Abstract
The large virulence plasmid pMYSH6000 of Shigella flexneri contains a determinant that is highly effective in stabilizing otherwise unstable plasmids in Escherichia coli. Expression of two small contiguous genes, mvpA and mvpT (formerly termed STBORF1 and STBORF2), was shown to be sufficient for stability. Mutations in mvpT abolished plasmid stability, and plasmids expressing only mvpT killed the cells unless mvpA was supplied from a separate plasmid or from the host chromosome. When replication of a plasmid carrying the minimal mvp region was blocked, growth of the culture stopped after a short lag and virtually all of the surviving cells retained the plasmid. Thus, the mvp system stabilizes by a highly efficient postsegregational killing (PSK) mechanism, with mvpT encoding a cell toxin and mvpA encoding an antidote. The regions that surround the mvp genes in their original context have an inhibitory effect that attenuates plasmid stabilization and PSK. The region encompassing the mvp genes also appears to contain an additional element that can aid propagation of a pSC101-based plasmid under conditions where replication initiation is marginal. However, this appears to be a relatively nonspecific effect of DNA insertion into the plasmid vector.
Enteroinvasive strains of shigella species and Escherichia coli contain related large plasmids which encode many of the factors required for virulence (11). The large virulence plasmid pMYSH6000 of Shigella flexneri contains a stability element (Stb) which can promote stable inheritance of plasmids carrying the replication region of pMYSH6000, or the P1 replicon in E. coli (11, 17). A 1.1-kb fragment of the plasmid was sufficient to promote plasmid stability in the absence of other pMYSH6000 sequences. It did so without any apparent increase in the plasmid copy number (12). The fragment contains three open reading frames (17). The 240-codon trbH open reading frame is a homolog of the trbH gene of the F plasmid of E. coli, a gene of unknown function that resides in the conjugal transfer region (12). The product of the pMYSH6000 trbH open reading frame could be interrupted by a nonsense mutation without impairing function, suggesting that a TrbH product is not needed for plasmid stability (17). The two other open reading frames, STBORF1 (75 codons) and STBORF2 (133 codons) lie in the opposite orientation and completely overlap trbH. Homologous open reading frame pairs are found in F trbH, in the chromosomes of certain pathogenic bacteria (17), and in plasmids of the pathogenic organisms Salmonella dublin and Dichelobacter nodosus (4, 15, 17). The STBORF1 and STBORF2 open reading frames show little similarity to any known plasmid stability element, but their general organization resembles that of postsegregational killing systems. Such systems encode a toxin and an unstable antidote. When the plasmid is lost from the cell, the antidote decays but the toxin persists, eventually killing most of the progeny of the plasmid-free cell. This promotes the maintenance of the plasmid in the growing population (5, 20). Here, we show evidence that the Stb element does encode a postsegregational killing (PSK) system and that the STBORF1 and STBORF2 open reading frames produce an antidote and toxin protein, respectively. We propose to name the genes mvpA and mvpT (for maintenance of virulence plasmid) and the protein products MvpA and MvpT.
MATERIALS AND METHODS
Media, chemicals, and DNA manipulations.
Media, reagents, enzymes, buffers, and chemicals used were as previously described (1). Enzymes were obtained from New England Biolabs (Beverly, Mass.) and Boehringer Mannheim (Indianapolis, Ind.), and used under conditions recommended by the manufacturers. Concentrations of antibiotics were as follows: ampicillin, 100 μg/ml; chloramphenicol, 10 μg/ml; and spectinomycin, 10 μg/ml. Cloning methods were as previously described (19), with strain BR2846 used for plasmid growth and transformation for DNA manipulations.
Map coordinates.
All map coordinates are given in base pairs and refer to the 1,127-bp sequence of the mvp (originally Stb) region (17).
Bacterial strains and plasmids.
The bacterial strains BR825 trp polA::Tn10 (13), BR2846 supE44 hsdR17 recA1 endA1 gyrA96 thi relA lacΔU169, a derivative of DH5 (8), and W3110 (2) were derived from E. coli K-12.
Plasmid pALA136 contains the pBR322 origin of replication, the P1 replication region, and a gene for chloramphenicol resistance, as previously described (17). Plasmid pHGB2 was a kind gift of V. François and C. Labie. It consists of the pSC101 derivative pGB2 (6), with the repA gene replaced by the equivalent region from the temperature-sensitive plasmid pHSG415 (9). Plasmid pALA1557 consists of the intact P1 par region inserted into the BamHI site of pALA136 (17), and pALA2518 consists of the 2.5-kb BamHI-HindIII fragment containing part of the P1 par region from pALA271 (7) inserted between the same sites of plasmid pHGB2. Plasmid pCS1367 consists of the SalI C (replicon) region and the SalI O (mvp) region of pMYSH6000 inserted into plasmid pBR322 (17). Plasmid pALA1196 consists of the 1,127-bp mvp region inserted into plasmid pALA136 (17). The 1,127-bp region was amplified by PCR with primers which generated the 1,127-bp region with a BamHI site at each end. The resulting fragment was inserted into the unique BamHI site of pALA136 to give plasmid pALA2515 (17). Plasmids pALA1585 and pALA1589 were made from pALA2515 by introducing mutations by modified strand overlap PCR with outside primers with BamHI sites and two complementary mutagenic primers (16). Plasmids pALA2534 and pALA2546 were produced similarly but used a BamHI-containing primer at one end and a SalI-containing primer at the other. Plasmid pALA1585 has a G-to-A change at base 333, creating a TGA stop codon in mvpA, and pALA1589 has an A-to-T mutation at base 465 creating a TAG stop codon in mvpT. Plasmid pALA2534 has a six-codon deletion in mvpA (mvpAΔ292–309) and pALA2546 has most of mvpA deleted (61 codons; mvpAΔ294–396).
The mvp region or portions of it present in all other plasmids were generated in a fashion similar to that of pALA2515, with PCR amplification from a pALA1196 template and suitable BamHI-SalI primers. The resulting fragments were inserted into the BamHI-SalI site of pALA136 to give the plasmids pALA2519-2529 and pALA2533 (see Fig. 1). Plasmids pALA2542 and pALA2543 have inserts identical to pALA2523 and pALA2528, respectively, but with the insert placed in the BamHI-SalI interval of plasmid pHGB2. The mvp open reading frames (or portions thereof) of all plasmids were oriented such that they run clockwise when the pBR322- or pHGB2-derived portions of the plasmids are displayed in their conventional orientations.
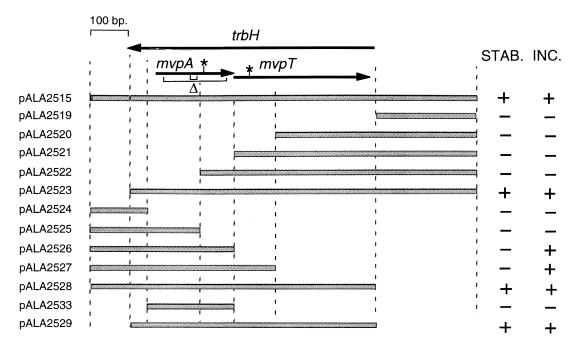
FIG. 1.
Physical map of the mvp region showing phenotypes of deletion derivatives. Shaded bars indicate regions present in the numbered plasmids. Arrows indicate the trbH and mvp open reading frames. Asterisks mark the positions of nonsense mutations in pALA1589 (TAG in mvpT) and pALA1585 (TGA in mvpA). Two brackets under the mvpA open reading frame (Δ) mark the extents of the deleted bases in the 6- and 61-codon in-frame deletions in pALA2534 and pALA2546, respectively. In stability tests (STAB.), a plus denotes >80% retention and a minus denotes <5% retention of plasmid in 25 generations of unselected growth. In incompatibility tests (INC.), a plus denotes >90% loss and a minus denotes <5% loss of resident plasmid pCS1367 (mvp+) in ca. 25 generations after the introduction of the incoming plasmids listed on the left.
Measurement of plasmid maintenance stability.
The ability of various inserts to stabilize the plasmid pALA136 when it is replicating as a mini-P1 plasmid at low copy number in strain BR825 polA was measured as previously described (17).
Measurement of plasmid copy number.
The copy numbers of pALA136 and its mvp derivatives were measured in Luria-Bertani (LB) broth at 30°C as previously described (17). This method compares the yield of plasmid DNA to that of the mini-P1 plasmid λ-P1:5RCm in a fixed quantity of cells that is mixed with the test sample prior to cell lysis and DNA extraction.
Measurement of transformation and survival frequencies.
Plasmid DNA of pALA2546 ΔmvpA mvpT+ was isolated from strain CC4451 which produces MvpA from the bacterial chromosome (see Results). Competent W3110 cells carrying the relevant resident pHGB2-based plasmid were prepared after growth at 30°C in the presence of spectinomycin (10 μg/ml). The cells were then transformed by the incoming plasmid to chloramphenicol resistance and the transformation frequency was determined by plating on agar with chloramphenicol and spectinomycin at 30°C. Pure lines of the transformants were grown in the presence of chloramphenicol and spectinomycin to an optical density at 600 nm (OD600) of 0.1, and dilutions were plated on prewarmed LB agar at 30 and 42°C to determine survival under conditions where the resident plasmid is lost.
Measurement of cell killing on loss of plasmids carrying the mvp genes.
Strain W3110 carrying the relevant pHGB2-based plasmid was grown overnight at 30°C in LB broth with spectinomycin. The culture was diluted 100-fold in the same medium and grown to an OD600 of 0.1. An aliquot was withdrawn, and the number of viable cells was determined by plating on LB agar with and without spectinomycin. The culture was diluted 50-fold into LB broth at 35 or 42°C, and exponential growth was continued for approximately 400 min. Periodically, the culture was diluted with prewarmed LB broth in order to keep the OD600 between 0.01 and 0.1. At the time points indicated, aliquots were withdrawn and the number of viable cells was determined by plating on LB agar with and without spectinomycin at 30°C. In some experiments, a second plasmid based on pALA136 was present. In these cases, the overnight cultures contained chloramphenicol in addition to spectinomycin. As these second plasmids contain the origin of plasmid pBR322, they are maintained as multiple copies in strain W3110 and are not lost from any of the cells during the course of the experiment (data not shown).
RESULTS
Deletion analysis of the 1,127-bp mvp region.
A series of deletions were constructed in the 1,127-bp mvp fragment present in the mini-P1 plasmid pALA2515 (Fig. 1). This plasmid relies on the function of the mvp region for its proper maintenance when present in a polA strain where the plasmid replicates at a low copy number under P1 control (17). Deletion analysis shows that a 714-bp region is necessary and sufficient to promote stable plasmid maintenance (pALA2529) (Fig. 1). This sequence includes the two short open reading frames, mvpA and mvpT, and a putative promoter region immediately upstream of them.
The 714-bp mvp fragment is very effective in promoting stable plasmid maintenance.
The stability conferred by the minimal 714-bp mvp fragment was consistently better than that conferred by its 1,127-bp progenitor (Table 1). Loss of the mini-P1 plasmid with the smaller fragment was undetectable, even after 100 generations of unselected growth, showing that the mvp system is highly efficient in promoting the maintenance of this construct. The mvp system proved more effective in stabilizing the mini-P1 plasmid construct in this assay than did the P1 par system (Table 1). P1 par promotes the active partition of daughter plasmids to daughter cells and is the system primarily responsible for the accurate maintenance of the P1 plasmid and its stable miniplasmid derivatives (14). As was the case with its 1,127-bp progenitor (12), the 714-bp mvp fragment confers stability without any apparent increase in plasmid copy number. The average copy numbers of the 714-bp derivative (pALA2529) and the pALA136 vector were not significantly different at 7.8 and 6.4 per cell, respectively, grown in LB broth at 30°C (see Materials and Methods).
TABLE 1.
The maintenance stability of mini-P1 plasmids carrying the mvp regiona
| Plasmid | Insert | % Retention (25 generations) |
|---|---|---|
| pALA136 | None | 5 |
| pALA1557 | P1 par+ | 94 |
| pALA2515 | bp 1–1127; mvpA+ mvpT+ | 95 |
| pALA2529 | bp 117–832; mvpA+ mvpT+ | 100b |
| pALA2528 | bp 1–832; mvpA+ mvpT+ | 100b |
| pALA2523 | bp 117–1127; mvpA+ mvpT+ | 100b |
| pALA1589 | bp 1–1127; mvpA+ mvpT (TAG) | 6 |
| pALA1585 | bp 1–1127; mvpA (TGA) mvpT+ | 5 |
| pALA2534 | bp 117–832; mvpAΔ292–309mvpT+ | 100 |
The plasmids were assayed in strain BR825 as described in Materials and Methods. The endpoints of the inserts and the positions of the nonsense mutations in pALA1589 and pALA1585 and the six-codon in-frame mvpA deletion in pALA2534 are illustrated in Fig. 1.
In a separate experiment, no loss of the plasmid was detected in 100 generations of unselected growth.
The increased stability conferred by the 714-bp fragment relative to its 1,127-bp progenitor suggests that inhibitory sequences exist in the regions upstream or downstream of the mvp genes which limit their effectiveness. Surprisingly, the inhibitory information appears to lie both upstream and downstream of the mvp genes, because deletion of excess information at either end of the genes results in highly efficient stabilization (pALA2523 and pALA2528) (Table 1). It seems probable that the efficiency of the mvp region is sensitive to the surrounding sequence context and that the surrounding bases from its natural context are not optimal for function, at least in E. coli.
The phenotypes of mutations in mvpA and mvpT.
A nonsense mutation (amber) at base 465 in mvpT abolished the stability phenotype (pALA1589) (Table 1). The amber mutation was designed such that it does not alter the coding of the trbH gene as it substitutes one leucine codon (CTT) for another (CTA). We conclude that the mvpT gene and likely its protein product are essential for the stability phenotype. The fact that truncation of a large carboxy-terminal segment of mvpT (pALA2527) (Fig. 1) also abolishes stability is consistent with this conclusion.
A G-to-A mutation was created at position 333, making a nonsense codon (UGA) in mvpA (Fig. 1). It does not affect the coding of the trbH open reading frame. The mutation abolished the stability phenotype (Table 1). As the only likely promoter in the region lies upstream of mvpA and as the open reading frames overlap, it is probable that the two genes form an operon and that translation of the two genes is coupled (17). Thus, the properties of the nonsense mutation do not necessarily imply an essential role for mvpA, as the mutation could have polar effects on mvpT expression. We therefore constructed in-frame deletions in mvpA to probe its function. A six-codon in-frame deletion within mvpA had no apparent effect on plasmid stability (Table 1). However, when a large in-frame deletion was used, the plasmid (pALA2546) could not be introduced into cells by transformation unless a second plasmid (pALA2547) containing an intact mvpA gene was present (Table 2). This suggests that MvpA is important and that mvpT expression is deleterious to the cells if no intact MvpA protein is present. The observation is consistent with the idea that mvp is a postsegregational killing system and that MvpT is a toxin and MvpA an antidote. Presumably the six amino acids deleted in the smaller mvpA in-frame deletion mutant leave MvpA function intact. The antidote proteins of the doc-phd and ccd systems of E. coli plasmids P1 and F also tolerate sizable deletions without loss of function (10). The ability of the plasmid containing the nonsense mutation in mvpA to transform cells is likely due to polarity exerted on MvpT synthesis such that neither protein is expressed.
TABLE 2.
Transformation and cell survival with a plasmid expressing MvpT in the absence of MvpAa
| Incoming plasmid and genotype | Temperature-sensitive resident plasmid | Transformation frequency | Frequency of survival at 42°C |
|---|---|---|---|
| pALA136 | pHGB2 | 1b | 1b |
| pALA2529 mvpA+ mvpT+ | pHGB2 | 0.4 | 1 |
| pALA2546 ΔmvpA mvpT+ | pHGB2 | <10−3 | N/A |
| pALA136 | pALA2547 mvpA+ ΔmvpT | 0.3 | 1 |
| pALA2529 mvpA+ mvpT+ | pALA2547 mvpA+ ΔmvpT | 0.3 | 1 |
| pALA2546 ΔmvpA mvpT+ | pALA2547 mvpA+ ΔmvpT | 0.05 | 10−3 |
Transformation and survival frequencies were determined at 30°C as described in Materials and Methods. N/A, not applicable.
Frequencies were normalized to the values obtained for this control strain.
Cells expressing MvpT die when MvpA expression is blocked.
Plasmid pALA2547 expresses MvpA from an insert in the temperature-sensitive plasmid pHGB2. Cells containing it could be transformed by pALA2546 ΔmvpA mvpT+ at 30°C, albeit at a somewhat reduced efficiency (Table 2). However, when the temperature was raised to 42°C in the presence of chloramphenicol so that the pALA2547 was lost from the cells but pALA2546 was retained, most of the cells died (Table 2). This confirms that mvpT expression in the absence of mvpA is lethal.
Of the minority of cells that survived at 42°C to form colonies in the presence of chloramphenicol, most (69 of 70) were spectinomycin resistant and contained both pALA2546 and a mutant form of pALA2547, whose replication is insensitive to temperature (data not shown). One colony was spectinomycin sensitive and contained pALA2546 but showed no evidence of a second plasmid. After these cells were cured of pALA2546, the resulting strain (CC4451) contained no plasmid DNA but retained a copy of the mvpA gene in the chromosome as detected by PCR amplification. This strain could readily be transformed by pALA2546 and provided a means of propagating the plasmid in the absence of other plasmid DNA (see Materials and Methods).
The mvpA gene acts as an incompatibility determinant against mvp-promoted plasmid stability.
The mini-pMYSH6000 plasmid pCS1367 is stably maintained at low copy number in a polA mutant strain due to the presence of the active mvp region (17). When a second plasmid containing mvp is introduced, pCS1367 becomes unstable (17). We mapped the mvp sequences responsible for exerting this incompatibility effect to the intact mvpA gene (Fig. 1). If mvpA produces an antidote, cells containing an extra copy of this gene would be immune to PSK when a plasmid carrying the mvp system is lost, thus explaining the incompatibility effect.
PSK by the mvp system.
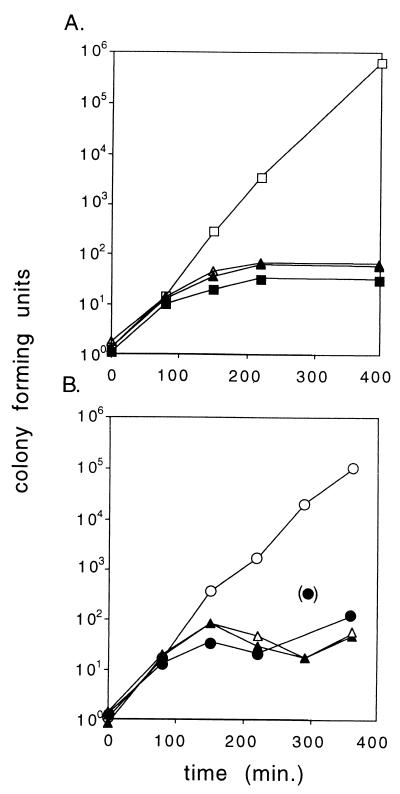
Killing or growth inhibition by MvpT in the absence of MvpA suggests that mvp is a PSK system with MvpT as the toxin and MvpA as the antidote. We introduced the 714-bp mvp region into the temperature-sensitive replicon pHGB2 and studied the fate of cells when plasmid replication was blocked at 42°C, so that the plasmid (pALA2530) could not be maintained. The block to plasmid replication appeared to shut off cell growth after a short lag. The result was a static population of viable cells, virtually all of which retain the plasmid (Fig. 2A). This result is consistent with highly efficient PSK (5). When the temperature of the culture is raised, the pHGB2 mvp+ plasmid (pALA2530) stops replicating and is presumably diluted from the growing cells until the copy number approaches a value of 1 per cell. Subsequent cell divisions give rise to plasmid-free cells, virtually all of which die.
FIG. 2.
Cell killing at 42°C upon loss of a temperature-sensitive plasmid with the 714-bp mvp region. Cultures were shifted from 30 to 42°C at time zero. Viable counts were normalized to a value of ca. 1 at time zero. Open symbols, unselected viable counts; solid symbols, counts on spectinomycin-selective agar. (A) Squares, pHGB2; triangles, pALA2530 (714-bp mvp region). (B) Triangles, pALA2530 in cells containing the vector pALA136; circles, pALA2530 in cells containing pALA2527 (mvpA+). The data are from one of several experiments which gave similar results.
The inviability of the plasmid-free cells when cured of pALA2530 is presumably due to the toxic effect of MvpT after decay of an unstable MvpA antidote activity. Consistent with this, we found that the presence of an additional copy of mvpA on a compatible plasmid (pALA2527) (Fig. 1) allowed efficient curing of pHGB2 mvp+ at 42°C with no deleterious effect on the cells (Fig. 2B).
Lack of PSK when the mvp genes are in their normal immediate context.
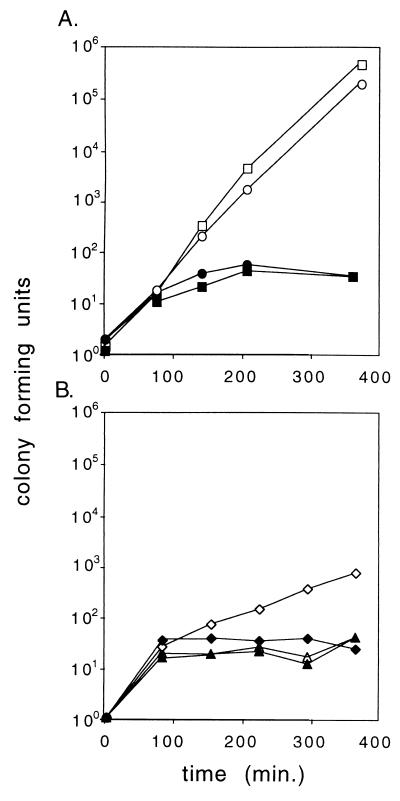
Surprisingly, the 1,127-bp stb fragment does not work in the PSK assay. Loss of the plasmid carrying it at 42°C had no measurable effect on the growth of the cells and a viable plasmid-free population was generated (Fig. 3). This lack of activity is not due to the temperature sensitivity of the mvp system or to an inadvertent mutation of sequences within the fragment (data not shown). Rather, it appears to be an extreme manifestation of the inhibitory context of the mvp genes when surrounded by the naturally occurring sequences. Removal of either of the sequences from the ends of the fragment that bracket the mvp genes restored most or all of the PSK in this assay (Fig. 3).
FIG. 3.
Lack of cell killing by the 1,127-bp mvp fragment is caused by bases surrounding the mvp genes. Cultures were shifted from 30 to 42°C at time zero. Viable counts were normalized to a value of ca. 1 at time zero. Open symbols, unselected viable counts; solid symbols, counts on spectinomycin-selective agar. (A) Squares, pHGB2; circles, pALA2511 (1,127-bp mvp region). (B) Plasmids derived from pALA2511 with the flanking regions deleted from upstream of the mvp genes (pALA2542) (triangles) or downstream of the mvp genes (pALA2543) (diamonds) are shown. The data are from one of several experiments which gave similar results.
Stabilization of a plasmid under conditions where DNA replication is compromised.
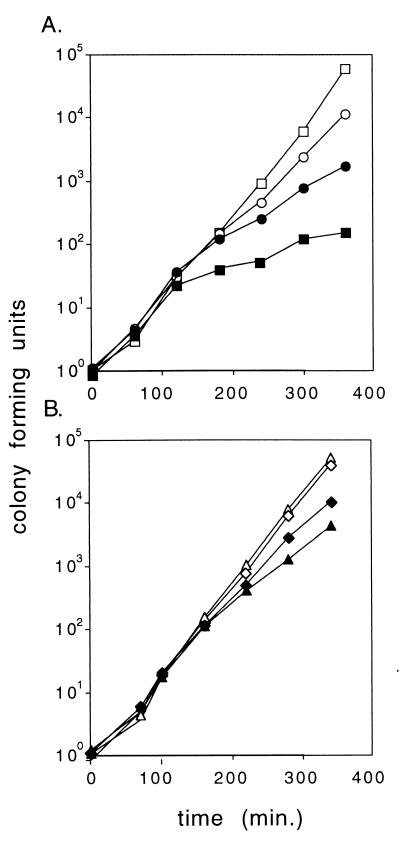
At 35°C, replication of pHGB2 is not shut off but is marginal. Some replication occurs, as shown by a significant and continuous increase in the number of plasmid-containing cells, but plasmid loss is frequent (Fig. 4). When the plasmid contained the 714-bp mvp fragment (pALA2530), the plasmid was substantially stabilized at 35°C (Fig. 4). However, not all of this added stability was due to the killing of plasmid-free cells. Otherwise, the growth curve of plasmid-containing cells (and the total cells) should mimic that of plasmid-containing cells in the pHGB2 vector control (Fig. 4). The additional stability does not appear to be due to the function of the mvp genes; a control fragment containing unrelated sequences also conferred some stability on pHGB2 at 35°C (pALA2518) (Fig. 4). This control fragment had no effect on the fate of the cell or the plasmid when replication was completely blocked at 42°C (data not shown). It appears that the insertion of nonspecific (or relatively common) sequences into pHGB2 can aid the stability of the plasmid but only when replication is marginal. The additional stability seen with the mvp fragment at 35°C is probably due to this nonspecific effect. Note that the 1,127-bp mvp fragment (pALA2511) also confers stability at 35°C (Fig. 4), despite the fact that, like the control fragment in pALA2518, it is inactive in PSK (Fig. 2). This stabilizing effect is insensitive to the incompatibility exerted by a second plasmid expressing MvpA (data not shown), again suggesting that it is a nonspecific effect. Maintenance of the pSC101 replicon employed in pHGB2 is sensitive to the degree of negative supercoiling induced by DNA gyrase (3). Perhaps the sequences that improve pHGB2 plasmid stability at 35°C contain gyrase binding sites that increase negative supercoiling.
FIG. 4.
The fate of cells carrying a temperature-sensitive mvp plasmid at 35°C. Cultures were shifted from 30 to 35°C at time zero. Viable counts were normalized to a value of ca. 1 at time zero. Open symbols, unselected viable counts; solid symbols, counts on spectinomycin selective agar. (A) Squares, pHGB2; circles, pALA2530 (714-bp mvp region). (B) Triangles, pALA2518 (control insert); diamonds, pALA2511 (1,127-bp mvp fragment). The data are from one of several experiments which gave similar results.
DISCUSSION
The properties of mutants of the mvp genes suggest that it is a PSK system, with MvpA as the antidote and MvpT as the toxin. Confirming this conclusion, we found that, when the replication of a plasmid containing the minimal mvp region was blocked, the number of viable cells stopped increasing and only plasmid-containing cells survived.
When the mvp system is embedded in an 1,127-bp fragment that includes surrounding sequences from the original pMYSH6000 plasmid, the system fails to kill cells when plasmid replication is blocked. Thus, the natural context of the system appears to inhibit function. The inhibitory sequences also appear to attenuate the ability to stabilize a mini-P1 plasmid. Removal of either the excess upstream or downstream sequences restores full mvp activity in either assay. The fact that extensions at both ends of the mvp region are required for this inhibitory effect makes it unlikely that some specific gene product is involved. For example, it is unlikely that mvp parallels the case of the pas system of plasmid pTF-FC2, where an additional 71-codon open reading frame (pasC) downstream of the antidote toxin genes attenuates pas activity (18). Rather, it seems that the naturally occurring sequences surrounding mvp have some fortuitous effect on gene expression which increases antidote levels or decreases toxin levels, rendering the system less effective at killing. Perhaps this contextual effect occurs in E. coli but not in the natural host, S. flexneri. In addition, it appears that the effect of context extends even to the vector sequences surrounding the 1,127-bp fragment. Thus, the mvp genes in the 1,127-bp fragment function in the mini-P1 vector to stabilize it (Table 1) but show no signs of cell killing in pHGB2 (Fig. 3). Our interpretation of this is that the genes function in one vector but not in another. This suggests that it is not just the immediate sequence context that can attenuate mvp function but rather some general property of the DNA region, such as supercoiling density or subcellular localization.
The minimal mvp system confers a high degree of stability on an unstable mini-P1 plasmid. The plasmid pALA136 is normally lost at a rate of ca. 5% per generation under the assay conditions, whereas no loss was seen during unselected growth of the minimal mvp derivative during many independent experiments. The apparent efficiency and small size of this element suggest that it may be useful for stabilizing plasmid vectors in practical applications.
The mvp locus is a member of a family of similar elements in a variety of disease-causing organisms. All examples so far described are linked to virulence genes on plasmids or host chromosomes (17). Perhaps mvp and its relatives play important roles in the maintenance of virulence in these organisms by efficiently killing cells which fortuitously lose blocks of virulence genes due to their deletion from the chromosome or loss of a plasmid which carries them.
ACKNOWLEDGMENTS
We are grateful to the expert assistance of Marilyn Powers for the operation of the automated sequencing machine.
This research was sponsored by the National Cancer Institute, DHHS, under contract with ABL.
REFERENCES
- 1.Abeles A. P1 plasmid replication. Purification and DNA-binding activity of the replication protein RepA. J Biol Chem. 1986;261:3548–3555. [PubMed] [Google Scholar]
- 2.Bachmann B J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972;36:525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaucage S L, Miller C A, Cohen S N. Gyrase-dependent stabilization of pSC101 plasmid inheritance by transcriptionally active promoters. EMBO J. 1991;10:2583–2588. doi: 10.1002/j.1460-2075.1991.tb07799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billington S J, Sinistaj M, Cheetham B F, Ayres A, Moses E K, Katz M E, Rood J I. Identification of a native Dichelobacter nodosus plasmid and implications for the evolution of the vap regions. Gene. 1996;172:111–116. doi: 10.1016/0378-1119(96)00032-7. [DOI] [PubMed] [Google Scholar]
- 5.Bugge-Jensen R, Gerdes K. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol Microbiol. 1995;17:205–210. doi: 10.1111/j.1365-2958.1995.mmi_17020205.x. [DOI] [PubMed] [Google Scholar]
- 6.Churchward G, Belin D, Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984;31:165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- 7.Friedman S A, Austin S J. The P1 plasmid-partition system synthesizes two essential proteins from an autoregulated operon. Plasmid. 1988;19:103–112. doi: 10.1016/0147-619x(88)90049-2. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 9.Hashimoto-Gotoh T, Franklin F C, Nordheim A, Timmis K N. Specific-purpose plasmid cloning vectors. I. Low copy number, temperature sensitive, mobilization-defective pSC101-derived containment vectors. Gene. 1981;16:227–235. doi: 10.1016/0378-1119(81)90079-2. [DOI] [PubMed] [Google Scholar]
- 10.Lehnherr H, Maguin E, Jafri S, Yarmolinsky M B. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J Mol Biol. 1993;233:414–428. doi: 10.1006/jmbi.1993.1521. [DOI] [PubMed] [Google Scholar]
- 11.Makino S-I, Sasakawa C, Yoshikawa M. Genetic relatedness of the basic replicon of the virulence plasmid in shigellae and enteroinvasive Escherichia coli. Microb Pathog. 1988;5:267–274. doi: 10.1016/0882-4010(88)90099-x. [DOI] [PubMed] [Google Scholar]
- 12.Maneewannakul K, Ippen-Ihler K. Construction and analysis of F plasmid traR, trbJ, and trbH mutants. J Bacteriol. 1993;175:1528–1531. doi: 10.1128/jb.175.5.1528-1531.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin K A, Davis M A, Austin S. Fine-structure analysis of the P1 plasmid partition site. J Bacteriol. 1991;173:3630–3634. doi: 10.1128/jb.173.12.3630-3634.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nordstrom K, Austin S J. Mechanisms that contribute to the stable segregation of plasmids. Annu Rev Genet. 1989;23:37–69. doi: 10.1146/annurev.ge.23.120189.000345. [DOI] [PubMed] [Google Scholar]
- 15.Pullinger G D, Lax A J. A Salmonella dublin virulence plasmid locus that affects bacterial growth under nutrient-limited conditions. Mol Microbiol. 1992;6:1631–1643. doi: 10.1111/j.1365-2958.1992.tb00888.x. [DOI] [PubMed] [Google Scholar]
- 16.Radnedge L, Davis M A, Austin S J. P1 and P7 plasmid partition: ParB protein bound to its partition site makes a separate discriminator contact with the DNA that determines species specificity. EMBO J. 1996;15:1155–1162. [PMC free article] [PubMed] [Google Scholar]
- 17.Radnedge L, Davis M A, Youngren B, Austin S J. Plasmid maintenance functions of the large virulence plasmid of Shigella flexneri. J Bacteriol. 1997;179:3670–3675. doi: 10.1128/jb.179.11.3670-3675.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rawlings D. Proteic toxin-antitoxin bacterial plasmid addiction systems and their evolution with special reference to the pas system of pTF-FC2. FEMS Microbiol Lett. 1999;176:269–277. doi: 10.1111/j.1574-6968.1999.tb13672.x. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 20.Yarmolinsky M B. Programmed cell death in bacterial populations. Science. 1995;267:836–837. doi: 10.1126/science.7846528. [DOI] [PubMed] [Google Scholar]