Abstract
The direct repeat region in Mycobacterium tuberculosis complex strains is composed of multiple direct variant repeats (DVRs), each of which is composed of a 36-bp direct repeat (DR) plus a nonrepetitive spacer sequence of similar size. It has been shown previously that clinical isolates show extensive polymorphism in the DR region by the variable presence of DVRs, and this polymorphism has been used in the epidemiology of tuberculosis. In an attempt to better understand the evolutionary scenario leading to polymorphic DR loci and to improve strain differentiation by spoligotyping, we characterized and compared the DNA sequences of the complete DR region and its flanking DNA of M. tuberculosis complex strains. We identified 94 different spacer sequences among 26 M. tuberculosis complex strains. No sequence homology was found between any of these spacers and M. tuberculosis DNA outside of the DR region or with any other known bacterial sequence. Although strains differed extensively in the presence or absence of DVRs, the order of the spacers in the DR locus was found to be well conserved. The data strongly suggest that the polymorphism in clinical isolates is the result of successive deletions of single discrete DVRs or of multiple contiguous DVRs from a primordial DR region containing many more DVRs than seen in present day isolates and that virtually no scrambling of DVRs took place during evolution. Because the majority of the novel spacer sequences identified in this study were confined to isolates of the rare Mycobacterium canettii taxon, the use of the novel spacers in spoligotyping led only to a slight improvement of strain differentiation by spoligotyping.
Bacterial isolates belonging to the Mycobacterium tuberculosis complex group of bacteria show an unusually high degree of conservation in housekeeping genes (19, 31). Because the mutation frequency of M. tuberculosis is similar to that of other bacteria, these observations have led to the speculation that the M. tuberculosis complex isolates presently seen have diverged from a common ancestor not more than 10,000 to 15,000 years ago (19). Much more DNA polymorphism in M. tuberculosis complex bacteria has been found to be associated with repetitive DNA, such as transposable elements, and short perfect or imperfect repeats (29, 32). Furthermore, the establishment of the genome sequence of M. tuberculosis (5) has led to the disclosure of variation in the presence of multiple regions carrying a variety of different genes (12).
The direct repeat (DR) locus is presently the only well-studied single locus in the genome of M. tuberculosis showing considerable strain-to-strain polymorphism (10, 14). This locus is composed of multiple 36-bp DR copies, which are interspersed by nonrepetitive short sequences of about equal length, the so-called spacers (16, 18). Clinical isolates of M. tuberculosis and Mycobacterium bovis generally differ in the presence or absence of one or more spacers and adjacent DRs. This polymorphism has been exploited to distinguish M. tuberculosis complex strains for epidemiological studies and to distinguish the taxons within the group of the M. tuberculosis complex, these being M. tuberculosis, M. bovis, M. microti, and M. canettii (9, 13, 15, 18, 22, 34, 37, 39, 40). The function of the DR locus in M. tuberculosis is presently unknown. The apparent omnipresence of this unusual region in M. tuberculosis complex and the strong sequence conservation of the DRs and spacers among clinical isolates may suggest a biological function of the DR region for the host and that, due to a selective advantage, this region has been maintained in the population. Although no significant homology has been reported between the M. tuberculosis DR sequence and DNA of other bacterial genera, loci with similar motifs composed of short repetitive and nonrepetitive sequences in other bacterial genera have been found (1, 4, 17, 20, 21, 24–26, 33). In Haloferax spp. the locus may be involved in replicon partitioning (26), but in other organisms the function has not been investigated.
Until now, DNA polymorphism in the DR locus has been investigated only in M. tuberculosis complex bacteria (10, 14) and in Streptococcus pyogenes (17). Isolates of M. tuberculosis complex differ in the presence or absence of one or more discrete DNA segments, each consisting of a DR plus the adjacent spacer, the so-called direct variant repeat (DVR). This suggests that homologous recombination between neighboring or distant DRs may lead to deletion of one or more discrete DVRs (10, 14). A similar polymorphism among clinical isolates was observed in the DR locus of S. pyogenes (17). Furthermore, the DR region in M. tuberculosis has been identified as a hot spot of integration of the insertion element IS6110 (10, 16), and IS6110-associated polymorphism in the DR region has been disclosed, also among outbreak strains (3, 10, 14, 23). To elucidate the mechanism of genetic variation in the DR locus and the evolutionary pathway of the DR locus in M. tuberculosis complex bacteria, we compared in this study the sequence of the DR locus and its flanking DNA from a variety of different strains, including pairs of isogenic strains with different spoligotypes. A second objective of this study was to improve the degree of strain differentiation based on DNA polymorphism in the DR region of M. tuberculosis by spoligotyping. In this method the whole DR region is amplified and labeled by PCR using DR-specific primers and the presence of any of a set of 43 different spacers is determined by hybridization of the amplified DNA to 43 spacer oligonucleotides, which are covalently linked to a membrane (18). Spoligotypes are expressed as the presence or absence of any of these 43 spacers. The method allows a high throughput, and no cultured cells are needed to differentiate strains (9, 30, 36, 40). A drawback of the presently used method of spoligotyping is the limited discriminative power, compared to other methods such as IS6110 fingerprinting (8, 13, 18, 22, 38, 39). The sequences disclosed in this study to characterize the nature of genetic variation in the DR locus allowed us to evaluate the novel spacers for their potential for improved strain differentiation.
MATERIALS AND METHODS
Bacterial strains.
Unless otherwise stated, all bacterial strains were clinical isolates of M. tuberculosis complex. Table 1 presents the strains for which the sequence of the DR region or part thereof was established. For spoligotyping, M. tuberculosis complex strains were selected from an RIVM culture collection comprising about 8,000 clinical isolates collected between 1993 and 1998. All strains in this collection were typed by IS6110 fingerprinting, and those with less than five IS6110 copies had been subtyped by polymorphic GC-rich sequence (PGRS) typing (39). A part of the strain collection (ca. 1,000 strains) has been spoligotyped as well. All M. tuberculosis complex strains, except the M. bovis BCG strains selected for hybridization with the novel spacers derived in this study differed in IS6110 restriction fragment length polymorphism (RFLP) and/or PGRS RFLP.
TABLE 1.
Mycobacterial strains used for DNA sequencing
| Strain no.a | Original designation | Species | Origin | Accession no.c | Source or reference |
|---|---|---|---|---|---|
| 1 | NLA009400768 | M. tuberculosis | Human | AF189746/AF189747 | This laboratory |
| 2 | NLA009400982 | M. tuberculosis | Human | AF189748/AF189749 | This laboratory |
| 3 | NLA000016314 | M. tuberculosis | Human (previously strain 103) | AF189750/AF189751 | Kremer et al. (22) |
| 4 | NLA009400319 | M. tuberculosis | Human | AF189752/AF189753 | This laboratory |
| 5 | NLA000016923 | M. tuberculosis | Human | AF189754/AF189755 | This laboratory |
| 6 | NLA009700584 | M. tuberculosis | Human | AF189756/AF189757 | This laboratory |
| 7 | NLA009401014 | M. tuberculosis | Human (previously strain 56) | AF189758/AF189759 | Kremer et al. (22) |
| 8 | H37Rv | M. tuberculosis | Laboratory strain, genome sequenced | Z81331 | Cole et al. (5) |
| 9 | DR strain 86 | M. tuberculosis | Human | Y14045NID | Fang et al. (10) |
| 10 | DR strain 257 | M. tuberculosis | Human | Y14047NID | Fang et al. (10) |
| 11 | DR strain 149 | M. tuberculosis | Human | Y14046NID | Fang et al. (10) |
| 12 | DR strain 191 | M. tuberculosis | Human | Y14048NID | Fang et al. (10) |
| 13 | DR strain 93 | M. tuberculosis | Human | Y14049NID | Fang et al. (10) |
| 14 | CSU#93 | M. tuberculosis | Human, genome being sequenced | B. B. Plikaytis | |
| 15 | NLA009400303 | M. tuberculosis | Human | AF189760 | This laboratory |
| 16 | NLA009400116 | M. tuberculosis | Human | AF189818/AF189761 | This laboratory |
| 17 | NLA000016319 | M. tuberculosis | Human (previously strain 111) | AF189762/AF189763 | Kremer et al. (22) |
| 18 | TN4937 | M. tuberculosis | Human | AF189819/AF189820 | This laboratory |
| 19 | NLA009402008 | M. tuberculosis | Human | AF189821/AF189822 | Kremer et al. (22) |
| 20 | NLA0000000P3 | M. bovis BCG | Vaccine strain, The Netherlands | X57835NID | This laboratory |
| 21 | NLA0000Russia | M. bovis BCG | Vaccine strain, Russia | AF189823/AF189824 | This laboratory |
| 22 | TMC401 | M. bovis | Bovine | U47864NID | Beggs et al. (2) |
| 23 | NLA009401854 | M. bovis | Bovine | AF189825/AF189826 | D. Brittain |
| 24 | NLA009502227 | M. tuberculosis complexb | Human | AF189827 | This laboratory |
| 25 | NLA000016240 | M. microti | Vole | AF189828/AF189829 | Van Soolingen et al. (36) |
| 26 | NLA000017727 | M. canettii | Human (previously strain SO93) | AF190853 | Van Soolingen et al. (37) |
| 27 | NLA000013667 | M. tuberculosis | Human, patient 1 | This laboratory | |
| 28 | NLA000013668 | M. tuberculosis | Human, patient 1 | This laboratory | |
| 29 | NLA009602019 | M. tuberculosis | Human, patient 2 | This laboratory | |
| 30 | NLA009602017 | M. tuberculosis | Human, patient 2 | This laboratory | |
| 31 | NLA000017877 | M. tuberculosis | Human | This laboratory | |
| 32 | NLA009401835 | M. tuberculosis | Human | This laboratory | |
| 33 | Strain A | M. bovis | Human | Blazquez et al. (3) | |
| 34 | Strain B | M. bovis | Human | Blazquez et al. (3) |
The complete DR regions of strains 1 to 26 were determined. Strains 27 to 34 comprise presumed isogenic pairs, and only part of the DR region was determined.
Phenotype between M. tuberculosis and M. bovis.
The sequences under two accession numbers were used to obtain the complete sequence of the DR locus.
Spoligotyping.
Culture and DNA isolation from mycobacteria was done as described previously (39). Spoligotyping using the previously published spacer oligonucleotides was done as described earlier (18). The incubation conditions of spoligotyping using the novel spacer oligonucleotides were slightly modified: hybridization was done for 45 min at 45°C, followed by two posthybridization washes for 10 min at 50°C and an incubation with strepavidin-peroxidase for 30 min at 42°C. All other conditions were as described by Kamerbeek et al. (18). Spoligotype designations were assigned to any unique hybridization pattern with the 43 different spacers used in traditional spoligotyping (18).
DNA sequence analysis.
The published sequences of the DR region of M. tuberculosis strains 8 to 14, M. bovis strain 22, and the partial sequence of strain 20, the BCG Pasteur strain P3, were used in this study (see Table 1). The DR region sequences of all other strains listed in Table 1 were determined in this study. Sequencing was done by using the fluorescence-labeled dideoxy nucleotide technology using PCR products obtained by amplifying 0.1- to 3.0-kb fragments of the DR region, purified with Qiaquick purification kits (Qiagen, Hilden, Germany). For this purpose primer oligonucleotides were used based on sequences of DNA flanking the DR region in strain H37Rv (5), known spacer sequences, and sequences of the IS6110 element. DNA sequencing was done using an Applied Biosystems Instruments automatic sequencers (Models 373 and 377; Perkin-Elmer, Applied Biosystems Division). The sequences obtained were assembled, edited, and analyzed with the DNAStar package (DNAStar, Inc., Madison, Wis.). The sequences derived in this study were deposited at GenBank; for the accession numbers, see Table 1. Spacers were numbered according to their location in the genome (Fig. 1). Because this numbering differs from the previously assigned numbers in the study of Kamerbeek et al. (18), the old and the new spacer designations are given in Table 2.
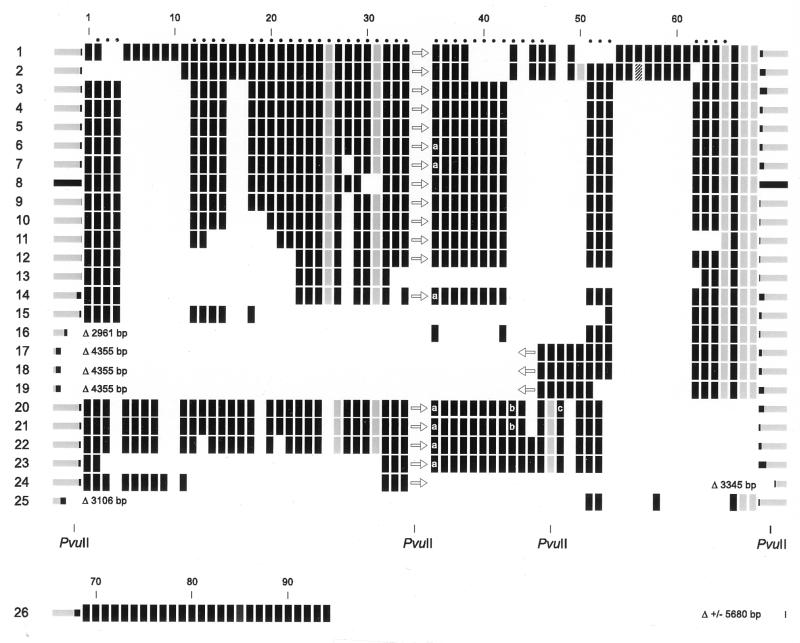
FIG. 1.
Genetic organization of the DR locus in 26 M. tuberculosis complex strains as follows: 1 to 19, M. tuberculosis; 20 to 23, M. bovis; 24, intermediate phenotype between M. tuberculosis and M. bovis; 25, M. microti; and 26, M. canettii. The rectangles depict individual DVRs, which are composed of a DR and the adjacent spacer. Except for the M. canettii strain, vertically aligned rectangles represent DVRs with identical spacers. The sequence of the DR part of the rectangles is identical, except for those in gray. These differ in one or a few nucleotides from the consensus sequence. The hatched spacer in strain 2 differs in a single nucleotide from that in strain 1. The numbers at the top correspond to the spacer numbers listed in Table 2. The presence and the orientation of IS6110 is depicted by an arrow. DNA flanking the DR region is depicted by the bars at the left and at the right. The black parts of these bars depict the stretches that have been sequenced. The size of the DNA sequence missing compared to strain 8 (M. tuberculosis H37Rv) is given in base pairs, after the triangle. DVRs occurring twice in the DR region are depicted as rectangles marked with a letter: a, a duplication of DVR 35 is present 3′ from spacer 45; b and c, duplicated DVRs 43 and 48, respectively, are present as tandem duplications.
TABLE 2.
List of spacer sequences in M. tuberculosis complex
Bold sequences used for oligonucleotide probes in spoligotyping.
The previously published sequence (18) contained a C at residue 17 erroneously.
The previously published sequence (18) contained two errors at residues 13 and 14 (both were A).
This spacer is polymorphic at position 21. Strain 1 is as shown, and strain 2 has a C at this position.
RESULTS
DNA sequence analysis of the DR locus in M. tuberculosis, M. bovis, M. microti, and M. canettii.
The DNA sequence of the complete DR region was determined for 12 M. tuberculosis strains, 2 M. bovis strains, 2 M. bovis BCG strains, 1 M. microti, and 1 M. canettii strain. Furthermore, we sequenced a few hundred base pairs of the DNA flanking the DR region on either side. The results are summarized in Fig. 1, together with the previously published sequences of seven M. tuberculosis strains and one M. bovis strain. The size of the DR region varied from six DVRs for M. microti 19 to 56 DVRs for M. tuberculosis 1. Except for three spacers, all spacers were found only once in the DR locus. Spacer 35 was duplicated as a part of a DVR in all four M. bovis strains (strains 20 to 23) and in three M. tuberculosis strains (strains 6, 7, and 14). This duplicated spacer was separated from spacer 35 by six DVRs and was invariantly located adjacent to spacer 41 (see Fig. 1). The two other duplicated spacers (spacers 43 and 49) were present as tandemly duplicated sequences. Spacer 43 was duplicated in both M. bovis BCG strains (strains 20 and 21), and spacer 49 was duplicated in one BCG strain (strain 20). Virtually no interstrain variation in the sequences of spacers was observed. Only one spacer, spacer 56, present in M. tuberculosis strain 2 differed by a single nucleotide from the corresponding spacer in strain 1 (depicted by a hatched rectangle in Fig. 1; see also Table 2). A small degree of intrastrain sequence variation was found among the DRs. In six DVRs we encountered DRs with single-base-pair mismatches from the consensus sequence. Furthermore, the DR in DVR26 lacked four residues (Table 3). Probably DVR26 has been deleted because the adjacent spacer (spacer 25) is only 25 bp long, which is shorter than any other DVR (see Table 2). Interestingly, in all strains carrying mutant DRs, these mutant DRs invariably were present at the same location within the DR region.
TABLE 3.
Variation in DR sequences
| DVR type | Sequencea |
|---|---|
| Consensus sequence | GTCGTCAGACCCAAAACCCCGAGAGGGGACGGAAAC |
| DVR26 | ----TCAGACCCAAAACCCCGAGAGGGGACGGAAAC |
| DVR31 | GTCGTCAGACCCAAAACCCCGAGAGAGGACGGAAAC |
| DVR47 | GTCGTCAGACCCAAAACCCCGAGAGGGGACGTAAAC |
| DVR50 | GTCGTCAGACCTAAAACCCCGAGAGGGGACGGAAAC |
| DVR65 | GTCGTCGGACCCAAAACCCCGAGAGGGGACGGAAAC |
| DVR67 | GTCCTCAGACCCAAAACCCCGAGAGGGGACGGAAAC |
| DVR68 | GTCGTCAGACCCAAAACCACGAGAGGGGACGGAAAC |
Polymorphisms are indicated in boldface.
For simplicity we numbered the DVRs and spacers according to their position in the DR locus, starting with number one at the left. Ninety-four different spacers were found among the 26 strains. Thirty-seven of these were not reported previously. Twenty-six of the novel spacers (spacers 69 to 94) were present only in M. canettii. The remaining 11 novel spacers were all found in M. tuberculosis 1, and nine of these spacers were present in only one other strain, M. tuberculosis 2. M. tuberculosis strains 1 and 2 are exceptional because of the presence of only a single copy of IS6110. The four M. bovis strains investigated differed from all M. tuberculosis strains in the presence of spacer 44 and in the absence of 16 spacers (spacers 53 and 54 to 68). These data confirm the previously observed M. bovis-characteristic signature in the DR region (18). The most remarkable finding is the strong conservation of the order of the DVRs in the various isolates. As depicted in Fig. 1, the polymorphism in the DR region appears to comprise mainly the presence or absence of single, discrete DVRs or stretches of contiguous DVRs. Except for the duplication of DVR35, no scrambling of DVRs appears to have taken place during the evolution of the DR region.
The DNA flanking the DR locus in most strains was identical to that in M. tuberculosis H37Rv (strain 8 in Fig. 1). Seven strains lacked 3- to 6-kb stretches 5′ or 3′ of the DR region. In all seven strains lacking DR-flanking DNA, IS6110 was either directly adjacent to the flanking DNA or IS6110 was absent from the DR region, suggesting the involvement of this insertion element in the deletion of chromosomal DNA. The DNA flanking the 3′ side of the DR region in the M. canettii strain shared no similarity with the sequence of H37Rv. Interestingly, this DNA was found to share 80% identity to the insertion element IS1096 from M. smegmatis (25). Strain 24 was the only other strain lacking sequences in the 3′-flanking region. This strain could not be classified as any of the known M. tuberculosis complex species because it shared biochemical characteristics of both M. tuberculosis and M. bovis. This may suggest that this strain belongs to a distinct, as-yet-unrecognized taxon within the M. tuberculosis complex group of bacteria. Because the origin of DRs and spacers in the DR region is unknown, we checked the GenBank database for sequences homologous to the DR and all of the spacers. No significant sequence similarity was found.
Rearrangements in the DR locus of presumed isogenic variants.
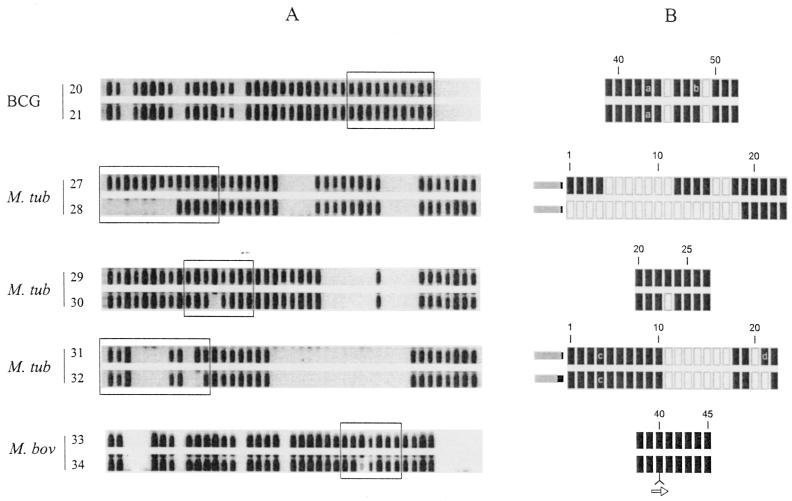
To investigate the nature of the rearrangements in the DR region, we attempted to collect isogenic strain pairs that differ in the DR locus. Only a single set of true isogenic strains was available. This set comprised the two M. bovis BCG strains investigated in this study. The Russian BCG strain, strain 21, differed from the Pasteur vaccine strain, strain 20, in the absence of a duplication of DVR49 as described above (Fig. 2).
FIG. 2.
Rearrangements in the DR locus of presumed isogenic variants. (A) Spoligotype of each of the five isogenic pairs determined by using the standard 43 spacers as a probe; the boxed areas have been sequenced and are depicted in panel B. (B) Arrangement of DVRs as determined by DNA sequencing; the numbers correspond to the spacer numbers as given in Fig. 1. Black rectangles depict DVRs that are present; gray rectangles represent DVRs that are absent. Numbers correspond to DVR numbering as shown in Fig. 1. The arrow represents the insertion element IS6110. Strain numbers are depicted on the left. a, b, and d, tandem duplications of DVR43; DVR48, and DVR21, respectively; c, a duplicated copy of DVR4 is located directly to the right of DVR19.
Unfortunately, we have been unable to derive in the laboratory subcultures which differ from parental strains in the DR region. Therefore, we have analyzed pairs of strains which are very likely to be isogenic, either because they were isolated during in a single outbreak or because they originated from the same patient. The results of partial sequencing of the DR region of these strains are depicted in Fig. 2. M. tuberculosis strains 27 and 28 were isolated from two different body sites of a 76-year-old tuberculosis patient, who probably reactivated from an infection acquired early in life. The IS6110 RFLP patterns of these isolates are related because 10 PvuII IS6110-containing restriction fragments are shared among the 12 IS6110 fragments in strain 27 and the 14 fragments in strain 28 (41). We assume that early in life the patient has been infected with a single strain and that the long period of dormancy allowed DNA rearrangements to take place. Strain 27 differed from strain 28 by the absence of 8 spacers in spoligotyping (Fig. 2A). Sequencing showed these two that strains differ in the absence of exactly nine discrete DVRs, of which eight are used in traditional spoligotyping using the set of 43 spacer probes (Fig. 2B). The third pair of strains comprised M. tuberculosis strains 29 and 30, isolated from a 73-year-old Dutch patient. These strains have identical IS6110 RFLP patterns (showing the presence of eight IS6110 copies) but differ in a single spacer in spoligotyping. Sequencing showed that strain 30 differed from strain 29 in the absence of a single DVR, DVR23. The fourth presumed isogenic pair comprised M. tuberculosis strains 31 and 32, isolated from two Somali immigrants, who probably were epidemiologically linked, although this link could not be confirmed by traditional contact tracing. Both strains have identical IS6110 RFLP patterns (showing 13 IS6110 copies) but differed in a single spacer reaction by spoligotyping. Sequencing showed that strain 31 differed from strain 32 in the presence of two DVRs, which are tandem duplications of DVR21. Finally, we investigated M. bovis strains 33 and 34, isolated during an epidemic of multidrug-resistant tuberculosis in Spain (3). These two strains differ in a single IS6110-containing restriction fragment and in a single spacer reaction by spoligotyping (Fig. 2A). Sequencing showed that strain 34 differed from 33 in the presence of a copy of IS6110 in the DR of DVR40 (Fig. 2B). We conclude that this insertion prevented amplification of DVR40 during the PCR and thus resulted in the apparent absence in the spoligotype pattern.
Although there is no direct evidence that four of the five strain pairs investigated are true isogenic pairs, the nature of the genetic variation found in the DR loci is consistent with this assumption. In four of the strain pairs the genetic rearrangements can be explained by a single genetic event comprising either the insertion or the deletion of a single, discrete DVR or a set of contiguous DVRs. In the remaining pair the variation is also explained by a single genetic event, namely, the insertion or deletion of IS6110 in the DR region.
Strain differentiation of M. tuberculosis complex strains by using novel spacers.
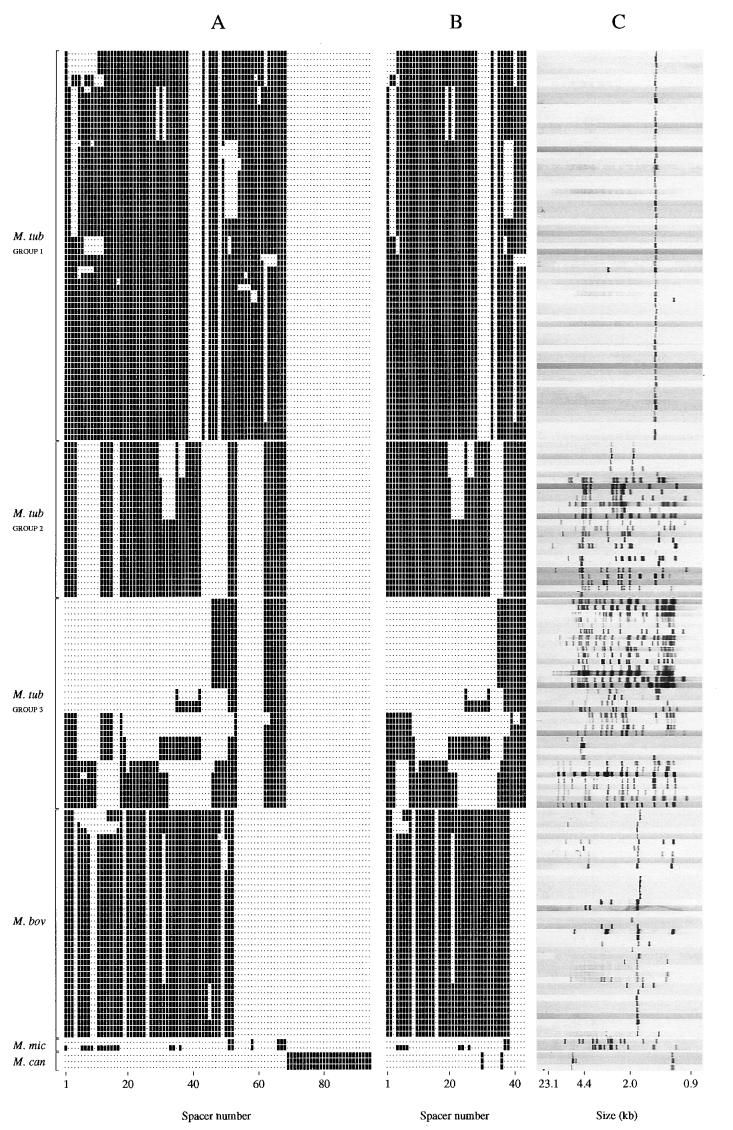
We investigated whether the degree of strain differentiation would be improved by typing using more spacers than the 43 spacers used in standard spoligotyping as described by Kamerbeek et al. (18). For this purpose we spoligotyped M. tuberculosis complex strains using the standard set of 43 spacers plus the 51 novel spacers. We analyzed 170 clinical isolates of M. tuberculosis complex. All of these isolates had previously been fingerprinted by IS6110 and, when fewer than five IS6110 copies were present, the PGRS subtypes were also known. Group 1 comprised 65 M. tuberculosis strains harboring a single IS6110 copy or only two IS6110 copies. Such strains are known to be hard to differentiate by IS6110 fingerprinting (32). By traditional spoligotyping these strains were differentiated into nine spoligotypes. By using the novel spacers, 23 strains were further subtyped, and the number of different hybridization patterns increased to 20 (Fig. 3). Interestingly, the group 1 strains displayed a characteristic spacer signature in the region of spacer 37 to 48 (Fig. 3).
FIG. 3.
Presence of spacers in and traditional spoligotypes and IS6110 RFLP patterns of 170 M. tuberculosis complex strains. (A) Presence or absence of any of the 94 spacers; the spacers are ordered as in Fig. 1, and this order corresponds to the presumed order in the genome. (B) Hybridization signals of the 43 spacers used in traditional spoligotyping. (C) IS6110 RFLP patterns. Strains of group 1 belong to the commonly found spoligotypes ST5, ST14, ST22, and ST38.
Group 2 strains comprised 26 M. tuberculosis strains that were selected because their spoligotypes corresponded to the most frequently encountered four spoligotypes among patients in The Netherlands. None of these strains were subdivided with the novel spacer set (Fig. 3).
Group 3 strains comprised 35 M. tuberculosis strains, which hybridized with relatively few spacers from the old set of 43 in standard spoligotyping. Fifteen of these strains were of the spoligotype, which is characteristic for strains of the Beijing genotype (38). By using the novel spacers only a single strain among the 35 strains tested was further differentiated (Fig. 3). Therefore, the use of additional spacers in spoligotyping led to a marginal increase in the level of strain differentiation. Compared to spoligotyping, IS6110 fingerprinting differentiated better for the strains harboring multiple IS6110 copies, and among the low-copy strains spoligotyping differentiated the strains slightly better.
A group of 38 M. bovis strains was investigated. M. bovis usually harbors only one or a few copies of IS6110, and therefore such strains are notoriously difficult to differentiate (6). The most common spoligotypes among M. bovis isolated in The Netherlands are the types 120 and 121 (unpublished observations). Therefore, we included 34 strains with these two spoligotypes for testing with the additional 51 novel spacer probes (Fig. 3). Seven M. bovis strains were subdivided, and the number of types increased from four to eight. Thus, similar to the low-IS6110-copy M. tuberculosis strains, the novel spacers contributed significantly to improving the strain differentiation of M. bovis.
The two M. microti strains shared the six spacers characterized by sequencing one strain. The other one contained, in addition, 15 other spacers (Fig. 3). Finally, we investigated three M. canettii strains, isolated in France, The Netherlands, and Switzerland (28, 37). All of these strains displayed a hybridization pattern identical to that of the 94 spacer probes. These strains did not share any spacers with those found in the other M. tuberculosis complex strains (Fig. 3).
DISCUSSION
In this study we compared the sequence of the complete DR locus and the bordering DNA in 26 M. tuberculosis complex strains in an attempt to better understand the mechanisms underlying the genesis and evolution of these peculiar genetic elements with unknown function. The size of the DR locus varied from 6 DVRs (0.6 kb) to 56 DVRs (6 kb), and both spacers and DRs showed little sequence interstrain variation. The most remarkable finding is the strong conservation of the order of the various DVRs among the strains. We did not find a single strain in which the order of the DVRs differed. This indicates that during evolution individual DVRs did not move within the DR region. In addition, the individual DRs also do not seem to move because the six single DRs, which varied slightly from the consensus sequence, were all found at the same position within the DR region. Except for occasional insertion-element-driven polymorphisms, we found that the major type of polymorphism comprised the presence or absence of single, discrete DVRs or stretches of contiguous DVRs. The data suggest that the DR loci in the clinical isolates we see today are the remnant of a primordial DR locus, which was composed of a large number, perhaps hundreds, of different DVRs. The most likely mechanism underlying the strain-to-strain variation is the successive deletion of single or multiple discrete DVRs from this archetypal DR region. We also observed a few rare duplications of discrete DVRs, mostly as tandem duplications. These deletions and duplications probably have been mediated by homologous recombination between neighboring or distant DRs and/or by slippage during DNA replication. The rearrangements observed in presumed isogenic strains are consistent with this view. These strains differed in the presence or absence of a single discrete DVR or a stretch of contiguous DVRs and the presence or absence of IS6110 at an uncommon site in the DR region. These observations are consistent with the evolutionary scenario recently proposed by Fang et al. (10) for a closely related group of M. tuberculosis strains isolated from different geographic areas. In this scenario strain variation was thought to be due to the deletion of discrete DVRs in the DR region and due to the transposition of IS6110.
In some of the strains we observed deletions in the DNA flanking the DR region. In these strains DVRs were missing DVRs, which in other strains were present at the left or the right border of the DR locus (see Fig. 1). This indicates that the deletions in DR flanks took place concurrently with deletion of the DVRs, which normally delimit the DR region. The lack of the IS6110 element in these strains strongly suggests that these deletions were IS6110 mediated. These observations are consistent with the idea of a primordial DR locus in which successive deletions led to the presently observed genetic variation.
Although variation by deletion from a primordial DR locus seems the easiest explanation for the genetic variation in the DR locus of present-day strains, the genesis of the hypothetical primordial DR locus remains enigmatic. Presently, the complete genome sequence of two M. tuberculosis strains is known. We have searched these genomes for sequence similarity with any of the 94 different spacer sequences, but except for sequences within the DR locus no significant sequence similarity was found. Therefore, it seems unlikely that in the present-day strains novel spacers in the DR locus are generated from a template of existing sequences elsewhere in the mycobacterial genome. At one time the DR perhaps had the capacity to multiply by replicative transposition or retroposition within a nonessential region of the genome. However, at present no examples in nature are known by which short pieces of DNA are duplicated in such a way that the repeats become separated by similarly sized nonrepetitive intervening sequences. Perhaps the DVRs evolved from directly repeated DNA without intervening spacer DNA. These repeats could have acquired a biological function, such as replicon partitioning, as has been found in Haloferax spp. (26). When the selective force was imposed on repeat length and part of the specific repeat sequence, such repeats could have diverged to the present-day DVR elements with a constant part and a variable part. It should be noted that in bacterial plasmids repeats have been identified which are involved in the regulation of replication and plasmid compatibility. These repeats or “iterons” have sizes similar to those of the DVRs, and also some sequence variation is found within the iterons on a single plasmid (7). The number of iterons per cell determines the copy number and stability. Perhaps DVRs have evolved from repeats with a similar function as iterons in plasmids.
Previous studies have revealed the existence of 57 different spacers. Forty-three of these have been used in standard spoligotyping for strain differentiation of M. tuberculosis complex isolates on the basis of the strain-dependent presence or absence of these spacers (18). In this study we disclosed 37 novel spacer sequences. The majority of these, 26 sequences, were found in M. canettii, a recently described taxon within the M. tuberculosis complex group of bacteria (37). M. canettii shared not a single spacer with other M. tuberculosis complex strains, and none of the M. canettii spacers were found in other M. tuberculosis complex strains. Thus, the DR locus in M. canettii differs greatly from the DR loci in other members of the M. tuberculosis complex. This difference confirms previous observations showing that M. canettii differs in many respects from the other species in this group of mycobacteria, such as the presence of multiple mutations in certain housekeeping genes, multiple chromosomal deletions, and differences in the cell wall composition (37). Other than the large number of M. canettii-specific spacers, we disclosed 11 novel spacer sequences in M. tuberculosis and M. bovis. The use of these novel spacers for strain typing improved the degree strain differentiation, in particular of strains harboring few IS6110 copies. From the very first publications on spoligotyping it has appeared that strains with certain spoligotypes are polymorphic when analyzed by other genetic markers such as IS6110 (13, 18, 22). In this study we have confirmed that strains with certain common spoligotypes encountered among clinical isolates of M. tuberculosis have an identical or almost identical DR region sequence in spite of their unrelated IS6110 and PGRS RFLP patterns. This indicates that the DR region remained unchanged during a long period of time during which genetic rearrangements took place at other chromosomal loci. Three mutually nonexclusive explanations are possible: (i) the DNA arrangement of the DR region in these strains is frozen because of an unknown structural property of the specific DR region sequence or because of a poor ability in homologous recombination or slipped strand mispairing during replication; (ii) the specific sequence of the DR region in these strains provides them with a selective advantage and therefore variants with DVR rearrangements do not persist in the population, or (iii) these strains acquired the DR region from other strains by horizontal DNA transfer. The latter possibility seems unlikely because other studies suggest that the population structure of M. tuberculosis is clonal rather than panmictic (11, 22). Furthermore, certain spacers in M. bovis and M. canettii were not found in M. tuberculosis, suggesting the absence of lateral transfer of DVR sequences. Presently, it is impossible to distinguish between the first two possibilities because the function of the DR region in M. tuberculosis is unknown and we have not been able to derive M. tuberculosis mutants with a rearranged DR region in the laboratory. The only bacterial species in which a function of a DR-like region has been proposed is Haloferax spp., in which the number of repeats seems to be involved in replicon partitioning (26).
In this study we confirmed the existence M. tuberculosis strains which are genetically divergent as measured with markers such as IS6110 and PGRS and which even so have an identical DR region. This observation complicates the use of spoligotyping for epidemiological analysis. Ideally, polymorphic genetic markers should have molecular clocks with equal paces for different strains. Our study strongly suggests that large differences among M. tuberculosis strains exist in the pace of the molecular clock of the DR region. Strains with spoligotypes such as type 38 often exhibit very different IS6110 RFLP patterns, although their spoligotypes are identical, indicating that molecular changes due to the mobility of IS6110 is quicker than that of rearrangements in the DR locus. We found the reverse situation in strains with a single or few IS6110 copies. In such strains the insertion element seems to be “frozen,” whereas they do exhibit DR-associated DNA polymorphism. The apparent strain-dependent pace of different molecular clocks of the various genetic markers used in the epidemiology of tuberculosis underlines the point that great care should be taken in the interpretation of strain typing in particular when only a single genetic marker is used.
ACKNOWLEDGMENTS
This work was financially supported by the Dutch Foundation for Technical Sciences and the European Union project on the development of novel standardized methodology and nomenclature for the identification of M. bovis strains.
We acknowledge Marjori Beggs, David Brittain, Solvig Roring, Robin Skuce, and Z. Fang for providing us with unpublished sequences of the DR region in M. bovis and M. tuberculosis.
REFERENCES
- 1.Bachellier S, Gilson E, Hofnung M, Hill C W. Repeated sequences. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B Jr, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: American Society for cellular and molecular biology. 2nd ed. Washingington, D.C.: Microbiology; 1996. pp. 2012–2040. [Google Scholar]
- 2.Beggs M L, Cave M D, Marlowe C, Cloney L, Duck P, Eisenach K D. Characterization of Mycobacterium tuberculosis complex direct repeat sequence for use in cycling probe reaction. J Clin Microbiol. 1996;34:2985–2989. doi: 10.1128/jcm.34.12.2985-2989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blazquez J, Espinoza de Los Monteros L E, Samper S, Martin C, Guerrero A, Cobo J, van Embden J, Baquero F, Gomez-Mampaso E. Genetic characterization of multidrug-resistant Mycobacterium bovis from a hospital outbreak involving human immunodeficiency virus-positive patients. J Clin Microbiol. 1997;35:1390–1393. doi: 10.1128/jcm.35.6.1390-1393.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J D, Geoghagen N S, Weidman J F, Fuhrmann J L, Nguyen D T, Utterback T, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Hurst M A, Roberts K M, Kaine B B, Borodovsky M, Klenk H P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 5.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver S, Osborne J, Quail M A, Rajandream M A, Rogers J, Rutter S, Seeger K, Skelton S, Squares S, Squares R, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 6.Cousins D, Williams S, Liebana E, Aranaz A, Bunschoten A, van Embden J D A, Ellis T. Evaluation of four DNA typing techniques in epidemiological investigations of bovine tuberculosis. J Clin Microbiol. 1998;36:168–178. doi: 10.1128/jcm.36.1.168-178.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DelSolar G, Giraldo R, RuizEchevarria M J, Espinosa M, DiazOrejas R. Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diaz R, Kremer K, de Haas P, Gomez R I, Marrero A, Valdivia J A, van Embden J D A, van Soolingen D. Molecular epidemiology of tuberculosis in Cuba outside of Havana, July 1994–June 1995: utility of spoligotyping versus IS6110 restriction fragment length polymorphism. Int J Tuberc Lung Dis. 1998;2:743–750. [PubMed] [Google Scholar]
- 9.Driscoll J R, McGarry M A, Taber H W. DNA typing of a nonviable culture of Mycobacterium tuberculosis in a homeless shelter outbreak. J Clin Microbiol. 1998;37:274–275. doi: 10.1128/jcm.37.1.274-275.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang Z, Morrison N, Watt B, Doig C, Forbes K J. IS6110 transposition and evolutionary scenario of the direct repeat locus in a group of closely related Mycobacterium tuberculosis strains. J Bacteriol. 1998;180:2102–2109. doi: 10.1128/jb.180.8.2102-2109.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frottingham R, Meeker-O'Connell W A. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology. 1998;144:1189–1196. doi: 10.1099/00221287-144-5-1189. [DOI] [PubMed] [Google Scholar]
- 12.Gordon S V, Brosch R, Billault A, Garnier T, Eiglmeijer K, Cole S T. Identification of variable regions in the genome of tubercle bacilli using artificial chromosome arrays. Mol Microbiol. 1999;32:643–655. doi: 10.1046/j.1365-2958.1999.01383.x. [DOI] [PubMed] [Google Scholar]
- 13.Goyal M, Saunders N A, van Embden J D A, Young D B, Shaw R J. Differentiation of Mycobacterium tuberculosis isolates by spoligotyping and IS6110 restriction fragment length polymorphism. J Clin Microbiol. 1997;35:647–651. doi: 10.1128/jcm.35.3.647-651.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groenen P M A, Bunschoten A E, van Soolingen D, van Embden J D A. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis: application for strain differentiation by a novel typing method. Mol Microbiol. 1993;10:1057–1085. doi: 10.1111/j.1365-2958.1993.tb00976.x. [DOI] [PubMed] [Google Scholar]
- 15.Gutiérrez M, Samper S, Jiménez M S, Embden J D A, García-Marín J F, Martin C. Identification by spoligotyping of a caprine genotype in Mycobacterium bovis strains causing human tuberculosis. J Clin Microbiol. 1997;35:3328–3330. doi: 10.1128/jcm.35.12.3328-3330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hermans P W M, van Soolingen D, Bik E M, de Haas P E W, Dale J W, van Embden J D A. The insertion element IS987 from Mycobacterium bovis BCG is located in a hot spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect Immun. 1991;59:2695–2705. doi: 10.1128/iai.59.8.2695-2705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoe N, Nakashima K, Grigsby D, Pan X, Dou S J, Naidich S, Garcia M, Kahn E, Bergmire-Sweat D, Musser J M. Rapid molecular genetic subtyping of serotype M1 group A Streptococcus strains. Emerg Infect Dis. 1999;5:254–263. doi: 10.3201/eid0502.990210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamerbeek J, Schouls L M, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten J E, Molhuizen H, Shaw R, Goyal M, van Embden J D A. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapur V, Whittam T S, Musser J M. Is Mycobacterium tuberculosis 15,000 years old? J Infect Dis. 1994;170:1348–1349. doi: 10.1093/infdis/170.5.1348. [DOI] [PubMed] [Google Scholar]
- 20.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, Sakai M, Ogura K, Otuka R, Nakazawa H, Takamiya M, Ohfuku Y, Funahashi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Nakamura Y, Robb T F, Horikoshi K, Masuchi Y, Shizuya H, Kikuchi H. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 21.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides B A, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L X, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, Dandrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 22.Kremer K, van Soolingen D, Frothingham R, Haas W H, Hermans P W M, Martin C, Palittapongarnpim P, Plikaytis B B, Riley L, Yakrus M A, Musser J M, van Embden J D A. Comparison of methods for typing of Mycobacterium tuberculosis complex strains for use in the epidemiology: an interlaboratory study on strain discriminatory power and reproducibility. J Clin Microbiol. 1999;37:2607–2618. doi: 10.1128/jcm.37.8.2607-2618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long R, Nobert E, Chomyc S, van Embden J D A, McNamee C, Duran R R, Talbot J, Fanning A. Transcontinental spread of multidrug-resistant Mycobacterium bovis. Am J Respir Crit Care Med. 1999;159:2014–2017. doi: 10.1164/ajrccm.159.6.9809076. [DOI] [PubMed] [Google Scholar]
- 24.Masepohl B, Gorlitz K, Bohme H. Long tandemly repeated repetitive (LTRR) sequences in the filamentous cyanobacterium Anabaena sp. PCC 7120 Biochim. Biophys Acta Gene Struct Expr. 1996;1307:26–30. doi: 10.1016/0167-4781(96)00040-1. [DOI] [PubMed] [Google Scholar]
- 25.McAdam R A, Weisbrod T R, Martin J, Scuderi J D, Brown A M, Cirillo J D, Bloom B R, Jacobs W R., Jr In vivo growth characteristics of leucine and methionine auxotrophic mutants of Mycobacterium bovis BCG generated by transposon mutagenesis. Infect Immun. 1995;63:1004–1012. doi: 10.1128/iai.63.3.1004-1012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moijica F J M, Ferrer G, Rodriguez-Valera F. Long stretches of short tandem repeats are present in the largest replicons of the archea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol Microbiol. 1995;17:85–93. doi: 10.1111/j.1365-2958.1995.mmi_17010085.x. [DOI] [PubMed] [Google Scholar]
- 27.Nakata A, Amemura M, Makino K. Unusual nucleotide arrangement with repeated sequences in the Escherichia coli K-12 chromosome. J Bacteriol. 1989;171:3553–3356. doi: 10.1128/jb.171.6.3553-3556.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfyffer G, Auckenthaler E R, van Embden J D A, van Soolingen D. Mycobacterium canettii, the smooth variant of M. tuberculosis, isolated from a Swiss patient exposed in Africa. Emerg Infect Dis. 1998;4:631–634. doi: 10.3201/eid0404.980414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poulet S, Cole S T. Repeated DNA sequences in mycobacteria. Arch Microbiol. 1995;163:79–86. doi: 10.1007/BF00381780. [DOI] [PubMed] [Google Scholar]
- 30.Qian L, Douglas J T, van der Zanden A, Duanmu H, van Embden J D A. Retrospective analysis of M. tuberculosis DNA types in preserved lung tissues. J Clin Microbiol. 1999;37:471–474. doi: 10.1128/jcm.37.2.471-474.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shreevatsan S, Escalante P, Pan X, Stockbauer K E, Conell N D, Kreiwirth B N, Whittam T S, Musser J M. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionary recent global dissemination. Proc Natl Acad Sci USA. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Small P M, van Embden J D A. Molecular epidemiology of tuberculosis. In: Bloom B, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C.: ASM Press; 1994. pp. 569–582. [Google Scholar]
- 33.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sola C, Devallois A, Horgen L, Maïsetti J, Filliol I, Legrand E, Rastogi N. Tuberculosis in the Caribbean: using spacer oligonucleotide typing to understand strain origin and transmission. Emerg Infect Dis. 1999;5:404–414. doi: 10.3201/eid0503.990311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor G M, Goyal M, Legge A J, Shaw R J, Young D. Genotypic analysis of Mycobacterium tuberculosis from medieval human remains. Microbiology. 1999;145:899–904. doi: 10.1099/13500872-145-4-899. [DOI] [PubMed] [Google Scholar]
- 36.van der Zanden A G M, Hoentjen A H, Heilmann F G C, Weltevreden E F, Schouls L M, van Embden J D A. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis complex in paraffin wax embedded tissues and in stained microscopic preparations. Mol Pathol. 1998;51:209–214. doi: 10.1136/mp.51.4.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Soolingen D, Hoogenboezem T, de Haas P E W, Hermans P W M, Koedam M A, Teppema K S, Brennan P J, Besra G S, Portaels F, Top J, Schouls L M, van Embden J D A. A novel pathogenic taxon of the Mycobacterium tuberculosis complex, Canetti: characterization of an exceptional isolate from Africa. Int J Syst Bacteriol. 1997;47:1236–1245. doi: 10.1099/00207713-47-4-1236. [DOI] [PubMed] [Google Scholar]
- 38.van Soolingen D, Qian L, de Haas P E W, Douglas J T, Traore H, Portaels F, Qing H Z, Enkhasaikan D, Nymadawa P, van Embden J D A. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J Clin Microbiol. 1995;33:3234–3238. doi: 10.1128/jcm.33.12.3234-3238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Soolingen D, de Haas P, Hermans P W M, Groenen P, van Embden J D A. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of M. tuberculosis. J Clin Microbiol. 1993;31:1987–1995. doi: 10.1128/jcm.31.8.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Soolingen D, van der Zanden A G M, de Haas P E W, Noordhoek G T, Kiers A, Foudraine N A, Portaels F, Kolk A H J, Kremer K, van Embden J D A. Diagnosis of Mycobacterium microti infections among humans by using novel genetic markers. J Clin Microbiol. 1998;36:1840–1845. doi: 10.1128/jcm.36.7.1840-1845.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Soolingen D, Hermans P W M. Epidemiology of tuberculosis by DNA fingerprinting. Eur Respir J. 1995;8:S649–S656. [PubMed] [Google Scholar]