbiol. 2018 Oct;45:189–194. doi: 10.1016/j.mib.2018.08.004. https://linkinghub.elsevier.com/retrieve/pii/S1369527417300851 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Górniak I., Bartoszewski R., Króliczewski J. Comprehensive review of antimicrobial activities of plant flavonoids. Phytochemistry Rev. 2019;18(1):241–272. doi: 10.1007/s11101-018-9591-z. [Internet] [DOI] [Google Scholar]
- 33.Gorlenko C.L., Kiselev HYu, Budanova E.V., Zamyatnin A.A., Ikryannikova L.N. Plant secondary metabolites in the battle of drugs and drug-resistant bacteria: new heroes or worse clones of antibiotics? Antibiotics. 2020 Apr 10;9(4):170. doi: 10.3390/antibiotics9040170. https://www.mdpi.com/2079-6382/9/4/170 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elmaidomy A.H., Shady N.H., Abdeljawad K.M., Elzamkan M.B., Helmy H.H., Tarshan E.A., et al. Antimicrobial potentials of natural products against multidrug resistance pathogens: a comprehensive review. RSC Adv. 2022;12(45):29078–29102. doi: 10.1039/d2ra04884a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kopel J., McDonald J., Hamood A. An assessment of the in vitro models and clinical trials related to the antimicrobial activities of phytochemicals. Antibiotics. 2022 Dec 17;11(12):1838. doi: 10.3390/antibiotics11121838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vargiu A.V., Pos K.M., Poole K., Nikaido H. Editorial: bad bugs in the XXIst century: resistance mediated by multi-drug efflux pumps in gram-negative bacteria. Front. Microbiol. 2016 May 31;7 doi: 10.3389/fmicb.2016.00833. http://journal.frontiersin.org/Article/10.3389/fmicb.2016.00833/abstract [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clifton L.A., Skoda M.W.A., Le Brun A.P., Ciesielski F., Kuzmenko I., Holt S.A., et al. Effect of divalent cation removal on the structure of gram-negative bacterial outer membrane models. Langmuir. 2015 Jan 13;31(1):404–412. doi: 10.1021/la504407v. https://pubs.acs.org/doi/10.1021/la504407v [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gutsmann T., Seydel U. Impact of the glycostructure of amphiphilic membrane components on the function of the outer membrane of Gram-negative bacteria as a matrix for incorporated channels and a target for antimicrobial peptides or proteins. Eur. J. Cell Biol. 2010 Jan;89(1):11–23. doi: 10.1016/j.ejcb.2009.10.011. https://linkinghub.elsevier.com/retrieve/pii/S0171933509003252 [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 39.Alderwick L.J., Harrison J., Lloyd G.S., Birch H.L. The mycobacterial cell wall—peptidoglycan and arabinogalactan. Cold Spring Harb Perspect Med. 2015 Aug;5(8):a021113. doi: 10.1101/cshperspect.a021113. http://perspectivesinmedicine.cshlp.org/lookup/doi/10.1101/cshperspect.a021113 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lehman M.K., Grabowicz M. Countering gram-negative antibiotic resistance: recent progress in disrupting the outer membrane with novel therapeutics. Antibiotics. 2019;8 doi: 10.3390/antibiotics8040163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cloeckaert A., Zygmunt M.S., Doublet B. Editorial: genetics of acquired antimicrobial resistance in animal and zoonotic pathogens. Front. Microbiol. 2017 Dec 5;8 doi: 10.3389/fmicb.2017.02428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munita J.M., Arias C.A. Mechanisms of antibiotic resistance. Microbiol. Spectr. 2016 Mar 25;4(2) doi: 10.1128/microbiolspec.VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mora-Ochomogo M., Lohans C.T. β-Lactam antibiotic targets and resistance mechanisms: from covalent inhibitors to substrates. RSC Med. Chem. 2021;12(10):1623–1639. doi: 10.1039/d1md00200g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zapun A., Contreras-Martel C., Vernet T. Penicillin-binding proteins and β-lactam resistance. FEMS Microbiol. Rev. 2008 Mar;32(2):361–385. doi: 10.1111/j.1574-6976.2007.00095.x. [DOI] [PubMed] [Google Scholar]
- 45.Tyson G.H., Li C., Hsu C.H., Bodeis-Jones S., McDermott P.F. Diverse fluoroquinolone resistance plasmids from retail meat E. coli in the United States. Front. Microbiol. 2019 Dec 5;10 doi: 10.3389/fmicb.2019.02826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Redgrave L.S., Sutton S.B., Webber M.A., Piddock L.J.V. Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 2014 Aug;22(8):438–445. doi: 10.1016/j.tim.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 47.Pitso L., Potgieter S., Van der Spoel van Dijk A. Prevalence of isoniazid resistance-conferring mutations associated with multidrug-resistant tuberculosis in Free State Province, South Africa. S. Afr. Med. J. 2019 Aug 28;109(9):659. doi: 10.7196/SAMJ.2019.v109i9.13730. [DOI] [PubMed] [Google Scholar]
- 48.Lempens P., Meehan C.J., Vandelannoote K., Fissette K., de Rijk P., Van Deun A., et al. Isoniazid resistance levels of Mycobacterium tuberculosis can largely be predicted by high-confidence resistance-conferring mutations. Sci. Rep. 2018 Feb 19;8(1):3246. doi: 10.1038/s41598-018-21378-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barozi V., Musyoka T.M., Sheik Amamuddy O., Tastan Bishop Ö. Deciphering isoniazid drug resistance mechanisms on dimeric Mycobacterium tuberculosis KatG via post-molecular dynamics analyses including combined dynamic residue network metrics. ACS Omega. 2022 Apr 19;7(15):13313–13332. doi: 10.1021/acsomega.2c01036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Darby E.M., Trampari E., Siasat P., Gaya M.S., Alav I., Webber M.A., et al. Molecular mechanisms of antibiotic resistance revisited. Nat. Rev. Microbiol. 2023 May 21;21(5):280–295. doi: 10.1038/s41579-022-00820-y. [DOI] [PubMed] [Google Scholar]
- 51.Cragg G.M., Newman D.J. Natural products: a continuing source of novel drug leads. Biochim. Biophys. Acta Gen. Subj. 2013 Jun;1830(6):3670–3695. doi: 10.1016/j.bbagen.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Radulovic N.S., Blagojevic P.D., Stojanovic-Radic Z.Z., Stojanovic N.M. Antimicrobial plant metabolites: structural diversity and mechanism of action. Curr. Med. Chem. 2013 Feb 1;20(7):932–952. http://www.eurekaselect.com/openurl/content.php?genre=article&issn=0929-8673&volume=20&issue=7&spage=932 [Internet] Available from: [PubMed] [Google Scholar]
- 53.Masyita A., Mustika Sari R., Dwi Astuti A., Yasir B., Rahma Rumata N., Emran T Bin, et al. Terpenes and terpenoids as main bioactive compounds of essential oils, their roles in human health and potential application as natural food preservatives. Food Chem. X. 2022 Mar;13 doi: 10.1016/j.fochx.2022.100217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cushnie T.P.T., Cushnie B., Lamb A.J. Alkaloids: an overview of their antibacterial, antibiotic-enhancing and antivirulence activities. Int. J. Antimicrob. Agents. 2014 Nov;44(5):377–386. doi: 10.1016/j.ijantimicag.2014.06.001. https://linkinghub.elsevier.com/retrieve/pii/S0924857914001885 [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 55.Park K.M., Choo J.H., Lee S.H., Hwang J.K. Antibacterial activity of panduratin A isolated from Kaempferia pandurata against Porphyromonas gingivalis. Food Sci. Biotechnol. 2005;14(2):286–289. [Google Scholar]
- 56.Rukayadi Y., Lee K., Han S., Yong D., Hwang J.K. In vitro activities of panduratin A against clinical Staphylococcus strains. Antimicrob. Agents Chemother. 2009 Oct;53(10):4529–4532. doi: 10.1128/AAC.00624-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rukayadi Y., Han S., Yong D., Hwang J.K. In vitro antibacterial activity of panduratin A against enterococci clinical isolates. Biol. Pharm. Bull. 2010;33(9):1489–1493. doi: 10.1248/bpb.33.1489. [DOI] [PubMed] [Google Scholar]
- 58.Song Min Soo, Shim Jae Seok, Gwon Song Hui, Lee Chan Woo, Kim Han Sung, Jkwan Hwang. Antibacterial activity of panduratin A and isopanduratin A isolated from Kaemfperia pandurata Roxb. against acne-causing microorganisms. Food Science and Biotechnology Biotechnol. 2008;17(6) [Google Scholar]
- 59.Rukayadi Y., Lee K.H., Hwang J.K. Activity of panduratin A isolated from Kaempferia pandurata Roxb. against multi-species oral biofilms in vitro. J. Oral Sci. 2009;51(1):87–95. doi: 10.2334/josnusd.51.87. [DOI] [PubMed] [Google Scholar]
- 60.Kanjanasirirat P., Suksatu, Ampa Kanjanasirirat P., Suksatu A., Manopwisedjaroen S., Munyoo B., Tuchinda P., Jearawuttanakul K., Seemakhan S., Charoensutthivarakul S., Wongtrakoongate P., Rangkasenee N., Pitiporn S., Waranuch N., Chabang N., Khemawoot P., Manopwisedjaroen S., Munyoo B., Tuchinda P., Jearawuttanakul K., et al. High-content screening of Thai medicinal plants reveals Boesenbergia rotunda extract and its component Panduratin A as anti-SARS-CoV-2 agents. Sci. Rep. 2020 Dec 17;10(1) doi: 10.1038/s41598-020-77003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kirana C., Jones G.P., Record I.R., McIntosh G.H. Anticancer properties of panduratin A isolated from Boesenbergia pandurata (Zingiberaceae) J. Nat. Med. 2007 Feb 28;61(2):131–137. [Google Scholar]
- 62.Reddy M., Gupta S., Jacob M., Khan S., Ferreira D. Antioxidant, antimalarial and antimicrobial activities of tannin-rich fractions, ellagitannins and phenolic acids from Punica granatum L. Planta Med. 2007 May;73(5):461–467. doi: 10.1055/s-2007-967167. [DOI] [PubMed] [Google Scholar]
- 63.Li Y., Guo C., Yang J., Wei J., Xu J., Cheng S. Evaluation of antioxidant properties of pomegranate peel extract in comparison with pomegranate pulp extract. Food Chem. 2006 May;96(2):254–260. [Google Scholar]
- 64.Nur Hanani Z.A., Aelma Husna A.B., Nurul Syahida S., Nor Khaizura M.A.B., Jamilah B. Effect of different fruit peels on the functional properties of gelatin/polyethylene bilayer films for active packaging. Food Packag. Shelf Life. 2018 Dec;18:201–211. [Google Scholar]
- 65.Andrade M.A., Lima V., Sanches Silva A., Vilarinho F., Castilho M.C., Khwaldia K., et al. Pomegranate and grape by-products and their active compounds: are they a valuable source for food applications? Trends Food Sci. Technol. 2019 Apr;86:68–84. https://linkinghub.elsevier.com/retrieve/pii/S0924224418308690 [Internet] Available from: [Google Scholar]
- 66.Dahham S.S., Ali M.N., Tabassum H., Khan M. Studies on antibacterial and antifungal activity of pomegranate. Am.-Eurasian J. Agric. Environ. Sci. 2010;9(3):273–281. [Google Scholar]
- 67.Barbieri M., Heard C.M. Isolation of punicalagin from Punica granatum rind extract using mass-directed semi-preparative ESI-AP single quadrupole LC-MS. J. Pharm. Biomed. Anal. 2019 Mar;166:90–94. doi: 10.1016/j.jpba.2018.12.033. https://linkinghub.elsevier.com/retrieve/pii/S0731708518309397 [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 68.Gosset-Erard C., Zhao M., Lordel-Madeleine S., Ennahar S. Identification of punicalagin as the bioactive compound behind the antimicrobial activity of pomegranate (Punica granatum L.) peels. Food Chem. 2021 Aug;352 doi: 10.1016/j.foodchem.2021.129396. https://linkinghub.elsevier.com/retrieve/pii/S0308814621004027 [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 69.Pelter A., Stainton P. The extractives from Derris scandens. Part II. The isolation of osajin and two new isoflavones, scandenone and scandinone. J. Chem. Soc. C Org. 1966:701. [Google Scholar]
- 70.Peleyeju G.B., Emmanuel T., Tata C.M., Djuidje Fotsing M.C., Niemann N., Rhyman L., et al. Crystal structure and antibacterial activity of scandenone (warangalone) from Erythrina plants. J. Mol. Struct. 2019 Sep;1191:43–51. https://linkinghub.elsevier.com/retrieve/pii/S0022286019304739 [Internet] Available from: [Google Scholar]
- 71.Peterson C., Fristad A., Tsao R., Coats J.R. Osajin and pomiferin, two isoflavones purified from osage orange fruits, tested for repellency to the maize weevil (Coleoptera: Curculionidae) Environ. Entomol. 2000 Dec 1;29(6):1133–1137. [Google Scholar]
- 72.Nkengfack A.E., Vouffo T.W., Vardamides J.C., Kouam J., Fomum Z.T., Meyer M., et al. Phenolic metabolites from Erythrina species. Phytochemistry. 1997 Oct;46(3):573–578. https://linkinghub.elsevier.com/retrieve/pii/S0031942297002914 [Internet] Available from: [Google Scholar]
- 73.Innok P., Rukachaisirikul T., Phongpaichit S., Suksamrarn A. Fuscacarpans A–C, new pterocarpans from the stems of Erythrina fusca. Fitoterapia. 2010 Sep;81(6):518–523. doi: 10.1016/j.fitote.2010.01.009. https://linkinghub.elsevier.com/retrieve/pii/S0367326X10000134 [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 74.Morris M.P., Pagán C. The isolation of the toxic principles of mamey. J. Am. Chem. Soc. 1953 Mar 1;75(6):1489. https://pubs.acs.org/doi/abs/10.1021/ja01102a514 [Internet] Available from: [Google Scholar]
- 75.Ouahouo B.M.W., Azebaze A.G.B., Meyer M., Bodo B., Fomum Z.T., Nkengfack A.E. Cytotoxic and antimicrobial coumarins from Mammea africana. Ann. Trop. Med. Parasitol. 2004 Oct 18;98(7):733–739. doi: 10.1179/000349804X3126. [DOI] [PubMed] [Google Scholar]
- 76.Lemus C., Smith-Ravin J., Marcelin O. Mammea americana: a review of traditional uses, phytochemistry and biological activities. J. Herb. Med. 2021 Oct;29 [Google Scholar]
- 77.Pájaro-González Y., Oliveros-Díaz A.F., Cabrera-Barraza J., Fernández-Daza E., Reyes N., Montes-Guevara O.A., et al. Mammea B/BA isolated from the seeds of Mammea americana L. (Calophyllaceae) is a potent inhibitor of methicillin-resistant Staphylococcus aureus. Front. Pharmacol. 2022 Mar 11;13 doi: 10.3389/fphar.2022.826404. https://www.frontiersin.org/articles/10.3389/fphar.2022.826404/full [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pino Pérez O., Jorge Lazo F., Morales T.E., Juan Khambay B.P.S. Isolation and characterization of active compounds from Mammea americana Lin. Rev. Cubana Quím. 2007;19(1):74–77. [Google Scholar]
- 79.Yang H., Protiva P., Gil R.R., Jiang B., Baggett S., Basile M.J., et al. Antioxidant and cytotoxic isoprenylated coumarins from Mammea americana. Planta Med. 2005 Sep;71(9):852–860. doi: 10.1055/s-2005-871257. [DOI] [PubMed] [Google Scholar]
- 80.Canning C., Sun S., Ji X., Gupta S., Zhou K. Antibacterial and cytotoxic activity of isoprenylated coumarin mammea A/AA isolated from Mammea africana. J. Ethnopharmacol. 2013 May;147(1):259–262. doi: 10.1016/j.jep.2013.02.026. [DOI] [PubMed] [Google Scholar]
- 81.Hill K.P., Gold M.S., Nemeroff C.B., McDonald W., Grzenda A., Widge A.S., et al. Risks and benefits of cannabis and cannabinoids in psychiatry. Am. J. Psychiatr. 2022 Feb;179(2):98–109. doi: 10.1176/appi.ajp.2021.21030320. [DOI] [PubMed] [Google Scholar]
- 82.Mechoulam R., Gaoni Y. Hashish—IV: the isolation and structure of cannabinolic cannabidiolic and cannabigerolic acids. Tetrahedron. 1965;21(5):1223–1229. doi: 10.1016/0040-4020(65)80064-3. [DOI] [PubMed] [Google Scholar]
- 83.Kabelik K., Krejci Z., Santavy F. Cannabis as a medicament. Bull Narcotics. 1960;5(12):5–23. https://www.unodc.org/unodc/en/data-and-analysis/bulletin/bulletin_1960-01-01_3_page003.html [Internet] Available from: [Google Scholar]
- 84.Appendino G., Gibbons S., Giana A., Pagani A., Grassi G., Stavri M., et al. Antibacterial cannabinoids from Cannabis sativa: a Structure−Activity study. J. Nat. Prod. 2008 Aug;71(8):1427–1430. doi: 10.1021/np8002673. https://pubs.acs.org/doi/10.1021/np8002673 [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 85.Farha M.A., El-Halfawy O.M., Gale R.T., MacNair C.R., Carfrae L.A., Zhang X., et al. Uncovering the hidden antibiotic potential of cannabis. ACS Infect. Dis. 2020 Mar 13;6(3):338–346. doi: 10.1021/acsinfecdis.9b00419. https://pubs.acs.org/doi/10.1021/acsinfecdis.9b00419 [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 86.Blaskovich M.A.T., Kavanagh A.M., Elliott A.G., Zhang B., Ramu S., Amado M., et al. The antimicrobial potential of cannabidiol. Commun. Biol. 2021 Dec 19;4(1):7. doi: 10.1038/s42003-020-01530-y. http://www.nature.com/articles/s42003-020-01530-y [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mahiou V., Roblot F., Hocquemiller R., Cavé A., Barrios A.A., Fournet A., et al. Piperogalin, a new prenylated diphenol from Peperomia galioides. J. Nat. Prod. 1995 Feb 1;58(2):324–328. doi: 10.1021/np50116a031. [DOI] [PubMed] [Google Scholar]
- 88.Jentsch N.G., Zhang X., Magolan J. Efficient synthesis of cannabigerol, grifolin, and piperogalin via alumina-promoted allylation. J. Nat. Prod. 2020 Sep 25;83(9):2587–2591. doi: 10.1021/acs.jnatprod.0c00131. [DOI] [PubMed] [Google Scholar]
- 89.Kitahara T., Kanda N. DB-2073, A new alkylresorcinol antibiotic. II. The chemical structure of DB-2073. J. Antibiot. (Tokyo) 1975;28(12):943–946. doi: 10.7164/antibiotics.28.943. [DOI] [PubMed] [Google Scholar]
- 90.Nachnani R., Raup-Konsavage W.M., Vrana K.E. The pharmacological case for cannabigerol. J. Pharmacol. Exp. Therapeut. 2021 Feb;376(2):204–212. doi: 10.1124/jpet.120.000340. [DOI] [PubMed] [Google Scholar]
- 91.Perucca E., Bialer M. Critical aspects affecting cannabidiol oral bioavailability and metabolic elimination, and related clinical implications. CNS Drugs. 2020 Aug 5;34(8):795–800. doi: 10.1007/s40263-020-00741-5. https://link.springer.com/10.1007/s40263-020-00741-5 [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 92.Butler M.S., Henderson I.R., Capon R.J., Blaskovich M.A.T. Antibiotics in the clinical pipeline as of December 2022. J. Antibiot. (Tokyo) 2023 Aug 8;76(8):431–473. doi: 10.1038/s41429-023-00629-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dahmer J., Marangon P., Adolpho L.O., Reis F.L., Maldaner G., Burrow R.A., et al. Alkaloids from the stem barks of Scutia buxifolia Reissek (Rhamnaceae): structures and antimicrobial evaluation. Phytochemistry. 2022 Apr;196 doi: 10.1016/j.phytochem.2021.113071. https://linkinghub.elsevier.com/retrieve/pii/S0031942221004209 [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 94.Habtemariam S. Berberine pharmacology and the gut microbiota: a hidden therapeutic link. Pharmacol. Res. 2020 May;155 doi: 10.1016/j.phrs.2020.104722. [DOI] [PubMed] [Google Scholar]
- 95.Gaba S., Saini A., Singh G., Monga V. An insight into the medicinal attributes of berberine derivatives: a review. Bioorg. Med. Chem. 2021 May;38 doi: 10.1016/j.bmc.2021.116143. [DOI] [PubMed] [Google Scholar]
- 96.Cai Y., Xin Q., Lu J., Miao Y., Lin Q., Cong W., et al. A new therapeutic candidate for cardiovascular diseases: berberine. Front. Pharmacol. 2021 Mar 17;12 doi: 10.3389/fphar.2021.631100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xia L., Luo M. Study progress of berberine for treating cardiovascular disease. Chronic Dis Transl Med. 2015 Dec;1(4):231–235. doi: 10.1016/j.cdtm.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang Y., Liu Y., Du X., Ma H., Yao J. pThe anti-cancer mechanisms of berberine: a review. Cancer Manag. Res. 2020 Jan;12:695–702. doi: 10.2147/CMAR.S242329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xu X., Yi H., Wu J., Kuang T., Zhang J., Li Q., et al. Therapeutic effect of berberine on metabolic diseases: both pharmacological data and clinical evidence. Biomed. Pharmacother. 2021 Jan;133 doi: 10.1016/j.biopha.2020.110984. [DOI] [PubMed] [Google Scholar]
- 100.Ye M., Fu S., Pi R., He F. Neuropharmacological and pharmacokinetic properties of berberine: a review of recent research. J. Pharm. Pharmacol. 2010 Jan 8;61(7):831–837. doi: 10.1211/jpp/61.07.0001. [DOI] [PubMed] [Google Scholar]
- 101.Wojtyczka R., Dziedzic A., Kępa M., Kubina R., Kabała-Dzik A., Mularz T., et al. Berberine enhances the antibacterial activity of selected antibiotics against coagulase-negative Staphylococcus strains in vitro. Molecules. 2014 May 22;19(5):6583–6596. doi: 10.3390/molecules19056583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xia S., Ma L., Wang G., Yang J., Zhang M., Wang X., et al. In vitro antimicrobial activity and the mechanism of berberine against methicillin-resistant Staphylococcus aureus isolated from bloodstream infection patients. Infect. Drug Resist. 2022 Apr;15:1933–1944. doi: 10.2147/IDR.S357077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Čerňáková M., Košťálová D. Antimicrobial activity of berberine—a constituent of Mahonia aquifolium. Folia Microbiol (Praha) 2002 Aug 19;47(4):375–378. doi: 10.1007/BF02818693. [DOI] [PubMed] [Google Scholar]
- 104.Wu S., Yang K., Hong Y., Gong Y., Ni J., Yang N., et al. A new perspective on the antimicrobial mechanism of berberine hydrochloride against Staphylococcus aureus revealed by untargeted metabolomic studies. Front. Microbiol. 2022 Jul 13:13. doi: 10.3389/fmicb.2022.917414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xia S., Ma L., Wang G., Yang J., Zhang M., Wang X., et al. In vitro antimicrobial activity and the mechanism of berberine against methicillin-resistant Staphylococcus aureus isolated from bloodstream infection patients. Infect. Drug Resist. 2022 Apr;15:1933–1944. doi: 10.2147/IDR.S357077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li X., Song Y., Wang L., Kang G., Wang P., Yin H., et al. A potential combination therapy of berberine hydrochloride with antibiotics against multidrug-resistant Acinetobacter baumannii. Front. Cell. Infect. Microbiol. 2021 Mar 25:11. doi: 10.3389/fcimb.2021.660431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li Y., Ge X. Role of berberine as a potential efflux pump inhibitor against MdfA from Escherichia coli: in vitro and in silico studies. Microbiol. Spectr. 2023 Apr 13;11(2) doi: 10.1128/spectrum.03324-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Morita Y., Nakashima K ichi, Nishino K., Kotani K., Tomida J., Inoue M., et al. Berberine is a novel type efflux inhibitor which attenuates the MexXY-mediated aminoglycoside resistance in Pseudomonas aeruginosa. Front. Microbiol. 2016 Aug 5;7 doi: 10.3389/fmicb.2016.01223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Peng L., Kang S., Yin Z., Jia R., Song X., Li L., et al. Antibacterial activity and mechanism of berberine against Streptococcus agalactiae. Int. J. Clin. Exp. Pathol. 2015;8(5):5217–5223. [PMC free article] [PubMed] [Google Scholar]
- 110.Boberek J.M., Stach J., Good L. Genetic evidence for inhibition of bacterial division protein FtsZ by berberine. PLoS One. 2010 Oct 29;5(10) doi: 10.1371/journal.pone.0013745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chu M., Zhang M bo, chen Liu Y., Kang J rui, yun Chu Z., Yin K lin, et al. Role of berberine in the treatment of methicillin-resistant Staphylococcus aureus infections. Sci. Rep. 2016 Apr 22;6(1) doi: 10.1038/srep24748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hu Q., Peng Z., Li L., Zou X., Xu L., Gong J., et al. The efficacy of berberine-containing quadruple therapy on Helicobacter pylori eradication in China: a systematic review and meta-analysis of randomized clinical trials. Front. Pharmacol. 2020 Feb 4;10 doi: 10.3389/fphar.2019.01694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xu Y., Liang B., Kong C., Sun Z. Traditional medicinal plants as a source of antituberculosis drugs: a system review. BioMed Res. Int. 2021 Sep 8;2021:1–36. doi: 10.1155/2021/9910365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bhagya N., Chandrashekar K.R. Tetrandrine – a molecule of wide bioactivity. Phytochemistry. 2016 May;125:5–13. doi: 10.1016/j.phytochem.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 115.Zhang Z., Yan J., Xu K., Ji Z., Li L. Tetrandrine reverses drug resistance in isoniazid and ethambutol dual drug-resistant Mycobacterium tuberculosis clinical isolates. BMC Infect. Dis. 2015 Dec 25;15(1):153. doi: 10.1186/s12879-015-0905-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Marealle A.I., Moyo A.A., Machumi F., Qwarse M., Chenyambuga Y.M., Heydenreich M., et al. Antimycobacterial activity of scopoletin from ethanolic extract of Hymenodictyon floribundum (Hochst. & Steud.) B.L.Rob. Stem bark. Sci Afr. 2023 Sep;21 [Google Scholar]
- 117.Mativandlela S.P.N., Meyer J.J.M., Hussein A.A., Lall N. Antitubercular activity of compounds isolated from Pelargonium sidoides. Pharm. Biol. 2007 Jan 7;45(8):645–650. [Google Scholar]
- 118.Chiang C.C., Cheng M.J., Peng C.F., Huang H.Y., Chen I.S. A novel dimeric coumarin analog and antimycobacterial constituents from Fatoua pilosa. Chem. Biodivers. 2010 Jul 15;7(7):1728–1736. doi: 10.1002/cbdv.200900326. [DOI] [PubMed] [Google Scholar]
- 119.Dandawate P., Vemuri K., Venkateswara Swamy K., Khan E.M., Sritharan M., Padhye S. Synthesis, characterization, molecular docking and anti-tubercular activity of Plumbagin–Isoniazid Analog and its β-cyclodextrin conjugate. Bioorg. Med. Chem. Lett. 2014 Nov;24(21):5070–5075. doi: 10.1016/j.bmcl.2014.09.032. [DOI] [PubMed] [Google Scholar]
- 120.Mathew R., Kruthiventi A.K., Prasad J.V., Kumar S.P., Srinu G., Chatterji D. Inhibition of mycobacterial growth by plumbagin derivatives. Chem. Biol. Drug Des. 2010 May 18;76(1):34–42. doi: 10.1111/j.1747-0285.2010.00987.x. [DOI] [PubMed] [Google Scholar]
- 121.Sarkar A., Ghosh S., Shaw R., Patra M.M., Calcuttawala F., Mukherjee N., et al. Mycobacterium tuberculosis thymidylate synthase (ThyX) is a target for plumbagin, a natural product with antimycobacterial activity. PLoS One. 2020 Feb 4;15(2) doi: 10.1371/journal.pone.0228657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mahdi F., Morgan J.B., Liu W., Agarwal A.K., Jekabsons M.B., Liu Y., et al. Sampangine (a copyrine alkaloid) exerts biological activities through cellular redox cycling of its quinone and semiquinone intermediates. J. Nat. Prod. (Lloydia) 2015 Dec 24;78(12):3018–3023. doi: 10.1021/acs.jnatprod.5b00819. [DOI] [PubMed] [Google Scholar]
- 123.Ibraheim Z.Z., Abdel-Mageed W.M., Dai H., Guo H., Zhang L., Jaspars M. Antimicrobial antioxidant daucane sesquiterpenes from Ferula hermonis Boiss. Phytother Res. 2012 Apr;26(4):579–586. doi: 10.1002/ptr.3609. [DOI] [PubMed] [Google Scholar]
- 124.Ozturk M., Chia J.E., Hazra R., Saqib M., Maine R.A., Guler R., et al. Evaluation of berberine as an adjunct to TB treatment. Front. Immunol. 2021 Oct 20;12 doi: 10.3389/fimmu.2021.656419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rukayadi Y., Lee K.H., Hwang J.K. Activity of panduratin A isolated from Kaempferia pandurata Roxb. against multi-species oral biofilms in vitro. J. Oral Sci. 2009;51(1):87–95. doi: 10.2334/josnusd.51.87. http://joi.jlc.jst.go.jp/JST.JSTAGE/josnusd/51.87?from=CrossRef [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 126.Dahmer J., Marangon P., Adolpho L.O., Reis F.L., Maldaner G., Burrow R.A., et al. Phytochemistry alkaloids from the stem barks of Scutia buxifolia reissek (Rhamnaceae): structures and antimicrobial evaluation. Phytochemistry. 2022;196(October 2021) doi: 10.1016/j.phytochem.2021.113071. [DOI] [PubMed] [Google Scholar]
- 127.Azimi G., Hakakian A., Ghanadian M., Joumaa A., Alamian S. Bioassay-directed isolation of quaternary benzylisoquinolines from Berberis integerrima with bactericidal activity against Brucella abortus. Res Pharm Sci. 2018;13(2):149–158. doi: 10.4103/1735-5362.223797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dong H., Yu P., Long B., Peng T., He Y., Xu B., et al. Total synthesis of kuwanons A and B and discovery of their antibacterial mechanism. J. Nat. Prod. 2023 Jul 27 doi: 10.1021/acs.jnatprod.3c00466. [DOI] [PubMed] [Google Scholar]
- 129.Hamoud R., Reichling J., Wink M. Synergistic antibacterial activity of the combination of the alkaloid sanguinarine with EDTA and the antibiotic streptomycin against multidrug resistant bacteria. J. Pharm. Pharmacol. 2014;67:264–273. doi: 10.1111/jphp.12326. [DOI] [PubMed] [Google Scholar]
- 130.Tan K.K., Khoo T.J., Rajagopal M., Wiart C. Antibacterial alkaloids from Artabotrys crassifolius Hook.f. & Thomson. Nat. Prod. Res. 2015;29(4):2346–2349. doi: 10.1080/14786419.2015.1013954. [DOI] [PubMed] [Google Scholar]
- 131.Gehrke I.T.S., Neto A.T., Pedroso M., Mostardeiro C.P., Da Cruz I.B.M., Silva U.F., et al. Antimicrobial activity of Schinus lentiscifolius (anacardiaceae) J. Ethnopharmacol. 2013;148(2):486–491. doi: 10.1016/j.jep.2013.04.043. [DOI] [PubMed] [Google Scholar]
- 132.Liu T., Wang S., Xu L., Fu W., Gibbons S., Mu Q. Sesquiterpenoids with anti-MDR Staphylococcus aureus activities from Ferula ferulioides. Chem. Biodivers. 2015;12:599–614. doi: 10.1002/cbdv.201400150. [DOI] [PubMed] [Google Scholar]
- 133.Zhao L yun, Liu H xin, Wang L., Xu Z fang, Tan H bo, Qiu S. Rhodomyrtosone B , a membrane-targeting anti-MRSA natural acylgphloroglucinol from Rhodomyrtus tomentosa. J. Ethnopharmacol. 2019;228:50–57. doi: 10.1016/j.jep.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 134.Shen F., Tang X., Wang Y., Yang Z., Shi X., Wang C., et al. Phenotype and expression profile analysis of Staphylococcus aureus biofilms and planktonic cells in response to licochalcone A. Applied Microbial and Cell Physiology. 2015;99:359–373. doi: 10.1007/s00253-014-6076-x. [DOI] [PubMed] [Google Scholar]
- 135.Zhang Z., Yan J., Xu K., Ji Z., Li L. Tetrandrine reverses drug resistance in isoniazid and ethambutol dual drug-resistant Mycobacterium tuberculosis clinical isolates. BMC Infect. Dis. 2015;15:1–7. doi: 10.1186/s12879-015-0905-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chiang C.C., Cheng M.J., Peng C.F., Huang H.Y., Chen I.S. A novel dimeric coumarin analog and antimycobacterial constituents from Fatoua pilosa. Chem. Biodivers. 2010 Jul 15;7(7):1728–1736. doi: 10.1002/cbdv.200900326. [DOI] [PubMed] [Google Scholar]
- 137.Claes P., Cappoen D., Mbala B.M., Jacobs J., Mertens B., Mathys V., et al. Synthesis and antimycobacterial activity of analogues of the bioactive natural products sampangine and cleistopholine. Eur. J. Med. Chem. 2013 Sep;67:98–110. doi: 10.1016/j.ejmech.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 138.Georgiev M.I., Weber J., Maciuk A. Bioprocessing of plant cell cultures for mass production of targeted compounds. Appl. Microbiol. Biotechnol. 2009 Jul 1;83(5):809–823. doi: 10.1007/s00253-009-2049-x. http://link.springer.com/10.1007/s00253-009-2049-x [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 139.Jeong S.H., Ryu Y.B., Curtis-Long M.J., Ryu H.W., Baek Y.S., Kang J.E., et al. Tyrosinase inhibitory polyphenols from roots of Morus lhou. J. Agric. Food Chem. 2009 Feb 25;57(4):1195–1203. doi: 10.1021/jf8033286. [DOI] [PubMed] [Google Scholar]
- 140.Bowen J.K., Chaparro J.M., McCorkle A.M., Palumbo E., Prenni J.E. The impact of extraction protocol on the chemical profile of cannabis extracts from a single cultivar. Sci. Rep. 2021 Dec 8;11(1) doi: 10.1038/s41598-021-01378-0. https://www.nature.com/articles/s41598-021-01378-0 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kuttruff C.A., Eastgate M.D., Baran P.S. Natural product synthesis in the age of scalability. Nat. Prod. Rep. 2014;31(4):419–432. doi: 10.1039/c3np70090a. http://xlink.rsc.org/?DOI=C3NP70090A [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 142.Zhu X., Liu X., Liu T., Wang Y., Ahmed N., Li Z., et al. Synthetic biology of plant natural products: from pathway elucidation to engineered biosynthesis in plant cells. Plant Commun. 2021 Sep;2(5) doi: 10.1016/j.xplc.2021.100229. https://linkinghub.elsevier.com/retrieve/pii/S2590346221001310 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tiwari P., Khare T., Shriram V., Bae H., Kumar V. Plant synthetic biology for producing potent phyto-antimicrobials to combat antimicrobial resistance. Biotechnol. Adv. 2021 May;48 doi: 10.1016/j.biotechadv.2021.107729. https://linkinghub.elsevier.com/retrieve/pii/S0734975021000355 [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 144.Pant B. Application of plant cell and tissue culture for the production of phytochemicals in medicinal plants. 2014. http://link.springer.com/10.1007/978-81-322-1774-9_3 Available from: [DOI] [PubMed]
- 145.Kametani T., Noguchi I., Saito K., Kaneda S. Studies on the syntheses of heterocyclic compounds. Part CCCII. Alternative total syntheses of (±)-nandinine, (±)-canadine, and berberine iodide. J. Chem. Soc. C. 1969;0(15):2036–2038. doi: 10.1039/j39690002036. [DOI] [PubMed] [Google Scholar]
- 146.Gatland A.E., Pilgrim B.S., Procopiou P.A., Donohoe T.J. Short and efficient syntheses of protoberberine alkaloids using palladium-catalyzed enolate arylation. Angew. Chem. Int. Ed. 2014 Dec 22;53(52):14555–14558. doi: 10.1002/anie.201409164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mori-Quiroz L.M., Hedrick S.L., De Los Santos A.R., Clift M.D. A unified strategy for the syntheses of the isoquinolinium alkaloids berberine, coptisine, and jatrorrhizine. Org. Lett. 2018 Jul 20;20(14):4281–4284. doi: 10.1021/acs.orglett.8b01702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tajiri M., Yamada R., Hotsumi M., Makabe K., Konno H. The total synthesis of berberine and selected analogues, and their evaluation as amyloid beta aggregation inhibitors. Eur. J. Med. Chem. 2021 Apr;215 doi: 10.1016/j.ejmech.2021.113289. [DOI] [PubMed] [Google Scholar]
- 149.Baran P.S. Natural product total synthesis: as exciting as ever and here to stay. J. Am. Chem. Soc. 2018 Apr 11;140(14):4751–4755. doi: 10.1021/jacs.8b02266. https://pubs.acs.org/doi/10.1021/jacs.8b02266 [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 150.Hetzler B.E., Trauner D., Lawrence A.L. Natural product anticipation through synthesis. Nat. Rev. Chem. 2022 Mar 14;6(3):170–181. doi: 10.1038/s41570-021-00345-7. https://www.nature.com/articles/s41570-021-00345-7 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jeong S.H., Ryu Y.B., Curtis-Long M.J., Ryu H.W., Baek Y.S., Kang J.E., et al. Tyrosinase inhibitory polyphenols from roots of Morus lhou. J. Agric. Food Chem. 2009 Feb 25;57(4):1195–1203. doi: 10.1021/jf8033286. [DOI] [PubMed] [Google Scholar]
- 152.Owens B. The professionalization of cannabis growing. Nature. 2019 Aug 29;572(7771):S10–S11. doi: 10.1038/d41586-019-02527-2. http://www.nature.com/articles/d41586-019-02527-2 [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 153.Monthony A.S., Page S.R., Hesami M., Jones A.M.P. The past, present and future of Cannabis sativa tissue culture. Plants. 2021 Jan 19;10(1):185. doi: 10.3390/plants10010185. https://www.mdpi.com/2223-7747/10/1/185 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ioannidis K., Dadiotis E., Mitsis V., Melliou E., Magiatis P. Biotechnological approaches on two high CBD and CBG Cannabis sativa L. (Cannabaceae) varieties: in vitro regeneration and phytochemical consistency evaluation of micropropagated plants using quantitative 1H-NMR. Molecules. 2020 Dec 15;25(24):5928. doi: 10.3390/molecules25245928. https://www.mdpi.com/1420-3049/25/24/5928 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Medema M.H., Kottmann R., Yilmaz P., Cummings M., Biggins J.B., Blin K., et al. Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol. 2015 Sep 18;11(9):625–631. doi: 10.1038/nchembio.1890. http://www.nature.com/articles/nchembio.1890 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Nützmann H.W., Osbourn A. Gene clustering in plant specialized metabolism. Curr. Opin. Biotechnol. 2014 Apr;26:91–99. doi: 10.1016/j.copbio.2013.10.009. https://linkinghub.elsevier.com/retrieve/pii/S0958166913006836 [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 157.Nützmann H.W., Huang A., Osbourn A. Plant metabolic clusters – from genetics to genomics. New Phytol. 2016;211(3):771–789. doi: 10.1111/nph.13981. https://nph.onlinelibrary.wiley.com/doi/abs/10.1111/nph.13981 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang H., Guo H., Wang N., Huo Y.X. Toward the heterologous biosynthesis of plant natural products: gene discovery and characterization. ACS Synth. Biol. 2021 Nov 19;10(11):2784–2795. doi: 10.1021/acssynbio.1c00315. [DOI] [PubMed] [Google Scholar]
- 159.Han J., Li S. De novo biosynthesis of berberine and halogenated benzylisoquinoline alkaloids in Saccharomyces cerevisiae. Commun. Chem. 2023 Feb 9;6(1):27. doi: 10.1038/s42004-023-00821-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Nagegowda D.A., Gupta P. Advances in biosynthesis, regulation, and metabolic engineering of plant specialized terpenoids. Plant Sci. 2020 May;294 doi: 10.1016/j.plantsci.2020.110457. https://linkinghub.elsevier.com/retrieve/pii/S0168945220300595 [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 161.Blin K., Shaw S., Kloosterman A.M., Charlop-Powers Z., van Wezel G.P., Medema M.H., et al. antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021 Jul 2;49(W1):W29–W35. doi: 10.1093/nar/gkab335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kautsar S.A., Suarez Duran H.G., Blin K., Osbourn A., Medema M.H. plantiSMASH: automated identification, annotation and expression analysis of plant biosynthetic gene clusters. Nucleic Acids Res. 2017 Apr 27;45(W1):W55–W63. doi: 10.1093/nar/gkx305. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Schläpfer P., Zhang P., Wang C., Kim T., Banf M., Chae L., et al. Genome-Wide prediction of metabolic enzymes, pathways, and gene clusters in plants. Plant Physiol. 2017 Apr;173(4):2041–2059. doi: 10.1104/pp.16.01942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Terlouw B.R., Blin K., Navarro-Muñoz J.C., Avalon N.E., Chevrette M.G., Egbert S., et al. MIBiG 3.0: a community-driven effort to annotate experimentally validated biosynthetic gene clusters. Nucleic Acids Res. 2023 Jan 6;51(D1):D603–D610. doi: 10.1093/nar/gkac1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yang J.Y., Sanchez L.M., Rath C.M., Liu X., Boudreau P.D., Bruns N., et al. Molecular networking as a dereplication strategy. J. Nat. Prod. 2013 Sep 27;76(9):1686–1699. doi: 10.1021/np400413s. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hubert J., Nuzillard J.M., Renault J.H. Dereplication strategies in natural product research: how many tools and methodologies behind the same concept? Phytochemistry Rev. 2017 Feb 19;16(1):55–95. http://link.springer.com/10.1007/s11101-015-9448-7 [Internet] Available from: [Google Scholar]
- 167.Johnson S.R., Lange B.M. Open-access Metabolomics databases for natural product research: present capabilities and future potential. Front. Bioeng. Biotechnol. 2015 Mar 4;3 doi: 10.3389/fbioe.2015.00022. http://www.frontiersin.org/Bioinformatics_and_Computational_Biology/10.3389/fbioe.2015.00022/abstract [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zani C.L., Carroll A.R. Database for rapid dereplication of known natural products using data from MS and fast NMR Experiments. J. Nat. Prod. 2017 Jun 23;80(6):1758–1766. doi: 10.1021/acs.jnatprod.6b01093. https://pubs.acs.org/doi/10.1021/acs.jnatprod.6b01093 [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 169.Smyth W.F., Smyth T.J.P., Ramachandran V.N., O'Donnell F., Brooks P. Dereplication of phytochemicals in plants by LC-ESI-MS and ESI-MSn. TrAC, Trends Anal. Chem. 2012 Mar;33:46–54. https://linkinghub.elsevier.com/retrieve/pii/S016599361200009X [Internet] Available from: [Google Scholar]
- 170.Sawada Y., Nakabayashi R., Yamada Y., Suzuki M., Sato M., Sakata A., et al. RIKEN tandem mass spectral database (ReSpect) for phytochemicals: a plant-specific MS/MS-based data resource and database. Phytochemistry. 2012 Oct;82:38–45. doi: 10.1016/j.phytochem.2012.07.007. https://linkinghub.elsevier.com/retrieve/pii/S003194221200310X [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 171.Alcover C.F., Bernadat G., Kabran F.A., Le Pogam P., Leblanc K., Fox Ramos A.E., et al. Molecular networking reveals serpentinine-related bisindole alkaloids from Picralima nitida, a previously well-investigated species. J. Nat. Prod. 2020 Feb 24 doi: 10.1021/acs.jnatprod.9b01247. [Internet] [DOI] [PubMed] [Google Scholar]
- 172.Wang M., Carver J.J., Phelan V.V., Sanchez L.M., Garg N., Peng Y., et al. Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotechnol. 2016;34(8):828–837. doi: 10.1038/nbt.3597. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nothias L.F., Nothias-Esposito M., da Silva R., Wang M., Protsyuk I., Zhang Z., et al. Bioactivity-based molecular networking for the discovery of drug leads in natural product bioassay-guided fractionation. J. Nat. Prod. 2018 Apr 27;81(4):758–767. doi: 10.1021/acs.jnatprod.7b00737. https://pubs.acs.org/doi/10.1021/acs.jnatprod.7b00737 [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 174.Ge Y., Ma Y., Zhao M., Wei J., Wu X., Zhang Z., et al. Exploring gabosine and chlorogentisyl alcohol derivatives from a marine-derived fungus as EcGUS inhibitors with informatic assisted approaches. Eur. J. Med. Chem. 2022 Nov;242 doi: 10.1016/j.ejmech.2022.114699. [DOI] [PubMed] [Google Scholar]
- 175.Azizah M., Pripdeevech P., Thongkongkaew T., Mahidol C., Ruchirawat S., Kittakoop P. UHPLC-ESI-QTOF-MS/MS-Based molecular networking guided isolation and dereplication of antibacterial and antifungal constituents of Ventilago denticulata. Antibiotics. 2020 Sep 15;9(9):606. doi: 10.3390/antibiotics9090606. https://www.mdpi.com/2079-6382/9/9/606 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Pontes M.C., Cavalcante N.B., Leal A.E.B.P., Oliveira de A.P., Coutinho H.D.M., Menezes de I.R.A., et al. Chemical constituents and antibacterial activity of Bromelia laciniosa (Bromeliaceae): Identification and structural characterization. Phytomedicine Plus. 2022;(1) https://linkinghub.elsevier.com/retrieve/pii/S266703132200001X [Internet] 100215. [Google Scholar]
- 177.Sheng X., Sun Y., Yin Y., Chen T., Xu Q. Cirsilineol inhibits proliferation of cancer cells by inducing apoptosis via mitochondrial pathway. J. Pharm. Pharmacol. 2008 Nov 1;60(11):1523–1529. doi: 10.1211/jpp/60.11.0014. http://openurl.ingenta.com/content/xref?genre=article&issn=0022-3573&volume=60&issue=11&spage=1523 [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 178.Ghane M., Babaeekhou L., Shams M. Antimicrobial activity of Rhus Coriaria L. and Salvia Urmiensis bunge against some food-borne pathogens and identification of active components using molecular networking and docking analyses. Food Sci. Technol. 2022;42 http://www.scielo.br/scielo.php?script=sci_arttext&pid=S0101-20612022000100907&tlng=en [Internet] Available from: [Google Scholar]
- 179.Hebra T., Elie N., Poyer S., Van Elslande E., Touboul D., Eparvier V. Dereplication, annotation, and characterization of 74 potential antimicrobial metabolites from Penicillium sclerotiorum using t-SNE molecular networks. Metabolites. 2021 Jul 8;11(7):444. doi: 10.3390/metabo11070444. https://www.mdpi.com/2218-1989/11/7/444 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Gomes P., Quirós-Guerrero L., Silva C., Pamplona S., Boutin J.A., Eberlin M., et al. Feature-based molecular network-guided dereplication of natural bioactive products from leaves of Stryphnodendron pulcherrimum. Willd.) Hochr. Metabolites. 2021 Apr 29;11(5):281. doi: 10.3390/metabo11050281. https://www.mdpi.com/2218-1989/11/5/281 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lap Balida, Regalado J.T.A., Teodosio J.J.R., Dizon K.A.H., Sun Z., Zhan Z.Q., et al. Antibiotic isoflavonoids, anthraquinones, and pterocarpanoids from pigeon pea (Cajanus cajan L.) seeds against multidrug-resistant Staphylococcus aureus. Metabolites. 2022 Mar 23;12(4):279. doi: 10.3390/metabo12040279. https://www.mdpi.com/2218-1989/12/4/279 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Babaeekhou L., Ghane M. Antimicrobial activity of ginger on cariogenic bacteria: molecular networking and molecular docking analyses. J. Biomol. Struct. Dyn. 2021 Mar 31;39(6):2164–2175. doi: 10.1080/07391102.2020.1745283. https://www.tandfonline.com/doi/full/10.1080/07391102.2020.1745283 [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 183.Méndez D., Escalona J.C., Pieters L., Cos P. The 24th International Electronic Conference on Synthetic Organic Chemistry. MDPI; Basel Switzerland: 2020. Chemical fingerprinting and antimicrobial evaluation of the methanolic extract of the leaves of the endemic Cuban plant Coccoloba cowellii; p. 124.https://www.mdpi.com/2673-4583/3/1/124 [Internet] Available from: [Google Scholar]
- 184.Egan J.M., van Santen J.A., Liu D.Y., Linington R.G. Development of an NMR-based platform for the direct structural annotation of complex natural products mixtures. J. Nat. Prod. 2021 Apr 23;84(4):1044–1055. doi: 10.1021/acs.jnatprod.0c01076. https://pubs.acs.org/doi/10.1021/acs.jnatprod.0c01076 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Flores-Bocanegra L., Al Subeh Z.Y., Egan J.M., El-Elimat T., Raja H.A., Burdette J.E., et al. Dereplication of fungal metabolites by NMR-based compound networking using MADByTE. J. Nat. Prod. 2022 Mar 25;85(3):614–624. doi: 10.1021/acs.jnatprod.1c00841. https://pubs.acs.org/doi/10.1021/acs.jnatprod.1c00841 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kleks G., Holland D.C., Porter J., Carroll A.R. Natural products dereplication by diffusion ordered NMR spectroscopy (DOSY) Chem. Sci. 2021;12(32):10930–10943. doi: 10.1039/d1sc02940a. http://xlink.rsc.org/?DOI=D1SC02940A [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Robinette S.L., Zhang F., Brüschweiler-Li L., Brüschweiler R. Web server based complex mixture analysis by NMR. Anal. Chem. 2008 May 1;80(10):3606–3611. doi: 10.1021/ac702530t. https://pubs.acs.org/doi/10.1021/ac702530t [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 188.Xia J., Bjorndahl T.C., Tang P., Wishart D.S. MetaboMiner – semi-automated identification of metabolites from 2D NMR spectra of complex biofluids. BMC Bioinf. 2008 Dec 28;9(1):507. doi: 10.1186/1471-2105-9-507. https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-9-507 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhang C., Idelbayev Y., Roberts N., Tao Y., Nannapaneni Y., Duggan B.M., et al. Small molecule accurate recognition Technology (SMART) to enhance natural products research. Sci. Rep. 2017 Oct 27;7(1) doi: 10.1038/s41598-017-13923-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Borges R.M., Ferreira G. de A., Campos M.M., Teixeira A.M., Costa F. das N., das Chagas F.O., et al. NMR as a tool for compound identification in mixtures. Phytochem. Anal. 2023 Jun 2;34(4):385–392. doi: 10.1002/pca.3229. [DOI] [PubMed] [Google Scholar]
- 191.Loftsson T. Essential Pharmacokinetics. Elsevier; 2015. Physicochemical properties and pharmacokinetics; pp. 85–104. [Google Scholar]
- 192.Franco V., Perucca E. Pharmacological and therapeutic properties of cannabidiol for epilepsy. Drugs. 2019 Sep 1;79(13):1435–1454. doi: 10.1007/s40265-019-01171-4. [DOI] [PubMed] [Google Scholar]
- 193.Huestis M.A., Solimini R., Pichini S., Pacifici R., Carlier J., Busardò F.P. Cannabidiol adverse effects and toxicity. Curr. Neuropharmacol. 2019 Sep 13;17(10):974–989. doi: 10.2174/1570159X17666190603171901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Huan Y., Kong Q., Mou H., Yi H. Antimicrobial peptides: classification, design, application and research progress in multiple fields. Front. Microbiol. 2020 Oct 16;11 doi: 10.3389/fmicb.2020.582779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Chen C.H., Lu T.K. Development and challenges of antimicrobial peptides for therapeutic applications. Antibiotics. 2020 Jan 13;9(1):24. doi: 10.3390/antibiotics9010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Sher Khan R., Iqbal A., Malak R., Shehryar K., Attia S., Ahmed T., et al. Plant defensins: types, mechanism of action and prospects of genetic engineering for enhanced disease resistance in plants. 3 Biotech. 2019 May 29;9(5):192. doi: 10.1007/s13205-019-1725-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Koo H.B., Seo J. Antimicrobial peptides under clinical investigation. Peptide Science. 2019 Sep 3;111(5) [Google Scholar]
- 198.Greco I., Molchanova N., Holmedal E., Jenssen H., Hummel B.D., Watts J.L., et al. Correlation between hemolytic activity, cytotoxicity and systemic in vivo toxicity of synthetic antimicrobial peptides. Sci. Rep. 2020 Aug 6;10(1) doi: 10.1038/s41598-020-69995-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Dijksteel G.S., Ulrich M.M.W., Middelkoop E., Boekema B.K.H.L. Review: lessons learned from clinical trials using antimicrobial peptides (AMPs) Front. Microbiol. 2021 Feb 22;12 doi: 10.3389/fmicb.2021.616979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Gaston T.E., Mendrick D.L., Paine M.F., Roe A.L., Yeung C.K. “Natural” is not synonymous with “Safe”: toxicity of natural products alone and in combination with pharmaceutical agents. Regul. Toxicol. Pharmacol. 2020 Jun;113 doi: 10.1016/j.yrtph.2020.104642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Günthardt B.F., Hollender J., Hungerbühler K., Scheringer M., Bucheli T.D. Comprehensive toxic plants–phytotoxins database and its application in assessing aquatic micropollution potential. J. Agric. Food Chem. 2018 Jul 25;66(29):7577–7588. doi: 10.1021/acs.jafc.8b01639. [DOI] [PubMed] [Google Scholar]
- 202.Othman L., Sleiman A., Abdel-Massih R.M. Antimicrobial activity of polyphenols and alkaloids in Middle eastern plants. Front. Microbiol. 2019 May 15:10. doi: 10.3389/fmicb.2019.00911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Cos P., Vlietinck A.J., Berghe D Vanden, Maes L. Anti-infective potential of natural products: how to develop a stronger in vitro ‘proof-of-concept’. J. Ethnopharmacol. 2006 Jul;106(3):290–302. doi: 10.1016/j.jep.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 204.Kok L.Y., Bannigan P., Sanaee F., Evans J.C., Dunne M., Regenold M., et al. Development and pharmacokinetic evaluation of a self-nanoemulsifying drug delivery system for the oral delivery of cannabidiol. Eur. J. Pharmaceut. Sci. 2022 Jan;168 doi: 10.1016/j.ejps.2021.106058. https://linkinghub.elsevier.com/retrieve/pii/S0928098721003602 [Internet] Available from: [DOI] [PubMed] [Google Scholar]
- 205.Hartung I.V., Huck B.R., Crespo A. Rules were made to be broken. Nat. Rev. Chem. 2023 Jan 1;7(1):3–4. doi: 10.1038/s41570-022-00451-0. [DOI] [PubMed] [Google Scholar]
- 206.Xiong G., Wu Z., Yi J., Fu L., Yang Z., Hsieh C., et al. ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021 Jul 2;49(W1):W5–W14. doi: 10.1093/nar/gkab255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017 Mar 3;7(1) doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ali J., Camilleri P., Brown M.B., Hutt A.J., Kirton S.B. Revisiting the general solubility equation: in silico prediction of aqueous solubility incorporating the effect of topographical polar surface area. J. Chem. Inf. Model. 2012 Feb 27;52(2):420–428. doi: 10.1021/ci200387c. [DOI] [PubMed] [Google Scholar]
- 209.Vidar W.S., Baumeister T.U.H., Caesar L.K., Kellogg J.J., Todd D.A., Linington R.G., et al. Interaction Metabolomics to discover synergists in natural product mixtures. J. Nat. Prod. (Lloydia) 2023 Apr 28;86(4):655–671. doi: 10.1021/acs.jnatprod.2c00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Stermitz F.R., Lorenz P., Tawara J.N., Zenewicz L.A., Lewis K. Synergy in a medicinal plant: antimicrobial action of berberine potentiated by 5′-methoxyhydnocarpin, a multidrug pump inhibitor. Proc. Natl. Acad. Sci. USA. 2000 Feb 15;97(4):1433–1437. doi: 10.1073/pnas.030540597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Tehrani K.H.M.E., Wade N., Mashayekhi V., Brüchle N.C., Jespers W., Voskuil K., et al. Novel cephalosporin conjugates display potent and selective inhibition of imipenemase-type metallo-β-lactamases. J. Med. Chem. 2021 Jul 8;64(13):9141–9151. doi: 10.1021/acs.jmedchem.1c00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Xie J., Pierce J.G., James R.C., Okano A., Boger D.L. A Redesigned Vancomycin Engineered for Dual <scp>d</scp> -Ala- <scp>d</scp> -Ala and <scp>d</scp> -Ala- <scp>d</scp> -Lac Binding Exhibits Potent Antimicrobial Activity against Vancomycin-Resistant Bacteria. J. Am. Chem. Soc. 2011 Sep 7;133(35):13946–13949. doi: 10.1021/ja207142h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Jelić D., Antolović R. From erythromycin to azithromycin and new potential ribosome-binding antimicrobials. Antibiotics. 2016 Sep 1;5(3):29. doi: 10.3390/antibiotics5030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sadri A. Is target-based drug discovery efficient? Discovery and “off-target” mechanisms of all drugs. J. Med. Chem. 2023 Sep 6 doi: 10.1021/acs.jmedchem.2c01737. [DOI] [PubMed] [Google Scholar]
- 215.Zhang H.W., Lv C., Zhang L.J., Guo X., Shen Y.W., Nagle D.G., et al. Application of omics- and multi-omics-based techniques for natural product target discovery. Biomed. Pharmacother. 2021 Sep;141 doi: 10.1016/j.biopha.2021.111833. [DOI] [PubMed] [Google Scholar]
- 216.Farha M.A., Brown E.D. Strategies for target identification of antimicrobial natural products. Nat. Prod. Rep. 2016;33(5):668–680. doi: 10.1039/c5np00127g. [DOI] [PubMed] [Google Scholar]
- 217.Huth S.W., Oakley J.V., Seath C.P., Geri J.B., Trowbridge A.D., Parker D.L., et al. μMap photoproximity labeling enables small molecule binding site mapping. J. Am. Chem. Soc. 2023 Aug 2;145(30):16289–16296. doi: 10.1021/jacs.3c03325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Tsou L.K., Lara-Tejero M., RoseFigura J., Zhang Z.J., Wang Y.C., Yount J.S., et al. Antibacterial flavonoids from medicinal plants covalently inactivate type III protein secretion substrates. J. Am. Chem. Soc. 2016 Feb 24;138(7):2209–2218. doi: 10.1021/jacs.5b11575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Wu Z., Li W., Liu G., Tang Y. Network-based methods for prediction of drug-target interactions. Front. Pharmacol. 2018 Oct 9;9 doi: 10.3389/fphar.2018.01134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Pinzi L., Lherbet C., Baltas M., Pellati F., Rastelli G. In silico repositioning of cannabigerol as a novel inhibitor of the enoyl acyl carrier protein (ACP) reductase (InhA) Molecules. 2019 Jul 15;24(14):2567. doi: 10.3390/molecules24142567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Sliwoski G., Kothiwale S., Meiler J., Lowe E.W. Computational methods in drug discovery. Pharmacol. Rev. 2014 Jan 31;66(1):334–395. doi: 10.1124/pr.112.007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Adewumi A.T., Oluyemi W.M., Adewumi N., Adekunle Y.A., Alahmdi M.I., Abo-Dya N.E., et al. Probing into the flap-dimer dynamics of the Mycobacterium tuberculosis kasa enzyme binding landscape provides the underlying inhibitory mechanisms of JSF-3285 and 5G. Curr. Top. Med. Chem. 2023 May;23(12):1065–1080. doi: 10.2174/1568026623666230125124433. [DOI] [PubMed] [Google Scholar]
- 223.Mayr F., Möller G., Garscha U., Fischer J., Rodríguez Castaño P., Inderbinen S.G., et al. Finding new molecular targets of familiar natural products using in silico target prediction. Int. J. Mol. Sci. 2020 Sep 26;21(19):7102. doi: 10.3390/ijms21197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Oluyemi W.M., Samuel B.B., Adewumi A.T., Adekunle Y.A., Soliman M.E.S., Krenn L. An allosteric inhibitory potential of triterpenes from Combretum racemosum on the structural and functional dynamics of Plasmodium falciparum lactate dehydrogenase binding landscape. Chem. Biodivers. 2022 Feb 4;19(2) doi: 10.1002/cbdv.202100646. [DOI] [PubMed] [Google Scholar]
- 225.Rathod S., Shinde K., Porlekar J., Choudhari P., Dhavale R., Mahuli D., et al. Computational exploration of anti-cancer potential of flavonoids against cyclin-dependent kinase 8: an in silico molecular docking and dynamic approach. ACS Omega. 2023 Jan 10;8(1):391–409. doi: 10.1021/acsomega.2c04837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Mehta J., Rolta R., Dev K. Role of medicinal plants from North Western Himalayas as an efflux pump inhibitor against MDR AcrAB-TolC Salmonella enterica serovar typhimurium: in vitro and in silico studies. J. Ethnopharmacol. 2022 Jan;282 doi: 10.1016/j.jep.2021.114589. [DOI] [PubMed] [Google Scholar]
- 227.Geethalakshmi R., Sundaramurthi J.C., Sarada D.V.L. Antibacterial activity of flavonoid isolated from Trianthema decandra against Pseudomonas aeruginosa and molecular docking study of FabZ. Microb. Pathog. 2018 Aug;121:87–92. doi: 10.1016/j.micpath.2018.05.016. [DOI] [PubMed] [Google Scholar]
- 228.Aliye M., Dekebo A., Tesso H., Abdo T., Eswaramoorthy R., Melaku Y. Molecular docking analysis and evaluation of the antibacterial and antioxidant activities of the constituents of Ocimum cufodontii. Sci. Rep. 2021 May 12;11(1) doi: 10.1038/s41598-021-89557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Snow Setzer M., Sharifi-Rad J., Setzer W. The search for herbal antibiotics: an in-silico investigation of antibacterial phytochemicals. Antibiotics. 2016 Sep 12;5(3):30. doi: 10.3390/antibiotics5030030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Theuretzbacher U., Outterson K., Engel A., Karlén A. The global preclinical antibacterial pipeline. Nat. Rev. Microbiol. 2020 May 19;18(5):275–285. doi: 10.1038/s41579-019-0288-0. http://www.nature.com/articles/s41579-019-0288-0 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Balakrishnan V.S. WHO's antibacterial pipeline reports. Lancet Infect. Dis. 2022 Oct;22(10):1424. doi: 10.1016/S1473-3099(22)00603-X. [DOI] [PubMed] [Google Scholar]
- 232.Reher R., Kim H.W., Zhang C., Mao H.H., Wang M., Nothias L.F., et al. A convolutional neural network-based approach for the rapid annotation of molecularly diverse natural products. J. Am. Chem. Soc. 2020 Mar 4;142(9):4114–4120. doi: 10.1021/jacs.9b13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Marks R.A., Hotaling S., Frandsen P.B., VanBuren R. Representation and participation across 20 years of plant genome sequencing. Nat. Plants. 2021 Nov 29;7(12):1571–1578. doi: 10.1038/s41477-021-01031-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Reidpath D.D., Allotey P. The problem of ‘trickle-down science’ from the Global North to the Global South. BMJ Glob. Health. 2019 Jul 24;4(4) doi: 10.1136/bmjgh-2019-001719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Reher R., Kim H.W., Zhang C., Mao H.H., Wang M., Nothias L.F., et al. A convolutional neural network-based approach for the rapid annotation of molecularly diverse natural products. J. Am. Chem. Soc. 2020 Mar 4;142(9):4114–4120. doi: 10.1021/jacs.9b13786. https://pubs.acs.org/doi/10.1021/jacs.9b13786 [Internet] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Hutchings M.I., Truman A.W., Wilkinson B. Antibiotics: past, present and future. Curr. Opin. Microbiol. 2019 Oct;51:72–80. doi: 10.1016/j.mib.2019.10.008. [DOI] [PubMed] [Google Scholar]