Abstract
Objectives
The objectives of this study were to determine the prevalence of feline coronavirus (FCoV) viremia, and its replication in peripheral blood using quantitative RT-PCR (qRT-PCR) methodology in a population of 205 healthy shelter cats in Southern California, as well as to assess any possible connection to longitudinal development of feline infectious peritonitis (FIP).
Methods
The study was performed on buffy-coat samples from EDTA-anticoagulated whole blood samples of 205 healthy shelter cats. From 50 of these cats, fecal samples were also examined. FCoV genomic and subgenomic RNA in the buffy coats was amplified by a total FCoV RNA qRT-PCR. Evidence for FCoV replication in peripheral blood and feces was obtained by M gene mRNA qRT-PCR.
Results
Nine of 205 cats (4.4%) were viremic by the total FCoV RNA qRT-PCR, and one of these cats had evidence of peripheral FCoV blood replication by an FCoV mRNA qRT-PCR. The single cat with peripheral blood replication had a unique partial M gene sequence distinct from positive controls and previously published FCoV sequences. Neither seven of the nine viremic cats with follow-up nor the single cat with replicating FCoV with positive qRT-PCR results developed signs compatible with FIP within 6 months of sample collection.
Conclusions and relevance
FCoV viremia and peripheral blood replication in healthy shelter cats have a low prevalence and do not correlate with later development of FIP in this study population, but larger case-control studies evaluating the prognostic accuracy of the qRT-PCR assays are needed.
Introduction
Feline infectious peritonitis (FIP), a lethal infectious disease of cats worldwide, is caused by feline coronavirus (FCoV) and characterized by multi-organ pyogranulomatous lesions and cavitary effusions. 1 FCoV is a positive-stranded RNA virus with a large genome of 29.2 kb. The genes encoding the membrane (M) and nucleocapsid (N) proteins are conserved among strains, representing well-suited targets for the molecular detection of FCoV. 2
FCoV is endemic in multi-cat environments, such as shelters and catteries, and normally replicates in feline enterocytes, causing mild, self-limiting gastroenteritis. This ubiquitous virus has been called the ‘feline enteric coronavirus (FECV) biotype’. 3 However, in a minority of cats (5–12%), the virus becomes highly pathogenic, causing the fatal disease FIP, and this viral biotype is named feline infectious peritonitis virus (FIPV). 4 While the exact mechanisms of this shift in biologic behavior are unknown, data in the literature primarily support an in vivo viral mutation in one or more genes such as those for the Spike protein or non-structural proteins 3c or 7a/b, with or without predisposing factors such as host immune deficiency, genetic susceptibility or a combination thereof.4–8 One study identified the presence of two distinct missense mutations (M1058L and S1060A) in the Spike protein relative to FECV in 113/118 (95.8%) FIPV isolates sequenced. 9 However, a follow-up study by a different research group did not show a correlation between the M1058L mutation and FIP development, but correlated with the presence of the virus outside the gastrointestinal tract. 10 A different study evaluating mutations that change susceptibility of the Spike protein to furin cleavage found an association between substitution mutations in the receptor-binding (S1) and fusion (S2) domains in this protein and the FIPV biotype. 8
Definitive ante-mortem diagnosis of FIP can be difficult, as it requires histopathologic evaluation with immunohistochemical/immunocytochemical detection of FCoV in macrophages in tissue or effusions.11,12 Routine serology is of limited value, as it cannot differentiate between antibodies induced by the benign FECV and virulent FIPV biotypes, and 80–90% of cats in multi-cat environments are seropositive for FCoV.13,14 RT-PCR assays are capable of detecting FCoV genomic RNA in a variety of samples, such as blood, feces and tissue with high sensitivity, although they cannot discriminate between FECV and FIPV biotypes. 3 In addition, previous studies have demonstrated that the detection of genomic FCoV RNA in blood of healthy cats by RT-PCR does not predict the development of FIP.15–18
However, a RT-PCR targeting the mRNA of the highly conserved M gene of FCoV (hereafter called M gene mRNA RT-PCR), with a forward primer specific for the 5’ ‘leader sequence’ that is inserted ahead of all coronavirus-coding mRNA transcripts, detected subgenomic mRNA generated during virus replication. 2 The hypothesis was that detecting replication outside of the gastrointestinal tract indicated a key pathogenic event in FCoV infection, and could discriminate between FECV and FIPV biotypes and potentially correlate with the development of FIP.2,7 Three epidemiologic studies in the Netherlands, Turkey, and Malaysia used this M gene mRNA RT-PCR assay and found 93–100% of cats with FIP were positive by this test. However, the number of positive results in cats that did not develop FIP varied widely from 5% to 52%.2,19,20
One recent case-control study evaluated cats in Germany with and without FIP (confirmed by histopathology and/or FCoV immunolabeling) using a quantitative RT-PCR for a region of the nucleocapsid N gene and membrane M genes (hereafter called total FCoV RNA quantitative RT-PCR [qRT-PCR]). PCRs of both peripheral blood mononuclear cells (PBMCs) and serum found high specificity (100% for both PBMCs and serum) for this assay but poor sensitivity (15.4–28.6%). 21 These conflicting results in widely different geographical regions using different methodologies and non-equivalent cat populations raise questions about the prevalence of both FCoV viremia and replication competency of the virus in peripheral blood of apparently healthy cats, the association (if any) between detection of replicating or genomic virus and later development of FIP, and the clinical utility of these molecular assays for FIP diagnosis.
The main objective of this study was to determine the frequency of FCoV viremia and replicating FCoV in circulating white blood cells in healthy cats from multi-cat environments in the USA. The primary hypothesis was that the frequency of FCoV viremia in healthy shelter cats would be low when compared with active FCoV replication in feces, and that the frequency of cats with replicating FCoV in blood would be a smaller subset of the cats with either replicating FCoV in feces or those with total FCoV RNA detected in blood. A secondary objective of the study was to track positive cats longitudinally, with the hypothesis that replicating FCoV in peripheral blood would be associated with the development of FIP.
Materials and methods
Study design
The study was designed as a cross-sectional molecular prevalence survey of a convenience sample of healthy shelter cats in Southern California. Two hundred and five cats enrolled in this study were housed at the San Gabriel Valley Humane Society, San Gabriel, CA (157 cats) or the Cats in Need rescue group, Pomona, CA (48 cats). Information about sex and approximate age were obtained from shelter records, and are presented in Table 1. Only healthy, adoptable cats of any age and sex currently residing in a shelter or multi-cat rescue environment were included. Cats with any sign of illness detected during initial examination by veterinary personnel (PPVD, FB, EJF, and multiple registered veterinary technicians) were excluded from enrollment in this study. This study was approved by the Western University Institutional Animal Care and Use Committee, under protocol number R10/IACUC/017.
Table 1.
Age and sex distribution of cats enrolled in the study
| Sex | n (%) | Age | n (%) |
|---|---|---|---|
| Male | 83 (40.5) | ⩽12 months | 91 (44.4) |
| Female | 77 (37.6) | >12 months | 80 (39.0) |
| No record | 45 (21.9) | No record | 34 (16.6) |
| Total | 205 (100) | Total | 205 (100) |
Sample collection and handling
All animals were humanely treated during sample collection. Two hundred and five whole-blood samples were collected in EDTA tubes, and from a subset of these 205 cats, 50 fecal samples from rectal swabs were collected in dry tubes during physical examination or while cats were under anesthesia for elective castration or ovariohysterectomy. Five hundred microliters to 2 ml blood were collected from each cat and centrifuged at 1500 g for 15 mins at 4°C. The buffy coat was immediately mixed 1:3 with Tri Reagent BD (Sigma-Aldrich), and stored at −80°C. Tips of rectal swabs for fecal specimens were stored at −80ºC. Samples from pyogranulomatous lesions located in the liver of a histopathology-confirmed FIP cat were aseptically collected during necropsy and used as positive controls for laboratory procedures.
RNA extraction
Total RNA was extracted from buffy coats by phenol–chloroform extraction (Sigma-Aldrich), and eluted with 50 μl DNAse/RNAse-free water. Total nucleic acids were extracted from fecal samples by glass fiber matrix binding and elution with the High-Pure PCR Template Preparation Kit (Roche) in 40 µl elution buffer.
Detection of total FCoV genomic and subgenomic RNA by qRT-PCR assay
cDNA of buffy-coat RNA was synthesized using the Protoscript First-Strand cDNA synthesis kit (New England Biolabs) and 25 ng standardized cDNA input was tested by a qRT-PCR for a 171-base pair (bp) segment of the FCoV genome containing both a portion of the membrane (M) and nucleocapsid (N) genes using a dual-labeled oligonucleotide TaqMan probe (Eurofins MWG Operon) as previously described. 22 This assay identifies the presence of both genomic FCoV RNA suggestive of viremia, as well as subgenomic mRNA, suggestive of active replication (assay hereafter referred to as total FCoV RNA qRT-PCR) and was determined in RNA of all buffy-coat samples. For positive controls, a quantified pGEM-T Easy (Promega) vector containing the 171-bp targeted region was used in serial dilutions. The detection limit of this assay were 10 copies of the M/N gene template.
Detection of replicating FCoV by subgenomic mRNA qRT-PCR assay
Replicating FCoV was determined in RNA of all buffy coat samples and four blinded controls, and in total nucleic acids of 50 fecal samples. The RT-PCR design followed the original approach reported by Simons et al, 2 with modifications in order to allow the use of qRT-PCR for better analytical sensitivity with less risk of laboratory contamination. 12 A proprietary and commercially available RT-PCR amplification method of the partial FCoV M gene subgenomic mRNA beginning at the 5’-end was performed by fluorescence resonance energy transfer RT-PCR. 23 Primers followed the published sequence but with the modification of degenerate bases designed to cover all known variants of the leader sequence and a highly conserved region of the M gene (assay hereafter referred to as FCoV mRNA qRT-PCR). Degenerate probes with 6-FAM and Bodipy 630/650 labels, respectively, targeted an intervening highly conserved region of the M gene. The assay was performed with 25 ng standardized cDNA input as one-step RT-PCR modeled on the proprietary Auburn University Molecular Diagnostics PCR thermal design (US patent 7,252,937). 23 The sensitivity of this assay was validated by serial dilution of cDNA standard templates. The limit of detection was a single mRNA copy per reaction as evident in the Poisson distribution of positive and negative amplification reactions at the limiting dilution. Validation of the specificity was performed by sequence determination of positive amplifications in this study.
DNA sequence analysis
The amplification product of the partial M gene of FCoV strain 55A was determined by automated Sanger DNA sequencing of both strands, using upstream and downstream primers. The sequence was deposited in GenBank under the accession number KX017227. The GenBank accession numbers of M gene sequences used for comparative analysis were: isolate 26M, KP143512; UCD15a, FJ917525.1; C1Je, DQ848678.1; UCD4, FJ943763.1; UCD14, FJ917524.1; UCD13, FJ943764.1; UCD6, FJ943771.1; RM, FJ938051.1; 79-1146, DQ010921.1; UCD1, AB086902.1; UU47, JN183882.1; Black AB086903.1. Sequence alignment was performed in the Vector NTI software package by use of the ClustalW algorithm, and phylogenetic reconstruction was achieved by the neighbor-joining algorithm in this package.
Statistical analysis
The percentage of infected cats was described with descriptive statistics. Confidence intervals and statistical tests were calculated using JMP Pro version 10 (SAS Institute). Regression coefficient (R2) and amplification efficiency (EFF%) were calculated for the total FCoV RNA qRT-PCR based on the detection of standard curves. Associations between signalment and PCR assay results from blood and fecal samples were calculated using Fisher’s exact test. Comparison between frequency of cats positive for FCoV mRNA qRT-PCR from blood and feces, as well as total FCoV RNA qRT-PCR from blood, were calculated using the McNemar test. The significance level for all statistical tests was set at P <0.05.
Results
Buffy coats from the 205 blood samples were tested by total FCoV RNA qRT-PCR as a measure of total FCoV viremia (positives potentially would include both non-viable and replicating virus, and FECV and FIPV biotypes), as well as by FCoV mRNA qRT-PCR, to detect samples with replicating FCoV. Of the subset of the enrolled population that had fecal samples tested by FCoV mRNA qRT-PCR, 56% were positive, with an average ± SD replicating viral load of 95,320 ± 310,960 copies/µg RNA (range: 180–1,600,160 copies/µg RNA). Only two of the cats with detectable replicating FCoV in feces were viremic by the total FCoV RNA qRT-PCR. These results are summarized in Tables 2 and 3.
Table 2.
Prevalence of feline coronavirus (FCoV) in clinical specimens from cats enrolled in this study
| Sample type | qRT-PCR assay | Presence of: | n/total (%) | 95% CI |
|---|---|---|---|---|
| Buffy coat | FCoV mRNA | Replicating FCoV | 1/205 (0.5) | 0.1–2.7 |
| Total FCoV RNA | FCoV viremia | 9/205 (4.4) | 2.3–8.1 | |
| Feces | FCoV mRNA | Replicating FCoV | 28/50 (56) | 42–69 |
CI = confidence interval; qRT-PCR = quantitative RT-PCR
Table 3.
RT-PCR prevalence of feline coronavirus (FCoV) in buffy coats and/or feces of 50 cats enrolled in this study
| FCoV mRNA from feces | Total FCoV RNA qRT-PCR in peripheral blood buffy coats | Total | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 2 (4) | 26 (52) | 28 (56) |
| Negative | 2 (4) | 20 (40) | 22 (44) |
| Total | 4 (8) | 46 (92) | 50 (100) |
Data are n (%)
qRT-PCR = quantitative RT-PCR
Out of the 205 peripheral blood buffy-coat samples tested by the total FCoV RNA qRT-PCR, nine cats (4.4%) were viremic, with an average ± SD viral load of 20,400 ± 15,900 copies/µg RNA (range: 480–44,000 copies/µg RNA). However, subgenomic mRNA suggestive of active FCoV replication was detected in only one of these total FCoV-positive cats enrolled in the study based on the FCoV mRNA qRT-PCR (replicating viral load of 520 copies/µg RNA in the cat positive by M gene qRT-PCR). The difference between the prevalence of overall viremia by total FCoV RNA qRT-PCR and evidence of FCoV replication by the FCoV mRNA qRT-PCR was statistically significant (P = 0.005, McNemar test). No statistically significant associations between age and sex were found when compared with the presence of FCoV genomic RNA in blood samples (by total FCoV RNA qRT-PCR) or replicating FCoV in feces samples (by FCoV mRNA qRT-PCR). The nine cats positive for FCoV in blood were adopted out before the laboratory results were available. Follow-up by telephone indicated that 7/9 cats did not develop FIP-related signs within 6 months of being tested, while the other two cats were lost to follow-up.
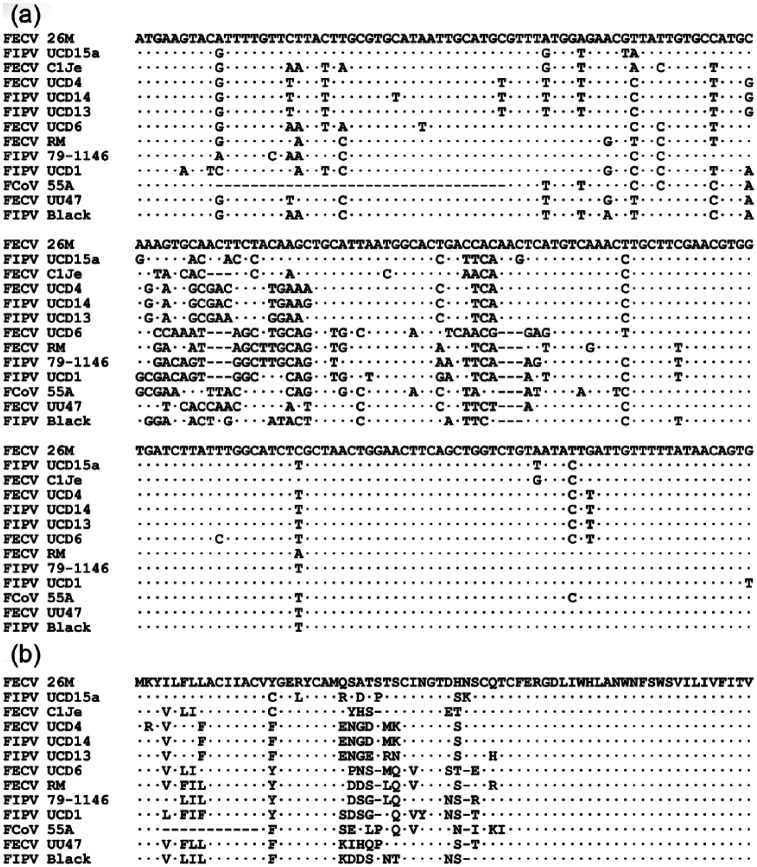
Cat 55A, which was positive by the FCoV mRNA qRT-PCR in buffy coat, was also positive when tested by the total FCoV RNA qRT-PCR, although the fecal sample tested negative for the FCoV mRNA qRT-PCR. Two separate partial M gene mRNA amplicons of this cat, originated from two distinct RNA extractions, were sequenced for both strands, and the sequences were identical. This mRNA sequence was aligned against the positive control and 11 additional published FCoV M gene sequences using ClustalW software (Figure 1). As evident in Figure 1(a), the M gene sequence amplified from this cat’s blood was distinct from both the positive control and those of published sequences of fecal and FIP FCoV isolates, including a unique upstream 33-bp in-frame deletion and multiple common and unique single nucleotide polymorphisms. Translation of the mRNA sequence reveals that this deletion uniquely eliminated amino acids 4–14 of the FCoV M gene open reading frame (Figure 1b). The unrooted partial M gene mRNA dendrogram in Figure 2 reveals the unique phylogenetic position of this isolate. In addition, it is clear from the complete intermixture of FECV and FIPV isolates in this dendrograms that this upstream portion of the M gene and M protein has no relation to the evolution of FIPV from FECV.
Figure 1.
Relationship of the partial M gene mRNA sequence of feline coronavirus (FCoV) strain 55A identified in this study with the main branches FCoV isolates. (a) mRNA sequences aligned with ClustalW. Strain feline infectious peritonitis virus (FIPV) 79-1146 corresponds to the positive control sample. Identical positions are indicated by dots and gaps by hyphens. Strain 55A shows a unique 33-base pair (bp) gap after position 9. (b) Deduced protein sequences. The 33-bp gap translates into an absence of 11 amino acids at positions 4–14
Figure 2.
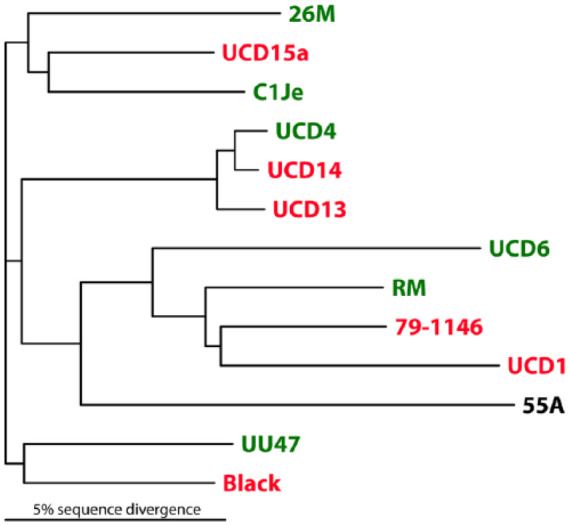
Phylogram of the partial feline coronavirus (FCoV) M gene mRNA sequences. Feline enteric coronavirus (FECV) strains are shown in green, and feline infectious peritonitis virus (FIPV) strains in red. The unknown status of FCoV strain 55A is indicated in black. A complete intermixture of FECV and FIPV isolates is evident
Cat 55A, positive in the peripheral blood buffy coat by FCoV mRNA qRT-PCR, was an 8-week-old female kitten, and did not develop FIP-related signs within 6 months of testing positive. During that time, the cat remained healthy at the shelter available for adoption, but was later euthanized owing to length of stay, as per the shelter’s policy on population control. The results of these laboratory tests were not available before this animal was euthanized; therefore, follow-up tests or necropsy evaluation were not performed.
Discussion
To our knowledge, this is the first field epidemiologic study evaluating the frequency of FCoV viremia and replicating FCoV in healthy shelter cats in the USA by comparing both total FCoV RNA qRT-PCR and FCoV mRNA qRT-PCR assays. Prior international studies found discrepant results, with the number of positive healthy cats ranging from 5% to 52%.2,19,20 Similar to those M gene assay studies, a low percentage (4.4%) of cats in this population were viremic for FCoV by total FCoV RNA qRT-PCR, confirming that FCoV may circulate in the bloodstream in a small number of cats. Out of these nine FCoV viremic cats, only a single one (11%) showed active replication of the virus. This rare occurrence of replication in the bloodstream may be due to replication inefficiency of FECV in the extraintestinal compartment. 17 This is supported by our data showing a very low copy number per reaction detected by the lone cat positive for FCoV mRNA in the buffy coat vs the typical replicating viral load by the M gene assay in feces. In contrast to the infrequent viremia, a much higher proportion (56%) of cats had active gastrointestinal FCoV infection, as detected by fecal FCoV mRNA qRT-PCR. These data suggest that only a small fraction of cats with gastrointestinal FCoV infection show a breakout of the virus into the extraintestinal compartment, and a small subset of that population of these breakout viruses actually manages to replicate outside the gastrointestinal tract; however, healthy cats have been shown to be intermittently viremic for FCoV, and single time-point sampling may have missed such cats. 15 Based on the data in this population, only a small fraction of apparently healthy FCoV-infected cats will develop FCoV replication in the bloodstream.
In our study population, replicating FCoV in the circulation was detected in only a single cat, and that cat did not develop FIP within 6 months of being tested. The sequence of the M gene mRNA of the single cat (55A) was distinct from the positive control and from any other FCoV strain, indicating that this was a true positive, and not due to laboratory contamination. The complete intermixture of FECV and FIPV strains along with the 55A M gene upstream partial mRNA sequence (Figure 2) suggests that upstream M gene polymorphisms do not associate with FECV to FIPV conversion; however, single nucleotide polymorphisms in multiple FCoV non-structural genes have been associated with conversion to FIPV biotype, and these minute differences may not show up in a sequence alignment phylogram.4,9
Previous studies using the FCoV mRNA qRT-PCR assay have not evaluated the later development of FIP in healthy cats that tested positive for replicating FCoV. The absence of FIP manifestation under extraintestinal FCoV replication in our study raises the question of whether a shift in FCoV tropism from enterocytes to leukocytes is necessary but not sufficient for development of FIP, with additional functional changes and/or mutations required. Another, possibly related, reason for the failure to precipitate FIP may be that marginal FECV replication in monocytes is not sufficient to provoke an inflammatory antiviral response, as previously suggested.15,20
Another possible explanation for the absence of FIP manifestation under extraintestinal FCoV replication is natural, presumably genetic, host resistance to FIP. While the mortality rate after experimental infection with FIPV and systemic spread has been reported to be as high as 100%, it depends on a number of variables (including viral strain, environment and comorbidities), and there is some evidence for innate resistance in cats. 16 For instance, Pedersen et al report that in experimental infection of 20 cats with a virulent FIPV strain, 19 succumbed to FIP within a short time, but a single cat survived until termination of the study. 24 Nevertheless, this cat seroconverted, indicating infection, but inability of the FIP virus to precipitate the disease in this cat. Genetic susceptibility, the corollary of resistance, has been suggested by the increased prevalence of FIP in pure-bred cats. 25
The low prevalence of FCoV viremia and blood replication in this study and the lack of association with development of FIP complement the existing literature and suggest that either the total FCoV RNA qRT-PCR or the FCoV mRNA qRT-PCR assays alone may be insufficient to diagnose or predict accurately the development of FIP. Methodological differences make direct comparisons between our results and previous FCoV mRNA qRT-PCR studies difficult.2,19,20 High analytical specificity of the two qRT-PCR assays used in this study is ensured by the use of internal sequence-specific probes for real-time detection of the amplification products. In contrast, previously published FCoV mRNA qRT-PCR assays rely on the sequence-independent agarose gel detection by amplicon size,2,19,20 and only a single study confirmed a subset of such positives by sequencing. 2 Such PCR assays without sequence-specific probes or sequencing of amplicons may give false-positive results, and depending on assay optimization, false-negative results because of insufficient analytical sensitivity.
This study has several important limitations. First, as only a single cat was positive by the FCoV mRNA qRT-PCR assay in this study, these findings must be replicated by additional studies. Such studies will likely require a very large population given the low prevalence in this study. Second, none of the healthy shelter cats in this study available for follow-up developed signs compatible with FIP. Therefore, necropsy with histopathology and immunohistochemistry was not performed on any cat. However, this would have been necessary to link definitively viremia or replicating FCoV to later microscopic or gross lesions consistent with FIP.
Future large-scale, longitudinal, prospective case-control studies are required to determine the significance of extraintestinal FCoV replication on development of FIP. Such studies should follow all cases of any clinically ill cats to necropsy with histopathology and FCoV immunohistochemistry for a gold-standard diagnosis of FIP in any applicable cases, and may reveal links between extraintestinal FCoV replication, mutations in other FCoV genes (eg, the spike protein), and subsequent development of FIP.
Conclusions
Our results indicate that both FCoV viremia and extraintestinal viral replication have a very low prevalence in apparently healthy shelter cats in a US multi-cat environment. In addition, the cats that tested positive for genomic or replicating FCoV in the bloodstream did not develop signs compatible with FIP (or any other illness) for 6 months until adoption with follow-up or euthanasia. Detection of FCoV viremia or replication in circulation in a cat without signs of FIP appears to be rare using the molecular methods described here, but the clinical significance is uncertain. A large-scale cross-sectional study with histologic evaluation of tissues as the gold standard for positive and negative FIP diagnosis is necessary to determine the relevance of extraintestinal FCoV genomes and replicating FCoV.
Acknowledgments
This study was made possible through the collaboration of the San Gabriel Valley Humane Society and the Cats in Need rescue groups. The authors would like to thank Maryam Tngrian, Colin Koziol and John Greenwood for their technical assistance.
Footnotes
BK is the director of the Auburn University Molecular Diagnostics Laboratory, and Y-CJ is a graduate research assistant in this laboratory. The FCoV mRNA qRT-PCR is offered as fee-for-service to veterinarians and was performed in this laboratory. All other authors report no conflict of interest.
Funding: This work was funded by the Winn Feline Foundation (grant number W 10-036). EF was supported by the Veterinary Student Scholars program from Morris Animal Foundation.
Accepted: 26 March 2017
References
- 1. Hartmann K. Feline infectious peritonitis and feline coronavirus infection. In: Ettinger SJ, Feldman EC. (eds). Textbook of veterinary internal medicine: diseases of the dog and the cat. St Louis, MO: Saunders Elsevier, 2010, pp 940–945. [Google Scholar]
- 2. Simons FA, Vennema H, Rofina JE, et al. A mRNA PCR for the diagnosis of feline infectious peritonitis. J Virol Methods 2005; 124: 111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pedersen NC, Allen CE, Lyons LA. Pathogenesis of feline enteric coronavirus infection. J Feline Med Surg 2008; 10: 529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vennema H, Poland A, Foley J, et al. Feline infectious peritonitis viruses arise by mutation from endemic feline enteric coronaviruses. Virology 1998; 243: 150–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chang HW, de Groot RJ, Egberink HF, et al. Feline infectious peritonitis: insights into feline coronavirus pathobiogenesis and epidemiology based on genetic analysis of the viral 3c gene. J Gen Virol 2010; 91: 415–420. [DOI] [PubMed] [Google Scholar]
- 6. Poland AM, Vennema H, Foley JE, et al. Two related strains of feline infectious peritonitis virus isolated from immunocompromised cats infected with a feline enteric coronavirus. J Clin Microbiol 1996; 34: 3180–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rottier PJ, Nakamura K, Schellen P, et al. Acquisition of macrophage tropism during the pathogenesis of feline infectious peritonitis is determined by mutations in the feline coronavirus spike protein. J Virol 2005; 79: 14122–14130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Licitra BN, Millet JK, Regan AD, et al. Mutation in spike protein cleavage site and pathogenesis of feline coronavirus. Emerg Infect Diseases 2013; 19: 1066–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chang HW, Egberink HF, Halpin R, et al. Spike protein fusion peptide and feline coronavirus virulence. Emerg Infect Diseases 2012; 18: 1089–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Porter E, Tasker S, Day MJ, et al. Amino acid changes in the spike protein of feline coronavirus correlate with systemic spread of virus from the intestine and not with feline infectious peritonitis. BMC Vet Res 2014; 45: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hartmann K. Feline infectious peritonitis. Vet Clin North Am Small Anim Pract 2005; 35: 39–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pedersen NC. An update on feline infectious peritonitis: diagnostics and therapeutics. Vet J 2014; 201: 133–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Giori L, Giordano A, Giudice C, et al. Performances of different diagnostic tests for feline infectious peritonitis in challenging clinical cases. J Small Anim Pract 2011; 52: 152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hartmann K, Binder C, Hirschberger J, et al. Comparison of different tests to diagnose feline infectious peritonitis. J Vet Intern Med 2003; 17: 781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gunn-Moore DA, Gruffydd-Jones TJ, Harbour DA. Detection of feline coronaviruses by culture and reverse transcriptase-polymerase chain reaction of blood samples from healthy cats and cats with clinical feline infectious peritonitis. Vet Microbiol 1998; 62: 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pedersen NC. A review of feline infectious peritonitis virus infection: 1963–2008. J Feline Med Surg 2009; 11: 225–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Desmarets LMB, Vermeulen BL, Theuns S, et al. Experimental feline enteric coronavirus infection reveals an aberrant infection pattern and shedding of mutants with impaired infectivity in enterocyte cultures. Sci Rep 2016; 6: 20022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vogel L, Van der Lubben M, te Lintelo EG, et al. Pathogenic characteristics of persistent feline enteric coronavirus infection in cats. Vet Res 2010; 41: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Can-Sahna K, Ataseven S, Pinar VD, et al. The detection of feline coronaviruses in blood samples from cats by mRNA RT-PCR. J Feline Med Surg 2007; 9: 369–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sharif S, Arshad SS, Hair-Bejo M, et al. Evaluation of feline coronavirus viraemia in clinically healthy and ill cats with feline infectious peritonitis. J Anim Vet Adv 2011; 10: 18–22. [Google Scholar]
- 21. Doenges SJ, Weber K, Dorsch R, et al. Comparison of real-time reverse transcriptase polymerase chain reaction of peripheral blood mononuclear cells, serum and cell-free body cavity effusion for the diagnosis of feline infectious peritonitis. J Feline Med Surg 2017; 19: 344–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dye C, Helps CR, Siddell SG. Evaluation of real-time RT-PCR for the quantification of FCoV shedding in the faeces of domestic cats. J Feline Med Surg 2008; 10: 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang C, Gao D, Vaglenov A, et al. One-step real-time duplex reverse transcription PCRs simultaneously quantify analyte and housekeeping gene mRNAs. BioTechniques 2004; 36: 508–516. [DOI] [PubMed] [Google Scholar]
- 24. Pedersen NC, Eckstrand C, Liu H, et al. Levels of feline infectious peritonitis virus in blood, effusions, and various tissues and the role of lymphopenia in disease outcome following experimental infection. Vet Microbiol 2015; 175: 157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Benetka V, Kubber-Heiss A, Kolodziejek J, et al. Prevalence of feline coronavirus types I and II in cats with histopathologically verified feline infectious peritonitis. Vet Microbiol 2004; 99: 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]