Abstract
A DNA library of pRJ28, a large linear plasmid encoding mercury resistance, was constructed, and the mercury resistance genes were cloned. The 5,921-bp sequence was analyzed and showed a high degree of similarity to the Streptomyces lividans 1326 mercury resistance operon. Genes merR, merT, merP, and orfIV were found in a similar order and in a single transcription unit. merA and merB were found to be transcribed in the opposite direction to genes merR, merT, merP, and orfIV, as in S. lividans 1326. A novel putative regulatory gene, orfX, was found 22 bp downstream of merA. orfX encodes a 137-amino acid protein with a potential helix-turn-helix motif in the N-terminal domain, characteristic of the MerR family of transcriptional regulators. Transcriptional studies showed that orfX is cotranscribed with merA and merB. It is hypothesized that orfX plays a role in the regulation of the mercury resistance operon, probably by binding at the MerR operator site.
Mercury resistance is widespread among prokaryotes, and resistance genes are often found on plasmids or transposons (11). The major mechanism of resistance is reductive detoxification of Hg(II) to elemental mercury Hg(0), which is extremely volatile and leaves the cell by diffusing through the cell membrane. The process is mediated intracellularly by a mercuric reductase (MerA). Mercuric ions are transported from outside the cell by a series of transporter proteins. MerP is an extracellular mercuric ion binding protein, and MerT is a membrane-anchored protein responsible for transporting Hg(II) into the cell. All gram-positive and some gram-negative systems are resistant to a broad range of mercuric compounds, including organomercurials like phenylmercuric acetate (PMA) (7). This ability is due to the presence of an organomercurial lyase (MerB) which cleaves the carbon-mercury bonds and releases Hg(II). Narrow-spectrum resistance is observed when the merB gene is missing (17). The systems are regulated by transcriptional regulator MerR. In all cases studied, with the exception of in Streptomyces lividans where MerR is a repressor (14, 16), MerR is an activator/repressor transcriptional regulator. In the presence of Hg(II), MerR binds Hg(II) and activates its own transcription as well as that of the other mer genes. In the absence of Hg(II), MerR binds tightly to an operator and represses the system (7). In a few mercury resistance operons, a second regulator gene, merD, is present and binds weakly to the MerR operator site. MerD has been shown to down-regulate the system (9, 10).
We have previously described mercury-resistant Streptomyces strain CHR28, in which mercury resistance genes are encoded by the large linear plasmid pRJ28 (330 kb) (13). CHR28 is an environmental strain isolated from a heavily polluted site in the Baltimore Harbor and might have developed resistance and/or regulation mechanisms adapted to its environment which differ from those of S. lividans 1326. Mercury resistance genes of the laboratory strain S. lividans 1326 have previously been cloned and sequenced (2, 16), and recently, the negatively regulated repressor MerR has been purified and characterized (14). In this study, we successfully constructed a DNA library of plasmid pRJ28 and cloned the mercury resistance genes. We report the analysis of a 5,921-bp sequence of the CHR28 mercury resistance operon and the discovery of a novel putative regulatory gene, orfX. Transcriptional analysis with a nuclease protection assay and reverse transcription-PCR (RT-PCR) is reported.
Cloning and sequence analysis of the Streptomyces sp. strain CHR28 mer operon.
Plasmid pRJ28 DNA was purified by electroelution from pulsed-field electrophoresis agarose gels. A library of plasmid pRJ28 was constructed in pBKSII. Screening of about 900 clones with insert sizes ranging from 2 to 4 kb by using probes MER-A, MER-B, and MER-RTP (12) allowed identification of five overlapping clones encoding mercury resistance genes. Each clone was sequenced on both strands by primer walking. The fragments were assembled into a 5,921-bp contiguous stretch of sequence, which is 840 bp longer than the sequence of S. lividans 1326 mercury resistance operon (16).
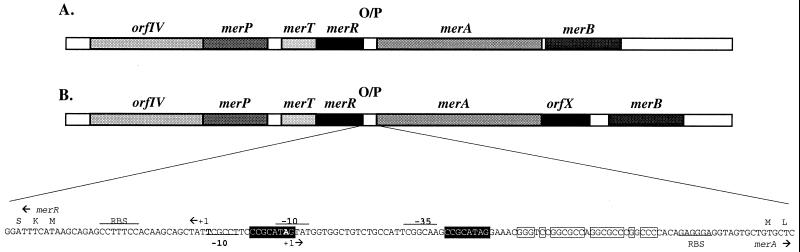
Seven open reading frames (ORFs) were found, and sequence comparison to the mercury resistance operon genes merA, merB, merR, merT, merP, and orfIV (a putative transporter gene) of S. lividans 1326 (16) and other mercury resistance genes permitted attribution of putative functions to each ORF. The analysis showed that all the genes found in the S. lividans 1326 mercury resistance operon were present in the same order in CHR28 (Fig. 1). The S. lividans 1326 mercury transporter genes, orfIV, merP, and merT, the regulator gene merR, the mercuric reductase gene merA, and the organomercurial lyase gene merB were aligned to the CHR28 sequences and were found to be highly similar (between 80 and 96% similarities at the nucleotide level and between 73 and 94% identities at the amino acid level). As in S. lividans 1326, merR and merT, as well as merP and orfIV, have overlapping start-stop codons. A 500-bp sequence was obtained downstream of merB, but no ORF was found in this region. Sequence comparison of the merA-merB region with S. lividans 1326 sequence revealed a 594-bp insert between the merA and merB genes (Fig. 1). In this insert, a new ORF was identified and termed orfX. This ORF is 411 bp and encodes a putative 137-amino-acid protein.
FIG. 1.
Structure of the mercury resistance operon of S. lividans 1326 (A) and Streptomyces sp. strain CHR28 (B). The promoter region is shown, and regulatory motifs are indicated, deduced by homology with those of the S. lividans 1326 mer operon (2, 14).
The mer operon in Streptomyces sp. strain CHR28 comprises two divergently oriented sets of four and three genes, respectively. Genes merR, merT, merP, and orfIV are transcribed leftward, and merA, orfX, and merB are transcribed from left to right. The promoter/operator region is 127 bp long and is located between genes merR and merA (Fig. 1). Upstream of merB, a 14-bp palindromic region identical to that in S. lividans 1326 indicated the presence of a transcriptional terminator (1). Another palindromic sequence was found in the region between merR and merA (Fig. 1). This region is also found in S. lividans 1326 and is probably involved in regulation (5). No specific structures were found in the 201-bp region between orfX and merB. The promoter/operator region is highly conserved when compared with the S. lividans 1326 promoter region. Two 8-bp direct repeats are also conserved in position but not in sequence between the two promoter regions and have no known function, although they are thought to be involved in regulation (Fig. 1). Delic et al. (4) described two Streptomyces promoters with 10-bp direct repeats and have implicated these repeats in regulation of chitinase genes.
Transcriptional analysis by RT-PCR and nuclease protection assay.
Transcriptional studies using RT-PCR to locate cotranscribed sets of genes indicated that merR and merA are not cotranscribed and that merA, orfX, and merB formed a contiguous transcript (Fig. 2). Primer sets spanning intergenic regions from merA to orfX and orfX to merB gave amplification products with cDNA synthesized from RNA prepared from cultures grown with HgCl2 and PMA (Fig. 2). No product was obtained for the primer set covering the divergently transcribed merR and merA genes (Fig. 2). The number of mRNA transcripts in cells grown without mercury compounds may be very low, explaining the lack of product with all primer sets in this case (Fig. 2).
FIG. 2.
RT-PCR analysis of RNA prepared from cultures grown with 0.05 mM HgCl2, 0.005 mM PMA, or without mercury with primer sets R2F-A1R, A2F-R3R, and R4F-B1R. +, thermoscript reverse transcriptase added; −, no Thermoscript reverse transcriptase added; C+, positive control (PCR performed with 100 ng of Streptomyces CHR28 genomic DNA); C−, negative control (PCR performed with no DNA added). A schematic representation of the Streptomyces CHR28 mercury resistance operon is shown with amplified regions indicated by thick lines.
A nuclease protection assay was performed by using the Multi-NPA kit (Ambion, Inc., Austin, Tex.). Primer probes were designed from the merA, merB, and merR genes to allow quantification of merA, merB, and merR mRNA amounts in cells grown in the presence or absence of HgCl2 and PMA and are as follows: MER-A (38 mer, position 3181 to 3216), 5 ′ - GC TCCAGGC C GAC G TAG C C GCCGCCGAGAACCAGCCAA-3′; MER-B (48 mer, position 4989 to 5033), 5′-GCGTGCCCAGGATGGCGGGGAAGATCAGGGTGTCCAGGGCGCACCCAA-3′; and MER-R (31 mer, position 2324 to 2351): 5′-CACGCGGGGCTGGAGATACCGGCGTGCCAA-3′ (the underlined sequence is nonhomologous to the target gene sequence in each case). Each oligonucleotide (10 pmol) was end labeled with T4 polynucleotide kinase with 10 pmol of [γ-32P]ATP (6,000 mCi/mmol) using the Ready-to-Go kit (Pharmacia BioChem). Each probe was gel purified to eliminate shorter-than-full-length oligonucleotides and was eluted in Probe Elution Buffer (Ambion). The amount of radioactive labeling of each probe was determined by scintillation counting, and specific activities were determined: MER-A, 1.0 × 106 cpm/pmol; MER-B, 9.8 × 105 cpm/pmol; and MER-R, 5.0 × 105 cpm/pmol. Total RNA extracts (5, 10, and 30 μg) from cultures grown with 0.05 mM HgCl2, 0.005 mM PMA, or no mercury were mixed with 5 fmol of each labeled probe (5- to 10-fold molar excess of probe over target mRNA) in a final reaction volume of 100 μl. RNA and probes were ethanol precipitated, and pellets were resuspended in 10 μl of Hybridization Buffer (Ambion). RNA and probes were incubated overnight at 37°C. Nuclease digestion treatment was carried out according to the manufacturer's recommendations (Ambion) for 45 min at room temperature with a nuclease mix diluted 1:100 in 1× Nuclease Digestion Buffer (Ambion). Nuclease consisted of a mixture of S1 nuclease-RNase A-RNase T1. Protected fragments were ethanol precipitated, dried pellets were resuspended in 10 μl of Gel Loading Buffer (Ambion), and protected fragments were separated on a 12% polyacrylamide gel. After electrophoresis, the gel was dried and exposed to a Phosphoscreen (Eastman-Kodak) for 48 h. The screen was scanned by using a Storm PhosphorImager (Molecular Dynamics). Digitized images were analyzed with the computer software ImageQuant (Molecular Dynamics).
Figure 3A shows that the signal intensity increased with the amount of total RNA, indicating that an excess of probe was present and validating the assay. The amount of merA mRNA was equal to that of merB mRNA under all conditions tested, thus confirming that merA and merB are cotranscribed (Fig. 3A). The amount of merR mRNA was three to four times higher than the amounts of merA or merB mRNA (Fig. 3A). The total mRNA levels of mer genes were two to three times higher when cells were grown in the presence of 0.05 mM HgCl2, compared with growth in 0.005 mM PMA (Fig. 3A). Numbers of mer transcripts were very low when cells were grown without mercuric compounds. In the absence of mercury, the amount of merR mRNA was four times higher than merA and merB but six times lower than merR mRNA from cells grown in the presence of 0.05 mM HgCl2. Analysis of these data reveals that the proportion of each transcript (merR, merA, or merB) remained constant under each condition tested (Fig. 3B). For each growth condition and amount of RNA tested, the proportion of merA and merB transcripts is consistently about 30% (Fig. 3B) and that of merR is about 70%. These results suggest that both sets of genes are coregulated. We hypothesize that as mercuric ion concentration increases, Hg(II) binds to the repressor MerR, conformational changes make MerR unable to bind to the promoter region, and transcription starts in both directions. The higher transcript levels of merR-orfIV may be due to that promoter being more efficient than the merA-merB promoter. Quantities of each of the three mRNAs were lower when cells were grown in the presence of 0.005 mM PMA than in the presence of 0.05 mM HgCl2. It appears that the system maintains a high basal level but that an increased concentration of Hg(II) increases transcription rates. The organic moiety of PMA is cleaved by MerB, and Hg(II) is released in the cell. However, the amounts of Hg(II) generated by this route are lower than the amounts of Hg(II) in cells grown in the presence of 0.05 mM HgCl2.
FIG. 3.
(A) Nuclease protection assay with probe MER-R (light gray bars), MER-A (dark gray bars), and MER-B (black bars), with 5, 10, and 30 μg of total RNA in each case. Quantitative analysis in relative intensity units (RIU) was done with the ImageQuant Software (Molecular Dynamics). (B) Intensity relative to a total intensity of 100%.
orfX: a putative second regulator gene.
Comparison of the CHR28 mercury resistance operon sequence to that of S. lividans 1326 revealed a 594-bp insert between gene merA and merB. An ORF encoding a putative 137-amino-acid protein named OrfX was found in the 5′ end of this insert. The start codon is located 22 bp downstream of the MerA stop codon. A putative ribosomal binding site is located in this region. Sequence analysis of OrfX showed homology with the MerR transcriptional regulator family (NolA, MerD, MerR [Bacillus and Tn501], and TipA) (Fig. 4). One major characteristic derived from amino acid alignments between members of this family is the high homology found at the N terminus of these proteins. This region is the DNA binding domain and contains a helix-turn-helix motif as indicated in Fig. 4. OrfX has the highest identity (42.2%) in this N-terminal region with NolA, a transcriptional regulator of nodulation in Bradyrhizobium japonicum (15). The function of OrfX is unknown, and analysis of the C-terminal domain indicates that it is unlikely to bind mercuric ions. Two cysteine residues are present in the predicted protein and only one is located in the C-terminal domain, the Hg(II) binding domain in MerR regulators, in which three highly conserved cysteine residues are located. However, it cannot be excluded that OrfX may be involved in the binding of organomercurial compounds. Figure 4 shows that OrfX has 32.6% identity with MerD, a protein which down-regulates the mercury resistance operon in Serratia marcescens (9, 10). Using a DNase footprinting assay, MerD has been shown to bind to a palindromic region located in the promoter/operator region (9). It is likely that OrfX also binds at the promoter/operator of CHR28 mer genes. orfX is located between merA and merB and is cotranscribed with these genes, as shown by RT-PCR (Fig. 2). This implies that as binding of Hg(II) to MerR derepresses the system, the amount of OrfX is likely to increase. OrfX then has the potential to bind to the promoter/operator region and activate or down-regulate the operon, as is the case with S. marcescens MerD (9). In this case, binding of MerD to the MerR operator was weak compared to the binding of MerR. It is possible that binding of OrfX at the promoter/operator represents a feedback inhibition mechanism for gene expression.
FIG. 4.
(A) Amino acid alignment of the N-terminal region of B. japonicum NolA (M58360), Bacillus cerus MerR (MerRb) (Y09027), Pseudomonas sp. Tn501 MerR (MerRt) (K02503), S. lividans TipA (M24524), and S. marcescens MerD (M15049) with OrfX from CHR28. The dashed line represents the helix-turn-helix (HTH) motif of the putative DNA binding site. (B) Table shows the similarity and identity between several proteins and OrfX over the region shown above.
Other transcriptional regulators of the MerR family have been described. However, like OrfX, their functions are unknown. These include a merR gene found on Tn5467, a Tn21-like transposon located on the Thiobacillus ferrooxidans plasmid pTF-FC2 (3). In this case, no other mer genes were found on the plasmid. However, a mercury resistance system is present on the chromosome and includes duplicated merR genes (3, 7, 8). This is also the case with gene yhdM in Escherichia coli, where adjacent ORFs have no similarity to any of the proteins usually associated with mercury resistance. Analysis of the complete sequence of the Haemophilus influenzae genome has revealed several candidate ORFs belonging to the MerR transcriptional regulator family (6). Our finding of OrfX in the mercury resistance operon present on a giant linear plasmid in Streptomyces sp. strain CHR28 adds an additional putative transcriptional regulator to this family.
Nucleotide sequence accession number.
The sequence of the Streptomyces sp. strain CHR28 mer operon has been deposited in GenBank under accession no. AF222792.
Acknowledgments
This study was supported by the Schering-Plough Research Institute and a NASA Biotechnology Grant (to J.D. and F.T.R.).
We thank Ann Horan for helpful discussions.
Footnotes
Contribution no. 517 from the Center of Marine Biotechnology.
REFERENCES
- 1.Bibb M J, Ward J M, Cohen S N. Nucleotide sequences encoding and promoting expression of three antibiotic resistance genes indigenous to Streptomyces. Mol Gen Genet. 1985;199:26–36. doi: 10.1007/BF00327505. [DOI] [PubMed] [Google Scholar]
- 2.Brünker P, Rother D, Sedlmeier R, Klein J, Mattes R, Altenbuchner J. Regulation of the operon responsible for broad-spectrum mercury resistance in Streptomyces lividans 1326. Mol Gen Genet. 1996;251:307–315. doi: 10.1007/BF02172521. [DOI] [PubMed] [Google Scholar]
- 3.Clennel A M, Johnston B, Rawlings D E. Structure and function of Tn5467, a Tn21-like transposon located on the Thiobacillus ferrooxidans broad-host-range plasmid pTF-FC2. Appl Environ Microbiol. 1995;61:4223–4229. doi: 10.1128/aem.61.12.4223-4229.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delic I, Robbins P, Westpheling J. Direct repeat sequences are implicated in the regulation of two Streptomyces chitinase promoters that are subject to carbon catabolite control. Proc Natl Acad Sci USA. 1992;89:1885–1889. doi: 10.1073/pnas.89.5.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eichenseer C, Altenbuchner J. The very large amplifiable element AUD2 from Streptomyces lividans 66 has insertion sequence-like repeats at its ends. J Bacteriol. 1994;176:7107–7112. doi: 10.1128/jb.176.22.7107-7112.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 7.Hobman J L, Brown N L. Bacterial mercury-resistance genes. Met Ions Biol Syst. 1997;34:527–568. [PubMed] [Google Scholar]
- 8.Inoue C, Sugawara K, Kusano T. The merR regulatory gene in Thiobacillus ferrooxidans is spaced apart from the mer structural genes. Mol Microbiol. 1991;5:2707–2718. doi: 10.1111/j.1365-2958.1991.tb01979.x. [DOI] [PubMed] [Google Scholar]
- 9.Mukhopadhyay D, Yu H R, Nucifora G, Misra T K. Purification and functional characterization of MerD. A coregulator of the mercury resistance operon in gram-negative bacteria. J Biol Chem. 1991;266:18538–18542. [PubMed] [Google Scholar]
- 10.Nucifora G, Silver S, Misra T K. Down regulation of the mercury resistance operon by the most promoter-distal gene merD. Mol Gen Genet. 1989;220:69–72. doi: 10.1007/BF00260858. [DOI] [PubMed] [Google Scholar]
- 11.Osborn A M, Bruce K D, Strike P, Ritchie D A. Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol Rev. 1997;19:239–262. doi: 10.1111/j.1574-6976.1997.tb00300.x. [DOI] [PubMed] [Google Scholar]
- 12.Ravel J, Amoroso M J, Colwell R R, Hill R T. Mercury-resistant actinomycetes from the Chesapeake Bay. FEMS Microbiol Lett. 1998;162:177–184. doi: 10.1111/j.1574-6968.1998.tb12996.x. [DOI] [PubMed] [Google Scholar]
- 13.Ravel J, Schrempf H, Hill R T. Mercury resistance is encoded by transferable giant linear plasmids in two Chesapeake Bay Streptomyces strains. Appl Environ Microbiol. 1998;64:3383–3388. doi: 10.1128/aem.64.9.3383-3388.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rother D, Mattes R, Altenbuchner J. Purification and characterization of MerR, the regulator of the broad-spectrum mercury resistance genes in Streptomyces lividans 1326. Mol Gen Genet. 1999;262:154–162. doi: 10.1007/s004380051070. [DOI] [PubMed] [Google Scholar]
- 15.Sadowsky M J, Cregan P B, Gottfert M, Sharma A, Gerhold D, Rodriguez-Quinones F, Keyser H H, Hennecke H, Stacey G. The Bradyrhizobium japonicum nolA gene and its involvement in the genotype-specific nodulation of soybeans. Proc Natl Acad Sci USA. 1991;88:637–641. doi: 10.1073/pnas.88.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sedlmeier R, Altenbuchner J. Cloning and DNA sequence analysis of the mercury resistance genes of Streptomyces lividans. Mol Gen Genet. 1992;236:76–85. doi: 10.1007/BF00279645. [DOI] [PubMed] [Google Scholar]
- 17.Walsh C T, Distefano M D, Moore M J, Shewchuk L M, Verdine G L. Molecular basis of bacterial resistance to organomercurial and inorganic mercuric salts. FASEB J. 1988;2:124–130. doi: 10.1096/fasebj.2.2.3277886. [DOI] [PubMed] [Google Scholar]