Abstract
Mutagenesis of Vibrio cholerae with TnphoA, followed by screening for fusions that were activated under low-iron conditions, led to the identification of seven independent fusion strains, each of which was deficient in the ability to utilize ferrichrome as a sole iron source for growth in a plate bioassay and had an insertion in genes encoding products homologous to Escherichia coli FhuA or FhuD. Expression of the gene fusions was independent of IrgB but regulated by Fur. We report here a map of the operon and the predicted amino acid sequence of FhuA, based on the nucleotide sequence. Unlike those of the E. coli fhu operon, the V. cholerae ferrichrome utilization genes are located adjacent and opposite in orientation to a gene encoding an ATP-binding cassette transporter homolog, but this gene, if disrupted, does not affect the utilization of ferrichrome in vitro.
Vibrio cholerae requires iron (0.5 to 1 μM) in a bioavailable form for growth and survival within the environment and within an animal host. Iron not only is necessary for bacterial multiplication within the host but also serves as an important environmental signal that regulates expression of other virulence determinants unrelated to iron acquisition (for reviews, see references 8, 13, 14, and 17). Many iron transport systems characterized to date involve iron-repressible outer membrane proteins (IROMPs) which bind a specific iron-containing compound and transport either free iron or the iron-bound ligand into the cell. Expression of many of these receptor proteins is mediated at the transcriptional level by the iron-binding repressor protein called Fur (ferric uptake regulator), which requires ferrous iron as a cofactor and acts as a repressor when environmental iron levels are high. Homologs of the Escherichia coli fur gene have been identified for at least 32 other bacterial species, including V. cholerae and Vibrio vulnificus (9, 11).
When V. cholerae is grown in vitro under iron-restricted conditions, the catechol siderophore vibriobactin is produced, as are six or more IROMPs. Only three of these IROMPs, ViuA, HutA, and IrgA, have been characterized. ViuA is the 74-kDa ferric vibriobactin receptor that allows internalization of iron from vibriobactin by an undefined mechanism (1, 20). HutA, HutB, and TonB1 have been characterized, and are all required for utilization of heme by V. cholerae (16). IrgA is a 77-kDa IROMP of unknown function that shares significant homology to TonB-dependent outer membrane proteins of gram-negative bacteria (4). Strains with a mutation in irgA show no defect in transport or utilization of iron from vibriobactin, heme, hemoglobin, ferrichrome, or ferric citrate, yet they show a 100-fold virulence defect in an infant mouse model of cholera (5). In vivo, a mutation in irgA leads to a more severe growth defect than a mutation in either hutA or viuA (21). The irgA gene is regulated by a positive transcriptional activator of the LysR family called IrgB (3). TonB1 is required not only for utilization of heme by V. cholerae but also for utilization of ferrichrome iron (16). The receptor for ferrichrome, however, has remained unidentified. We now present evidence that V. cholerae contains an operon consisting of genes homologous to those in the E. coli ferrichrome utilization system and that the products of these genes are required for the utilization of ferrichrome as a sole iron source in vitro.
The strains used in this study are listed in Table 1. TnphoA fusions to iron-regulated genes of classical V. cholerae strain O395 were constructed and screened as described previously (5). Mutant strains containing TnphoA fusions activated under low-iron conditions were individually screened for the ability to grow with ferrichrome as a sole iron source, using a growth stimulation assay (20). Ferrichrome is a siderophore produced by the rust fungus Ustilago sphaerogena and was purchased from Sigma Chemical Co. (no. F8014; St. Louis, Mo.). Ethylenediamine-di(o-hydroxyphenylacetic acid) (EDDA) was deferrated as described previously (20) and incorporated into Luria-Bertani (LB) agar in such a way that there was no growth of the indicator strain in the absence of a usable exogenous iron source placed on the surface of the agar. This usually required 30 μg of deferrated EDDA/ml, with the indicator strain seeded at 105 CFU/ml. Indicator plates were spotted with 10 μl of various iron sources, and after 24 h, zones of growth were measured. Unlike the parent strain, O395, seven TnphoA fusion strains were found to produce no zone of growth specifically around either 1 or 10 mM ferrichrome but were able to utilize vibriobactin (produced by the O395 strain), 1 mM hemin, 0.233 mM hemoglobin, or 36 mM ferric sulfate normally. Results for three representative fusion strains and controls are listed in Table 2. MBG14, deficient in the vibriobactin receptor, was unable to utilize vibriobactin but could grow in the presence of all other iron sources tested, including ferrichrome, as reported previously (20). CA40130, a vibriobactin biosynthetic mutant, was able to grow on all five iron sources tested, as was MBG40, a strain with a mutation in irgA.
TABLE 1.
Bacterial strains used in this study
| V. cholerae strain | Relevant characteristicsa | Reference |
|---|---|---|
| O395 | Classical; Smr | 15 |
| MBG14 | O395 viuA::TnphoA Smr Knr | 5 |
| MBG40 | O395 irgA::TnphoA Smr Knr | 5 |
| CML19 | O395 fur::Knr Smr Knr | 10 |
| CA40130 | Classical CA401 vibriobactin synthesis mutant | 19 |
| BE | O395 fhuA::TnphoA Smr Knr | This study |
| BG | O395 fhuA::TnphoA Smr Knr | This study |
| JAS73 | O395 fhuA::TnphoA Smr Knr | This study |
| JAS73 fur | JAS73 fur::pCML13 Smr Knr Apr | This study |
| JAS73 irgB | JAS73 irgB::pMBG111 Smr Knr Apr | This study |
| PAC6b (MBG18) | O395 fhuD::TnphoA Smr Knr | 5 |
| PAC6 fur | PAC6 fur::pCML13 Smr Knr Apr | This study |
| PAC6 irgB | PAC6 irgB::pMBG111 Smr Knr Apr | This study |
| PAC10 (MBG39) | O395 fhuD::TnphoA Smr Knr | 5 |
| PAC12 (BD) | O395 fhuA::TnphoA Smr Knr | 5 |
| PAC13 (MBG27) | O395 fhuA::TnphoA Smr Knr | 5 |
Abbreviations: Ap, ampicillin; Gn, gentamicin; Kn, kanamycin; Sm, streptomycin.
PAC strains were derived from the previously isolated iron-regulated TnphoA insertion strains shown (5), by curing of the Gnr delivery plasmid pPH1JI.
TABLE 2.
Utilization of various iron sources by wild-type and mutant V. choleraea
| Indicator strain | Growth on:
|
|||||
|---|---|---|---|---|---|---|
| O395 supernatant (vibriobactin) | Hemin | Hemoglobin | Ferrichrome
|
FeSO4 | ||
| 1 mM | 10 mM | |||||
| O395 | + | + | + | + | + | + |
| CA40130 | + | + | + | + | + | + |
| MBG14 | − | + | + | + | + | + |
| MBG40 | + | + | + | + | + | + |
| PAC6 | + | + | + | − | − | + |
| PAC12 | + | + | + | − | − | + |
| JAS73 | + | + | + | − | − | + |
Zones of growth (+) were all >5 mm in diameter. Indicator strains were seeded into low-iron LB agar containing 30 μg of deferrated EDDA/ml at a density of approximately 105 CFU/ml. Iron sources (10 μl), including culture supernatant of O395 containing vibriobactin, 1 mM hemin, 0.233 mM hemoglobin, 1 and 10 mM ferrichrome, and 36 mM ferric sulfate, were spotted onto the surface of the agar.
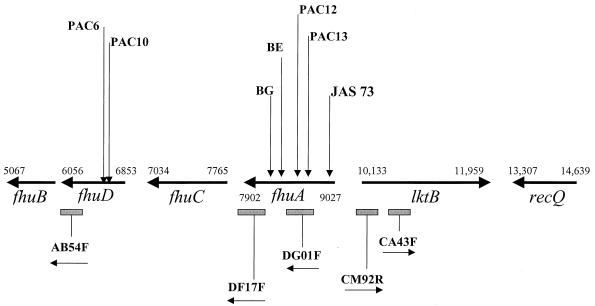
Chromosomal DNA was isolated from the seven fusion strains and was digested with XbaI and EcoRV, which do not cut within TnphoA, and Southern analysis was performed as previously described (5), using a probe internal to TnphoA, to demonstrate that each strain contained only a single insertion (data not shown). Inverse PCR was used to obtain DNA flanking the TnphoA insertions as follows. Three to four micrograms of chromosomal DNA from each mutant strain was digested with various enzymes, including XhoI, PstI, SacII, SfuI, and TaqI, followed by phenol-chloroform extraction and ethanol precipitation. Aliquots (25 to 50 ng) of this digested DNA were self ligated in 50-μl reaction mixtures with 1 U of T4 DNA ligase (Boehringer Mannheim), followed by heat inactivation at 70°C for 10 min. Aliquots of the ligation reaction product were then used in PCRs with 200 to 250 pmol of each primer (divergently oriented and both binding to either the 5′ or 3′ end of TnphoA). Flanking DNA both upstream (SfuI derived) and downstream (TaqI derived) of the PAC12 insertion revealed homology to a 626-bp fragment (gvc.dg01f) in the nonannotated genomic DNA sequence collection of V. cholerae El Tor N16961 at The Institute for Genomic Research (TIGR). This collection of 5,523 sequences was downloaded from the TIGR website into a local server in the Department of Molecular Biology, Massachusetts General Hospital, in August 1997 and used for additional analyses described below. BLASTX analysis of this fragment in turn displayed 23% identity and 43% similarity to the E. coli FhuA protein, amino acids 245 to 415 (of 747 total). Similarly, inverse-PCR products from fusion strain PAC13 also indicated that TnphoA had been inserted into the region encompassed by gvc.dg01f (Fig. 1).
FIG. 1.
Map (not to scale) of the V. cholerae O395 gene cluster with homology to fhuACDB of E. coli. Shown are bp 5067 to 14639 (of 48,695) from V. cholerae N16961 TIGR contig asm818 (see URL in the text). Locations of the seven TnphoA fusions are indicated with arrows, and original TIGR fragments are indicated with boxes. Open reading frame designations other than fhuA are presumptive, based on TIGR contig asm818 regions of similarity to the E. coli fhu genes (see the text).
We then used a λZAP II phagemid library containing 5- to 10-kbp fragments from V. cholerae O395, derived by partial Sau3A1 digestion (courtesy of Shelley Trucksis), to isolate larger genomic fragments with homology to inverse-PCR fragments from the remaining five fusions, which showed no similarity to any of the 5,523 TIGR fragments available at that time. Restriction mapping of overlapping phage clones was combined with PCR analysis using gvc.dg01f as a reference point. Ordering of the TnphoA fusions was accomplished using PCR analysis with a primer to the 3′ end of TnphoA and an opposing primer to the 3′ end of TIGR fragment gvc.ab54f, resulting in the map shown in Fig. 1. This provided the first suggestion of an operon structure, with the JAS73 fusion as the most proximal fusion relative to the orientation of the fhuA ortholog. In addition, the fact that all seven TnphoA fusions were inserted into either fhuA or fhuD homologs is consistent with the requirement for the N terminus of the fusion protein to provide a signal sequence for the export of PhoA to the periplasm or outer membrane to yield an active fusion protein in our screen. It is thus not surprising that no fusions were isolated from fhuC or fhuB, which are localized to the cytoplasmic membrane.
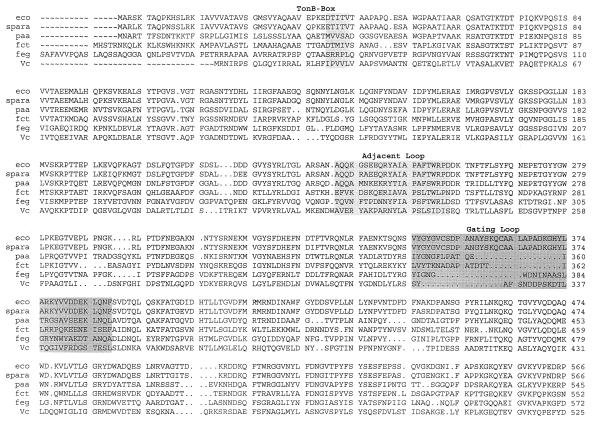
Appropriate regions of phage clones encompassing the fhuA ortholog were amplified by PCR and sequenced. The DNA sequence of this gene from strain O395 was very similar to that of V. cholerae El Tor strain N16961, now available in the TIGR database, bp 229508 to 231607 of contig 1752 (chromosome 1). There were two areas of substantial difference in otherwise virtually identical predicted proteins, however: amino acids (aa) 193 to 200 (ITRIKTVP) in O395 were DYANQDGS in the N16961 sequence in the TIGR database, and aa 214 to 223 (WAVERYAKPA) in O395 were GQLNG*TQTS in the TIGR database (the asterisk denotes a stop codon). Each of these areas of substantial difference resulted from frameshift mutations in the sequences in N16961 compared to sequences in O395. It is uncertain if these frameshifts (and the resulting stop codon at amino acid 219 of FhuA in N16961) are correct or might represent sequencing errors yet to be corrected in the TIGR database. The fhuA open reading frame from strain O395 was 2,100 bp, encoding a protein of 700 aa. The predicted FhuA protein sequence shared homologies with a variety of siderophore receptors from many bacteria but was most similar to E. coli FhuA (AE000124) (33% identity; 53% similarity), Bradyrhizobium japonicum FegA (U61401) (34% identity; 53% similarity), the ferrichrome receptors for Pantoea (Enterobacter) agglomerans (Y14026) and Salmonella enterica serovar Paratyphi (Y14067) (33% identity; 52% similarity), and a hydroxamate-type ferrisiderophore receptor for Pseudomonas aeruginosa (AF051691) (36% identity; 54% similarity). The peptide sequence is shown aligned to known FhuA homologs in Fig. 2. Certain features were conserved, but the largest differences were seen in a region known to form the “gating loop” (6); the significance of these differences for FhuA function in V. cholerae is uncertain. It also appeared that the V. cholerae FhuA protein may contain an “adjacent loop” that may be an alternate binding site that allows the transport of ferrichrome and albomycin (6).
FIG. 2.
Amino acid sequence alignment of E. coli FhuA (eco), S. enterica serovar Paratyphi B FhuA (spara), P. agglomerans FhuA (paa) (6), Erwinia chrysanthemi ferrichrysobactin receptor (fct) (18), B. japonicum FegA (feg) (7), and V. cholerae O395 FhuA (Vc), using the Pileup program in the Genetics Computer Group package with the default settings. Amino acids representing the adjacent loop and the gating loop are shaded, as are the TonB box sequences, as in reference 6. Only the first 525 to 572 residues of each protein are shown.
We examined whether expression of the fhu operon was regulated by Fur in an iron-dependent fashion (as in other organisms) and analyzed whether the V. cholerae transcriptional activator IrgB played a role in expression of this operon. Fusion strains JAS73 (which has the most proximal insertion in fhuA) and PAC6 (which has the most distal insertion in fhuD) were chosen for this analysis. For each strain, either irgB or fur was disrupted by allelic exchange with pMBG111 or pCML13, respectively, as described previously (3, 9), and these mutations were confirmed by Southern hybridization (data not shown). Alkaline phosphatase assays were performed using strains grown overnight in LB medium with or without 2,2-dipyridyl (200 μM) to limit iron availability. Results shown in Table 3 clearly indicate that both fusions were induced 12- and 16-fold under low-iron conditions but were more constitutively expressed if fur was disrupted. Disruption of irgB had no effect on the expression of the fusions. In addition, Northern blot analysis of fhuA expression was performed using total RNA isolated from O395 grown in LB medium (high iron) and LB medium with 200 μM 2,2-dipyridyl (low iron), probed with a 593-bp PCR fragment internal to TIGR fragment gvc.dg01f (bp 23 to 615). As a control, total RNA was isolated from a fur mutant derivative, CML19, under high- and low-iron conditions and probed in parallel. A hybridization signal was detected in wild-type cells under low-iron conditions only but under both low- and high-iron conditions in CML19 (data not shown).
TABLE 3.
Alkaline phosphatase activities of representative fusion strains and their corresponding regulatory mutant strains under high- and low-iron conditions
| Strain | Activity (U/OD600) ina:
|
Avg fold inductionb | |
|---|---|---|---|
| High-iron medium | Low-iron medium | ||
| JAS73 | 21 (2.5) | 223 (18) | 11 |
| JAS73::irgB | 25 (3.7) | 252 (84) | 10 |
| JAS73::fur | 123 (16) | 320 (42) | 2.6 |
| PAC6 | 3 (0.8) | 40 (5.0) | 13 |
| PAC6::irgB | 3 (0) | 52 (34) | 17 |
| PAC6::fur | 15 (2.1) | 36 (1.3) | 2.4 |
Values are the means of at least five separate experiments, with standard errors of the means in parentheses. OD600, optical density at 600 nm. High-iron medium is LB broth and low-iron medium is LB with 200 μM 2,2-dipyridyl.
Between low- and high-iron media.
More recent searching of the nonannotated V. cholerae N16961 genomic sequence at the TIGR website (http://www.tigr.org/cgi-bin/BlastSearch/blastcgi?organism=v_cholerae) revealed a much larger, 48,695-bp contig, asm818, that contained the entire ferrichrome uptake operon and neighboring genes. This region (bp 5067 to 14639) is shown in Fig. 1 (not to scale). The V. cholerae genome can now be searched as two contigs: 1752, which is 2,962,721 bp, and 1741, which is 1,072,915 bp (see URL above); the fhu operon is in contig 1752. BLASTX searches (GenBank release 112.0) using portions of contig asm818, bp 5067 to 14639, as a query allowed us to establish the positions and orientations of the remainder of the fhu operon. Interestingly, bp 10133 to 11959 shared 50% identity with and 65% similarity to H. influenzae LktB aa 5 to 612 (of 614 aa). An internal 1,167-bp portion of lktB was amplified by PCR, cleaved with HincII (this encompasses the region encoding aa 125 to 511), blunt end ligated into the EcoRV site of suicide vector pGP704 (3), and integrated into the O395 chromosome by a single crossover as previously described (9); integration was confirmed by Southern analysis (data not shown). Insertion into this gene did not abrogate the ability to use ferrichrome in a growth stimulation assay (data not shown).
In summary, multiple TnphoA fusions to iron-regulated genes were isolated. Inverse PCR was used to determine that seven fusions had been inserted into genes with similarity to E. coli fhuA and fhuD. Larger genomic fragments isolated from a phagemid library showed that these genes were clustered into an operon structure. The seven fusions were mapped and ordered with respect to the most distal known TIGR sequence at the time (gvc.ab54f) by PCR. All TnphoA insertions prevented the utilization of ferrichrome (but not other substances) as an iron source in a plate growth stimulation assay. The ability to bind and transport vibriobactin was not required for ferrichrome utilization. The most proximal fusion in fhuA and the most distal fusion in fhuD were shown to be regulated by Fur (but not IrgB) at the level of transcription. The gene for an interesting ABC transporter protein homologous to LktB was identified immediately upstream of and opposite in orientation to fhuA in V. cholerae. Disruption of this gene, however, did not affect ferrichrome utilization in a plate bioassay. The sequence of the 700-aa V. cholerae FhuA protein adds to the large amount of information on this multifunctional outer membrane protein, a paradigm for ligand-specific gated channel proteins (2, 6, 12).
Nucleotide sequence accession number.
The sequence of O395 fhuA determined here has been submitted to GenBank (accession no. AF203702).
Acknowledgments
This work was supported by NIAID grant R01AI34968 (S.B.C.) and NIDDK grant F32 DK09651 (M.B.R.).
We thank Shelley Trucksis for the packaged λZAP II O395 library. Preliminary sequence data were obtained from the TIGR website at http://www.tigr.org. Sequencing of V. cholerae N16961 by TIGR was accomplished with support from NIAID.
REFERENCES
- 1.Butterton J R, Stoebner J A, Payne S M, Calderwood S B. Cloning, sequencing, and transcriptional regulation of viuA, the gene encoding the ferric vibriobactin receptor of Vibrio cholerae. J Bacteriol. 1992;174:3729–3738. doi: 10.1128/jb.174.11.3729-3738.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferguson A D, Hofmann E, Coulton J W, Diederichs K, Welte W. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science. 1998;282:2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- 3.Goldberg M B, Boyko S A, Calderwood S B. Positive transcriptional regulation of an iron-regulated virulence gene in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88:1125–1129. doi: 10.1073/pnas.88.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldberg M B, Boyko S A, Butterton J R, Stoebner J A, Payne S M, Calderwood S B. Characterization of a Vibrio cholerae virulence factor homologous to the family of TonB-dependent proteins. Mol Microbiol. 1992;6:2407–2418. doi: 10.1111/j.1365-2958.1992.tb01415.x. [DOI] [PubMed] [Google Scholar]
- 5.Goldberg M B, DiRita V J, Calderwood S B. Identification of an iron-regulated virulence determinant in Vibrio cholerae, using TnphoA mutagenesis. Infect Immun. 1990;58:55–60. doi: 10.1128/iai.58.1.55-60.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Killmann H, Herrmann C, Wolff H, Braun V. Identification of a new site for ferrichrome transport by comparison of the FhuA proteins of Escherichia coli, Salmonella paratyphi B, Salmonella typhimurium, and Pantoea agglomerans. J Bacteriol. 1998;180:3845–3852. doi: 10.1128/jb.180.15.3845-3852.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LeVier K, Guerinot M L. The Bradyrhizobium japonicum fegA gene encodes an iron-regulated outer membrane protein with similarity to hydroxamate-type siderophore receptors. J Bacteriol. 1996;178:7265–7275. doi: 10.1128/jb.178.24.7265-7275.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Litwin C M, Calderwood S B. Role of iron in regulation of virulence genes. Clin Microbiol Rev. 1993;6:137–149. doi: 10.1128/cmr.6.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Litwin C M, Calderwood S B. Cloning and genetic analysis of the Vibrio vulnificus fur gene and construction of a fur mutant by in vivo marker exchange. J Bacteriol. 1993;175:706–715. doi: 10.1128/jb.175.3.706-715.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Litwin C M, Calderwood S B. Analysis of the complexity of gene regulation by Fur in Vibrio cholerae. J Bacteriol. 1994;176:240–248. doi: 10.1128/jb.176.1.240-248.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Litwin C M, Boyko S A, Calderwood S B. Cloning, sequencing, and transcriptional regulation of the Vibrio cholerae fur gene. J Bacteriol. 1992;174:1897–1903. doi: 10.1128/jb.174.6.1897-1903.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Locher K P, Rees B, Koebnik R, Mitschler A, Moulinier L, Rosenbusch J P, Moras D. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell. 1998;95:771–778. doi: 10.1016/s0092-8674(00)81700-6. [DOI] [PubMed] [Google Scholar]
- 13.Martinez J L, Delgado-Iribarren A, Baguero F. Mechanisms of iron acquisition and bacterial virulence. FEMS Microbiol Rev. 1990;75:45–56. doi: 10.1111/j.1574-6968.1990.tb04085.x. [DOI] [PubMed] [Google Scholar]
- 14.Mekalanos J J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mekalanos J J, Swartz D J, Pearson G D N. Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature. 1983;306:551–557. doi: 10.1038/306551a0. [DOI] [PubMed] [Google Scholar]
- 16.Occhino D A, Wyckoff E E, Henderson D P, Wrona T J, Payne S M. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol Microbiol. 1998;29:1493–1507. doi: 10.1046/j.1365-2958.1998.01034.x. [DOI] [PubMed] [Google Scholar]
- 17.Payne S M. Iron and virulence in the family Enterobacteriaceae. Crit Rev Microbiol. 1988;16:81–111. doi: 10.3109/10408418809104468. [DOI] [PubMed] [Google Scholar]
- 18.Sauvage C, Franza T, Expert D. Analysis of the Erwinia chrysanthemi ferrichrysobactin receptor gene: resemblance to the Escherichia coli fepA-fes bidirectional promoter region and homology with hydroxamate receptors. J Bacteriol. 1996;178:1227–1231. doi: 10.1128/jb.178.4.1227-1231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sigel S P, Stoebner J A, Payne S M. Iron-vibriobactin transport system is not required for virulence of Vibrio cholerae. Infect Immun. 1985;47:360–362. doi: 10.1128/iai.47.2.360-362.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stoebner J A, Butterton J R, Calderwood S B, Payne S M. Identification of the vibriobactin receptor of Vibrio cholerae. J Bacteriol. 1992;174:3270–3274. doi: 10.1128/jb.174.10.3270-3274.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tashima K T, Carroll P A, Rogers M B, Calderwood S B. Relative importance of three iron-regulated outer membrane proteins for in vivo growth of Vibrio cholerae. Infect Immun. 1996;64:1756–1761. doi: 10.1128/iai.64.5.1756-1761.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]