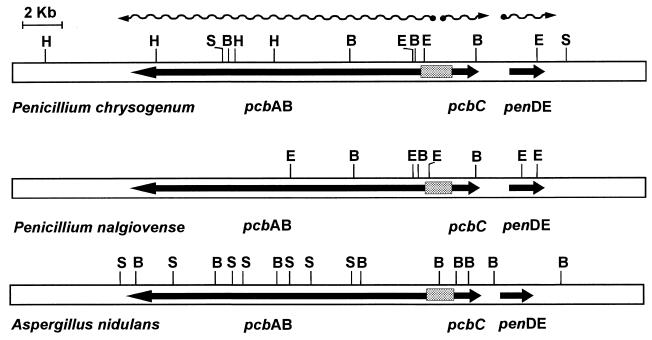
Genes for the biosynthesis of secondary metabolites are usually arranged in clusters (15, 59) together with genes for resistance to the toxic action of secondary metabolites on the producer organisms (20) and sometimes with genes for biosynthesis of antibiotic precursors (54). The penicillin biosynthesis cluster consists of three genes pcbAB, pcbC, and penDE (58) and are arranged the same in Penicillium chrysogenum, Aspergillus nidulans (1), and Penicillium nalgiovense (51) (Fig. 1). The pcb genes encode enzymes involved in penicillin and cephalosporin biosynthesis, whereas pen genes are specific for the penicillin pathway. The pcbAB and pcbC genes are expressed from a 1.16-kb bidirectional promoter region in opposite directions (6, 7, 21, 63). The expression of these genes is subject to sophisticated controls by both nutritional and developmental factors (1, 8, 9, 55, 60).
FIG. 1.
Penicillin gene clusters in P. chrysogenum, A. nidulans, and P. nalgiovense. The pcbAB, pcbC, and penDE genes encode ACV synthetase, IPN synthase, and IPN acyltransferase, respectively. The arrows indicate the orientation of the genes. The bidirectional pcbAB-pcbC promoter region is stippled. The transcripts of the three genes are indicated by black wavy lines at the top of the figure. Restriction site abbreviations: S, SalI; B, BamHI; H, HindIII; E, EcoRI.
Multiple DNA-binding proteins appear to bind to different regions of the pcbAB-pcbC bidirectional promoter. Proteins that interact with the pcbAB-pcbC intergenic control region have been found by DNA mobility shift and DNA footprinting assays (17, 29; J. F. Martín, K. Kosalková, A. T. Marcos, F. Fierro, F. J. Fernández, and S. Gutiérrez, Abstr. 8th Int. Symp. Genet. Ind. Microorg., p. 22, 1998). Penicillin biosynthesis is sensitive to nitrogen source repression; the global-acting regulatory protein NIT2 of Neurospora crassa binds strongly to a site in the bidirectional promoter containing two close GATA sequences (29). Glucose strongly represses penicillin biosynthesis by preventing expression of the three penicillin biosynthesis genes (26, 75); the glucose effect is not due to a decrease in pH and is not reversed by alkaline pHs (37). Glucose repression of penicillin biosynthesis does not seem to be exerted by the global carbon catabolite regulator CreA (86) but appears to be mediated by complex regulatory protein interactions (48). Cooperation between these DNA-binding proteins modulate penicillin gene expression. Differences in the interacting proteins may explain the widely different levels of penicillin production in P. chrysogenum and A. nidulans.
THE PENICILLIN GENE CLUSTER PCBAB-PCBC-PENDE IS AMPLIFIED IN TANDEM REPEATS IN HIGH-LEVEL PENICILLIN-PRODUCING STRAINS
The three genes encoding δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine (ACV) synthetase, isopenicillin N (IPN) synthase (cyclase), and IPN acyltransferase, named pcbAB, pcbC, and penDE, respectively (21, 83), are clustered in a 15-kb DNA region in all known penicillin-producing fungi (51, 62, 81, 82) (Fig. 1). The conserved arrangement of the penicillin biosynthesis genes in different species is undoubtedly important for coordinated regulation of expression of the three genes (57). The three genes occur only in a few evolutionarily related fungal species (1, 36).
The penicillin biosynthesis genes of P. chrysogenum AS-P-78 are amplified in tandem repeats of a 106.5-kb DNA region (five or six copies) linked by conserved TTTACA sequences (31, 68). The wild-type strains P. chrysogenum NRRL 1951 and Penicillium notatum ATCC 9478 (Fleming's isolate) contain a single copy of the 106.5-kb region. This region is flanked by the same TTTACA hexanucleotide found between tandem repeats in strain AS-P-78. A penicillin-overproducing strain, P. chrysogenum E1, contains a large number of copies in tandem with a 57.9-kb DNA fragment (corresponding to the righthand side moiety of the 106.5-kb region that includes the three penicillin genes) linked by the same hexanucleotide (or its reverse complementary TGTAAA) sequence. The amplification has occurred within chromosome I, the largest P. chrysogenum chromosome, which is 11.0 Mb long in strain AS-P-78 (31).
Several mutants unable to produce penicillin have been shown to have a deletion of the penicillin gene cluster (33). The P. chrysogenum npe-10 deletion mutant showed a deletion of 57.9 kb that corresponds exactly to the DNA fragment that is amplified in strain E1. The conserved hexanucleotide TTTACA sequence was reconstituted at the deletion site. The tandem reiterations and deletions appear to arise by mutation-induced site-specific recombination at the conserved hexanucleotide sequences (30). No other gene, either structural or regulatory, that may affect penicillin biosynthesis is present in the amplified DNA region that occurs in the high-level penicillin-producing strains (32, 36).
PACC-MEDIATED PH CONTROL OF PENICILLIN GENE EXPRESSION
A pH regulatory circuit which controls extracellular enzymes, permeases, and other cellular processes is known in A. nidulans and other fungi (13, 88). Espeso and coworkers (27) showed that transcription of the A. nidulans gene encoding isopenicillin N synthase is under the control of the pH regulatory system mediated by the PacC protein and proposed that external alkaline pH overrides carbon regulation. PacC is a transcriptional factor that contains three putative Cys2His2 zinc fingers (88). The intergenic pcbAB-pcbC region of A. nidulans contains at least five PacC binding sites GCCARG. However, the PacC binding sites did not correspond to the cis-acting region involved in carbon catabolite regulation (88).
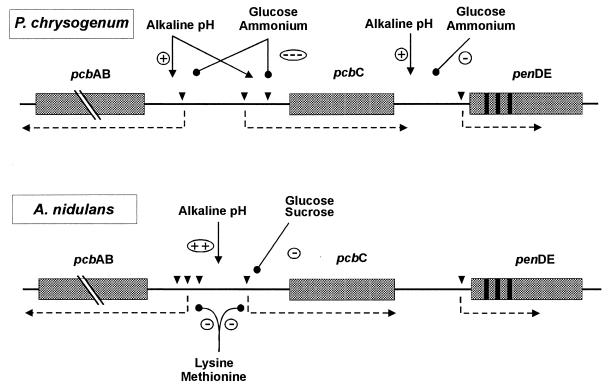
The effect of pH on regulation of penicillin biosynthesis was also observed in P. chrysogenum (18) although the effect of alkaline pH values seems to be weaker in this fungus than in A. nidulans (37). Alkaline pH values stimulate only slightly the expression of the penicillin biosynthesis genes, particularly of the pcbC and penDE genes (Fig. 2).
FIG. 2.
Regulatory circuits controlling expression of the pcbAB-pcbC bidirectional promoter and the penDE promoter in P. chrysogenum and A. nidulans. Induction of gene expression is indicated by an arrow with ⊕, and repression of gene expression is indicated by —• with ⊝. The intensity of the induction or repression effect is shown by the number of + or − symbols. Transcription initiation sites (▾) are also shown.
CARBON CATABOLITE REGULATION OF PENICILLIN BIOSYNTHESIS
Penicillin biosynthesis in P. chrysogenum is strongly regulated by glucose and sucrose and to a lower extent by other sugars (maltose, fructose, and galactose), but not by lactose (74). Lactose is the classically used carbon source for penicillin production, although continuous slow feeding of glucose, thus avoiding glucose repression, is frequently used in industrial processes.
High glucose concentrations prevent formation of ACV (the first intermediate of the pathway) and depress isopenicillin N synthase and (only to a low extent) acyl-coenzyme A:isopenicillin N acyltransferase (10, 75). Glucose-grown cultures showed reduced α-aminoadipic acid pools (41), apparently caused by stimulating lysine biosynthesis and growth (75).
In A. nidulans, penicillin biosynthesis is repressed by sucrose and to a lower extent by glucose (10, 26). Some strains of A. nidulans (e.g., strain G191) seem to be less sensitive to glucose repression than others (46).
The repressive effect of glucose on penicillin biosynthesis is strongly dependent on the presence of inorganic phosphate in the culture medium. In a phosphate-limited complex medium (containing Pharmamedia as the only source of phosphate and nitrogen), the reduction in the penicillin levels due to the repressive effect of glucose (139 mM) is about 13% when the sugar is added at inoculation time, whereas it increases to 59% when the medium is supplemented with 100 mM inorganic phosphate (56; F. Antequera and J. F. Martín, unpublished results). Inorganic phosphate (100 mM) has no effect per se on penicillin production under glucose-limited conditions or in lactose-based medium.
Interestingly, penicillin biosynthesis in P. chrysogenum is also repressed by 2-deoxyglucose (73). This glucose analogue is phosphorylated to 2-deoxyglucose-6-phosphate but appears not to be metabolized further in glycolysis. This result suggests that signal transduction leading to glucose repression of penicillin biosynthesis genes proceeds by modification (perhaps phosphorylation) of a penicillin regulatory protein exerted by glucose or 2-deoxyglucose or a phosphorylated derivative of these sugars. The signal transduction cascade may involve a two-component regulatory system including a sensor protein and a partner response regulator protein. A glucokinase-deficient mutant (glk-1) of P. chrysogenum is simultaneously derepressed in carbon catabolite regulation of β-galactosidase and isopenicillin N synthase (5), suggesting that a common step of the signal transduction cascade (probably a sensor kinase) is involved in carbon catabolite regulation of both sugar utilization and penicillin biosynthesis. Hexokinase II has been implicated in carbon catabolite regulation of sugar utilization in yeasts (24, 35), but its involvement in signal transduction in filamentous fungi is obscure (76).
TRANSCRIPTION OF THE PCBAB, PCBC, AND PENDE GENES OF P. CHRYSOGENUM IS STRONGLY REPRESSED BY GLUCOSE
Recent results in our laboratory showed that glucose repressed transcription of the three penicillin biosynthesis genes pcbAB, pcbC, and penDE (Fig. 2) when added at inoculation time to cultures of P. chrysogenum AS-P-78, but it had little repressive effect when added at 12 h of incubation and no effect when added after 24 or 36 h (37). Most of the pcbAB and pcbC transcripts are accumulated in the initial 36 to 48 h of the culture, while the penDE transcript (expressed from a different promoter) is synthesized later. The time window for the glucose repression effect was longer (up to 36 h) when the cultures were inoculated directly with conidia (Antequera and Martín, unpublished).
These results are consistent with the hypothesis that glucose (when added early to the culture) triggers a signal transduction cascade that modifies a carbon regulatory protein that interacts with the pcbAB, pcbC, and penDE promoters (Fig. 3). Repression by glucose of the three penicillin biosynthesis genes was also observed using the lacZ reporter gene coupled to each of the three promoters in single-copy transformants with the constructions integrated at the pyrG locus (37). Although the drastic decrease in the steady-state levels of the pcbAB, pcbC, and penDE transcripts might also be due to an increased degradation of the transcripts in glucose-supplemented cells, an effect of glucose on preventing transcription from the penicillin promoters is supported by the severe reduction in reporter β-galactosidase activity observed in single-copy constructions in which the lacZ gene was coupled to each of the three promoters (37). Similarly, repression of pcbAB and penDE was also established by the use of the uidA reporter system (28).
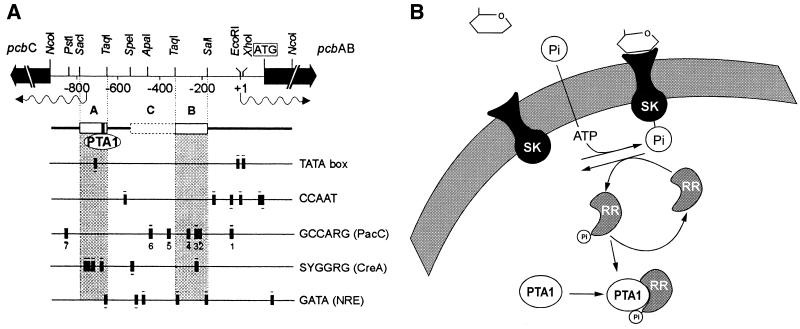
FIG. 3.
(A) Scheme showing the locations of the TATA and CCAAT boxes and the PacC, CreA, and NRE consensus motifs in the P. chrysogenum pcbAB-pcbC bidirectional promoter. The regions corresponding to box A and box B are shaded. The PTA1 protein is shown bound to box A (see text). The transcripts of pcbAB and pcbC are indicated by wavy lines under the map at the top of the figure. (B) Model showing glucose signal transduction in P. chrysogenum. When glucose is absent, interaction of PTA1 with the modified (phosphorylated?) response regulator is proposed to activate penicillin gene expression. SK, sensor kinase; RR, response regulator; PTA1, penicillin transcriptional activator 1 (see text).
The residual expression of the pcbC and penDE genes in glucose-grown cultures was slightly increased at pH 6.8, 7.4, and 8.0, but alkaline pH values did not override the strong repression exerted by glucose. Transcription of the actin gene used as a control was not significantly affected by glucose or alkaline pH (37).
IS CARBON CATABOLITE REGULATION OF PENICILLIN BIOSYNTHESIS MEDIATED BY CREA?
All available evidence suggests that carbon catabolite regulation of penicillin biosynthesis is exerted by a carbon regulatory protein that transduces the nutritional signal (high or low glucose levels) to the penicillin gene promoters (Fig. 3). This carbon regulatory protein appears to be formed (or modified) in a short time window following inoculation of the penicillin fermentation, since the repressive effect is clearly lower when glucose is added after 12 to 24 h of incubation.
In A. nidulans, glucose repression of structural genes required for utilization of other alternative carbon sources is mediated by a transcriptional regulatory protein encoded by the creA gene (2–4, 22, 50). A similar creA gene in Aspergillus niger has also been reported (23), and there have been other recent reports of similar genes in Trichoderma resei and Trichoderma harzianum (42), Sclerotinia sclerotiorum (92), and Metharhizium anisopliae (78). The CreA protein of filamentous fungi contains two Cys2-His2 zinc fingers which are similar to those of the yeast repressor MIG1, although the overall amino acid identity of CreA and MIG1 is not very high (67).
Two other proteins SSN6 (CYC8) and TUP1 are required for repression by MIG1 (89). Gene repression by glucose is relieved via a mechanism that requires the SNF1 protein kinase complex (84). When glucose is absent, the repressing activity of MIG1 is inhibited by protein kinase SNF1, which is activated upon removal of glucose. The MIG1 protein is readily phosphorylated in vitro by the SNF1 kinase under these conditions (69, 90). These results indicate that the MIG1 protein may be regulated by phosphorylation; these results are in good agreement with the observed involvement of phosphate in glucose repression of penicillin biosynthesis and the possible modification of a response regulator (Fig. 3).
The CreA and MIG1 proteins are closely related to a family of GC box binding proteins that includes the Wilms tumor and SP1 proteins (67). The CreA and MIG1 transcriptional repressors mediate carbon catabolite repression in A. nidulans and Saccharomyces cerevisiae by interaction of the two Cys2-His2 zinc fingers of the protein with the consensus 5′-SYGGGG-3′ nucleotide sequence (S = C or G; Y = C or T; R = A or G) as shown by DNase I protection analysis (50, 66). Cubero and Scazzochio (19) expanded the consensus recognition sequence to 5′-SYGGRG-3′. However, in all A. nidulans creA mutants tested, glucose still represses the ipnA (=pcbC) mRNA (11, 26). Deletion of a 29-bp region which is protected by the CreA protein did not alter sucrose repression in A. nidulans (27). Mutations in creB and creC also have very minor effect on carbon regulation of penicillin biosynthesis (25). These results suggest that in A. nidulans a second creA-independent mechanism of carbon repression is involved in penicillin biosynthesis.
DIFFERENCES IN CARBON CATABOLITE REGULATION AND PH CONTROL OF PENICILLIN BIOSYNTHESIS IN P. CHRYSOGENUM AND A. NIDULANS
Sucrose (or glucose) repression of the ipnA (=pcbC) gene of A. nidulans is known (10, 26, 72), but Litzka and coworkers reported that expression of the A. nidulans pcbAB gene using reporter gene fusions was not repressed by the carbon source (53). The basic regulatory mechanisms of carbon and nitrogen regulation were initially believed to be similar in both fungi (28, 47). However, Suárez and Peñalva (86) proposed that the mechanisms of control by pH and carbon catabolite regulation are different in P. chrysogenum and A. nidulans. Our results support that conclusion (37).
The extracellular pH affects penicillin production in A. nidulans (79) and to a much lower extent in P. chrysogenum (18, 37). Espeso and coworkers (27) showed that external alkaline pH in A. nidulans overrides sucrose regulation of ipnA. By contrast, in P. chrysogenum, alkaline extracellular pH produces a small stimulation of transcription of pcbAB, pcbC, and penDE, but it does not override the strong catabolite regulation exerted by glucose (37). Brakhage (8, 9) has proposed that carbon source regulation of the ipnA gene in A. nidulans is mediated by a control mechanism independent of pH regulation. There are differences in the fine control of penicillin gene expression in P. chrysogenum and A. nidulans (86) that might be related to the higher expression of the penicillin genes in P. chrysogenum than in A. nidulans. The high expression of these genes in P. chrysogenum is due to the presence of a transcription-activating protein that binds to a specific sequence in the pcbAB-pcbC intergenic region (48).
PROTEIN BINDING SITES IN THE UPSTREAM REGION OF THE PENICILLIN BIOSYNTHESIS GENES
Several proteins in crude extracts of P. chrysogenum are able to bind to the pcbAB-pcbC intergenic region. Chu et al. (17) showed that proteins in partially purified extracts recognize the TGCCAAG sequence. In parallel, Feng et al. (29) reported a nuclear factor (NF-A) that recognizes a slightly different binding sequence, GCCAAGCC. Both of these sites contain the CAAG motif and most likely correspond to the PacC binding sites (Fig. 3) that have been identified in A. nidulans [consensus sequence GCCA(A/G)G] (86, 88). Indeed, this motif is also located in the promoter region of the pH-regulated acid phosphatase phoA gene of P. chrysogenum (40).
The level of expression of the P. chrysogenum pcbAB gene was monitored by measuring the β-galactosidase activity of strains carrying sequential deletion derivatives of the pcbAB promoter fused to the Escherichia coli lacZ gene (Martín et al., Abstr. 8th Int. Symp. Genet. Ind. Microorg.). These fused constructions were targeted to the chromosomal pyrG locus of P. chrysogenum (37). Transformants integrated in single copy at the pyrG locus were selected. Analysis of the regulatory effect of deleting different DNA fragments of the pcbAB-pcbC intergenic region revealed two important promoter regions (boxes A and B) for expression of the pcbAB gene (Fig. 3). Using protein extracts from mycelia grown under carbon catabolite-repressing or -derepressing conditions, we have found DNA-binding proteins that specifically shift both promoter fragments. These proteins form DNA-protein complexes for each box: AG1 complex with box A in glucose-grown cells and AL1 in lactose-grown cells. Similarly, box B forms two complexes, BG1 and BG2, in glucose-grown cells and BL1 in lactose-grown cells (48).
Uracil interference assays showed that a protein in P. chrysogenum cell extracts interacts with the thymines in a palindromic heptanucleotide 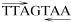 . The effects of point mutations and deletion of the entire TTAGTAA sequence supported the involvement of this sequence in the binding of a transcriptional activator that was named PTA1. The results of in vivo studies using constructions carrying point mutations in the TTAGTAA sequence or a deletion of the complete heptanucleotide confirmed that this intact sequence is required for protein binding and high-level expression of the pcbAB gene (48). The TTAGTAA sequence resembles the target sequence of BAS2 (PHO2), a factor required for expression of several genes in yeasts.
. The effects of point mutations and deletion of the entire TTAGTAA sequence supported the involvement of this sequence in the binding of a transcriptional activator that was named PTA1. The results of in vivo studies using constructions carrying point mutations in the TTAGTAA sequence or a deletion of the complete heptanucleotide confirmed that this intact sequence is required for protein binding and high-level expression of the pcbAB gene (48). The TTAGTAA sequence resembles the target sequence of BAS2 (PHO2), a factor required for expression of several genes in yeasts.
The pcbAB-pcbC intergenic region binding proteins were purified by heparin agarose chromatography, ammonium sulfate precipitation, gel filtration chromatography, and ion-exchange chromatography (Martín et al., Abstr. 8th Int. Symp. Genet. Ind. Microorg.). Formation of the DNA-PTA1 complex using extracts of glucose-grown P. chrysogenum cells is prevented by extracts of lactose-grown (glucose-limited) cells; i.e., a protein present in glucose-limited cells interacts with PTA1 or modifies this regulator, preventing formation of the AG1 DNA-repressor protein complex that occurs in glucose-grown cells (K. Kosalková and J. F. Martín, unpublished results) (Fig. 3). In summary, catabolite regulation of penicillin biosynthesis is mediated by protein-protein and protein-DNA interactions.
THE CCAAT BOX-BINDING PROTEIN COMPLEX PENR1 OF A. NIDULANS
Based on the results of a moving-window analysis of the pcbAB-pcbC intergenic region of A. nidulans and supported by band shift and methyl interference assays, a CCAAT-containing DNA motif that is recognized by a protein complex designated PENR1 was located 409 bp upstream of the A. nidulans pcbAB gene ATG initiation codon (72, 87). A 4-bp deletion within this motif led to an eightfold increase of pcbAB expression and to a simultaneous reduction of ipnA expression to about 30% of the original level (87). Another CCAAT box that is also recognized by the PENR1-protein complex is located 250 bp upstream of the transcriptional start site of penDE. Substitution of the CCAAT sequence by GATCC resulted in a fourfold reduction of expression of penDE (53). Similar CCAAT boxes are present in the promoters of many eukaryotic genes (43, 65).
A CCAAT-binding complex (the HAP complex) has been characterized in S. cerevisiae. It consists of at least four subunits, HAP2, HAP3, and HAP5 (forming a heterotrimeric complex essential for DNA binding) and HAP4, an acidic protein acting as a transcriptional activator. An A. nidulans gene designated hapC, similar to S. cerevisiae HAP3, was reported recently (70). In an A. nidulans strain in which hapC had been deleted, binding of the transcriptional factor AnCF (44) to the CCAAT boxes of the amdS and gatA genes did not occur, and indeed, ΔhapC strains grow very poorly on acetamide as the sole nitrogen source. These results indicate that the HapC protein is a functional part of the AnCF complex that binds to the CCAAY motif (45).
Further studies by Brakhage and coworkers indicated that the binding consensus sequence of the PENR1 complex is RRCCAAT(C/A)RCR (reviewed in reference 9), which matches the CCAAT site in the amdS promoter recognized by AnCF. Therefore, it is likely that PENR1 and AnCF share at least some protein components (85). Immunoprecipitation and supershift experiments with anti-HapC antibodies provided evidence that HapC is a component of the PENR1 complex (52).
Deletion or mutagenesis of the PENR1 binding sites increased eightfold the expression of the pcbAB gene, while expression of pcbC was reduced (87). In summary, CCAAT box I mediates a negative effect on pcbAB and a positive effect on pcbC. Similarly, mutation of CCAAT box II upstream of the penDE gene reduced expression of this gene (53). There are other CCAAT boxes in the A. nidulans pcbAB-pcbC bidirectional promoter, but they do not appear to bind PENR1 (53).
The real contribution of PENR1 to the control of penicillin gene expression is unclear. The lack of PENR1 in the PENR1 deletion mutant should lead to higher pcbAB transcript levels and to a large increase in penicillin production, since expression of the pcbAB gene in A. nidulans is known to be limiting for penicillin biosynthesis (46). However, expression of a pcbAB-uidA gene fusion was not significantly affected in a ΔhapC A. nidulans mutant (nonfunctional PENR1) and the penicillin titers were reduced by about 30% in this mutant (52). Brakhage (9) proposed that in addition to PENR1, a repressor protein binds closely to or overlaps with the PENR1 binding site (CCAAT); this hypothesis would explain why the lack of PENR1 in the ΔhapC mutant did not result in increased pcbAB expression. Indeed, binding of proteins to sites adjacent to CCAAT sequences has been reported in A. nidulans (91) and in animal cells.
In summary, it seems that PENR1 represents a HAP-like protein which shares the HapC core protein with AnCF. However, both complexes may differ in ancillary proteins what would explain the different regulation of the penicillin biosynthesis and amidase formation.
AREA MEDIATES NITROGEN REGULATION OF PENICILLIN BIOSYNTHESIS
For many years, it has been known that ammonium ions decrease penicillin biosynthesis in P. chrysogenum (55, 77, 80). Ammonium concentrations above 40 mM repressed expression of the uidA reporter gene when fused to the pcbAB and pcbC promoters in P. chrysogenum (28).
A general mechanism of nitrogen regulation occurs in filamentous fungi and other microorganisms to ensure the proper utilization of different nitrogen sources. Ammonium ions or glutamine are favored and thus repress the permeases and enzymes required for the utilization of alternative nitrogen sources (nitrate, proteins, other amino acids, etc.). Nitrogen regulation is mediated in N. crassa by the nit-2 gene product (34) and in A. nidulans by the areA gene product (49). Both genes encode positive regulatory factors with a single Cys-X2-Cys-X17-Cys-X2-Cys zinc finger.
The areA gene of P. chrysogenum (named nre) was cloned by PCR and complementation of a nit-2 mutant of N. crassa (38). These transcriptional factors recognize a consensus GATA sequence (61). Sometimes interaction occurs with two GATA elements separated by a varying number of base pairs (16). The Nit-2 protein of N. crassa and AreA of A. nidulans and P. chrysogenum have 98% homology in the GATA-recognizing domains, although the rest of the protein shows only about 30% overall identity.
There are six GATA sequences in the P. chrysogenum bidirectional pcbAB-pcbC promoter (Fig. 3). Interaction of the zinc finger fragment of the Nre protein of P. chrysogenum occurs specifically with a site containing two GATA sequences separated by 27 bp (38, 39). In separate experiments, the same GATA boxes were recognized by the N. crassa Nit-2 protein (28).
Although clear in vivo studies correlating alterations in GATA sequences or modifications of the Nre protein with changes in nitrogen regulation of penicillin biosynthesis have still not been done, it is likely that nitrogen regulation of penicillin biosynthesis in P. chrysogenum is mediated by the same regulatory protein involved in the nitrogen regulatory circuit.
Very little is known about the molecular mechanism of nitrogen regulation of penicillin biosynthesis in A. nidulans. When the bidirectional pcbAB-pcbC promoter region of P. chrysogenum was introduced into A. nidulans, it was sensitive to nitrogen regulation, indicating that the A. nidulans Nre protein acts on the P. chrysogenum GATA boxes (47). However, there are not tandem GATA boxes in the A. nidulans intergenic pcbAB-pcbC promoter (there is a single GATA sequence), making it more unlikely that it might be recognized by Nre and perhaps explaining the lack of evidence of nitrogen regulation of penicillin biosynthesis in A. nidulans.
It is also worth mentioning that the intergenic pcbAB-pcbC region of the cephalosporin producer Acremonium chrysogenum contains 15 GATA sequences (64). Cephalosporin biosynthesis is known to be regulated by nitrogen, but no studies have been performed on the role of Nre in this fungus.
MUTATIONS AFFECTING EXPRESSION OF THE PENICILLIN BIOSYNTHESIS GENES IN TRANS
There are probably several domain-wide or pathway-specific regulatory genes affecting the cascade of penicillin gene expression. These have been hard to identify. Based on a search of clones expressing low levels of a chromogenic reporter gene fused to the A. nidulans pcbC promoter integrated at a specific locus, three mutations prgA, prgB (penicillin regulation), and npeE1 (nonproducer of penicillin) were found (12, 71). These prg mutants exhibited reduced levels of penicillin biosynthesis. However, cloning of the corresponding genes complementing the prgA, prgB, and npeE mutations is required to support the involvement of these genes in penicillin biosynthesis.
During a screening of mutants blocked in penicillin biosynthesis, we isolated 10 mutants impaired in penicillin biosynthesis, two of which (npe-2 and npe-3) showed reduced levels of the penicillin intermediate α-aminoadipyl-cysteinyl-valine and low activities of all the penicillin biosynthesis enzymes (14). It was proposed that these strains correspond to mutants with alterations in penicillin regulatory genes (prg). These strains provide a useful tool for further characterization of the penicillin regulatory mechanisms.
ACKNOWLEDGMENTS
This work was supported in part by grants from the European Union (BIO-CT94-2100 and BIO4-CT96-0535) and the CICYT (Madrid) (BIO97-0289-CO2-02).
I thank S. Gutiérrez, F. Fierro, J. Casqueiro, K. Kosalková, and O. Bañuelos for fruitful scientific discussions and M. Corrales for excellent technical assistance.
REFERENCES
- 1.Aharonowitz Y, Cohen G, Martín J F. Penicillin and cephalosporin biosynthetic genes: structure, organization, regulation and evolution. Annu Rev Microbiol. 1992;46:461–496. doi: 10.1146/annurev.mi.46.100192.002333. [DOI] [PubMed] [Google Scholar]
- 2.Arst H N, Jr, MacDonald D W. A gene cluster in Aspergillus nidulans with an internally located cis-acting regulatory region. Nature. 1975;254:26–31. doi: 10.1038/254026a0. [DOI] [PubMed] [Google Scholar]
- 3.Arst H N, Jr, Tollervey D, Dowzer C E A, Kelly J M. An inversion truncating the creA gene of Aspergillus nidulans results in carbon catabolite derepression. Mol Microbiol. 1990;4:851–854. doi: 10.1111/j.1365-2958.1990.tb00656.x. [DOI] [PubMed] [Google Scholar]
- 4.Bailey C, Arst H N., Jr Carbon catabolite repression in Aspergillus nidulans. Eur J Biochem. 1975;51:573–577. doi: 10.1111/j.1432-1033.1975.tb03958.x. [DOI] [PubMed] [Google Scholar]
- 5.Barredo J L, Alvarez E, Cantoral J M, Díez B, Martín J F. A glucokinase-deficient mutant of Penicillium chrysogenum is derepressed in glucose catabolite regulation of both β-galactosidase and penicillin biosynthesis. Antimicrob Agents Chemother. 1988;32:1061–1067. doi: 10.1128/aac.32.7.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barredo J L, Cantoral J M, Alvarez E, Díez B, Martín J F. Cloning, sequence analysis and transcriptional study of the isopenicillin N synthase of Penicillium chrysogenum AS-P-78. Mol Gen Genet. 1989;216:91–98. doi: 10.1007/BF00332235. [DOI] [PubMed] [Google Scholar]
- 7.Barredo J L, van Solingen P, Díez B, Alvarez E, Cantoral J M, Kattevilder A, Smaal E B, Groenen M A M, Veenstra A E, Martín J F. Cloning and characterization of acyl-CoA:6-APA acyltransferase gene of Penicillium chrysogenum. Gene. 1989;83:291–300. doi: 10.1016/0378-1119(89)90115-7. [DOI] [PubMed] [Google Scholar]
- 8.Brakhage A A. Molecular regulation of penicillin biosynthesis in Aspergillus (Emericella) nidulans. FEMS Microbiol Lett. 1998;148:1–10. doi: 10.1111/j.1574-6968.1997.tb10258.x. [DOI] [PubMed] [Google Scholar]
- 9.Brakhage A A. Molecular regulation of β-lactam biosynthesis in filamentous fungi. Microbiol Mol Biol Rev. 1998;62:547–585. doi: 10.1128/mmbr.62.3.547-585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brakhage A A, Browne P, Turner G. Regulation of Aspergillus nidulans penicillin biosynthesis and penicillin biosynthesis genes acvA and ipnA by glucose. J Bacteriol. 1992;174:3789–3799. doi: 10.1128/jb.174.11.3789-3799.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brakhage A A, Turner G. Biotechnical genetics of antibiotic biosynthesis. In: Kück U, editor. The Mycota II. Genetics and biotechnology. Berlin, Germany: Springer-Verlag KG; 1995. pp. 263–285. [Google Scholar]
- 12.Brakhage A A, van den Bralle J. Use of reporter genes to identify recessive trans-acting mutations specifically involved in the regulation of Aspergillus nidulans penicillin biosynthesis genes. J Bacteriol. 1995;177:2781–2788. doi: 10.1128/jb.177.10.2781-2788.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caddick M X, Brownlee A G, Arst H N., Jr Regulation of gene expression by pH of the growth medium in Aspergillus nidulans. Mol Gen Genet. 1986;203:346–353. doi: 10.1007/BF00333978. [DOI] [PubMed] [Google Scholar]
- 14.Cantoral J M, Gutiérrez S, Fierro F, Gil-Espinosa S, van Liempt H, Martín J F. Biochemical characterization and molecular genetics of nine mutants of Penicillium chrysogenum impaired in penicillin biosynthesis. J Biol Chem. 1993;268:737–744. [PubMed] [Google Scholar]
- 15.Chater K, Bibb M. Regulation of bacterial antibiotic production. In: Kleinkauf H, von Döhren H, editors. Biotechnology: products of secondary metabolism. Weinheim, Germany: VCH Verlag; 1997. pp. 59–105. [Google Scholar]
- 16.Chiang T Y, Marzluf G A. DNA recognition by the NIT2 nitrogen regulatory protein: importance of the number, spacing, and orientation of GATA core elements and their flanking sequences upon NIT2 binding. Biochemistry. 1994;33:576–582. doi: 10.1021/bi00168a024. [DOI] [PubMed] [Google Scholar]
- 17.Chu Y-W, Renno D, Saunders G. Detection of a protein which binds specifically to the upstream region of the pcbAB gene in Penicillium chrysogenum. Curr Genet. 1995;27:184–189. doi: 10.1007/BF00315786. [DOI] [PubMed] [Google Scholar]
- 18.Chu Y-W, Renno D, Saunders G. Extracellular pH affects regulation of the pcbAB gene in Penicillium chrysogenum. Appl Microbiol Biotechnol. 1997;47:250–254. doi: 10.1007/s002530050922. [DOI] [PubMed] [Google Scholar]
- 19.Cubero B, Scazzochio C. Two different, adjacent and divergent zinc finger binding sites are necessary for CREA-mediated carbon catabolite repression in the proline gene cluster of Aspergillus nidulans. EMBO J. 1994;13:407–415. doi: 10.1002/j.1460-2075.1994.tb06275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cundliffe E. How antibiotic-producing organisms avoid suicide. Annu Rev Microbiol. 1989;43:207–233. doi: 10.1146/annurev.mi.43.100189.001231. [DOI] [PubMed] [Google Scholar]
- 21.Díez B, Gutiérrez S, Barredo J L, van Solingen P, van deer Voort L H M, Martín J F. The cluster of penicillin biosynthetic genes. Identification and characterization of the pcbAB gene encoding the α-aminoadipyl-cysteinyl-valine synthetase and linkage to the pcbC and penDE genes. J Biol Chem. 1990;265:16358–16365. [PubMed] [Google Scholar]
- 22.Dowzer C E A, Kelly J M. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol Cell Biol. 1991;11:5701–5709. doi: 10.1128/mcb.11.11.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drysdale M R, Kolze S E, Kelly J M. The Aspergillus niger carbon catabolite repressor encoding gene, creA. Gene. 1993;130:241–245. doi: 10.1016/0378-1119(93)90425-3. [DOI] [PubMed] [Google Scholar]
- 24.Entian K-D, Zimmermann F K. New genes involved in carbon catabolite repression and derepression in the yeast Saccharomyces cerevisiae. J Bacteriol. 1982;151:1123–1128. doi: 10.1128/jb.151.3.1123-1128.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Espeso E A, Fernández-Cañón J M, Peñalva M A. Carbon regulation of penicillin biosynthesis in Aspergillus nidulans: a minor effect of mutations in creB and creC. FEMS Microbiol Lett. 1995;126:63–68. doi: 10.1016/0378-1097(94)00527-x. [DOI] [PubMed] [Google Scholar]
- 26.Espeso E A, Peñalva M A. Carbon catabolite repression can account for temporal pattern of expression of a penicillin biosynthetic gene in Aspergillus nidulans. Mol Microbiol. 1992;6:1457–1465. doi: 10.1111/j.1365-2958.1992.tb00866.x. [DOI] [PubMed] [Google Scholar]
- 27.Espeso E A, Tilburn J, Arst H N, Jr, Peñalva M A. pH regulation is a major determinant in expression of a fungal penicillin biosynthetic gene. EMBO J. 1993;12:3947–3956. doi: 10.1002/j.1460-2075.1993.tb06072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng B, Friedlin E, Marzluf G A. A reporter gene analysis of penicillin biosynthesis gene expression in Penicillium chrysogenum and its regulation by nitrogen and glucose catabolite repression. Appl Environ Microbiol. 1994;60:4432–4439. doi: 10.1128/aem.60.12.4432-4439.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng B, Friedlin E, Marzluf G A. Nuclear DNA-binding proteins which recognize the intergenic control region of penicillin biosynthetic genes. Curr Genet. 1995;27:351–358. doi: 10.1007/BF00352104. [DOI] [PubMed] [Google Scholar]
- 30.Fierro F, Barredo J L, Díez B, Gutiérrez S, Fernández F J, Martín J F. The penicillin gene cluster is amplified in tandem repeats linked by conserved hexanucleotide sequences. Proc Natl Acad Sci USA. 1995;92:6200–6204. doi: 10.1073/pnas.92.13.6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fierro F, Gutiérrez S, Díez B, Martín J F. Resolution of four large chromosomes in penicillin-producing filamentous fungi: the penicillin gene cluster is located on chromosome II (9.6 Mb) in Penicillium notatum and chromosome I (10.4 Mb) in Penicillium chrysogenum. Mol Gen Genet. 1993;241:573–579. doi: 10.1007/BF00279899. [DOI] [PubMed] [Google Scholar]
- 32.Fierro F, Martín J F. Molecular mechanism of chromosomal rearrangement in fungi. Crit Rev Microbiol. 1999;25:1–17. doi: 10.1080/10408419991299185. [DOI] [PubMed] [Google Scholar]
- 33.Fierro F, Montenegro E, Gutiérrez S, Martín J F. Mutants blocked in penicillin biosynthesis show a deletion of the entire penicillin gene cluster at a specific site within a conserved hexanucleotide sequence. Appl Microbiol Biotechnol. 1996;43:597–604. doi: 10.1007/BF00172491. [DOI] [PubMed] [Google Scholar]
- 34.Fu Y H, Marzluf G A. nit-2, the major positive-acting nitrogen regulatory gene of Neurospora crassa, encodes a sequence-specific DNA-binding protein. Proc Natl Acad Sci USA. 1990;87:5331–5335. doi: 10.1073/pnas.87.14.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gancedo J M. Yeast carbon catabolite repression. Microbiol Mol Biol Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gutiérrez S, Fierro F, Casqueiro J, Martín J F. Gene organization and plasticity of the β-lactam genes in different filamentous fungi. Antonie Leewenhoek. 1999;75:81–94. doi: 10.1023/a:1001861025070. [DOI] [PubMed] [Google Scholar]
- 37.Gutiérrez S, Marcos A T, Casqueiro J, Kosalková K, Fernández F J, Velasco J, Martín J F. Transcription of the pcbAB, pcbC and penDE genes of Penicillium chrysogenum AS-P-78 is repressed by glucose and the repression is not reversed by alkaline pHs. Microbiology. 1999;145:317–324. doi: 10.1099/13500872-145-2-317. [DOI] [PubMed] [Google Scholar]
- 38.Haas H, Bauer B, Redl B, Stöffler G, Marzluf G A. Molecular cloning and analysis of nre, the major nitrogen regulatory gene of Penicillium chrysogenum. Curr Genet. 1995;27:150–158. doi: 10.1007/BF00313429. [DOI] [PubMed] [Google Scholar]
- 39.Haas H, Marzluf G A. NRE, the major nitrogen regulatory protein of Penicillium chrysogenum binds specifically to elements in the intergenic promoter regions of nitrate assimilation and penicillin biosynthetic gene clusters. Curr Genet. 1995;28:177–183. doi: 10.1007/BF00315785. [DOI] [PubMed] [Google Scholar]
- 40.Haas H, Redl B, Friedlin E, Stöffler G. Isolation and analysis of the Penicillium chrysogenum phoA gene encoding a secreted phosphate-repressible acid phosphatase. Gene. 1992;113:129–133. doi: 10.1016/0378-1119(92)90680-n. [DOI] [PubMed] [Google Scholar]
- 41.Hönlinger C, Kubicek C P. Regulation of δ-(L-α-aminoadipyl)-L-cysteinyl-D-valine and isopenicillin N biosynthesis in Penicillium chrysogenum by the α-aminoadipate pool size. FEMS Microbiol Lett. 1989;65:71–76. doi: 10.1016/0378-1097(89)90368-6. [DOI] [PubMed] [Google Scholar]
- 42.Ilmen M, Thrane C, Penttilä M. The glucose repressor gene cre1 of Trichoderma: isolation and expression of a full-length and a truncated mutant form. Mol Gen Genet. 1996;251:451–460. doi: 10.1007/BF02172374. [DOI] [PubMed] [Google Scholar]
- 43.Johnson P F, McKnight S L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- 44.Kato M, Aoyama A, Naruse F, Kobayashi T, Tsukagoshi N. An Aspergillus nidulans nuclear protein, AnCP, involved in enhancement of Taka-amylase A gene expression, binds to the CCAAT-containing taaG2, amdS, and gatA promoters. Mol Gen Genet. 1997;254:119–126. doi: 10.1007/s004380050399. [DOI] [PubMed] [Google Scholar]
- 45.Kato M, Aoyama A, Naruse F, Tateyama Y, Hayashi K, Miyazaki M, Papagiannopoulos P, Davis M A, Hynes M J, Kobayashi T, Tsukagoshi N. The Aspergillus nidulans CCAAT-binding factor, AnCP/AnCF, is a heteromeric protein analogous to the HAP complex of Saccharomyces cerevisiae. Mol Gen Genet. 1998;257:404–411. doi: 10.1007/s004380050664. [DOI] [PubMed] [Google Scholar]
- 46.Kennedy J, Turner G. δ-(L-α-aminoadipyl)-L-cysteinyl-D-valine synthetase is a rate limiting enzyme for penicillin production in Aspergillus nidulans. Mol Gen Genet. 1996;253:189–197. doi: 10.1007/s004380050312. [DOI] [PubMed] [Google Scholar]
- 47.Kolar M, Holzmann K, Weber G, Leitner E, Schwab H. Molecular characterization and functional analysis in Aspergillus nidulans of the 5′-region of the Penicillium chrysogenum isopenicillin N synthetase gene. J Biotechnol. 1991;17:67–80. doi: 10.1016/0168-1656(91)90027-s. [DOI] [PubMed] [Google Scholar]
- 48.Kosalková K, Marcos A T, Fierro F, Hernando-Rico V, Gutiérrez S, Martín J F. A novel heptameric sequence (TTAGTAA) is the binding site for a protein required for high level expression of pcbAB, the first gene of the penicillin biosynthesis in Penicillium chrysogenum. J Biol Chem. 2000;275:2423–2430. doi: 10.1074/jbc.275.4.2423. [DOI] [PubMed] [Google Scholar]
- 49.Kudla B, Caddick M X, Langdon T, Martinez-Rossi N M, Benett S E, Silbey S, Davis R W, Arst H N., Jr The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 1990;9:1355–1365. doi: 10.1002/j.1460-2075.1990.tb08250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kulmburg P, Mathieu M, Dowzer C, Kelly J, Felenbok B. Specific binding sites in the ethanol regulon for the CREA repressor mediating carbon catabolite repression in Aspergillus nidulans. Mol Microbiol. 1993;7:847–857. doi: 10.1111/j.1365-2958.1993.tb01175.x. [DOI] [PubMed] [Google Scholar]
- 51.Laich F, Fierro F, Cardoza R E, Martín J F. Organization of the gene cluster for biosynthesis of penicillin in Penicillium nalgiovense and antibiotic production in cured dry sausages. Appl Environ Microbiol. 1999;65:1236–1240. doi: 10.1128/aem.65.3.1236-1240.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Litzka O, Papagiannopoulos P, Davis M A, Hynes M J, Brakhage A A. The penicillin regulator PENR1 of Aspergillus nidulans is a HAP-like transcriptional complex. Eur J Biochem. 1998;251:758–767. doi: 10.1046/j.1432-1327.1998.2510758.x. [DOI] [PubMed] [Google Scholar]
- 53.Litzka O, Then Bergh K, Brakhage A A. The Aspergillus nidulans penicillin biosynthesis gene aat (penDE) is controlled by a CCAAT containing DNA element. Eur J Biochem. 1996;238:675–682. doi: 10.1111/j.1432-1033.1996.0675w.x. [DOI] [PubMed] [Google Scholar]
- 54.Martín J F. New aspects of genes and enzymes for β-lactam antibiotic biosynthesis. Appl Microbiol Biotechnol. 1998;50:1–15. doi: 10.1007/s002530051249. [DOI] [PubMed] [Google Scholar]
- 55.Martín J F, Aharonowitz Y. Regulation of biosynthesis of β-lactam antibiotics. In: Demain A L, Solomon N A, editors. Antibiotics containing the β-lactam structure. New York, N.Y: Springer; 1983. pp. 189–228. [Google Scholar]
- 56.Martín J F, Casqueiro J, Kosalková K, Marcos A T, Gutiérrez S. Penicillin and cephalosporin biosynthesis: mechanism of carbon catabolite regulation of penicillin production. Antonie Leeuwenhoek. 1999;75:21–31. doi: 10.1023/a:1001820109140. [DOI] [PubMed] [Google Scholar]
- 57.Martín J F, Gutiérrez S. Genes for β-lactam antibiotic biosynthesis. Antonie Leeuwenhoek. 1995;67:181–200. doi: 10.1007/BF00871213. [DOI] [PubMed] [Google Scholar]
- 58.Martín J F, Gutiérrez S, Demain A L. β-Lactams. In: Anke T, editor. Fungal biotechnology. Weinheim, Germany: Chapman & Hall; 1997. pp. 91–127. [Google Scholar]
- 59.Martín J F, Liras P. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu Rev Microbiol. 1989;43:173–206. doi: 10.1146/annurev.mi.43.100189.001133. [DOI] [PubMed] [Google Scholar]
- 60.Martín J F, López-Nieto M J, Castro J M, Cortés J, Romero J, Ramos F R, Cantoral J M, Alvarez E, Domínguez M G, Barredo J L, Liras P. Enzymes involved in β-lactam biosynthesis controlled by carbon and nitrogen regulation. In: Kleinkauf H, editor. Regulation of secondary metabolite formation. Basel, Switzerland: Verlag Chemie; 1986. pp. 41–47. [Google Scholar]
- 61.Marzluf G A. Genetic regulation of nitrogen metabolism in the fungi. Microbiol Mol Biol Rev. 1997;61:17–32. doi: 10.1128/mmbr.61.1.17-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCabe A P, Riach M B R, Unkles S E, Kinghorn J R. The Aspergillus nidulans npeA locus consists of three contiguous genes required for penicillin biosynthesis. EMBO J. 1990;9:279–287. doi: 10.1002/j.1460-2075.1990.tb08106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCabe A P, van Liempt H, Palissa H, Unkles S E, Riach M B R, Pfeifer E, von Döhren H, Kinghorn J R. δ-(L-α-aminoadipyl)-L-cysteinyl-D-valine synthetase from Aspergillus nidulans—molecular characterisation of the acvA gene encoding the first enzyme of the penicillin biosynthetic pathway. J Biol Chem. 1991;266:12646–12654. [PubMed] [Google Scholar]
- 64.Menne S, Walz M, Kück U. Expression studies with the bidirectional pcbAB-pcbC promoter region from Acremonium chrysogenum using reporter gene fusions. Appl Microbiol Biotechnol. 1994;42:57–66. doi: 10.1007/BF00170225. [DOI] [PubMed] [Google Scholar]
- 65.Mitchel P J, Tjian R. Transcriptional regulation in mammalian cells by sequence specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 66.Nehlin J O, Carlberg M, Ronne H. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 1991;10:3373–3377. doi: 10.1002/j.1460-2075.1991.tb04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nehlin J O, Ronne H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J. 1990;9:2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Newbert R W, Barton B, Greaves P, Harper J, Turner G. Analysis of a commercially improved Penicillium chrysogenum strain series: involvement of recombinogenic regions in amplification and deletion of the penicillin biosynthesis gene cluster. J Ind Microbiol Biotechnol. 1997;19:18–27. doi: 10.1038/sj.jim.2900411. [DOI] [PubMed] [Google Scholar]
- 69.Ostling J, Carlberg M, Ronne H. Functional domains in the Mig1 repressor. Mol Cell Biol. 1996;16:753–761. doi: 10.1128/mcb.16.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Papagiannopoulos P, Andrianopoulos A, Sharp J A, Davis M A, Hynes M J. The hapC gene of Aspergillus nidulans is involved in the expression of CCAAT-containing promoters. Mol Gen Genet. 1996;251:412–421. doi: 10.1007/BF02172369. [DOI] [PubMed] [Google Scholar]
- 71.Pérez-Esteban B, Gómez-Pardo E, Peñalva M A. A lacZ reporter fusion method for the genetic analysis of regulatory mutations in pathways of fungal secondary metabolism and its application to the Aspergillus nidulans penicillin pathway. J Bacteriol. 1995;177:6069–6076. doi: 10.1128/jb.177.21.6069-6076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pérez-Esteban B, Orejas M, Gómez-Pardo E, Peñalva M A. Molecular characterization of a fungal secondary metabolism promoter: transcription of the Aspergillus nidulans isopenicillin N synthetase gene is modulated by upstream negative elements. Mol Microbiol. 1993;9:881–895. doi: 10.1111/j.1365-2958.1993.tb01746.x. [DOI] [PubMed] [Google Scholar]
- 73.Revilla G. Regulation of penicillin biosynthesis in Penicillium chrysogenum. Ph.D. thesis. Salamanca, Spain: University of Salamanca; 1983. [Google Scholar]
- 74.Revilla G, López-Nieto M J, Martín J F. Carbon catabolite repression of penicillin biosynthesis by Penicillium chrysogenum. J Antibiot. 1984;37:781–789. doi: 10.7164/antibiotics.37.781. [DOI] [PubMed] [Google Scholar]
- 75.Revilla G, Ramos F R, López-Nieto M J, Alvarez E, Martín J F. Glucose represses formation of δ-(l-α-aminoadipyl)-l-cysteinyl-d-valine and isopenicillin N synthase but not penicillin acyltransferase in Penicillium chrysogenum. J Bacteriol. 1986;168:947–952. doi: 10.1128/jb.168.2.947-952.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ruijter G J, Visser J. Carbon repression in aspergilli. FEMS Microbiol Lett. 1997;151:103–114. doi: 10.1111/j.1574-6968.1997.tb12557.x. [DOI] [PubMed] [Google Scholar]
- 77.Sánchez S, Flores M E, Demain A L. Nitrogen regulation of penicillin and cephalosporin fermentation. In: Sanchez-Esquivel S, editor. Nitrogen source control of microbial processes. Boca Raton, Fla: CRC Press, Inc.; 1988. pp. 121–136. [Google Scholar]
- 78.Screen S, Bailey A, Charnley K, Cooper R, Clarkson J. Carbon regulation of the cuticle-degrading enzyme PR1 from Metarhizium anisopliae may involve a trans-acting DNA-binding protein CRR1, a functional equivalent of the Aspergillus nidulans CREA protein. Curr Genet. 1997;31:511–518. doi: 10.1007/s002940050238. [DOI] [PubMed] [Google Scholar]
- 79.Shah A J, Tilburn J, Adlard M W, Arst H N., Jr pH regulation of penicillin production in Aspergillus nidulans. FEMS Microbiol Lett. 1991;77:209–212. doi: 10.1016/0378-1097(91)90553-m. [DOI] [PubMed] [Google Scholar]
- 80.Shen Y-Q, Heim J, Solomon N A, Wolfe S, Demain A L. Repression of β-lactam production in Cephalosporium acremonium by nitrogen source. J Antibiot. 1984;37:503–511. doi: 10.7164/antibiotics.37.503. [DOI] [PubMed] [Google Scholar]
- 81.Smith D J, Burnham M K R, Bull J H, Hodgson J E, Ward J M, Browne P, Brown J, Barton B, Earl A J, Turner G. β-Lactam antibiotic biosynthetic genes have been conserved in clusters in prokaryotes and eukaryotes. EMBO J. 1990;9:741–747. doi: 10.1002/j.1460-2075.1990.tb08168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smith D J, Burnham M R K, Edwards J, Earl A J, Turner G. Cloning and heterologous expression of the penicillin biosynthetic gene cluster from Penicillium chrysogenum. Bio/Technology. 1990;8:39–41. doi: 10.1038/nbt0190-39. [DOI] [PubMed] [Google Scholar]
- 83.Smith D J, Earl A J, Turner G. The multifunctional peptide synthetase performing the first step of penicillin biosynthesis is a 421 073 dalton protein similar to Bacillus brevis peptide antibiotic synthetases. EMBO J. 1990;9:2743–2750. doi: 10.1002/j.1460-2075.1990.tb07461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Smith F C, Davies S P, Wilson W A, Carling D, Hardie D G. The SNF1 kinase complex from Saccharomyces cerevisiae phosphorylates the transcriptional repressor protein Mig1p in vitro at four sites within or near regulatory domain 1. FEBS Lett. 1999;453:219–223. doi: 10.1016/s0014-5793(99)00725-5. [DOI] [PubMed] [Google Scholar]
- 85.Steidl S, Papagiannopoulos P, Litzka O, Andrianopoulos A, Davis M A, Brakhage A A, Hynes M J. AnCT, the CCAAT binding complex of Aspergillus nidulans, contains products of the hapB, hapC, and hapE genes and is required for activation by the pathway-specific regulatory gene amdR. Mol Cell Biol. 1999;19:99–106. doi: 10.1128/mcb.19.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Suárez T, Peñalva M A. Characterisation of a Penicillium chrysogenum gene encoding a PacC transcription factor and its binding sites in the divergent pcbAB-pcbC promoter of the penicillin biosynthetic cluster. Mol Microbiol. 1996;20:529–540. doi: 10.1046/j.1365-2958.1996.5421065.x. [DOI] [PubMed] [Google Scholar]
- 87.Then Berg K, Litzka O, Brakhage A A. Identification of a major cis-acting DNA element controlling the bidirectionally transcribed penicillin biosynthesis genes acvA (pcbAB) and ipnA (pcbC) of Aspergillus nidulans. J Bacteriol. 1996;178:3908–3916. doi: 10.1128/jb.178.13.3908-3916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tilburn J, Sarkar S, Widdick D A, Espeso E A, Orejas M, Mungroo J, Peñalva M A, Arst H N., Jr The Aspergillus PacC zinc finger transcription factor mediates regulation of both acidic- and alkaline-expressed genes by ambient pH. EMBO J. 1995;14:779–790. doi: 10.1002/j.1460-2075.1995.tb07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Treitel M A, Carlson M. Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc Natl Acad Sci USA. 1995;92:3132–3136. doi: 10.1073/pnas.92.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Treitel M A, Kuchin S, Carlson M. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:6273–6280. doi: 10.1128/mcb.18.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Van Heeswick R, Hynes M J. The amdR product and a CCAAT-binding factor bind to adjacent, possibly overlapping DNA sequences in the promoter region of the Aspergillus nidulans amdS gene. Nucleic Acids Res. 1991;19:2655–2660. doi: 10.1093/nar/19.10.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vautard G, Cotton P, Fèvre M. The glucose repressor CRE1 from Sclerotinia sclerotiorum is functionally related to CREA from Aspergillus nidulans but not to the Mig proteins from Saccharomyces cerevisiae. FEBS Lett. 1999;453:54–58. doi: 10.1016/s0014-5793(99)00691-2. [DOI] [PubMed] [Google Scholar]