Abstract
South Asia, making up around 25% of the world’s population, encompasses a wide range of individuals with tremendous genetic and environmental diversity. This region, which spans eight countries, is home to over 4,500 anthropologically defined groups that speak numerous languages and have an array of religious beliefs and cultures, making it one of the most diverse places in the world. Much of the region’s rich genetic diversity and structure is the result of a complex combination of population history, migration patterns, and endogamous practices. Despite the overwhelming size and diversity, South Asians have often been underrepresented in genetic research, making up less than 2% of the participants in genetic studies. This has led to a lack of population specific understanding of genetic disease risks. We aim to raise awareness about underlying genetic diversity in this ancestry group, call attention to the lack of representation of the group, and to highlight strategies for future studies in South Asians.
Keywords: South Asian, Diversity, Equity, Inclusion
Introduction
South Asia is home to nearly two billion individuals, eight countries, hundreds of languages, and the largest populations of five of the twelve major world religions (Hindus, Muslims, Sikhs, Jains, and Zoroastrians). However, despite making up about 25% of the world’s population, the South Asian ancestry group has been severely underrepresented in genetic research overall, making up less than 2% of human study participants in genome wide association studies (GWAS) (Morales et al. 2018). This lack of representation and inclusion has critical consequences for genetic applications in this population, equity in genomic medicine, and for the generalizability and advancement of genetic research.
Although numerous papers have discussed the need for diversity in human genetic research in general (Li and Keating 2014; Popejoy and Fullerton 2016; Bentley et al. 2017; Sirugo et al. 2019), occasionally championing the inclusion of South Asian participants specifically (Chambers et al. 2014; Nakatsuka et al. 2017), South Asians continue to be one of the least represented groups in human genetics overall and in behavior genetics specifically. Further exacerbating these representational disparities, researchers often fail to explicate the degree of South Asian missingness across papers and genetic repositories by using inappropriate labels based on continentally derived racial categories (such as “Asian”) rather than on stringent, genetically informative ancestral categories (such as “South Asian” or “East Asian”) (Popejoy and Fullerton 2016; Sirugo et al. 2019). Across the spectrum of single ancestry GWAS, trans-ethnic GWAS, cross-ancestry GWAS, and papers calling for diversity in GWAS, the South Asian ancestry group has been continually overlooked in the data and discussions about the representativity in the data. This article aims to raise awareness about research already done in this ancestry group, to emphasize the scientific and ethical importance of including South Asian populations in genetic research, and to provide avenues for better inclusion in the future.
Who are South Asians?
South Asia, as defined by the United Nations, includes Afghanistan (sometimes considered part of Central Asia or the Middle East/Southwest Asia), Bangladesh, Bhutan, India, Iran (frequently considered a part of the Middle East or Western Asia), the Maldives, Nepal, Pakistan, and Sri Lanka (United Nations (UN) 2017). Home to three of the ten most populous countries in the world, South Asia is the most populated sub-region within Asia (and, by extension, the world). In addition to the massive population within the sub-continent, the South Asian diaspora, comprising nearly 44 million individuals outside of the region, is one of the fastest-growing groups worldwide (United Nations Publications 2021). With substantial populations in over 30 countries, South Asians currently make up the largest ethnic minority groups in Canada and the UK (Rangaswamy 2005).
The enormity of this population means that South Asians account for a disproportionately high degree of global burden of disease, even when the prevalence estimates for a given condition are lower in South Asia than what is observed in other populations. For example, age is a primary risk factor for many diseases (MacNee et al. 2014). By 2050, the number of individuals aged 65 and older in India alone will exceed 200 million people, making up about 14% of its total population; meanwhile, despite having a higher proportion of individuals of the same age range (about 21%), only about 84 million individuals in the US will be over the age of 65 by the same year (He et al. 2016). The incomparability of prevalence and resultant number of affected individuals is major reason to increase South Asian inclusion in future studies. Without a proper characterization of the risk factors impacting the largest sub population in the world, the global impact of most research is going to be largely limited and disparate.
Migration History
The first modern humans (Homo sapiens) out of Africa migrated to South Asia approximately 60,000 to 50,000 years ago (Teixeira and Cooper 2019) (Figure 1). In the following 20,000 years, individuals from this wave migrated south towards Sri Lanka, crossing the Indian Ocean, and peopling the Andaman and Nicobar Islands (Tamang et al. 2012). Secondary migrations into South Asia likely include at least two main sets of migrations that are largely responsible for the admixture observed in contemporary South Asian populations. The first migration from the Iranian plateau occurred approximately 12,000 years ago (Figure 1). An analysis of an ancient Harappan–otherwise known as the mature Indus Valley Civilization (IVC)--genome suggests a genetic gradient resulting from gene flow between the Ancient Iranians and Andaman Hunter Gatherers (AHG), the early inhabitants of South Asia (Shinde et al. 2019).
Figure 1.
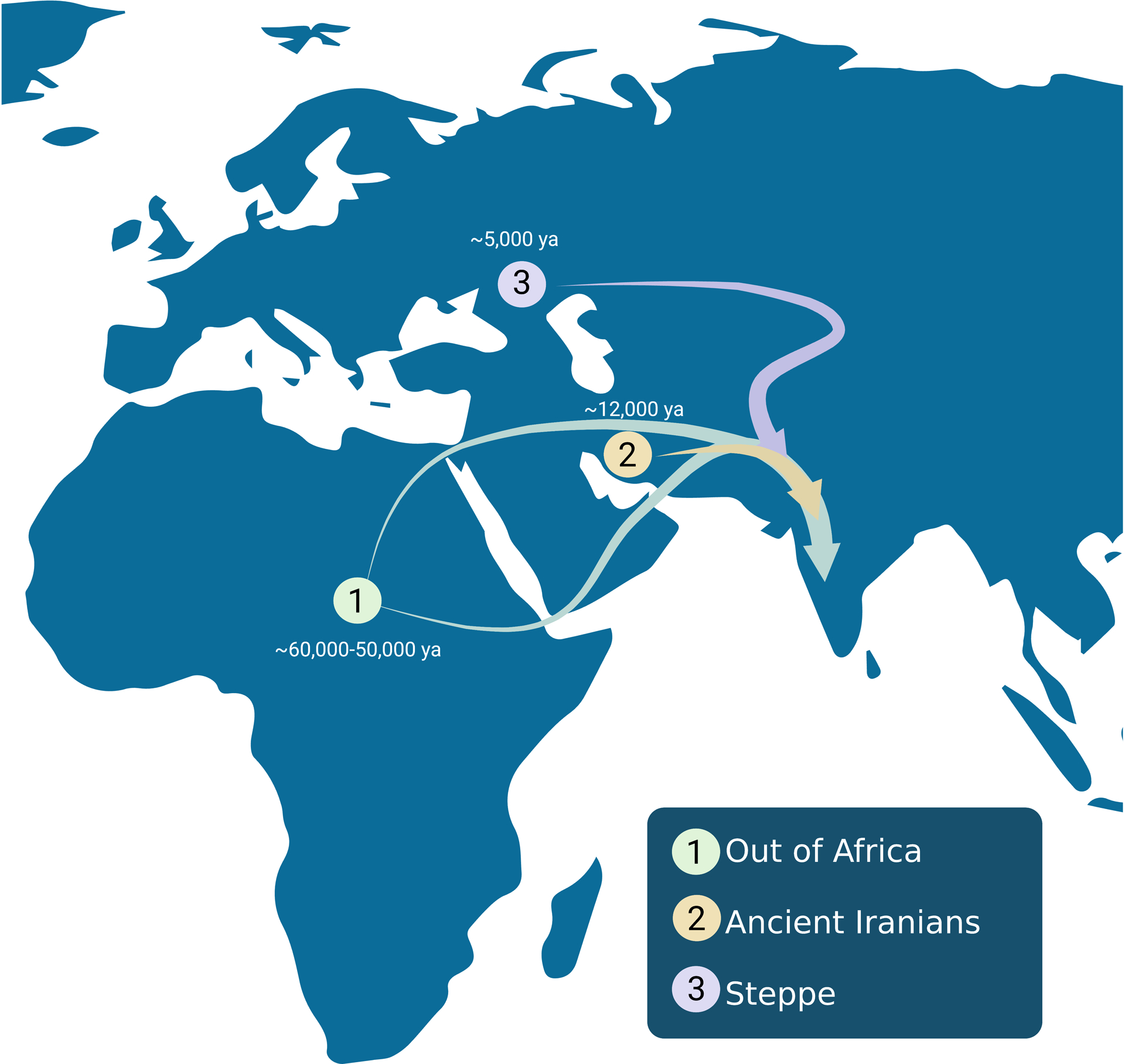
Major Migrations into South Asia. The first major migration into South Asia was the out of Africa migration estimated to be around 60,000 to 50,000 years ago that peopled the Indian subcontinent. The second main migration occurred nearly 12,000 years ago with the arrival of individuals from the Iranian plateau. The third major migration into the subcontinent was the influx of the Steppe after the decline of the Indus Valley Civilization approximately 5,000 years ago.
The second major migration began about 5,000 years ago with the arrival of eastern-moving Yamnaya Steppe people, who were likely responsible for introducing Proto-Indo-Aryan language to South Asia (Figure 1). By about 2000 BCE, the ensuing mixture of the post-IVC population with AHG in the South gave rise to the Ancestral South Indian (ASI) population, while gene flow between the post-IVC and the Yamnaya Steppe group in the North resulted in the Ancestral North Indian (ANI) population (Narasimhan et al. 2019). The following 2 to 3,000 years included a high degree of movement and gene flow across the subcontinent, and as a result, mainland South Asians today are an admixed population with varying proportions of all three ancestries (AHG, Ancient Iranians, and the Steppe) (Shinde et al. 2019). Additional models hypothesize that Ancestral Austro-Asiatic individuals from Southeast Asia and Ancestral Tibeto-Burman individuals fromEast Asia and/or Southeast Asia likely migrated into eastern South Asia in the last 10,000 years, but additional work is needed to accurately determine the exact migratory patterns that occurred in this region (Basu et al. 2016).
Language as a Catalyst of Diversity
In the years following the Steppe migration, population substructures in South Asia co-evolved with the development of language in the area (Moorjani et al. 2013). Languages in South Asia belong to at least seven linguistics families. Understanding the evolution of language in this region is vital to understanding the population structure that has resulted likely due to potential social homogamy based on language (Nagoshi et al. 1990; Moorjani et al. 2013; Metspalu et al. 2018; GenomeAsia100K Consortium 2019). Figure 2 (see below) demonstrates how four of South Asia’s major linguistic families: Indo-European, Dravidian, Austro-Asiatic, and Tibeto-Burman, map onto genetic clusters of modern-day Indians (Metspalu et al. 2011). Currently, most of the genetic literature studying South Asians focuses largely on Indo-European and Dravidian populations due to the sociolinguistic groups that happen to be represented in existing samples and reference panels (Cavalli-Sforza 2005; International HapMap 3 Consortium et al. 2010; 1000 Genomes Project Consortium et al. 2015; Taliun et al. 2021). This lack of sufficiently representative samples from other South Asian groups demonstrates just one way in which the current approximation of the variation in the human genome is largely incomplete (GenomeAsia100K Consortium 2019).
Figure 2.
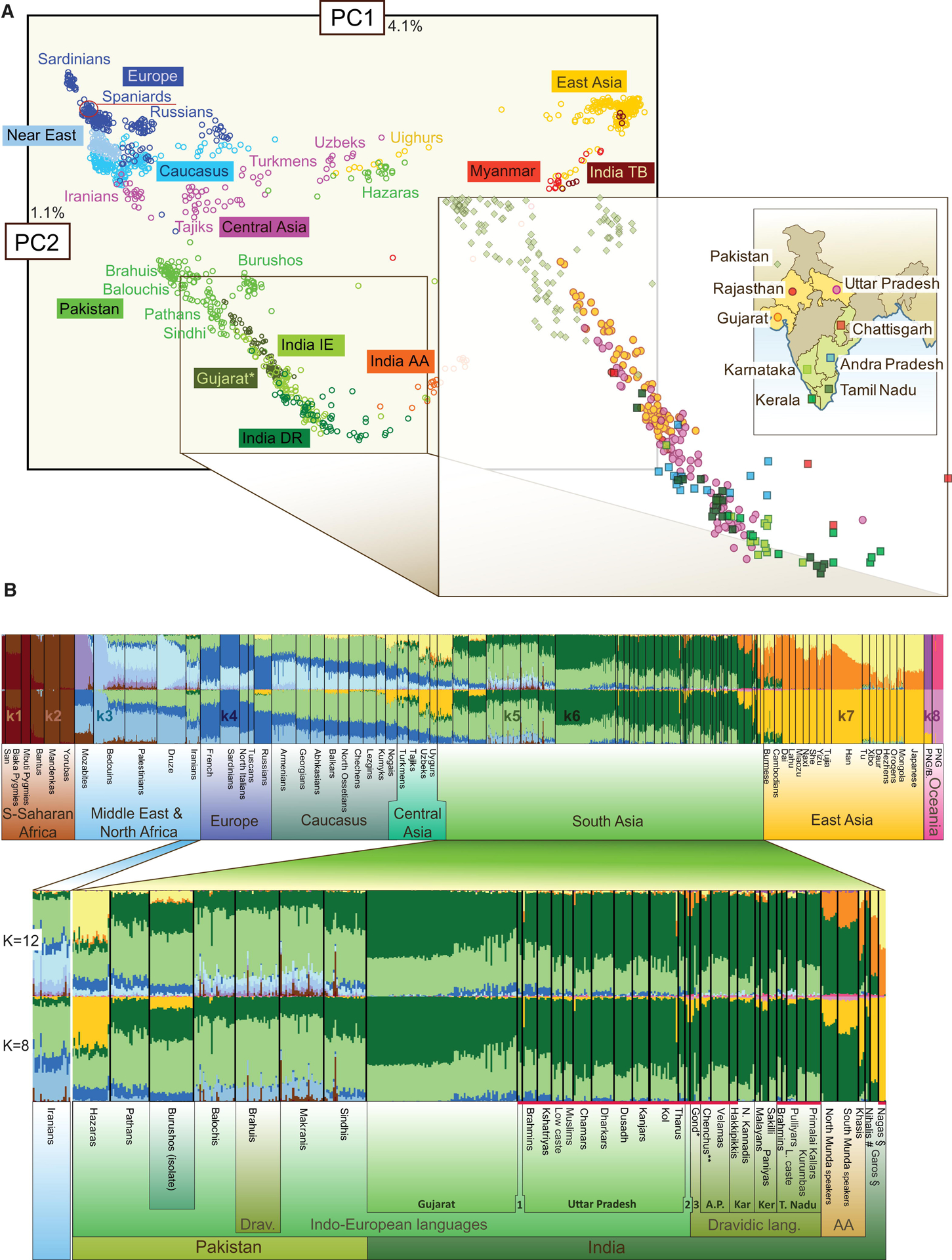
Reprinted from Metspalu 2011 with permission from Elsevier. Principal component analysis (PCA) of the Eurasian populations. The following abbreviations are used: IE, Indo European speakers; DR, Dravidic speakers; AA, Austroasiatic speakers; TB, Tibeto Burman speakers;?using data from Hapmap
Evolution of Social Structure
Along with language, the rise of complex social structure has completely transformed the cultural landscape of South Asia in the last few millennia (Mastana 2014). The social classification system, colloquially known as caste (originating from the Portuguese word “casta” meaning lineage) refers to a sociological construct that identifies groups that interact socially and economically but are separated by the practice of endogamy, marrying within a select group (Berreman 1960). While the presence of social structures likely date back as far as the Indus Valley Civilization (Sen 1992), the practice of endogamy likely began around third century AD (Moorjani et al. 2013).
In addition to endogamy practices, consanguinity patterns can also have important implications for population substructure and kinship analyses (Arciero et al. 2021; Wall et al. 2023). Differing customs around intrafamilial marriage has resulted in heterogeneity of consanguinity trends across sub-regions and religions (Bittles and Black 2010; Hamamy 2012; Acharya and Sahoo 2021; Iqbal et al. 2022). Within the context of South Asia’s complex history, the cultural practices around endogamy and consanguinity are just one element that profoundly impacts the diverse environmental and genetic patterns observed in South Asians.
Genetic Diversity
Today, the various influences of language, caste, and cultural history have resulted in over 4,500 anthropologically defined groups in South Asia (Mastana 2014). Together, these groups constitute a complex gradient both culturally and genetically. Nakatsuka et al. (2017) assessed 263 of these unique South Asian groups and found that 81 of these groups showed higher identity-by-descent (IBD) scores, a measure of stronger founder event strength, than the Finnish and Ashkenazi Jews--two populations that also have a history of strong founder events. A similar follow-up analysis performed by the GenomeAsia consortium using various groups across South Asia and other groups around the world showed that some South Asian groups demonstrate amongst the strongest genetic founder effects identified (GenomeAsia100K Consortium 2019).
While founder effects may have minimal impact on high frequency common variants, alleles in the low, rare, and ultra-rare frequency range of the original source population can reach much higher frequencies in the subsequent bottlenecked populations due to genetic drift (Lim et al. 2014). As a result, even deleterious alleles can increase in frequency in the new populations (Slatkin 2004). The South Asian ancestry today is enriched for putative loss of function (pLOF) variants, with number of novel homozygous protein truncating variants identified varying largely across the various South Asian groups (Figure 3) (GenomeAsia100K Consortium 2019; Wall et al. 2023).
Figure 3.
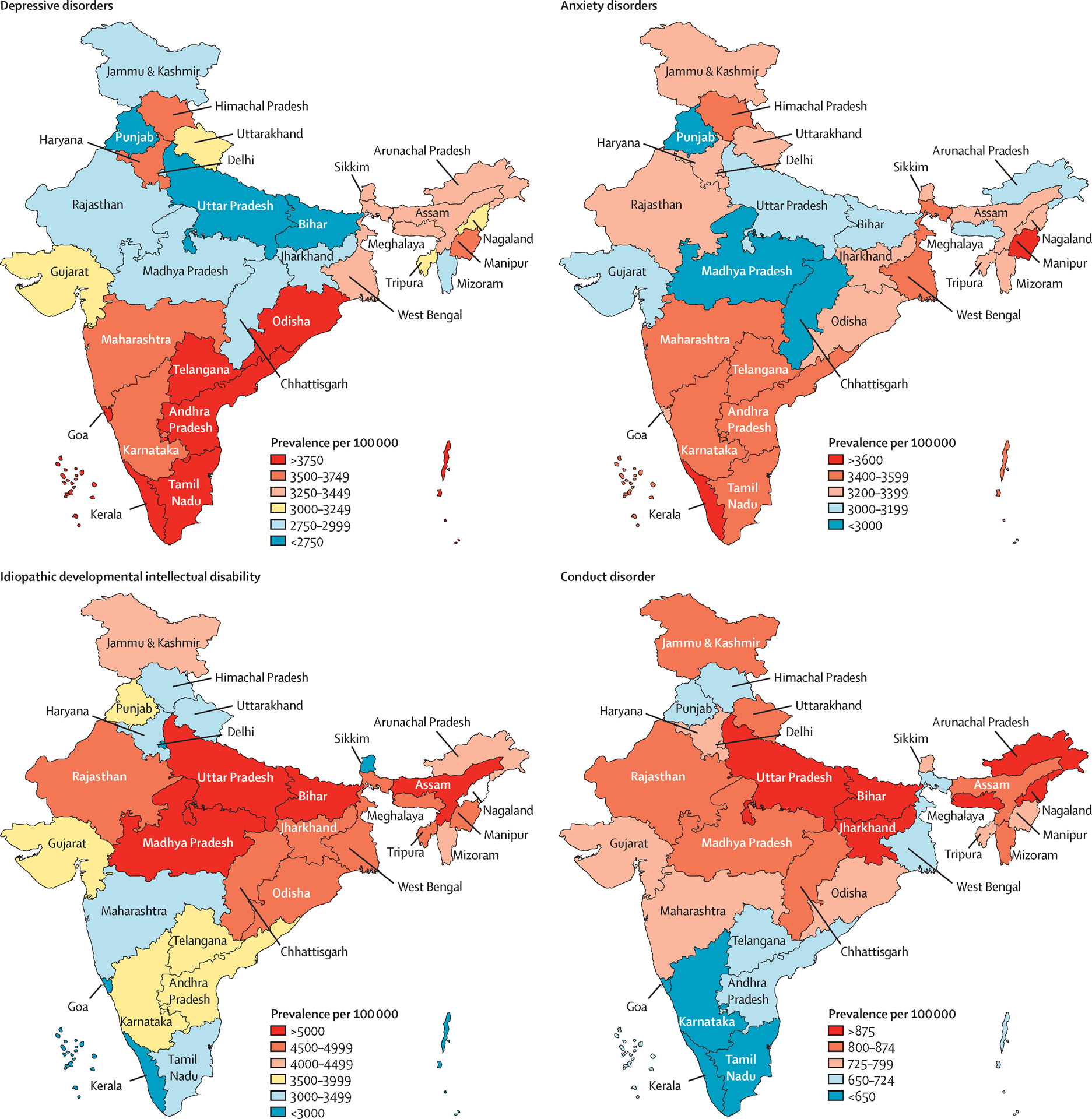
Open Access. Creative Commons Attribution IGO (CC BY 3.0 IGO) 2017 crude prevalence estimates of major mental health disorder across states in India
The increased prevalence of founder effects and observed genetic diversity across groups in South Asia dictates the need for representation from diverse groups in the region (at minimum from all the ethnolinguistic groups) to accurately characterize the genetic architecture of any complex trait in South Asians (Figure 4) (Chambers et al. 2014; Sengupta et al. 2016). However, to determine what groups are still needed, it is first important to identify–and include, wherever possible–the groups that are already represented.
Figure 4.
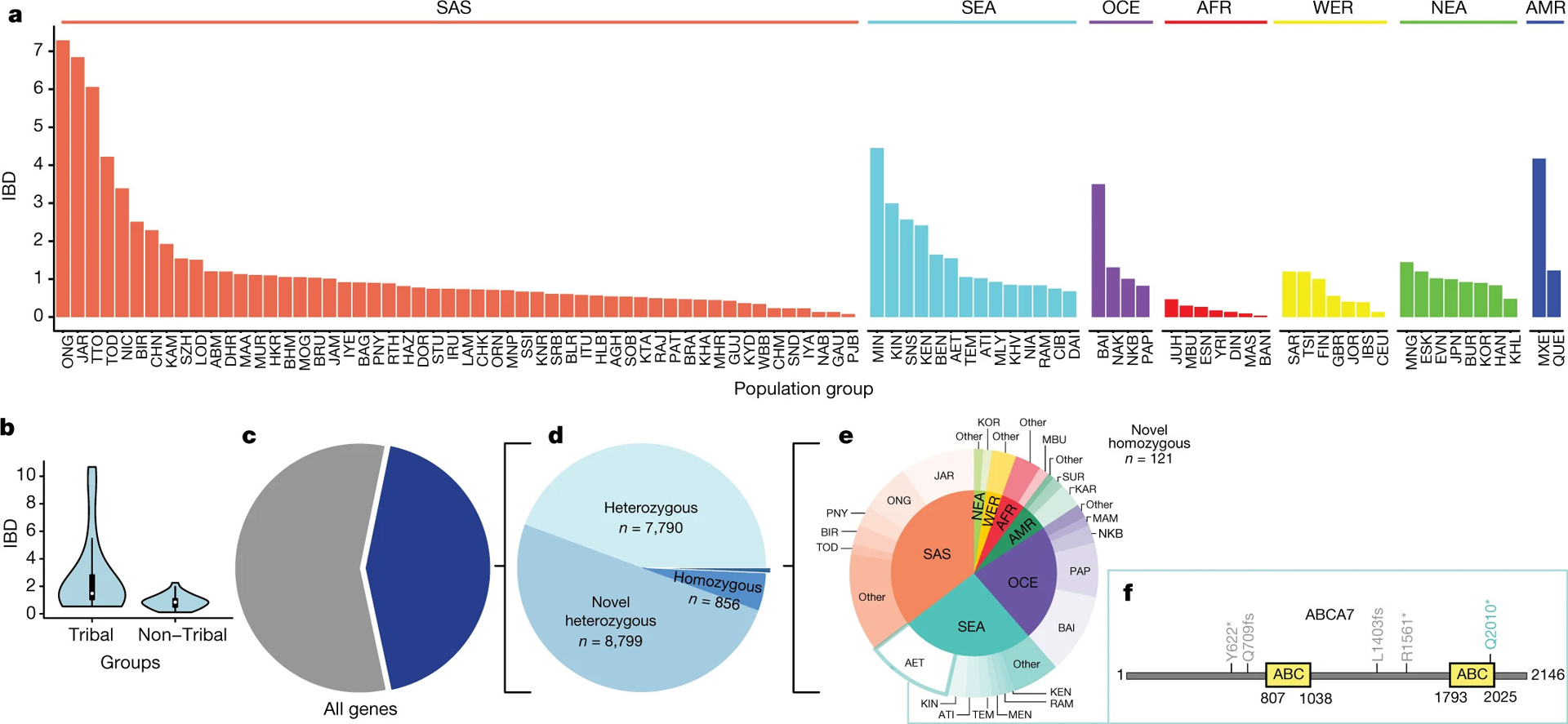
Open Access Creative Commons Attribution 4.0 (CC BY 4.0). a. IBD scores relative to the Finnish group across different population groups across the world. b. Violin plot of the IBD scores in tribal groups versus non-tribal groups. c. Proportion of genes with at least one high-confidence protein truncating variant (PTV) d. Proportion of novel, known, heterozygous, and homozygous PTVs in the Genome Asia pilot dataset. e. Pie chart of novel homozygous PTVs plotted by region (inner circle) and population group (outer circle). Groups with less than two PTVs were grouped as other. f. Novel homozygous PTV Q2010* (green) found in ABCA7 localizes to the C-terminal ABC domain. Previously reported PVTs are shown in grey
Currently Represented Groups
The rise of GWAS has immensely transformed our understanding of human genetics over the last decade. However, a critical component of GWAS is an appropriate imputation of the participating samples using ancestry-based reference panels. To determine the appropriate imputation panel necessary for a population, it is important to determine which groups are currently represented in existing reference panels. The International HapMap Project, has one South Asian reference panel, Gujaratis in Houston (labeled as GIH), that was generated in the third phase of the project, HapMap3 (International HapMap 3 Consortium et al. 2010). While the Human Genome Diversity Project (HGDP) offers a more expanded representation of South Asia, it is critical to note that almost all of the South Asian samples in this panel have come from diverse groups in Pakistan, which are cumulatively classified as “Central and South Asian” despite only measuring genetic diversity captured from one country of origin with its own unique cline (Cavalli-Sforza 2005). Similarly, the only South Asian representation in TOPMed comes from the Pakistani Risk of Myocardial Infarction Study (PROMIS) (Taliun et al. 2021). While there were no South Asian groups in the pilot phase of the 1000 genomes project, the most recent 1000 genomes data set has 5 South Asian groups. Of the 5 groups, 2 of the population additions, Indian Telugus in the UK (ITU) and Sri Lankan Tamils in the UK (STU), were the first South Asian reference populations that captured genetic ancestry from the Dravidian language speaking group from the southern part of the Indian subcontinent (important to note that both populations were captured from the UK diaspora and may not capture the full diversity of the region) (1000 Genomes Project Consortium et al. 2015). In addition to samples from the South, one eastern reference population, Bengalis in Bangladesh (BEB) was also added.
Most GWASs today use a combination of HGDP and 1000 genomes populations to determine ancestry of samples. Here we recommend the additional inclusion of the reference populations generated in the Simons Genome Diversity Project (SGDP) to determine ancestry (Mallick et al. 2016). The SGDP includes a comprehensive set of genomes that better span the Indian subcontinent in comparison to previous efforts. Additionally, when possible, we recommend using the GenomeAsia imputation panel for imputing South Asian samples (available on the Michigan Imputation Server) (GenomeAsia100K Consortium 2019). GenomeAsia is a coordinated initiative from scientific groups across Asia to better characterize the genetic architecture of diverse populations within Asia. The imputation panel is generated from thousands of samples across hundreds of groups in Asia and specifically South Asia, it spans diverse regions, language groups, and social groups (GenomeAsia100K Consortium 2019). Future genetic efforts should also aim to capture genetic diversity from other countries in South Asia that are currently still underrepresented or missing, including Afghanistan, Bhutan, Nepal, Sri Lanka, and the Maldives.
Unique Environmental Considerations
The genetic diversity and complexity of South Asia is rivaled only by the rich diversity of its environment. Like in any other population, outcomes for nearly every trait are contextually driven by multiple layers of environmental influences such as geographical factors (crops that do and do not grow in the area, proximity to water bodies, elevation, etc.) and socio-cultural factors (caste, family structures, religion, etc.). Currently, the norms around genetic and genomic modeling from relatedness thresholds to assumptions about homogeneity, stratification, and rearing environments are based on standards derived from data from individuals of European ancestry. Thus, it is important to re-evaluate assumptions about such standards as human genetics integrates increasingly diverse ancestries.
Figure 3 demonstrates how even traits such as psychiatric disorders can show geographical trends in India, likely due to complex interactions between environmental and genetic variability (India State-Level Disease Burden Initiative Mental Disorders Collaborators 2020). With environmental and genetic structures historically co-evolving, disentangling the two components from one another will prove to be more difficult in this population without sufficient representation from diverse groups across the region. It is extremely important to check when conducting a GWAS in South Asians that the phenotypes in participants are not ethnolinguistically stratified. If there are sufficient data, samples should also be assessed for other cultural or group-based stratification (such as religion or caste), as not doing so may result in false positives relating to population structure (or cultural differences) rather than identification of actual functional loci.
Another significant factor to consider in South Asian populations is concerning the impact of differential family structures on patterns of familial similarities in phenotypic values. In recent years, there is growing interest in understanding how gene-environment correlations (such as “genetic nurture”) within families can influence or inform trait values. In addition to the classical twin design, many of the family-based models used to explore these topics inherently stress the framework of two (biological or adoptive) parents within a nuclear family structure as the primary influencers of their offspring’s’ “shared” rearing environment. However, among most South Asian families, more collectivist family structures are more normative and often include larger households in which grandparents, parents, uncles, aunts, nieces, and nephews may live together (Chadda and Deb 2013). Given these factors and other genetic factors (such as endogamy and consanguinity), even more biologically distant relations such as first cousin pairs or avuncular pairs can/may show a greater degree of phenotypic similarity when compared to analogous relations in European-ancestry individuals (Arciero et al. 2021; Wall et al. 2023).
Climate patterns and geography could also act as major environmental pressures that can contribute to a differential trait architecture unique to South Asians. For example, much of the Indian subcontinent’s agriculture is largely dependent on the temperamental El Niño Southern Oscillation. Some studies show that before 5,000 years ago, this phenomenon was largely absent or infrequent, leading to periodic famines (Pomeroy et al. 2019). This temporal phenomenon could have in turn led to the selection for low lean mass (lower lean tissue mass relative to height) and shorter stature to improve metabolic efficiency as on average increased body size and higher muscle composition require higher energy expenditure (Westerterp 2017). Analysis of ancient skeletons shows that this feature of low lean mass has likely characterized the South Asian physique since at least the early Holocene period and could therefore be a long-term environmental adaptation in this population (Pomeroy et al. 2019). Such an adaptation can have clinical implications in areas such as the use of (body mass index) BMI to predict risk for other disorders. A recent study in the UK showed that a BMI cut off of 23.9 kg/m2 in South Asians is associated with the same Type 2 diabetes incidence rate as the group referred to by the authors as “Whites” in the UK with a cut off of 30.0 kg/m2 (Caleyachetty et al. 2021). Such indications of differential underlying risk could explain the variability in the degree of transferability of polygenic scores from European to South Asians across different traits (Huang et al. 2022). If some traits have ancestry specific risk factors and investigating these differences can help us better characterize the unique underlying genetic architecture and develop effective interventions for diverse groups. Failing to identify these factors could be detrimental to providing equitable care.
The inclusion of more diverse populations can also encourage new avenues of research not only into traits that may present differently from other populations, but also into traits that are unique to specific populations. For example, despite the massive interest in research around substance use and substance use disorders, there has been minimal work done in humans or animal models to understand the genetic or environmental factors associated with areca nut (betel nut) use and/or abuse. While the use of areca nut is relatively uncommon in the US and Europe, it is highly prevalent across populations in East Africa, South Asia, and Southeast Asia, where the plant grows readily (Garg et al. 2014). With over 600 million users worldwide, the habitual chewing of the areca nut (the preparation of which is sometimes called paan) is believed to bethe fourth-most common form of self-administered psychoactive substance in humans after alcohol, caffeine, and nicotine (Garg et al. 2014). The areca nut, often used as a digestive aid, is a psychoactive substance that contains the compound arecoline (a partial agonist of muscarinic acetylcholine receptors), a stimulant with similar psychoactive properties to nicotine (Liu et al. 2016). The inclusion of diverse groups can offer a new outlook into the phenotypic diversity that exists across the globe for the array of traits commonly studied in behavior genetics.
What should we change?
Inclusivity and the Need for Diversity
As a global scientific society, including individuals from diverse parts of the world in any conversations on diversity is a minimum courtesy we must extend. Despite numerous calls from researchers in South Asia and collaborators worldwide advocating for the need to study this group, South Asians have been repeatedly left out of genetic research and seminal papers summarizing the current state of human genetics. Additionally, researchers frequently gloss over South Asian representation, even when explicitly discussing the importance of diversity and representation in genetics research. Despite being the largest subpopulation in the world, there are few precise statistics summarizing the degree of representation of individuals from this region in human genetic and genomic work. Often, when diversity statistics are reported in papers, South Asians, Southeast Asians, Middle Easterners (also sometimes known as West Asians), and other unspecified Asians are collectively lumped into a group called “Other Asian”. Morales et al. (2018) offers a framework for a somewhat more detailed, specific, and appropriate set of ancestry groups that could be used in human genetics; however, even that paper falls prey to categorical oversimplification in the graphics, by using “Other Asian” to encapsulate all Asians not of East Asian descent. All these groups are genetically and socially different and require, at minimum, the dignity of separate categories so that the research community can better understand where deficiencies in representation exist and to contextualize findings more accurately.
Based on 26 populations sampled from around the world, the 1000 Genomes Project has developed five main super populations: Africans (AFR), Admixed Americans (AMR), East Asians (EAS), Europeans (EUR), and South Asians (SAS). Although the number of trans-ancestral and cross-ancestral GWAS is rising, including South Asian ancestry is the exception, not the norm, even when every other super population is included. Furthermore, it is rare that such studies even mention the lack of inclusion in the limitations of the paper. For example, the most recent GWAS from the GWAS and Sequencing Consortium of Alcohol and Nicotine use (GSCAN), a meta-analysis of ancestrally diverse GWASs on tobacco and alcohol use (Saunders et al. 2022) was a massive stride in incorporating diverse groups with the inclusion of AFR, AMR, EAS, and EUR GWASs. However, the study fell short by failing to point out lack of South Asians representativity in the limitations. While this is one example, there are many studies across the genetic literature that neither include nor discuss the lack of South Asian samples. While performing a genetic analysis, the inclusion of every major ancestry group is not always feasible (either due to data and sample size limitations) or appropriate (because of issues with heterogeneity). However, at a minimum, it is vital for researchers working with human data to mention groups not included in the limitations of the papers to raise awareness of the issue and to speak about gaps that need to be addressed in future data collection efforts. Failure to address these voids will delay future solutions and exacerbate existing disparities resulting from discrepancies in research on human genetic data across ancestries.
Labels matter: What is “Asian”?
In the absence of genetic data, self-reported race/ethnicity categories are often used to categorize individuals into groups. In the United States, the Office of Management and Budget (OMB) currently offers five main categorical racial designations: White, Black or African American, American Indian or Alaska Native, Asian, and Native Hawaiian or Other Pacific Islander, along with only one category for ethnicity: Hispanic or Latino (Holup et al. 2007). According to the Revisions to the Standards for the Classification of Federal Data on Race and Ethnicity, the OMB defines Asian as a “person having origins in any of the original peoples of the Far East, Southeast Asia, or the Indian subcontinent including, for example, Cambodia, China, India, Japan, Korea, Malaysia, Pakistan, the Philippine Islands, Thailand, and Vietnam” (Holup et al. 2007). Moreover, health data often groups Native Hawaiian/Pacific Islander populations into the broader Asian-American demographic. Many of these racial/ethnic labels were developed before there was a methodfor parsing and determining genetic ancestry. It is widely accepted that the use of the phrase “race and/or ethnicity” falsely implies that these terms are interchangeable and can be assumed as an indicator of a “racial ancestry.” Today, we have a much better understanding of the implications of these terms, so we must advocate changing these outdated groupings. Although the Census Bureau specifically states that race and ethnicity are socially constructed categories that do not aim to biologically, anthropologically, or genetically group individuals (Bennett 1997), these terms continue to be utilized in health and societal settings, such as when evaluating demographics for clinical trials, consortia diversity, and systematic disparities.
The definitions below are commonly used definitions across the biological and social sciences for race, ethnicity, and genetic ancestry.
Race: is a socio-political construct that groups people based on perceived physical differences (Messer and Gonzalez 2021)
Ethnicity: is a cultural construct based on common cultural characteristics including language, religion, dietary practices, and nationality (Messer and Gonzalez 2021)
Genetic Ancestry: a construct based on DNA about biological origins and reflects individuals who share a demographic history(Jorde and Bamshad 2020)
Given these definitions, it does not follow that 60% of the human population should be collectively categorized under the term “Asian” based on continent of origin (Holup et al. 2007; Lee and Ramakrishnan 2020; Chan et al. 2022). The over 4 billion Asians do not share the same perceived physical features, nor do they all share common cultural characteristics or common demographic history. Although researchers must work within the confines of society and bureaucratic processes, the scientific community must demand appropriate group labels. Alongside this, we need to educate ourselves and our trainees so that society moves toward an improved understanding of the diversity and complexity of ancestry. Contemporary human genetics research stresses the use of ancestral groups (i.e., genetically meaningful categories indicating shared descent) (Khan et al. 2022), so it is unacceptable to continue to employ categorical oversimplifications that likely have little-to-no merit within the genetic sphere (such as purely continental categories). Researchers must be clear about their intentions and motivations for using such an umbrella term when utilizing a term that encompasses so many people with highly diverse backgrounds. If deemed inappropriate for the line of question, it is necessary to break down umbrella termsinto comprehensible parts to ensure sound science. For example, global platforms such as the GWAS Diversity monitor, TOPMed diversity monitor, NHGRI Diversity monitor, and GTEx have condensed ancestral categories into continental categories of American, Asian, African, and European (Mills and Rahal 2020; GTEx Consortium 2020; Taliun et al. 2021). Individuals of East Asian ancestry are the second-most represented group of humans in genetic research (after individuals of European descent). While the rise in East Asian samples represents an essential stride in diverse human genome-wide research, collapsing across groups with less representation (i.e., “South Asian”, “Southeast Asians”) into one group (i.e., “Asians”), provides a false sense of progress across all groups and compromises scientific rigor. Lumping of ancestral groups in human genetics research is not unique to research on those of South Asian descent. Still, such over-broad classifications, likely intended as heuristic solutions to statistical power, diminish our ability to recognize underrepresentation and generalize findings. The usage of large overarching terms can have negative repercussions not only on the representation of data, but also in the use of data in an effective and equitable way. It deprives clinicians, researchers, drug developers, study participants, and the general public of accurate and population specific health information, an oversight that could have deadly consequences.
History: Tales of Caution
In addition to better characterizing diverse groups in health data, as researchers, we must aim to integrate the scientific history from diverse groups around the world to bring awareness to historical events that may contribute to hesitancies around research involvement and provide caution for the future. A relatively unknown fact is that much of the ideology in early scientific racism, the pseudoscientific rationale to justify racism with “empirical data” such as craniometry (Hudson 1996; Jackson and Weidman 2005),was built upon the radical misinterpretation and misappropriation of a historical and religious Hindu text (the Rigveda) for the purposes of fueling the notion of a superior “Aryan race” (Thapar 1996). The identification of linguistic similarities between Sanskrit and other European languages, made the Rigveda one of the earliest texts in the Indo-European language family. In a time when ethnography was coming to rise (Hudson 1996), there was a need to determine where individuals in South Asia fit into the relatively static concepts of “race”. The term “Arya” meaning “noble” within the Indo-Iranian language branch was used as a term of self-identification in the Rigveda. The definition of nobility here, however, wasn’t one of superiority, but rather nobility in one’s actions. Unfortunately, this idea was radicalized to represent an imaginary race of people who had supposedly subjugated the indigenous people of South Asia under the guise of an endogamous caste system (which in reality did not arise until a few thousand years after the composition of the Rigveda) (Thapar 1996). This idea provided the basis for many atrocities that were committed and many of the terms and symbols from the text were misappropriated as symbols of racist ideology. Some of these continue to be associated with this negative history rather than the culture from which they were originally derived.
The pathway from this text to subsequent negative consequences (mainly the conceptualization of race) that continue to permeate today is another illustration of the importance of understanding the history and evolution of diverse language, geography, and culture (Weaver 2022). According to The Races of Mankind Before European Expansion, published by Charles Scribner’s Sons in 1891, probably due to the shared linguistic components, much of South Asia was considered part of the “Aryan race”. Around the same time, in 1885, according to the Meyers Konversations-Lexikon released in Germany, the northernmost part of South Asia was considered “Indo-Arier” which came under the “Caucasoid race”, while the southern part was called “Dravida u. Singhalesen” (a race of unknown status). The rest of the subcontinent was considered to be a mix of these two “races”. Within the next few years, by 1909, according to a text called People of India by Herbert Hope Risley,South Asia was divided in 7 “races”: “Mongloid”, “Indo-Aryan”, “Dravidian”, “Monglo-Dravidian”, “Aryo-Dravidian”, “Scytho-Dravidian”, and “Turko Iranian”. In addition to the linguistic and geographically determined “races”, it was determined that caste could be integrated with the designations of “race” (Thapar 1996), such that, as one went down the caste strata, the features moved from “Aryan” to “Dravidian” (Risley 1999; Weaver 2022). While this notion has been completely debunked as baseless and racist by many scientists across a diverse array of fields, the effect of such concepts has left major scars on people’s notions of self and identity. The impact of colonialist ideas about racial superiority and implications on the modern caste system persists to the present day in the form of discrimination and colorism (Vijaya and Bhullar 2022). Additionally, to reinforce the fickle nature of the concept of “race”, South Asians in the US have also experienced a gamut of changes in racial designation, where they have been historically classified as “Hindu”, “White”, “Other”, and now most recently “Asian” (Morning 2001). While Asian is the most accurate among the terms in describing South Asians based on at least geographical terms, none of these categories capture the essence of South Asia as its own separate entity. So, while “race” can sometimes be an indicator of genetic ancestry, it is important to note that it is not always the case, and its misuse could have detrimental effects. Understanding why and how racism has evolved is necessary to ensure we do not inadvertently allow it to perpetuate and that we do not promote any harmful claims that may intensify downstream disparities.
History is unfortunately riddled with cases of abuse and violation, so taking the time to understand and respect the history and hesitations of a population around participating in science are the first steps to a more conscientious approach to diversity, inclusion, and equity initiatives within the realm of research. Behavior genetics sits at the juncture of the natural and social sciences with the potential to begin shifting some of these narratives that arose from the eugenic period. It will take time and effort to educate health care providers, educators, and the public about accurate information regarding appropriate use of race, ethnicity, and ancestry. The goal is to shift perceptions, dialogue, and policies in a manner that will be beneficial to the rapidly rising diverse communities of today. However, for behavior genetics to inspire such global change it will first require active dialogue within the community to first understand the use and implications of these terms and then arrive at a global consensus of appropriate future use.
How can we get involved?
While the availability of South Asian data remains minimal, the table below lists some of the currently available datasetsthat include individuals with South Asian ancestry. Additionally, institutions around South Asia are currently designing prospective datasets that are likely to become available in upcoming years. In the meantime, researchers around the world should aim not only to develop more diverse genetic datasets but also to form active collaborations with a diverse array of scientists globally to receive input on ways to collectively improve analysis of this diversity.
Moving forward, the inclusion of multiple populations should become the standard in the field of human behavior genetics. Individual populations should not have to justify the value of their inclusion, as it should be a given. Given our deeper understanding and recognition of past limitations in diversity, equity, and inclusion, we must pause and think critically to identify the missing groups. In some cases, past research has failed to recognize an ancestry group, and in others it has failed to incorporate diverse groups from a particular ancestry group. For example, even the minimal representation of South Asian data present thus far is largely from specific subgroups of India, Pakistan, and Bangladesh. Inclusion and equity in data will take time but expanding the diversity in the individuals involved in the conversations of diversity and inclusion will facilitate advancement of these efforts more effectively and equitably. Although this paper has aimed to shed light on the identities of many groups present in and descended from the area, it has still missed many, along with other populations across the world not yet part of genetics research. Research lays the foundation for novel scientific discoveries, advancement in our understanding of human diversity, and progressive medical care. Gaps in inclusion within this foundational work can have serious repercussions when it comes to current and future applications of ensuing results in healthcare and other societal changes.
Table 1:
Examples of Currently Available South Asian Cohorts
| Study | Sample Size | Descriptive Publication |
|---|---|---|
| Colombo Twin and Singleton Study (CoTASS) | 2934 twins and 1035 singletons | Siribaddana et al. 2008 |
| Pakistan Risk of Myocardial Infarction Study (PROMIS) | 5,000 cases confirmed acute myocardial infarction over 5,000 matched controls | Saleheen et al. 2009 |
| Genomics for Understanding Rare Diseases: India Alliance Network (GUaRDIAN) | No available numbers | GUaRDIAN Consortium et al. 2019 |
| Longitudinal Aging Study in India (LASI) + LASI-DAD The Harmonized Diagnostic Assessment of Dementia for the Longitudinal Aging Study in India | Pilot-1,600 individuals aged 45 and older Wave 1: 73,396 adults age 45 and older (no genetic data yet) LASI-DAD-3,000 individuals aged 60 and older |
Bloom et al. 2021
Lee et al. 2019 |
| East London Genes & Health | 38,899 British Bangladeshi and British Pakistani individuals from East London | Finer et al. 2020 |
| London Life Sciences Population Study (LOLIPOP) | 17,606 Indian Asians (defined as all four grandparents born on the Indian subcontinent) |
Kooner et al. 2011
Chambers et al. 2015 |
| UKBiobank | 5,716 Indian 1,748 Pakistani 221 Bangladeshi |
Bycroft et al. 2018 |
References
- 1000 Genomes Project Consortium, Auton A, Brooks LD, et al. (2015) A global reference for human genetic variation. Nature 526:68–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya S, Sahoo H (2021) Consanguineous Marriages in India: Prevalence and Determinants. J Health Manag 23:631–648 [Google Scholar]
- Arciero E, Dogra SA, Malawsky DS, et al. (2021) Fine-scale population structure and demographic history of British Pakistanis. Nat Commun 12:7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A, Sarkar-Roy N, Majumder PP (2016) Genomic reconstruction of the history of extant populations of India reveals five distinct ancestral components and a complex structure. Proc Natl Acad Sci U S A 113:1594–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett T (1997) “Racial” and ethnic classification: two steps forward and one step back? Public Health Rep 112:477–480 [PMC free article] [PubMed] [Google Scholar]
- Bentley AR, Callier S, Rotimi CN (2017) Diversity and inclusion in genomic research: why the uneven progress? J Community Genet 8:255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berreman GD (1960) Caste in India and the United States. Am J Sociol 66:120–127 [Google Scholar]
- Bittles AH, Black ML (2010) Consanguinity, human evolution, and complex diseases. Proceedings of the National Academy of Sciences 107:1779–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom DE, Sekher TV, Lee J (2021) Longitudinal Aging Study in India (LASI): new data resources for addressing aging in India. Nature Aging 1:1070–1072 [DOI] [PubMed] [Google Scholar]
- Bycroft C, Freeman C, Petkova D, et al. (2018) The UK Biobank resource with deep phenotyping and genomic data. Nature 562:203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caleyachetty R, Barber TM, Mohammed NI, et al. (2021) Ethnicity-specific BMI cutoffs for obesity based on type 2 diabetes risk in England: a population-based cohort study. Lancet Diabetes Endocrinol 9:419–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL (2005) The Human Genome Diversity Project: past, present and future. Nat Rev Genet 6:333–340 [DOI] [PubMed] [Google Scholar]
- Chadda RK, Deb KS (2013) Indian family systems, collectivistic society and psychotherapy. Indian J Psychiatry 55:S299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JC, Abbott J, Zhang W, et al. (2014) The South Asian genome. PLoS One 9:e102645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JC, Loh M, Lehne B, et al. (2015) Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinol 3:526–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SH, Bylstra Y, Teo JX, et al. (2022) Analysis of clinically relevant variants from ancestrally diverse Asian genomes. Nat Commun 13:6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finer S, Martin HC, Khan A, et al. (2020) Cohort Profile: East London Genes & Health (ELGH), a community-based population genomics and health study in British Bangladeshi and British Pakistani people. Int J Epidemiol 49:20–21i [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A, Chaturvedi P, Gupta PC (2014) A review of the systemic adverse effects of areca nut or betel nut. Indian J Med Paediatr Oncol 35:3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GenomeAsia100K Consortium (2019) The GenomeAsia 100K Project enables genetic discoveries across Asia. Nature 576:106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium (2020) The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369:1318–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUaRDIAN Consortium, Sivasubbu S, Scaria V (2019) Genomics of rare genetic diseases-experiences from India. Hum Genomics 14:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamy H (2012) Consanguineous marriages : Preconception consultation in primary health care settings. J Community Genet 3:185–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Goodkind D, Kowal PR (2016) An Aging World: 2015. https://www.researchgate.net/profile/Paul-Kowal/publication/299528572_An_Aging_World_2015/links/56fd4be108ae17c8efaa1132/An-Aging-World-2015.pdf. Accessed 11 Apr 2023
- Holup JL, Press Nancy, Vollmer WM, et al. (2007) Performance of the US Office of Management and Budget’s revised race and ethnicity categories in Asian populations. Int J Intercult Relat 31:561–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang QQ, Sallah N, Dunca D, et al. (2022) Transferability of genetic loci and polygenic scores for cardiometabolic traits in British Pakistani and Bangladeshi individuals. Nat Commun 13:4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson N (1996) From “Nation to “Race”: The Origin of Racial Classification in Eighteenth-Century Thought. Eighteenth Century Stud 29:247–264 [Google Scholar]
- India State-Level Disease Burden Initiative Mental Disorders Collaborators (2020) The burden of mental disorders across the states of India: the Global Burden of Disease Study 1990–2017. Lancet Psychiatry 7:148–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap 3 Consortium, Altshuler DM, Gibbs RA, et al. (2010) Integrating common and rare genetic variation in diverse human populations. Nature 467:52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal S, Zakar R, Fischer F, Zakar MZ (2022) Consanguineous marriages and their association with women’s reproductive health and fertility behavior in Pakistan: secondary data analysis from Demographic and Health Surveys, 1990–2018. BMC Womens Health 22:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JP, Weidman NM (2005) The Origins of Scientific Racism. The Journal of Blacks in Higher Education 66–79 [Google Scholar]
- Jorde LB, Bamshad MJ (2020) Genetic Ancestry Testing: What Is It and Why Is It Important? JAMA 323:1089–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AT, Gogarten SM, McHugh CP, et al. (2022) Recommendations on the use and reporting of race, ethnicity, and ancestry in genetic research: Experiences from the NHLBI TOPMed program. Cell Genom 2:. 10.1016/j.xgen.2022.100155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooner JS, Saleheen D, Sim X, et al. (2011) Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet 43:984–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Banerjee J, Khobragade PY, et al. (2019) LASI-DAD study: a protocol for a prospective cohort study of late-life cognition and dementia in India. BMJ Open 9:e030300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Ramakrishnan K (2020) Who counts as Asian. Ethn Racial Stud 43:1733–1756 [Google Scholar]
- Li YR, Keating BJ (2014) Trans-ethnic genome-wide association studies: advantages and challenges of mapping in diverse populations. Genome Med 6:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim ET, Würtz P, Havulinna AS, et al. (2014) Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet 10:e1004494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-J, Peng W, Hu M-B, et al. (2016) The pharmacology, toxicology and potential applications of arecoline: a review. Pharm Biol 54:2753–2760 [DOI] [PubMed] [Google Scholar]
- MacNee W, Rabinovich RA, Choudhury G (2014) Ageing and the border between health and disease. Eur Respir J 44:1332–1352 [DOI] [PubMed] [Google Scholar]
- Mallick S, Li H, Lipson M, et al. (2016) The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 538:201–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastana SS (2014) Unity in diversity: an overview of the genomic anthropology of India. Ann Hum Biol 41:287–299 [DOI] [PubMed] [Google Scholar]
- Messer RH, Gonzalez GDS (2021) Relationship Between Culture and Race. In: Shackelford TK, Weekes-Shackelford VA (eds) Encyclopedia of Evolutionary Psychological Science. Springer International Publishing, Cham, pp 6538–6540 [Google Scholar]
- Metspalu M, Mondal M, Chaubey G (2018) The genetic makings of South Asia. Curr Opin Genet Dev 53:128–133 [DOI] [PubMed] [Google Scholar]
- Metspalu M, Romero IG, Yunusbayev B, et al. (2011) Shared and unique components of human population structure and genome-wide signals of positive selection in South Asia. Am J Hum Genet 89:731–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills MC, Rahal C (2020) The GWAS Diversity Monitor tracks diversity by disease in real time. Nat Genet 52:242–243 [DOI] [PubMed] [Google Scholar]
- Moorjani P, Thangaraj K, Patterson N, et al. (2013) Genetic evidence for recent population mixture in India. Am J Hum Genet 93:422–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales J, Welter D, Bowler EH, et al. (2018) A standardized framework for representation of ancestry data in genomics studies, with application to the NHGRI-EBI GWAS Catalog. Genome Biol 19:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morning A (2001) The racial self-identification of South Asians in the United States. J Ethn Migr Stud 27:61–79 [Google Scholar]
- Nagoshi CT, Johnson RC, Danko GP (1990) Assortative mating for cultural identification as indicated by language use. Behav Genet 20:23–31 [DOI] [PubMed] [Google Scholar]
- Nakatsuka N, Moorjani P, Rai N, et al. (2017) The promise of discovering population-specific disease-associated genes in South Asia. Nat Genet 49:1403–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan VM, Patterson N, Moorjani P, et al. (2019) The formation of human populations in South and Central Asia. Science 365:. 10.1126/science.aat7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeroy E, Mushrif-Tripathy V, Cole TJ, et al. (2019) Ancient origins of low lean mass among South Asians and implications for modern type 2 diabetes susceptibility. Sci Rep 9:10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popejoy AB, Fullerton SM (2016) Genomics is failing on diversity. In: Nature Publishing Group UK. 10.1038/538161a. Accessed 5 Feb 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaswamy P (2005) South Asian Diaspora. In: Ember M, Ember CR, Skoggard I (eds) Encyclopedia of Diasporas: Immigrant and Refugee Cultures Around the World. Springer; US, Boston, MA, pp 285–296 [Google Scholar]
- Risley H (1999) The People of India. Asian Educational Services [Google Scholar]
- Saleheen D, Zaidi M, Rasheed A, et al. (2009) The Pakistan Risk of Myocardial Infarction Study: a resource for the study of genetic, lifestyle and other determinants of myocardial infarction in South Asia. Eur J Epidemiol 24:329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders GRB, Wang X, Chen F, et al. (2022) Genetic diversity fuels gene discovery for tobacco and alcohol use. Nature 612:720–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen R (1992) Formation of state and the Indus Valley Civilization. Indian Anthropologist 22:25–40 [Google Scholar]
- Sengupta D, Choudhury A, Basu A, Ramsay M (2016) Population Stratification and Underrepresentation of Indian Subcontinent Genetic Diversity in the 1000 Genomes Project Dataset. Genome Biol Evol 8:3460–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde V, Narasimhan VM, Rohland N, et al. (2019) An Ancient Harappan Genome Lacks Ancestry from Steppe Pastoralists or Iranian Farmers. Cell 179:729–735.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siribaddana SH, Ball HA, Hewage SN, et al. (2008) Colombo Twin and Singleton Study (CoTASS): a description of a population based twin study of mental disorders in Sri Lanka. BMC Psychiatry 8:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirugo G, Williams SM, Tishkoff SA (2019) The Missing Diversity in Human Genetic Studies. Cell 177:26–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M (2004) A population-genetic test of founder effects and implications for Ashkenazi Jewish diseases. Am J Hum Genet 75:282–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliun D, Harris DN, Kessler MD, et al. (2021) Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 590:290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamang R, Singh L, Thangaraj K (2012) Complex genetic origin of Indian populations and its implications. J Biosci 37:911–919 [DOI] [PubMed] [Google Scholar]
- Teixeira JC, Cooper A (2019) Using hominin introgression to trace modern human dispersals. Proc Natl Acad Sci U S A 116:15327–15332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapar R (1996) The Theory of Aryan Race and India: History and Politics. Social Scientist 24:3–29 [Google Scholar]
- United Nations Publications (2021) International Migration 2020: Highlights. UN [Google Scholar]
- United Nations (UN) (2017) World economic situation and prospects 2017
- Vijaya RM, Bhullar N (2022) Colorism and employment bias in India: an experimental study in stratification economics. Review of Evolutionary Political Economy 3:599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Sathirapongsasuti JF, Gupta R, et al. (2023) South Asian medical cohorts reveal strong founder effects and high rates of homozygosity. Nat Commun 14:3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LJ (2022) The Laboratory of Scientific Racism: India and the Origins of Anthropology. Annu Rev Anthropol 51:67–83 [Google Scholar]
- Westerterp KR (2017) Control of energy expenditure in humans. Eur J Clin Nutr 71:340–344 [DOI] [PubMed] [Google Scholar]


