Abstract
The complexity of CRISPR machinery is a challenge to its application for nonviral in vivo therapeutic gene editing. Here, we demonstrate that proteins, regardless of size or charge, efficiently load into porous silicon nanoparticles (PSiNPs). Optimizing the loading strategy yields formulations that are ultrahigh loading—>40% cargo by volume—and highly active. Further tuning of a polymeric coating on the loaded PSiNPs yields nanocomposites that achieve colloidal stability under cryopreservation, endosome escape, and gene editing efficiencies twice that of the commercial standard Lipofectamine CRISPRMAX. In a mouse model of arthritis, PSiNPs edit cells in both the cartilage and synovium of knee joints, and achieve 60% reduction in expression of the therapeutically relevant MMP13 gene. Administered intramuscularly, they are active over a broad dose range, with the highest tested dose yielding nearly 100% muscle fiber editing at the injection site. The nanocomposite PSiNPs are also amenable to systemic delivery. Administered intravenously in a model that mimics muscular dystrophy, they edit sites of inflamed muscle. Collectively, the results demonstrate that the PSiNP nanocomposites are a versatile system that can achieve high loading of diverse cargoes and can be applied for gene editing in both local and systemic delivery applications.
Keywords: porous silicon nanoparticles, CRISPR, arthritis, Duchenne’s muscular dystrophy, ribonucleoprotein
Graphical Abstract

INTRODUCTION
The modularity of the CRISPR system, in which the guide RNA (gRNA) can be readily designed to target any genomic locus,1 has simplified and generated explosive growth in the use of gene editing.2 Previous methods, such as zinc finger nucleases, required tedious engineering of a new protein for each specific genomic site.3 The popularity of the CRISPR system is largely because new gRNA sequences can be synthesized quickly, inexpensively, and with more reliable outcomes.4 This modularity also attracts commercial interest, as the platform nature of CRISPR greatly expands the opportunity to pursue various disease applications. Exploration of the CRISPR system for therapeutic use is further justified by the recent FDA approval of several RNA-based drugs including 4 short interfering RNA drugs5 and the 2 mRNA-based COVID-19 vaccines.6-8
Current methods of CRISPR delivery can be broadly categorized as either viral or nonviral. Viruses are generally the most efficient gene delivery modality, but manufacturing, immunogenicity, and safety risks have motivated a search for alternatives.9 Among nonviral approaches, lipid nanoparticles (LNPs) have garnered much attention after the success of LNP gene silencing drug Onpattro and COVID-19 vaccines.10 In the context of gene editing, LNPs are usually engineered to codeliver the gRNA and mRNA encoding Cas9, but they have also been successfully adapted to deliver the combined gRNA and Cas9 ribonucleoprotein (RNP) complex.11 Delivery of the preformed RNP complex has several advantages over that of mRNA platforms. First, mRNA is over 9× the mass of the protein it encodes. Thus, the smaller protein should require lower quantity of carrier material and be associated with fewer nonspecific carrier effects. With delivery of mRNA, the cargo must be translated into the active Cas9 enzyme within the cell prior to editing activity; the ideal the mRNA:gRNA stoichiometry is difficult to define, as there is a risk that the active gRNA may be degraded during the lag time required for protein expression. mRNA also carries the risk of over-expression, which can result in problematic immunogenicity and off-target editing.12
RNP delivery offers the ability to overcome the uncertainties of mRNA formulations but also presents challenges. Whereas mRNA and gRNA have similar physicochemical properties, and thus similar packaging considerations, Cas9 RNP is a combination of protein and RNA. RNPs thus present challenges due to a more diverse charge distribution and Cas9 enzyme sensitivity to denaturation. It has been recently shown that LNP formulations should therefore be independently optimized for RNP delivery rather than directly using systems previously optimized for RNA delivery.11,13 Newer gene-editing strategies may require even more complex mixtures of additional proteins or large donor-DNA sequences, which require additional optimization.14 There is, therefore, a significant need for modular, high-capacity, cargo-agnostic delivery systems for gene editing.
Porous silicon nanoparticles (PSiNPs) are an attractive solution for delivery of complex gene-editing cargos.15 PSiNPs are honeycomb-like, rigid silicon structures with tunable pore and overall size.16 Their structurally rigid pores enable loading of diverse cargos such as small-molecule drugs,17-20 siRNA,17,21-24 peptide nucleic acids,25-27 proteins,28-31 and even large plasmid DNA.32,33 For all cargo types, PSiNPs typically load 10–25 wt % (% w/w), which (due to the high-density of silicon) corresponds to all the pore volume being completely filled with cargo; impressively, this equates to production of formulations where approximately 50 volume percent of the nanoparticle (% v/v) comprises cargo. PSiNPs, therefore, often achieve loading capacities that are >10-fold higher than other systems. For example, Waggoner et al. improved loading of a therapeutic protein by >50-fold 13% w/w (~50% v/v) in PSiNPs vs <1% w/w (<1% v/v) in PLGA].28 Although no one has yet demonstrated Cas9 RNP delivery with PSiNPs, silica nanoparticles have been demonstrated for gene editing.34-39 While the route of synthesis, geometry, and chemical structure are very different between PSi and silica, both can achieve high drug loading (~10% w/w).
Beyond their impressive loading, PSiNPs are nontoxic, biodegradable,40 and easily surface-modified.15,41,42 Furthermore, they can be tailored with optical,43,44 magnetic,45-47 or sonic30,48 properties to enable imaging and theranostic applications.49 For example, not only are PSiNPs inherently fluorescent in the tissue-penetrative NIR window, but they also have emission lifetimes ~104 longer than typical fluorophores (like Cy5), allowing for ultrasensitive detection in vivo.44
The many potential benefits of PSiNPs motivated our exploration of their use for the delivery of gene-editing components. This application requires (1) loading without denaturing the Cas9 enzyme, (2) stabilizing the nanoparticles from agglomeration, and (3) delivery across the plasma and endosomal membranes to reach the nucleus. Previous work has demonstrated the in vitro and in vivo utility of forming a polymeric coating on PSiNPs using poly(ethylene glycol)-b-poly(N,N-dimethylaminomethacrylate-stat-n-butyl methacrylate) (PEGDB).26,27 PEG, which forms the hydrophilic corona-block, endows the PSiNPs with colloidal stability while the DB-block confers pH-responsiveness and endosomal membrane disruptive properties for cytosolic delivery of cargo.50,51
In this work, we devised a three-step process for easily and repeatedly preparing protein or RNP-loaded, PEGDB-coated PSiNP nanocomposites. After the formulation protocol was optimized, PSiNPs were tested for gene editing activity in vitro and in vivo. Specifically, we first explored the ability of PSiNPs to deliver two gene editing tools of distinct size and charge: Cre Recombinase and Cas9 in its RNP form. We then optimized the PEGDB coating procedure to yield PSiNP nanocomposites for in vitro gene editing. Finally, we demonstrated in vivo editing after local administration in models of arthritis and muscular dystrophy as well as systemic biodistribution and gene editing after intravenous administration.
RESULTS AND DISCUSSION
PSiNP Design and Synthesis.
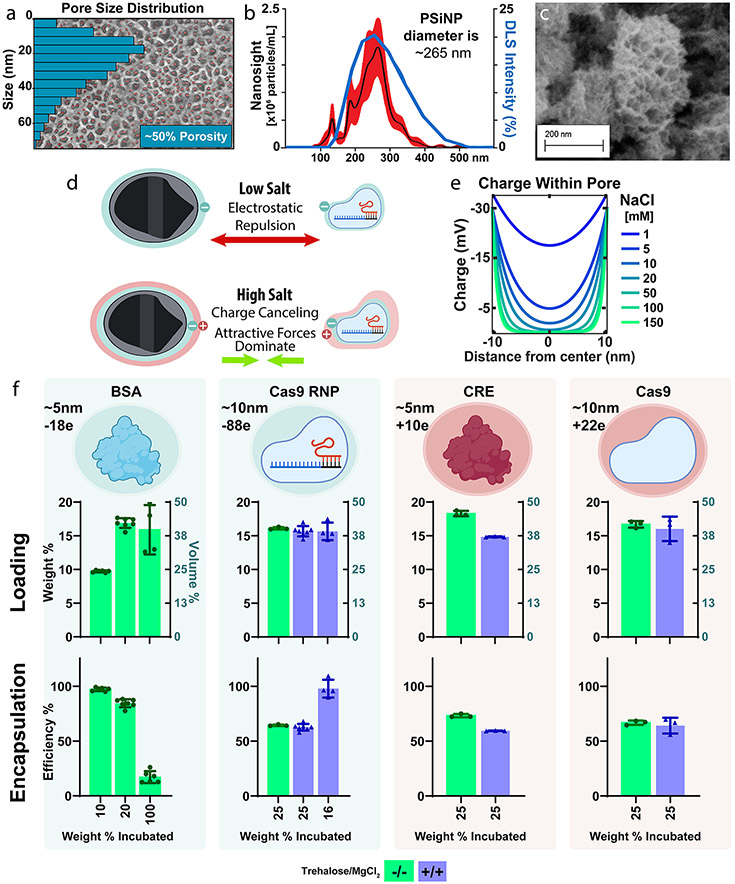
PSi was formed with a mean pore size diameter of ~20 nm; this pore size was selected to provide enough space for loading of ~10 nm Cas9 RNP cargo. Pore size and porosity were determined by scanning electron microscopy and quantified in ImageJ (Figure 1a, Figure S2) which confirmed the porous silicon wafer to have a porosity of ~50% and a median pore size of ~20 nm.
Figure 1.
Porous silicon nanoparticles load a diversity of protein and ribonucleoprotein cargos efficiently and independent of charge. (a) (b) Dynamic light scattering (blue line) and particle-tracking (NanoSight, Black line ± SEM in red) of bare PSiNPs. (c) SEM of porous silicon nanoparticles. (d) Graphical representation of the hypothesized mechanism for efficient protein loading into PSiNPs. (e) Poisson–Boltzmann based modeling of electric potential within a pore as a function of NaCl concentration. (f) Quantification of PSiNP loading weight percent and encapsulation efficiency for protein cargoes of different charge. Each point overlaid onto the bar graph represents an independently prepared replicate. All error bars are the standard deviation.
Our manufacturing protocol yielded PSiNPs with an average size of ~267 nm(Figure 1b,c), as confirmed by both dynamic light scattering (DLS) and NanoSight. DLS found a Z-average diameter of 250 nm and an intensity peak of ~255 nm, while NanoSight illustrated a mean of 250 nm and a mode of 265 nm (Figure S1). The DLS and NanoSight distribution curves are in agreement, and NanoSight, the higher-resolution method, found that 80% of particles fell between 180 and 320 nm (D10–D90, Figure S1).
Intermolecular Forces and PSiNP Loading.
Whereas polymer- or lipid-based nanoparticles designed for RNA delivery generally rely on electrostatic interactions for loading, the PSiNPs’ rigid geometry enables charge-independent loading. Physically, this is due to counterions like sodium or magnesium shielding electrostatic charges and allowing attractive forces, including van der Waals interactions, to dominate (Figure 1d).52-54
We first wanted to optimize the loading conditions for maximum Cas9-RNP loading and activity. Previous work demonstrated that a high (1.5 M) concentration of divalent cations form a stable silicate that traps cargo within the pores.52 Practically, this yields a slower and more controlled cargo release profile. The high salt concentration used in the prior studies may, however, be incompatible with maintaining the stability and activity of enzymes. In contrast to these high MgCl2 concentrations, work by Givens et al. established that 10 mM NaCl was sufficient to shield charge repulsion between anionic albumin and spherical SiO2 nanoparticles.4 However, we would expect the confined geometry of the pore to concentrate the electric field within, as all force vectors converge upon the same physical space (Figure S3). Therefore, a higher counterion concentration may be necessary for PSiNPs than would be the case for spherical silica.
We thus sought to estimate the charge within the pore of silicon as a function of the NaCl concentration by mathematically modeling the pore as two parallel plates, a well-established norm (Figure 1e, Figure S3).55 Our calculations suggest that NaCl concentrations greater than 100 mM should be sufficient to completely passivate the electrostatic effects (Figure 1e, Figure S3). Indeed, modifying the parallel-plate equation to model the more concentrated electric field within a cylinder also confirms ~100 mM NaCl as the minimum salinity (Figure S3d). We therefore predict that, at physiologic salinity, protein cargo should readily load into PSiNPs regardless of size or charge. This treatment of the modeling is simplistic, as it ignores protein–protein interactions as well as the breakdown of DLVO theory in such small dimensions. However, it serves as a useful gauge to inform the design and interpretation of our complementary experimental approaches.
We next sought to predict the maximum possible loading capacity of the PSiNPs using the method of Mughal et al. for calculating the maximal packing factor of a sphere within a cylinder.56 We estimated (Figure S4) that if the pores were maximally filled with protein, the final mass of the protein, relative to PSiNP mass, would be ~ 16.3 wt % for the smaller proteins like BSA (Bovine Serum Albumin, 66 kDa), and ~ 12.9 wt % for larger proteins like Cas9 RNP (Cas9 with sgRNA, 200 kDa).56 We tested this prediction experimentally (Figure 1f) with the two positively charged proteins (CRE Recombinase and Cas9 without sgRNA) and two negatively charged cargos (BSA and Cas9 RNP). PSiNPs were incubated with various amounts of protein, ranging from below to above their maximum theoretical loading (see Figure S4 for more detail). In these measurements, PSiNP mass was known to be within ~2% error (Figure S4b). This study showed that PSiNPs have high loading that was close to and even slightly exceeded the estimated maximum for all proteins (BSA 16.9 wt %, Cas9 RNP 16.1 wt %, CRE 18.3 wt %, Cas9 16.7 wt %).
We next wanted to test the roles of addition of both magnesium, which has been previously studied for charge-shielding,54 and trehalose, which is kosmotropic and increases protein stability.57 We explored whether adding both of these excipients at 150 mM each would affect loading (Figure 1f). These studies showed no significant effect on loading of Cas9 or Cas9 RNP (unpaired t test), but there was a 19% decrease in CRE loading (p < 0.0001, unpaired t test).
Finally, we sought to assess the limits of loading and encapsulation efficiency of Cas9 RNP using varied RNP to PSiNP formulation ratios. Impressively, adding exactly the limit of PSiNPs loading capacity (16 wt %) resulted in nearly 100% encapsulation efficiency and no reduction in final wt % of cargo loaded into the formulation vs when excess cargo is added during the loading process (Figure 1f).
Discussion: PSiNP Design and Synthesis.
Overall, these data demonstrate that PSiNPs can be controllably fabricated and enable, due to their rigid geometry and colloidal forces, ultrahigh loading of proteins of diverse size and charge. Protein loading slightly exceeded our estimated maximum, suggesting that the pore volume of the nanoparticle is completely saturated with cargo. This equates to ~18.3 wt % for CRE recombinase and ~15.7% for Cas9 RNP. Based on our understanding of the system, the slightly higher CRE wt % loading is not due to charge but is instead due to the its smaller size more efficiently packing into the cylindrical pores. These are impressively high cargo weight percentages, especially considering that silicon is denser than lipid- or polymer-based carriers. An alternative consideration is to look at the percentage of the nanoparticle volume comprising the loaded cargo. By this metric, PSiNPs achieve 40–45 vol % loading (Figure 1f). For comparison, RNP-Loaded LNPs achieve ~1.5 wt %,11 which is ~1.5 vol %.58 Therefore, PSiNPs achieve ~25-fold greater % vol loading than LNP systems. Additionally, no Cas9 RNP is wasted (~100% encapsulation efficiency), which is of practical benefit for expensive protein-based therapies. This improvement over a clinically established class of carriers (LNPs) is critical and is in line with the field-leading loading efficiency achieved with porous silica-nanoparticles.34-39
Cargo Release, Timing, and Excipient Effects on PSiNP Gene Editing Activity In Cellulo.
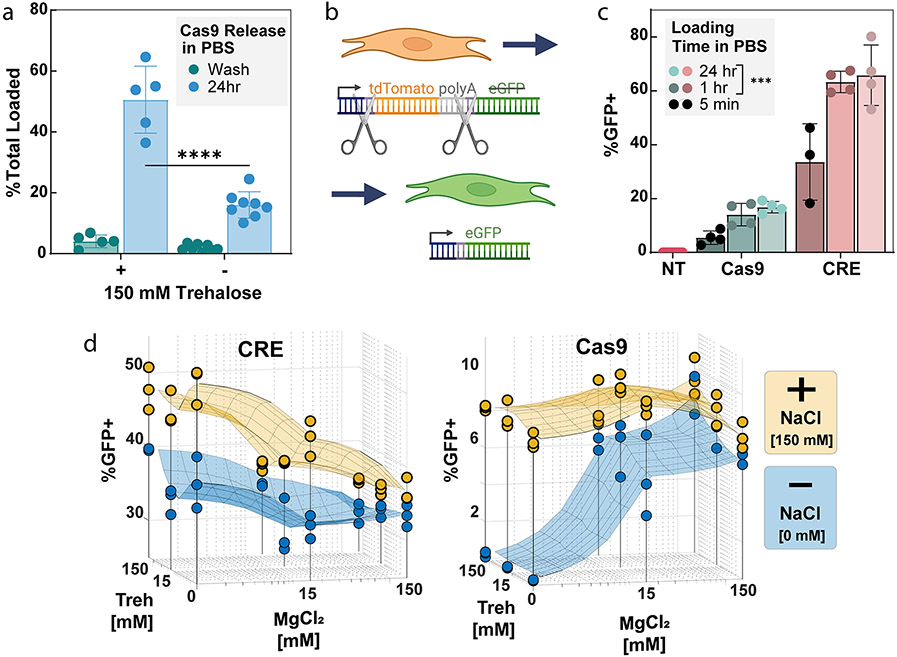
As trehalose is known to stabilize protein structure, we hypothesized that including trehalose during the loading steps may improve Cas9 stability and prevent denaturation within the confined space of the PSiNP pore, thus improving later Cas9 release and activity. To study this, we tested the addition of 150 mM trehalose in the loading buffer (which also contained 10 mM HEPES, 150 mM NaCl, and 15 mM MgCl2) and evaluated the release of Cas9 from PSiNPs after 24 h (Figure 2a). The results demonstrate that Cas9 was stably loaded (less than 10% was released during a centrifugal wash step) even with trehalose addition. However, nearly 3 times (p < 0.00001, t test) as much Cas9 released over 24 h in PBS from the PSiNPs loaded using trehalose. Further measurement of release kinetics using BSA as a model protein showed that protein loaded in trehalose buffer is mostly released during the first 6 h (Figure S5).
Figure 2.
Effects of excipient and salts in loading buffer and loading time on editing activity. (a) Cargo loading is stable based on negligible release during a wash, spin-down, and resuspension of the PSiNPs, with or without addition of 150 mM trehalose in the loading buffer. However, addition of trehalose during Cas9 loading increases release of Cas9 RNP from the PSiNPs over 24 h (****p ≤ 0.0001, Šídák’s multiple comparisons test). Each point on the graph represents an independent replicate. (b) Schematic of the mTmG reporter NIH 3T3 fibroblasts. The mTmG reporter cassette was inserted into the Rosa26 genomic locus. Cas9 or CRE recombinase targets LoxP sites within the cassette. Successful editing removes tdTomato and the polyadenylation tail, turning on eGFP expression. (c) Effect of time of cargo incubation during the PSiNP loading effect on gene editing activity. NT = No Treatment, statistical comparison vs 5 min loading time group, ***p ≤ 0.001, Tukey’s multiple comparisons test. (d) Effect of NaCl, MgCl2, and trehalose concentration in loading buffer on positively charged CRE or negatively charged RNP editing activity in cells. In sections c and d, each point represents a separate treatment well, with percent positive measured by flow cytometry. All error bars are standard deviation.
We next sought to test the effects of loading time and excipient presence on cargo loading and gene editing in vitro. To quantify the level of gene editing in cells we used NIH 3T3 fibroblasts, a relatively difficult to transfect cell line.59 A fluorescent mTmG reporter (Figure 2b) was inserted into the genomic DNA of the NIH 3T3s at the Rosa26 locus. Upon DNA editing, the mTmG reporter changes from tdTomato to eGFP expression. PSiNPs were coated with PEGDB, a polymer which aids in colloidal stability and endosome escape of PSiNPs, as we have previously established for intracellular delivery of peptide nucleic acids as micro-RNA antagonists.26,27 The Cas9-loaded, PEGDB-coated PSiNP nanocomposites used in Figure 2 were formed using the protocol described in the Methods section and the optimized 50B-250 polymer described below in Figure 4.
Figure 4.
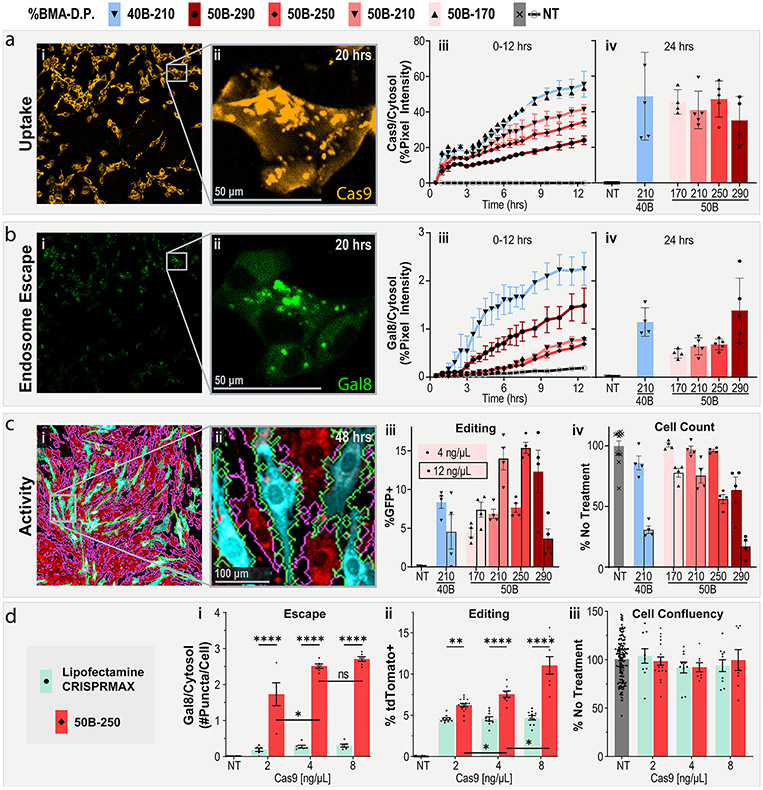
PEGDB molecular weight affects in vitro RNP delivery and editing. (a) Cell internalization and (b) Gal8 endosomal escape reporter imaging of cells treated with PSiNP-PEGDB nanocomposites. (i) Full-field and (ii) inset confocal microscopy representative images. (iii) Image quantification from 0 to 12 and at (iv) 24 h. (c) Gene editing activity of PSiNP-PEGDB nanocomposites in NIH 3T3 mTmG (tdTomato in red, GFP in teal) reporter cell line. Cell outlines are shown to demonstrate image cytometry recognition of cell area (GFP+ in green, tdTomato+ in pink. (i) Full-field and (ii) inset confocal microscopy example images. (iii) Percentage of cells positive for GFP expression. (iv) Relative cell area quantified from images, a proxy for cell viability. (d) Comparison of RNP delivery with PSiNP-PEGDB nanocomposites vs Lipofectamine CRISPRMAX in NIH 3T3 Ai9 reporter cell line. Quantification of (i) relative Gal8 endosomal escape, (ii) percentage of cells positive for tdTomato expression, and (iii) relative cell area as a proxy for cell viability. All error bars are SEM. All data points represent one well. All statistics result from ANOVA with Tukey’s multiple comparison’s test using GraphPad Prism (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001).
We first explored the time needed to load cargo into PSiNPs for effective editing. For both CRE and Cas9 RNP, we found that the 5 min loading groups were significantly less active (p < 0.001, Tukey’s multiple comparisons) than the 1 and 24 h loading groups (Figure 2c). Therefore, we concluded that one h of incubation ensures complete cargo loading of the PSiNPs.
Although MgCl2 and trehalose had negligible effects on protein loading, we wanted to test if it affects in vitro activity. PSiNPs were loaded with CRE Recombinase or Cas9 RNP (Figure 2d) over a range of NaCl, MgCl2, and trehalose concentrations. Full PSiNP PEGDB nanocomposites were then made and treated onto the mTmG reporter cell line. The editing results confirmed the physical theory and mirrored the loading and release results. For both proteins, 150 mM NaCl resulted in better editing, likely due to greater protein stability. For the positively charged CRE, increasing MgCl2 concentrations decreased activity, likely due to competition between CRE and Mg2+ to bind to the negatively charged PSiNP surface, consistent with the slightly decreased CRE loading seen in Figure 1f at 150 mM MgCl2 and 150 mM Trehalose concentrations. Conversely, MgCl2 improved Cas9 RNP editing in the no-NaCl group, likely due to both enzyme stabilization and charge shielding. Trehalose also slightly, but insignificantly, improved editing in the no-NaCl Cas9 group. In the 150 mM NaCl Cas9 group, the excipients had little to no effect on editing percentages. However, since 15 mM MgCl2 and Trehalose performed well in the Cas9 no-NaCl group, we concluded to use a low (15 mM) concentration of these excipients in conjunction with 150 mM NaCl going forward. Subsequent experiments used the ideal loading conditions derived from these 3-dimensional plots, i.e., Cas9 RNPs were loaded in 150 mM NaCl, 15 mM trehalose, and 15 mM MgCl2 whereas CRE was loaded in 150 mM NaCl.
The PEGDB–PSiNP Nanocomposite System.
We sought to optimize the PEGDB–PSiNP nanocomposite system for the formulation and intracellular delivery of active nuclease enzymes. The diblock copolymer PEGDB comprises a hydrophilic “stealth” block of polyethylene-glycol (PEG) and a pH-responsive, endosomolytic block of statistically copolymerized 50 mol % DMAEMA and 50 mol % BMA (“DB”).50 The DMAEMA subunits contain an amine group that is protonatable at pH ~ 6.7, while the BMA subunits are hydrophobic and allow for membrane interaction/disruption. The DB block is multifunctional and serves to stabilize and neutralize the surface of the nanoparticles while also providing cytosolic delivery via endosome escape.
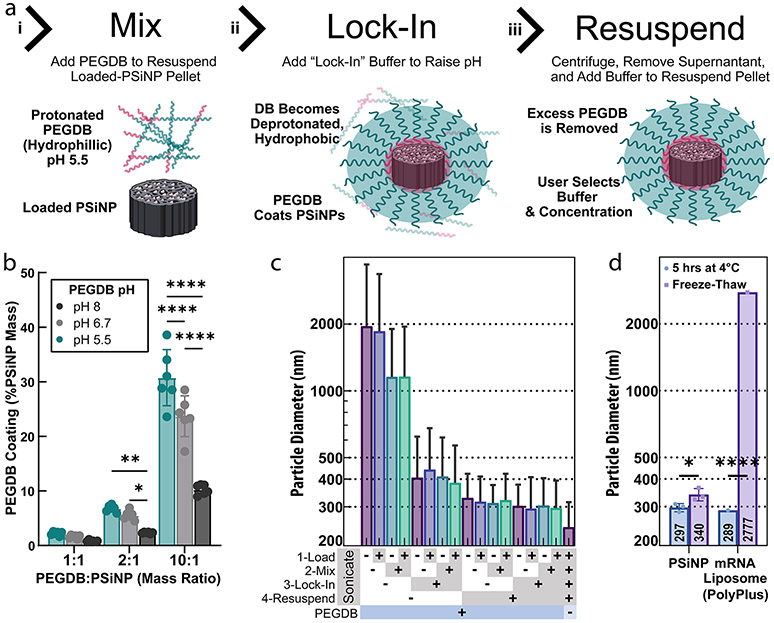
Prior work by Beavers et al. demonstrated that the PEGDB can coat oxidized PSiNPs by applying a solution of PEGDB to the PSiNPs in 100% ethanol.26 While effective for PNA encapsulation, enzymes are not typically compatible with 100% ethanol, motivating our development of an aqueous formulation process. We therefore used an acidic pH 5.5 solution to protonate and better solubilize the DB polymer block in water, anticipating that this would reduce PEGDB micellization and allow the DB block to interact with and coat the PSiNPs (Figure 3a.i). By next shifting the pH to 7.4, above the pKa of DMAEMA, the DB becomes more hydrophobic and “locks” onto the PSiNPs (Figure 3a.ii). Measuring the PEGDB concentration at each step confirmed this modified method as effective (Figure S6a).
Figure 3.
Optimizing the PEGDB coating of PSiNPs to form nanocomposites. (a) Steps used to form polymer and PSiNP nanocomposites: (i) PEGDB is mixed with PSiNP to “coat” the PSiNP based on electrostatic interactions; (ii) the DB block is deprotonated by raising the solution pH to “lock in” the polymer surface coating around the PSiNP; (iii) the PSiNPs are pelleted by centrifugation, and the excess PEGDB is removed with the supernatant. (b) Relative amount of PEGDB coated onto PSiNPs using different loading buffer pHs. Each data point is an independently prepared replicate, and error bars indicate standard deviation. (c) Z-average diameter of PSiNPs as a function of sonication at each preparation step. Bar colors are for ease of viewing. Each bar is a single sample, and error bars represent standard deviation, calculated from polydispersity index. (d) The stability of PSiNP nanocomposites vs PolyPlus mRNA lipid nanoparticles after incubation at 4 °C for 5 h or flash-freezing and thawing at room temperature. All statistics result from ANOVA with Dunnett’s or Tukey’s multiple comparisons test using GraphPad Prism (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001).
Prior work has illustrated that PSiNP surface saturation with the maximal amount of PEGDB coating is optimal for resultant composite NP stability and activity.26,27 To test if protonating the DB facilitates PSiNP coating, PEGDB was solubilized in unbuffered water (pH adjusted to ~6.7 with 0.1 M HCl and NaOH), 10 mM sodium acetate (pH adjusted to ~5.5), or 10 mM Tris buffer (pH adjusted to ~8). Mixing of PSiNPs and PEGDB at varied ratio evinced that increasing the protonation of the DB block (i.e., lower pH) improved polymer association with the PSiNPs (Figure 3b, pH 5.5 > 6.7 > 8, p < 0.0001, Tukey’s multiple comparisons).
To maximize the coating, we next tested a range of PEGDB concentrations and PEGDB:PSi mass ratios during the mixing step (Figure S6b). At a 1:1 PEGDB:PSi ratio, only a 2.5 mg/mL solution was tested, for practical purposes. The 1:1 ratio obtained only ~1/3 the coating achieved by the 10:1 PEGDB:PSi ratio (p < 0.0001, Dunnett’s multiple comparisons test). To ensure the coating is a function of PEGDB:PSi mass ratio and not PEGDB concentration, we tested coating with 2.5, 10, and 20 mg/mL PEGDB solutions at the 10:1 mass ratio (Figure S6b, middle). Coating was invariant to concentration through this range (ns, ordinary one-way ANOVA). Therefore, we concluded that a 10:1 ratio of PEGDB:PSi was the best for saturating the PSiNP surface at concentrations of at least 2.5 mg/mL of PEGDB.
After the PSiNPs are mixed with PEGDB, a “lock-in” buffer is added to raise the pH, deprotonating the DB to stabilize the coating around the PSiNPs. To test the importance of the “lock-in” step, we compared unbuffered water and PBS, demonstrating that the buffer created a significant (p < 0.01, t test) improvement in coating (Figure S6c). To ensure that salinity was not the key factor, we tested PBS vs an equivalently buffered isosmotic trehalose solution and found no significant difference (Figure S6c, p = 0.11, t test). Additionally, trehalose is of interest because it acts as a cryoprotectant, and because it may reduce particle aggregation compared to salt solutions with equivalent osmolarity because salts shield charge-repulsion between particles.
After “lock-in”, PSiNP nanocomposites are centrifuged and resuspended in the final treatment vehicle to purify away any free PEGDB not coated onto the particles. We measured “free” PEGDB as a function of the PEGDB:PSiNP mass ratio, determining that the 10:1 ratio was optimal in terms of maximal coating and minimal “free” PEGDB (Figure S6d). Based on these PEGDB coating studies, all subsequent experiments used a 10:1 PEGDB:PSiNP ratio, with coating done at pH 5.5 buffer and lock-in performed using pH 7.4 trehalose or NaCl solutions.
Sonication and Stability of PSiNP Nanocomposites.
We sought for our nanoparticles to be both fully polymer coated and as small and monodispersed as possible. During the centrifugation steps, PSiNPs are compacted together and thus may become agglomerated. Conveniently, PSiNPs act as nucleation points for cavitation bubbles from ultrasound, providing a facile method for dispersion of PSiNP agglomerates.30,60 We thus sought to determine the optimal sonication protocol for PSiNPs during preparation (Figure 3c). We either sonicated or did not sonicate PSiNPs at each stage of preparation, i.e., (1) after loading the cargo for 1 h, (2) while being resuspended in the PEGDB coating solution, (3) while being diluted in the trehalose lock-in buffer, or (4) while being resuspended in the final trehalose treatment buffer. The combinatorics (24) of this yielded 16 different samples to test for size. Samples were left suspended in trehalose buffer for 1 h at room temperature before dilution in water and size measurement to ensure stability and experimental consistency. The results indicate that sonication in the resuspension buffer (4) is critical for yielding small, monodisperse nanoparticles (p < 0.05 paired t test). Also sonicating at the lock-in step (3) makes a small but significant difference (p < 0.05 paired t test). Sonication at the coating step (2) does not affect size but is still beneficial in practice because it helps to resuspend the pelleted PSiNPs more quickly.
As additional evidence that the PEGDB forms a well-ordered coating on the nanoparticles, we compared the size of uncoated PSiNPs (Figure 3c, far-right column) with that of our polymer nanocomposites. Compared to uncoated PSiNPs, PEGDB-coated PSiNPs increase in hydrodynamic diameter from ~250 to ~300 nm (Z-avg). The ~50 nm increase is likely the result of a single layer of PEGDB stably coating the PSiNP based on simple estimations. First, assuming ~1.26 Å per repeat unit, the polymer used in this experiment (250 repeat units of DB + 5 kDa PEG) should be ~42 nm if perfectly linear. Second, treating the system as a sphere with a radius of 125 nm surrounded by a 25 nm spherical shell means the total volume of PEGDB per particle should be ~44% of the PSiNP volume. Assuming the density of PSiNPs is ~ 1.66 g/mL (50% Si, 50% water) and the density of the PegDB is ~1 g/mL, this yields an estimated mass ratio of ~27%, which matches experimental observations across independent experiments in Figures 3b and S6b-d. Measurement of particle surface charge also indicates effective coating, the polymer coating causing a shift in zeta potential from −19 to +14.5 mV for all sonication conditions (Figure S6e). To accentuate the differences before and after coating, we recorded these zeta potentials in a trehalose vehicle containing negligible salts.
Stability of PSiNPs to Freeze–Thaw Process.
For translational considerations, NP samples can ideally be stored frozen or freeze-dried. Additionally, freezing in aliquots allows for facile and thorough quality control of formulations used across multiple preclinical studies. We, therefore, evaluated the stability of PSiNP nanocomposites after being flash-frozen in liquid nitrogen and then thawed at room temperature. In parallel, we also prepared commercial mRNA liposomes (PolyPlus in vivo-jetRNA). jetRNA was prepared according to the manufacturer’s protocol with Cas9 mRNA and sgRNAs at a 1:1 mass ratio. PSiNPs were prepared containing either Cas9 RNP or CRE Recombinase. DLS was performed on the nanoparticles after 5 h on ice (to further verify stability), or were frozen immediately after preparation, and then thawed 1 h before DLS measurement (Figure 3d). In studies shown later in this article, both frozen and fresh PSiNPs were administered intramuscularly to Ai9 mice.
For all DLS measurements of PSiNPs (Figure 3d), the protein (Cas9 RNP or CRE) loading did not affect the particle size. This observation further suggests the pore loading and cargo-agnostic aspects of our system. PSiNP size remained the same after 5 h of 4 °C incubation, and only a small (309 to 340 nm), although significant (p < 0.05, t test), Z-avg diameter increase was observed after freeze–thawing. The jetRNA liposomes, in contrast, were essentially destroyed by freeze–thawing (p < 0.0001), with DLS reading a Z-average diameter of ~3000 nm, a 10-fold increase. This measurement yielded poor size-quality report and high PDI, however, so the average size is irrelevant given knowledge of their instability.
This set of experiments suggests that PSiNP formulations can be frozen in aliquots and used as needed at future dates without requiring additional formulations to be made with each experiment. This enables better quality control, enhances experimental consistency, and contributes to the potential for clinical translation.
PSiNP-PEGDB Nanocomposite Uptake and Endosomal Escape.
To be active, the Cas9 RNP must be taken up by the cells and escape endolysosomal vesicles to eventually reach the nucleus. Here, we tracked fluorescent Cas9 RNP in our fluorescent Gal8 reporter cell line. The Gal8 reporter has diffuse cytosolic YFP expression that becomes concentrated around disrupted endosomes. This shift from diffuse to punctate is a useful method for quantifying endosomal escape of therapeutic formulations.61,62
We chose to test uptake and endosome escape effects as a function of both the number of polymeric repeat units and the %BMA of the DB block of PEGDB. Prior work by Kelly et al. demonstrated that the ratio of DMAEMA (cationic) to BMA (hydrophobic) affected both the stability and activity of porous silicon nanoparticles, highlighting 40% BMA (40B) and 50% BMA (50B) as the lead candidates.27 The number of repeat units in the DB block has been shown to be another important variable in the PEGDB system62 but, until this study, had not been explored in PSiNP nanocomposites. We chose to test several different DB lengths, presented here as the number of repeat units (170, 210, 250, and 290) of DB monomers.
Uptake of the RNP is readily apparent in confocal microscopy of cells treated with RNP-loaded PSiNP nanocomposites (Figure 4a.i). RNP signal rapidly associated with the cell surface upon application of treatment. Over time, the RNPs were concentrated within the intracellular puncta (Figure 4a.ii). These puncta are highly, but not totally, colocalized with Gal8 puncta, as can be seen by comparing Figure 4a.ii and Figure 4b.ii, which are the same field of view. Points where Cy5 and Gal8 do not overlay are likely locations where the endosome has not yet been disrupted or where the cargo already escaped.
Quantifying the total cellular Cy5 signal yields further insight into the PSiNP nanocomposite formulations (Figure 4a.iii-iv). Uptake increases linearly over time, but at different rates for different polymer compositions. Uptake rate increases with shorter DB (170 > 210 > 250 > 290), or a lower percent BMA (40B-210 > 50B-210). This is consistent with the PEGDB system in previous works and general results with cationic polymers wherein more cationic (Lower %B) and less-stable (shorter DB block) nanoparticles tend to associate with cells more readily. While the formulations show different rates of uptake early on (Figure 4a.iii), the differences in uptake between formulations are less apparent at the 24-h time point (Figure 4a.iv).
Overall, by utilizing time course, live-cell confocal microscopy, and automated computational image analysis, we can draw useful conclusions about PSiNP nanocomposite formulations. PSiNPs are well dispersed, stable, and internalized by cells. The rate of association with the cell membrane can be slowed by decreasing the percentage of the cationic monomer (lower percentage DMAEMA, higher percentage BMA) or by increasing the length of the core-forming DB block of PEGDB.
Endosomal disruption is apparent in microscopy of MDA 231 (human epithelial mammary adenocarcinoma) cells expressing the Gal8 endosomal-disruption reporter after treatment with PSiNP nanocomposites (Figure 4b.i-ii). Whereas there is an obvious trend between uptake and length of the DB block within PEGDB, endosomal disruption is consistent over time for three shorter polymers (170 = 210 = 250, Figure 4b.iii). The 40B-210 and 50B-290, however, had higher levels of endosomal disruption more quickly, with increases of ~4-fold and ~2-fold, respectively, during the first 12 h. This is because the pKa of 40B is higher than 50B, activating its endosomal disruption behavior at a lower pH.27 For 50B, we also know that its endosomolytic potency is molecular weight dependent,62 explaining the higher Gal8 recruitment activity of 50B-290 than the smaller molecular weight analogues.
In parallel to the Gal8-cell treatment, nanocomposites were treated onto the mTmG reporter cell line, which expresses tdTomato before editing, and GFP after successful gene editing (Figure 2b). This change in fluorescence is then quantified by flow or image cytometry. For Cas9 gene editing, 100% editing efficiency results in ~33% of the cells becoming positive due to differences in the DNA repair mechanisms.63
In vitro studies measured editing in NIH 3T3 fibroblasts, a relatively difficult cell line to transfect/edit; the Lipofectamine CRISPRMAX product literature suggests that these cells are of similar difficulty to edit as induced pluripotent stem cells (iPSCs).59 We also tested a more difficult “forward” transfection approach in full-serum media, using a 24 h treatment time to better simulate in vivo conditions. Finally, by using dual-cut "turn-on" systems, we decreased the chance of false positives inherent to turn-off systems.
Cells were treated with PSiNPs at either 4 or 12 ng/μL Cas9 RNP (~24 or 72 ng/μL PSiNP) in 100 μL of treatment medium in a 96-well plate. GFP expression was imaged by confocal microscopy, and cell area was masked using an open-source MATLAB image analysis program (Figure 4c.i-ii). 50B-290 and 40B-210 were found to be more toxic than the other PEGDB polymers without a corresponding increase in peak editing activity. At the high dose, both yielded a decrease in editing activity—likely due to toxicity. However, at the lower dose, 50B-290 had an activity/toxicity profile that was competitive with those of the 50B-250 and 50B-210 formulations.
The 50B-170, 210, and 250 formulations had similar efficacy/toxicity trends, with the 210 and 250 lengths performing best. At the highest dose of 12 ng/μL 50B-250 achieved nearly 15% GFP+ (which should correspond to ~45% editing efficiency, based on literature). Cumulatively considering the uptake, Gal8, editing, and toxicity results, we selected 50B-250 as our lead formulation. 50B-290 had similar efficacy/toxicity performance and may be interesting to pursue in future work, but we prioritized 50B-250 due to its better cytocompatibility.
Comparison with Lipofectamine CRISPRMAX.
We next compared editing activity to Lipofectamine CRISPRMAX, a popular and effective reagent for Cas9 RNP.59 For this experiment, we used an NIH-3T3 cell line with the Ai9 reporter instead of the mTmG reporter. This was done in preparation for forthcoming in vivo studies in Ai9 reporter mice and because it requires two distinct sgRNA sequences, like what is necessary for the exon-skipping strategy for treating DMD. In systems which can only include a single sgRNA per particle, like many AAV systems, the need for two sgRNA would be expected to lower editing efficiencies.9,64
Treating the Ai9-NIH 3T3 with either PSiNP nanocomposites (50B-250) or Lipofectamine CRISPRMAX demonstrated significant differences between the transfection reagents (Figure 4d). At all dosages, PSiNPs significantly outperformed CRISPRMAX. Approximately 10-fold more Gal8 puncta were seen per cell, and PSiNPs more than doubled the editing efficiency of CRISPRMAX at 8 ng/μL Cas9 (Figure 4d.ii). Interestingly, the PSiNPs showed a linear trend of increased editing with dosage, whereas increasing the Lipofectamine CRISPRMAX dosage beyond the recommended 2 ng/μL of Cas9 did not increase editing. Our proxy for toxicity (relative cell count at 48 h) showed no significant difference in cell area versus no treatment at any dosage for any group (Figure 4d.iii).
This experiment also demonstrated the highly reproducible nature of the PSiNP system. Although the experiments were performed months apart, with different sgRNAs and reportergenes (mTmG vs Ai9), the 4 ng/μL 50B-250 group yielded the same percent-positive cells (~7.5%). The equivalent editing with a single or double sgRNA system suggests that PSiNPs may be amenable to multiplexing—a highly valuable trait for therapeutic strategies that require dual-sgRNA, like DMD exon skipping. This equivalent activity is likely a result of the high loading capacity.
In Vivo Editing.
Encouraged by the in vitro editing results, we sought proof of concept data for in vivo editing. We chose to investigate two local administration routes, intraarticular and intramuscular, corresponding to disease targets of post-traumatic osteoarthritis and muscular dystrophy, respectively. Additionally, to achieve data more relevant to the clinical needs of muscular dystrophy patients, we tested systemic (intravenous) administration of PSiNP nanocomposites in an inflamed-muscle model.
Intraarticular Editing in Ai9Mice.
To assess future potential for an antiarthritic therapy, we tested intraarticular administration into the knees of a mouse model of arthritis (Figure 5a).65,66 This model involves subjecting the knees of adult mice to mechanical loading for 2 weeks (Figure 5b), which has been previously shown to induce the desired PTOA phenotype. The day after the final loading and again after 7 days, formulations were injected locally into the intraarticular space. Two treatments per knee were chosen to maximize the cumulative impact. Two weeks after the second injection, hindlimbs were harvested for analysis.
Figure 5.
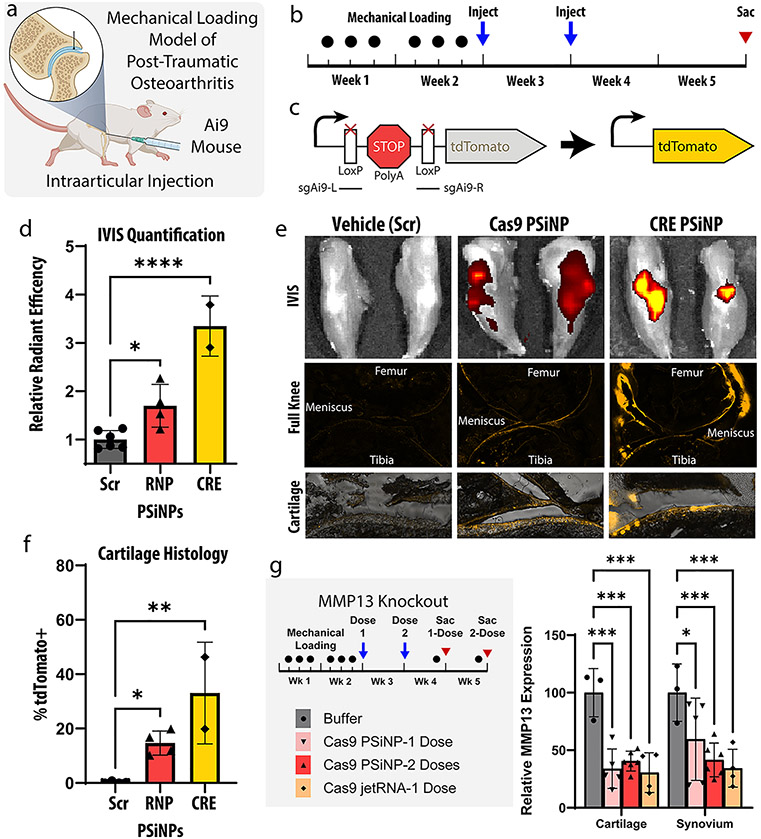
Intraarticular gene-editing in a post-traumatic osteoarthritis model. (a) A mouse mechanical loading model of post-traumatic osteoarthritis (PTOA) was used in these studies assessing editing within the knee joint following intraarticular PSiNP administration. (b) Experimental timeline for study. (c) Schematic representation of the Ai9 reporter cassette gene. (d) IVIS (total radiant efficiency) quantification from mouse knees after sacrifice. (*p ≤ 0.05, ****p ≤ 0.0001, Holm-Šídák’s multiple comparisons test). (e) Representative IVIS and cryohistology images from each group. The bottom row is higher magnification images of articular cartilage, demonstrating reporter turn-on as a marker for gene editing in chondrocytes. (f) Quantification of the percentage of tdTomato positive cartilage area from histological images of the Ai9 mouse knees. (*p ≤ 0.05, **p ≤ 0.01, Holm-Šídák’s multiple comparisons test). (g) Experimental scheme and qPCR results from MMP13 knockout experiment (*p ≤ 0.05, ***p ≤ 0.001, Holm-Šídák’s multiple comparisons test). Throughout the entire figure, all error bars are standard deviation and each data point represents one knee.
In this study, a well-established Ai9 reporter mouse was chosen. Briefly, these mice are engineered to contain a stably integrated reporter gene in the Rosa26 safe harbor site of the genome (Figure 5c). Cells in these mice express tdTomato fluorescent protein when gene was edited by either CRE recombinase or Cas9 ribonucleoprotein. Notably, due to reliance on the cell machinery to repair the DNA into the correct reading frame, only about 1 in 3 Cas9 editing events result in proper tdTomato expression. Thus, the Cas9 editing signal is only expected to be about 1/3 as bright as for CRE. The tdTomato fluorescence is visible on IVIS imaging, which allows for fast and facile assessment of efficacy. Additionally, tdTomato fluorescence can be seen in cryohistology, which elucidates the location of the edited cells. Localization of edited cells can be difficult to deduce by other methods and can be important to disease outcomes.
In the Ai9 study, mice were administered either saline or PSiNP composites loaded with one of three cargos: (1) Cas9+scrambled sgRNA, (2) Cas9+Ai9 sgRNA, or (3) CRE. The control groups (PBS, Cas9+scrambled sgRNA) showed no difference from one another by IVIS. They were therefore pooled into one statistical group to minimize the animal count needed (Figure 5d, Figure 5e, and Figure 5f). At the macro level, both treatment groups showed a significant increase in IVIS fluorescence compared to no treatment (p < 0.05, p < 0.0001, Holm-Šídák’s multiple comparisons test). The RNP group had a 70 ± 22% (mean ± SEM) increase in fluorescent signal, while the CRE group had a 235 ± 44% increase. This is consistent with the expected 1:3 signal ratios if equivalent gene editing was achieved.
Cryohistology from both the Cas9 and CRE groups (Figure 5e) indicates that the signal is coming from edited cells in all three target tissues: the cartilage, meniscus, and synovium. Quantifying the percentage of cartilage tissue positive for tdTomato signal in all groups indicated the Cas9+Ai9 sgRNA group yielded 14.7 ± 2.2 (mean ± SEM) percent positive cartilage area while the CRE group yielded 33.1 ± 13.2.
Intraarticular Editing of the Therapeutically Relevant MMP13 Gene.
Because the results in Ai9 mice indicated that approximately 33% of the cells along the interior surface of the target tissues—cartilage, synovium, and meniscus—were gene edited, we next pursued a potential therapeutic gene target. Matrix Metallopeptidase 13 (MMP13) is a secreted enzyme which breaks down extracellular matrix and is highly overexpressed in rheumatoid and osteoarthritis.67 MMP13 activity can create a positive feedback mechanism that propagates injury induced cartilage degradation and inflammation.67 Inhibition of MMP13 has thus long been a therapeutic target, but so far drug trials of small molecules have failed due to specificity issues and side effects.67-69 Therefore, work using siRNA or Cas9 to reduce MMP13 levels in the knees have drawn interest.69
To assess MMP13 knockout, we repeated a similar mechanical loading model but decided to test both single- and two-dose treatment protocol. We chose for both groups to receive treatment 12 h after the last loading injury to model the clinical realities of a patient receiving treatment after acute injury or surgery. In the two-dose group, a second administration was given 1 week later to simulate a follow-up appointment. The three intraarticular treatments tested were: (1) the resuspension buffer used with the PSiNPs (Buffer) as a negative control, (2) PSiNPs loaded with Cas9 RNP targeting MMP13 (Cargo concentration 1 mg/mL, Silicon concentration 6 mg/mL), or (3) PolyPlus in vivo jetRNA targeting MMP13 (0.1 mg/mL cargo concentration, as indicated by manufacturer).
Knockdown of MMP13 was quantified with qPCR and found to be ~60% in both the cartilage and synovium in all groups (Figure 5g). MMP13 expression was significantly reduced in all groups versus the buffer control.
Discussion: Intraarticular Gene Editing.
Overall, intra-articular administration of PSiNP nanocomposites achieved editing levels consistent with a promising future therapeutic for PTOA. The signal by both IVIS and quantification of histology suggests that the system is cargo agnostic—editing levels are consistent between Cas9 and CRE. The ~60% knockdown of MMP13 shown in Figure 5g is consistent with prior publications which demonstrated therapeutic efficacy.65,66,68-70 The greater MMP13 knockdown percentage (~60%, Figure 5g) than surface-cell editing percentage (~30%, Figure 5f) may be attributed to the positive-feedback nature of MMP13 expression.
Muscle Editing.
We next sought to prove editing in skeletal muscle, toward proof of concept relevant to the treatment of Duchenne Muscular Dystrophy (DMD), a disease that has been a major focus for application of gene editing technology.71 Early publications using Cas9 in mice were applied to DMD models using AAVs (Adeno Associated Virus), which are difficult to manufacture, difficult to dose, and cause immune responses which prevent repeated dosing.72-74 The current standard for genetic DMD treatment is an exon-skipping strategy, which uses two different gRNAs to remove a region of mutated DNA.71 The Ai9 cassette is a good model of this strategy, as it also requires a large deletion by two different sgRNAs, and is thus frequently utilized.75
Stability of PSiNP Nanocomposites.
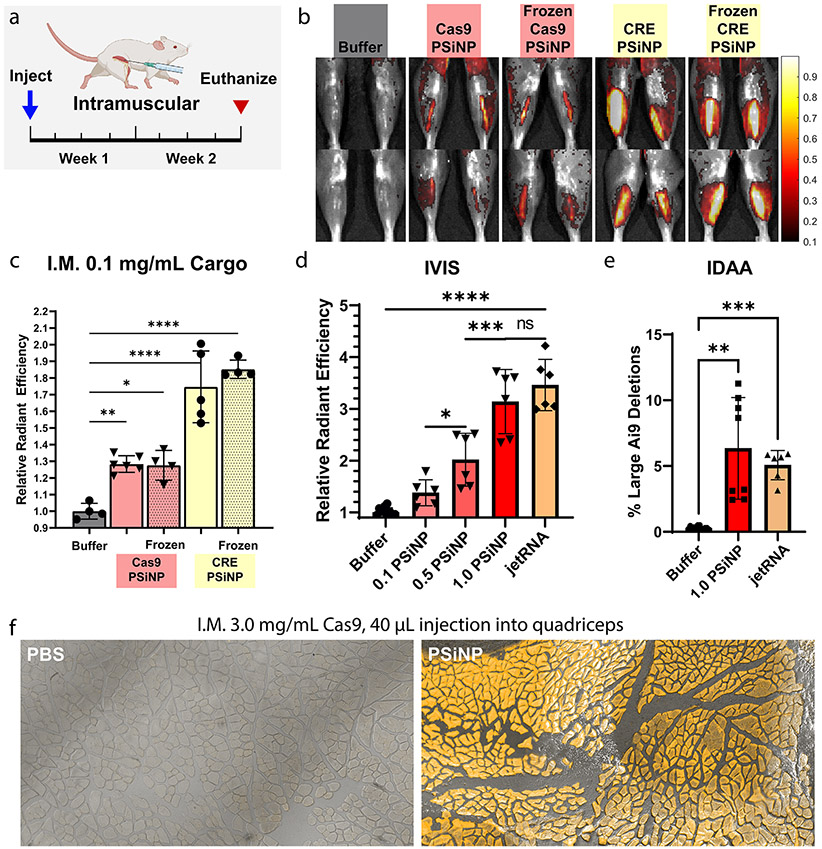
To complement earlier in vitro particle stability analyses (Figure 3d), we sought to test whether the PSiNP nanocomposite system is amenable to in vivo activity after freeze–thaw using the IM delivery route. We administered the PSiNPs at a relatively low concentration (0.1 mg/mL cargo) and volume (20 μL). Two weeks after injection, mice were sacrificed (Figure 6a) and no difference in IVIS signal between the frozen and fresh PSiNPs (Figure 6b, Figure 6c) was found. Even at this low dose, an ~30% increase in signal over no treatment was seen for the Cas9 groups and an ~80% increase for CRE. All groups were significantly different from PBS control injection (Šídák multiple comparisons).
Figure 6.
Editing in Ai9 mice following intramuscular administration of PSiNPs. (a) Experimental timeline for direct intramuscular injection studies. Muscle was injected with PSiNPs or PBS and harvested 2 weeks after the injection. (b) Representative IVIS images of legs from each group. (c) IVIS (average radiant efficiency) data from mouse muscle in freeze–thaw study. (d) IVIS data from mouse muscle in Cas9-PSiNP dose escalation study. (e) Indel detection by amplicon analysis (IDAA) data from the highest dose used in the dose escalation study. (f) Cryohistology to visualize tdTomato fluorescence in edited muscle fibers in separate study done with injection into the larger quadriceps muscle. All statistics result from ANOVA with the Šídák multiple comparisons test using GraphPad Prism (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001). All error bars are standard deviation and each data point represents one tibialis anterior muscle.
Intramuscular Dose-Escalation and Benchmarking vs Liposomes.
With intramuscular activity established, we next pursued dose escalation studies in Ai9 mice (Figure 6d). PEGDB–PSiNP nanocomposites loaded with Cas9 RNP targeting the Ai9 site were injected intramuscularly (20 μL at 0.1, 0.5, or 1.0 mg/mL Cas9 concentration), and the mice were sacrificed after 2 weeks. PSiNP resuspension buffer (Buffer) and PolyPlus in vivo jetRNA loaded with Cas9 mRNA and sgRNA were administered as controls. Dose response was linear (Figure S7, r2 = 0.81) across the range tested. No adverse health effects were noticed at any dosage. At the highest PSiNP dosage, the fluorescent signal matched that of the PolyPlus in vivo jetRNA mRNA liposome control, with no significant difference observed in the IVIS signal. To assess gene editing at the DNA level, indel detection by amplicon analysis (IDAA, Figure 6e and Figure S8) was performed. This method also indicated roughly equivalent editing between the high-dose PSiNP and jetRNA groups, both groups having ~5% indel efficiency at the DNA level. Finally, to pilot the potential scalability of these solutions, we tested a larger 40 μL injection into the quadriceps of Ai9 mice at a Cas9 concentration of 3.0 mg/mL. Again, no adverse safety events were observed. At the injection site, nearly 100% positive muscle fibers were observed over a large area (Figure 6f, Figure S9d). IVIS signal indicated that gene-editing was located at the injection site and not present over the entire muscle (Figure S9c).
Biodistribution of Intravenous PEGDB—PSiNP Nanocomposites in BaCl2 Muscle-Inflammation Model.
Localized, direct intramuscular administration is not a clinically viable strategy to treat muscular dystrophy patients. Ideally, a single intravenous injection would be sufficient to ameliorate the disease. DMD causes substantial inflammation of the skeletal muscle, the severity of which is not matched by dystrophin knockout (mdx) mice due to physiological differences between mice and humans. Here, we used injection of 20 μL of 1.2% barium chloride (BaCl2) into the tibialis anterior muscle as a method to model muscle inflammation of DMD.
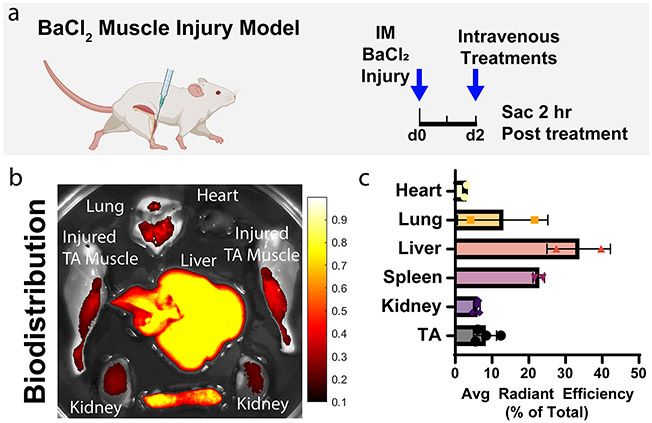
To measure biodistribution, PEGDB–PSiNP nanocomposites were loaded with RNPs comprising Cy5-labeled tracrRNA. These were administered intravenously in the BaCl2 model at a concentration of 0.1 mg/mL Cas9 (0.6 mg/mL PSiNP) in 150 μL of trehalose freezing buffer. Two hours after injection, mice were sacrificed, and major organs/tissues were excised (Figure 7a). Tissues (heart, lung liver, spleen, kidneys, and legs) were imaged for Cy5 fluorescence on IVIS, indicating the localization of Cas9 RNP (Figure 7b,c, S10). Unsurprisingly, a substantial percentage of the cargo is cleared in the liver, but these studies confirmed that a measurable amount of biodistribution occurred to the inflamed skeletal muscle, a very high bar and significant challenge for nonviral therapies.
Figure 7.
Biodistribution of intravenously administered Cas9-loaded PEGDB–PSiNPs. (a) Experimental scheme. Muscles were injured with barium chloride 2 days before intravenous treatment with PSiNPs containing fluorescently labeled Cas9. (b) Qualitative image of biodistribution of Cas9 loaded within PSiNPs. (c) Quantification of Cas9 fluorescent signal distribution by percentage of total signal. All error bars are standard deviation. Each data point represents one organ.
Intravenous Administration of PSiNP Nanocomposite.
Observation of biodistribution to inflamed muscle with intravenous delivery motivated us to assess gene editing in BaCl2 injured muscle after the intravenous administration of PSiNPs.
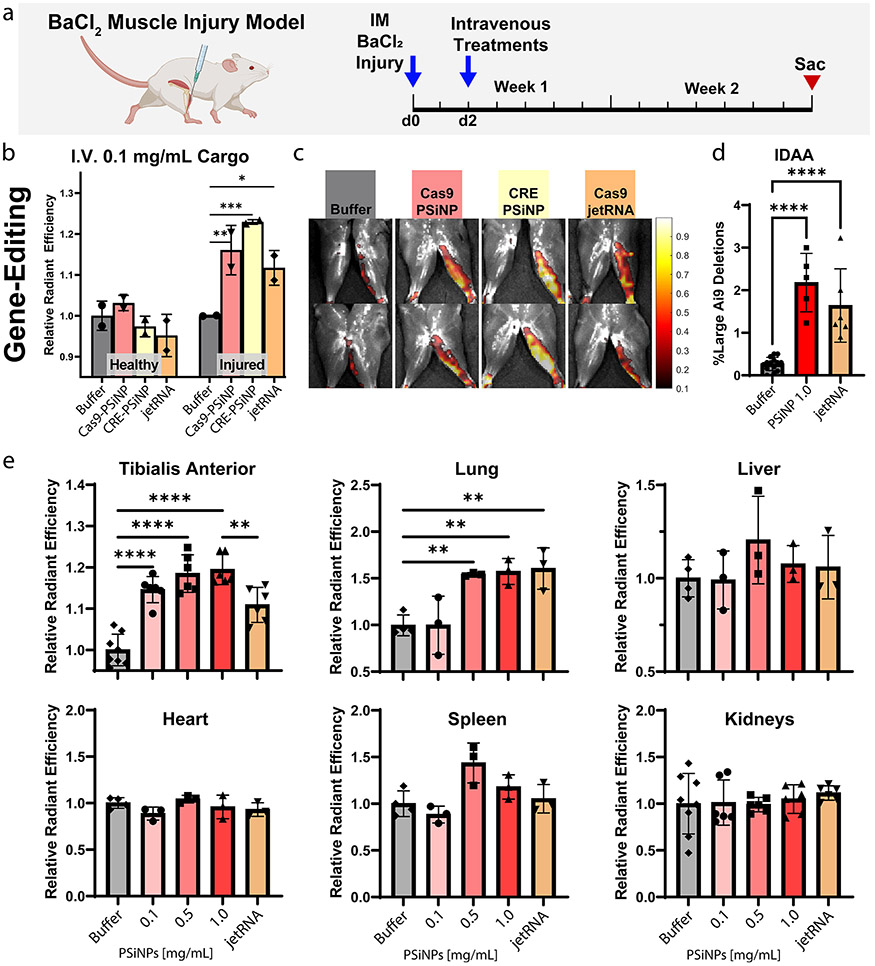
BaCl2 was administered into the left leg of Ai9 mice, leaving the right leg as a contralateral control. 48 h later, treatments were administered intravenously, and mice were sacrificed 2 weeks after treatment (Figure 8a). As a positive control, we administered gold-standard, commercially available in vivo jetRNA liposomes (PolyPlus) containing Cas9 mRNA and Ai9-targetting sgRNA. As a negative control, the PSiNP resuspension buffer (“Buffer”) was administered intravenously. For experimental groups, PEGDB–PSiNP nanocomposites were formed loaded with either Cas9 RNP or CRE and administered at a cargo concentration of 0.1 mg/mL (0.6 mg/mL PSiNP). For all groups,150 μL was injected via tail vein. Notably, this is well below the maximum tolerated dose of PSiNP nanocomposites, which have been shown to be nontoxic up to at least 5 mg/kg of cargo (>6-fold our ~0.75 mg/kg dose).26
Figure 8.
Systemically administered PSiNPs gene edit in BaCl2 injured muscle. (a) Experimental strategy outline. (b) Quantification of tdTomato signal (gene editing) from healthy and BaCl2-injured TA muscle and (c) representative IVIS images. (d) Indel detection by amplicon analysis (IDAA) data from the high dose in the intravenous dose response study. (e) Quantification of tdTomato IVIS signal (gene editing) from major organs excised from Ai9 mice in the intravenous dose response study. All error bars are standard deviation, and each data point represents one organ. Statistical comparisons using Šídák multiple comparisons, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
Even at this low dose, all formulations generated significant reporter turn-on (Figure 8b, Figure 8c, Šídák multiple comparison). The Cas9-mRNA liposome increased average IVIS signal over the TA muscle by ~10%, and the Cas9-PSiNP nanocomposite increased signal by ~15%, although these groups were not significantly different. CRE-PSiNP Nanocomposites had an increase of ~20%. Results indicate editing in the inflamed but not contralateral TA. The contrast between healthy vs injured legs suggests improved targeting to the inflamed muscle, likely due to induction of vascular leakiness, a phenomenon known to occur during muscular dystrophy, and in the BaCl2 model.76,77
In a follow-up study, we sought to test the dose response and add more replicates while also confirming gene-editing at the DNA level. Both tibialis anterior muscles were injected with BaCl2 on each mouse to increase sample size. To confirm gene-editing at the DNA level, DNA was extracted from the tibialis anterior of mice from three groups, those injected with either: (1) trehalose resuspension buffer (Buffer), (2) Cas9-loaded PSiNPs at the highest dose of 1.0 mg/mL Cas9, 6 mg/mL PSiNPs (1.0 PSiNPs), or (3) jetRNA liposomes (Figure 8d). IDAA was run on these samples, indicating editing percentages of 2.2 ± 0.3% for PSiNPs, and 1.6 ± 0.4% (mean ± SEM) for jetRNA. Both groups significantly differed from the Buffer control (p < 0.0001, Tukey’s multiple comparisons test), but did not significantly differ from each other.
Quantification of IVIS imaging of tdTomato expression paralleled the IDAA results; the 1.0 mg/mL Cas9 PSiNP group and jetRNA group exhibited an increase over baseline of 19.5 ± 1.5% and 10.9 ± 1.7% (mean ± SEM), respectively, with the PSiNP group significantly outperforming jetRNA (p < 0.01, Figure 8e). In the lung, the 0.5 and 1.0 mg/mL Cas9 PSiNP groups, as well as the jetRNA group, had significantly increased fluorescence signal over the no treatment control (p < 0.01, Figure 8e). No other organ demonstrated significantly increased fluorescence that would be indicative of gene-editing.
Overall, these data from systemically administering Cas9 gene therapies in a BaCl2 muscle inflammation model are promising when considered in a historical context. An intravenous administration achieving ~2% indels and ~20% increase in fluorescence is comparable to early studies with AAVs.72,73,75 While 2.2% deletion efficiency may seem low, prior work has demonstrated that this level of editing in the mdx model may be close to the therapeutic level, yielding ~8% of wild-type dystrophin levels by Western blot and ~67% of positive muscle fibers by immunostaining. Our results were also promising relative to jetRNA, as PSiNPs outperformed jetRNA by about 40% (2.2% vs 1.6% deletion efficiency).
CONCLUSIONS
This work demonstrates that porous silicon and polymer nanocomposites are a modular system for loading diverse protein cargoes, including Cas9 RNP complexes for gene editing. The rigid 20 nm pores of porous silicon, in contrast to more common “soft” nanoparticles, are ideal for loading—achieving ~16 wt % loading and nearly 100% loading efficiency regardless of the protein’s charge or size. The density of silicon allows for facile separation from unloaded cargo or other excipients used in formulation. In vitro, this system yielded twice the gene editing efficacy of a commercial transfection reagent. In vivo, we demonstrated gene editing after local administration in disease-relevant models of Duchenne’s muscular dystrophy and post-traumatic osteoarthritis. In a muscle-inflammation model of muscular dystrophy, intravenously administered PSiNPs localize to and gene-edit in injured muscle better than a commercial liposomal reagent—achieving ~2% editing by IDAA analysis. Considering prior publications in which PSiNPs deliver DNA, RNA, and small molecules, the universal protein loading appliations demonstrated in this work suggests that future work may combine modalities for targeted genomic insertion of DNA, base editing, and other manifestations of CRISPR technology.
METHODS
PSiNP Manufacturing.
Porous silicon was formed by electrochemical perforation etching and sonication as performed in previously published methods.26,27 Briefly, silicon wafers were etched in a repeating sequence of high and low voltage to create fractioning layers. Pore size distribution was determined by scanning electron microscopy (Vanderbilt VINSE Zeiss Merlin) and quantified in ImageJ (Figure S1). The porous silicon was then lifted from the wafer by scraping with a razor blade in absolute ethanol.
Nanoparticles were formed by successive sonication and centrifugal size-selection steps. First, the PS-ethanol solution was placed in a nuclease-free Erlenmeyer flask in a large ice and salt bath to dissipate heat. The porous silicon was sonicated into nanoparticles using a probe tip sonicator for 30 min at a time. After each 30 min step, a size selection step was performed. The solution was first centrifuged at 2000 RCF for 3 min to pellet particles > ~300 nm, which were then resonicated. This process was repeated until the yield plateaued—approximately 5 times. All supernatants (the smaller particles) were pooled, pelleted at maximum RCF (rotor dependent) for enough time to pellet 100% PSiNPs > 50 nm (21k RCF, 10 min, vesicles.niifhm.ru). Particles were resuspended in absolute ethanol, sonicated an additional 30 min, and then the entire process was repeated with only 10 min of sonication of the pellet at each step. This repeated sonication and centrifugation optimizes particle size, quality, and yield because pelleting 100% of >400 nm particles will also pellet a significant percentage of correctly sized particles. Also, less-porous silicon will be denser, therefore should pellet with the oversized particles.
The size selected particles were dried and thermally oxidized at 400 °C for 3 h. Eight etches yielded ~300 mg of PSiNPs. The particles were suspended in molecular biology grade ethanol to 5 mg/mL and sonicated in a water bath for 1 h. Concentration was confirmed via thermogravimetric analysis (Figure S4). Particles were diluted to 0.1 mg/mL in molecular biology grade ethanol and stored at −80 °C. Hydrodynamic diameter was determined by both NanoSight (Malvern Panalytical NS300) and dynamic light scattering (Malvern ZS nano) to be ~250 nm.
Thermogravimetric Analysis (TGA).
Thermogravimetric analysis was performed on a TGA Instrument Specialists TGA 1000. To determine PSiNP concentration, a known volume of dilute PSiNPs (~2 mL) was pelleted and then resuspended in 20 μL of absolute ethanol. This was pipetted onto a tared, flame-dried platinum pan within the TGA. The temperature was raised to the following temperatures and held constant until the mass plateaued: 73 °C (evaporate Ethanol without boiling), 105 °C (remove residual ethanol and trace Water). The temperature was then raised linearly to 600 °C, but the mass was found to be invariant to this temperature range over 4 samples. The mass remaining on the platinum pan was taken as the total mass silicon in the original 2 mL solution. Samples with ethanol only were run to ensure the validity of the method. The concentration by TGA was found to be consistent with a standard made from a known mass of dried particles measured by analytical balance, but TGA allows for accurate measurements using much smaller amounts of particles (micrograms instead of milligrams).
DLS and NanoSight.
Dynamic light scattering and Zeta Potential measurements were performed on a Zetasizer nano ZS using a disposable zeta cell and standard parameters. The machine selected an attenuation of 7, and all results shown passed the size quality report (except, of course, for some of the agglomerated particles in Figure 3). PSiNPs were suspended in absolute ethanol (bare particles) or in trehalose injection buffer which was then diluted 1:10 in molecular biology grade water (coating and sonication study). NanoSight was performed on a Malvern NS300 instrument following manufacturer recommendations (Figure S1).
SEM (scanning electron microscopy) and ImageJ.
SEM was performed on a MERLIN SEM instrument with the GEMINI II column in the Vanderbilt Institute for Nanoscale Science and Engineering (VINSE) core facility. The "Analyze Particle" function of ImageJ was used to segment pores vs walls, and pore size distribution was estimated by summing the distribution of Feret Max, Feret Min, EQPC (diameter of a circle of equal projection of area), and PED (diameter of a circle of equal perimeter) (Figure S2).
DLVO Modeling.
Charge within pore as a function of NaCl concentration was modeled using Poisson–Boltzmann theory as espoused in Chapter 4 of Surface and Interfacial Forces by Butt and Kappl,55 adapted to parallel plate conditions. We used the linearized Poisson–Boltzmann equation (eqs 4.7 and 4.8 in the textbook) to calculate the Debye length in a system with low Zeta Potential voltage (<25 mV). According to our calculations (Figure S3e), the zeta potential of PSiNPs was within the linear range. Physically, the Debye length is a measure of the exponential decay length of electric field away from a charged surface. As salt concentration increases, more counterions are available to shield a charged surface. Thus, the Debye length shortens with an increase in salt (Figure S3a).
The Linear Superposition Approximation (SLA, eq 4.57 in Butt and Kappl)55 is the assumption that overlapping electric fields add linearly. This is acceptable for minimally overlapping fields. This study seeks to use a high enough salt concentration to have zero charge in the center of the pore, and thus, the SLA is appropriate (Figure S3c). To model the electric field within a pore, we sought to convert from a parallel plate system to a cylindrical system. Our simplified way of doing this was to consider relative areas. Between plates, the area of the Gaussian surface does not change with distance. Within a cylinder, the relative area decreases toward the center. We thus normalized the data in Figure S3c to the relative area of a cylinder to generate Figure S3d.
We realize this modeling is simplified—several of the assumptions used in these foundational models do not apply to our system, particularly with the small length scales and presence of divalent ions—but we believe the modeling is still useful in understanding the system. Better models of similar systems have been explored in literature, and support the same general conclusions.78
Calculations were performed in MATLAB. See Figure S3 for more detail. For the script, see github.com/BrockFletcher/Cas9_Silicon.
Protein Loading and Release Studies.
PSiNP concentration was known based on analytical balance and confirmed by thermogravimetric analysis (Figure S4b). Protein concentration was known from the manufacturer and confirmed by A280 and BCA assay. PSiNPs and cargo were suspended in PBS for 1 h at 4 °C with mild agitation. PSiNPs were then pelleted via centrifugation at 21k RCF for 15 min. Particles were then resuspended in molecular biology grade water to wash, pelleted again, and then resuspended in 0.1% SDS solution and sonicated for 1 h to strip out the cargo. For the release studies, instead of resuspension in 0.1% SDS, PSiNPs were suspended in DPBS for the prescribed time period and then pelleted and the protein concentration in the supernatant measured.
Cargo concentration in each solution was measured by gel densitometry (Bio-Rad ImageLab), micro-BCA assay, or CBQCA assay. All three methods were found to be consistent, thus confirming the accuracy of the results.
Cas9 Ribonucleoprotein Preparation and sgRNA Sequences.
Cas9 was purchased from IDT (Alt-R spCas9 Nuclease), and guide RNAs were purchased from Synthego (Synthetic sgRNA with 2′-O-Me and phosphorothioate modifications). Guide RNA target sequences are listed in Table S1. Cas9 was stored at 10 mg/mL at −80 °C in 10 μL aliquots. Guide RNA was stored at 100 μM at −80 °C in Synthego’s provided TE buffer. Before RNP formation, guide RNAs were “annealed” in PCR tubes on a thermocycler programmed to heat to 95 °C for 5 s, then drop stepwise by 10 °C, stopping for 2 s at each step, down to 35 °C. Cas9 protein was then mixed with the freshly annealed sgRNA at a 4:1 mass ratio of Cas9:sgRNA (1:1.25 molar ratio) in molecular biology grade PBS with added NaCl. Care was taken so that Cas9 never experienced NaCl concentrations below 150 mM to prevent any potential instability. The Cas9-sgRNA mix was then placed on a thermocycler at 37 °C for 10 min to aid in RNP formation. RNPs were then immediately used, stored at 4 °C for up to 8 h, or stored at −80 °C for later use. For the multiguide Ai9 and Rosa26 strategies, RNPs were formed separately for each guide and then mixed at equimolar ratios before use.
CRE Recombinase Production.
Expression plasmid for CRE Recombinase production was obtained from Addgene (Plasmid #62730) and transformed into competent E. coli (Rosetta 2(DE3) singles, Novagen 71400) by heat shock following manufacturer protocol. Cells were plated onto agar plates containing 100 μg/mL Ampicillin and 34 μg/mL Chloramphenicol overnight at 37 °C. A single colony was selected to grow a 10 mL starter culture overnight, and this was diluted 1:100 into 1 L of Terrific Broth (TB) the next day. All growth media contained freshly prepared 100 μg/mL Ampicillin and 34 μg/mL Chloramphenicol. Once OD reached 0.6, temperature was reduced to 20 °C,and expression of Cre recombinase was induced using 0.5 mM IPTG. Approximately 16 h after induction, media was pelleted in 1 L centrifugation jars at 4 °C at 8000g RCF for 15 min. Pellets were flash frozen in liquid nitrogen and stored at −80 °C until purification. To purify CRE, pellets were resuspended in cold BugBuster Lysis buffer with 1 mM PMSF and 100 μg/mL Lysozyme. Pellets were lysed by probe-tip sonication (6W output) on ice with 1 s pulses for 15 min. Lysates were then treated with RNase A (10 μg/mL) and DNase I (5 μg/mL) on ice for 15 min, until a change in consistency was observed. Lysates were then clarified via centrifugation for 30 min at 20,000g at 4 °C. Clarified lysates were then purified on a Cobalt NTA column (Cytiva HiTrap TALON crude) on an FPLC and eluted with imidazole. A second purification was then performed by FPLC using cation exchange chromatography (Cytiva HiTrap SP). Finally, the buffer was exchanged into a CRE freezing buffer (50% Glycerol, 250 mM NaCl, 10 mM HEPES, pH 7.4) using Vivaspin 20 MWCO 3000 spin filters. CRE recombinase was concentrated to 1 mg/mL and flash frozen in liquid nitrogen.
Salt Loading Study.
For the salt-loading activity study, all PSiNP-composites were prepared under the exact same conditions except were loaded with a variety of Cas9 RNP or CRE recombinase at a variety of Trehalose, MgCl2, and NaCl concentrations. These particles were treated onto mT/mG reporter cells. The percentage of positive cells was measured by flow cytometry after 48 h.
Cell Line Creation.
MDA-231 human epithelial mammary adenocarcinoma Gal8 reporter cells were used from frozen stocks from Kilchrist et al.62 Ai9 and mTmG plasmids were purchased from Addgene (Addgene #22799 and Addgene #17787, respectively). Plasmid was maximum prepared and then transfected into NIH 3T3 fibroblasts with Lipofectamine 3000. Cas9 RNPs were simultaneously transfected using Lipofectamine CRISPRMAX and gRNAs targeting the Rosa26 left and right arms of the plasmids.79,80 Cells were selected with G418 at 500 μg/mL for at least 3 weeks, at least 1 week after untransfected controls wells were completely dead from G418 selection. mTmG cells were then also flow sorted for the top 10% highest-performance tdTomato-expressing cells. Both cell lines were cultured with a maintenance antibiotic concentration of 200 mg/mL until 48 h before experimentation.
Nanocomposite Preparation.
Unless otherwise indicated, PSiNP composites were prepared as follows:
Briefly, PSiNPs were sonicated in water to disperse and then mixed with cargo solution for 1 h to allow loading of the cargo into the PSiNPs. Then, PSiNPs were pelleted by centrifugation and resuspended in a protonated PEGDB polymer solution. A “lock-in” buffer was added to raise the pH to above the pKa of the DB-block of PEGDB. The particles were pelleted again and then finally resuspended in the final buffer of choice.
To provide more detail for those seeking to utilize PSiNP nanocomposites:
First, the 0.1 mg/mL EtOH-Stored PSiNPs stocks are pelleted at 21k RCF for 15 min and the supernatant removed. Then, PSiNPs are resuspended in molecular-biology grade water and sonicated for 10 min. The pH of this solution should be checked and adjusted to pH ~ 7–8 with NaOH or HCl if necessary, to ensure the silica surface is sufficiently charged.81 The charge and low tonicity should ensure that the PSiNPs are optimally dispersed. For all studies here, PSiNPs were suspended to 1 mg/mL.
The Loading Step:
A solution containing the protein cargo is added to the PSiNP particles in a 2 mL microcentrifuge tube and well-mixed via pipetting. The tube is sonicated in an iced (<10 °C) water-bath sonicator in half second pulses for 1 min. This is then incubated on a rocker at 4 °C for 1 h to allow loading. The system is amenable to a range of concentrations, if optimal conditions for the particular cargo (pH, salinity, etc.) are maintained. Care should be taken that the cargo is pure, as many proteins are sold with albumin to help stabilize. For all studies here published, the cargo volume was >10× that of the PSiNP volume, to minimize dilution of the cargo buffer. Unless otherwise specified, we recommend and use the following as the final concentration in the loading step: 0.1 mg/mL PSiNPs, with cargo concentration at 100% theoretical loading (18.3 μg/mL CRE, for example), 10 mM HEPES Buffer, 150 mM NaCl. For negatively charged proteins like Cas9 RNP, we also recommend 15 mM Trehalose and 15 mM MgCl2. It is vital that all reagents should be RNase, DNase, and Protease Free (DEPC or RNA-Secure treat Trehalose, for example).
The Coating Step:
After at least 1 h of loading (and up to 24 h has been proven successful, Figure 2C), the nanoparticles are pelleted via centrifugation (>10k RCF for >5 min should be sufficient, although we recommend >21k RCF for 15 min to be safe). PSiNPs are then resuspended in cold 10 mg/mL PEGDB solution (pH 5.5, 30 mM sodium acetate) by vigorous pipetting and brief vortexing. Particles are visually inspected to ensure there is no pellet or aggregates remaining before water-bath sonication for 1 min with half second pulses at 4 °C. A cold “lock-in” buffer is added immediately after raising the pH of the solution, deprotonating the PEGDB. The specifics of this buffer should be flexible, as long as it fully raises the pH, but we recommend the following: Add lock-in buffer at >5× the volume of PEGDB solution, lock-in buffer consists of 50 mM buffering capacity at pH ~ 7.5 (we used HEPES, but tris and phosphates may also be amenable) and ~300 mOsm osmotic pressure (we recommend/used 250 mM Trehalose, although PBS with added HEPES may be passable).
The Resuspension Step:
Particles are then resuspended in the needed buffer to the desired concentration. We used 10 mM HEPES with 270 mM Trehalose for all of our studies. PSiNPs were either used immediately in this buffer or flash-frozen in this buffer in liquid nitrogen. Frozen PSiNPs were thawed at room temperature on the bench.
PEGDB Synthesis.
PEGDB was synthesized, as previously described, by RAFT polymerization from a PEG-linked chain transfer agent and then purified by ether precipitation.27,50,51,62 Polymer composition and length were confirmed by NMR and GPC.
PEGDB Coating Assay.
PEGDB was synthesized as previously described, with rhodamine (TAMRA) fluorophore incorporated into the RAFT polymerization.26 PSiNP nanocomposites were prepared by the standard protocol, then pelleted and resuspended in 0.1%SDS/95% EtOH, in order to “strip” off the PEGDB coated onto the PSiNPs. Finally, the particles were pelleted again and resuspended in 0.1%SDS/95% EtOH to test the efficacy of the stripping step. All supernatants were measured for fluorescence on a plate reader and normalized to an appropriate standard curve. Confirmation of this protocol is included in Figure S6a, as well as published by Kelly et al.27
High Content Imaging, Uptake, and Endosome Escape.
Images were acquired on a Nikon C1 Confocal microscope using Nikon Elements software and a Ti Eclipse main unit with a 20x Plan Apo Fluor nosepiece. The acquisition was automated to take multiple images per well and, for the time course studies, images of the exact same field of view over time. Care was taken not to oversaturate the images. Acquired images were analyzed using the open-source Baxter Algorithms github.com/klasma/BaxterAlgorithmsoftware and Gal8 analysis program published by Kilchrist et al.62 Briefly, the cytosol and Gal8 puncta are segmented out, and the intensity of Gal8 regions is quantified and normalized to the cytosolic pixel intensity. To track uptake, the signal was quantified from the Cy5-labeled cas9 tracrRNA over the cytosolic area and normalized to the cytosolic intensity to account for cell number and any focal aberrations.
The mTmG images were analyzed by first segmenting regions positive for either GFP or tdTomato. The two segmentations are overlaid to yield the total cytosolic area, and then the area GFP+ is quantified relative to the total cytosolic area to yield the percentage of cells GFP positive. The relative cell area for each well was also used as a proxy for toxicity, as toxic formulations should either impede cell growth or cause cell loss from the plate.
Preparation of PolyPlus In Vivo-jetRNA Liposome-Based mRNA Delivery Reagent.
PolyPlus in vivo-jetRNA was prepared following the manufacturer’s protocols in all experiments. For example, for a 200 μL injection, 20 μg of RNA (1 μg/μL) is diluted in 160 μL of the provided mRNA buffer to a final concentration. Then 20 μL of in vivo-jetRNA is added to the tube and mixed by gently pipetting. This yields a final RNA concentration of 0.1 μg/μL. After a 15 min incubation, the mix is injected. A minor modification was incorporated to allow codelivery. mRNA and sgRNA were mixed in a 1:1 mass ratio before incorporation into the experiment. For example, the protocol calls for 20 μg of mRNA for a 200 μL injection volume. Instead, we loaded 10 μg of mRNA and 10 μg of sgRNA for a total RNA mass of 20 μg.
Animal Work.
All mouse procedures were reviewed and approved by Vanderbilt University’s IACUC. Housing was standard (12 h light-dark cycle, 40–60% humidity, 18–23 °C). Ai9 mice were purchased from the Jackson laboratory and then used to start a colony. Ai9 mice are homozygous, yet PCR was still performed regularly to confirm the presence of the Ai9 cassette.
Mechanical Loading PTOA Model and Intraarticular Gene Editing.
The post-traumatic osteoarthritis (PTOA) model was performed as previously published.65 Briefly, mechanical loading was performed on knee joints of mice three times per week for 2 weeks. Each loading consisted of 250 cycles of compressive mechanical loading at 9N of force. The day after the last loading, the mice were administered 30 μL of PSiNP nanocomposites intraarticularly at 1 mg/mL protein cargo concentration.
In the Ai9 mouse studies, the mice were then redosed 7 days later and sacrificed 2 weeks after this final injection. Hindlimbs were skinned, imaged by IVIS, and fixed following the histology protocol enumerated in another section of the methods.
In the MMP13 studies, mice were split into two treatment groups: either injected once or twice. Two weeks after each group’s last injection, one final mechanical loading was performed to induce inflammation and MMP13 expression. Mice were sacrificed 12 h after the final mechanical loading. Hindlimbs were skinned and stored in RNAlater.
Muscle Editing Models.
In the initial intramuscular editing study, 20 μL of PSiNPs was injected into the tibialis anterior of healthy Ai9 mice at a concentration of 0.1 mg/mL Cargo (0.6 mg/mL PSiNP). Mice were sacrificed after 2 weeks and imaged on IVIS. In the subsequent tibialis anterior injections, the same protocol was followed, but dosage increased. For the quadriceps study, 40 μL of PSiNPs (3 mg/mL cargo, 18 mg/mL PSiNP concentration) was administered by direct injection. Fluorescence of tdTomato was imaged by IVIS and then legs were fixed and frozen for cryohistology.
For the systemic muscle-editing study, 20 μL of 1.2% BaCl2 was administered intramuscularly into the tibialis anterior to induce muscle damage and inflammation. 48 h after BaCl2 injury, 150 μL of treatment was administered by tail vein. The “Buffer” treatment was the PSiNP resuspension buffer alone, with no PSiNPs. PSiNPs were administered at a concentration of 0.1, 0.5, or 1.0 mg/mL cargo (0.6, 3.0, or 6.0 mg/mL PSiNP).
Histology for Knees and Muscle.
Histology was performed with the goal of maintaining tdTomato fluorescence while also keeping tissue morphology. 4% PFA was prepared from boiling paraformaldehyde. 10× PBS was added to create 1× PBS in the PFA solution. NaOH and/or HCl were added to adjust the pH to 7.4, and then the solution was sterile filtered and frozen at −20 °C for less than 6 months before use. After IVIS imaging, all legs were placed in the thawed ice-cold PFA for 2 h. Then, legs were placed in 50 mL tubes full of 4 °C 30% sucrose overnight until they sunk to the bottom of the tube.
Legs were then positioned in the OCT in cryomolds, frozen in liquid nitrogen, and stored at −80 °C. Before cryomicrotome sectioning, the OCT blocks were cut with a razor blade close to the desired location. Slices were collected with prolonged gold, coverslipped, and sealed with fingernail polish. The entire area with tissue on each slide was then imaged using a Nikon Ti-Eclipse/C1 Confocal using the large-image stitching automation program within the NIS Elements software.
IVIS Imaging.
IVIS images were acquired on an IVIS Lumina. For Figure 5 and Figure 6, images were acquired at 520 and 570 Ex/Em and analyzed using Living Image software. For Figure 8b and c, supply the highest quality data, hyperspectral imaging was performed across broad excitation/emission wavelengths. MATLAB code was written to analyze the images (github.com/brockfletcher). The script normalizes the fluorescence signal at an optimal wavelength for tdTomato expression to a nonoptimal tdTomato wavelength. This serves to remove noise and false positive signal, which may result from tissue autofluorescence or uneven lighting. For all figures, the quantitative data was acquired by placing identical ROIs centered on the tissue of interest, and the radiant efficiency measured.
Indel Detection by Amplicon Analysis (IDAA).
IDAA was performed following standard protocols published in Nature Protocols by Lonowski et al.82 Briefly, a triprimer PCR was performed (sequences in Table S2) using NEB Q5 universal master mix and two custom primers which surround the entire Ai9 deletion. Either the forward or reverse primer was extended to allow incorporation of a FAM-labeled universal primer. As prescribed, a 10:1:10 molar ratio of FamFwd:Fwd:Rev primer was used. Touchdown PCR was performed, starting at 72 °C, and then dropping by 1 °C per cycle to the melt temperature calculated for Q5 polymerase on the NEB Web site. ~30 cycles were then performed to result in a total of 35 PCR cycles, including the touchdown cycles. Importantly, extension times were set to 1 min (twice the necessary time for the 1.2 kb amplicon) to reduce potential amplicon size bias. Three different primer combinations were used to reduce any potential bias, specifically (1) Ai9_1.FWD and IDAA_Ai9_4.REV, (2) IDAA_Ai9_2.FWD and Ai9_5.REV, and (3) IDAA_Ai9_3.FWD and Ai9_5.REV. Primer sequences listed in Table S2.
DNA and RNA Extraction from Tissues.
Tissues were individually lysed and homogenized using a rotor-stator homogenizer for 10 s per samples, and visually inspected to ensure complete tissue destruction. DNA and RNA extractions were performed using the AllPrep Universal kit from Qiagen, following all steps as prescribed.
qPCR.
cDNA was formed from RNA using iScript Reverse Transcription Supermix (Bio-Rad), and qPCR was performed using Luna qPCR Mastermix (NEB) following the standard protocol. qPCR was performed with two control genes, ActB and Rpl13a, and one target gene, MMP13. Four technical replicates were performed for each gene. Primer sequences listed in Table S2. The entire process was repeated to confirm the knockdown results, and the two experiments were consistent. The graphed values represent the average of the two experiments.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the Vanderbilt Institute for Nanoscale Science and Engineering (VINSE) and the Vanderbilt High Throughput Screening Facility for technical support and equipment sharing. We acknowledge the National Institutes of Health (NIH) for funding provided under contracts R01CA224241, R38HL143619, and 1R21AR078636-01. R.B.F. acknowledges Biorender.com and Adobe Illustrator for figure prep.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.2c12261.
Tables of sgRNA and primer DNA sequences, NanoSight data, SEM images, electrostatic modeling, protein loading calculations, method-supporting data, additional in vivo biodistribution and gene editing data (PDF)
The authors declare no competing financial interest.
Contributor Information
R. Brock Fletcher, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37235-1631, United States.
Larry D. Stokes, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37235-1631, United States
Isom B. Kelly, III, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37235-1631, United States.
Katelyn M. Henderson, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37235-1631, United States
Isabel C. Vallecillo-Viejo, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37235-1631, United States
Juan M. Colazo, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37235-1631, United States
Benjamin V. Wong, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37235-1631, United States
Fang Yu, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37235-1631, United States.
Richard d’Arcy, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37235-1631, United States.
Morgan N. Struthers, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37235-1631, United States
Brian C. Evans, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37235-1631, United States
Jacob Ayers, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37235-1631, United States.
Matthew Castanon, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37235-1631, United States.
Michael J. Weirich, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37235-1631, United States
Sarah K. Reilly, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37235-1631, United States
Shrusti S. Patel, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37235-1631, United States
Yoanna I. Ivanova, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37235-1631, United States
Carlos A. Silvera Batista, Department of Chemical and Biomolecular Engineering, Vanderbilt University, Nashville, Tennessee 37235-1631, United States.
Sharon M. Weiss, Department of Electrical Engineering, Vanderbilt University, Nashville, Tennessee 37235-1631, United States
Charles A. Gersbach, Department of Biomedical Engineering, Duke University, Durham, North Carolina 27708, United States
Jonathan M. Brunger, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37235-1631, United States
Craig L. Duvall, Department of Biomedical Engineering, Vanderbilt University, Nashville, Tennessee 37235-1631, United States
REFERENCES
- (1).Jiang F; Doudna JA CRISPR-Cas9 Structures and Mechanisms. Annu. Rev. Biophys 2017, 46 (1), 505–529. [DOI] [PubMed] [Google Scholar]
- (2).Nelson CE; Gersbach CA Engineering Delivery Vehicles for Genome Editing. Annu. Rev. Chem. Biomol. Eng 2016, 7 (1), 637–662. [DOI] [PubMed] [Google Scholar]
- (3).González Castro N; Bjelic J; Malhotra G; Huang C; Alsaffar SH Comparison of the Feasibility, Efficiency, and Safety of Genome Editing Technologies. Int. J. Mol. Sci 2021, 22 (19), 10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Gaj T; Gersbach CA; Barbas CF ZFN, TALEN, and CRISPR/Cas-Based Methods for Genome Engineering. Trends Biotechnol. 2013, 31 (7), 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Mullard A. FDA Approves Fifth RNAi Drug — Alnylam’s next-Gen HATTR Treatment. Nat. Rev. Drug Discovery 2022, 21 (8), 548–549. [DOI] [PubMed] [Google Scholar]
- (6).Walsh EE; Frenck RW; Falsey AR; Kitchin N; Absalon J; Gurtman A; Lockhart S; Neuzil K; Mulligan MJ; Bailey R; Swanson KA; Li P; Koury K; Kalina W; Cooper D; Fontes-Garfias C; Shi P-Y; Türeci O; Tompkins KR; Lyke KE; Raabe V; Dormitzer PR; Jansen KU; Şahin U; Gruber WC Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med 2020, 383 (25), 2439–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Jackson LA; Anderson EJ; Rouphael NG; Roberts PC; Makhene M; Coler RN; McCullough MP; Chappell JD; Denison MR; Stevens LJ; Pruijssers AJ; McDermott A; Flach B ; Doria-Rose NA; Corbett KS; Morabito KM; O’Dell S; Schmidt SD; Swanson PA; Padilla M; Mascola JR; Neuzil KM; Bennett H; Sun W; Peters E; Makowski M; Albert J; Cross K; Buchanan W; Pikaart-Tautges R; Ledgerwood JE; Graham BS; Beigel JH An MRNA Vaccine against SARS-CoV-2 — Preliminary Report. N. Engl. J. Med 2020, 383 (20), 1920–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Kenjo E; Hozumi H; Makita Y; Iwabuchi KA; Fujimoto N; Matsumoto S; Kimura M; Amano Y; Ifuku M; Naoe Y; Inukai N; Hotta A Low Immunogenicity of LNP Allows Repeated Administrations of CRISPR-Cas9MRNA into Skeletal Muscle in Mice. Nat. Commun 2021, 12 (1), 7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).He X; Urip BA; Zhang Z; Ngan CC; Feng B Evolving AAV-Delivered Therapeutics towards Ultimate Cures. J. Mol. Med 2021, 99 (5), 593–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Paunovska K; Loughrey D; Dahlman JE Drug Delivery Systems for RNA Therapeutics. Nat. Rev. Genet 2022, 23 (5), 265–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Wei T; Cheng Q; Min Y-L; Olson EN; Siegwart DJ Systemic Nanoparticle Delivery of CRISPR-Cas9 Ribonucleoproteins for Effective Tissue Specific Genome Editing. Nat. Commun 2020, 11 (1), 3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Liang X; Potter J; Kumar S; Zou Y; Quintanilla R; Sridharan M; Carte J; Chen W; Roark N; Ranganathan S; Ravinder N; Chesnut JD Rapid and Highly Efficient Mammalian Cell Engineering via Cas9 Protein Transfection. J. Biotechnol 2015, 208, 44–53. [DOI] [PubMed] [Google Scholar]
- (13).Cheng Q; Wei T; Farbiak L; Johnson LT; Dilliard SA; Siegwart DJ Selective Organ Targeting (SORT) Nanoparticles for Tissue-Specific MRNA Delivery and CRISPR-Cas Gene Editing. Nat. Nanotechnol 2020, 15 (4), 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Tenjo-Castaño F; Montoya G; Carabias A Transposons and CRISPR: Rewiring Gene Editing. Biochemistry 2022, DOI: 10.1021/acs.biochem.2c00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Li W; Liu Z; Fontana F; Ding Y; Liu D; Hirvonen JT; Santos HA Tailoring Porous Silicon for Biomedical Applications: From Drug Delivery to Cancer Immunotherapy. Adv. Mater 2018, 30 (24), 1703740. [DOI] [PubMed] [Google Scholar]
- (16).Qin Z; Joo J; Gu L; Sailor MJ Size Control of Porous Silicon Nanoparticles by Electrochemical Perforation Etching. Part. Part. Syst. Charact 2014, 31 (2), 252–256. [Google Scholar]
- (17).Tieu T; Wojnilowicz M; Huda P; Thurecht KJ; Thissen H; Voelcker NH; Cifuentes-Rius A Nanobody-Displaying Porous Silicon Nanoparticles for the Co-Delivery of SiRNA and Doxorubicin. Biomater. Sci 2021, 9 (1), 133–147. [DOI] [PubMed] [Google Scholar]
- (18).Luo M; Lewik G; Ratcliffe JC; Choi CHJ; Mäkilä E; Tong WY; Voelcker NH Systematic Evaluation of Transferrin-Modified Porous Silicon Nanoparticles for Targeted Delivery of Doxorubicin to Glioblastoma. ACS Appl. Mater. Interfaces 2019, 11 (37), 33637–33649. [DOI] [PubMed] [Google Scholar]
- (19).Xia B; Zhang Q; Shi J; Li J; Chen Z; Wang B Co-Loading of Photothermal Agents and Anticancer Drugs into Porous Silicon Nanoparticles with Enhanced Chemo-Photothermal Therapeutic Efficacy to Kill Multidrug-Resistant Cancer Cells. Colloids Surf. B Biointerfaces 2018, 164, 291–298. [DOI] [PubMed] [Google Scholar]
- (20).Shahbazi M-A; Almeida PV; Correia A; Herranz-Blanco B ; Shrestha N; Mäkilä E; Salonen J; Hirvonen J; Santos HA Intracellular Responsive Dual Delivery by Endosomolytic Polyplexes Carrying DNA Anchored Porous Silicon Nanoparticles. J. Controlled Release 2017, 249, 111–122. [DOI] [PubMed] [Google Scholar]
- (21).Tieu T; Dhawan S; Haridas V; Butler LM; Thissen H; Cifuentes-Rius A; Voelcker NH Maximizing RNA Loading for Gene Silencing Using Porous Silicon Nanoparticles. ACS Appl. Mater. Interfaces 2019, 11 (26), 22993–23005. [DOI] [PubMed] [Google Scholar]
- (22).Kim B; Sun S; Varner JA; Howell SB; Ruoslahti E; Sailor MJ Securing the Payload, Finding the Cell, and Avoiding the Endosome: Peptide-Targeted, Fusogenic Porous Silicon Nanoparticles for Delivery of SiRNA. Adv. Mater 2019, 31 (35), 1902952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Kim B; Park J-H; Sailor MJ Rekindling RNAi Therapy: Materials Design Requirements for In Vivo SiRNA Delivery. Adv. Mater 2019, 31 (49), 1903637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Tong WY; Alnakhli M; Bhardwaj R; Apostolou S; Sinha S; Fraser C; Kuchel T; Kuss B; Voelcker NH Delivery of SiRNA in Vitro and in Vivo Using PEI-Capped Porous Silicon Nanoparticles to Silence MRP1 and Inhibit Proliferation in Glioblastoma. J. Nanobiotechnology 2018, 16 (1), 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Neri M; Kang J; Zuidema JM; Gasparello J; Finotti A; Gambari R; Sailor MJ; Bertucci A; Corradini R Tuning the Loading and Release Properties of MicroRNA-Silencing Porous Silicon Nanoparticles by Using Chemically Diverse Peptide Nucleic Acid Payloads. ACS Biomater. Sci. Eng 2022, 8 (10), 4123–4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Beavers KR; Werfel TA; Shen T; Kavanaugh TE; Kilchrist KV; Mares JW; Fain JS; Wiese CB; Vickers KC; Weiss SM; Duvall CL Porous Silicon and Polymer Nanocomposites for Delivery of Peptide Nucleic Acids as Anti-MicroRNA Therapies. Adv. Mater 2016, 28 (36), 7984–7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Kelly IB; Fletcher RB; McBride JR; Weiss SM; Duvall CL Tuning Composition of Polymer and Porous Silicon Composite Nanoparticles for Early Endosome Escape of Anti-MicroRNA Peptide Nucleic Acids. ACS Appl. Mater. Interfaces 2020, 12 (35), 39602–39611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Waggoner LE; Kang J; Zuidema JM; Vijayakumar S; Hurtado AA; Sailor MJ; Kwon EJ Porous Silicon Nanoparticles Targeted to the Extracellular Matrix for Therapeutic Protein Delivery in Traumatic Brain Injury. Bioconjugate Chem. 2022, 33 (9), 1685–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Kumeria T; Wang J; Kim B; Park J-H; Zuidema JM; Klempner M; Cavacini L; Wang Y; Sailor MJ Enteric Polymer-Coated Porous Silicon Nanoparticles for Site-Specific Oral Delivery of IgA Antibody. ACS Biomater. Sci. Eng 2022, 8 (10), 4140–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Qi S; Zhang P; Ma M; Yao M; Wu J; Mäkilä E; Salonen J; Ruskoaho H; Xu Y; Santos HA; Zhang H Cellular Internalization-Induced Aggregation of Porous Silicon Nanoparticles for Ultrasound Imaging and Protein-Mediated Protection of Stem Cells. Small 2019, 15 (1), 1804332. [DOI] [PubMed] [Google Scholar]
- (31).Turner CT; McInnes SJP; Melville E; Cowin AJ; Voelcker NH Delivery of Flightless I Neutralizing Antibody from Porous Silicon Nanoparticles Improves Wound Healing in Diabetic Mice. Adv. Healthc. Mater 2017, 6 (2), 1600707. [DOI] [PubMed] [Google Scholar]
- (32).Chaix A; Cueto-Diaz E; Delalande A; Knezevic N; Midoux P; Durand J-O; Pichon C; Cunin F Amino-Acid Functionalized Porous Silicon Nanoparticles for the Delivery of PDNA. RSC Adv. 2019. 9 (55), 31895–31899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Wareing N; Szymanski K; Akkaraju GR; Loni A; Canham LT; Gonzalez-Rodriguez R; Coffer JL In Vitro Gene Delivery with Large Porous Silicon Nanoparticles Fabricated Using Cost-Effective, Metal-Assisted Chemical Etching. Small 2017, 13 (3), 1602739. [DOI] [PubMed] [Google Scholar]
- (34).LaBauve AE; Saada EA; Jones IKA; Mosesso R; Noureddine A; Techel J; Gomez A; Collette N; Sherman MB; Serda RE; Butler KS; Brinker CJ; Schoeniger JS; Sasaki D; Negrete OA Lipid-Coated Mesoporous Silica Nanoparticles for Anti-Viral Applications via Delivery of CRISPR-Cas9 Ribonucleoproteins. Sci. Rep 2023, 13 (1), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Wang Y; Shahi PK; Wang X; Xie R; Zhao Y; Wu M; Roge S; Pattnaik BR; Gong S In Vivo Targeted Delivery of Nucleic Acids and CRISPR Genome Editors Enabled by GSH-Responsive Silica Nanoparticles. J. Controlled Release 2021, 336, 296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Wang Y; Shahi PK; Xie R; Zhang H; Abdeen AA; Yodsanit N; Ma Z; Saha K; Pattnaik BR; Gong S A PH-Responsive Silica-Metal-Organic Framework Hybrid Nanoparticle for the Delivery of Hydrophilic Drugs, Nucleic Acids, and CRISPR-Cas9 Genome-Editing Machineries. J. Controlled Release 2020, 324, 194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Wang Y; Wang X; Xie R; Burger JC; Tong Y; Gong S Overcoming the Blood-Brain Barrier for Gene Therapy via Systemic Administration of GSH-Responsive Silica Nanocapsules. Adv. Mater 2023, 35 (6), 2208018. [DOI] [PubMed] [Google Scholar]
- (38).Salekdeh PR; Ma’mani L; Tavakkoly-Bazzaz J; Mousavi H; Modarressi MH; Salekdeh GH Bi-Functionalized Amino-guanidine-PEGylated Periodic Mesoporous Organosilica Nanoparticles: A Promising Nanocarrier for Delivery of Cas9-SgRNA Ribonucleoproteine. J. Nanobiotechnology 2021, 19 (1), 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Noureddine A; Maestas-Olguin A; Saada EA; LaBauve AE; Agola JO; Baty KE; Howard T; Sabo JK; Espinoza CRS; Doudna JA; Schoeniger JS; Butler KS; Negrete OA; Brinker CJ; Serda RE Engineering of Monosized Lipid-Coated Mesoporous Silica Nanoparticles for CRISPR Delivery. Acta Biomater. 2020, 114, 358–368. [DOI] [PubMed] [Google Scholar]
- (40).Gongalsky MB; Sviridov AP; Bezsudnova Yu. I.; Osminkina LA Biodegradation Model of Porous Silicon Nanoparticles. Colloids Surf. B Biointerfaces 2020, 190, 110946. [DOI] [PubMed] [Google Scholar]
- (41).Jung Y; Huh Y; Kim D Recent Advances in Surface Engineering of Porous Silicon Nanomaterials for Biomedical Applications. Microporous Mesoporous Mater. 2021, 310, 110673. [Google Scholar]
- (42).Jung Y; Kim D Recent Advances in Hybrid System of Porous Silicon Nanoparticles and Biocompatible Polymers for Biomedical Applications. Biomed. Eng. Lett 2021, 11 (3), 171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Park J-H; Gu L; von Maltzahn G; Ruoslahti E; Bhatia SN; Sailor MJ Biodegradable Luminescent Porous Silicon Nanoparticles for in Vivo Applications. Nat. Mater 2009, 8 (4), 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Gu L; Hall DJ; Qin Z; Anglin E; Joo J; Mooney DJ; Howell SB; Sailor MJ In Vivo Time-Gated Fluorescence Imaging with Biodegradable Luminescent Porous Silicon Nanoparticles. Nat. Commun 2013, 4 (1), 2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Kim J; Jo D; Yang S-H; Joo C-G; Whiting N; Pudakalakatti S; Seo H; Son HY; Min S-J; Bhattacharya P; Huh Y-M; Shim JH; Lee Y 29Si Isotope-Enriched Silicon Nanoparticles for an Efficient Hyperpolarized Magnetic Resonance Imaging Probe. ACS Appl. Mater. Interfaces 2021, 13 (48), 56923–56930. [DOI] [PubMed] [Google Scholar]
- (46).Seo H; Choi I; Whiting N; Hu J; Luu QS; Pudakalakatti S; McCowan C; Kim Y; Zacharias N; Lee S; Bhattacharya P; Lee Y Hyperpolarized Porous Silicon Nanoparticles: Potential Theragnostic Material for 29Si Magnetic Resonance Imaging. ChemPhysChem 2018, 19 (17), 2143–2147. [DOI] [PubMed] [Google Scholar]
- (47).Jin JH; Um H; Oh JH; Huh Y; Jung Y; Kim D Gadolinium Silicate-Coated Porous Silicon Nanoparticles as an MRI Contrast Agent and Drug Delivery Carrier. Mater. Chem. Phys 2022, 287, 126345. [Google Scholar]
- (48).Kang J; Kim D; Wang J; Han Y; Zuidema JM; Hariri A; Park J-H; Jokerst JV; Sailor MJ Enhanced Performance of a Molecular Photoacoustic Imaging Agent by Encapsulation in Mesoporous Silicon Nanoparticles. Adv. Mater 2018, 30 (27), 1800512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Osminkina LA; Timoshenko VY Porous Silicon as a Sensitizer for Biomedical Applications. Open Mater. Sci 2016, DOI: 10.1515/mesbi-2016-0005. [DOI] [Google Scholar]
- (50).Nelson CE; Kintzing JR; Hanna A; Shannon JM; Gupta MK; Duvall CL Balancing Cationic and Hydrophobic Content of PEGylated SiRNA Polyplexes Enhances Endosome Escape, Stability, Blood Circulation Time, and Bioactivity in Vivo. ACS Nano 2013, 7 (10), 8870–8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Werfel TA; Jackson MA; Kavanaugh TE; Kirkbride KC; Miteva M; Giorgio TD; Duvall C Combinatorial Optimization of PEG Architecture and Hydrophobic Content Improves Ternary SiRNA Polyplex Stability, Pharmacokinetics, and Potency in Vivo. J. Controlled Release 2017, 255, 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Kang J; Joo J; Kwon EJ; Skalak M; Hussain S; She Z-G; Ruoslahti E; Bhatia SN; Sailor MJ Self-Sealing Porous Silicon-Calcium Silicate Core-Shell Nanoparticles for Targeted SiRNA Delivery to the Injured Brain. Adv. Mater 2016, 28 (36), 7962–7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Wang J; Kumeria T; Bezem MT; Wang J; Sailor MJ Self-Reporting Photoluminescent Porous Silicon Microparticles for Drug Delivery. ACS Appl. Mater. Interfaces 2018, 10 (4), 3200–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Hyung Kang R; Oh Jung K; Kim D Self-Sealing Chemistry of Calcium/Magnesium Silicate on Porous Silicon Nanoparticles for Enhanced Drug-Loading and Slowed Drug-Releasing. Mater. Lett 2022, 324, 132719. [Google Scholar]
- (55).Butt HJ; Kappl M Electrostatic Double-Layer Forces. In Surface and Interfacial Forces; John Wiley & Sons, Ltd., 2010; pp 93–125. . [Google Scholar]
- (56).Mughal A; Chan HK; Weaire D; Hutzler S Dense Packings of Spheres in Cylinders: Simulations. Phys. Rev. E 2012, 85 (5), 051305. [DOI] [PubMed] [Google Scholar]
- (57).Jain NK; Roy I Trehalose and Protein Stability. Curr. Protoc. Protein Sci 2010, 59 (1), 4.9.1–4.9.12. [DOI] [PubMed] [Google Scholar]
- (58).Henrickson A; Kulkarni JA; Zaifman J; Gorbet GE; Cullis PR; Demeler B Density Matching Multi-Wavelength Analytical Ultracentrifugation to Measure Drug Loading of Lipid Nanoparticle Formulations. ACS Nano 2021, 15 (3), 5068–5076. [DOI] [PubMed] [Google Scholar]
- (59).Yu X; Liang X; Xie H; Kumar S; Ravinder N; Potter J; de Mollerat du Jeu X; Chesnut JD Improved Delivery of Cas9 Protein/GRNA Complexes Using Lipofectamine CRISPRMAX. Biotechnol. Lett 2016, 38 (6), 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Sviridov AP; Osminkina LA; Nikolaev AL; Kudryavtsev AA; Vasiliev AN; Timoshenko V. Yu. Lowering of the Cavitation Threshold in Aqueous Suspensions of Porous Silicon Nanoparticles for Sonodynamic Therapy Applications. Appl. Phys. Lett 2015, 107 (12), 123107. [Google Scholar]
- (61).Patel SS Core polymer optimization of ternary siRNA nanoparticles enhances in vivo safety, pharmacokinetics, and tumor gene silencing. Biomaterials 2023, 297, 122098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Kilchrist KV; Dimobi SC; Jackson MA; Evans BC; Werfel TA; Dailing EA; Bedingfield SK; Kelly IB; Duvall CL Gal8 Visualization of Endosome Disruption Predicts Carrier-Mediated Biologic Drug Intracellular Bioavailability. ACS Nano 2019, 13 (2), 1136–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Yang F; Liu C; Chen D; Tu M; Xie H; Sun H; Ge X; Tang L; Li J; Zheng J; Song Z; Qu J; Gu F CRISPR/Cas9-LoxP-Mediated Gene Editing as a Novel Site-Specific Genetic Manipulation Tool. Mol. Ther. - Nucleic Acids 2017, 7, 378–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Chen F; Alphonse M; Liu Q Strategies for Nonviral Nanoparticle-Based Delivery of CRISPR/Cas9 Therapeutics. WIREs Nanomedicine Nanobiotechnology 2020, 12 (3), No. e1609. [DOI] [PubMed] [Google Scholar]
- (65).Bedingfield SK; Colazo JM; Yu F; Liu DD; Jackson MA; Himmel LE; Cho H; Crofford LJ; Hasty KA; Duvall CL Amelioration of Post-Traumatic Osteoarthritis via Nanoparticle Depots Delivering Small Interfering RNA to Damaged Cartilage. Nat. Biomed. Eng 2021, 5 (9), 1069–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Bedingfield SK; Colazo JM; Di Francesco M; Yu F; Liu DD; Di Francesco V; Himmel LE; Gupta MK; Cho H; Hasty KA; Decuzzi P; Duvall CL Top-Down Fabricated MicroPlates for Prolonged, Intra-Articular Matrix Metalloproteinase 13 SiRNA Nanocarrier Delivery to Reduce Post-Traumatic Osteoarthritis. ACS Nano 2021, 15 (9), 14475–14491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Hu Q; Ecker M Overview of MMP-13 as a Promising Target for the Treatment of Osteoarthritis. Int. J. Mol. Sci 2021, 22 (4), 1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Liang Y; Xu X; Xu L; Iqbal Z; Ouyang K; Zhang H; Wen C; Duan L; Xia J Chondrocyte-Specific Genomic Editing Enabled by Hybrid Exosomes for Osteoarthritis Treatment. Theranostics 2022, 12 (11), 4866–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Zhao L; Huang J; Fan Y; Li J; You T; He S; Xiao G; Chen D Exploration of CRISPR/Cas9-Based Gene Editing as Therapy for Osteoarthritis. Ann. Rheum. Dis 2019, 78 (5), 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Colazo JM; Hoogenboezem EN; Shah V; Yu F; Lo JH; Sorets AG; Francini N; Cho H; Michell DL; Vickers KC; Gibson-Corley KN; Hasty KA; Crofford L; Cook RS; Duvall CL Albumin-Binding RNAi Conjugate for Carrier Free Treatment of Arthritis. bioRxiv, June 8, 2023. (accessed June 14, 2023). [Google Scholar]
- (71).Choi E; Koo T CRISPR Technologies for the Treatment of Duchenne Muscular Dystrophy. Mol. Ther 2021, 29 (11), 3179–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Nelson CE; Wu Y; Gemberling MP; Oliver ML; Waller MA; Bohning JD; Robinson-Hamm JN; Bulaklak K; Castellanos Rivera RM; Collier JH; Asokan A; Gersbach CA Long-Term Evaluation of AAV-CRISPR Genome Editing for Duchenne Muscular Dystrophy. Nat. Med 2019, 25 (3), 427–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Nelson CE; Hakim CH; Ousterout DG; Thakore PI; Moreb EA; Rivera RMC; Madhavan S; Pan X; Ran FA; Yan WX; Asokan A; Zhang F; Duan D; Gersbach CA In Vivo Genome Editing Improves Muscle Function in a Mouse Model of Duchenne Muscular Dystrophy. Science 2016, 351 (6271), 403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).FDA Briefing Document Delandistrogene Moxeparvovec Sarepta Therapeutics, Inc. Cellular, Tissue and Gene Therapies Advisory Committee Meeting, 2023. https://www.fda.gov/media/168021/download.
- (75).Tabebordbar M; Zhu K; Cheng JKW; Chew WL; Widrick JJ; Yan WX; Maesner C; Wu EY; Xiao R; Ran FA; Cong L; Zhang F; Vandenberghe LH; Church GM; Wagers AJ In Vivo Gene Editing in Dystrophic Mouse Muscle and Muscle Stem Cells. Science 2016, 351 (6271), 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Kwon JB; Ettyreddy AR; Vankara A; Bohning JD; Devlin G; Hauschka SD; Asokan A; Gersbach CA In Vivo Gene Editing of Muscle Stem Cells with Adeno-Associated Viral Vectors in a Mouse Model of Duchenne Muscular Dystrophy. Mol. Ther. - Methods Clin. Dev 2020, 19, 320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Pessina P; Cabrera D; Morales MG; Riquelme CA; Gutiérrez J; Serrano AL; Brandan E; Muñoz-Cánoves P Novel and Optimized Strategies for Inducing Fibrosis in Vivo: Focus on Duchenne Muscular Dystrophy. Skelet. Muscle 2014, 4 (1), 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Prakash S; Zambrano HA; Rangharajan KK; Rosenthal-Kim E; Vasquez N; Conlisk AT Electrokinetic Transport of Monovalent and Divalent Cations in Silica Nanochannels. Microfluid. Nanofluidics 2016, 20 (1), 8. [Google Scholar]
- (79).Muzumdar MD; Tasic B; Miyamichi K; Li L; Luo L A Global Double-Fluorescent Cre Reporter Mouse. genesis 2007, 45 (9), 593–605. [DOI] [PubMed] [Google Scholar]
- (80).Madisen L; Zwingman TA; Sunkin SM; Oh SW; Zariwala HA; Gu H; Ng LL; Palmiter RD; Hawrylycz MJ; Jones AR; Lein ES; Zeng H A Robust and High-Throughput Cre Reporting and Characterization System for the Whole Mouse Brain. Nat. Neurosci 2010, 13 (1), 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Kim S; Stébé M-J; Blin J-L; Pasc A PH-Controlled Delivery of Curcumin from a Compartmentalized Solid Lipid Nanoparticle@mesostructured Silica Matrix. J. Mater. Chem. B 2014, 2 (45), 7910–7917. [DOI] [PubMed] [Google Scholar]
- (82).Bennett EP; Petersen BL; Johansen IE; Niu Y; Yang Z; Chamberlain CA; Met O; Wandall HH; Frödin M INDEL Detection, the ‘Achilles Heel’ of Precise Genome Editing: A Survey of Methods for Accurate Profiling of Gene Editing Induced Indels. Nucleic Acids Res. 2020, 48 (21), 11958–11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.