Abstract
The integration of diverse chemical tools like small-molecule inhibitors, activity-based probes (ABPs), and proteolysis targeting chimeras (PROTACs) advances clinical drug discovery and facilitates the exploration of various biological facets of targeted proteins. Here, we report the development of such a chemical toolbox for the human Parkinson disease protein 7 (PARK7/DJ-1) implicated in Parkinson’s disease and cancers. By combining structure-guided design, miniaturized library synthesis, and high-throughput screening, we identified two potent compounds, JYQ-164 and JYQ-173, inhibiting PARK7 invitro and in cells by covalently and selectively targeting its critical residue, Cys106. Leveraging JYQ-173, we further developed a cell-permeable Bodipy probe, JYQ-196, for covalent labeling of PARK7 in living cells and a first-in-class PARK7 degrader JYQ-194 that selectively induces its proteasomal degradation in human cells. Our study provides a valuable toolbox to enhance the understanding of PARK7 biology in cellular contexts and opens new opportunities for therapeutic interventions.
Introduction
Human Parkinson disease protein 7 (PARK7), also known as DJ-1, is a small (∼20 kDa) multifunctional protein,1,2 which is associated with various types of cancer and Parkinson’s disease.3−5 Throughout the years, PARK7 has been found to play a major role in protecting cells from stress conditions, especially oxidative stress, via its enzymatic and nonenzymatic functions.6 The key element for PARK7 functioning is the highly conserved cysteine residue at position 106. Cys106 serves as the active-site residue for the enzymatic glyoxalase activity of PARK7, while oxidation of Cys106 is essential for PARK7 to accomplish nonenzymatic functions, including antioxidant, chaperone, cotranscription factor, and antiapoptotic/ferroptotic functions.7 On the other hand, the excessive oxidation of the Cys106 residue leads to the loss of its neuroprotective activity and the development of neurodegenerative diseases.8
Its critical roles in a plethora of biological processes, including cell protective/survival activities and promoting tumorigenesis, demonstrate PARK7 to be an attractive therapeutic target. However, exactly how PARK7 fulfills its multifarious functions remains to be explored. This highlights the need for potent inhibitors and degraders that specifically target PARK7 in cells to explore PARK7 biology and advance drug discovery. A few small-molecule inhibitors were initially identified to bind PARK7,9,10 and this was later extended by a group of inhibitors based on the endogenous metabolite isatin, which bound PARK7 and reacted with Cys106 in a covalent manner.11−13 Amino-epoxycyclohexanones were also reported to covalently modify Cys106, and the inclusion of an alkyne moiety in these compounds allowed for in situ profiling of PARK7 by two-step labeling.14 We recently reported the development of cyanimide-containing inhibitor JYQ-88,15 along with two fluorescent probe variants, which covalently react with PARK7 Cys106, and demonstrated the successful application of these compounds in cell lysate. However, only a few compounds developed so far have been shown to engage with PARK7 in the context of live cells,11,12,14 which encouraged us to develop highly potent, cell-permeable small-molecule compounds specifically targeting the Cys106 residue of PARK7.
Taking JYQ-88 as a starting point, we here applied combined strategies of structure-guided design, miniaturized synthesis, and high-throughput screening to obtain improved PARK7 inhibitors JYQ-164 and JYQ-173 having submicromolar potency in cells. Both compounds specifically bind PARK7 and react to the Cys106 residue in cells as evidenced from a streamlined cysteine activity-based protein profiling (SLC-ABPP) experiment,16 with JYQ-173 being the most potent one. Moreover, we report a cell-permeable fluorescent probe JYQ-196 with a Bodipy dye, which was shown to covalently label PARK7 activity in both HEK293T and A549 cells. Finally, we report a first-in-class PARK7 degrader JYQ-194, which induced PARK7 degradation in different tumor cell lines. Altogether, we have developed a complete chemical toolbox of PARK7 to boost further studies on its diverse functions and future drug development.
Results and Discussion
Combining Structure-Guided Design and High-Throughput Synthesis to Discover Improved PARK7 Inhibitors
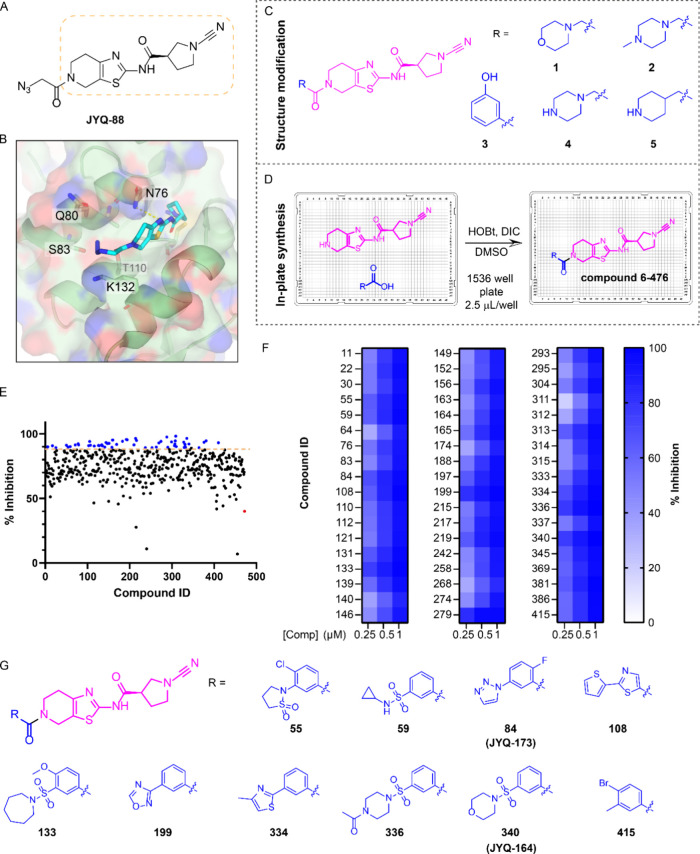
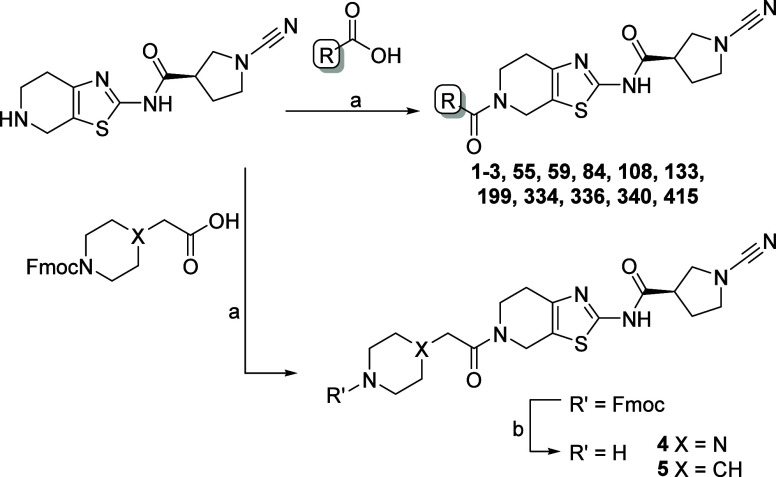
The PARK7 inhibitor JYQ-88 (Figure 1A) that we recently reported potently inhibits PARK7 in cell lysates.15 However, administering the compound to intact HEK293T cells revealed that it showed poor cellular engagement (Supporting Information Figure S1). Therefore, taking JYQ-88 as a starting point, we conducted an optimization to further improve the molecule in terms of inhibitory potency, specificity, and cellular uptake. Our previously generated crystal structure of the PARK7-JYQ-88 complex (PDB 7PA3) showed that the azidoacetyl moiety of JYQ-88 does not interact with PARK7, thereby leaving the mainly hydrophilic pocket surrounding this moiety largely unoccupied (Figure 1B). We therefore opted to improve the inhibitor by introducing different substituents replacing the azidomethyl moiety using two strategies. First, a small tailored library was designed and synthesized to investigate whether the introduction of larger, hydrophilic substituents on JYQ-88 would enhance the interaction with the PARK7 pocket. We introduced five different hydrophilic moieties, including morpholine, 4-methyl-piperazine, 3-hydroxyphenyl, piperazine, and piperidine replacing the azidomethyl moiety (compounds 1–5 in Figure 1C, Scheme 1). The second approach involved the generation of a compound library containing diverse substituents at the azidomethyl position. The crude compound library was prepared using Echo acoustic dispensing and in-plate synthesis, where we performed an amidation reaction in DMSO with the cyanimide amine moiety of JYQ-88 (Figure 1A, yellow box) and 471 diverse carboxylic acids using N,N′-diisopropylcarbodiimide (DIC) and hydroxybenzotriazole (HOBt) in a 1536-well plate (compound 6-476 in Figure 1D, Supplementary Data S1). We next performed a high-throughput screen of this crude compound library using our in-house developed PARK7 fluorescence polarization (FP) assay.15 The compounds, at 1 μM final concentration, were incubated with recombinant PARK7, followed by incubation with carboxyrhodamine-tagged FP reagent JYQ-107, which labels all residual active PARK7, and monitoring of the FP signal. Overall, most of the compounds showed over 50% inhibition, and we specifically selected the 54 hits showing over 90% inhibition (Figure 1E). All of these hits could be validated at 1, 0.5, and 0.25 μM final concentrations in the same FP assay (Figure 1F, Supplementary Data S1). As all compounds exist as crude mixtures in the library plate, we selected the top 10 hits, which were resynthesized and purified for further biochemical characterization (Figure 1G, Scheme 1, Supplementary Data S1). Of note here is that the cyanimide amine precursor for all compounds (Figure 1D, left) also inhibits PARK7. An inhibition of ∼40% at 1 μM of crude mixture was found in the HTS (Figure 1E), but inhibition potency of the pure compound was strongly reduced to 3 and 20% at 0.25 and 0.5 μM, respectively (Supporting Information Figure S2).
Figure 1.
Discovery of improved PARK7 inhibitors. (A) Structure of PARK7 inhibitor JYQ-88. (B) Crystal structure of PARK7-JYQ-88 complex (PDB 7PA3) showing an unoccupied pocket around the azidoacetyl moiety. (C) Chemical structures of designed compounds with a hydrophilic substituent. (D) Schematic illustration of in-plate synthesis to build a compound library. (E) Screening results using an FP assay at 1 μM compound concentration. Blue color represents compounds showing over 90% inhibition. Red color shows inhibition data for the amine precursor compound. (F) Heatmap displaying validation of the screening hits at 0.25, 0.5, and 1 μM using the FP assay. (G) Chemical structures of the resynthesized top 10 compounds.
Scheme 1. Synthesis of PARK7 Inhibitors.
Reagents and conditions: (a) carboxylic acid, HCTU, DIPEA, DCM; (b) DBU, DCM.
JYQ-164 and JYQ-173 Potently Bind with PARK7 in Cells
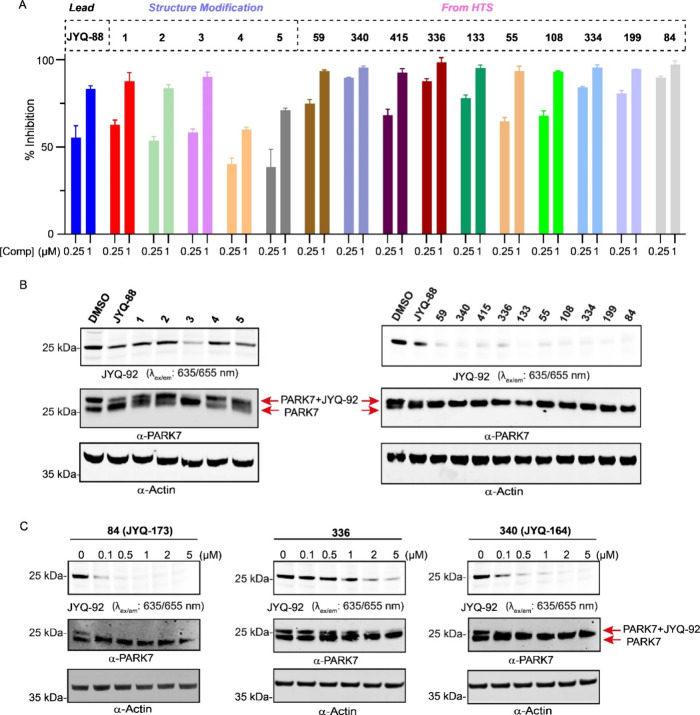
The 15 pure compounds from the two different approaches (5 compounds from structure modification and 10 compounds from HTS) were assessed for their potency to inhibit PARK7 at 0.25 and 1 μM final concentrations using the FP assay (Figure 2A). Overall, 11 compounds showed improved potency compared to JYQ-88, all of which contain an aromatic substituent. Among these, compounds 84, 336, and 340, which were discovered via HTS, displayed the highest potency with close to 100% inhibition at 0.25 μM, while compound 3, the only compound from the designed structural modification, showed a slight improvement in potency compared to JYQ-88. By contrast, the compounds with polar groups, such as 4-methyl-piperazine, piperazine, and piperidine, showed decreased potency compared to JYQ-88.
Figure 2.
Assessment of potency and cell permeability of the selected compounds. (A) Inhibition of PARK7 at 0.25 and 1 μM final concentration of 5 compounds designed from structural modification and top 10 compounds from HTS, determined using the PARK7 FP assay.15 (B) Fluorescent probe labeling of PARK7 remaining activity after inhibitor treatment to investigate cellular engagement. HEK293T cells were treated with the indicated compounds for 24 h. After cell lysis and incubation with the fluorescent PARK7 probe JYQ-92 for 1 h, the samples were analyzed by SDS-PAGE, fluorescence scanning, and immunoblot against PARK7 and β-actin. β-Actin was used as a loading control. (C) Target engagement of compounds 84, 336, and 340 in HEK293T cells in a dose–response manner. HEK293T cells were incubated with the compounds at the indicated final concentrations for 24 h, prior to cell lysis and incubation with PARK7 probe JYQ-92. The samples were analyzed by SDS-PAGE, fluorescence scanning, and immunoblot against PARK7 and β-actin. β-Actin was used as a loading control.
Since JYQ-88 is a potent inhibitor in vitro but did not show inhibition of PARK7 in cells (Supporting Information Figure S1), we next evaluated the cellular target engagement of all 15 compounds in a cell-based competition assay. Live HEK293T cells were treated with 5 μM of compounds for 24 h, followed by cell lysis and incubation with our previously developed SulfoCy5 PARK7 probe JYQ-92.15 Samples were resolved by SDS-PAGE and analyzed by fluorescence scanning and Western blotting (Figure 2B). The target engagement of cellular PARK7 is reflected by the disappearance of the PARK7 labeling band in the fluorescence gel scan and by the loss of the shifted probe-labeled PARK7 band in the Western blot. This revealed that all of the 11 aforementioned compounds with improved potency were able to bind to PARK7 in cells, indicating improved cellular target engagement. Remarkably, the remaining 4 compounds (1, 2, 4, 5) with decreased potency did not bind with PARK7 in cells.
Cyanimide-based compounds are reported to potently inhibit deubiquitinating enzymes (DUBs).17−19 We therefore investigated the inhibitory potential of our compounds against DUBs in cells using the fluorescent activity-based DUB probe Rho-Ub-PA (Supporting Information Figure S3).20 For this purpose, we used the HEK293T cell line for which the activity-based DUB profiling has been well-established and used to assess DUB inhibitors.17−19,21 Only 4 out of the 11 compounds (108, 199, 334, and 415) showed inhibition of one or more DUBs, mainly UCHL1, while the others, including our most potent compounds 84, 336, and 340, did not show interference with the activity of any DUB. Based on the potency and selectivity results, we selected compounds 84, 336, and 340 and further assessed their binding to cellular PARK7 in a dose-dependent manner. HEK293T cells were treated with a 0.1–5 μM dilution series of these inhibitors for 24 h, followed by cell lysis and incubation with probe JYQ-92. Fluorescence scanning and Western blotting revealed that compounds 84 and 340 engaged PARK7 from 0.1 μM, while compound 336 was less potent and engaged PARK7 activity from 1 μM (Figure 2C). Hence, it was decided to continue with the most potent, cell-permeable compounds 84 and 340, which were renamed JYQ-173 and JYQ-164, respectively, for further experiments and to ease future reference (Figure 1G). As expected, based on our previous data for JYQ-88, both JYQ-164 and JYQ-173 bind covalently with PARK7 as evidenced by intact protein mass spectrometry (Supporting Information Figure S4). The ability of JYQ-164 and JYQ-173 to inhibit PARK7 enzymatic activity was assessed using the DiFMUAc assay reagent,13 along with JYQ-88 and the previously reported isatin-based PARK7 inhibitor STK793590.11,13 This assay relies on the PARK7 active-site cysteine106-dependent deacetylation of the fluorogenic substrate 6,8-difluoro-4-methylumbelliferyl, thereby mimicking the glyoxalase activity of PARK7. JYQ-164 and JYQ-173 potently inhibited PARK7 activity with IC50 values of 21 and 19 nM, respectively (Supporting Information Figure S5A), showing a 5-fold improved potency compared with JYQ-88 (IC50 120 nM) and STK793590 (IC50 130 nM). These inhibitors also showed increased selectivity toward PARK7 compared to UCHL1 with a nearly 1000-fold difference (Supporting Information Figure S5B and Table S1). After showing the selectivities and potencies of JYQ-164 and JYQ-173 toward PARK7 in HEK293T cells, we conducted further experiments in A549 cells, a model cell line of lung adenocarcinoma and used as a model in previous PARK7 studies.14,22−26 Both compounds did not show any cytotoxicity in A549 cells up to 5 μM after 72 h incubation but still showed complete inhibiton of PARK7 engagement with the JYQ-92 probe (Supporting Information Figure S6).
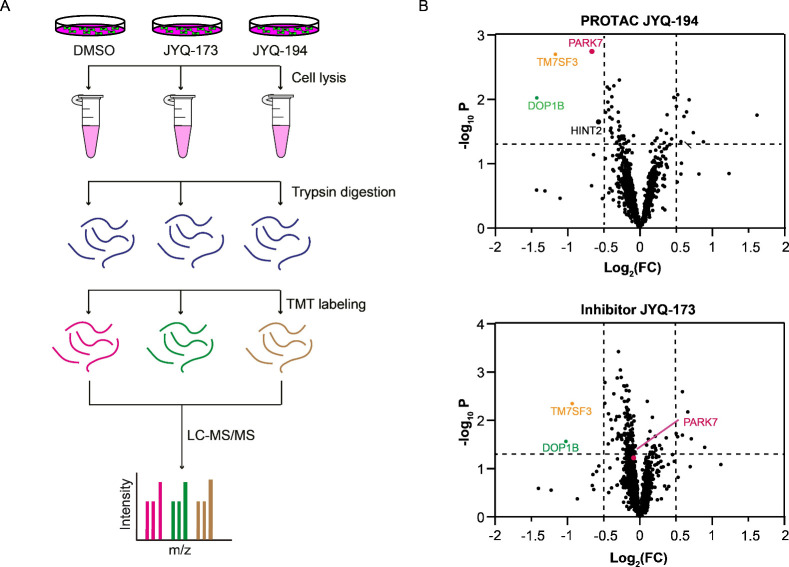
JYQ-164 and JYQ-173 Are Highly Selective Inhibitors for PARK7
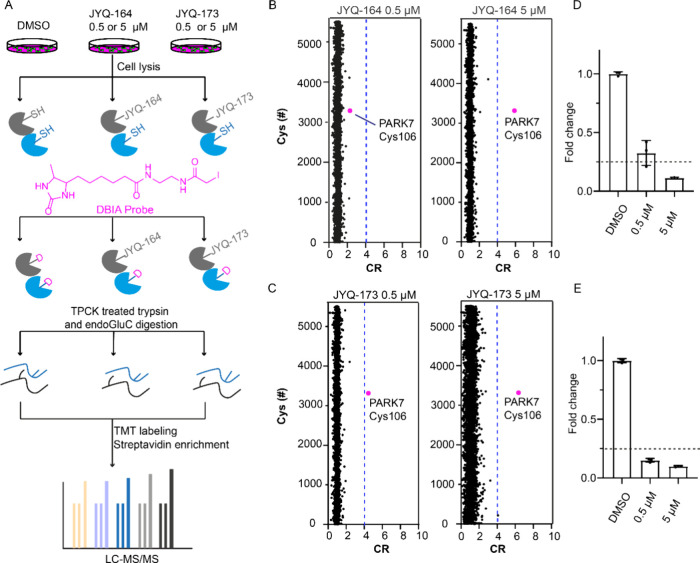
To investigate the selectivities of JYQ-164 and JYQ-173 in cells, we performed a streamlined cysteine activity-based protein profiling (SLC-ABPP) that can be used to profile and quantify the reactive cysteines binding to covalent inhibitors.16 A549 cells were treated with 0.5 or 5 μM of each inhibitor for 4 h along with a DMSO control, followed by cell lysis and incubation with a desthiobiotin iodoacetamide (DBIA) probe that is used to differentiate and enrich reactive cysteine sites that are not bound to JYQ-164 or JYQ-173. Further, the samples were digested with tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin and endoGluC to increase the coverage of protein sequences, especially the PARK7 peptide containing Cys106 residue. Peptides generated by digestion from replicates of each sample were labeled using TMT16-plex to perform tandem mass tag (TMT)-based quantitative proteomics profiling. Following the TMT labeling, all samples were pooled and the peptides conjugated to the DBIA probe were enriched by streptavidin beads and analyzed by LC-MS/MS (Figure 3A). A total of 13,492 peptides containing cysteine sites were detected and listed according to their abundance ratios for all the conditions (Supplementary Data S2). With the aim of representing our data, competition ratio (CR) values for each modified peptide were first calculated as the abundance ratios between each replicate of different concentrations of JYQ-164 and JYQ-173 and the average abundance ratios of the DMSO samples for the same peptide (DMSO/inhibitor ratio). All cysteines were represented with the shortest peptide by using the average CR values of all the peptides with the same modified cysteine from the same protein in the same condition.16,27,28 A total of 5512 unique cysteine sites were quantified in this analysis (Supplementary Data S3). Only the cysteines with a competitive ratio ≥4 (named CR threshold), corresponding to ≥75% reduction of DBIA probe alkylation at the cysteine by JYQ-164 or JYQ-173, were considered as targeted (Figure 3B–E). Following treatment with 5 μM JYQ-164, PARK7 Cys106 was identified as the only targeted site, while no targeted site that showed ≥75% reduction of DBIA probe alkylation was identified with the treatment of 0.5 μM JYQ-164 (Figure 3B,D). In addition, we identified PARK7 Cys106 as the only targeted cysteine that showed ≥75% reduction of DBIA probe alkylation in JYQ-173-treated cells (at 0.5 and 5 μM) (Figure 3C,E). Together, these data demonstrate that JYQ-164 and JYQ-173 are highly selective to PARK7 Cys106, and JYQ-173 has a higher potency than JYQ-164 toward PARK7 in cells.
Figure 3.
Investigation of the cellular selectivities of JYQ-164 and JYQ-173. (A) Schematic illustration of the workflow for the SLC-ABPP experiment. (B, C) SLC-ABPP profiling of PARK7 inhibitor JYQ-164 or JYQ-173 in A549 cells using two different concentrations (0.5 and 5 μM) quantified >5500 cysteine sites. All experiments were performed in triplicates. Data are represented as means ± s.d. Dotted lines represent a CR threshold of 4 (75% reduction in DBIA probe binding). (D, E) Fold changes of annotated peptide for PARK7 Cys106 residue in inhibitor-treated samples compared to a DMSO control are represented as a column graphic where the 0.25 fold change corresponding to 75% reduction in DBIA probe binding is highlighted with dotted lines.
JYQ-164 and JYQ-173 Turn into Cell-Permeable Activity-Based Probes
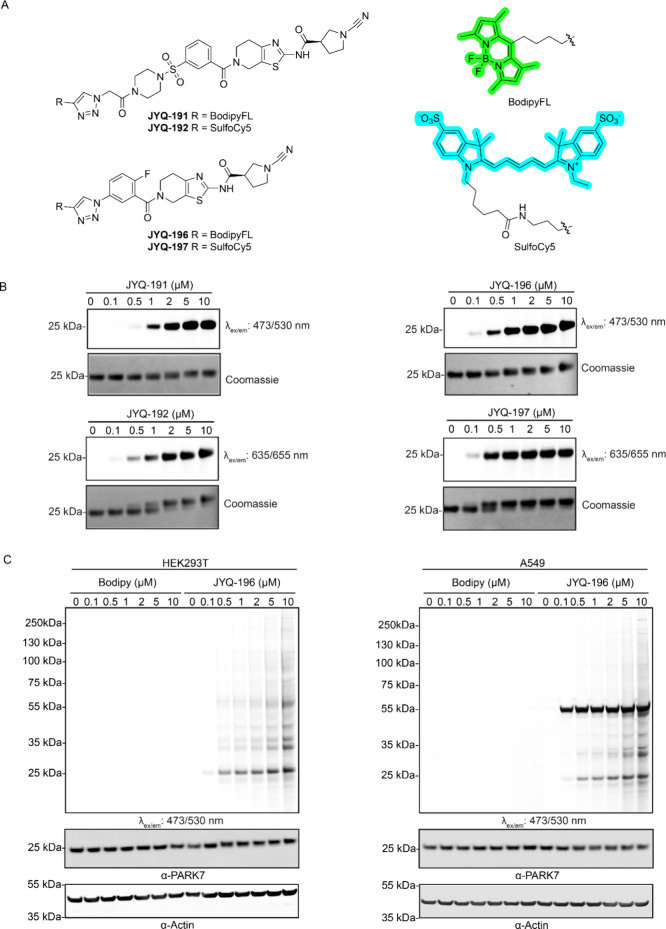
With the confirmation that JYQ-164 and JYQ-173 are highly potent and selective for cellular PARK7, we opted to convert our inhibitors into fluorescent, cell-permeable activity-based probes (ABPs), by installing a fluorescent dye. Because of our successful experience previously, we chose to install SulfoCy5-alkyne and BodipyFL-alkyne via the copper(I)-catalyzed azide–alkyne cycloaddition (or click) reaction. We took advantage of the triazole group already present in JYQ-173. A new compound was synthesized, where we replaced the triazole moiety in JYQ-173 with an azide, which was converted into the corresponding triazole after the click reaction with both alkynes, resulting in probes JYQ-196 and JYQ-197 (Figure 4A, Scheme 2). A convenient point of attachment was not readily available for inhibitor JYQ-164. We therefore substituted its morpholine moiety with 4-azidoacetyl piperazine and reacted this with both alkynes to obtain probes JYQ-191 and JYQ-192 (Figure 4A, Scheme 2). The ability of these probes to label and visualize PARK7 activity in vitro was evaluated by treating recombinant human PARK7 protein (1 μM) with a concentration series of the probes for 1 h at 37 °C, followed by SDS-PAGE analysis under nondenaturing conditions (without boiling and β-mercaptoethanol). Fluorescence scanning of the gel clearly showed that all four probes could label recombinant PARK7 in a dose-dependent manner. PARK7 labeling with probes JYQ-196 and JYQ-197 based on inhibitor JYQ-173 was more efficient than the probes JYQ-191 and JYQ-192 based on inhibitor JYQ-164 (Figure 4B). Taking the advantage of a band shift caused by the Bodipy and SulfoCy5 probes, we could also follow the complete labeling of recombinant PARK7 by these probes. Coomassie staining showed that JYQ-196 and JYQ-197 completely labeled recombinant PARK7 protein at 2 and 1 μM concentration, respectively. On the other hand, JYQ-191 reached complete labeling at 5 μM and JYQ-192 at 2 μM concentration, which implies a higher labeling efficiency of the JYQ-173-based probes than the JYQ-164-based probes (Figure 4B).
Figure 4.
(A) Chemical structures of activity-based probes JYQ-191, JYQ-192, JYQ-196, and JYQ-197. (B) Labeling of purified recombinant human PARK7 with the four probes. Recombinant human PARK7 was incubated with indicated concentrations of the probes for 1 h followed by SDS-PAGE, fluorescence scanning, and coomassie staining. (C) Fluorescence labeling of PARK7 activity in HEK293T and A549 cells with JYQ-196. HEK293T and A549 cells were incubated with indicated final concentration of JYQ-196 for 4 h, followed by cell lysis, SDS-PAGE, fluorescence scanning, and immunoblotting against PARK7 and β-actin. β-Actin was used as a loading control.
Scheme 2. Synthesis of Fluorescent PARK7 Probes.
Reagents and conditions: (a) azidoacetic acid, HCTU, DIPEA, DCM; (b) (i) DBU, DCM; (ii) 3-(chlorosulfonyl)benzoic acid, DIPEA, DCM; (c) (R)-1-cyano-N-(4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl)pyrrolidine-3-carboxamide,15 HCTU, DIPEA, DCM; (d) CuSO4·5H2O, sodium ascorbate, BodipyFL-alkyne or sulfoCy5-alkyne, DMF; (e) NaNO2, NaN3, H2SO4, H2O, 0 °C.
To further investigate the ability of these probes to visualize PARK7 in cells, HEK293T and A549 cells were incubated with 5 μM of these probes for 24 h, followed by cell lysis, SDS-PAGE, fluorescence scanning, and immunostaining (Supporting Information Figure S7). A clear band around 25 kDa was observed for both SulfoCy5 probes (JYQ-192 and JYQ-197) and BodipyFL probe JYQ-196, corresponding to the expected mass of ABP-labeled PARK7. We also observed a band shift of the PARK7 protein in the samples treated with JYQ-192, JYQ-196, and JYQ-197 by immunoblot against PARK7 in A549 cells. On the other hand, we did not observe any shift of the PARK7 protein in the sample treated with Bodipy probe JYQ-191, indicating inefficient labeling of cellular PARK7 by this probe. Therefore, we further investigated JYQ-192, JYQ-196, and JYQ-197. To characterize their cell permeability, A549 cells were incubated with 5 μM final concentration of these probes for the indicated time points, followed by confocal microscopy and by cell lysis, SDS-PAGE, fluorescence scanning, and immunostaining. All three probes already entered the cells after 1 h incubation (Supporting Information Figure S8A). However, only Bodipy probe JYQ-196 showed complete labeling of cellular PARK7 after 4 h incubation, while SulfoCy5 probes JYQ-192 and JYQ-197 labeled a small portion of cellular PARK7 even after 24 h incubation (Supporting Information Figure S8B), suggesting poor target engagement of JYQ-192 and JYQ-197 in intact cells. Besides PARK7, we observed extra bands in the fluorescence scan for both HEK293T and A549 cells and a very strong band labeling between 55 and 75 kDa in A549 cells with a 5 μM final concentration of all probes.
Despite the fact that probe JYQ-196 outperformed SulfoCy5 probes JYQ-192 and JYQ-197 in terms of the cellular PARK7 labeling efficiency, this probe still lacked specificity toward PARK7. PARK7 is known to form dimers, trimers, and even high-molecular-weight complexes under nonreducing conditions.29,30 Hence, to examine whether these extra bands are labeling of different forms of PARK7, PARK7 was depleted in HEK293T and A549 cells via siRNA transfection for 48 h and PARK7-depleted and control cells were subsequently incubated with 5 μM final concentration of JYQ-196 for an adiditional 4 h. Disapparence of a clear band corresponding to PARK7 was observed in the PARK7-depleted samples, while other bands did not show any changes compared to control samples (Supporting Information Figure S9), suggesting unspecific labeling of other proteins with JYQ-196. To investigate further whether lower concentrations of JYQ-196 can decrease unspecific labeling, HEK293T and A549 cells were incubated with increased concentrations (0–10 μM) of JYQ-196 for 4 h, followed by cell lysis, SDS-PAGE, fluorescence scanning, and immunostaining (Figure 4C). This showed that a lower concentration of JYQ-196 can still efficiently label PARK7 while showing less unspecific labeling.
PROTAC Derived from JYQ-173 Provides a Functional Cellular PARK7 Degrader
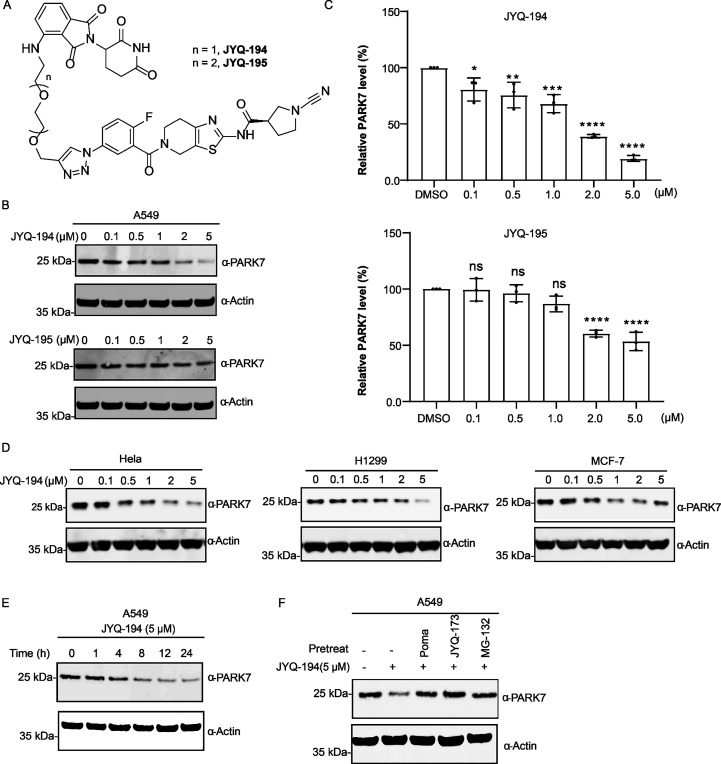
After having successfully converted our inhibitors into cell-permeable ABPs, we investigated the option of generating proteolysis targeting chimeras (PROTACs). PROTACs are heterobifunctional molecules that bind with an E3 ligase and protein of interest (POI) to form a ternary complex, enabling the degradation of the POI by targeting it to the proteasome.31,32 As such, we designed and synthesized a small set of PROTACs, JYQ-187 and JYQ-188 (based on inhibitor JYQ-164), and JYQ-194 and JYQ-195 (based on inhibitor JYQ-173), in a similar approach as for the above-described ABPs, yet this time conjugating the cereblon ligand pomalidomide, via two PEG linkers of different length (Figure 5A, Scheme 3, Supporting Information Figure S10A). We assessed their ability to induce PARK7 degradation in cells by treating A549 cells with increasing concentrations (from 0.1 to 5 μM) of the PROTACs for 8 h. Of them, JYQ-194 stood out as the most potent PARK7 degrader by inducing degradation of PARK7 starting at 0.1 μM and reaching a Dmax of ∼80% at a concentration of 5 μM (Figure 5B,C). JYQ-195 was less efficient than JYQ-194 as it degraded PARK7 starting at 2 μM. No degradation of PARK7 was observed for JYQ-187 and JYQ-188, both of which are based on JYQ-164 (Supporting Information Figure S10B). Therefore, JYQ-194 was selected for further evaluation.
Figure 5.
(A) Structures of PROTACs JYQ-194, JYQ-195. (B) PARK7 degradation efficacy with PROTACs JYQ-194, JYQ-195. A549 cells were incubated with the indicated concentration of PROTACs JYQ-194 and JYQ-195 for 8 h, followed by cell lysis, SDS-PAGE, and immunoblot analysis. (C) Quantification of Western blot of JYQ-194, JYQ-195 in panel B. Total PARK7 protein levels at each concentration of JYQ-194 were normalized to a DMSO control. Quantified data represent mean ± SD for three independent biological replicates. All significance was calculated using standard Student’s t test: *p <0.05, **p <0.01, ***p <0.001. (D) JYQ-194 induces PARK7 degradation in multiple tumor cell lines. Cells were treated with indicated concentrations of JYQ-194 for 8 h, followed by cell lysis and immunoblot analysis. (E) Time-course experiment for JYQ-194 (5 μM). A549 cells were treated with JYQ-194 (5 μM) for the indicated time, followed by cell lysis, running SDS-PAGE gel, and immunoblot analysis. (F) PARK7 degradation relies on ternary complex formation and proteasomal degradation. A549 cells were pretreated with 5 μM CRBN binder pomalidomide (POMA), 5 μM PARK7 inhibitor JYQ-173, or proteasome inhibitor 10 μM MG-132 for 4 h, followed by treatment with JYQ-194 (5 μM) for additional 8 h. Following cell lysis, the samples were run in SDS-PAGE gel, and immunoblot analysis was performed. All immunoblots performed in this figure were against PARK7 and β-actin. β-Actin was used as a loading control.
Scheme 3. Synthesis of PARK7 PROTACs.
Reagents and conditions: (a) NaOAc, AcOH, reflux; (b) TEA, DMF; (c) CuSO4·5H2O, sodium ascorbate, DMF.
Next, we determined the efficiency of JYQ-194-induced PARK7 degradation in multiple tumor cell lines, including Hela, H1299, and MCF7. PARK7 degradation was observed in all these cells in a dose-dependent manner (Figure 5D). To explore JYQ-194-induced PARK7 degradation kinetics in cells, we performed a time-course experiment in A549 cells using a fixed concentration of 5 μM JYQ-194, which had shown to induce maximal degradation among the tested concentrations in all cell lines we tested. Degradation of PARK7 was observed starting from 4 h, and the maximal degradation of PARK7 was reached by 8 h and remained until 24 h (Figure 5E). To test the functionality of each component of the JYQ-194 PROTAC and to investigate whether PARK7 degradation indeed depends on the ubiquitin-proteasome system, we pretreated A549 cells with the CRBN binder pomalidomide,33 the PARK7 inhibitor JYQ-173, or the proteasome inhibitor MG-132 for 4 h,34 followed by treatment with JYQ-194 for additional 8 h. The degradation of PARK7 is abolished by each inhibitor targeting an individual component of the functional PROTAC and the proteasome (Figure 5F), suggesting ubiquitin/proteasome system-dependent PARK7 degradation via JYQ-194.
JYQ-194 Selectively Induces PARK7 Degradation
To explore the degradation selectivity of PROTAC JYQ-194 toward PARK7, we performed a quantitative TMT-based total proteome profiling by incubating A549 cells with 5 μM JYQ-194 for 8 h, along with 5 μM JYQ-173 and DMSO as a control (Figure 6A). Over 6000 proteins were identified in the samples (Supplementary Data S4). Notably, PROTAC JYQ-194 induced the degradation of PARK7. As expected, JYQ-173 did not change the PARK7 protein level (Figure 6B). The proteomic data also revealed TM7SF3 (transmembrane 7 superfamily member 3) and DOP1B (DOP1 leucine zipper-like protein B) to be decreased by PROTAC JYQ-194, while these proteins were also downregulated by inhibitor JYQ-173 (Figure 6B), suggesting that downregulation of these two proteins is associated with Cys106-dependent functions of PARK7. In addition, PROTAC JYQ-194, unlike inhibitor JYQ-173, partly decreased HINT2 (histidine triad nucleotide-binding protein 2), implying that HINT2 might be an off-target of PROTAC JYQ-194 or might be regulated by scaffold function of PARK7. PARK7 still remains the major target of JYQ-194. Collectively, these findings revealed that JYQ-194 is a selective degrader of PARK7.
Figure 6.
(A) Schematic illustration of the workflow of TMT-based total proteomics experiments. A549 cells were incubated with DMSO, JYQ-173, or JYQ-164 for 8 h. Upon cell lysis and trypsin digestion, the peptides were labeled with TMTpro label reagents, and samples were pooled for LC-MS/MS analysis. (B) Proteomics analysis of proteins differentially expressed in PROTAC JYQ-194 and inhibitor JYQ-173-treated A549 cells compared to DMSO-treated A549 cells. Volcano plots of the −log10 (p-value) versus the log2 fold change (FC) for JYQ-194 versus DMSO (top panel) and JYQ-173 versus DMSO (bottom panel). All the samples were prepared as n = 3 biological replicates. Proteins with −log10 (p-value) > 1.3 and log2 FC < −0.5 or log2 FC > 0.5 were considered to be significantly changed in abundance. p-Values were calculated using standard Student’s t-test.
Conclusions
Besides serving as potential therapeutic agents, small-molecule inhibitors are great tools to investigate the biological function of specific proteins, especially enzymes, by binding to them via catalytic or allosteric mechanisms and affecting their cellular functions.34 These inhibitors have also provided a foundation to design and extend chemical tools such as activity-based probes (ABPs) and protein degraders (PROTACs). Combining these chemical tools derived from similar structures can help study diverse perspectives of target proteins in vitro, in living cells, and even in complex animal models.35−37 By using our recently developed high-throughput screening compatible FP assay,15 herein, we report the development of highly potent and selective inhibitors JYQ-164 and JYQ-173 for an attractive therapeutic target, PARK7.1,38−42 Both compounds potently inhibited PARK7 enzymatic activity with IC50 values of 19 and 21 nM, respectively, showing a 5-fold improved potency compared to our previously reported inhibitor JYQ-88. We identified these compounds in a chemical optimization study, which involved the replacement of the 2-azidoacetyl moiety in JYQ-88 by a large variety of substituents with the goal to fill the largely unoccupied protein pocket surrounding this moiety. The in vitro FP assay on our panel of 15 purified compounds showed that the introduction of an aromatic substituent was beneficial for the inhibitory potency, whereas a nonaromatic, hydrophilic moiety did not improve (compounds 1 and 2) or even lowered (compounds 4 and 5) the potency. Interestingly, all compounds that showed improved inhibition contain a meta-substituted aromatic moiety, which provides some possible insights into the binding mode within the protein pocket of these compounds. Notably, we showed direct target engagement of inhibitors JYQ-164 and JYQ-173 in live cells in a cell-based competition assay. In-cell SLC-ABP profiling revealed that these compounds specifically target PARK7’s highly conserved Cys106 residue that is essential for the biological functions of PARK7 and sensitive to oxidative stress.2
With these potent and selective inhibitors in hand, we designed and synthesized four activity-based probes, JYQ-191, JYQ-192, JYQ-196, and JYQ-197, by installment of Bodipy or a SulfoCy5 dyes onto them. Probe JYQ-196, derived from JYQ-173 by installing Bodipy dye, outperformed other probes by presenting efficient labeling of PARK7 activity in living cells. However, JYQ-196 also showed unspecific labeling of other proteins in the labeling experiment, while the original compound, JYQ-173, showed high selectivity toward PARK7 in the SLC-ABPP experiment. The reason for the disagreement between the fluorescent labeling and SLC-ABPP experiments could be due to the changes in the chemical composition of JYQ-173 by installment of dye as the similar effect observed in the previous studies for UCHL1-selective probes.17,18,43 Previously, Sieber and co-workers reported an alkyne-equipped probe to monitor PARK7 in different cell lines after a postlysis click reaction with rhodamine- or biotin-azide.14 The presented probes here provide great advantages because of the highly efficient one-step labeling of cellular PARK7 without a need for an intermediate click chemistry reaction, thus omitting all disadvantages associated with two-step labeling. Therefore, our probes could potentially serve as diagnostic tools with further endeavors, as PARK7 is a potential biomarker for various cancers and Parkinson’s diseases.44−47 In addition, we further developed a first-in-class selective PARK7 degrader JYQ-194 that induced PARK7 degradation in different tumor cell lines. It should be noted that the covalent, irreversible nature of JYQ-194 may limit its potency as it cannot be recycled upon PARK7 degradation, which possibly lowers its efficacy in an in vivo setting.31 Despite its limitation, the creation of JYQ-194 PROTAC provides new opportunities for targeting PARK7 in cancer therapy since PARK7 is overexpressed in various types of cancer.1,5,48
In summary, we have largely expanded the chemical toolbox for PARK7 by the development of two cell-permeable, potent, and specific PARK7 inhibitors, two activity-based probes to monitor or visualize PARK7 activity in cells, and a selective degrader to induce PARK7 degradation. The potent and selective inhibitors, ABPs, and PROTAC provided by our study can be further investigated for their therapeutic potential, and these chemical tools would be helpful to shed light on the biological function of PARK7 in health and diseases and to explore possible future therapeutic agents for PARK7.
Experimental Section
Chemistry
All reagents and solvents were purchased from commercially available sources and used as received unless indicated otherwise. All reaction progress was monitored by thin-layer chromatography (TLC) under UV light or by using a solution of KMnO4 (7.5 g L–1) and K2CO3 (50 g L–1) in H2O and liquid chromatography–mass spectrometry (LC-MS). Compounds were purified by Büchi flash column chromatography (unless indicated otherwise) using GraceResolv Davisil silica with indicated eluents. NMR spectra (1H, 13C) were recorded on a Bruker Ultrashield 300 MHz spectrometer at 298 K. Resonances are indicated with symbols “d” (doublet), “s” (singlet), “t” (triplet), and “m” (multiplet). Chemical shifts (δ) are given in ppm relative to CDCl3, DMSO-d6, or CD3OD as an internal standard, and coupling constants (J) are quoted in hertz (Hz). LC-MS measurements were performed on an LC-MS system equipped with a Waters 2795 Separation Module (Alliance HT), a Waters 2996 Photodiode Array Detector (190–750 nm), an Xbridge C18 column (2.1 × 100 mm, 3.5 μm), and an LCT ESI-Orthogonal Acceleration Time of Flight Mass Spectrometer. Samples were run using 2 mobile phases: A = 1% CH3CN and 0.1% formic acid in H2O and B = 1% H2O and 0.1% formic acid in CH3CN. Data processing was performed using Waters MassLynx Mass Spectrometry Software 4.1. High-resolution mass spectra (HR-MS) were recorded on a Waters Acquity H-class UPLC with a UPLC BEH C18 column (1.7 μm, 2.1 × 50 mm) coupled to a Xevo G2-XS Qtof mass spectrometer with ESI. Preparative HPLC was performed on a Waters preparative automated HPLC with mass detection. Samples were run on an Xbridge PREP C18 column (5 μm, 19 × 150 mm) using base (NH4OH)-modified CH3CN/H2O gradients. Gradient: 0–2.5 min: 95% H2O, 5% CH3CN; 2.5–17.5 min: 5–40% CH3CN; 17.5–20.90 min: 40–95% CH3CN; 20.90–21.00 min: 95–5% CH3CN; 1 mL min–1 CH3CN (with 1% 4 M NH4OH) mixed through the eluent. All final compounds had a purity of >95% as confirmed by LC-MS and NMR.
General Procedure A
To a solution of (R)-1-cyano-N-(4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl)pyrrolidine-3-carboxamide15 (1.0 equiv) in DCM were added HCTU (1.2 equiv) and DIPEA (3.6 equiv). The reaction mixture was stirred for 10 min. The indicated carboxylic acid (1.2 equiv) was then added, and the stirring was continued for 2 h at rt. The solvents were evaporated under reduced pressure, and the resulting residue was taken up in EtOAc. The organic layer was washed with 1 M HCl, sat. aq. NaHCO3, and brine, followed by drying with Na2SO4 and evaporating under reduced pressure. The resulting residue was purified by Büchi flash chromatography to obtain the desired product.
General Procedure B
The first step was performed as general procedure A. The obtained product from general procedure A and DBU (0.5 equiv) were dissolved in DCM, and the reaction mixture was stirred at rt for 2 h. After the complete removal of the Fmoc group, the organic layer was evaporated under reduced pressure. The resulting residue was purified by Büchi flash chromatography.
(R)-1-Cyano-N-(5-(2-morpholinoacetyl)-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl)pyrrolidine-3-carboxamide (1)
This compound was prepared according to the general procedure A using 2-morpholinoacetic acid (17.4 mg, 120 μmol, 1.2 equiv) as starting material. Purification after Büchi flash chromatography (DCM to 8% CH3OH/DCM) yielded 1 as a white solid (30 mg, 74.2 μmol, 74%). 1H NMR (300 MHz, DMSO-d6) δ 12.13 (d, J = 2.9 Hz, 1H), 4.58 (d, J = 44.9 Hz, 2H), 3.71 (dt, J = 11.4, 5.9 Hz, 2H), 3.58–3.28 (m, 9H), 3.20 (d, J = 7.3 Hz, 2H), 2.73–2.48 (m, 3H), 2.34 (s, 3H), 2.21–1.87 (m, 2H). 13C NMR (75 MHz, DMSO-d6) δ 170.8, 156.4, 143.3, 118.9, 117.5, 66.5, 53.4, 53.2, 52.5, 50.4, 43.5, 43.4, 29.7, 27.6, 26.4. HR-MS calculated for C18H24N6O3S [M + H]+ 405.1709, found 405.1703.
(R)-1-Cyano-N-(5-(2-(4-methylpiperazin-1-yl)acetyl)-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl)pyrrolidine-3-carboxamide (2)
This compound was prepared according to the general procedure A using 4-methylpiperazine-1-carboxylic acid (18.9 mg, 120 μmol, 1.2 equiv) as starting material. Purification after Büchi flash chromatography (DCM to 10% CH3OH/DCM) yielded 2 as a white solid (20 mg, 47.9 μmol, 48%). 1H NMR (300 MHz, DMSO-d6) δ 4.65 (d, J = 43.7 Hz, 2H), 3.79–3.75 (m, 3H), 3.52–3.47 (m, 2H), 3.42 (ddt, J = 9.2, 5.9, 2.8 Hz, 3H), 3.36–3.22 (m, 3H), 2.67 (dt, J = 44.7, 5.9 Hz, 2H), 2.44 (d, J = 12.9 Hz, 6H), 2.23 (d, J = 11.5 Hz, 3H), 2.19–1.98 (m, 2H). 13C NMR (75 MHz, DMSO) δ 170.9, 168.7, 156.5, 143.4, 119.3, 119.0, 117.6, 60.9, 54.6, 52.5, 52.2, 50.4, 45.4, 43.1, 29.7, 27.5. HR-MS calculated for C19H27N7O2S [M + H]+ 418.2025, found 418.2019.
(R)-1-Cyano-N-(5-(4-hydroxybenzoyl)-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl)pyrrolidine-3-carboxamide (3)
This compound was prepared according to the general procedure A using 3-hydroxybenzoic acid (16.6 mg, 120 μmol, 1.2 equiv) as starting material. Purification after Büchi flash chromatography (DCM to 6% CH3OH/DCM) yielded 3 as a white solid (22 mg, 55.3 μmol, 55%). 1H NMR (300 MHz, DMSO-d6) δ 12.24 (s, 1H), 9.91 (s, 1H), 7.38–7.23 (m, 2H), 6.92–6.76 (m, 2H), 4.66 (s, 2H), 3.75 (s, 2H), 3.56 (ddd, J = 29.7, 9.5, 6.9 Hz, 2H), 3.43 (td, J = 7.4, 5.0 Hz, 2H), 3.29 (d, J = 7.1 Hz, 1H), 2.73 (d, J = 7.5 Hz, 2H), 2.11 (ddt, J = 37.2, 12.8, 6.6 Hz, 2H). 13C NMR (75 MHz, DMSO) δ 170.8, 170.4, 159.5, 156.6, 143.5, 129.7, 126.5, 118.9, 117.6, 115.5, 52.5, 50.4, 43.5, 29.7. HR-MS calculated for C19H19N5O3S [M + H]+ 398.1287, found 398.1283.
(R)-1-Cyano-N-(5-(2-(piperazin-1-yl)acetyl)-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl)pyrrolidine-3-carboxamide (4)
This compound was prepared according to the general procedure B using 4-(((9H-fluoren-9-yl)methoxy)carbonyl)piperazine-1-carboxylic acid (44.0 mg, 120 μmol, 1.2 equiv) as starting material. Purification after Büchi flash chromatography yielded 4 as a white solid (5.1 mg, 12.6 μmol, 13% over 2-steps). 1H NMR (300 MHz, CD3OD) δ 4.68 (d, J = 30.1 Hz, 2H), 3.87–3.77 (m, 2H), 3.62–3.55 (m, 2H), 3.53–3.38 (m, 3H), 3.31 (d, J = 12.7 Hz, 3H), 2.93 (d, J = 19.4 Hz, 3H), 2.77 (s, 1H), 2.65 (s, 1H), 2.61–2.49 (m, 4H), 2.16 (ddt, J = 20.1, 13.0, 6.3 Hz, 2H). 13C NMR (75 MHz, CD3OD) δ 187.5, 142.8, 119.0, 60.1, 52.2, 51.5, 51.2, 50.0, 44.2, 44.1, 43.6, 43.1, 42.8, 39.9, 39.6, 29.4, 26.9, 25.8. HR-MS calculated for C18H25N7O2S [M + H]+ 404.1869, found 404.1863.
(R)-1-Cyano-N-(5-(2-(piperidin-4-yl)acetyl)-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl)pyrrolidine-3-carboxamide (5)
This compound was prepared according to the general procedure B using 1-(((9H-fluoren-9-yl)methoxy)carbonyl)piperidine-4-carboxylic acid (43.8 mg, 120 μmol, 1.2 equiv) as starting material. Purification after Büchi flash chromatography yielded 5 as white solid (4.8 mg, 11.9 μmol, 12% over 2-steps). 1H NMR (300 MHz, CD3OD) δ 4.64 (d, J = 2.2 Hz, 2H), 3.81 (dt, J = 16.5, 5.9 Hz, 2H), 3.59 (d, J = 7.0 Hz, 2H), 3.56–3.37 (m, 2H), 3.15 (s, 1H), 2.86–2.62 (m, 4H), 2.40 (dd, J = 11.2, 6.9 Hz, 2H), 2.26–2.05 (m, 2H), 1.98 (s, 1H), 1.84 (d, J = 13.2 Hz, 2H), 1.30 (s, 2H), 1.21 (s, 1H). HR-MS calculated for C19H26N6O2S [M + H]+ 403.1916, found 403.1911.
(R)-1-Cyano-N-(5-(3-(N-cyclopropylsulfamoyl)benzoyl)-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl)pyrrolidine-3-carboxamide (59)
This compound was prepared according to the general procedure A using 3-(N-cyclopropyl sulfamoyl)benzoic acid (17.4 mg, 72 μmol, 1.2 equiv) as starting material. Purification after Büchi flash chromatography (DCM to 5% CH3OH/DCM) yielded 59 as a white solid (15.1 mg, 30.2 μmol, 50%). 1H NMR (300 MHz, CD3OD) δ 8.11–7.92 (m, 2H), 7.75 (d, J = 7.4 Hz, 2H), 4.61 (s, 2H), 4.12 (s, 1H), 3.80–3.41 (m, 6H), 2.85 (d, J = 18.3 Hz, 2H), 2.41–2.12 (m, 3H), 1.48–1.29 (m, 1H), 0.54 (dt, J = 12.1, 3.7 Hz, 4H). 13C NMR (75 MHz, DMSO-d6) δ 170.9, 168.8, 156.7, 141.1, 137.1, 131.2, 130.3, 128.5, 125.5, 118.6, 117.6, 52.5, 50.4, 43.5, 29.7, 24.6, 5.6. HR-MS calculated for C22H24N6O4S2 [M + H]+ 501.1379, found 501.1373.
(R)-1-Cyano-N-(5-(3-(morpholinosulfonyl)benzoyl)-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl)pyrrolidine-3-carboxamide (340, JYQ-164)
This compound was prepared according to the general procedure A using 3-(morpholino sulfonyl)benzoic acid (19.5 mg, 72 μmol, 1.2 equiv) as starting material. Purification after Büchi flash chromatography (DCM to 5% CH3OH/DCM) yielded 340 as a light yellow solid (6.5 mg, 12.3 μmol, 20%). 1H NMR (300 MHz, DMSO-d6) δ 12.26 (s, 1H), 7.83 (dd, J = 21.7, 7.5 Hz, 4H), 4.79 (s, 1H), 4.57 (s, 1H), 3.63 (ddd, J = 9.7, 5.9, 3.5 Hz, 6H), 3.55–3.40 (m, 3H), 3.15 (qd, J = 7.3, 4.2 Hz, 2H), 2.95–2.88 (m, 4H), 2.80–2.68 (m, 2H), 2.30–2.00 (m, 2H). 13C NMR (75 MHz, DMSO-d6) δ 170.9, 156.7, 137.5, 135.4, 131.8, 130.5, 129.3, 126.3, 118.5, 117.5, 65.7, 52.5, 50.4, 46.4, 43.5, 42.3, 29.7. HR-MS calculated for C23H26N6O5S2 [M + H]+ 531.1484, found 531.1478.
(R)-N-(5-(4-Bromo-3-methylbenzoyl)-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl)-1-cyanopyrrolidine-3-carboxamide (415)
This compound was prepared according to the general procedure A using 4-bromo-3-methylbenzoic acid (15.5 mg, 72 μmol, 1.2 equiv) as starting material. Purification after Büchi flash chromatography (DCM to 5% CH3OH/DCM) yielded 415 as a white solid (18.2 mg, 38.4 μmol, 64%). 1H NMR (300 MHz, DMSO-d6) δ 12.25 (s, 1H), 7.67 (d, J = 8.1 Hz, 1H), 7.46 (s, 1H), 7.21 (s, 1H), 4.66 (d, J = 52.4 Hz, 2H), 3.96–3.38 (m, 6H), 3.27–3.10 (m, 1H), 2.81–2.68 (m, 2H), 2.39 (s, 3H), 2.12 (ddd, J = 44.5, 12.9, 6.5 Hz, 2H). 13C NMR (75 MHz, DMSO-d6) δ 170.8, 156.6, 138.3, 136.0, 132.7, 129.9, 126.6, 126.1, 118.6, 117.5, 52.5, 50.4, 43.5, 42.3, 38.1, 29.7, 23.8, 22.8. HR-MS calculated for C20H20BrN5O2S [M + H]+ 474.0599, found 474.0595.
(R)-N-(5-(3-((4-Acetylpiperazin-1-yl)sulfonyl)benzoyl)-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl)-1-cyanopyrrolidine-3-carboxamide (336)
This compound was prepared according to the general procedure A using 3-((4-acetylpiperazin-1-yl)sulfonyl)benzoic acid (20.3 mg, 65 μmol, 1.2 equiv) as starting material. Purification after Büchi flash chromatography (DCM to 5% CH3OH/DCM) yielded 336 as a white solid (11.7 mg, 20.4 μmol, 38%). 1H NMR (300 MHz, DMSO-d6) δ 12.25 (s, 1H), 7.84 (s, 2H), 7.78 (d, J = 7.0 Hz, 2H), 4.67 (d, J = 66.5 Hz, 2H), 3.97 (s, 1H), 3.67–3.39 (m, 9H), 2.94 (d, J = 15.8 Hz, 4H), 2.83–2.67 (m, 2H), 2.29–2.01 (m, 2H), 1.96 (s, 3H). 13C NMR (75 MHz, DMSO-d6) δ 170.9, 168.8, 166.0, 137.6, 135.8, 132.1, 130.6, 129.2, 126.2, 118.5, 117.6, 52.6, 50.4, 46.4, 46.2, 45.4, 43.5, 29.7, 21.6.HR-MS calculated for C25H29N7O5S2 [M + H]+ 572.1750, found 572.1754.
(R)-N-(5-(3-(Azepan-1-ylsulfonyl)-4-methoxybenzoyl)-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl)-1-cyanopyrrolidine-3-carboxamid (133)
This compound was prepared according to the general procedure A using 3-(azepan-1-ylsulfonyl)-4-methoxybenzoic acid (20.4 mg, 65 μmol, 1.2 equiv) as starting material. Purification after Büchi flash chromatography (DCM to 5% CH3OH/DCM) yielded 133 as a white solid (10.5 mg, 18.4 μmol, 34%). 1H NMR (300 MHz, DMSO-d6) δ 12.26 (s, 1H), 7.82 (d, J = 1.6 Hz, 1H), 7.79–7.68 (m, 1H), 7.32 (d, J = 8.6 Hz, 1H), 4.69 (s, 2H), 3.96 (s, 3H), 3.91–3.36 (m, 6H), 3.26 (t, J = 5.7 Hz, 5H), 2.81–2.69 (m, 2H), 2.30–1.97 (m, 2H), 1.72–1.51 (m, 8H). 13C NMR (75 MHz, DMSO-d6) δ 170.8, 168.7, 157.9, 156.7, 133.9, 130.2, 127.7, 118.7, 117.6, 113.5, 56.9, 52.5, 50.4, 48.3, 43.5, 29.7, 29.4, 27.0. HR-MS calculated for C26H32N6O5S2 [M + H]+ 573.1954, found 573.1958.
(R)-N-(5-(3-(Azepan-1-ylsulfonyl)-4-methoxybenzoyl)-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl)-1-cyanopyrrolidine-3-carboxamide (55)
This compound was prepared according to the general procedure A using 4-chloro-3-(1,1-dioxidoisothiazolidin-2-yl)benzoic acid (17.9 mg, 65 μmol, 1.2 equiv) as starting material. Purification after Büchi flash chromatography (DCM to 5% CH3OH/DCM) yielded 55 as a white solid (6.5 mg, 12.2 μmol, 23%). 1H NMR (300 MHz, DMSO-d6) δ 11.95 (s, 1H), 7.38 (t, J = 7.6 Hz, 2H), 7.24–7.10 (m, 1H), 4.45 (s, 1H), 4.28 (s, 1H), 3.48–3.38 (m, 6H), 3.12 (t, J = 7.3 Hz, 4H), 2.51 (s, 1H), 2.47–2.39 (m, 2H), 2.18–2.10 (m, 2H), 1.97–1.69 (m, 2H). 13C NMR (75 MHz, DMSO-d6) δ 171.0, 156.7, 135.7, 131.3, 128.5, 118.5, 117.6, 52.5, 50.3, 49.7, 47.3, 43.5, 40.6, 40.3, 29.7, 19.6. HR-MS calculated for C22H23ClN6O4S2 [M + H]+ 535.0989, found 535.0987.
(R)-1-Cyano-N-(5-(2-(thiophen-2-yl)thiazole-5-carbonyl)-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl)pyrrolidine-3-carboxamide (108)
This compound was prepared according to the general procedure A using 2-(thiophen-2-yl)thiazole-5-carboxylic acid (13.7 mg, 65 μmol, 1.2 equiv) as starting material. Purification after Büchi flash chromatography (DCM to 5% CH3OH/DCM) yielded 108 as a white solid (5.7 mg, 12.1 μmol, 22%). 1H NMR (300 MHz, DMSO-d6) δ 12.27 (s, 1H), 8.19 (s, 1H), 7.83–7.78 (m, 2H), 7.22 (dd, J = 5.0, 3.8 Hz, 1H), 4.86 (s, 2H), 3.96 (t, J = 5.7 Hz, 2H), 3.65–3.40 (m, 4H), 3.31–3.11 (m, 1H), 2.78 (d, J = 27.1 Hz, 2H), 2.29–1.98 (m, 2H). 13C NMR (75 MHz, DMSO-d6) δ 170.9, 163.7, 161.0, 156.8, 143.4, 136.3, 130.7, 129.2, 118.5, 117.6, 52.5, 50.4, 43.5, 42.3, 29.7. HR-MS calculated for C20H18N6O2S3 [M + H]+ 471.0732, found 471.0718.
(R)-1-Cyano-N-(5-(3-(4-methylthiazol-2-yl)benzoyl)-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridine-2-yl)pyrrolidine-3-carboxamide (334)
This compound was prepared according to the general procedure A using 3-(4-methylthiazol-2-yl) benzoic acid (14.2 mg, 65 μmol, 1.2 equiv) as starting material. Purification after Büchi flash chromatography (DCM to 5% CH3OH/DCM) yielded 334 as a white solid (6.3 mg, 13.2 μmol, 24%). 1H NMR (300 MHz, DMSO-d6) δ 12.25 (s, 1H), 8.02 (dt, J = 7.4, 1.7 Hz, 1H), 7.94 (s, 1H), 7.60 (t, J = 7.5 Hz, 2H), 7.39 (d, J = 1.1 Hz, 1H), 4.70 (d, J = 55.5 Hz, 2H), 3.97 (s, 1H), 3.72–3.39 (m, 6H), 2.84–2.67 (m, 2H), 2.44 (dd, J = 3.2, 1.0 Hz, 3H), 2.29–1.96 (m, 2H). 13C NMR (75 MHz, DMSO-d6) δ 170.9, 165.8, 153.9, 137.3, 133.9, 130.1, 127.6, 124.7, 118.7, 117.6, 115.8, 52.5, 50.4, 45.3, 43.5, 29.7, 17.4. HR-MS calculated for C23H22N6O2S2 [M + H]+ 479.1324, found 479.1319.
(R)-N-(5-(3-(1,2,4-Oxadiazol-3-yl)benzoyl)-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl)-1-cyanopyrrolidine-3-carboxamide (199)
This compound was prepared according to the general procedure A using 3-(1,2,4-oxadiazol-3-yl)benzoic acid (13.7 mg, 72 μmol, 1.2 equiv) as starting material. Purification after Büchi flash chromatography (DCM to 5% CH3OH/DCM) yielded 199 as a white solid (5.9 mg, 13.2 μmol, 22%). 1H NMR (300 MHz, DMSO-d6) δ 12.26 (s, 1H), 9.77 (s, 1H), 8.19–8.11 (m, 1H), 8.07 (s, 1H), 7.71 (d, J = 6.0 Hz, 2H), 4.70 (d, J = 56.2 Hz, 2H), 3.98 (s, 1H), 3.68–3.38 (m, 5H), 3.30–3.13 (m, 1H), 2.83–2.65 (m, 2H), 2.31–1.95 (m, 2H). 13C NMR (75 MHz, DMSO-d6) δ 170.8, 168.2, 166.8, 156.7, 137.4, 130.3, 128.9, 126.7, 126.0, 118.6, 117.6, 52.5, 50.4, 43.5, 29.7. HR-MS calculated for C21H19N7O3S [M+H]+ 450.1348, found 450.1345.
(R)-1-Cyano-N-(5-(2-fluoro-5-(1H-1,2,3-triazol-1-yl)benzoyl)-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl)pyrrolidine-3-carboxamide (84, JYQ-173)
This compound was prepared according to the general procedure A using 2-fluoro-5-(1H-1,2,3-triazol-1-yl)benzoic acid (14.9 mg, 72 μmol, 1.2 equiv) as starting material. Purification after Büchi flash chromatography (DCM to 5% CH3OH/DCM) yielded 84 as a white solid (7.8 mg, 16.7 μmol, 28%). 1H NMR (300 MHz, DMSO-d6) δ 12.26 (s, 1H), 8.86 (dd, J = 6.7, 1.2 Hz, 1H), 8.09 (dddd, J = 11.4, 5.6, 4.4, 2.8 Hz, 2H), 7.99 (dd, J = 3.3, 1.2 Hz, 1H), 7.70–7.51 (m, 1H), 4.70 (d, J = 92.9 Hz, 2H), 4.01 (t, J = 5.8 Hz, 1H), 3.68–3.49 (m, 5H), 3.35–3.25 (m, 1H), 2.73 (d, J = 25.2 Hz, 2H), 2.29–1.95 (m, 2H). 13C NMR (75 MHz, DMSO-d6) δ 170.9, 163.8, 157.4 (d, J = 247.3 Hz), 156.7, 143.5, 143.1, 135.1, 133.9, 125.7 (d, J = 20.5 Hz), 124.0, 123.7, 120.8 (d, J = 12.9 Hz), 118.3, 118.0, 117.6, 52.5, 50.3, 44.9, 43.5, 29.7, 27.3, 26.5. HR-MS calculated for C21H19FN8O2S [M + H]+ 467.1414, found 467.1417.
(9H-Fluoren-9-yl)methyl 4-(2-azidoacetyl)piperazine-1-carboxylate (477)
To a solution of N-(9-fluorenylmethoxycarbonyl)piperazine hydrochloride (1 g, 2.90 mmol, 1.0 equiv) in DCM were added HCTU (1.44 g, 3.48 mmol, 1.2 equiv) and DIPEA (1.72 mL, 10.44 mmol, 3.6 equiv). The reaction mixture was stirred for 10 min. Azidoacetic acid (262 μL, 3.48 mmol, 1.2 equiv) was then added, and stirring was continued for 2 h at rt. The solvents were evaporated under reduced pressure, and the resulting residue was taken up in EtOAc. The organic layer was washed with 1 M HCl, sat. aq. NaHCO3, and brine (50 mL), followed by drying with Na2SO4 and evaporating under reduced pressure. The resulting residue was purified by Büchi flash chromatography (DCM to 2% MeOH/DCM) to yield the title compound as a white solid (1.02 g, 2.61 mmol, 90%). 1H NMR (300 MHz, CDCl3) δ 7.80 (dt, J = 7.6, 1.0 Hz, 2H), 7.58 (dd, J = 7.4, 1.1 Hz, 2H), 7.48–7.40 (m, 2H), 7.39–7.32 (m, 2H), 4.56 (d, J = 6.2 Hz, 2H), 4.26 (t, J = 6.2 Hz, 1H), 3.96 (s, 2H), 3.64–3.18 (m, 8H). 13C NMR (75 MHz, CDCl3) δ 143.8, 141.4, 127.8, 127.1, 124.8, 120.1, 67.3, 50.8, 47.4, 45.0, 43.4, 41.7.
3-((4-(2-Azidoacetyl)piperazin-1-yl)sulfonyl)benzoic Acid (478)
To a solution of compound 477 (1.0 g, 2.55 mmol, 1.0 equiv) in DCM (5 mL) was added DBU (190 μL, 1.23 mmol, 0.5 equiv), and the reaction mixture was stirred at rt for 2 h. After complete removal of the Fmoc group, 3-(chlorosulfonyl) benzoic acid (563.4 mg, 2.55 mmol, 1.0 equiv) and DIPEA (920 μL, 5.78 mmol, 2.2 equiv) were added. The resulting reaction mixture was stirred at rt for 0.5 h, followed by removing the solvents under reduced pressure. The crude material was taken up in EtOAc (50 mL), and the organic layer was washed with 1 M HCl (3 × 25 mL). The organic layer was dried over Na2SO4 and evaporated under reduced pressure. The resulting residue was used as such without further purification.
(R)-N-(5-(3-((4-(2-Azidoacetyl)piperazin-1-yl)sulfonyl)benzoyl)-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl)-1-cyanopyrrolidine-3-carboxamide (479)
To a solution of (R)-1-cyano-N-(4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl) pyrrolidine-3-carboxamide (138.5 mg, 0.5 mmol, 1.0 equiv) in DCM were added HCTU (247.6 mg, 0.6 mmol, 1.2 equiv) and DIPEA (297 μL, 1.8 mmol, 3.6 equiv). The reaction mixture was stirred for 10 min. Compound 478 (212 mg, 0.6 mM, 1.2 equiv) was then added, and stirring was continued for 2 h at rt. The solvents were evaporated under reduced pressure, and the resulting residue was taken up in EtOAc (20 mL). The organic layer was washed with 1 M HCl (2 × 10 mL), sat. aq. NaHCO3 (3 × 10 mL), and brine (10 mL), followed by drying with Na2SO4 and evaporating under reduced pressure. The resulting residue was purified by Büchi flash chromatography (DCM to 5% MeOH/DCM) to yield the title compound as a white solid (90 mg, 0.15 mmol, 25%). 1H NMR (300 MHz, DMSO-d6) δ 7.86 (d, J = 8.5 Hz, 2H), 7.78 (d, J = 7.0 Hz, 2H), 4.66 (d, J = 70.8 Hz, 2H), 4.10 (s, 3H), 3.57 (q, J = 11.5, 10.1 Hz, 8H), 3.35–3.21 (m, 2H), 2.96 (t, J = 4.9 Hz, 4H), 2.86–2.62 (m, 2H), 2.31–1.94 (m, 2H). 13C NMR (75 MHz, DMSO-d6) δ 171.0, 168.2, 166.5, 156.7, 137.5, 135.7, 132.1, 130.6, 129.2, 126.2, 118.5, 117.6, 52.6, 50.4, 50.1, 46.0, 43.7, 43.5, 29.7.
BodipyFL Probe JYQ-191
A solution of compound 479 (20.0 mg, 32.6 μmol, 1.0 equiv) and BodipyFL-alkyne (12.9 mg, 39.1 μmol, 1.2 equiv) in dry DMF (2 mL) was degassed by argon for 30 min. Aqueous solutions of sodium ascorbate (0.5 M) and CuSO4·5H2O (0.5 M) were prepared in 1 mL volume and degassed for 30 min with argon bubbling. The degassed sodium ascorbate (131 μL, 39.2 μmol, 1.2 equiv) and CuSO4·5H2O (112 μL, 32.6 μmol, 1.0 equiv) solutions were added to the reaction mixture, followed by stirring for 2 h. The resulting crude material was purified by preparative HPLC to yield JYQ-191 as a red solid (7.5 mg, 8.0 μmol, 24%). 1H NMR (300 MHz, DMSO-d6) δ 12.25 (s, 1H), 7.82 (dt, J = 26.2, 7.6 Hz, 5H), 7.67 (s, 1H), 6.22 (s, 2H), 5.36 (s, 2H), 4.68 (d, J = 66.5 Hz, 2H), 3.97 (s, 1H), 3.56 (d, J = 17.6 Hz, 7H), 3.14–2.89 (m, 7H), 2.83–2.60 (m, 4H), 2.39 (s, 6H), 2.37 (s, 6H), 2.29–1.92 (m, 3H), 1.86–1.72 (m, 2H), 1.68–1.51 (m, 2H). 13C NMR (75 MHz, DMSO-d6) δ 170.9, 165.0, 153.5, 147.1, 146.6, 141.4, 137.6, 135.8, 132.1, 131.1, 130.6, 129.2, 126.2, 124.1, 122.1, 117.6, 52.5, 50.9, 50.4, 46.1, 43.9, 43.5, 31.2, 29.9, 29.6, 28.1, 25.0, 16.3, 14.5. HR-MS calculated for C44H51BF2N12O5S2 [M + H]+ 941.3694, found 941.3714.
SulfoCy5 Probe JYQ-192
This compound was prepared according to the procedure of JYQ-191 using compound 479 (10.0 mg, 16.3 μmol, 1.0 equiv) and SulfoCy5-alkyne (14.2 mg, 19.6 μmol, 1.2 equiv) as starting materials. Purification by preparative HPLC yielded JYQ-192 as a blue solid (6.2 mg, 4.7 μmol, 28.5%). 1H NMR (300 MHz, DMSO-d6) δ 12.25 (s, 1H), 8.35 (t, J = 13.1 Hz, 2H), 7.91–7.75 (m, 7H), 7.70–7.60 (m, 3H), 7.37–7.23 (m, 3H), 7.10 (s, 1H), 6.93 (s, 1H), 6.58 (t, J = 12.3 Hz, 1H), 6.30 (dd, J = 13.7, 2.3 Hz, 2H), 5.37 (s, 2H), 4.79 (s, 1H), 4.57 (s, 1H), 4.10 (q, J = 7.7 Hz, 4H), 3.66–3.50 (m, 7H), 3.10–2.96 (m, 6H), 2.80–2.70 (m, 2H), 2.60–2.54 (m, 2H), 2.17 (s, 1H), 2.08–1.98 (m, 3H), 1.73–1.65 (m, 15H), 1.54 (t, J = 7.3 Hz, 2H), 1.25 (t, J = 7.2 Hz, 4H). 13C NMR (75 MHz, DMSO-d6) δ 173.5, 173.1, 172.3, 170.9, 165.0, 156.7, 154.8, 145.7, 142.5, 142.0, 137.6, 135.8, 130.6, 129.2, 126.6, 126.2, 120.5, 117.6, 110.6, 110.4, 103.9, 103.6, 52.5, 50.9, 50.4, 49.4, 46.1, 43.5, 38.4, 35.6, 29.7, 29.5, 27.5, 27.4, 26.2, 25.4, 23.1, 12.6. HR-MS calculated for C63H75N13O12S4 [M + H]2+ 667.7349, found 667.7340.
5-Azido-2-fluorobenzoic Acid (480)
To a solution of 5-amino-2-fluorobenzoic acid (300 mg, 1.93 mmol, 1.0 equiv) in a mixture of H2SO4 (2 mL) and H2O (10 mL) was added NaNO2 (133.4 mg, 1.93 mmol, 1.0 equiv) solution in H2O (1 mL) at 0 °C. The reaction mixture was stirred for 15 min. NaN3 (150.9 mg, 2.32 mmol, 1.2 equiv) solution in H2O (1 mL) was then added dropwise, and stirring was continued for 2 h at 0 °C. After the reaction was complete, the reaction mixture was extracted with EtOAc. The combined organic layer was washed with brine, followed by drying with Na2SO4 and evaporating under reduced pressure to yield the title compound as a white solid (285 mg, 1.57 mmol, 81%). 1H NMR (300 MHz, DMSO-d6) δ 7.25–7.21 (m, 1H), 7.14 (d, J = 1.7 Hz, 1H), 7.12 (d, J = 2.7 Hz, 1H).
(R)-N-(5-(5-Azido-2-fluorobenzoyl)-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl)-1-cyanopyrrolidine-3-carboxamide (481)
To a solution of (R)-1-cyano-N-(4,5,6,7-tetrahydrothiazolo[5,4-c]pyridin-2-yl) pyrrolidine-3-carboxamide (138.5 mg, 0.5 mmol, 1.0 equiv) in DCM were added HCTU (247.6 mg, 0.6 mmol, 1.2 equiv) and DIPEA (297 μL, 1.8 mmol, 3.6 equiv). The reaction mixture was stirred for 10 min. Compound 480 (108.7 mg, 0.6 mmol, 1.2 equiv) was then added, and stirring was continued for 2 h at rt. The solvents were evaporated under reduced pressure, and the resulting residue was taken up in EtOAc (20 mL). The organic layer was washed with 1 M HCl (2 × 10 mL), sat. aq. NaHCO3 (3 × 10 mL), and brine (10 mL), followed by drying with Na2SO4 and evaporating under reduced pressure. The resulting residue was purified by Büchi flash chromatography (DCM to 5% MeOH/DCM) to yield the title compound as a white solid (180 mg, 0.40 mmol, 81%). 1H NMR (300 MHz, DMSO-d6) δ 12.26 (s, 1H), 7.43–7.32 (m, 1H), 7.32–7.12 (m, 2H), 4.85–4.43 (m, 2H), 3.97 (t, J = 5.9 Hz, 1H), 3.66–3.58 (m, 2H), 3.58–3.54 (m, 2H), 3.42 (d, J = 2.0 Hz, 1H), 3.36–3.07 (m, 1H), 2.82–2.59 (m, 2H), 2.28–1.97 (m, 2H). 13C NMR (75 MHz, DMSO-d6) δ 170.9, 164.1, 155.2 (d, J = 243.2 Hz), 143.6, 143.1, 136.8, 125.8 (d, J = 20.5 Hz), 122.7, 119.5, 118.3, 118.0 (d, J = 23.5 Hz), 117.6, 52.5, 50.4, 44.7, 43.5, 42.3, 29.7, 27.2.
BodipyFL Probe JYQ-196
This compound was prepared according to the procedure of JYQ-191 using compound 481 (20 mg, 45.4 μmol, 1.0 equiv) and BodipyFL-alkyne (17.9 mg, 54.5 μmol, 1.2 equiv) as starting materials. Purification by preparative HPLC yielded JYQ-196 as a red solid (8.2 mg, 10.7 μmol, 24%). 1H NMR (300 MHz, DMSO-d6) δ 12.17 (s, 1H), 8.66 (d, J = 7.8 Hz, 1H), 8.11–7.92 (m, 2H), 7.65–7.53 (m, 1H), 6.23 (d, J = 2.9 Hz, 2H), 4.85 (s, 1H), 4.54 (s, 1H), 4.00 (d, J = 5.9 Hz, 1H), 3.67–3.59 (m, 2H), 3.58–3.39 (m, 4H), 3.00 (d, J = 15.3 Hz, 2H), 2.91–2.63 (m, 5H), 2.55 (s, 1H), 2.40 (s, 12H), 2.30–1.97 (m, 3H), 1.90 (t, J = 7.3 Hz, 2H), 1.67 (s, 2H). 13C NMR (75 MHz, DMSO-d6) δ 170.9, 163.9, 157.3 (d, J = 247.1 Hz), 153.6, 148.4, 147.1, 141.3, 134.0, 131.2, 125.7 (d, J = 20.3 Hz), 122.2, 121.1, 118.3, 117.6, 52.6, 50.4, 44.8, 31.2, 29.6, 28.1, 27.3, 26.5, 25.0, 16.3, 14.5. HR-MS calculated for C38H40BF3N10O2S [M + H]+ 769.3187, found 769.3191.
SulfoCy5 Probe JYQ-197
This compound was prepared according to the procedure of JYQ-191 using compound 481 (10 mg, 22.7 μmol, 1.0 equiv) and SulfoCy5-alkyne (19.7 mg, 27.2 μmol, 1.2 equiv) as starting materials. Purification by preparative HPLC yielded JYQ-197 as a blue solid (6.1 mg, 5.3 μmol, 23%). 1H NMR (300 MHz, DMSO-d6) δ 12.24 (d, J = 13.9 Hz, 1H), 8.63 (d, J = 5.9 Hz, 1H), 8.35 (t, J = 13.1 Hz, 2H), 8.08–7.90 (m, 2H), 7.90–7.78 (m, 3H), 7.69–7.51 (m, 3H), 7.32 (dd, J = 8.2, 1.3 Hz, 2H), 7.28 (s, 1H), 7.11 (s, 1H), 6.94 (s, 1H), 6.58 (t, J = 12.4 Hz, 1H), 6.30 (d, J = 13.7 Hz, 2H), 4.84 (s, 1H), 4.53 (s, 1H), 4.19–3.97 (m, 5H), 3.67–3.58 (m, 2H), 3.57–3.46 (m, 3H), 3.10 (q, J = 6.6 Hz, 2H), 2.81–2.64 (m, 4H), 2.28–2.13 (m, 1H), 2.08–1.96 (m, 3H), 1.76 (dd, J = 5.9, 3.6 Hz, 2H), 1.68 (s, 12H), 1.55 (p, J = 7.3 Hz, 2H), 1.39–1.29 (m, 2H), 1.25 (t, J = 7.1 Hz, 3H). 13C NMR (75 MHz, DMSO-d6) δ 173.4, 173.1, 172.4, 170.9, 163.9, 163.7, 157.3 (d, J = 247.4 Hz), 156.9, 156.7, 154.8, 148.3, 145.7, 145.6, 143.6, 143.1, 142.5, 142.0, 141.1, 141.0, 134.0, 126.6, 126.3, 125.7 (d, J = 20.5 Hz), 121.0, 120.4, 118.4, 117.6, 110.6, 110.4, 103.9, 103.6, 52.5, 50.4, 49.3, 44.9, 43.9, 43.5, 38.4, 35.6, 29.7, 29.2, 27.4, 27.1, 26.2, 25.4, 23.0, 12.5. HR-MS calculated for C57H64FN11O9S3 [M + H]2+ 581.7095 found 581.7094.
2-(2,6-Dioxopiperidin-3-yl)-4-fluoroisoindoline-1,3-dione (482)
To a solution of 4-fluoroisobenzofuran-1,3-dione (1.0 g, 6.02 mmol, 1.0 equiv) and 2,6-dioxopiperidine-3-ammonium chloride (0.99 g, 6.02 mmol, 1.0 equiv) in AcOH (50 mL), NaOAc (0.59 g, 7.22 mmol, 1.2 equiv) was added. The resulting reaction mixture was refluxed and stirred for 3 h, followed by pouring into water (300 mL) and extraction with EtOAc (3 × 150 mL). The organic layers were combined, dried with Na2SO4, and evaporated under reduced pressure. The resulting residue was purified by Büchi flash chromatography (DCM to 3.5% MeOH/DCM) to yield the title compound as a white solid (1.3 g, 4.76 mmol, 78%). 1H NMR (300 MHz, DMSO-d6) δ 11.12 (s, 1H), 7.92 (ddd, J = 8.3, 7.3, 4.6 Hz, 1H), 7.80–7.66 (m, 2H), 5.13 (dd, J = 13.0, 5.4 Hz, 1H), 2.94–2.77 (m, 1H), 2.63–2.49 (m, 2H), 2.10–1.98 (m, 1H). 13C NMR (75 MHz, DMSO-d6) δ 173.2, 170.2, 166.6, 164.4, 156.8 (d, J = 262.2 Hz), 138.1 (d, J = 8.0 Hz), 133.9, 123.0 (d, J = 19.6 Hz), 120.5, 117.1 (d, J = 12.5 Hz), 49.5, 31.4, 22.3.
2-(2,6-Dioxopiperidin-3-yl)-4-((2-(2-(prop-2-yn-1-yloxy)ethoxy)ethyl)amino)isoindoline-1,3-dione (483a)
To a solution of compound 482 (200 mg, 0.72 mmol, 1.0 equiv) in DMF (5 mL) were added 2-(2-(prop-2-yn-1-yloxy)ethoxy)ethan-1-amine (134.6 mg, 0.94 mmol, 1.3 equiv) and TEA (195 μL, 1.44 mmol, 2.0 equiv). The reaction mixture was stirred for 24 h at 95 °C. After the reaction was completed, the solvents were evaporated under reduced pressure and the resulting residue was taken up in EtOAc (10 mL). The organic layer was washed with brine (5 mL), followed by drying with Na2SO4 and evaporating under reduced pressure. The resulting residue was purified by Büchi flash chromatography (heptane to 70% heptane/EtOAc) to yield the title compound as a yellow solid (70 mg, 0.18 mmol, 24%). 1H NMR (300 MHz, CDCl3) δ 8.34 (s, 1H), 7.41 (dd, J = 8.5, 7.1 Hz, 1H), 7.02 (dd, J = 7.1, 0.6 Hz, 1H), 6.85 (dd, J = 8.5, 0.6 Hz, 1H), 4.91–4.78 (m, 1H), 4.13 (d, J = 2.4 Hz, 2H), 3.67–3.54 (m, 6H), 3.41 (t, J = 5.5 Hz, 2H), 2.85–2.63 (m, 3H), 2.36 (t, J = 2.4 Hz, 1H), 2.12–1.98 (m, 1H). 13C NMR (75 MHz, C DCl3) δ 171.3, 169.3, 168.5, 167.6, 146.8, 136.0, 132.5, 116.9, 111.7, 110.3, 74.6, 70.5, 69.6, 69.2, 58.5, 48.9, 42.4, 31.4, 22.8.
2-(2,6-Dioxopiperidin-3-yl)-4-((2-(2-(2-(prop-2-yn-1-yloxy)ethoxy)ethoxy)ethyl)amino)isoindoline-1,3-dione (483b)
This compound was prepared according to the procedure for compound 483a using 482 (200 mg, 0.72 mmol, 1.0 equiv) and 2-(2-(2-(prop-2-yn-1-yloxy)ethoxy)ethoxy)ethan-1-amine (176.2 mg, 0.94 mmol, 1.3 equiv) as the starting materials. Purification after Büchi flash chromatography (heptane to 70% heptane/EtOAc) yielded the title compound as a yellow solid (65 mg, 0.14 mol, 19%). 1H NMR (300 MHz, CDCl3) δ 7.42 (dd, J = 8.5, 7.1 Hz, 1H), 7.07–6.99 (m, 1H), 6.86 (d, J = 8.4 Hz, 1H), 4.92–4.80 (m, 1H), 4.13 (d, J = 2.4 Hz, 2H), 3.64 (d, J = 9.3 Hz, 10H), 3.41 (q, J = 5.5 Hz, 2H), 2.90–2.56 (m, 3H), 2.36 (t, J = 2.4 Hz, 1H), 2.13–1.95 (m, 1H). 13C NMR (75 MHz, CDCl3) δ 171.0, 169.3, 168.3, 167.6, 146.9, 136.1, 132.5, 116.8, 111.7, 110.3, 74.6, 70.7, 70.7, 70.5, 69.5, 69.1, 58.4, 48.9, 42.4, 31.4, 22.8.
PROTAC JYQ-187
This compound was prepared according to the procedure of JYQ-191 using azide 479 (10 mg, 22.7 μmol, 1.0 equiv) and alkyne 483a (19.7 mg, 27.2 μmol, 1.2 equiv) as starting materials. After the reaction was complete, the reaction mixture was poured into water (20 mL) and extracted with DCM (3 × 20 mL). The combined organic layer was washed with brine, followed by drying with Na2SO4 and evaporating under reduced pressure. The resulting residue was purified by Büchi flash chromatography (DCM to 6% MeOH/DCM) to yield JYQ-187 as a yellow solid (8.5 mg, 8.4 μmol, 26%). 1H NMR (300 MHz, DMSO-d6) δ 12.27 (s, 1H), 11.10 (s, 1H), 7.94–7.65 (m, 5H), 7.62–7.55 (m, 1H), 7.13 (d, J = 8.6 Hz, 1H), 7.03 (d, J = 6.9 Hz, 1H), 6.60 (t, J = 5.8 Hz, 1H), 5.41 (s, 2H), 5.05 (dd, J = 12.9, 5.4 Hz, 1H), 4.79 (s, 1H), 4.51 (s, 3H), 3.97 (s, 1H), 3.70–3.51 (m, 14H), 3.01 (d, J = 22.2 Hz, 4H), 2.90–2.52 (m, 8H), 2.27–1.93 (m, 3H). 13C NMR (75 MHz, DMSO-d6) δ 173.3, 170.9, 170.6, 169.4, 167.8, 164.9, 146.8, 144.1, 137.6, 136.7, 135.7, 132.5, 132.2, 130.6, 129.2, 126.2, 126.0, 118.5, 117.9, 117.6, 111.1, 109.7, 70.1, 69.3, 63.9, 52.5, 51.0, 50.4, 49.0, 46.1, 44.0, 43.5, 42.1, 41.2, 31.4, 29.6, 22.6. HR-MS calculated for C45H49N13O11S2 [M + H]+ 1012.3194, found 1012.3198.
PROTAC JYQ-188
This compound was prepared according to the procedure for JYQ-187 using azide 479 (20.0 mg, 32.6 μmol, 1.0 equiv) and alkyne 483b (17.4 mg, 39.2 μmol, 1.2 equiv) as starting materials. Purification after Büchi flash chromatography (DCM to 6% MeOH/DCM) yielded JYQ-188 as a yellow solid (10.8 mg, 10.2 μmol, 31%). 1H NMR (300 MHz, DMSO-d6) δ 12.26 (s, 1H), 11.10 (s, 1H), 7.99–7.63 (m, 5H), 7.57 (dd, J = 8.6, 7.1 Hz, 1H), 7.12 (d, J = 8.6 Hz, 1H), 7.03 (d, J = 7.0 Hz, 1H), 6.58 (t, J = 5.7 Hz, 1H), 5.40 (s, 2H), 5.04 (dd, J = 12.9, 5.4 Hz, 1H), 4.78 (s, 1H), 4.49 (s, 3H), 3.96 (s, 1H), 3.78–3.57 (m, 15H), 3.43–3.25 (m, 5H), 3.01 (d, J = 22.2 Hz, 4H), 2.90–2.60 (m, 3H), 2.60–2.54 (m, 1H), 2.31–1.94 (m, 3H). 13C NMR (75 MHz, DMSO-d6) δ 173.4, 171.0, 170.6, 169.4, 167.8, 164.9, 146.9, 144.1, 137.5, 136.8, 135.7, 132.5, 132.1, 117.9, 117.6, 111.2, 109.6, 70.2, 69.3, 63.9, 52.5, 51.0, 50.4, 49.0, 46.1, 45.9, 43.5, 42.1, 31.4, 29.4, 22.6. HR-MS calculated for C47H53N13O12S2 [M + H]+ 1056.3456, found 1056.3456.
PROTAC JYQ-194
This compound was prepared according to the procedure for JYQ-187 using azide 481 (20.0 mg, 45.5 μmol, 1.0 equiv) and alkyne 483a (21.8 mg, 54.5 μmol, 1.2 equiv) as starting materials. Purification after Büchi flash chromatography (DCM to 5% MeOH/DCM) yielded JYQ-194 as a yellow solid (8.1 mg, 9.7 μmol, 21%). 1H NMR (300 MHz, DMSO-d6) δ 12.25 (d, J = 11.8 Hz, 1H), 11.09 (s, 1H), 8.78 (d, J = 8.5 Hz, 1H), 8.11–7.92 (m, 2H), 7.63–7.50 (m, 2H), 7.13 (d, J = 8.6 Hz, 1H), 7.03–6.98 (m, 1H), 6.59 (t, J = 5.7 Hz, 1H), 5.02 (dd, J = 12.9, 5.4 Hz, 1H), 4.84 (s, 1H), 4.61 (d, J = 2.4 Hz, 2H), 4.00 (t, J = 5.9 Hz, 1H), 3.61 (tt, J = 7.6, 4.3 Hz, 10H), 3.44–3.24 (m, 6H), 2.94–2.72 (m, 2H), 2.71–2.53 (m, 2H), 2.29–2.00 (m, 3H). 13C NMR (75 MHz, DMSO-d6) δ 173.4, 171.0, 170.6, 169.4, 167.8, 163.9, 157.5 (d, J = 247.7 Hz), 156.7, 146.8, 145.7, 143.5, 143.1, 136.7, 133.8, 132.5, 125.7 (d, J = 20.7 Hz), 123.6, 122.9, 120.7 (d, J = 11.3 Hz), 118.3, 117.9, 117.6, 111.2, 109.6, 70.1, 69.6, 69.4, 63.8, 52.5, 50.4, 49.0, 44.8, 43.5, 42.1, 31.4, 29.7, 22.6. HR-MS calculated for C39H38FN11O8S [M + H]+ 840.2688, found 840.2691.
PROTAC JYQ-195
This compound was prepared according to the procedure for JYQ-187 using azide 481 (20.0 mg, 45.5 μmol, 1.0 equiv) and alkyne 483b (24.2 mg, 54.5 μmol, 1.2 equiv) as starting materials. Purification after Büchi flash chromatography (DCM to 5% MeOH/DCM) yielded JYQ-195 as a yellow solid (6.5 mg, 7.4 μmol, 16%). 1H NMR (300 MHz, DMSO-d6) δ 12.25 (d, J = 12.4 Hz, 1H), 11.10 (s, 1H), 8.80 (s, 1H), 8.11–7.93 (m, 2H), 7.65–7.52 (m, 2H), 7.11 (d, J = 8.6 Hz, 1H), 7.02 (d, J = 6.9 Hz, 1H), 6.57 (t, J = 5.8 Hz, 1H), 5.03 (dd, J = 12.8, 5.4 Hz, 1H), 4.84 (s, 1H), 4.60 (d, J = 2.4 Hz, 2H), 4.53 (s, 1H), 4.00 (t, J = 5.8 Hz, 1H), 3.71–3.56 (m, 15H), 3.46–3.25 (m, 6H), 2.96–2.54 (m, 2H), 2.30–2.10 (m, 1H), 2.10–1.91 (m, 2H). 13C NMR (75 MHz, DMSO-d6) δ 173.4, 171.0, 170.6, 169.4, 167.8, 163.9, 163.7, 157.1 (d, J = 248.1 Hz), 156.8, 146.8, 145.8, 143.6, 143.1, 136.7, 133.8, 132.5, 125.7 (d, J = 20.7 Hz), 123.6, 118.3, 117.9, 117.6, 111.2, 109.6, 70.2, 69.6, 69.3, 63.8, 52.5, 50.3, 49.0, 44.8, 43.5, 42.1, 31.4, 29. 7, 27.2, 26.4, 22.6. HR-MS calculated for C41H42FN11O9S [M + H]+ 884.2950, found 884.2953.
Echo-Mediated High-Throughput Synthesis
Stock solutions of the cyanimide amine in DMSO (200 mM), carboxylic acids in DMSO (100 mM), and a mixture of HOBt and DIC in DMSO (111.1 mM) were prepared. The stock solutions of the amine (125 nL, final concentration 10 mM) and carboxylic acids (1250 nL, final concentration 50 mM) were transferred to an Echo-ready 1536-well LDV plate (Labcyte LP-0400) by using a Labcyte Echo550 acoustic dispenser, followed by adding the mixture of HOBt and DIC (1125 nL, final concentration 10 mM). The plate was sealed and kept overnight at room temperature. In total, 471 compounds were synthesized as crude reaction mixtures in a total volume of 2.5 μL per well with a maximum concentration of 10 mM, assuming 100% reaction conversion. LC-MS analysis of a representative number of compounds (38 in total) was used to confirm compound formation and purity (Supporting Information Figure S11). A 1 mM daughter plate was made by diluting the compounds 10x in DMSO (300 nL compound solution plus 2700 nL DMSO) in an Echo-ready 1536-well LDV plate (Labcyte LP-0400) by using a Labcyte Echo550 acoustic dispenser.
High-Throughput Screening
The screen was performed in a buffer containing 50 mM Tris-HCl, 150 mM NaCl, 2 mM TCEP, pH 7.5, 1 mg/mL CHAPS, and conducted in a 1536-well plate (Corning 3724) with a reaction volume of 8 μL per well. Stock solutions of 0.26 μM PARK7 and 80 nM JYQ-107 were prepared. Using a Labcyte Echo550 acoustic dispenser, 10 nL of 1 mM DMSO stock solutions of the library compounds was transferred from the source plates into the empty 1536-well screening plates to obtain a 1.25 μM final compound concentration. Next, PARK7 (6 μL, final concentration 0.2 μM) was dispensed using a Biotek MultiflowFX liquid dispenser and incubated for 2 h, followed by dispensing the probe JYQ-107 (2 μL, final concentration 20 nM). After 2 h incubation, the FP signal was recorded on a BMG Labtech PHERAstar plate reader (λex/em 480/520 nm). The percentage inhibition of each compound was calculated from the FP values, normalized to the positive (5 mM N-ethylmaleimide, 100% inhibition) and negative (DMSO, 0% inhibition) controls.
Hit Picking and Validation
The validation was performed using the conditions described above in a 384-well plate (Corning 3820) with a reaction volume of 20 μL per well in triplicate. Fifty four compounds showing inhibition above 90% were picked and checked at three different concentrations. Using a Labcyte Echo550 acoustic dispenser, 20, 10, and 5 nL of 1 mM compound stock solutions were transferred from the source plates into an empty 384-well plate to obtain 1, 0.5, 0.25 μM final concentrations. Next, PARK7 (15 μL, final concentration 0.2 μM) was dispensed with a Biotek MultiFlowFX liquid dispenser and incubated for 1 h, followed by dispensing the probe JYQ-107 (5 μL, final concentration 20 nM). The FP signal was monitored on a BMG Labtech PHERAstar plate reader (λex/em 480/520 nm) for 2 h. The relative loss of FP signal compared with reference controls (5 mM NEM, 100% inhibition, and DMSO, 0% inhibition) was used to calculate the remaining enzyme activity. The inhibition percentage of these 54 compounds was plotted as heatmaps using Graphpad Prism 9.0.1 software.
Cell Lines and Cell culture
HEK293T and HeLa cells were originally obtained from the American Type Culture Collection (ATCC). A549 cells and MCF7 cells were kindly provided by Prof. Dr. Peter ten Dijke (LUMC), while H1299 cells were kindly provided by Dr. A.G. Jochemsen (LUMC). All cell lines were cultured in Dulbecco’s modified Eagles’ medium (DMEM) (Gibco) supplemented with 7.5% fetal bovine serum (FBS) at 37 °C and 5% CO2.
Cell Viability Assay
5 × 103 A549 cells were seeded into 96-well plates. The following day, the indicated final concentrations of JYQ-164 and JYQ-173 were added to the cells, and DMSO was used as a control. Inhibitors were renewed at 36 h, and cells were incubated with them an additional 36 h. Cell viability was measured using the CellTiter-Blue viability assay (Promega). Relative survival was normalized to the DMSO-treated sample and corrected for background signal.
siRNA Transfection
Oligos used to knockdown PARK7 were purchased from Dharmacon (Cat#: MQ-005984-00-0002). Silencing was performed in HEK293T and A549 cells as follows: for a 24-well plate format, 50 μL of siRNA (500 nM stock) was incubated with 1 μL of Dharmafect reagent 1 (Dharmacon) diluted in 50 μL of medium without supplements (total volume of 100 μL of transfection mix) with gentle shaking for 20 min at room temperature (RT). A total of 30 × 103 HEK293T or 50 × 103 cells were added to transfection mixes to a total volume of 500 μL per well and cultured for 48 h prior to incubation with 5 μM final concentration of JYQ-196 for 4 h.
Antibodies and Fluorescent Dyes
The following antibodies were used for the detection of endogenous protein by immunoblot analysis in a 1:1000 dilution: rabbit anti-PARK7 (Abcam, Cat# ab18257), rabbit anti-UCHL1 (Abcam, Cat# ab27053), Mouse anti-β-actin (Sigma-Aldrich, Cat# A5441) was used as a loading control in a 1:10,000 dilution for Western blot. Secondary IRDye 800CW goat antirabbit IgG (H + L) (Li-COR, Cat# 926-32211, 1:5000) and IRDye 680LT goat antimouse IgG (H + L) (Li-COR, Cat# 926-68020, 1:20,000) were used for detection using the Odyssey Classic imager (LI-COR).
SDS-PAGE and Immunoblotting
Samples were separated by 12 or 4–12% SDS-PAGE. After proteins were transferred to a nitrocellulose membrane at 300 mA for 2.5 h, the membranes were blocked with 5% skim milk in PBS and incubated with a primary antibody diluted in 5% skim milk in 0.1% PBS-Tween 20 (PBST) for 1 h at RT. After washing with 0.1% PBST three times for 10 min, proteins were incubated with secondary antibodies diluted in 0.1% PBST for 30 min and washed three times again in 0.1% PBST. The signal was detected using direct imaging by the Odyssey Classic imager (LI-COR).
Cell Lysate Preparation
Cell pellets were suspended in the cell lysis buffer (50 mM Tris, 150 mM NaCl, 0.5% Triton X-100, and 2 mM TCEP at pH 7.5) supplemented with protease inhibitor cocktail (11836145001, Roche). The samples were kept on ice and sonicated using 10 cycles of 30 s pulse on, 30 s pulse off (Bioruptor, Diagenode). The cell lysate was centrifuged at 14,000 rpm with Eppendorf Centrifuge 5430 R for 20 min at 4 °C, and supernatant fractions were collected.
Cell-Based Competition Assay
HEK293T cells were treated with the indicated concentrations of indicated inhibitors for indicated incubation time. The cells were then washed with PBS and collected, followed by cell lysis. The prepared cell lysate of each sample was incubated by JYQ-92 (1 μM) for 30 min, followed by adding NuPAGE LDS sample buffer (4X). Samples were resolved by SDS-PAGE using a 12% Bis-Tris gel with MES SDS running buffer and visualized by fluorescence scanning on a Typhoon FLA 9500 using a Cy5 channel (λex/em 635/655 nm), followed by protein transfer to nitrocellulose membranes and immunoblot analysis.
IC50 Determination
The assay was performed in PBS buffer and conducted in a 384-well plate (Corning 3820) with a reaction volume of 20 μL in triplicate. Stock solutions of compounds of 0.01, 0.1, and 1 mM were prepared. Compounds were transferred using a Labcyte Echo550 acoustic dispenser to obtain a 12-point serial dilution of 0.001–100 μM. Next, PARK7 (15 μL, final concentration 0.1 μM) was dispensed with a Biotek MultiFlowFX liquid dispenser and incubated for 1 h, followed by dispensing the substrate DiFMUAc (5 μL, final concentration 300 μM). The fluorescence intensity (FI) signal was monitored on a BMG Labtech PHERAstar plate reader (λex/em 350/450 nm) for 1 h. All samples were normalized to the positive and negative controls and plotted against the inhibitor concentrations (in μM) using the built-in equation “[inhibitor] vs response–variable slope (four parameters), least-squares fit” with constraints “Bottom = 0” and “Top = 100” in GraphPad Prism 9.0.1 software to obtain the IC50 values.
Covalent Complex Formation Mass Spectrometry Analysis
The stock concentration of 20 μM PARK7 was prepared in Tris-buffer (50 mM Tris-HCl, 150 mM NaCl, 2 mM TCEP, pH 7.5), followed by incubating with DMSO or 30 μM indicated compounds in buffer (10 μM PARK7 and 15 μM indicated compounds final concentration) for 1 h. Samples were then diluted 10-fold with acetonitrile:water (1:1), and 1 μL of sample was injected to LC-MS analysis. LC-MS analysis was performed on Waters Acquity H-class UPLC with a UPLC protein BEH C4 column (1.7 μm, 2.1 × 50 mm) coupled to a Xevo G2-XS Qtof mass spectrometer with ESI. Samples were run using CH3CN/H2O gradients equipped with 0.1% formic acid for 7 min (2–100% CH3CN). Deconvoluted mass was obtained from convolution spectra (50–1200 m/z) using with MaxEnt1 function of Waters MassLynx mass spectrometry software 4.1.
Probe Labeling of Purified Recombinant PARK7
The assay was conducted in Tris-buffer (50 mM Tris-HCl, 150 mM NaCl, 2 mM TCEP, pH 7.5). PARK7 (1 μM final concentration) was incubated with different concentrations (0, 0.1, 0.5, 1, 2, 5, and 10 μM) of the indicated probes for 1 h at 37 °C. After completing the incubation time, all of the reactions were stopped by adding NuPAGE LDS sample buffer (4x). These samples were resolved by SDS-PAGE using precast 12% Bis-Tris gels (Invitrogen, NuPAGE) with MES SDS running buffer (Novex, NuPAGE). The gels were visualized by Typhoon FLA 9500 (GE Healthcare Life Sciences) fluorescence scanning: PARK7-JYQ-191 and PARK7-JYQ-196 adducts were visualized with a Rhodamine channel (λex/em 473/530 nm); PARK7-JYQ-192 and PARK7-JYQ-197 adducts were visualized with a Cy5 channel (λex/em 635/655 nm), followed by staining with Instant Blue Coomassie protein stain (Expedeon) and scanning on an Amersham Imager 600 (GE Healthcare Life Sciences).
Probe Labeling of Endogenous PARK7 in Cells
HEK293T or A549 cells were incubated with indicated concentration of the probes for the indicated time points, followed by cell lysis and adding loading buffer (4x). Samples were resolved by SDS-PAGE using a 12% Bis-Tris gel with MES SDS running buffer and visualized by fluorescence scanning on a Typhoon FLA 9500 using a Cy5 channel (λex/em 635/655 nm), followed by transferring to nitrocellulose membranes and immunoblot analysis.
Confocal Microscopy
A549 cells were seeded into 24-well plates (Costar, Cat# 3524) containing glass coverslips (Menzel Gläser, Cat# MENZCB00130RAC) and incubated with 5 μM of final concentration of probes JYQ-192, JYQ-196, and JYQ-197 for fluorescence confocal microscopy of fixed samples. Fixation was performed in 3.7% formaldehyde (acid-free, Merck Millipore) in phosphate-buffered saline (PBS) for 20 min. After washing 3× with PBS, cells were mounted using a ProLong Gold antifade Mounting medium with DAPI (Life Technologies, Cat# P36941). Samples were imaged using a Leica SP8 microscope equipped with appropriate solid-state lasers, HCX PL 63 times magnification oil emersion objectives and HyD detectors. Data were collected using a digital zoom in 1 in 1024 by 1024 scanning format with line averaging. Postcollection image processing was performed using the Fiji software.
Activity Evaluation of PROTACs in Cells
A549 cells were seeded into 6-well plates. The next day after seeding, cells were incubated with the indicated concentrations of indicated PROTACs for 8 h. The cells were then washed with PBS and collected. The collected cell pellets were lysed with Tris-triton buffer as described above. The prepared cell lysate of each sample (10 μL) was added 5 μL 3× LDS + 5% β-mercaptoethanol, and boiled for 10 min. Samples were resolved by SDS-PAGE using a 4–12% Bis-Tris gel with MOPS SDS running buffer, followed by transferring to nitrocellulose membranes and immunoblot analysis.
Streamlined Cysteine Activity-Based Protein Profiling
A549 cells were seeded into 10 cm plates in 10 mL of media. The day after seeding, cells were incubated with the indicated concentrations of JYQ-164, and JYQ-173 for 4 h. The cells were then washed two times with cold PBS (1 mL per 10 cm plate) and collected by scraping them with a cell scraper. Cells were lysed by sonication in ice-cold PBS and centrifuged at 15,000 rpm for 2 min to remove cell debris. The BCA Gold protein assay (Thermo Fisher Scientific) was performed to determine protein concentration. Each sample (100 μg) was labeled with 500 μM DBIA probe for 1 h in the dark at room temperature (RT). Excess DBIA and disulfide bonds were quenched and reduced, respectively, using 5 mM dithiothreitol (DTT) for 30 min in the dark at RT. Reduced disulfide bonds were alkylated using 20 mM iodoacetamide for 30 min in the dark at RT. Proteins were precipitated using chloroform/methanol, resolubilized in 40 mM HEPES (pH 8.4), and digested using TPCK-treated trypsin and endoGluC (1:12.5 enzyme/protein ratio) overnight at 37 °C. Digested peptides were labeled with TMTpro16-plex reagents (Thermo Fisher Scientific) in a 1:4 ratio by mass (peptides/TMT reagents) for 1 h at RT. Excess TMT reagent was quenched with 5 μL 6% hydroxylamine for 15 min at RT. All samples were then pooled and lyophilized.
Lyophilized samples were reconstituted in 1 mL of PBS and enriched with Pierce streptavidin magnetic beads (Catalog number: 88816, Thermo Scientific) by rotating end-over-end for 4 h at RT. Nonspecific binding peptides were washed away using the following procedure: 3 × 1 mL of PBS pH 7.4, 2 × 1 mL of PBS with 0.1% SDS pH 7.4, and 3 × 1 mL of HPLC-grade water. DBIA probe-containing peptides were eluted using 700 μL of 50% acetonitrile in 0.1% TFA, lyophilized using a Speedvac, and desalted using a handmade mini-SPE column (10% sorbent material of a Waters, OASIS 1 cm3 HLB 30 mg cartridge). Column was equilibrated with 200 μL of 90% acetonitrile and 3 × 200 μL of 10 mM NH4HCO3 pH 8.4. The dried sample was dissolved in 200 μL of 10 mM NH4HCO3 pH 8.4, loaded onto the column, washed 3x with 200 μL of 10 mM NH4HCO3 pH 8.4, and eluted into 3 fractions with 150 μL of 10, 20, and 50% acetonitrile (in 10 mM NH4HCO3). Samples were then lyophilized.
TMT-Based Global Proteomic Profiling
A549 cells were seeded into 10 cm plates in 10 mL of media. Two days after seeding, cells were incubated with 5 μM JYQ-173 or JYQ-194 for 8 h. The cells were then washed two times with cold PBS (1 mL per 10 cm plate) and collected by scraping them with a cell scraper. Cells were lysed using a 5% SDS lysis buffer (100 mM Tris-HCl, pH 7.6) and were incubated at 95 °C for 4 min. Protein determination was performed using the Pierce BCA Gold protein assay. 100 μg of protein of each sample was reduced and alkylated, and excess iodoacetamide was quenched using 5 mM TCEP, 15 mM iodoacetamide, and 10 mM DTT, respectively. The protein was pelleted down using chloroform/methanol precipitation, and the resulting pellets were resolubilized in 40 mM HEPES pH 8.4. Trypsin (1:12.5 enzyme/protein ratio) was added to digest protein overnight at 37 °C. The Pierce BCA Gold protein assay was used to determine the peptide concentration. The peptides were labeled with TMTpro Label Reagents in a 1:4 ratio by mass (peptides/TMT reagents) for 1 h at RT. Five μL of 6% hydroxylamine was added to quench excess TMT reagent and incubated for 15 min at RT. Samples were pooled and lyophilized. The sample was subsequently fractionated on an Agilent 1200 series HPLC system (Agilent Technologies). 75 μg of the pooled sample was dissolved in solvent A (10 mM NH4HCO3, pH 8.4), injected onto, and eluted from an Agilent Eclipse Plus C18 2.1 × 150 mm 3.5 μm column (Agilent Technologies). The gradient was run from 2 to 90% solvent B (10 mM NH4HCO3, pH 8.4 final concentration, 20/80 water/acetonitrile v/v) in 30 min at a flow rate of 200 μL/min. Twelve fractions were made; every 30 s a fraction was collected in a vial, and the eluate of the next 30 s was collected in the next vial and so on until it loops back to the first vial (5 loops of 6 min each). Afterward, the samples were lyophilized.
Mass Spectrometry
Lyophilized peptides were dissolved in 0.1% formic acid (FA) and analyzed by online C18 nano-HPLC MS/MS with a system consisting of an Ultimate3000nano gradient HPLC system (Thermo, Bremen, Germany), and an Exploris480 mass spectrometer (Thermo). Fractions were injected onto a cartridge precolumn (300 μm × 5 mm, C18 PepMap, 5 μm, 100 A) and eluted via a homemade analytical nano-HPLC column (50 cm × 75 μm; Reprosil-Pur C18-AQ 1.9 um, 120 A (Dr. Maisch, Ammerbuch, Germany)). The gradient was run from 2 to 36% solvent B (20/80/0.1 water/acetonitrile/FA v/v) in 120 min. The nano-HPLC column was drawn to a tip of ∼10 μm and acted as the electrospray needle of the MS source. The mass spectrometer was operated in data-dependent MS/MS mode for a cycle time of 3 s, with a HCD collision energy at 36% and recording of the MS2 spectrum in the Orbitrap, with a quadrupole isolation width of 1.2 Da. In the master scan (MS1), the resolution was 120,000, the scan range 350–1600, at standard AGC target @maximum fill time of 50 ms. A lock mass correction on the background ion m/z = 445.12 was used. Precursors were dynamically excluded after n = 1 with an exclusion duration of 45 s, and with a precursor range of 20 ppm. Charge states 2–5 were included. For MS2, the first mass was set to 110 Da, and the MS2 scan resolution was 45,000 at an AGC target of 200% with a maximum fill time set to auto.
Data Analysis
For SLC-ABBP, in a postanalysis process, raw data were first converted to peak lists using Proteome Discoverer version 2.4 (Thermo Electron) and then submitted to the Uniprot Homo sapiens minimal database (20296 entries), using Mascot v. 2.2.04 (www.matrixscience.com) for protein identification. Mascot searches were done with 10 ppm and 0.02 Da deviation for the precursor and fragment mass, respectively, and the enzymes trypsin and EndoGluC was specified, up to three missed cleavages were allowed. Methionine oxidation, acetyl (protein N-term), and DBIA on Cys were set as a variable modification. Carbamidomethyl on Cys and TMTpro on Lys and N-term were set as a fixed modification. Peptides with an FDR <1% were accepted. Peptides were only retained, if they were quantified and if there was exactly one DBIA modification on the peptide. The values for each TMT channel were normalized to the average of the values for the DMSO samples for the same peptide, and these values were named CR. For each peptide, the first entry of the Master Protein Accessions column was chosen as the Master Protein Accession, and the residue number of the cysteine modified by DBIA within the entire protein sequence was determined. Using this information, each peptide was assigned an identifier consisting of the Master Protein Accession and the residue number of the modified cysteine to assign each peptide to the respective modified cysteine residue. Within each replicate, CR values for all peptides with the same identifier (and thus the same modified cysteine residue) were combined by averaging and only the shortest Annotated Sequence was kept as the final Annotated Sequence. The CR values of all replicates for the same identifier under the same condition were averaged to give the “Average CR” for the respective cysteine under that condition.
For TMT-based global proteomics, in a postanalysis process, raw data were first converted to peak lists using Proteome Discoverer version 2.5 (Thermo Electron) and submitted to the Uniprot database (Homo sapiens, 20596 entries), using Mascot v. 2.2.07 (www.matrixscience.com) for protein identification. Mascot searches were with 10 ppm and 0.02 Da deviation for precursor and fragment mass, respectively. Enzyme specificity was set to trypsin. Up to two missed cleavages were allowed. Methionine oxidation and acetyl on protein N-terminus were set as variable modifications. Carbamidomethyl on Cys and TMTpro on Lys and N-terminus were set as fixed modifications. Protein FDR of 1% was set. Normalization was on total peptide amount. The values for each TMT channel were normalized to the average of the values for the DMSO samples for the same peptide, and these values were named FC. The log2 FC values of all replicates for the same master protein were averaged to give the “Average log2 FC”. A one-sample t test was performed against the null hypothesis of log2 FC = 0, and the resulting −log10p values were reported for each protein. The volcano plot was plotted using GraphPad Prism v.9.01.
Acknowledgments
We thank Bjorn van Doodewaerd for LC-MS measurement and compound purification, Angeliki Moutsiopoulou for help with protein purification, and Vito Pol for chemical synthesis. This project is funded by the Institute for Chemical Immunology (grant no. ICI00026 to A.S.) and the Innovative Medicines Initiative 2 (IMI2) Joint Undertaking under grant agreement no. 875510 (EUbOPEN project). Y.J. is supported by the China Scholarship Council.
Glossary
Abbreviations
- ABP
activity-based probe
- CHAPS
(3-((3-cholamidopropyl) dimethylammonio)-1-propanesulfonate)
- CR
competition ratio
- CRBN
cereblon
- DBIA
desthiobiotin iodoacetamide
- DIC
N,N′-diisopropylcarbodiimide
- DiFMUAc
6,8-difluoro-4-methylumbelliferyl acetate
- DIPEA
N,N-diisopropylethylamine
- DOP1B
DOP1 leucine zipper-like protein B
- DUBs
deubiquitinating enzymes
- FC
fold change
- FP
fluorescence polarization
- HCTU
O-(1H-6-chlorobenzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate
- HINT2
histidine triad nucleotide-binding protein 2
- HOBt
1-hydroxybenzotriazole
- LDV
low dead volume
- PARK7
Parkinson Disease Protein 7
- POI
protein of interest
- POMA
pomalidomide
- PROTACs
proteolysis targeting chimeras
- SLC-ABPP
streamlined cysteine activity-based protein profiling
- TCEP
tris(2-carboxyethyl)phosphine
- TEA
triethylamine
- TM7SF3
transmembrane 7 superfamily member 3
- TMT
tandem mass tag
- TPCK
tosylsulfonyl phenylalanyl chloromethyl ketone
- UCHL1
ubiquitin C-terminal hydrolase L1.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.3c02410.
Assessment of the cell permeability and covalent bond formation, inhibition of PARK7, activity evaluation of inhibitors, IC50 determination, inhibition values and selectivity ratios, cytotoxicity assay, PARK7 labeling, cell permeability and cellular PARK7 labeling efficiency, off-target validation, degradation evaluation, and plate lay-out and LC-MS analysis (PDF)
Supplementary Data S1 including high-throughput screening and validation data (XLSX)
Supplementary Data S2 including SLC-ABPP data analysis of all cysteine sites (XLSX)
Supplementary Data S3 including SLC-ABPP data analysis of unique cysteine sites (XLSX)
Supplementary Data S4 including quantitative TMT-based total proteome profiling data analysis of PROTACs (XLSX)
Molecular formula strings for all compounds (CSV)
The authors declare no competing financial interest.
Notes
The mass spectrometry proteomics data have been deposited with the ProteomeXchange Consortium via the PRIDE partner repository with data set identifier PXD047093.
Supplementary Material
References
- Jin W. Novel Insights into PARK7 (DJ-1), a Potential Anti-Cancer Therapeutic Target, and Implications for Cancer Progression. J. Clin. Med. 2020, 9 (5), 1256. 10.3390/jcm9051256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. A. The role of cysteine oxidation in DJ-1 function and dysfunction. Antioxid Redox Signal 2011, 15 (1), 111–22. 10.1089/ars.2010.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repici M.; Straatman K. R.; Balduccio N.; Enguita F. J.; Outeiro T. F.; Giorgini F. Parkinson’s disease-associated mutations in DJ-1 modulate its dimerization in living cells. J. Mol. Med. (Berl) 2013, 91 (5), 599–611. 10.1007/s00109-012-0976-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen F. L.-H.; Scafidi A.; Poli A.; Michelucci A. PARK7/DJ-1 in microglia: implications in Parkinson’s disease and relevance as a therapeutic target. J. Neuroinflammation 2023, 20 (1), 95. 10.1186/s12974-023-02776-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivo E.; La Chimia M.; Ceramella J.; Catalano A.; Chiaradonna F.; Sinicropi M. S.; Cuda G.; Iacopetta D.; Scumaci D. Moving beyond the Tip of the Iceberg: DJ-1 Implications in Cancer Metabolism. Cells 2022, 11 (9), 1432. 10.3390/cells11091432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss R.; Zhu M.; Jójárt B.; Czajlik A.; Solti K.; Fórizs B.; Nagy É.; Zsila F.; Beke-Somfai T.; Tóth G. Structural features of human DJ-1 in distinct Cys106 oxidative states and their relevance to its loss of function in disease. Biochim. Biophys. Acta Gen. Subj. 2017, 1861 (11 Pt A), 2619–2629. 10.1016/j.bbagen.2017.08.017. [DOI] [PubMed] [Google Scholar]
- Sun M. E.; Zheng Q. The Tale of DJ-1 (PARK7): A Swiss Army Knife in Biomedical and Psychological Research. Int. J. Mol. Sci. 2023, 24 (8), 7409. 10.3390/ijms24087409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solti K.; Kuan W. L.; Forizs B.; Kustos G.; Mihaly J.; Varga Z.; Herberth B.; Moravcsik E.; Kiss R.; Karpati M.; Mikes A.; Zhao Y.; Imre T.; Rochet J. C.; Aigbirhio F.; Williams-Gray C. H.; Barker R. A.; Toth G. DJ-1 can form beta-sheet structured aggregates that co-localize with pathological amyloid deposits. Neurobiol Dis 2020, 134, 104629 10.1016/j.nbd.2019.104629. [DOI] [PubMed] [Google Scholar]
- Miyazaki S.; Yanagida T.; Nunome K.; Ishikawa S.; Inden M.; Kitamura Y.; Nakagawa S.; Taira T.; Hirota K.; Niwa M.; Iguchi-Ariga S. M.; Ariga H. DJ-1-binding compounds prevent oxidative stress-induced cell death and movement defect in Parkinson’s disease model rats. J. Neurochem 2008, 105 (6), 2418–34. 10.1111/j.1471-4159.2008.05327.x. [DOI] [PubMed] [Google Scholar]
- Kitamura Y.; Watanabe S.; Taguchi M.; Takagi K.; Kawata T.; Takahashi-Niki K.; Yasui H.; Maita H.; Iguchi-Ariga S. M.; Ariga H. Neuroprotective effect of a new DJ-1-binding compound against neurodegeneration in Parkinson’s disease and stroke model rats. Mol. Neurodegener. 2011, 6, 48. 10.1186/1750-1326-6-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro S.; Caaveiro J. M. M.; Nakakido M.; Tanabe A.; Nagatoishi S.; Tamura Y.; Matsuda N.; Liu D.; Hoang Q. Q.; Tsumoto K. Discovery and Optimization of Inhibitors of the Parkinson’s Disease Associated Protein DJ-1. ACS Chem. Biol. 2018, 13 (9), 2783–2793. 10.1021/acschembio.8b00701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. B.; Zhu H. Y.; Bao K.; Jiang L.; Zhu H.; Ying M. D.; He Q. J.; Yang B.; Sheng R.; Cao J. Bis-isatin derivatives: design, synthesis, and biological activity evaluation as potent dimeric DJ-1 inhibitors. Acta Pharmacol Sin 2021, 42 (7), 1160–1170. 10.1038/s41401-020-00600-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimovic I.; Finkin-Groner E.; Fukase Y.; Zheng Q.; Sun S.; Michino M.; Huggins D. J.; Myers R. W.; David Y. Deglycase-activity oriented screening to identify DJ-1 inhibitors. RSC Med. Chem. 2021, 12 (7), 1232–1238. 10.1039/D1MD00062D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsel J.; Mandl F. A.; Sieber S. A. Chemical Probe To Monitor the Parkinsonism-Associated Protein DJ-1 in Live Cells. ACS Chem. Biol. 2018, 13 (8), 2016–2019. 10.1021/acschembio.8b00633. [DOI] [PubMed] [Google Scholar]
- Jia Y.; Kim R. Q.; Kooij R.; Ovaa H.; Sapmaz A.; Geurink P. P. Chemical Toolkit for PARK7: Potent, Selective, and High-Throughput. J. Med. Chem. 2022, 13288. 10.1021/acs.jmedchem.2c01113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuljanin M.; Mitchell D. C.; Schweppe D. K.; Gikandi A. S.; Nusinow D. P.; Bulloch N. J.; Vinogradova E. V.; Wilson D. L.; Kool E. T.; Mancias J. D.; Cravatt B. F.; Gygi S. P. Reimagining high-throughput profiling of reactive cysteines for cell-based screening of large electrophile libraries. Nat. Biotechnol. 2021, 39 (5), 630–641. 10.1038/s41587-020-00778-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooij R.; Liu S.; Sapmaz A.; Xin B. T.; Janssen G. M. C.; van Veelen P. A.; Ovaa H.; Dijke P. T.; Geurink P. P. Small-Molecule Activity-Based Probe for Monitoring Ubiquitin C-Terminal Hydrolase L1 (UCHL1) Activity in Live Cells and Zebrafish Embryos. J. Am. Chem. Soc. 2020, 142 (39), 16825–16841. 10.1021/jacs.0c07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panyain N.; Godinat A.; Lanyon-Hogg T.; Lachiondo-Ortega S.; Will E. J.; Soudy C.; Mondal M.; Mason K.; Elkhalifa S.; Smith L. M.; Harrigan J. A.; Tate E. W. Discovery of a Potent and Selective Covalent Inhibitor and Activity-Based Probe for the Deubiquitylating Enzyme UCHL1, with Antifibrotic Activity. J. Am. Chem. Soc. 2020, 142 (28), 12020–12026. 10.1021/jacs.0c04527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W. C.; Liu X.; Magin R. S.; Girardi N. M.; Ficarro S. B.; Hu W.; Guzman M. I. T.; Starnbach C. A.; Felix A.; Adelmant G.; Varca A. C.; Hu B.; Bratt A. S.; DaSilva E.; Schauer N. J.; Maisonet I. J.; Dolen E. K.; Ayala A. X.; Marto J. A.; Buhrlage S. J. Accelerating inhibitor discovery for deubiquitinating enzymes. Nat. Commun. 2023, 14 (1), 686. 10.1038/s41467-023-36246-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekkebus R.; van Kasteren S. I.; Kulathu Y.; Scholten A.; Berlin I.; Geurink P. P.; de Jong A.; Goerdayal S.; Neefjes J.; Heck A. J.; Komander D.; Ovaa H. On terminal alkynes that can react with active-site cysteine nucleophiles in proteases. J. Am. Chem. Soc. 2013, 135 (8), 2867–70. 10.1021/ja309802n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altun M.; Kramer H. B.; Willems L. I.; McDermott J. L.; Leach C. A.; Goldenberg S. J.; Kumar K. G.; Konietzny R.; Fischer R.; Kogan E.; Mackeen M. M.; McGouran J.; Khoronenkova S. V.; Parsons J. L.; Dianov G. L.; Nicholson B.; Kessler B. M. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem. Biol. 2011, 18 (11), 1401–12. 10.1016/j.chembiol.2011.08.018. [DOI] [PubMed] [Google Scholar]
- Zhou J.; Qi C.; Fang X.; Wang Z.; Zhang S.; Li D.; Song J. DJ-1 modulates Nrf2-mediated MRP1 expression by activating Wnt3a/beta-catenin signalling in A549 cells exposed to cigarette smoke extract and LPS. Life Sci. 2021, 276, 119089 10.1016/j.lfs.2021.119089. [DOI] [PubMed] [Google Scholar]
- Niki T.; Endo J.; Takahashi-Niki K.; Yasuda T.; Okamoto A.; Saito Y.; Ariga H.; Iguchi-Ariga S. M. M. DJ-1-binding compound B enhances Nrf2 activity through the PI3-kinase-Akt pathway by DJ-1-dependent inactivation of PTEN. Brain Res. 2020, 1729, 146641 10.1016/j.brainres.2019.146641. [DOI] [PubMed] [Google Scholar]
- Bahmed K.; Boukhenouna S.; Karim L.; Andrews T.; Lin J.; Powers R.; Wilson M. A.; Lin C. R.; Messier E.; Reisdorph N.; Powell R. L.; Tang H. Y.; Mason R. J.; Criner G. J.; Kosmider B. The effect of cysteine oxidation on DJ-1 cytoprotective function in human alveolar type II cells. Cell Death Dis. 2019, 10 (9), 638. 10.1038/s41419-019-1833-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi-Niki K.; Ganaha Y.; Niki T.; Nakagawa S.; Kato-Ose I.; Iguchi-Ariga S. M. M.; Ariga H. DJ-1 activates SIRT1 through its direct binding to SIRT1. Biochem. Biophys. Res. Commun. 2016, 474 (1), 131–136. 10.1016/j.bbrc.2016.04.084. [DOI] [PubMed] [Google Scholar]
- Chang Y. H.; Lee S. H.; Chang H. C.; Tseng Y. L.; Lai W. W.; Liao C. C.; Tsay Y. G.; Liao P. C. Comparative secretome analyses using a hollow fiber culture system with label-free quantitative proteomics indicates the influence of PARK7 on cell proliferation and migration/invasion in lung adenocarcinoma. J. Proteome Res. 2012, 11 (11), 5167–85. 10.1021/pr300362g. [DOI] [PubMed] [Google Scholar]
- Zanon P. R. A.; Lewald L.; Hacker S. M. Isotopically Labeled Desthiobiotin Azide (isoDTB) Tags Enable Global Profiling of the Bacterial Cysteinome. Angew. Chem., Int. Ed. Engl. 2020, 59 (7), 2829–2836. 10.1002/anie.201912075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backus K. M.; Correia B. E.; Lum K. M.; Forli S.; Horning B. D.; González-Páez G. E.; Chatterjee S.; Lanning B. R.; Teijaro J. R.; Olson A. J.; Wolan D. W.; Cravatt B. F. Proteome-wide covalent ligand discovery in native biological systems. Nature 2016, 534 (7608), 570–574. 10.1038/nature18002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulac S.; LaVoie M. J.; Strahle J.; Schlossmacher M. G.; Xia W. Dimerization of Parkinson’s disease-causing DJ-1 and formation of high molecular weight complexes in human brain. Mol. Cell. Neurosci. 2004, 27 (3), 236–46. 10.1016/j.mcn.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Kobayashi M.; Muramatsu K.; Haruyama T.; Uesugi H.; Kikuchi A.; Konno H.; Noguchi N.; Saito Y. Polymerization of Oxidized DJ-1 via Noncovalent and Covalent Binding: Significance of Disulfide Bond Formation. ACS Omega 2019, 4 (6), 9603–9614. 10.1021/acsomega.9b00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond M. J.; Chu L.; Nalawansha D. A.; Li K.; Crews C. M. Targeted Degradation of Oncogenic KRAS(G12C) by VHL-Recruiting PROTACs. ACS Cent Sci. 2020, 6 (8), 1367–1375. 10.1021/acscentsci.0c00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimster N. P. Covalent PROTACs: the best of both worlds?. RSC Med. Chem. 2021, 12 (9), 1452–1458. 10.1039/D1MD00191D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Shao X.; Zhong T.; Wu Y.; Xu A.; Sun X.; Gao H.; Liu Y.; Lan T.; Tong Y.; Tao X.; Du W.; Wang W.; Chen Y.; Li T.; Meng X.; Deng H.; Yang B.; He Q.; Ying M.; Rao Y. Discovery of a first-in-class CDK2 selective degrader for AML differentiation therapy. Nat. Chem. Biol. 2021, 17 (5), 567–575. 10.1038/s41589-021-00742-5. [DOI] [PubMed] [Google Scholar]
- Lu J.; Qian Y.; Altieri M.; Dong H.; Wang J.; Raina K.; Hines J.; Winkler J. D.; Crew A. P.; Coleman K.; Crews C. M. Hijacking the E3 Ubiquitin Ligase Cereblon to Efficiently Target BRD4. Chem. Biol. 2015, 22 (6), 755–63. 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijdeven R. H.; Neefjes J.; Ovaa H. How chemistry supports cell biology: the chemical toolbox at your service. Trends Cell Biol. 2014, 24 (12), 751–60. 10.1016/j.tcb.2014.07.002. [DOI] [PubMed] [Google Scholar]
- Bekes M.; Langley D. R.; Crews C. M. PROTAC targeted protein degraders: the past is prologue. Nat. Rev. Drug Discov 2022, 21 (3), 181–200. 10.1038/s41573-021-00371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaz A.; Rasul A.; Sarfraz I.; Nawaz J.; Sadiqa A.; Zara R.; Gul Khan S.; Selamoglu Z., Chemical Biology Toolsets for Drug Discovery and Target Identification. In Cheminformatics and its Applications, Stefaniu A.; Razul A.; Hussain G., Eds. IntechOpen: Rijeka, 2020; p Ch. 9. [Google Scholar]
- Dolgacheva L. P.; Berezhnov A. V.; Fedotova E. I.; Zinchenko V. P.; Abramov A. Y. Role of DJ-1 in the mechanism of pathogenesis of Parkinson’s disease. J. Bioenerg. Biomembr. 2019, 51 (3), 175–188. 10.1007/s10863-019-09798-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S.; Tan J.; Qin L.; Lv L.; Yan W.; Zhang H.; Tang B.; Wang C. Molecular chaperones and Parkinson’s disease. Neurobiol Dis 2021, 160, 105527 10.1016/j.nbd.2021.105527. [DOI] [PubMed] [Google Scholar]
- Tsoporis J. N.; Drosatos I. A.; Gupta S.; Amatullah H.; Izhar S.; Dos Santos C. C.; Salpeas V.; Rigopoulos A. G.; Toumpoulis I. K.; Triantafyllis A. S.; Sakadakis E.; Kavantzas N.; Marshall J. C.; Rizos I. K.; Parker T. G. Cytoprotective Mechanisms of DJ-1: Implications in Cardiac Pathophysiology. Molecules 2021, 26 (13), 3795. 10.3390/molecules26133795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.; Marion T. N.; Cao X.; Wang W.; Cao Y. Park 7: A Novel Therapeutic Target for Macrophages in Sepsis-Induced Immunosuppression. Front. Immunol. 2018, 9, 2632. 10.3389/fimmu.2018.02632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Wang J.; Wang J.; Yang B.; He Q.; Weng Q. Role of DJ-1 in Immune and Inflammatory Diseases. Front. Immunol. 2020, 11, 994. 10.3389/fimmu.2020.00994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M.; Grethe C.; Recknagel S.; Kipka G. M.; Klink N.; Gersch M. N-Cyanopiperazines as Specific Covalent Inhibitors of the Deubiquitinating Enzyme UCHL1. Angew. Chem., Int. Ed. 2024, 63 (12), e202318849 10.1002/anie.202318849. [DOI] [PubMed] [Google Scholar]
- Wang W.; Wang H.; Xiang L.; Ni T.; Jin F.; Deng J.; Zhang Y.; Shintaro I.; Zhou Y.; Liu Y. DJ1 is a new prognostic marker and predicts chemotherapy efficacy in colorectal cancer. Oncol. Rep. 2020, 44 (1), 77–90. 10.3892/or.2020.7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Kang M.; Lu W.; Guo Q.; Zhang B.; Xie Q.; Wu Y. DJ-1, a novel biomarker and a selected target gene for overcoming chemoresistance in pancreatic cancer. J. Cancer Res. Clin Oncol 2012, 138 (9), 1463–74. 10.1007/s00432-012-1205-3. [DOI] [PubMed] [Google Scholar]
- Gao H.; Niu Y.; Li M.; Fang S.; Guo L. Identification of DJ-1 as a contributor to multidrug resistance in human small-cell lung cancer using proteomic analysis. Int. J. Exp Pathol 2017, 98 (2), 67–74. 10.1111/iep.12221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y. DJ-1 as a Biomarker of Parkinson’s Disease. Adv. Exp. Med. Biol. 2017, 1037, 149–171. 10.1007/978-981-10-6583-5_10. [DOI] [PubMed] [Google Scholar]
- Cao J.; Lou S.; Ying M.; Yang B. DJ-1 as a human oncogene and potential therapeutic target. Biochem. Pharmacol. 2015, 93 (3), 241–50. 10.1016/j.bcp.2014.11.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.