Abstract
Pseudomonas aeruginosa OprM is a protein involved in multiple-antibiotic resistance as the outer membrane component for the MexA-MexB-OprM efflux system. Planar lipid bilayer experiments showed that OprM had channel-forming activity with an average single-channel conductance of only about 80 pS in 1 M KCl. The gene encoding OprM was subjected to insertion mutagenesis by cloning of a foreign epitope from the circumsporozoite form of the malarial parasite Plasmodium falciparum into 11 sites. In Escherichia coli, 8 of the 11 insertion mutant genes expressed proteins at levels comparable to those obtained with the wild-type gene and the inserted malarial epitopes were surface accessible as assessed by indirect immunofluorescence. When moved to a P. aeruginosa OprM-deficient strain, seven of the insertion mutant genes expressed proteins at variable levels comparable to that of wild-type OprM and three of these reconstituted MIC profiles resembling those of the wild-type protein, while the other mutant forms showed variable MIC results. Utilizing the data from these experiments, in conjunction with multiple sequence alignments and structure predictions, an OprM topology model with 16 β strands was proposed.
One general mechanism that contributes to intrinsic multiple antibiotic resistance in gram-negative bacteria is multidrug efflux. Resistance-nodulation-division efflux systems are usually composed of three components: a cytoplasmic membrane pump protein, an outer membrane channel, and a cytoplasmic membrane-anchored membrane fusion protein which helps to link the other two components. A large number of such resistance-nodulation-division efflux system homologues have been identified in a wide variety of species (24). Antimicrobial agents that enter the cell or the cytoplasmic membrane are captured by the pump and extruded directly into the medium. Bacteria expressing these multidrug efflux systems are resistant to several chemical classes of antibiotics and are thus causing great concern. It is important to understand the molecular mechanism of these efflux systems in order to design methods of inhibition.
In Pseudomonas aeruginosa, a number of multidrug efflux systems have been identified to date (19, 26, 31, 32). The mexA-mexB-oprM system is expressed constitutively in wild-type P. aeruginosa and contributes to the intrinsic resistance and, when overexpressed, to the mutational resistance of this microorganism to a wide variety of structurally unrelated antimicrobial agents (18, 21, 22). In addition to multiple-antibiotic resistance, the MexA-MexB-OprM system was also shown to affect quorum sensing, possibly through the export of the P. aeruginosa autoinducer N-(3-oxo)-dodecanoyl-l-homoserine lactone (9). OprM has been assumed to be a porin-like protein and is homologous to the outer membrane components of other efflux systems in P. aeruginosa and in other bacteria. Similar to the Escherichia coli TolC protein, OprM was shown to be able to act as the outer membrane component of other efflux systems (38, 46) and was important for efficient efflux of multiple antibiotics. However, of the efflux outer membrane proteins, only the distantly related protein TolC has been shown to have porin activity (5). Although the proton motive force-driven pump proteins and the membrane fusion proteins anchored in the cytoplasmic membrane have been shown to be responsible for substrate selectivity (38), they do not work in the absence of an outer membrane component, which must therefore be important for direct passage of the wide range of antimicrobial agents into the medium. Understanding the mechanism whereby OprM is involved in facilitating the extrusion of antibiotics will help to develop appropriate inhibitory compounds for these multidrug efflux systems. A knowledge of the molecular structure of OprM is also a starting point for studying its structure-function relationships.
The structures of a number of porins have been determined by mutagenesis and crystallography (1, 8, 37, 41). They consist of three identical monomers, each of which is a 16- to 18-stranded β barrel whose β strands are tilted with respect to the plane of the membrane. The β strands, which often comprise alternating hydrophobic and hydrophilic amino acids and are relatively conserved in sequence and length among homologous proteins, are connected by longer, more flexible loops exposed to the cell surface and shorter loops exposed to the periplasmic space. OprM does not show very high homology to either general porins or substrate-specific porins. However, it was possible here to identify conserved regions of amino acid residues from its multiple-sequence alignment with various highly homologous outer membrane proteins that are also involved in efflux. In fact, there are approximately 20 homologues of OprM in the P. aeruginosa genome (our web page, http://www.cmdr.ubc.ca/bobh/OprMfamily.html). Applying this information with structure prediction methods such as β-turn prediction (30) and hydrophobicity calculation (10) for outer membrane proteins, together with the results from insertion mutagenesis of OprM in this study, a topology model for an OprM monomer is proposed here to provide a basis for understanding the structure-function relationship of this efflux system component.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli DH5α was used as the primary host for expressing oprM mutant plasmids. Strain CE1248 (3), deficient in the expression of OmpC, OmpF, and PhoE, was used for the expression experiments. For expression experiments and antimicrobial susceptibility assays with P. aeruginosa, OprM-deficient mutant strain OCR03T (12) was utilized. The initially cloned oprM gene (termed here oprM*) carried on plasmid pT7-7 (40) was used as the source for creation of the insertion mutants. Plasmid pUC4KAPA (obtained from John Smit, University of British Columbia) was used as the source of the kanamycin resistance cassette contained on a HincII fragment. The oprM gene and its various insertion mutant forms were then cloned into plasmid pVLT35 or its derivative pVLT31 (23) for expression and antimicrobial susceptibility studies. Plasmids pXZL33 and pXZL34, with the native oprM gene cloned into pT7-7 and pVLT31, respectively, were obtained from Keith Poole.
Media.
Bacterial strains were routinely grown with shaking at 37°C in Luria broth (LB) medium (1% tryptone–0.5% yeast extract–0.5% NaCl for E. coli, 0.05% NaCl for P. aeruginosa strains) or on LB agar with the addition of 2% (wt/vol) Bacto Agar. The following antimicrobials were used in selective media: HgCl2 (15 μg/ml) for OprM-deficient P. aeruginosa strain OCR03T; ampicillin (100 μg/ml), spectinomycin (30 μg/ml), and tetracycline (10 μg/ml) for E. coli with constructs made from pT7-7, pVLT35, and pVLT31, respectively; and streptomycin (45 μg/ml) and tetracycline (50 μg/ml) for OCR03T with constructs made from pVLT35 or pVLT31, respectively.
For expression of insertion mutant forms of oprM and oprM, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to mid-log-phase cell cultures to a final concentration of 0.1 mM and cells were induced for 2 h before harvest.
DNA methodology.
Restriction endonucleases and T4 DNA ligase purchased from Gibco/BRL and New England Biolabs, Inc., were used in accordance with the protocols supplied by the manufacturers. Plasmid DNA was prepared by the alkaline lysis protocol as previously described (35). Transformations of E. coli and P. aeruginosa with plasmid DNA or ligation products were performed by the CaCl2 and MgCl2 protocols, respectively (35).
Insertion mutagenesis.
Insertion of 12-bp linkers into the oprM gene was accomplished as described previously (43). Briefly, pT7-7::oprM* was partially digested with frequently cutting blunt-end restriction endonuclease RsaI or HaeIII. After digestion, the singly cut plasmid was purified and ligated to a 1.3-kb HincII fragment isolated from the kanamycin resistance plasmid pUC4KAPA. After transformation of the ligation products to E. coli DH5α, clones were selected for both ampicillin and kanamycin resistance. The resultant colonies were screened by PCR assay for insertion of the Kanr gene into oprM, using an internal primer for the Kanr gene and a forward primer annealing to the 5′ end and a reverse primer annealing to the 3′ end of oprM*. The kanamycin resistance gene was then removed by PstI digestion, leaving a 12-bp linker sequence containing the unique PstI site in oprM*. After religation of the plasmids, these were transformed back into DH5α and sequenced. The linker mutant plasmids were then linearized by PstI and ligated to synthetic oligonucleotides encoding the malarial epitope with the corresponding reading frame to ensure correct translation. The resultant insertion mutant forms of oprM* were then excised by XbaI and HindIII and cloned into pVLT35 for expression experiments.
When pXZL33 became available, PCR was used for site-directed insertion of the malarial epitope into oprM. Primers containing a DNA region encoding the amino acid sequence KRKNPNANPNANPN were designed to introduce the malarial epitope (NANP) at the chosen insertion sites. The KRK motif was used to ensure that insertion into the amphipathic β strands would interrupt the correct folding of the protein. The PCR products were then phosphorylated, religated, and transformed into E. coli DH5α cells, and the oprM insertion mutant forms were eventually cloned into pVLT35 or pVLT31 for expression experiments.
Outer membrane preparation, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and Western immunoblotting.
Outer membranes were isolated by passage through a French press and sucrose density gradients or Sarkosyl solubilization, and samples were subjected to SDS-PAGE as previously described (13, 34). Proteins were stained with Coomassie brilliant blue. Protein concentrations of the isolated outer membrane samples were determined by a modified Lowry assay (36).
For Western immunoblotting, proteins from unstained gels were transferred to Immobilon polyvinylidene difluoride membranes (Millipore, Bedford, Mass.) in cold transfer buffer (25 mM Tris, 0.2 M glycine, 20% [vol/vol] methanol) at 100 V for 1 h. Proteins were then detected as previously described (29), using an OprM-specific murine monoclonal antibody (kindly provided by N. Gotoh) or a monoclonal antibody against the malarial epitope (pf2A.10, obtained from R. Wirtz) as the primary antibody and a goat anti-mouse immunoglobulin G alkaline phosphatase-conjugated secondary antibody. The bound antibodies were detected with 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium.
Indirect immunofluorescence.
Surface-exposed proteins were detected by the method of Hofstra et al. (14). Briefly, aliquots of cells were incubated with an anti-OprM or anti-malarial epitope monoclonal antibody and then a fluorescein isothiocyanate-conjugated secondary antibody (Gibco/BRL). Washings were done with phosphate-buffered saline. Cells were spread and air dried on poly-l-lysine-coated slides (Sigma) and covered with mounting reagent and a coverslip, and fluorescence was monitored with a Zeiss microscope fitted with a halogen lamp and filters set for emission at 525 nm.
DNA sequencing.
Primers annealing to different regions of oprM were synthesized on an ABI DNA synthesizer. Template DNA was prepared with a QIAprep spin miniprep kit (Qiagen), and PCRs were done in accordance with the protocols provided by Applied Biosystems Inc. DNA sequencing was performed on an ABI model 373 sequencer using the manufacturer's protocols.
MIC determinations.
MICs were determined by serial twofold dilution in LB using the method described by Amsterdam (2). Results were determined after incubation at 37°C for 24 h.
Solubilization and purification of protein.
E. coli strain CE1248 carrying oprM on pVLT35 (pKW35TM) was used to isolate OprM. Outer membrane proteins isolated from sucrose density gradients were solubilized subsequently in 0.5 and 3% (vol/vol) n-octyl-polyoxyethylene (Bachem Bioscience Inc.) in 10 mM Tris (pH 8). This fraction was dialyzed into buffer A (10 mM Tris [pH 8], 1% [vol/vol] n-octyl-polyoxyethylene) and then passed through an anion-exchange MonoQ column and eluted with buffer B (buffer A with 1 M NaCl) by fast protein liquid chromatography. A fraction containing OprM was subjected to SDS-PAGE without heating of the sample, and OprM was excised from the gel and eluted in 10 mM Tris (pH 8) with 0.1% SDS at 4°C overnight.
Planar lipid bilayer experiments.
Analysis of the pore-forming ability of proteins was done with the planar lipid bilayer technique as previously described (4). Membranes were composed of 1.5% oxidized cholesterol in n-decane. Purified protein was diluted into 0.1% (vol/vol) Triton X-100 (1:100) prior to addition to the solution of 1 M KCl bathing the planar bilayer membrane.
RESULTS
Prediction of OprM topology.
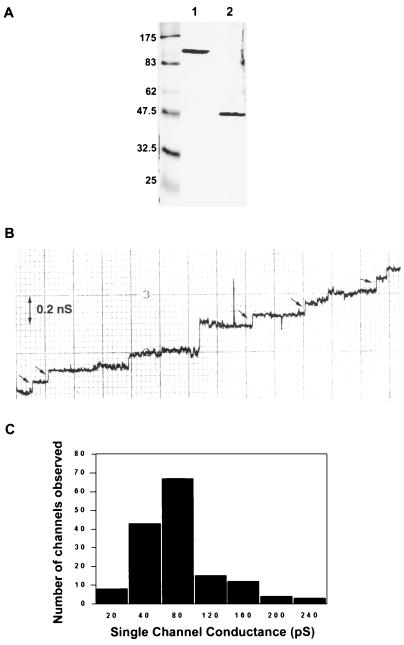
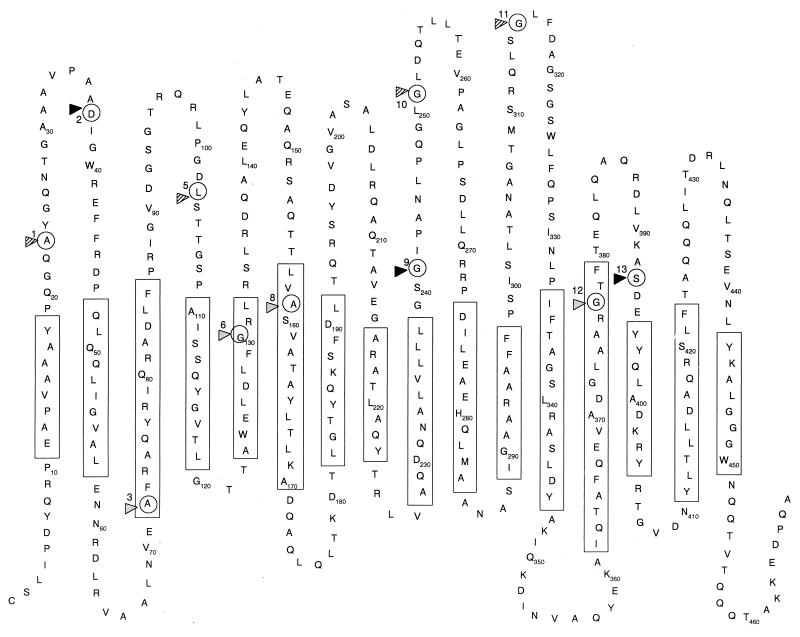
OprM has been presumed to form a channel in the outer membrane of P. aeruginosa to assist in the efflux of multiple antibiotics (24, 28). OprM showed some homology to and thus was predicted to function similarly to the E. coli outer membrane protein TolC, which had been previously shown to form channels (5) and was predicted, based on studies with two-dimensional crystals, to form a trimer of β-barrel monomers (20). Data from planar lipid bilayer experiments using the gel-purified oligomeric form of OprM showed that it had channel-forming activity with a single-channel conductance of 82 ± 3.8 pS (mean ± standard error of 152 events) in 1 M KCl (pH 7) at an applied membrane potential of 50 mV (Fig. 1), which was similar to that obtained from oligomeric TolC in the same salt concentration (5). Hence, a topology model of OprM was predicted based on the β-barrel motif evident in previously published porin models. Multiple-sequence alignment with several homologous proteins was performed to identify conserved regions. In addition, possible β turns were identified and the amphipathicity profile of OprM was calculated using previously published methods (10, 15). Using these methods, an OprM topology model with 16 β strands connected by long, surface-exposed loops and shorter, periplasmic loops was proposed. The identified amphipathic and conserved regions were mostly contained in the transmembrane β strands, and residues with both hydrophobic and hydrophilic properties, such as tyrosine, were localized mainly to the water-lipid interface. This preliminary model was tested by insertion mutagenesis to determine the flexible (loop region) portions of OprM, and the model was revised accordingly (Fig. 2).
FIG. 1.
(A) SDS-PAGE of purified OprM. The sample was untreated (lane 1) or heated at 100°C for 10 min (lane 2). The molecular masses (in kilodaltons) of the prestained markers in the leftmost lane are indicated on the left. (B) Step increases in single-channel conductance after OprM was added to the aqueous phase (1 M KCl) bathing the lipid bilayer membrane. Each small arrow indicates an increase of 80 pS. (C) Histogram of single-channel conductance increases from more than 150 membrane insertion events. Conductance increases showed a mean distribution (± standard error) of 82 ± 3.8 pS.
FIG. 2.
Predicted membrane topology model of the OprM monomer. Rectangles enclose the proposed transmembrane β strands. Circles indicate insertion sites ME1 to -3, -5, -6, and -8 to -13 (numbered correspondingly) for the malarial epitope. Permissive insertions are indicated by solid triangles, partially permissive ones are indicated by shaded triangles, and nonpermissive ones are indicated by gray triangles.
Epitope insertion mutagenesis.
Epitope insertion mutagenesis was done to determine if insertion of a large stretch of amino acids was tolerated in various regions of OprM. The malarial epitope from the circumsporozoite form of Plasmodium falciparum was chosen because previous studies in this laboratory have shown that this epitope is permissive and antigenic when inserted into the loop regions of the outer membrane proteins OprF (44) and OprP (39). Nine oprM insertion mutant forms were created by random insertion of the malarial epitope (NANP) repeats into oprM*, and these constructs were named pKWIN plasmids (Table 1). The protein encoded by oprM* contained a substitution of 14 unrelated amino acids for the 22 C-terminal residues of native OprM but was capable of reconstituting OprM function (42). To supplement these mutants, a PCR approach was used to create four additional epitope insertion mutant forms of oprM with the native 3′ sequence when pXZL33 became available (Table 1). When the whole oprM gene from these mutants was sequenced, some PCR-introduced errors were discovered. Except for the Y18C and Q20C changes in pKWIN2, the other three plasmids contained conserved changes (A110V in pKWIN5, S152T in pKWIN9, and A400V in pKWIN13). We retained pKWIN2 for our studies, since subsequent analysis demonstrated efficient expression in both E. coli and P. aeruginosa, indicating the relatively benign effects of the observed mutations. The insertion sites were named ME1 to ME13 from the N to the C terminus, corresponding to the pKWIN plasmid number (i.e., pKWIN1 carries an insertion at site ME1).
TABLE 1.
Expression and characterization of OprM epitope insertion mutant proteins in E. coli
| Plasmida | Insertion site (amino acid)b | Amino acids inserted | Protein expression (by SDS-PAGE)c | Surface reactivityd (antimalarial) |
|---|---|---|---|---|
| pVLT35 | None (control vector) | − | − | |
| pKW35TMe | None (wild-type OprM) | +++ | − | |
| pKWIN1 | ME1 (23) | GPAPNA(NPNA)2GHAGP | +++ | +++ |
| pKWIN2f | ME2 (37) | KRK(NPNA)2NPN | +++ | +++ |
| pKWIN3 | ME3 (72) | GPAPNA(NPNA)2GHAGP | + | − |
| pKWIN4 | ME4 (77) | None (stop) | − | − |
| pKWIN5f | ME5 (103) | KRKNPNANPN | +++ | +++ |
| pKWIN6 | ME6 (130) | TC(NPNA)3CRS | +++ | +++ |
| pKWIN7 | ME7 (143) | None (stop) | − | − |
| pKWIN8 | ME8 (159) | GPAPNA(NPNA)2GHAGP | ++ | +++ |
| pKWIN9f | ME9 (241) | KRK(NPNA)2NPN | + | + |
| pKWIN10 | ME10 (251) | GTC(NPNA)3CRS | ++++ | +++ |
| pKWIN11 | ME11 (315) | GTC(NPNA)3CRS | +++ | +++ |
| pKWIN12 | ME12 (377) | DLQ(NPNA)2NALDVQV | ++ | + |
| pKWIN13f | ME13 (393) | KRKNPNAPNANPN | + | − |
Plasmids pKWIN1 to pKWIN12 contain insertion mutation oprM cloned into pVLT35; pKWIN13 contains insertion mutation oprM cloned into pVLT31 (a derivative of pVLT35).
Position 1 is the N-terminal amino acid of the mature OprM amino acid sequence.
These results were obtained using outer membranes of E. coli CE1248 containing the various plasmids. Expression levels ranged from undetectable (−) to strong (++++). Expression was confirmed by Western immunoblot assay using a murine monoclonal antibody against OprM.
Fluorescence levels ranged from no (−) to strong (+++) fluorescence as assessed by indirect immunofluorescence assay.
pKW35TM contains wild-type oprM with the native sequence cloned into pVLT35.
This plasmid contains the native oprM sequence at the 3′ end; the other plasmids contain an insertion into the previously published oprM sequence (termed oprM*).
Expression of OprM insertion mutant proteins in E. coli.
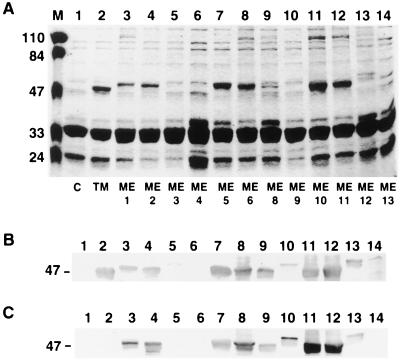
The various pKWIN plasmids were transformed into porin-deficient E. coli strain CE1248 to allow overexpression of the cloned insertion oprM mutant forms. Outer membranes were isolated for analysis by SDS-PAGE and Western immunoblotting with both anti-OprM and anti-malarial epitope antibodies. Plasmid pKW35TM (pVLT35::oprM with the native sequence) and the control vector pVLT35 were also transformed into E. coli strain CE1248 as controls.
Insertions at sites ME4 and ME7 generated translational stop sites and, as expected, did not produce mutant proteins (Fig. 3A to C; Table 1). Three of the mutant plasmids, with insertion sites at ME3, ME9, and ME13, led to expression of the mutant proteins at greatly reduced levels in comparison with wild-type OprM. Mutant proteins with insertions at ME8 and ME12 were expressed at intermediate levels but were strongly detected by the anti-OprM antibody. The other six plasmids directed expression of the mutant proteins at levels similar to or higher than that from pKW35TM, as determined by SDS-PAGE and Western immunoblotting with an anti-OprM antibody (Fig. 3A and B). Most of these mutant proteins exhibited slightly lower electrophoretic mobility compared to that of wild-type OprM. Similar results were obtained from Western immunoblotting with an anti-malarial epitope antibody, except that the mutant proteins with insertions at ME12 and ME13 became less detectable and undetectable, respectively (Fig. 3C).
FIG. 3.
(A) SDS-PAGE of outer membranes of E. coli CE1248 carrying control vector pVLT35 (lane 1; C), native OprM-expressing plasmid pKW35TM (lane 2; TM), and OprM-encoding plasmids with malarial epitope insertions at sites ME1 to ME6 (lanes 3 to 8) and ME8 to ME13 (lanes 9 to 14). Each lane was loaded with 20 μg of protein, treated with 5% (vol/vol) β-mercaptoethanol, and heated at 100°C for 10 min. The molecular masses (in kilodaltons) for the prestained markers in lane M are indicated on the left. Expression of oprM and the insertion mutants was confirmed by Western immunoblotting with an anti-OprM antibody (B) and an anti-malarial epitope antibody (C). Samples were in the same order as in the gel shown in panel A, and the corresponding regions of the blots are shown, with the relevant molecular mass indicated on the left.
Surface exposure of insertion mutant proteins.
To determine if the OprM mutant proteins were properly inserted into the outer membrane of the E. coli host cells, clones were subjected to indirect immunofluorescence for surface exposure analysis of the mutant proteins. Cells carrying the pKWIN plasmids or the control plasmids pVLT35 and pKW35TM were incubated with anti-malarial epitope antibodies, followed by a goat anti-mouse fluorescein isothiocyanate-conjugated antibody, and examined under a fluorescence microscope. Those seven mutant proteins detected strongly in a Western immunoblot with an antibody specific for the malarial epitope also bound the antibody on the surface of the cell, demonstrating that these mutant proteins were surface exposed and that insertions in these sites were similarly surface exposed (Table 1). Mutant proteins with insertions at ME9 and ME12 fluoresced very weakly. The other two mutant proteins, with insertions at ME3 and ME13, did not fluoresce. These clones also showed reduced or undetectable levels of the mutant proteins. Thus, we assumed that these sites, ME3, -9, -12, and -13, were not well exposed on the surface of E. coli CE1248.
Expression of OprM insertion mutant proteins in P. aeruginosa.
To determine if the OprM mutant proteins were also expressed in similar ways in the native host for OprM, the pKWIN plasmids and the negative control plasmids pVLT35 and pVLT31 and the native OprM-expressing plasmid pKW35TM were transformed into P. aeruginosa. Strain OCR03T, an OprM-deficient mutant of P. aeruginosa, was utilized to ensure that the modest level of OprM constitutively produced in wild-type cells would not interfere with detection of the mutant proteins. Interestingly, the expression pattern of the OprM mutant proteins in P. aeruginosa strain OCR03T was somewhat different from that obtained in E. coli strain CE1248.
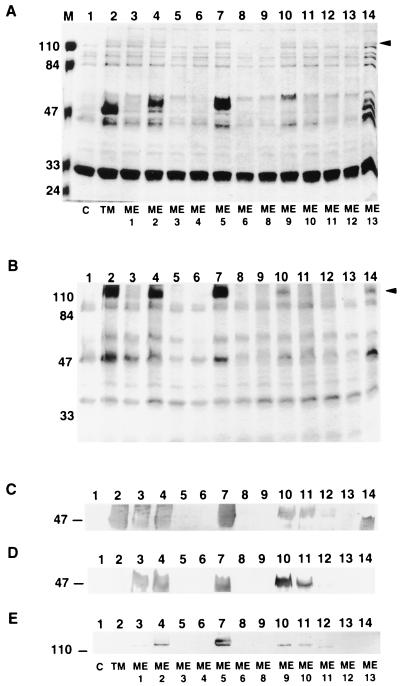
Among the three plasmids which produced reduced levels of mutant proteins in E. coli, pKWIN3 did not produce any of the mutant protein in P. aeruginosa OCR03T cells while plasmids pKWIN9 and pKWIN13 led to medium expression levels of the oprM mutant forms they carried (Fig. 4A to C). The result for these latter two plasmids could reflect the ability of MexAMexB, present only in P. aeruginosa, to stabilize these mutant forms of OprM or could reflect different proteases that degrade mutant proteins in these two organisms. On the other hand, three other plasmids (pKWIN6, pKWIN8, and pKWIN12) led to undetectable expression of their oprM mutant forms in P. aeruginosa (Fig. 4A to C; Table 2). We assume that this reflects the enhanced susceptibility of these mutant OprM forms to proteolytic degradation in P. aeruginosa. P. aeruginosa OCR03T cells carrying the remaining five plasmids (pKWIN1, pKWIN2, pKWIN5, pKWIN10, and pKWIN11) showed various levels of expression of the oprM mutant forms (Fig. 4A to C; Table 2).
FIG. 4.
(A) SDS-PAGE of outer membranes of P. aeruginosa OCR03T carrying control vector pVLT35 (lane 1; C), native OprM-expressing plasmid pKW35TM (lane 2; TM), and OprM-encoding plasmids with malarial epitope insertions at sites ME1 to ME6 (lanes 3 to 8) and ME8 to ME13 (lanes 9 to 14). Each lane was loaded with 20 μg of protein, treated with 5% β-mercaptoethanol, and heated at 100°C for 10 min. The molecular masses (in kilodaltons) of the prestained markers in lane M are indicated on the left. The arrowhead on the right indicates the position of the oligomeric forms of the proteins. (B) SDS-PAGE of the same samples unheated (room temperature for 10 min) and loaded in the same order as in panel A. The arrowhead on the right indicates the position of the oligomeric forms of the proteins. Relative molecular masses (in kilodaltons) are indicated on the left. Expression of insertion oprM mutant forms was confirmed by Western immunoblotting with an anti-OprM antibody (C) and an anti-malarial epitope antibody (D and E). The corresponding regions of the blots are shown, with the relevant molecular mass (in kilodaltons) indicated on the left. Samples are in the same order as in the gel shown in panel A. Proteins for the Western immunoblots were untreated (E) or treated with 5% (vol/vol) β-mercaptoethanol and heated at 100°C for 10 min (C and D).
TABLE 2.
Expression and characterization of OprM epitope insertion mutant proteins in P. aeruginosa
| Plasmida | Insertion site (amino acid)b | Western immunoblot resultc obtained with:
|
|
|---|---|---|---|
| Anti-OprM antibody | Antimalarial epitope antibody | ||
| pVLT35 | None (control vector) | − | − |
| pKW35TMd | None (wild-type OprM) | +++ | − |
| pKWIN1 | ME1 (23) | ++ | +++ |
| pKWIN2e | ME2 (37) | +++ | +++ |
| pKWIN3 | ME3 (72) | − | − |
| pKWIN4 | ME4 (77) | − | − |
| pKWIN5e | ME5 (103) | +++ | +++ |
| pKWIN6 | ME6 (130) | − | − |
| pKWIN7 | ME7 (143) | − | − |
| pKWIN8 | ME8 (159) | − | − |
| pKWIN9e | ME9 (241) | ++ | +++ |
| pKWIN10 | ME10 (251) | ++ | +++ |
| pKWIN11 | ME11 (315) | + | ++ |
| pKWIN12 | ME12 (377) | − | − |
| pKWIN13e | ME13 (393) | ++ | − |
pKWIN1 to pKWIN12 contain insertion mutant gene oprM cloned into pVLT35, and pKWIN13 contains insertion mutant gene oprM cloned into pVLT31 (a derivative of pVLT35).
Position 1 is the N-terminal amino acid of the mature OprM amino acid sequence.
These results were obtained using outer membranes of P. aeruginosa OprM-deficient strain OCR03T containing the various plasmids. Expression levels ranged from undetectable (−) to strong (+++).
pKW35TM contains wild-type oprM cloned into pVLT35.
This plasmid contains the native oprM sequence at the 3′ end, and the other plasmids contain an insertion into oprM*, the previously published oprM sequence.
When reacted with the anti-malarial epitope antibody, most of the results obtained with the OprM mutant proteins were similar to that obtained with the anti-OprM antibody, except for the mutant OprM protein from pKWIN13, which became undetectable (Fig. 4D). This was probably due to inaccessibility of the epitope when inserted at site ME13. Wild-type OprM is heat modifiable and partially runs on SDS-PAGE as an oligomeric form of about 100 kDa (25). This oligomeric form was also observed for the mutant forms of OprM that were expressed (Fig. 4A and B). When reacted with the antimalarial epitope antibody, these expressed mutant proteins gave results similar to those given by their monomeric forms (Fig. 4E).
Antimicrobial susceptibilities of P. aeruginosa cells carrying the OprM insertion mutants.
The P. aeruginosa clones carrying the various pKWIN plasmids were subjected to antimicrobial susceptibility assays to determine if the malarial epitope insertions affected the function of the OprM mutants. MICs of various antimicrobial agents were determined for the P. aeruginosa clones (Table 3). Antimicrobial susceptibility assays performed on clones carrying the two oprM sequences in a P. aeruginosa oprM-deficient background strain did not show any significant differences (Table 3). Apparently, variation at the 3′ end of oprM did not affect the function of the protein and so should not have affected the results in this study.
TABLE 3.
Antimicrobial susceptibilities of P. aeruginosa OprM-deficient strain OCR03T carrying plasmids expressing wild-type or insertion mutant forms of oprM
| Plasmid | OprM/mutant protein expressiona | MICb (μg/ml) of:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TET | CM | NA | NFX | MERO | CTAX | CFP | CFSD | CARB | ||
| pVLT35 | − (vector) | 0.20 | 0.10 | 1.56 | <0.05 | <0.01 | 1.00 | <0.03 | <0.05 | <0.20 |
| pKW35TM | +++ | 3.13 | 6.25 | 25.00 | 0.20 | 0.20 | 4.00 | 0.25 | 0.78 | 25.00 |
| pKWIN1 | ++ | 1.56 | 3.13 | 12.50 | 0.39 | 0.10 | 1.00 | 0.06 | 0.20 | 50.00 |
| pKWIN2 | +++ | 3.13 | 6.25 | 25.00 | 0.20 | 0.20 | 2.00 | 0.13 | 0.39 | 12.50 |
| pKWIN5 | +++ | 1.56 | 3.13 | 12.50 | 0.10 | 0.10 | 2.00 | 0.13 | 0.20 | 12.50 |
| pKWIN9 | ++ | 1.56 | 3.13 | 12.50 | 0.10 | 0.10 | 2.00 | 0.13 | 0.39 | 12.50 |
| pKWIN10 | ++ | 0.78 | 1.56 | 6.25 | <0.05 | 0.02 | 0.50 | 0.06 | 0.20 | 6.25 |
| pKWIN11 | + | 0.39 | 0.39 | 3.13 | <0.05 | 0.02 | 0.25 | <0.03 | 0.10 | 1.56 |
| pXZL34c | +++ | —d | 12.50 | 50.00 | 0.20 | 0.39 | 4.00 | 0.25 | 1.56 | 25.00 |
| pKPM-2c | +++ | —d | 25.00 | 50.00 | 0.20 | 0.39 | 8.00 | 0.50 | 0.78 | 50.00 |
| pKWIN13c | ++ | —d | 25.00 | 200.00 | 0.39 | 0.39 | 16.00 | 0.50 | 1.56 | 100.00 |
Expression levels ranged from undetectable (−) to strong (+++), and results were obtained from outer membranes of strain OCR03T carrying the various plasmids.
These results were obtained from three identical experiments. Abbreviations: TET, tetracycline; CM, chloramphenicol; NA, nalidixic acid; NFX, norfloxacin; MERO, meropenem; CTAX, ceftriaxone; CFP, cefepime; CFSD, cefsulodin; CARB, carbenicillin. Results in boldface indicate a reduction of at least fourfold compared with cells carrying wild-type OprM.
pXZL34 contains wild-type oprM with the native sequence cloned into pVLT31. pKPM-2 contains oprM* cloned into pVLT31. pKWIN13 was also constructed using pVLT31.
This plasmid carries a tetracycline resistance gene.
All nonexpressing clones failed to influence antibiotic resistance (data not shown). In contrast, P. aeruginosa cells carrying the expressed mutant proteins showed variable patterns in their resistance profiles. Cells with pKWIN2, pKWIN9, and pKWIN13 had resistance profiles identical to those obtained with wild-type OprM. Cells carrying pKWIN1 and pKWIN5 had resistance profiles very similar to that of the control with wild-type OprM, except for only partial restoration of resistance to some of the β-lactams tested. Cells with pKWIN10 and pKWIN11 showed only partially restored resistance to all of the various antimicrobials tested, with at least a fourfold reduction in MICs compared with those obtained for cells carrying wild-type OprM (Table 3). The variation in expression levels of oprM mutant forms was not solely responsible for this result (cf. pKWIN9 and pKWIN10 in Table 3). Therefore, this variation in antimicrobial susceptibility caused by the different mutant proteins was likely due to different effects asserted by insertion of the malarial epitope at specific sites. These experimental data were used to generate the revised OprM topology model shown in Fig. 2.
DISCUSSION
OprM is the outer membrane component of the MexA-MexB-OprM multiple-antibiotic efflux system in P. aeruginosa and had been predicted to function like a porin (24, 28). Results from planar lipid bilayer experiments showed that a gel-purified oligomeric form of OprM had channel activity with a single-channel conductance of about 80 pS in 1 M KCl (pH 7). This suggested that OprM might indeed function as a channel in the outer membrane, like porins. OprM was thus shown to function like the E. coli outer membrane protein TolC and could, in principle, associate with other efflux systems (38, 46). However, it is interesting that the P. aeruginosa genome sequence contains a different protein, with the highest homology to TolC, that is phylogenetically distinct from the OprM subfamily of efflux outer membrane proteins (our web page, http://www.cmdr.ubc.ca/bobh/OprMfamily.html). From two-dimensional crystals, TolC was predicted to have a β-barrel structure (20). Therefore, based on the previously published porin models, multiple-sequence alignment with homologous proteins, β-turn prediction (30), hydrophobicity calculation (10), and the insertion mutagenesis results of this study, we have proposed a topology model for the OprM monomer here (Fig. 2). A 16-β-strand motif of many previously published porin models (8, 41) was predicted. The β strands contained mostly amphipathic stretches of amino acid residues and the majority of the conserved regions from multiple-sequence alignment of OprM with highly homologous proteins such as P. aeruginosa OprJ (31) and P. putida SrpC (17). OprM has been predicted to have a putative lipoprotein signal peptidase cleavage site (33), and the homologous OpcM protein in Burkholderia cepacia was experimentally shown to be a lipoprotein (6). Therefore, the OprM topology was shown as that of a mature protein after modification by signal peptidase at the N terminus (Fig. 2) but without any possible lipid modification.
The last 22 amino acids at the carboxy-terminal end of native OprM were different from the sequence previously published (termed OprM* here) but consistent with the genomic sequence (http://www.pseudomonas.com). This variation (including 8 extra residues and 14 different residues) apparently did not affect the expression and function of the proteins. Both oprM sequences were expressed similarly in P. aeruginosa OprM-deficient cells and restored resistance to various antimicrobial agents without any noticeable difference (Table 3). In addition, the C terminus of native OprM contains a large stretch of hydrophilic residues and is presumably located in the periplasm, where variations are usually tolerable. Therefore, results obtained with our OprM insertion mutant proteins should not have been affected by the presence of the native or the incorrect OprM C-terminal sequence.
On the other hand, a deletion of the last 70 amino acid residues at the C terminus completely abolished the function of the protein and apparently affected the proper folding since no oligomeric or surface-expressed form was obtained from the truncated monomer (data not shown). Therefore, it seems that amino acid residues 399 to 468 of OprM are indispensable for the expression of OprM.
Insertion mutagenesis of PhoE and LamB has shown that insertion sites located in the loop regions are permissive (1, 7). Mutant forms of oprM with insertions of the malarial epitope at sites ME1, ME2, ME5, ME9, ME10, ME11, and ME13 were expressed in P. aeruginosa, and the proteins were detected by the anti-OprM antibody (Fig. 4C). These mutant proteins also maintained their abilities to form oligomers which were reduced to monomers, as previously shown for wild-type OprM (25), by heating and treatment with β-mercaptoethanol (Fig. 4A and B). These insertion sites seemed to be permissive for OprM and were placed in the surface loops. However, there were variations in antimicrobial susceptibilities from cells carrying the pKWIN plasmids with these insertions (Table 3). It was not entirely clear what gave rise to this variation in MIC profiles. Although it has been shown that the cytoplasmic membrane-associated proteins (MexA and MexB) in this efflux system were involved in determining the substrate specificity including β-lactams (38), an outer membrane component was indispensable for efflux of antimicrobials. The fact that efflux systems with chimeric outer membrane components were less effective suggested that these outer membrane proteins more readily associate with their native cytoplasmic membrane-associated components (38). Conversely, the outer membrane components might also have some influence on substrate passage. For instance, it was shown that OprM could contribute to resistance to certain agents in the absence of MexAB (48), although it was unclear if other cellular components were involved. Consequently, the insertion of a long stretch of amino acids into these particular sites might have caused sufficient disruption of the conformation of the mutant OprM proteins, such as changes in the size and charge distribution of the pore, to affect the passage of antibiotics. It was previously shown that a TonB homologue in P. aeruginosa influenced efflux-mediated multidrug resistance, and it was suggested OprM might be a gated channel (47). However, the fact that we obtained channel activity with OprM indicates that it is not simply a gated channel. This conductance (80 pS) is much lower than that observed for E. coli OmpC monomers (500 pS) that have been known to restrict β-lactam passage (16). Therefore, it is possible that the OprM channel under the influence of MexA+MexB and a substrate increases its channel diameter through movement of one or more of its loop or turn regions, like a snake opening its jaws wider for larger prey. In the crystal structures of E. coli OmpF and LamB, loop 3 is folded entirely into the β barrel to form the eyelet and some other loops also fold over the channel to different extents (8, 37). Similarly, some loop regions in OprM might fold partly into the channel to restrict the channel diameter at the surface, and to provide partial gating or some selectivity on substrate passage. In such a model, insertions at ME10 and ME11 in predicted loops 5 and 6 would be proposed to partially impair the normal function of such loops and thus influence antibiotic passage. Further studies, such as deletion and site-directed mutagenesis, would help to further resolve the involvement of these loops.
P. aeruginosa cells carrying plasmids with insertions at four sites (ME3, ME6, ME8, and ME12) did not express the mutant forms of oprM and did not show any increase in resistance to the various antimicrobial agents tested. These insertion sites were proposed to be localized within β strands. Incorporation of the malarial epitope repeats into the β strands would disrupt the folding patterns of the proteins, rendering them more susceptible to proteolytic degradation in the periplasm (27, 45), and thus they could not be inserted into the outer membrane to form complete functional efflux complexes with MexA and MexB.
Some of the proposed periplasmic loops of OprM were longer. Since OprM apparently interacts with the membrane fusion proteins of different multidrug efflux systems (38, 46), these longer loops may be involved to facilitate the interaction. A periplasmic domain was also observed for the homologous E. coli TolC protein by two-dimensional crystals (20). Two large hydrophilic loops predicted in the MexB topology model were suggested to transmit cellular energy to OprM (11). The longer periplasmic loops proposed in the OprM model here might also be involved. These interactions could be quite important and specific, making chimeric pump systems less effective than the native ones (38).
There were some differences in expression of the oprM mutants between the E. coli cells and the P. aeruginosa cells carrying the same plasmids. OprM mutant proteins with malarial epitope insertions at ME6, ME8, and ME12 were expressed in an E. coli CE1248 background but not in P. aeruginosa strain OCR03T. The proposed locations of these three sites are within β strands but very close to the cell surface. The alternating hydrophobic and hydrophilic residues being inserted at these sites might be compatible with extension of the β strands and still be accommodated in E. coli without extensive disruption of the β barrel, and the mutant proteins would thus be recovered in the outer membranes. Presumably, these less stable mutant proteins were more readily degraded in P. aeruginosa by its different proteolytic enzymes and therefore these three OprM mutant proteins were not recovered. Since P. aeruginosa is the native host for the MexA-MexB-OprM system, we based our model more on results obtained in the P. aeruginosa background. For the insertion at ME13, the mutant protein was expressed and detected by an anti-OprM antibody in both E. coli and P. aeruginosa and the protein also reconstituted the function of wild-type OprM. However, it was not detected by the antimalarial epitope antibody either in Western immunoblotting or indirect immunofluorescence. Insertion at this particular site was therefore tolerable but might not contain sufficient malarial epitope (NANP) repeats for detection, as observed in some OprF insertion mutants (44).
The proposed topology model for an OprM monomer will serve as a starting point from which we can begin to define the structure of the protein, its involvement in multiple-antibiotic efflux in terms of mechanism, and its interaction with other components of the efflux system. Further testing of the model would help to better resolve the structure-function relationships of the outer membrane protein.
ACKNOWLEDGMENTS
This work was supported by a grant from the Medical Research Council of Canada (MRC). Kendy Wong is supported by a studentship from the Canadian Cystic Fibrosis Foundation. R. E. W. Hancock is a recipient of the MRC Distinguished Scientist Award.
We thank Keith Poole for the pKPM-2 (pVLT31::oprM), pXZL33, and pXZL34 (pVLT31::native oprM) constructs and OprM-deficient strain K613, Victor de Lorenzo for the pVLT31 and pVLT35 vectors, Thilo Köhler for the OCR03T strain, and Naomasa Gotoh for the monoclonal antibody against OprM.
REFERENCES
- 1.Agterberg M, Adriaanse H, Lankhof H, Meloen R, Tommassen J. Outer membrane PhoE protein of Escherichia coli K-12 as an exposure vector: possibilities and limitations. Gene. 1990;88:37–45. doi: 10.1016/0378-1119(90)90057-x. [DOI] [PubMed] [Google Scholar]
- 2.Amsterdam D. Susceptibility testing of antimicrobials in liquid media. In: Lorian V, editor. Antibiotics in laboratory medicine. 3rd ed. Baltimore, Md: The Williams & Wilkins Co.; 1991. pp. 72–78. [Google Scholar]
- 3.Bauer K, Struyve M, Bosch D, Benz R, Tommassen J. One single lysine residue is responsible for the special interaction between polyphosphate and the outer membrane porin PhoE of Escherichia coli. J Biol Chem. 1989;264:16393–16398. [PubMed] [Google Scholar]
- 4.Benz R, Jando K, Boos W, Langer P. Formation of large ion-permeable membrane channels by the matrix protein (porin) of Escherichia coli. Biochim Biophys Acta. 1978;511:305–319. doi: 10.1016/0005-2736(78)90269-9. [DOI] [PubMed] [Google Scholar]
- 5.Benz R, Maier E, Gentschev I. TolC of Escherichia coli functions as an outer membrane channel. Zentbl Bakteriol. 1993;278:187–196. doi: 10.1016/s0934-8840(11)80836-4. [DOI] [PubMed] [Google Scholar]
- 6.Burns J L, Wadsworth C D, Barry J J, Goodall C P. Nucleotide sequence analysis of a gene from Burkholderia (Pseudomonas) cepacia encoding an outer membrane lipoprotein involved in multiple antibiotic resistance. Antimicrob Agents Chemother. 1996;40:307–313. doi: 10.1128/aac.40.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charbit A, Molla A, Saurin W, Hofnung M. Versatility of a vector for expressing foreign polypeptides at the surface of Gram-negative bacteria. Gene. 1988;70:181–189. doi: 10.1016/0378-1119(88)90116-3. [DOI] [PubMed] [Google Scholar]
- 8.Cowan S W, Schirmer T, Rummel G, Steiert M, Ghosh R, Paupitt P A, Janosonius J N, Rosenbursh J P. Crystal structures explain functional properties of two Escherichia coli porins. Nature (London) 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 9.Evans K, Passador L, Srikumar R, Tsang E, Nezezon J, Poole K. Influence of the MexAB-OprM multidrug efflux system on quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1998;180:5443–5447. doi: 10.1128/jb.180.20.5443-5447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gromiha M M, Majumdar R, Ponnuswamy P K. Identification of membrane spanning beta strands in bacterial porins. Protein Eng. 1997;10:497–500. doi: 10.1093/protein/10.5.497. [DOI] [PubMed] [Google Scholar]
- 11.Guan L, Ehrmann M, Yoneyama H, Nakae T. Membrane topology of the xenobiotic-exporting subunit, MexB, of the MexA,B-OprM extrusion pump in Pseudomonas aeruginosa. J Biol Chem. 1999;274:10517–10522. doi: 10.1074/jbc.274.15.10517. [DOI] [PubMed] [Google Scholar]
- 12.Hamzehpour M M, Pechere J C, Plesiat P, Köhler T. OprK and OprM define two genetically distinct multidrug efflux systems in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:2392–2396. doi: 10.1128/aac.39.11.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancock R E W, Carey A M. Outer membrane of Pseudomonas aeruginosa: heat- and 2-mercaptoethanol-modifiable proteins. J Bacteriol. 1979;140:902–910. doi: 10.1128/jb.140.3.902-910.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofstra H, vanTol M J D, Dankert J. Immunofluorescent detection of the major outer membrane protein II in Escherichia coli O26K60. FEMS Microbiol Lett. 1979;6:147–150. [Google Scholar]
- 15.Huang H, Jeanteur D, Pattus F, Hancock R E. Membrane topology and site-specific mutagenesis of Pseudomonas aeruginosa porin OprD. Mol Microbiol. 1995;16:931–941. doi: 10.1111/j.1365-2958.1995.tb02319.x. [DOI] [PubMed] [Google Scholar]
- 16.Jaffe A, Chabbert Y A, Semonin O. Role of porin proteins OmpF and OmpC in the permeation of beta-lactams. Antimicrob Agents Chemother. 1982;22:942–948. doi: 10.1128/aac.22.6.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kieboom J, Dennis J J, de Bont J A, Zylstra G J. Identification and molecular characterization of an efflux pump involved in Pseudomonas putida S12 solvent tolerance. J Biol Chem. 1998;273:85–91. doi: 10.1074/jbc.273.1.85. [DOI] [PubMed] [Google Scholar]
- 18.Köhler T, Kok M, Michea-Hamzehpour M, Plesiat P, Gotoh N, Nishino T, Curty L K, Pechere J C. Multidrug efflux in intrinsic resistance to trimethoprim and sulfamethoxazole in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1996;40:2288–2290. doi: 10.1128/aac.40.10.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Köhler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechere J C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 20.Koronakis V, Li J, Koronakis E, Stauffer K. Structure of TolC, the outer membrane component of the bacterial type I efflux system, derived from two-dimensional crystals. Mol Microbiol. 1997;23:617–626. doi: 10.1046/j.1365-2958.1997.d01-1880.x. [DOI] [PubMed] [Google Scholar]
- 21.Li X Z, Nikaido H, Poole K. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X Z, Zhang L, Poole K. Role of the multidrug efflux systems of Pseudomonas aeruginosa in organic solvent tolerance. J Bacteriol. 1998;180:2987–2991. doi: 10.1128/jb.180.11.2987-2991.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorenzo V, Eltis L, Kessler B, Timmis K N. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene. 1993;123:17–24. doi: 10.1016/0378-1119(93)90533-9. [DOI] [PubMed] [Google Scholar]
- 24.Ma D, Cook D N, Hearst J E, Nikaido H. Efflux pumps and drug resistance in Gram-negative bacteria. Trends Microbiol. 1994;2:489–493. doi: 10.1016/0966-842x(94)90654-8. [DOI] [PubMed] [Google Scholar]
- 25.Masuda N, Sakagawa E, Ohya S. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:645–649. doi: 10.1128/AAC.39.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mine T, Morita Y, Kataoka A, Mizushima T, Tsuchiya T. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:415–417. doi: 10.1128/aac.43.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Misra R, Peterson A, Ferenci T, Silhavy F J. A genetic approach for analyzing the pathway of LamB assembly into the outer membrane of Escherichia coli. J Biol Chem. 1991;266:13592–13597. [PubMed] [Google Scholar]
- 28.Nikaido H. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science. 1994;264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 29.O'Connor G G, Ashman L K. Application of the nitrocellulose transfer technique and alkaline phosphatase conjugated anti-immunoglobulin for determination of the specificity of monoclonal antibodies to protein mixtures. J Immunol Methods. 1982;54:267–271. doi: 10.1016/0022-1759(82)90068-0. [DOI] [PubMed] [Google Scholar]
- 30.Paul C, Rosenbusch J P. Folding patterns of porin and bacteriorhodopsin. EMBO J. 1985;4:1593–1597. doi: 10.1002/j.1460-2075.1985.tb03822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J, Li X Z, Nishino T. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 32.Poole K, Heinrichs D E, Neshat S. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol Microbiol. 1993;10:529–544. doi: 10.1111/j.1365-2958.1993.tb00925.x. [DOI] [PubMed] [Google Scholar]
- 33.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poxton I R, Bell G T, Barclay G R. The association on SDS-polyacrylamide gels of lipopolysaccharide and outer membrane proteins of Pseudomonas aeruginosa as revealed by monoclonal antibodies and Western blotting. FEMS Microbiol Lett. 1985;27:247–251. [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Sandermann H, Strominger J L. Purification and properties of C55-isoprenoid alcohol phosphokinase from Staphylococcus aureus. J Biol Chem. 1972;247:5123–5131. [PubMed] [Google Scholar]
- 37.Schirmer T, Keller T A, Wang Y F, Rosenbusch J P. Structural basis for sugar translocation through maltoporin channels at 3.1 Å resolution. Science. 1995;267:512–514. doi: 10.1126/science.7824948. [DOI] [PubMed] [Google Scholar]
- 38.Srikumar R, Li X Z, Poole K. Inner membrane efflux components are responsible for beta-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J Bacteriol. 1997;179:7875–7881. doi: 10.1128/jb.179.24.7875-7881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sukhan A, Hancock R E. Insertion mutagenesis of the Pseudomonas aeruginosa phosphate-specific porin OprP. J Bacteriol. 1995;177:4914–4920. doi: 10.1128/jb.177.17.4914-4920.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiss M S, Wacker T, Weckesser J, Welte W, Schulz G E. The three-dimensional structure of porin from Rhodobacter capsulatus at 3 Å resolution. FEBS Lett. 1990;267:268–272. doi: 10.1016/0014-5793(90)80942-c. [DOI] [PubMed] [Google Scholar]
- 42.Wong K K Y, Poole K, Gotoh N, Hancock R E W. Influence of OprM expression on multiple antibiotic resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:2009–2012. doi: 10.1128/aac.41.9.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong R S Y, Jost H, Hancock R E W. Linker-insertion mutagenesis of Pseudomonas aeruginosa outer membrane protein OprF. Mol Microbiol. 1993;10:283–292. [PubMed] [Google Scholar]
- 44.Wong R S Y, Wirtz R A, Hancock R E W. Pseudomonas aeruginosa outer membrane OprF as a presentation vector for foreign epitopes: the effects of positioning and length on the antigenicity of the epitope. Gene. 1995;158:55–60. doi: 10.1016/0378-1119(95)00155-y. [DOI] [PubMed] [Google Scholar]
- 45.Wulfing C, Pluckthun A. Protein folding in the periplasm of Escherichia coli. Mol Microbiol. 1994;12:685–692. doi: 10.1111/j.1365-2958.1994.tb01056.x. [DOI] [PubMed] [Google Scholar]
- 46.Yoneyama H, Ocaktan A, Gotoh N, Nishino T, Nakae T. Subunit swapping in the Mex-extrusion pumps in Pseudomonas aeruginosa. Biochem Biophys Res Commun. 1998;244:898–902. doi: 10.1006/bbrc.1998.8351. [DOI] [PubMed] [Google Scholar]
- 47.Zhao Q, Li X Z, Mistry A, Srikumar R, Zhang L, Lomovskaya O, Poole K. Influence of the TonB energy-coupling protein on efflux-mediated multidrug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:2225–2231. doi: 10.1128/aac.42.9.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao Q, Li X Z, Srikumar R, Poole K. Contribution of outer membrane efflux protein OprM to antibiotic resistance in Pseudomonas aeruginosa independent of MexAB. Antimicrob Agents Chemother. 1998;42:1682–1688. doi: 10.1128/aac.42.7.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]