Abstract
We have isolated several Saccharomyces cerevisiae mutants resistant to calcofluor that contain mutations in the PBS2 or HOG1 genes, which encode the mitogen-activated protein kinase (MAPK) and MAP kinases, respectively, of the high-osmolarity glycerol response (HOG) pathway. We report that blockage of either of the two activation branches of the pathway, namely, SHO1 and SLN1, leads to partial resistance to calcofluor, while simultaneous disruption significantly increases resistance. However, chitin biosynthesis is independent of the HOG pathway. Calcofluor treatment also induces an increase in salt tolerance and glycerol accumulation, although no activation of the HOG pathway is detected. Our results indicate that the antifungal effect of calcofluor depends on its binding to cell wall chitin but also on the presence of a functional HOG pathway. Characterization of one of the mutants isolated, pbs2-14, revealed that resistance to calcofluor and HOG-dependent osmoadaptation are two different physiological processes. Sensitivity to calcofluor depends on the constitutive functionality of the HOG pathway; when this is altered, the cells become calcofluor resistant but also show very low levels of basal salt tolerance. Characterization of some multicopy suppressors of the calcofluor resistance phenotype indicated that constitutive HOG functionality participates in the maintenance of cell wall architecture, a conclusion supported by the antagonism observed between the protein kinase and HOG signal transduction pathways.
Chitin is a polymer of the Saccharomyces cerevisiae cell wall that has been shown to be essential for cell viability (8, 36). Chitin is assumed to confer strength to the cell wall, allowing cells to grow in nonstabilized media. In vivo, calcofluor, a fluorescent brightener that interacts specifically with chitin (30), acts as an antifungal agent by killing fungal cells (30, 31). This antifungal effect is associated with an increase in the rate of chitin synthesis, although the polymer synthesized is abnormal and does not confer enough strength on the cell wall to guarantee cell survival. Based on these observations, it has been assumed that chitin is the primary target of the drug, a hypothesis that was reinforced by the isolation of several mutants resistant to calcofluor, all of them showing severe reductions in the levels of chitin synthesis (32, 38). The first mutants resistant to calcofluor that do not have apparent defects in chitin synthesis have been described only very recently (19). However, the characterization of these mutants was preliminary, and from the report it is difficult to deduce the existence of other cellular targets for the action of calcofluor. The effect of calcofluor is also characterized by an increase in chitin synthesis that is dependent on chitin synthase III activity and on de novo protein synthesis (31). These results suggest that calcofluor induces some intracellular responses that are triggered by the drug binding to the cell wall chitin.
S. cerevisiae contains several mitogen-activated protein kinase (MAPK) cascades that translate physiological modifications into intracellular responses (see reference 11 for a recent review). The protein kinase C (PKC) cascade is involved in cell wall integrity and morphogenesis (17) by participating in the control of β-glucan synthesis (29), although recently it has been shown that this pathway also plays a role in the transcriptional control of CHS1, CHS2, and CHS3, the genes for the catalytic subunits of chitin synthases (13). Despite this, the relationship between the PKC pathway and chitin synthesis is not clear because the regulation of chitin synthesis in S. cerevisiae mostly occurs by posttranslational modification of Chs3p (7, 9). In theory, a hitherto-uncharacterized relationship might exist between other MAPK pathways, i.e., those involved in mating or sporulation, and cell wall synthesis because S. cerevisiae requires cell wall reorganization at different stages of its biological cycle (8).
S. cerevisiae contains a fourth MAPK pathway, the high-osmolarity glycerol response (HOG) pathway. It is basically involved in the adaptation of cells to high-osmolarity environments and is required for S. cerevisiae growth in media supplemented with high NaCl (0.9 M) or sorbitol (1.5 M) concentrations (11). The HOG pathway is well known in yeast and has been shown to be made up of three typical kinases (MAPKKK, MAPKK, and MAPK), which are products of the SSK2-22, PBS2, and HOG1 genes, respectively (11). Activation of this route in response to high osmolarity can take place through two independent routes: a two-component regulatory sensor (SLK1-SLN1), which activates the first kinase (21), or a single sensor (SHO1), which transfers the signal to the PBS2 kinase (22) through the MAPKKK protein Ste11p (26). The timing and intensity of the response varies, depending on the activation route (22), but in both cases an increase occurs in the intracellular glycerol concentration that allows the cells to grow in high-osmolarity media. This response is mediated by an increase in the transcription of GPD1 (1), which encodes the enzymatic activity that controls glycerol production. In addition, the expression of several other genes dependent upon this pathway is also increased, among them CTT1 (35) and GLO1 (14). The major transcription factor(s) controlled by the HOG pathway has not yet been identified in S. cerevisiae, although there is clear evidence that suggests that part of the transcriptional induction of the GPD1 gene is the result of relief of the repression caused by the Tup1p-Ssn6p complex (24).
In addition to its role in osmoregulation, the HOG pathway has also been implicated in the architecture of the S. cerevisiae cell wall through its control over β-glucan synthesis. PBS2 has been implicated in the transcriptional regulation of EXG1, a gene that encodes the major yeast glucanase. This regulation is translated into significant changes in β(1-6)glucan contents, depending on the deletion or overexpression of the PBS2 gene (15). Similarly, deletion or overexpression of this gene leads to significant differences in β(1-3)glucan synthase and β(1-3)glucan levels through an unknown mechanism (16). This involvement of the HOG pathway in cell wall architecture together with the activation of the PKC pathway under hypo-osmotic conditions (10) suggests an interconnection between these two signaling pathways. Based on this and other evidence, a model has very recently been proposed in which the PKC and HOG pathways would have opposite roles in the balancing of cell wall plasticity (28).
The present work focuses on the characterization of the resistance to calcofluor of S. cerevisiae mutants affected in the HOG pathway and shows that the antifungal effect of calcofluor depends not only on chitin, as previously suspected, but also, at least partially, on the presence of this signal transduction pathway. In addition, we show that the HOG pathway maintains its functionality under noninducing conditions, guaranteeing cellular protection against sudden increases in osmolarity (osmotic tolerance) and contributing to the maintenance of the cell wall architecture.
MATERIALS AND METHODS
Yeast strains, media, and general methods.
The yeast strains used in this work are described in Table 1. pbs2 null mutants in different genetic backgrounds were obtained by gene disruption as described elsewhere (35). Typical yeast media were used: yeast extract-peptone-dextrose (YEPD) as a complex medium and synthetic dextrose (SD) as a minimal medium, to which the corresponding supplements were added. Typically, experiments involving different NaCl concentrations were carried out on YEPD medium, while resistance to calcofluor was always measured on SD solid medium supplemented with 50 mM biphthalate buffer, pH 6.1, and different concentrations of calcofluor (31). Unless specified, all experiments were carried out with logarithmically growing cells (1 × 107 to 2.5 × 107/ml).
TABLE 1.
S. cerevisiae strains
| Strain | Genotype | Source or reference |
|---|---|---|
| W3031A | MATa ade2-1 his3-11,15 leu2-3,115 trp1-1 ura3-1 canr1-100 | Laboratory stock |
| TC1A | W3031A chs3::LEU2 | 9 |
| YSH 392 | W3031A gpd1::TRP1 | 1 |
| YSH788-1A | W3031B gpd2::URA3 | 2 |
| YSH788-2D | W3031B gpd1::TRP1 gpd2::URA3 | 2 |
| LJY1 | W3031A pbs2::LEU2 | This work |
| LJY2 | W3031A CTT1-7x-Leu2-LacZ | This work |
| LJY3 | W3031A pHV7 | This work |
| LJYE14 | W3031A pbs2-14 | This work |
| LJYE8 | W3031A hog1-E8 | This work |
| YPH499 | MATa leu2 ura3 his3 trp1 ade2 lys2 | 5 |
| JBY10 | YPH499 hog1::TRP1 | 5 |
| TM141 | MATa ura3 leu2 trp1 his3 | 22 |
| TM257 | TM141 ssk2::LEU2 ssk22::LEU2 | 22 |
| TM285 | TM141 MATα sho1::TRP1 | 22 |
| TM310 | TM141 ssk2::URA3 ssk22::LEU2 sho1::TRP1 | 22 |
| LJY4 | TM141 pbs2::LEU2 | This work |
| 15Dau | MATα ade1 his2 leu2-3,112 trp1-1a ura3ΔNS | 23 |
| 15Dau pkc1ts | 15Dau pkc1ts | 23 |
| LJY5 | 15Dau pkc1ts pbs2::LEU2 | This work |
Mutant selection was carried out in strain LJY3 (Table 1) by plating cells directly onto SD medium supplemented with 0.5 mg of calcofluor/ml. Two different aliquots were plated: one from a log-phase culture, allowing the isolation of spontaneous mutants, and the other containing cells mutagenized with ethyl methanesulfonate as described previously (32), with a survival rate of 45%. Approximately 5 × 106 cells were plated for each aliquot. Clones able to grow were retested in the same medium and chosen for further study. Complementation groups were obtained by crossing individual mutants with previously characterized mutants and testing the resistance of the corresponding diploids to calcofluor. When required, the diploids were sporulated and tetrad analysis was carried out (33).
All techniques involving the manipulation of yeast cells have been previously described (33). Molecular techniques were used as described elsewhere (34).
Plasmids.
Plasmid pCTT1-18/7x (35) was integrated into the chromosome after linearization with NcoI. Plasmids pLJ1 (pRS316::PBS2) and pLJ2 (pRS426::PBS2) were obtained by direct cloning of PBS2 as a SpeI/SacI fragment obtained from plasmid L42 (16). pLJ3 (pRS316::PBS2-14) and pLJ4 (pRS426::PBS2-14) contain a mutated copy of PBS2 generated by site-directed mutagenesis. The cloning of PCR products was accomplished in the pGEM-T plasmid (Promega Corporation) following the manufacturer's instructions.
Assays for tolerance to osmotic dehydration.
Tolerance to osmotic dehydration (osmotic tolerance) (3) is defined as the capacity of a culture to grow in dehydration medium (YEPD agar supplemented with 1.4 M NaCl) and is expressed as the percentage of cells that grow in this medium compared to that grown in YEPD agar. Alternatively, sorbitol-supplemented (YEPD agar–2.2 M sorbitol) plates were used as a dehydration medium. Typically, a culture growing logarithmically was diluted, and after one generation time, the culture was assayed for its osmotic tolerance after appropriate dilution by plating different aliquots onto YEPD or dehydration medium. Colonies were counted after 48 h (YEPD medium) or 7 days (dehydration medium) of growth. The osmotic tolerance of log-phase cells is defined as basal osmotic tolerance. When required, different aliquots of this log-phase culture were supplemented with the compound to be tested and osmotic tolerance was measured at different times (see Results for details of specific protocols). Osmotic tolerance in early-stationary-phase cells was assayed after 7 h of growth in YEPD medium. Cells containing plasmids were pregrown in selective medium, but dilution and further incubation were carried out in YEPD medium.
Analytical determinations.
Chitin contents were measured as the amount of N-acetyl-glucosamine present in the alkali-insoluble cellular fraction after its complete digestion with chitinase. The whole protocol has been described previously (38).
β-Galactosidase activity from the CTT1 7x Leu2 LacZ chimera was assayed as described previously (33) in early-log-phase cells to avoid expression of the CTT1 promoter under derepression conditions (35).
Glycerol was determined enzymatically with a commercial glycerol determination kit (Boehringer Mannheim) following the manufacturer's specifications. Intracellular glycerol was determined as follows. Cells (1.5 ml) were recovered by 5 min of centrifugation at 13,000 × g, resuspended in the same volume of boiling water, and heat treated at 95°C for 10 min in tubes with a pear-drop condenser (3). After cooling, the cells were decanted by centrifugation, and the supernatant was assayed for its glycerol concentration. The cells were counted after boiling in a Thoma chamber, and intracellular glycerol was always expressed as nanomoles/107 cells.
Immunological procedures.
Pbs2p and Pbs2-14p were detected in total cellular extracts as follows. Total-protein samples were obtained as described previously (9), quantified by absorbance at 280 nm, and loaded in equivalent amounts into sodium dodecyl sulfate–7.5% polyacrylamide gels. After electrophoresis, the proteins were transferred to an Immobilon-P membrane (Millipore), incubated with anti-Pbs2p goat polyclonal antibody (PBS2-yN-19; Santa Cruz Biotechnology), and immunodetected by the ECL method (Amersham Life Science).
Total Hog1p protein was detected in total cellular extract as described above by using anti-Hog1p goat polyclonal antibody (HOG1-yC-20; Santa Cruz Biotechnology). Phosphorylated Hog1p was detected in total cellular extract as described previously (26) but using phosphospecific p38 MAPK rabbit polyclonal antibody (New England Biolabs).
Isolation of multicopy suppressors of the pbs2-14 mutation.
Multicopy suppressors of the pbs2-14 mutation were isolated as follows. Strain EJY14 was transformed with an S. cerevisiae library constructed in the multicopy plasmid YEp24 (6). Transformants were grown on selective medium, and individual clones were screened for resistance to calcofluor. The clones that were sensitive to 0.5 mg of calcofluor/ml were subjected to plasmid loss by growing them on 5-fluoro-orotic acid plates. All the clones in which sensitivity to calcofluor was associated with the presence of a plasmid were selected for further study. We isolated five clones that might contain bona fide suppressors. In these cases, the plasmids were isolated and both ends of the cloned regions were sequenced using primers that anneal onto vector sequences. Comparison of the region sequenced with databases allowed us to pinpoint the exact restriction map of the clones, and a specific subcloning strategy was therefore designed for each suppressor. The open reading frame contained in the minimum DNA fragment able to complement the calcofluor resistance of strain EJY14 was considered to be the suppressor.
RESULTS
Isolation of mutants resistant to calcofluor.
The strategy of isolating mutants resistant to calcofluor has been very useful in S. cerevisiae for the identification of genes involved in chitin biosynthesis (8). However, this strategy has not been fully exploited because it is time-consuming in the sense that more than 90% of mutants arise because of mutations in the CHS3 gene (38). To avoid this problem, we designed a new screening procedure using a strain, LJY3, that contains two copies of CHS3: the chromosomal copy and another carried by pHV7 (39). In addition, selection was carried out in synthetic complete medium supplemented with only 0.5 mg of calcofluor/ml. Using this strategy, we isolated 63 new mutants resistant to calcofluor that, after rechecking, were back-crossed with all previously known chs− mutants. Analysis of the diploids formed indicated that 48 of the mutants were affected in previously defined loci (38), and their study was therefore abandoned (Table 2). The 15 remaining mutants could be classified in two groups in terms of septum enlargement after calcofluor treatment (Table 2, last column). Previous studies had indicated that the absence of septum enlargement is usually associated with a defect in chitin synthesis (9, 32). This work focuses on the group of calcofluor-resistant mutants without apparent defects in chitin synthesis.
TABLE 2.
Characterization of mutants resistant to calcofluor
| Mutanta | Complementation group | Growth on YEPD supplemented with NaClb
|
Septum enlargementbc | ||
|---|---|---|---|---|---|
| 0.9 M | 1.2 M | 1.4 M | |||
| W303 | None | +++ | +++ | +++ | +++ |
| Several (10) | CHS4 | NT | NT | NT | − |
| Several (3) | CHS5 | NT | NT | NT | − |
| One (1) | CHS6 | NT | NT | NT | − |
| Several (24) | CHS7 | NT | NT | NT | − |
| E8 | HOG1 | − | − | − | +++ |
| E10 | PBS2 | − | − | − | +++ |
| E11 | PBS2 | +++ | ++ | − | +++ |
| E14 | PBS2 | +++ | +++ | +++ | +++ |
| E5, E6, E9, E13, M9 | Not yet determined | +++ | +++ | +++ | +++ |
| E3, E12, E15, E16, M25, M26, M27 | Not yet determined | +++ | +++ | +++ | − |
Mutant nomenclature is as follows: E, spontaneous mutants; M, ethyl methanesulfonate induced mutants. The number in parentheses is the number of independent isolates.
Relative levels of growth or septum enlargement are indicated by the + symbols. −, absence of growth or septum enlargement. NT, not tested.
Septum enlargement was observed under a fluorescence microscope after growth for 3 to 4 h in medium supplemented with 0.075 mg of calcofluor/ml.
Preliminary characterization of the newly isolated mutants indicated that some of them were unable to grow at high NaCl concentrations, which prompted us to back-cross all these mutants with known HOG pathway mutants. The diploids formed after all the new mutants were crossed with pbs2 and hog1 null mutants were tested for salt and calcofluor sensitivity (Table 2). This complementation analysis indicated that strain E8 contains a mutation in the HOG1 gene, while strains E10, E11, and E14 contain mutations in PBS2. The remaining mutants do not seem to be affected in any of these genes. All the mutants affected in the HOG pathway, as well as others as yet uncharacterized, contained apparently normal levels of cellular chitin and had enlarged septa after calcofluor treatment. Further genetic characterization of these mutants indicated that resistance to calcofluor segregated in a Mendelian fashion in the E8, E10, E11, and E14 mutants, and this character cosegregated with salt sensitivity in strains E8 and E11. However, mutant E10 contained a second, unlinked mutation that was also related to salt sensitivity (data not shown). Our study focused on strain E14, which apparently contains a pbs2 mutant allele able to confer resistance to calcofluor but also to sustain full growth at 1.4 M NaCl.
HOG pathway mutants show different degrees of resistance to calcofluor but do not have alterations in chitin synthesis.
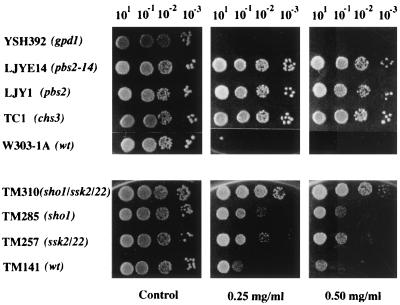
In order to understand the mechanism(s) involved in the resistance to calcofluor observed in the above-mentioned mutants, we tested the resistance levels of several HOG pathway mutants (Fig. 1). Clearly, the pbs2 null mutant can be classified as calcofluor-resistant, since it was able to grow at 0.5 mg of Calcofluor/ml (Fig. 1) and even up to 0.75 mg/ml (data not shown). However, growth at 1.0 mg/ml, a concentration at which the chs3 mutant can grow, was severely impaired (data not shown). sho1 and ssk2 ssk22 mutants also showed partial resistance to calcofluor, and this resistance increased significantly in the triple sho1 ssk2 ssk22 mutant. Unfortunately, we were unable to compare the absolute resistance values due to significant differences in the genetic backgrounds of the strains (Fig. 1, compare the respective wild types, W303 and TM141). By contrast, mutants with mutations in the GPD1 gene, a known target of the HOG pathway, were calcofluor sensitive (Fig. 1), as were the gpd2 and gpd1 gpd2 null mutants (data not shown).
FIG. 1.
S. cerevisiae growth on plates supplemented with different calcofluor concentrations. Serial dilutions of each strain as indicated above the panels were plated onto complete SD medium supplemented with different calcofluor concentrations as indicated below the panels, incubated for 48 h at 28°C, and photographed. Note the different genetic backgrounds used: W303 (upper) and TM141 (lower). wt, wild type.
This resistance prompted us to analyze the possible involvement of the HOG pathway in chitin synthesis. As shown in Table 3, different mutants in this pathway contain wild-type levels of chitin (approximately 0.2 to 0.3 nmol of N-acetyl-glucosamine/100 mg of cells). Additionally, osmotic treatment (0.4 M NaCl), which induced the HOG pathway, did not increase the chitin content in any of the strains. Calcofluor treatment increased the rate of chitin synthesis by two- to threefold, an effect that was clearly visible in the enlargement of septa (30). Although pbs2 and hog1 null mutants showed enlarged septa after calcofluor treatment, we measured the exact amount of chitin in these strains grown in the presence of calcofluor. As expected, they showed increases in chitin synthesis similar to that observed in wild-type strains but absent in the chs3 mutant, as previously described (32).
TABLE 3.
Effect of osmoregulation cascade on chitin biosynthesis
| Strain | Amt of chitina
|
||
|---|---|---|---|
| Control | Calcofluor | NaCl | |
| W303 (wild type) | 0.280 ± 0.04 | 0.710 ± 0.05 | 0.280 ± 0.04 |
| LJY1(pbs2::LEU2) | 0.210 ± 0.04 | 0.700 ± 0.19 | NT |
| JBY10 (hog1::TRP1) | 0.280 ± 0.05 | 0.850 ± 0.35 | 0.247 ± 0.06 |
| TC1(chs3::LEU2) | 0.022 ± 0.01 | 0.020 ± 0.00 | 0.028 ± 0.02 |
Chitin was measured as described in Materials and Methods and is expressed as nanomoles of GlcNAc per 100 mg of cells (wet weight). The cells were harvested after 3 h in YEPD medium (Control) or YEPD medium supplemented with calcofluor (0.075 mg/ml) or NaCl (0.4 M). NT, not tested.
In conclusion, the HOG pathway is not involved in chitin synthesis during vegetative growth or in the regulation of chitin synthesis during calcofluor treatment.
Calcofluor induces osmotic tolerance and increased glycerol concentrations but does not induce HOG pathway activation.
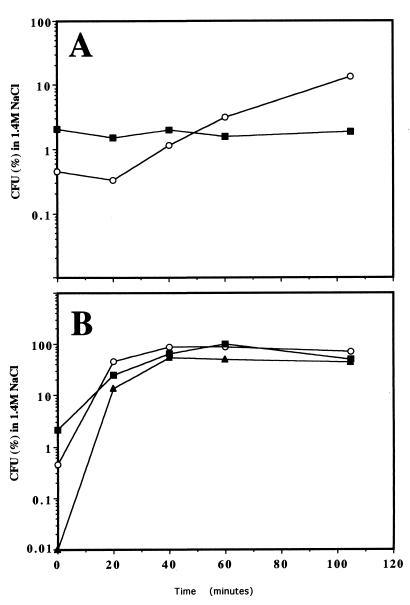
Activation of the HOG pathway by increased external osmolarity induces two clear physiological effects, an increase in the concentration of intracellular glycerol (1) and an increase in osmotic tolerance (20). To test the effect of calcofluor on osmotic tolerance, we incubated early logarithmically growing cells in YEPD supplemented with 0.075 mg of calcofluor/ml and determined their osmotic tolerances (see Materials and Methods) after different treatment times. Calcofluor treatment induced osmotic tolerance in the wild-type strain in a time-dependent fashion, since the basal levels of osmotic tolerance (zero time) were increased by approximately 30-fold after 105 min of treatment (Fig. 2 and Table 4). A similar increase in osmotic tolerance was obtained when plates supplemented with sorbitol were used (data not shown). The increase in osmotic tolerance due to calcofluor treatment was not observed in the chs3 mutant, although the basal levels in this strain were considerably higher (Fig. 2A and Table 4). This increase was very significant, although it was considerably less than that observed after induction of the HOG pathway by NaCl treatment (20) (Fig. 2B and Table 4). Apparently, the binding of calcofluor to chitin is required for the observed increase in osmotic tolerance.
FIG. 2.
Osmotic tolerance of different S. cerevisiae strains. (A) Osmotic tolerance of a logarithmically growing culture after incubation in the presence of 0.075 mg of calcofluor/ml for the indicated times. (B) Osmotic tolerance after osmotic (0.4 M NaCl) treatment for the indicated times. Osmotic tolerance is expressed as the percentage of cells able to grow on dehydration medium compared to those growing on YEPD. See Materials and Methods for details. ○, W303 (wild type); ■, TC1 (chs3−; ▴, LJYE14 (pbs2-14).
TABLE 4.
Osmotic tolerancea of S. cerevisiae strains grown under different conditions
| Strain | Osmotic tolerance (%)
|
|||
|---|---|---|---|---|
| Log phaseb | Stationaryc | NaCld | Calcofluore | |
| W303 (wild type) | 3.9 × 10−2 | 18.1 | 90.1 | 1.2 |
| TC1A (chs3−) | 2.9 × 10−1 | 31.0 | 100.0 | 2.6 × 10−1 |
| LJYE14 (pbs2-14) | 1.2 × 10−3 | 1.2 | 49.6 | 7.2 × 10−3 |
| LJYE14 (pRS426:: pbs2-14) | 7.9 × 10−2 | NDf | ND | ND |
Osmotic tolerance was measured as described in Materials and Methods in cells growing under different conditions.
Early logarithmically growing cells.
Early stationary-phase cells.
Early log-phase cells grown for 1 h in 0.4 M NaCl.
Early log-phase cells grown for 105 min in 0.075 mg of calcofluor/ml.
ND, not determined.
Calcofluor also caused a modest but reproducible increase (2.19-fold) in intracellular glycerol levels (Table 5), although this was considerably less than that observed after salt treatment (approximately 25-fold [data not shown]). This increase was not observed in the chs3− mutant or, more importantly, in the pbs2 or hog1 mutant (Table 5). The increase in intracellular glycerol levels observed after calcofluor treatment was apparently not due to the action of the GPD1 and GPD2 genes, since it was also observed in gpd1, gpd2, or gpd1 gpd2 mutants, although these strains had lower absolute levels of glycerol (Table 5).
TABLE 5.
Intracellular glycerol concentrations in different S. cerevisiae strains
| Strain | Intracellular glycerola
|
||
|---|---|---|---|
| YEPD | YEPD + calcofluor | ΔGlycerolb | |
| W303 (wild type) | 9.33 ± 2.2 | 20.43 ± 2.4 | 2.19 |
| TC1A (chs3−) | 18.2 ± 0.3 | 18.74 ± 2.4 | 1.03 |
| LJY1 (pbs2−) | 9.27 ± 1.1 | 10.74 ± 2.2 | 1.15 |
| LJYE14 (pbs2-14) | 6.01 ± 1.8 | 7.22 ± 2.4 | 1.20 |
| LJYE8 (hog1-E8) | 15.97 ± 1.6 | 15.82 ± 4.2 | 0.99 |
| YSH392 (gpd1) | 8.24 ± 2.4 | 16.72 ± 0.5 | 2.03 |
| YSH188-1A (gpd2) | 6.56 ± 0.5 | 15.63 ± 3.7 | 2.38 |
| YSH788-2D (gpd1 gpd2) | 4.16 ± 0.7 | 9.33 ± 1.1 | 2.24 |
Intracellular glycerol concentrations were measured as described in Materials and Methods and are expressed in nanomoles/107 cells. Glycerol levels were measured in cells growing for 105 min in YEPD medium without or with 0.075 mg of calcofluor/ml.
Ratio of glycerol increase after calcofluor treatment.
These results indicate that calcofluor triggers some of the responses of hyperosmotic shock, although at much lower levels. To confirm this hypothetical relationship between calcofluor treatment and hyperosmotic shock, we also measured the induction of CTT1 expression after calcofluor treatment. The CTT1 gene encodes the protein responsible for catalase T activity, which has been implicated in the osmotic response, and accordingly, its expression is strongly induced after hyperosmotic shock (35). To measure CTT1 induction, we used the GG18 strain, which contains the LacZ reporter gene under the control of the CTT1 promoter (35). While salt treatment increased β-galactosidase activity by approximately 30-fold, β-galactosidase activity in calcofluor-treated cells was only 10 to 15% higher than that seen in the controls, a value that does not correspond to the increase in osmotic tolerance or even to the increase in glycerol accumulation (data not shown). We also determined the phosphorylation status of the Hog1p protein (see Materials and Methods) after calcofluor treatment. We were unable to detect any tyrosine-phosphorylated form of Hog1p after 5, 15, 30, 60, or even 120 min of calcofluor treatment. Under similar conditions, salt-induced phosphorylation of Hog1p was detected from 5 to 15 min after hyperosmotic shock (data not shown). Taken together, these results clearly indicate that calcofluor treatment does not activate the HOG pathway.
Characterization of the pbs2-14 mutant.
We have shown above that strain E-14 contains a mutation that confers resistance to calcofluor but also supports growth at high salt concentrations (Fig. 1 and Table 2) or at 1.5 M sorbitol (data not shown). Diploid strain E14/LJY1 (pbs2::LEU2) is resistant to calcofluor, as are the parent strains, indicating that E14 contains a mutation in the PBS2 gene. To confirm this, segregation analysis was carried out on this diploid, resulting in a calcofluor resistance segregation of 4:0. In addition, the resistance of strain E14 to calcofluor was reversed after transformation with a centromeric plasmid containing the PBS2 gene. Therefore, strain E14 must contain a new pbs2 mutant allele; we call it pbs2-14.
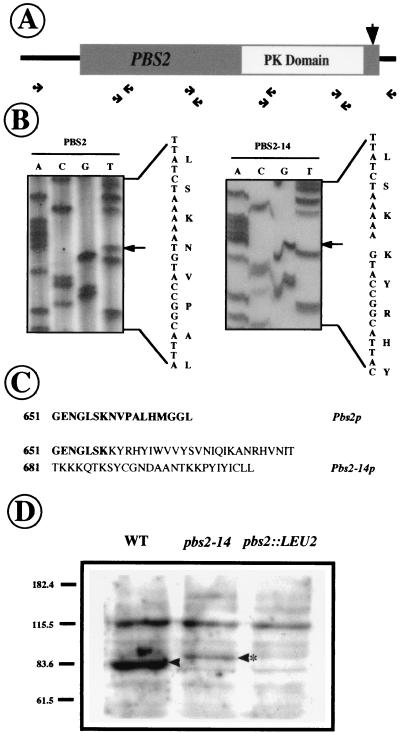
This mutant allele was cloned by PCR using the strategy outlined in Fig. 3A. The strategy basically consists of cloning the mutant allele in small (600-bp) overlapping fragments. These fragments were cloned and sequenced, and their sequences were compared to that of the wild-type PBS2 gene. To avoid Taq polymerase errors, at least three different clones from each fragment were sequenced and any difference in sequence was confirmed in a totally independent PCR amplification of the same region. Following this, we found that pbs2-14 contains a deletion of the thymidine (T) at position 1977 of the open reading frame (Fig. 3B). This deletion changes the reading frame and increases the size of the protein (Fig. 3C). The mutation is at the very end of the C region, altering only the last 10 amino acids, and it is therefore not expected to cause alterations in the protein kinase domain, which is located in the middle of the protein (Fig. 3A). To confirm the nature of the altered protein, we carried out the Western immunoblotting experiment shown in Fig. 3D. While the Pbs2p wild-type protein ran with its expected molecular size, Pbs2-14p showed a higher molecular weight that matched that predicted by the sequence data. However, the amount of Pbs2-14p was considerably lower than that observed for Pbs2p, suggesting greater instability of the mutated protein.
FIG. 3.
Molecular characterization of the PBS2-14 mutation. (A) Schematic representation of PBS2-14 PCR cloning. Primer pairs and the conserved protein kinase (PK) domain are indicated. The arrow shows the approximate position of the E14 mutation. (B) Sequence discrepancy between wild type and mutant; the deleted base is indicated by an arrow. The protein translation is shown on the right side of each picture. (C) C-terminal sequence of Pbs2 and Pbs2-14 proteins. (D) Immunodetection of Pbs2p and Pbs2-14p in Western blot using anti-Pbs2p antibodies (see Materials and Methods). The arrowheads indicate the Pbs2 protein that is absent in the pbs2 null mutant. The altered Pbs2-14 protein is indicated by an asterisk. WT, wild type.
The most relevant characteristic of the pbs2-14 mutant is its ability to grow at high salt concentrations, despite its resistance to calcofluor. However, when we determined the basal tolerance levels of this mutant we observed a severe reduction in this parameter. The basal tolerance of the pbs2-14 mutant was more than 30-fold smaller than that of the wild-type strains (Table 4). This difference was not only observed in log-phase cells but also in stationary-phase cells, despite the overall higher tolerance (Table 4). Consistent with its ability to grow at high salt concentrations, pretreatment of the pbs2-14 mutant with 0.4 M NaCl increased its osmotic tolerance to values close to those of the controls (Table 4), and its induction kinetics were also similar (Fig. 2B). This treatment also induced a normal HOG response in pbs2-14 mutants, as determined by the expression levels of STRE-LacZ fusions or the intracellular increase in glycerol concentrations (data not shown). Apparently, the pbs2-14 mutation impairs basal osmotic tolerance but not the induction of the HOG pathway due to high external osmolarity.
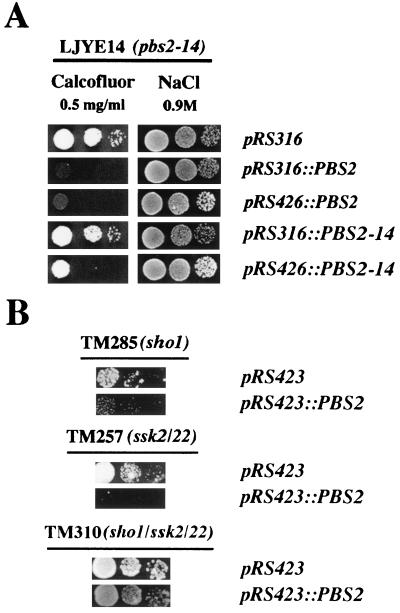
In order to confirm the relationship between the pbs2-14 mutation and the observed phenotypes, we constructed monocopy and multicopy plasmids expressing the Pbs2p or Pbs2-14p protein (see Materials and Methods). These plasmids were transformed into W303 (wild-type), LJY1 (pbs2::LEU2) and LJYE14 (pbs2-14) isogenic strains. The phenotypes of some of these strains are shown in Fig. 4A. As expected, overexpression of these genes had no apparent effect on the wild-type strain (data not shown). However, PBS2, either in monocopy or multicopy plasmids, complemented the resistance of the pbs2-14 mutant to calcofluor (Fig. 4A) and the calcofluor resistance and salt sensitivity of the pbs2::LEU2 mutant (data not shown). PBS2-14 only weakly complemented the salt concentration-dependent growth defect of the pbs2 null mutant when expressed in monocopy plasmids. However, its overexpression completely abolished the salt sensitivity of this strain (data not shown). Similarly, only the overexpression of PBS2-14 in the pbs2 null mutant partially complemented the calcofluor resistance of this strain (not shown). PBS2-14 overexpression in the pbs2-14 mutant led to a significant reduction in calcofluor resistance (Fig. 4A). PBS2-14 overexpression also elicited a significant increase in the basal osmotic tolerance of the pbs2-14 mutant (Table 4). Taken together, these data suggest that all the pbs2-14 mutant phenotypes observed are due to lower levels of the Pbs2p protein, which can be compensated for by the overexpression of mutated protein.
FIG. 4.
Mutant growth in different media after transformation with different plasmids. Serial dilutions of each strain were plated onto SD medium supplemented with 0.5 mg of calcofluor/ml or YEPD medium supplemented with 0.9 M NaCl. The plates were photographed after 48 h of growth at 28°C. The strains are indicated above each picture. The plasmids carried by each strain are indicated at the right. (A) Calcofluor and salt plates are shown (W303 background). (B) Only calcofluor plates are shown. Note that TM285, TM257, and TM310 strains are in the TM141 background.
In addition to these data, we observed that overexpression of PBS2 was able to alleviate the calcofluor resistance phenotype associated with sho1 or ssk2 ssk22 mutations. However, it had no apparent effect on the resistance of the sho1 ssk2 ssk22 triple mutant (Fig. 4B).
Isolation of multicopy suppressors of the pbs2-14 mutation.
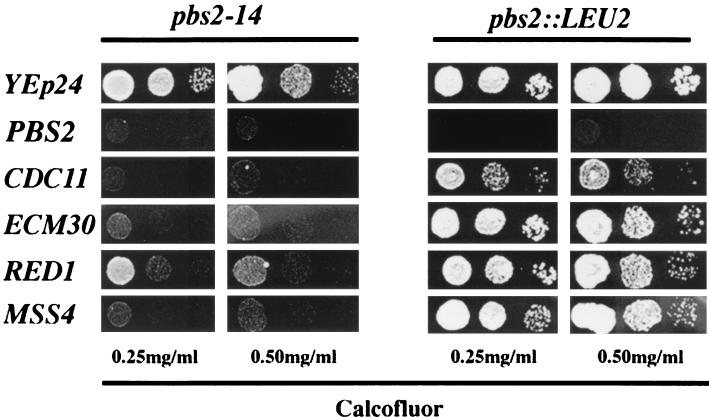
In an attempt to identify some of the genes involved in calcofluor sensitivity and resistance through the HOG pathway, we searched for multicopy suppressors of the pbs2-14 mutation. To do so, we transformed strain LJYE14 with an S. cerevisiae gene bank constructed in the episomic vector YEp24 and looked for transformants sensitive to calcofluor. We selected several transformants that were more sensitive to calcofluor than the parent strain (Fig. 5). After standard procedures (plasmid loss, end sequencing, and subcloning), we were able to assign the specific suppressors to four independently isolated genes, ECM30, CDC11, MSS4, and RED1, none of which were previously associated with the HOG pathway. The ECM30 gene has been identified in a screening for insertion mutants hypersensitive to calcofluor (19). CDC11 is a member of the septin family and hence is involved in the organization of the septum during cell division (18). MSS4 encodes a phosphatidylinositol-4-phosphate 5-kinase that is required for cell morphogenesis (12). Finally, RED1 encodes a protein required for chromosome segregation during meiosis, although its exact function is unknown (37). The four genes clearly suppressed the calcofluor resistance phenotype associated with the pbs2-14 mutation but also weakly suppressed the resistance observed in the null pbs2 mutant (Fig. 5). In addition, these multicopy suppressors also conferred weak hypersensitivity to calcofluor when introduced in wild-type strains (data not shown).
FIG. 5.
Effects of different multicopy suppressors on calcofluor resistance. Serial dilutions of each strain were plated onto SD selective medium supplemented with different calcofluor concentrations (shown at bottom). The recipient strains (top) were transformed with multicopy plasmids containing the genes shown on the left.
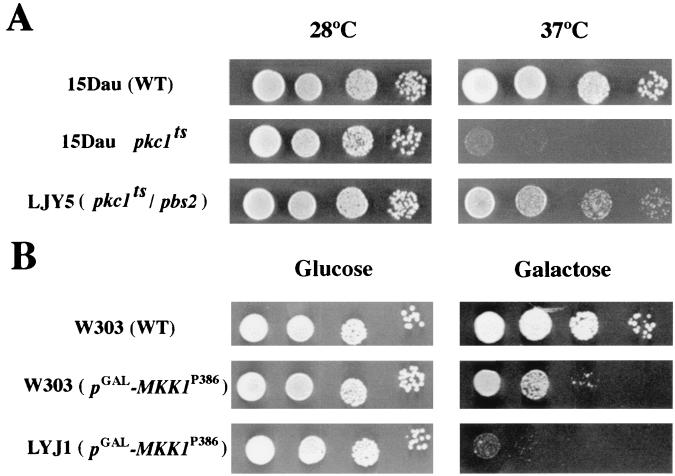
There is no obvious relationship among these four genes, but at least three of them, ECM30, CDC11, and MSS4, can be associated with cell wall or morphogenetic processes. These results suggest that the effect of the HOG pathway in resistance and sensitivity to calcofluor could be mediated through the participation of this pathway in the control of cell wall architecture. To test this possibility, we investigated the epistatic relationships between the PKC and HOG signal transduction pathways. Figure 6A shows that disruption of the PBS2 gene clearly alleviates the temperature-sensitive phenotype of the pck1 mutation (23). Overexpression of an activated form of Mkk1p kinase (40) leads to a highly activated PKC pathway that significantly reduces cell growth (Fig. 6B), a deleterious effect that is increased in the absence of the PBS2 gene (Fig. 6B). Thus, the PKC and HOG pathways seem to play opposite roles in cell physiology.
FIG. 6.
Epistatic relationship between the PKC and HOG signal transduction pathways. (A) Serial dilutions of each strain (15Dau background) were plated onto YEPD medium, and replica plates were incubated at the indicated temperatures for 48 h. (B) Isogenic W3031A or LJY1 (pbs2::LEU2) strains were transformed with plasmid pNV7-MKK1P386 in which the hyperactive form of the MKK1 gene is under the control of the GAL1 promoter (40). Serial dilutions of each strain were plated onto selective medium with either glucose or galactose as a carbon source. Growth was scored after 2 (glucose) or 4 (galactose) days of growth.
DISCUSSION
The HOG pathway is involved in S. cerevisiae resistance and sensitivity to calcofluor.
Previous attempts to isolate S. cerevisiae mutants resistant to calcofluor have been successful in the identification of genes involved in chitin synthesis (32, 38). The fact that all the complementation groups identified are deficient in chitin synthesis prompted us to assume that chitin is the primary and only target of this drug (8). However, in addition to the chitin defect, it should be possible to obtain resistance by blocking the increase in chitin synthesis induced by calcofluor treatment. This hypothesis is sustained by earlier reports that indicate that calcofluor binds chitin, increases its synthesis, and disturbs the cell wall (30, 31). Very recently, new mutants partially resistant to calcofluor but with no apparent defect in chitin synthesis have been isolated (19). However, this study focused on the isolation strategy and not on mutant characterization, and it would therefore be unwise to attempt to draw any conclusions about the mechanism(s) of action of calcofluor based on these data alone.
The present report addresses the isolation and characterization of new S. cerevisiae mutants resistant to calcofluor, using a strategy that circumvents the problem of resistance saturation by mutation in the CHS3 gene (38). We obtained several new mutants affected in previously described chs complementation groups. It is interesting that the statistical distribution of these mutants is quite different from those reported in previous studies, probably due to the lower concentration of calcofluor used. Our strategy was successful, since 16 new mutants were isolated. Preliminary characterization indicated that they belong to several new complementation groups. The most intriguing result of this screening was the isolation of some mutants sensitive to high salt concentration that turned out to be mutant alleles of the two best-characterized kinases of the HOG pathway: PBS2 and HOG1. The null mutants for these genes were also resistant to calcofluor, although their resistance levels were lower (0.5 to 0.75 mg/ml) than those observed for different chs− mutants (1.0 mg/ml). HOG activation in S. cerevisiae occurs through two independent mechanisms mediated by the SHO1 and SLN1-SLK1 genes, respectively (11). This dual activation route clearly explains the partial resistance of sho1 or ssk2 ssk22 mutants to calcofluor, while the resistance of the sho1 ssk2 ssk22 triple mutant is similar to that of pbs2 null mutant (Fig. 1).
Is there any relationship between the HOG pathway and chitin synthesis? The results obtained here seem to show that this is not the case. Neither PBS2 nor HOG1 is involved in chitin synthesis, and neither of them mediates the increase in chitin levels observed during calcofluor treatment (Table 3). In addition, induction of the HOG pathway by salt does not affect chitin synthesis (Table 3). These results indicate that the presence of chitin is not enough to explain sensitivity or resistance to calcofluor. The lower resistance to calcofluor observed in the pbs2 null mutant compared to that in different chs mutants suggests that chitin is absolutely required for calcofluor action but that the HOG pathway is also partially involved in the process.
Figure 2 and Table 5 indicate that calcofluor induces certain intracellular responses which could be related to osmoregulation; the drug increased intracellular glycerol concentrations and osmotic tolerance. When it was possible to carry out the test, we observed that these effects were dependent on a functional HOG pathway, because they were absent in pbs2 and hog1 mutants (Table 5). These effects were not observed in chs3 (Fig. 2 and Table 5) or in other chs mutants (data not shown). Glycerol accumulation, however, does not depend on the function of GPD genes, since different gpd mutants were seen to increase intracellular glycerol levels after calcofluor treatment (Table 5). Therefore, the increased glycerol levels, and possibly the increase in osmotic tolerance, could reflect general changes in cell physiology dependent on the HOG pathway (see below). However, it is clear that the absolute levels of glycerol cannot be directly related to calcofluor resistance or sensitivity, since the gpd mutants, which have very low levels of intracellular glycerol (Table 5), were sensitive to calcofluor.
Interestingly, the increase in osmotic tolerance or glycerol accumulation due to calcofluor treatment was significantly different from that observed after salt treatment, not only in the size of the response but also in its kinetics. In addition, we were unable to detect any calcofluor-dependent phosphorylation of Hog1p. In view of these results, the small increase observed in the expression of the STRE-LacZ fusion after calcofluor treatment is not surprising. Taken together, these results indicate that calcofluor treatment does not induce activation of the HOG pathway. Accordingly, the isolation of mutant E14, which is resistant to calcofluor but also to high salt concentrations, is a clear indication that HOG-dependent osmoadaptation and calcofluor resistance are two physiologically different phenomena.
Physiological role of the HOG pathway under noninducing conditions.
Sequencing of the pbs2-14 mutation indicated that Pbs2-14p is only altered in its C-terminal domain; therefore, the mutation should not affect the catalytic domain of the kinase (4). This is in agreement with the growth of the pbs2-14 mutant at high salt concentrations. Figure 2 shows that osmoadaptation after salt treatment is normal in this mutant and follows kinetics similar to those of the wild-type strain. In addition, induction of the STRE-LacZ fusion and the increase in intracellular glycerol levels by salt in this mutant are comparable to those observed in the wild-type strain (data not shown). All these observations are clear indications that activation of the HOG pathway by increased osmolarity occurs normally in the pbs2-14 mutant.
Like the pbs2 null mutant, the pbs2-14 mutant is resistant to calcofluor. Are there any differences between this mutant and the wild-type apart from their sensitivity or resistance to calcofluor? We observed that basal tolerance in strain LJYE14 (pbs2-14) was only 2 to 3% of that of the wild-type. This difference persisted in early-stationary-phase cells despite higher overall levels. These results indicate that the decrease in basal osmotic tolerance is intrinsic to the pbs2-14 mutation. However, the difference in osmotic tolerance was not significant after salt treatment (Fig. 2). Apparently, the HOG pathway could have some uncharacterized function under noninducing conditions; this function would be severely impaired in the pbs2-14 mutant. To date, resistance to calcofluor and low basal tolerance have been linked to the pbs2-14 allele.
After these results one would have to address the issue of why the pbs2-14 mutant is unable to maintain the steady-state level of the HOG pathway. A possible explanation for this can be found in Fig. 3: Pbs2-14p levels were much lower than those of Pbs2p, and such low levels of protein would not be enough for biological function, despite the functionality of the mutated protein. Phosphorylation of Pbs2-14p by the induction of the HOG pathway would increase Pbs2p-14 kinase activity, overcoming the defects associated with this low protein level. At this point we cannot be sure of the reasons at the molecular level for the low abundance of this protein, but the differences observed suggest low Pbs2p-14 stability. In addition, we cannot exclude the possibility that the described mutation could have some minor effect on kinase activity. Regardless of the molecular reasons, if this hypothesis is correct an alleviation of phenotypes associated with the pbs2-14 mutation after overexpression of this protein would be expected. This indeed seems to be the case: overexpression of PBS2-14 in a multicopy plasmid significantly decreased not only the resistance phenotype observed in strain LJYE14 (pbs2-14) (Fig. 4) but also its basal osmotic tolerance (Table 4). Similarly, complementation of the salt sensitivity phenotype of the pbs2 null mutant by PBS2-14 was only complete when the mutated protein was overexpressed (data not shown).
We have already shown that the HOG pathway is required for osmotic tolerance and calcofluor sensitivity, but it is also important to note that the functionality of this pathway under noninducing conditions depends not only on the kinases but also on the activation routes. This conclusion is based on several lines of evidence. (i) Mutations in the SHO1 or SSK2-22 branches only conferred partial calcofluor resistance, whereas the triple mutant was significantly more resistant (Fig. 2). (ii) PBS2 overexpression suppressed the calcofluor resistance phenotype observed in sho1 and ssk2 ssk22 mutants but not that found in the sho1 ssk2 ssk22 triple mutant (Fig. 4). (iii) Overexpression of PBS2 in wild-type strains leads to calcofluor hypersensitivity (data not shown).
We have been able to show the effect of the removal of the HOG pathway on resistance to calcofluor and osmotic tolerance. However, we do not yet know the molecular reasons for these cellular effects. It has previously been reported that PBS2 elimination causes increases in β(1-6)glucan levels (15) and decreases in β(1-3) levels (16); the opposite effects on glucan synthesis are observed after PBS2 overexpression. The effects on β(1-6)glucan are due to an altered regulation of EXG1, the gene that encodes the major exoglucanase. However, the alteration in β-glucan synthesis through PBS2 cannot account for calcofluor resistance and sensitivity, since deletion or overexpression of EXG1 does not alter the calcofluor sensitivity of wild-type strains (data not shown). In addition, a decrease in the rate of β(1-3)glucan synthesis, such as that observed in fks1 mutants, leads to calcofluor hypersensitivity (27).
The data presented in this paper clearly suggest a scenario in which calcofluor resistance and sensitivity would depend, in addition to the absence or presence of chitin, on a physiological state rather than on the existence of a unique intracellular target for calcofluor. This hypothesis is favored by the nature of the suppressors of the pbs2-14 mutation that were isolated. Although they were isolated as suppressors of the calcofluor resistance of the pbs2-14 mutant, all of them caused calcofluor hypersensitivity in the wild-type strain (data not shown), clearly indicating that we had not isolated true suppressors but rather genes whose overexpression confers calcofluor hypersensitivity. However, there must be some kind of direct relationship between the effects of these suppressors and the HOG pathway, because their overexpression elicited only minor effects on pbs2 null mutants (Fig. 5). The most likely explanation regarding these suppressors is that they would have pleiotropic effects on cell physiology, and these effects would depend on a functional HOG pathway. All the suppressors—with the exception of RED1, whose exact function is not yet clear—have been related more or less directly to cell wall synthesis or assembly (12, 18, 19). Therefore, some of their effects would be expected to affect cell wall synthesis and/or assembly, even though no effect on chitin synthesis was detected (data not shown). This hypothesis seems to be supported by the study of the epistatic relationships between the PKC and HOG signal transduction pathways, the results of which (Fig. 6) clearly indicate that the pathways act antagonistically. Very recently it has been proposed that these pathways play opposite roles in the cell, balancing the plasticity of the cell wall during growth (28). However, to our knowledge this is the first time that a direct relationship between these two signal transduction pathways has been shown in S. cerevisiae. The molecular link between these pathways is not yet known, although it has been reported that Ptp2 and Ptp3 phosphatases can act as global regulators of MAPK signaling, including the HOG and PKC pathways (25). Additionally, the hypothetical participation of the HOG pathway in the control of cell wall plasticity could be also envisioned as the molecular link between this pathway and resistance and sensitivity to calcofluor.
The results reported here clearly support the current hypothesis about the coordinated contribution of the HOG and PKC pathways to cell wall architecture, offering a new approach that could open new areas of study in the field.
ACKNOWLEDGMENTS
We thank C. Sculler, H. Saito, F. Posas, S. Homman, and J. Ramos for providing us with plasmids and strains. Special thanks are due to S. Homman, who, in addition to plasmids and strains, provided us with very helpful comments at the beginning of this work. Thanks are also due to J. A. Trilla, M. H. Valdivieso, and Y. Sanchez for comments on the manuscript and to the rest of A. Duran's lab for scientific criticism.
This work has been supported by a MEC predoctoral fellowship (to L.J.G.R.) and CICYT grants BIO95-0500 and BIO98-0814 (to C.R.).
REFERENCES
- 1.Albertyn J, Hohmann S, Thevelein J M, Prior B A. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol Cell Biol. 1994;14:4135–4144. doi: 10.1128/mcb.14.6.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansell R, Granath K, Hohmann S, Thevelein J M, Adler L. The two isoenzymes for yeast NAD+-dependent glycerol 3-phosphate dehydrogenase encoded by GPD1 and Gpd2 have distinct roles in osmoadaptation and redox regulation. EMBO J. 1997;16:2179–2187. doi: 10.1093/emboj/16.9.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blomberg A, Larsson C, Gustafsson L. Microcalorimetric monitoring of growth of Saccharomyces cerevisiae: osmotolerance in relation to physiological state. J Bacteriol. 1988;170:4562–4568. doi: 10.1128/jb.170.10.4562-4568.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boguslawski G, Polazzi J O. Complete nucleotide sequence of a gene conferring polymyxin B resistance on yeast: similarity of the predicted polypeptide to protein kinases. Proc Natl Acad Sci USA. 1987;84:5848–5852. doi: 10.1073/pnas.84.16.5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brewster J L, de Valoir T, Dwyer N D, Winter E, Gustin M C. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 6.Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted and intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- 7.Chuang J S, Schekman R W. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J Cell Biol. 1996;135:597–610. doi: 10.1083/jcb.135.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cid V J, Duran A, del Rey F, Snyder M P, Nombela C, Sanchez M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cos T, Ford R A, Trilla J A, Duran A, Cabib E, Roncero C. Molecular analysis of Chs3p participation in chitin synthase III activity. Eur J Biochem. 1998;256:419–426. doi: 10.1046/j.1432-1327.1998.2560419.x. [DOI] [PubMed] [Google Scholar]
- 10.Davenport K R, Sohaskey M, Kamada Y, Levin D E, Gustin M C. A second osmosensing signal transduction pathway in yeast. J Biol Chem. 1995;270:30157–30161. doi: 10.1074/jbc.270.50.30157. [DOI] [PubMed] [Google Scholar]
- 11.Gustin M V, Albertyn J, Alexander M, Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Homma K, Terui S, Minemura M, Qadota H, Anraku Y, Kanaho Y, Ohya Y. Phosphatidylinositol-4-phosphate 5-kinase localized on the plasma membrane is essential for yeast cell morphogenesis. J Biol Chem. 1998;273:15779–15786. doi: 10.1074/jbc.273.25.15779. [DOI] [PubMed] [Google Scholar]
- 13.Igual J C, Johnson A L, Johnston L H. Coordinated regulation of gene expression by the cell cycle transcription factor SWI4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 1996;15:5001–5013. [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue Y, Tsujimoto Y, Kimura A. Expression of the glyoxalase I gene of Saccharomyces cerevisiae is regulated by high glycerol mitogen-activated protein kinase pathway in osmotic stress response. J Biol Chem. 1998;273:2977–2983. doi: 10.1074/jbc.273.5.2977. [DOI] [PubMed] [Google Scholar]
- 15.Jiang B, Ram A F, Sheraton J, Klis F M, Bussey H. Regulation of cell wall beta-glucan assembly: PTC1 negatively affects PBS2 action in a pathway that includes modulation of EXG1 transcription. Mol Gen Genet. 1995;248:260–269. doi: 10.1007/BF02191592. [DOI] [PubMed] [Google Scholar]
- 16.Lai M H, Silverman S J, Gaughran J P, Kirsch D R. Multiple copies of PBS2, MHP1 or LRE1 produce glucanase resistance and other cell wall effects in Saccharomyces cerevisiae. Yeast. 1997;13:199–213. doi: 10.1002/(SICI)1097-0061(19970315)13:3<199::AID-YEA76>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 17.Levin D E, Bartlett-Heubusch E. Mutants in the S. cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J Cell Biol. 1992;116:1221–1229. doi: 10.1083/jcb.116.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Longtine M S, DeMarini D J, Valencik M L, Al-Awar O S, Fares H, De Virgilio C, Pringle J R. The septins: roles in cytokinesis and other processes. Curr Opin Cell Biol. 1996;6:106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- 19.Lussier M, et al. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics. 1997;147:435–450. doi: 10.1093/genetics/147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackenzie K F, Blomberg A, Brown A D. Water stress plating hypersensitivity of yeast. J Gen Microbiol. 1986;132:2053–2056. doi: 10.1099/00221287-132-7-2053. [DOI] [PubMed] [Google Scholar]
- 21.Maeda T, Wurgler-Murphy S M, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 22.Maeda T, Takekawa M, Saito H. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science. 1995;269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- 23.Marini N J, Meldrum E, Buehrer B, Hubberstey A W, Stone D E, Traynor-Kaplan A, Reed S I. A pathway in the yeast cell division cycle linking protein kinase C (Pkc1) to activation of Cdc28 at START. EMBO J. 1996;15:3040–3052. [PMC free article] [PubMed] [Google Scholar]
- 24.Marquez J A, Pascual-Ahuir A, Proft M, Serrano R. The Ssn6-Tup1 repressor complex of Saccharomyces cerevisiae is involved in the osmotic induction of HOG-dependent and -independent genes. EMBO J. 1998;17:2543–2553. doi: 10.1093/emboj/17.9.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mattison C P, Spencer S S, Kresge K A, Lee J, Ota I M. Differential regulation of the cell wall integrity mitogen-activated protein kinase pathway in budding yeast by the protein tyrosine phosphatases Ptp2 and Ptp3. Mol Cell Biol. 1999;19:7651–7660. doi: 10.1128/mcb.19.11.7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 27.Ram A F J, Brekelmans S S C, Oehlen L J W M, Klis F M. Identification of two cell-cycle regulated genes affecting the b1,3-glucan content of cell walls in Saccharomyces cerevisiae. FEBS Lett. 1995;358:165–170. doi: 10.1016/0014-5793(94)01418-z. [DOI] [PubMed] [Google Scholar]
- 28.Reynolds T B, Hopkins B D, Lyons M R, Graham T R. The high osmolarity glycerol response (HOG) MAP kinase pathway controls localization of a yeast golgi glycosyltransferase. J Cell Biol. 1998;143:935–946. doi: 10.1083/jcb.143.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roemer T, Paravicini G, Payton M A, Bussey H. Characterization of the yeast (1→6)-beta-glucan biosynthetic components, Kre6p and Skn1p, and genetic interactions between the PKC1 pathway and extracellular matrix assembly. J Cell Biol. 1994;127:567–579. doi: 10.1083/jcb.127.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roncero C, Duran A. Effect of Calcofluor white and Congo red on fungal wall morphogenesis: in vivo activation of chitin polymerization. J Bacteriol. 1985;163:1180–1185. doi: 10.1128/jb.163.3.1180-1185.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roncero C, Valdivieso M H, Ribas J C, Duran A. Effect of Calcofluor white on chitin synthases from Saccharomyces cerevisiae. J Bacteriol. 1988;170:1945–1949. doi: 10.1128/jb.170.4.1945-1949.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roncero C, Valdivieso M H, Ribas J C, Duran A. Isolation and characterization of Saccharomyces cerevisiae mutants resistant to Calcofluor white. J Bacteriol. 1988;170:1950–1954. doi: 10.1128/jb.170.4.1950-1954.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Schuller C, Brewster J L, Alexander M R, Gustin M C, Ruis H. The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene. EMBO J. 1994;13:4382–4389. doi: 10.1002/j.1460-2075.1994.tb06758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaw J A, Mol P C, Bowers B, Silverman S J, Valdivieso M H, Duran A, Cabib E. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1991;114:111–123. doi: 10.1083/jcb.114.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson E A, Roeder S G. Expression and DNA sequence of RED1, a gene required for meiosis I chromosome segregation in yeast. Mol Gen Genet. 1989;218:293–301. doi: 10.1007/BF00331281. [DOI] [PubMed] [Google Scholar]
- 38.Trilla J A, Duran A, Roncero C. Chs7p, a new protein involved in the control of protein export from the endoplasmic reticulum that is specifically engaged in the regulation of chitin synthesis in Saccharomyces cerevisiae. J Cell Biol. 1999;145:1153–1163. doi: 10.1083/jcb.145.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valdivieso M H, Mol P C, Shaw J A, Cabib E, Duran A. CAL1, a gene required for activity of chitin synthase 3 in Saccharomyces cerevisiae. J Cell Biol. 1991;114:101–109. doi: 10.1083/jcb.114.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watanabe Y, Irie K, Matsumoto K. Yeast RLM1 encodes a serum response factor-like protein that may function downstream of the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol Cell Biol. 1995;15:5740–5749. doi: 10.1128/mcb.15.10.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]