Abstract
Artificial Intelligence (AI) in oncology is advancing beyond algorithm development to integration into clinical practice. This review describes the current state of the field, with a specific focus on clinical integration. AI applications are structured according to cancer type and clinical domain, focusing on the four most common cancers and tasks of detection, diagnosis, and treatment. These applications encompass various data modalities, including imaging, genomics, and medical records. We conclude with a summary of existing challenges, evolving solutions, and potential future directions for the field.
Introduction
Artificial Intelligence (AI) is increasingly being applied to all aspects of oncology. These applications have been set in motion by two fundamental shifts. The first is the development of new computational models and tools. In particular, advances in deep learning1 over the past decade have made it feasible to learn complex patterns directly from real-world data, making it the core driver of AI advances in and out of healthcare2. These advances in deep learning have been accompanied with rapid advances in graphical processing units (GPUs) and cloud computing that enable the development of increasingly large models trained on massive data sets. The second shift is the advancing digital landscape of oncology itself. This includes the storage of patient data in electronic medical record (EMR) systems, the digitization of radiology and pathology imaging3, and the increasing adoption of routine genomic profiling4. While this digitization is non-uniform across data modalities and clinical sites, there is increasing availability of detailed, longitudinal information for cancer patients. This data can be used to build and train AI models, and critically, the real-time availability of this data can enable individualized, clinically relevant AI predictions to further the goal of precision oncology5.
Within this review, we aim to summarize the current landscape of AI in oncology. While the field can encompass numerous applications, including biological and drug discovery6, this review specifically focuses on AI use cases targeting direct integration into clinical practice. This focus is motivated by the accelerated advancement of AI applications from development to clinical use. Additionally, we focus on AI approaches based on modern, deep learning methods, rather than other machine learning or rule-based methods. Finally, given the numerous possible applications and breadth of AI work, the review focuses on the four most common cancer types: Breast, Prostate, Lung, and Colorectal, accounting for 50% of all new cancer cases in 20237. AI applications for these cancer types serve as a good representation of the current state of the field, especially as their prevalence facilitates large-scale data collection and encourages clinical application.
To first motivate the focus on applications targeting clinical integration and illustrate the pathway to clinical deployment, we highlight the use of AI for breast cancer detection in mammography (Figure 1). To develop such a system, one typically curates thousands of mammograms (x-ray images of the breast) along with ground truth annotations of whether breast cancer is present. A deep artificial neural network is then trained to detect the presence of breast cancer based on this data (Figure 1A). Initial testing of the resulting model often consists of pilot, retrospective studies. Towards clinical integration, commercialization may follow, consisting of steps such as product engineering, more rigorous testing, and regulatory clearance (Figure 1B). In the case of mammography, there are now multiple commercial AI devices cleared by the US Federal Drug Administration (FDA) and/or the European Union (EU) that are designed to aid radiologists in breast cancer detection. There are also multiple ongoing clinical trials evaluating the real-world effectiveness of such systems.
Figure 1: Stages of AI development and clinical translation: Example of AI in mammography.
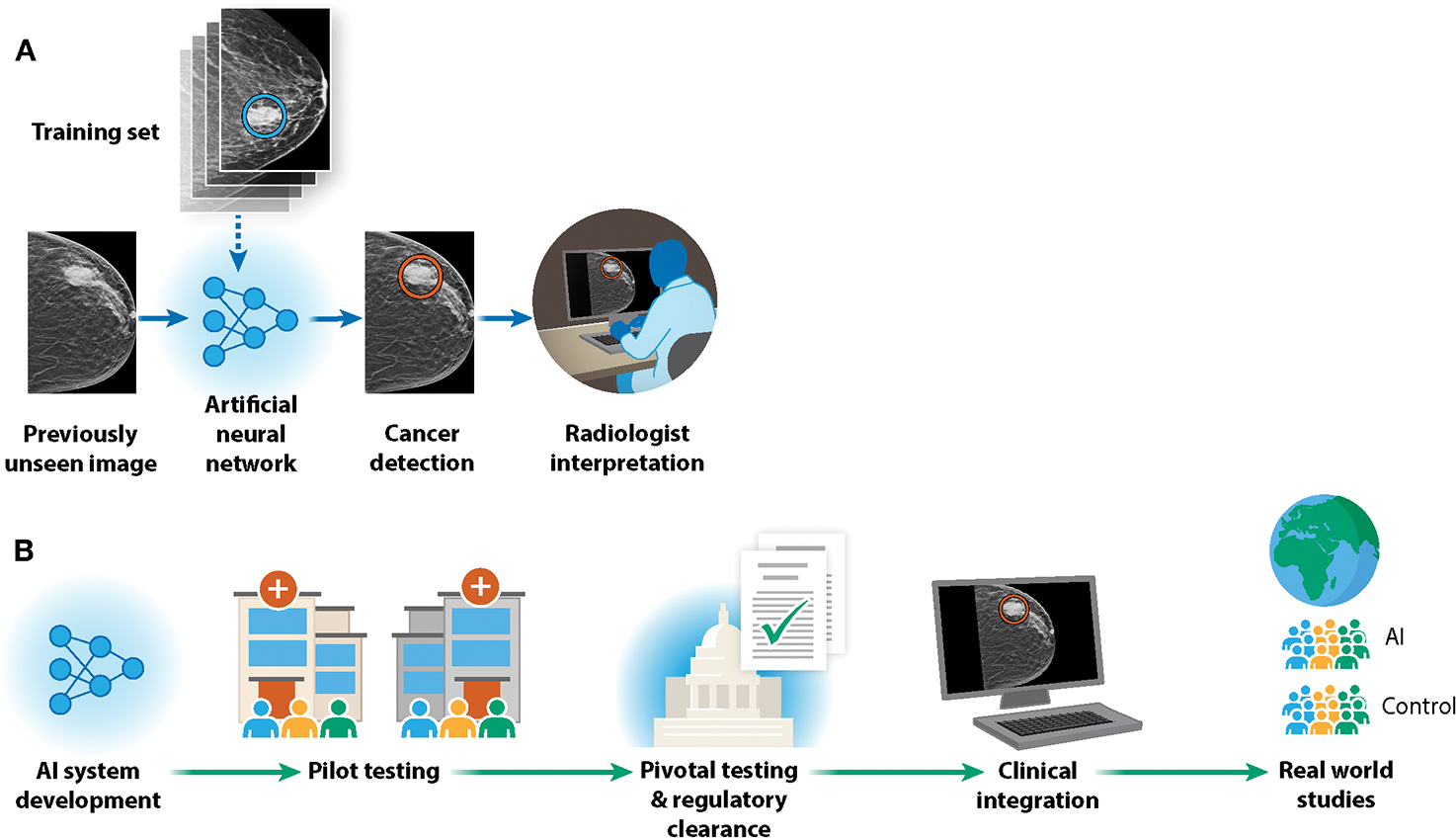
We consider the development of AI for breast cancer detection in mammography as a lens to understand the broader landscape of AI in oncology. A) An artificial neural network is trained on thousands of labeled mammography images. The performance of the network in detecting breast cancer is iteratively improved, and then the network can be used to make predictions on previously unseen images. These predictions can then be used to assist radiologists in their workflow. B) AI in mammography has transitioned from research on retrospective samples to regulatory clearance, clinical integration, and real-world clinical evaluation.
While there are many application-specific considerations and the trajectory is not always linear, the use of AI in mammography illustrates both the core steps and challenges for developing AI applications in oncology. In addition to cancer detection applications like mammography, AI is being heavily applied all along the cancer care continuum using different types of data (Figure 2). Below, we provide a structured summary of these applications categorized along a clinical trajectory divided into three stages: detection, diagnosis, and treatment, with subdivisions by cancer type. We then summarize overarching challenges, such as data curation, fairness, and regulation, along with efforts currently underway to address these challenges. We conclude with forward looking principles for guiding the development and deployment of clinically-effective, equitable AI in oncology.
Figure 2: Overview of AI in Oncology, with specific examples highlighted.
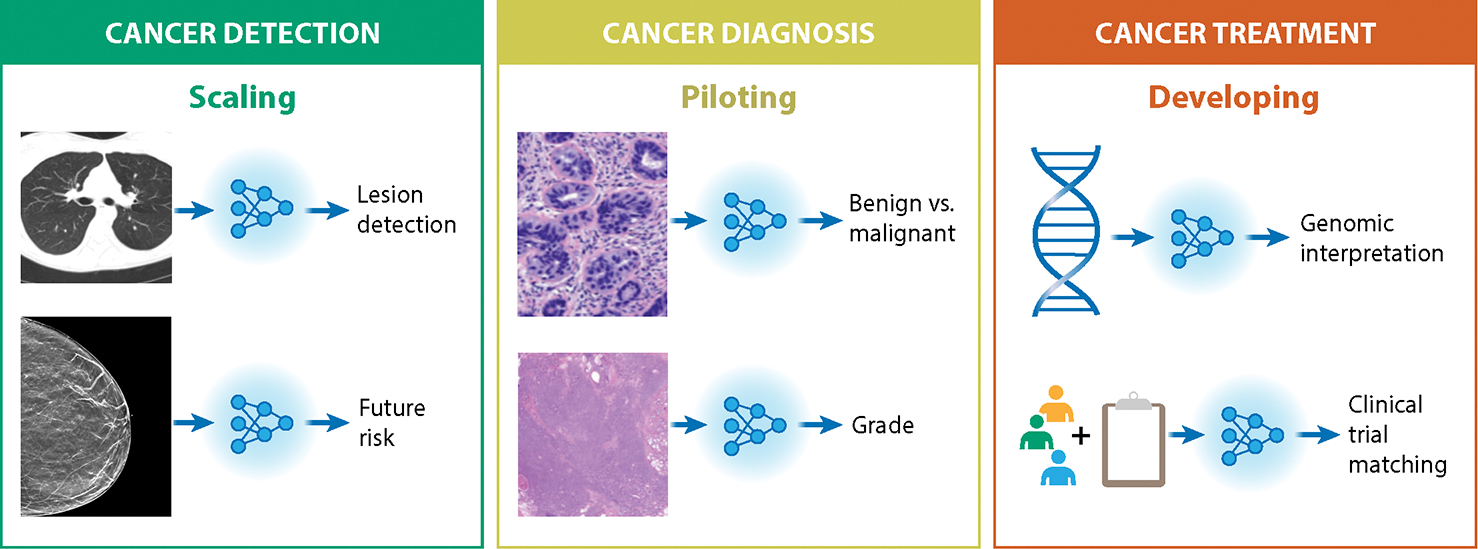
AI is being applied across the patient care trajectory, where this review groups applications into three main categories across this trajectory. Detection applications tend to currently have the highest level of clinical maturity, where several applications have regulatory clearances and published clinical trials, which we denote as “Scaling”. Diagnosis applications tend to be less mature, but regulatory clearances exist and validation studies are underway (“Piloting”). Prognosis and treatment applications are generally furthest from maturity with much emerging research (“Developing”). The review highlights AI applications in each of the clinical categories, with a specific focus on Breast, Prostate, Lung, and Colorectal cancers.
Cancer Detection
Large-scale data generated through current and expanding screening programs present a unique opportunity for the development and deployment of AI applications. Screening focuses on early cancer detection, where, especially in the context of breast and lung cancer, radiology-based medical imaging techniques are used. Outside of screening, imaging techniques are still often a core component of initial cancer detection, either in symptomatic patients or via incidental detection. Although not yet uniformly adopted across imaging providers or cancer hospitals, there are multiple FDA-cleared and/or EU-certified commercial devices in the cancer detection domain, and multiple reported clinical trials.
Breast Cancer:
As described above, the use of AI for breast cancer detection in X-ray mammography is one of the most active areas of development and clinical translation. An early catalyst in this space was the Digital Mammography DREAM Challenge, an open AI competition where groups were able to benchmark and compare different AI approaches8. A later study led by Google in 2020 garnered much attention in reporting evidence of expert-level breast cancer detection by an AI system9. A subsequent study presented an AI approach that encompasses both 2D and 3D mammography, with evidence of detecting breast cancer 1–2 years earlier than standard radiographic review for some cases10. There are now several FDA-cleared AI products with an intended use of aiding radiologists in detecting breast cancer from mammograms (K220105, K211541, K200905 RRID:SCR_012945). FDA-cleared AI algorithms have also been developed to aid in the interpretation of magnetic resonance imaging (MRI) (DEN1700, RRID:SCR_012945) and breast ultrasound exams (K190442, K210670, P150043 RRID:SCR_012945). Prospective studies are also underway to evaluate such products in clinical care. This includes the Mammography Screening with Artificial Intelligence (MASAI) clinical trial in Sweden, which recently reported that an AI-assisted workflow led to a 44% reduction in radiologist workload with comparable clinical performance metrics11. Several studies have also examined the use of AI to predict future breast cancer risk. For instance, the Mirai system developed by Yala et al.12 is designed to predict 5-year cancer risk directly from mammograms and has been retrospectively evaluated across multiple hospitals. These AI risk prediction algorithms have been shown to outperform traditional risk models13,14, and could be used to identify women who would benefit from supplemental screening. For example, the ScreenTrustMRI clinical trial will be using AI risk prediction models to select women for supplemental MRIs, and is expected to report results in 2025 (NCT04832594, RRID:SCR_002309).
Colorectal Cancer:
In the context of colorectal cancer screening, deep learning has been applied extensively to colonoscopy images and video. For example, Zhou et al.15 have reported on CRCNet, a deep learning model for detecting the presence of colorectal cancer within endoscopy images, with reported high performance across three independent data sets. Multiple companies have received FDA clearance or EU certification for computer-aided detection (CADe) systems in identifying polyps from colonoscopy images (K211951, K223473 RRID:SCR_012945), and multiple randomized controlled trials have also reported promising results16–22. Several of the clinical studies have reported an increased Adenoma Detection Rate (ADR) with AI, a central criteria in evaluating the effectiveness of colonoscopy procedures. However, most of these same studies also found an increased detection rate of diminutive adenomas (defined as polyps ≤5 mm in size), which are very rarely malignant23,24. Future research is therefore needed to determine whether these AI tools result in long term patient benefit, a core challenge across AI applications. To that end, a longer term, prospective clinical trial is now seeking to assess the connection between colonoscopy CADe systems and incidence and mortality from colorectal cancer after a 10 year follow-up period25.
Lung Cancer:
Since 2013, the US Preventive Services Task Force has recommended low-dose computed tomography (LDCT) to screen for lung cancer in high risk populations26. These recommendations are based on multiple studies27,28 including the National Lung Cancer Screening Trial (NLST), which has become a key public data resource for machine learning research. Multiple groups have now leveraged NLST imaging data to develop lung cancer detection models. For example, Ardila et al.29 developed an approach to localize lung nodules and predict their likelihood of malignancy, where this model is now licensed by the AI company Aidence30. In addition to the use of computed tomography (CT) exams, there have been efforts to detect lung nodules on chest x-rays using deep learning. For example, Nam et al.31 have published promising results from a single center, open-label randomized control trial that utilized an AI based system for chest x-rays from the company Lunit, and reported that the AI arm had an improved detection rate of actionable nodules compared to the non-AI arm. Beyond detecting the current presence of lung cancer, Mikhael et al.32 have recently reported on an AI model that predicts 6-year lung cancer risk. The model, referred to as Sybil, was trained on NLST data and subsequently validated using two independent data sets.
Prostate Cancer:
Current US and European guidelines recommend prostate cancer detection via MRI prior to biopsy33,34. There have been many AI efforts in this domain, including those aimed at directly classifying clinically significant prostate cancer and approaches to quantify clinically-relevant features such as segmenting and measuring the volume of the prostate gland35. This has resulted in numerous academic studies and regulatory-cleared commercial software devices36. For example, a commercial prototype from Siemens was shown to increase the diagnostic accuracy of clinically significant prostate cancer in a retrospective reader study involving seven radiologists37. More recently, Hamm et al.38 presented an explainable AI model that goes beyond classification to output AI predictions in terms of PI-RADS criteria39, a standard set of criteria used by radiologists to evaluate multiparametric prostate MRIs. Multiple open AI data challenges are also underway. For instance, the Prostate Imaging - Cancer AI (PI-CAI) challenge enables participants to train on a multi-center, multi-vendor data set of 1500 MRI prostate exams36. These challenges provide a transparent mechanism for evaluating and benchmarking AI models, paving the way for future algorithmic improvements.
Cancer Diagnosis
Once cancer is suspected, definitive diagnosis usually requires histopathologic examination. Beyond a determination of malignancy, histopathology techniques are used to classify the disease into standard clinical categories such as cancer subtype, grade, and stage. AI has been applied to each of these tasks, though these applications are generally further from clinical adoption with fewer FDA or EU clearances than the imaging-based detection applications described above. In addition to augmenting standard histochemical diagnostic workflows, several studies have demonstrated the potential of using AI to facilitate intra-operative diagnosis (e.g., during surgical resection) using label-free40,41 and cryosectioned42,43 tissue samples.
Breast Cancer:
AI has been applied extensively to the analysis of Hematoxylin and Eosin (H&E) images for aiding in breast cancer diagnosis. A core line of work in this domain has focused on the detection of metastatic lesions in sentinel lymph nodes. Treatment decisions in breast cancer frequently hinge on the detection of such lesions44, but pathology review remains labor intensive and subject to significant inter-reader variability (arXiv:1703.02442). Beyond the clinical significance, a key driving force in this application has been the Cancer Metastases in Lymph Nodes Challenge (CAMELYON), an open competition hosted in 2016 (CAMELYON16) and 2017 (CAMELYON17)45. This AI challenge became an early model for many AI projects in cancer, as it provided a publicly-available, expertly curated set of images and a transparent method for evaluating and comparing AI models. This evaluation includes comparison to a reference panel of pathologists working on the same data set with predefined time constraints designed to mimic routine pathology workflows, where many of the models in the challenge exhibited higher performance than the panel of pathologists. The CAMELYON16/17 data has subsequently become a standard benchmark for deep learning models in pathology. For example, Google developed LYmph Node Assistant (LYNA)46, and reported that pathologists who used the system were able to detect significantly more micrometastases with shorter review time. Multiple commercial platforms for breast cancer lymph node metastasis are also clinically-available or in development. For instance, Challa et al.47 evaluated the Visiopharm Integrator System (VIS) metastasis AI algorithm in pathology workflows in a retrospective study at Ohio State University with promising results. The AI company Paige has also recently been granted FDA Breakthrough Device Designation status for Paige Lymph Node, providing a path for accelerated FDA clearance48.
Beyond metastases detection, groups have reported on the use of deep learning for breast cancer diagnosis and subtyping from H&E slides49–51, and quantifying other clinically-relevant features such as mitotic cell counting52,53. For example, Sandbank et al.50 have recently presented a deep learning model for classifying invasive and non-invasive breast cancer subtypes and generating 51 predicted clinical and morphological features. The model was validated with external data sets from the Institut Curie, and then piloted as a second reader diagnostic system at Maccabi Healthcare Services in Israel. The model is now being further developed and Conformité Européenne (CE)-marked as Galen Breast54 by Ibex Medical Analytics. Towards intra-operative diagnosis of breast cancer during surgical resection, You et al.40 have presented an approach that combines intra-operative microscopy with a deep learning model to classify malignant vs. benign breast tissue. Beyond histological subtyping, CE-marked products exist for quantifying estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), and Ki67 status from digital pathology images55–57. As another example, Wang et al.58 have developed DeepGrade, a deep learning model that aims to improve on the Nottingham grading system. Much of this work has now been integrated into Stratipath59, a Swedish based AI start-up focused on digital pathology.
Prostate Cancer:
Multiple academic groups and companies have developed AI models for prostate cancer diagnosis and grading from H&E images. For instance, Raciti et al.60 reported on Paige Prostate Alpha, a commercial decision support system trained to detect prostate cancer from core needle biopsies. In a retrospective study, pathologists demonstrated improved sensitivity in diagnosing prostate cancer when using the AI platform, along with a reduction in review time and no statistically significant change in specificity. The platform was subsequently validated in an external cohort61 and eventually cleared by the FDA (DEN200080, RRID:SCR_012945). Along with using AI to aid in the diagnosis of prostate cancer from H&E images resulting from biopsies, AI is being used to guide biopsies themselves through prostate segmentation and quantitative feature extraction on MRI images (K193283, RRID:SCR_012945). Beyond diagnosis, multiple groups have reported on Gleason scoring predictions from prostate cancer H&E images62–64. For example, Nagpal et al.62 used a deep learning model to predict Gleason scores using H&E images from The Cancer Genome Atlas (TCGA), and reported significantly higher diagnostic accuracy compared to a cohort of 29 board-certified pathologists. There are now several CE-marked products for Gleason grading55,65,66. More recently, Kartasalo et al.67 have developed an AI model using H&E images to detect perineural invasion – a key prognostic marker associated with poor outcomes in prostate cancer.
Lung and Colorectal Cancers:
Significant efforts have also been made in using AI for histopathological diagnosis in lung and colorectal cancers, though FDA-cleared devices for these tasks do not exist at the time of writing. However, there are several CE-marked devices for treatment-related biomarker quantification for lung and colorectal cancers, which are described in the Treatment section below. In terms of histologic diagnosis, a 2018 study by Coudray et al.68 trained a deep learning model on TCGA H&E images to classify non-small cell lung cancer (NSCLC) subtypes. The paper was also notable as one the first studies to predict mutation status for key driver genes directly from H&E images. More recently, Lu et al.69 used a weakly supervised deep learning method – dubbed clustering-constrained-attention multiple-instance learning (CLAM) – to predict subtypes in renal cell carcinoma and NSCLC. A study by Ozyoruk et al.43 also demonstrated the potential of using AI to facilitate intra-operative subtyping of NSCLC by transforming cryosectioned tissue slides into the style of standard formalin-fixed and paraffin-embedded slides. Histopathological classification studies have been reported in colorectal cancer70,71. For instance, Korbar et al.72 developed a deep learning model to classify colorectal polyps into one of five common types based on H&E whole slide images.
Cancer Treatment
While cancer diagnosis and classification strongly inform patient treatment, AI algorithms are also being developed to directly improve treatment itself. These approaches include using AI to aid in treatment selection, design personalized treatments, and provide guidance during treatment delivery. We summarize advances in these applications below. We start by describing cancer-specific applications, where, in particular, there are increasing efforts of using digital pathology images to guide treatment selection by predicting patient prognosis73–75 and/or existing treatment biomarkers76–79 using AI. While there are some clinically-available devices for these tasks, they tend to be further from clinical maturity than cancer detection and diagnosis tasks. We then describe several specific treatment applications that are more fitting to be described in a pan-cancer setting: radiotherapy, molecular oncology, clinical trial matching, treatment decision support, and optimizing administrative workflows and patient engagement. The last three sections highlight the rapid advancement in the application of Large Language Models (LLMs) to healthcare. Models such as BERT80, GPT81, and ChatGPT (arXiv:2303.08774) are rapidly diffusing into industry, and healthcare specific models are also being developed82 (arXiv:2106.03598, arXiv:1904.05342). These increasingly large models have attracted attention for their improving ability to perform “zero-shot” and “in-context” learning, which can enable them to perform a myriad set of downstream tasks without necessarily being fine-tuned on those tasks directly, as described in the applications below.
Breast Cancer:
A recent study by Ogier du Terrail et al.83 presented a federated learning approach for predicting response to neoadjuvant chemotherapy in triple-negative breast cancer. The approach included weakly supervised training to predict response from H&E whole slide images, with subsequent application of interpretability approaches to identify features that correlate with higher response predictions, such as the presence of tumor-infiltrating lymphocytes (TILs) and necrosis. In another recent study, Amgad et al.84 explicitly enforce interpretability by using deep learning to extract human interpretable features, and subsequently use these features to generate a risk score for breast cancer prognosis. Other efforts have used AI on H&E images to directly predict TIL presence 85,86, a notable prognostic and predictive biomarker in breast cancer, and programmed cell death ligand 1 (PD-L1) expression status, a predictive biomarker for immunotherapies87.
Colorectal Cancer:
Prognostic AI models for colorectal cancer have been developed for various data modalities, including MRI88, histopathology89–91, and multiplex imaging92,93. In terms of histopathology, DoMore Diagnostics has a CE-marked product that predicts colorectal cancer prognosis from H&E slides94. There have additionally been several validation studies of histopathology-based prognostic models95,96, including a study demonstrating that a prognostic feature previously identified through AI analysis can be learned and used by pathologists95. Along with directly predicting prognosis, there have been several studies using AI to predict previously identified prognostic/predictive biomarkers. For instance, a line of work97–99 has focused on developing and validating AI for predicting microsatellite instability (MSI), a feature associated with clinical outcomes and a biomarker for immunotherapy100. This application is beginning to be commercialized, such as the CE-marked product101 developed by Owkin to predict MSI from H&E images.
Lung Cancer:
The relatively poor prognosis of NSCLC and the variable success of immunotherapies for NSCLC have motivated many efforts in using AI to predict prognosis, treatment response, and existing biomarkers for these patients. These efforts include histology102 and CT-based103 models for survival prediction, and the correlation of CT imaging features with histology risk factors104. Numerous studies have demonstrated the potential for using AI to predict immunotherapy biomarkers from H&E images of NSCLC patients, including tumor mutation burden (TMB)76, PD-L1 expression105, and TILs106,107. For instance, a study106 by AI startup Lunit used Lunit-Scope, a product currently available for research only, to categorize tumors into inflamed, immune-excluded, or immune-desert based on estimated TIL distributions in H&E images. These AI-classified phenotypes were then shown to differentially correlate with immune checkpoint inhibitor response. A study by Vanguri et al.108 demonstrated that a multimodal model that included CT images, digitized PD-L1 immunohistochemistry (IHC) slides, and molecular alteration data performed significantly better than unimodal models in predicting response to immunotherapy in NSCLC. Additionally, CE-marked products exist for quantifying PD-L1 expression from IHC slides109–111.
Prostate Cancer:
Efforts are also underway to use AI to improve prostate cancer prognostication. For instance, P-NET112 is a sparse deep learning model that processes molecular profiling data in a biologically-informed, pathway-driven fashion to predict prostate disease state such as metastases, where the resulting prediction scores were shown to independently correlate with prognosis. AI company ArteraAI has developed and tested a multimodal AI approach113,114 that uses histology images and clinical data to predict prostate cancer prognosis. This technology has been commercialized as the ArteraAI Prostate Test, which is now clinically available through a Clinical Laboratory Improvement Amendments (CLIA)-certified laboratory115.
Radiotherapy:
Radiotherapy is a critical therapeutic approach for multiple cancer types, and an intense area of AI research, development, and commercialization. This includes the use of AI at all steps of radiotherapy, including image segmentation, treatment planning, outcome prediction, quality assurance, and AI-guided radiosurgery116. Of these, a particularly clinically-advanced area is the use of deep learning to automatically contour Organs at Risk (OARs) within clinical images such as CT, positron emission tomography and computed tomography (PET-CT), and MRI. Multiple companies have now received FDA clearances and/or EU certification for these approaches (K201232, K223774, K211881 RRID:SCR_012945). A recent study by Radici et al.117 evaluated the FDA-cleared and CE-marked Limbus Contour (LC) AI platform for auto-contouring within a single European tertiary cancer hospital, and found that the technology significantly reduced the procedure time and interobserver variability. MD Anderson Cancer Center is also creating a comprehensive AI-based Radiation Planning Assistant for auto-contouring and radiotherapy planning, and will primarily target low- and middle-income countries, where AI can provide scalable assistance118. AI applications are also now advancing in the field of image-guided radiotherapy, adaptive radiotherapy, stereotactic radiosurgery119, and surgery in general, where auto-contouring is similarly helpful120. For a more comprehensive overview of recent advances, we refer readers to an excellent review by Huynh et al121.
Molecular Oncology:
Rich molecular data are increasingly critical for personalized cancer treatment across cancer types. This includes the use of AI in nearly every step of next generation sequencing (NGS) processing and interpretation, including mutation identification122,123, artifact filtering124 and driver gene identification125. The interpretation of germline and somatic mutations observed in cancer continues to be a challenge, and new AI tools are also emerging to automatically annotate and interpret variants. For example, a recent study investigated the use of deep learning for pathogenic germline variant detection in cancer126. The approach was evaluated in prostate cancer and melanoma, and outperformed standard genetic analysis approaches, providing a foundation for integration into clinical practice. “Dig” is a deep learning framework that identifies somatic mutations that are found to be under positive selection when compared to a genome-wide neutral mutation rate model127. The method outperformed multiple other methods for driver identification in accuracy and power and ran multiple orders of magnitude faster. Applied to 37 cancer types from the Pan-Cancer Analysis of Whole Genomes data set128, Dig identified several potential cryptic splice sites, recurrent variants in non-coding regions, as well as mutations in rarely mutated genes. Most recently, AlphaMissense, a deep learning model that builds on the protein structure prediction tool AlphaFold129, was introduced130. AlphaMissense predicts 32% of all possible missense variants in the human proteome to be pathogenic, but which subset is oncogenic in the setting of cancer remains to be evaluated. Additionally, CancerVar131 is a AI platform that uses a deep learning framework to predict oncogenicity of somatic variants using both functional and clinical features, and may provide a robust alternative to manually curated databases, such as CiVIC132 and OncoKB133.
An additional longstanding diagnostic challenge is cell-of-origin prediction, which is particularly relevant for cancers of unknown primary where accurate origin information could significantly impact treatment strategy. Multiple AI methods have been developed to perform these predictions. This includes genomics-based algorithms, where targeted next generation sequencing algorithms have been rigorously evaluated and are currently being deployed and assessed for clinical utility134,135. Transcriptomic-based algorithms have also been developed136,137, although these approaches will require expansion of clinical transcriptome profiling for assessments of generalizability. Additional approaches that integrate multiple molecular inputs for refined cell of origin prediction may further enhance performance. A study by Lu et al.138 has also demonstrated the potential of using AI to predict tumor origin from routine histology slides.
Deep learning methods are also having an impact on emerging novel therapeutic approaches involving molecular oncology. For example, personalized cancer vaccines are being tested in a wide variety of clinical indications, and recent trial data indicates their potential role in combination with existing immune checkpoint blockade139. Increasingly, detection of cancer specific neoantigens and related immune microenvironmental cell types is being informed by deep learning strategies. These include neoantigen discovery methods, e.g. NetMHCPan140 and HLAthena141, as well as associated T-cell receptor (TCR) discovery methods, e.g. DeepTCR142 and pMTnet143. Ultimately, assessments of these approaches will be performed in clinical trial settings that may guide the ongoing use of these immunogenomics and deep learning algorithms in clinical settings.
Clinical trial matching and automated pre-screening:
Clinical trials are the primary mechanism for evaluating and advancing new cancer therapeutics. However, only 6% of cancer patients enroll in clinical trials144 and there continue to be race-, sex- and age-based disparities within clinical trials145,146. The clinical trial system also suffers from overall inefficiencies, where 20% of trials are prematurely terminated, many due to insufficient accrual of patients147. Properly deployed, AI could increase efficiencies and patient diversity within the clinical trial system and assist physicians in matching patients to clinical trials. From a computer science perspective, clinical trial matching is usually framed as a natural language processing (NLP) challenge, where patient medical records — usually only available as unstructured clinical notes — are matched against inclusion and exclusion clinical trial criteria148. Multiple non-deep learning clinical trial matching applications now exist. A 2022 meta-analysis reported on ten published studies across multiple commercial and academic platforms, and found that most applications offered comparable, if not superior performance to manual screening, with dramatic improvements in efficiency149. The advent of LLMs now offers a renewed opportunity to improve NLP trial matching. For example, multiple groups have now reported on the use of LLMs to effectively extract structured data from clinical notes150 and to match patients to clinical trials (arXiv:2303.16756, arXiv:2306.02077, arXiv:2304.07396, arXiv:2308.02180). All of these efforts will benefit from the development of reference data sets, ideally derived from multiple hospitals, and prospective studies evaluating clinical integration across diverse hospital settings.
Treatment decision support:
The rapid advancement of LLMs has sparked interest in their potential for treatment decision support in oncology. Several recent studies have explored the feasibility of using LLMs to assist with tasks such as identifying personalized treatment options for patients with complex cancer cases. For instance, a recent study evaluated the performance of four LLMs in suggesting treatment options for fictional patients with advanced cancer. While the LLMs generated a wider range of treatment options compared to human experts, their recommendations often deviated from expert consensus. Despite these limitations, LLMs were able to correctly identify several important treatment strategies and even suggested some reasonable options that were not readily apparent to experts151. Similarly, a second study investigated the use of ChatGPT for providing first-line treatment recommendations for advanced solid tumors. Performance was generally promising, though there were cases in which the model arguably provided outdated information, such as investigational drug names152. Overall, these recent endeavors suggest that LLMs offer promising applications for clinical decision support in oncology, but further research is necessary to address limitations in accuracy, reliability, integration with existing clinical workflows, and regulatory requirements.
Optimizing administrative workflows and patient engagement:
LLMs also have the potential to significantly improve administrative workflows and patient engagement across the patient care trajectory, including treatment. Documentation and coding requirements in modern EMRs are extensive, imposing a significant time burden on clinicians153 and contributing to burnout154 that risks driving providers away from clinical practice and impairing access to care. Several companies have developed AI-assisted scribing technologies to generate documentation based on transcriptions of a clinical encounter155,156. LLMs could also be leveraged to unify disparate medical records into concise summaries157 of a patient’s case, allowing clinicians to focus on treatment planning together with patients. In terms of patient engagement, the complexity of modern oncology and the emotionally laden nature of dealing with cancer creates an environment in which patients’ understanding of their diseases may be limited. Many patients with advanced cancer, for example, do not fully grasp the palliative intent of their treatment158. Advances in LLMs that can generate patient-facing summaries of complex and multimodal information could facilitate patients’ understanding of their condition without significant additional burden on clinicians’ time. It would be critical for such summaries to be accurate and interpretable across diverse populations in order to prevent misunderstanding. A recent study has demonstrated promise towards these goals, where ChatGPT was shown to exhibit good performance at answering questions about cancer myths and misconceptions159.
Challenges and Opportunities
While there has been much progress in AI for oncology, significant challenges persist. These challenges include obtaining and sharing robust medical data, risks to patient privacy, and ensuring fairness in AI performance across all patients. As more AI models reach the clinic, there are also challenges in effectively integrating AI applications into clinical workflows and assessing the ultimate impact on patients and healthcare systems. Here, we summarize core challenges as they relate to AI in oncology (Figure 3), and describe multiple approaches already taken by the community to address these challenges.
Figure 3: Challenges and Opportunities.
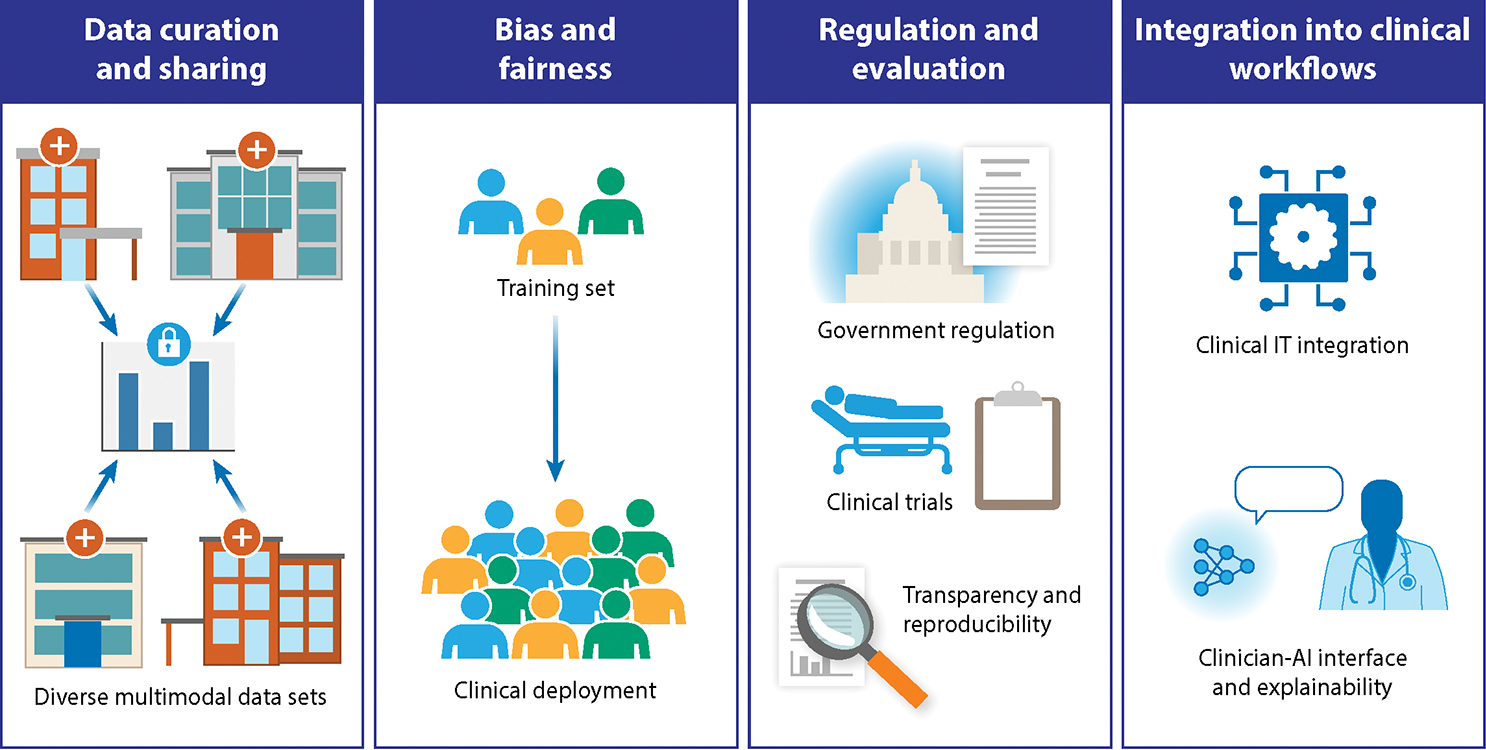
Like many other domains in medicine, AI in oncology faces significant challenges to effective development and clinical translation. These challenges pertain to curating and sharing data across institutions, guarding against biases from training through to deployment, ensuring appropriate regulation and evaluation, and integrating into clinical workflows across diverse clinical settings.
Data Curation and Sharing
Robust, generalizable AI models require vast amounts of training data, ideally sourced from multiple, independent sources160. This is particularly important in cancer, where each site is limited in patient size, patient diversity, and its own set of embedded clinical practices. It is even more acutely important for rare cancers, where individual sites may only treat a small number of such patients, and multi-site data integration is necessary to establish meaningful scientific or clinical insights161. Beyond training, AI models must also be tested across multiple independent sites, and evaluated for overall accuracy and potential bias162. Despite the need for multi-site training and evaluation, significant obstacles to cancer data sharing persist163. This includes protection of intellectual property, data interoperability challenges, and protection of patient privacy, as codified by multiple regulatory frameworks such as the Health Insurance Portability and Accountability Act (HIPAA), the Federal Policy for the Protection of Human Subjects, and the EU General Data Protection Regulation (GDPR)163,164.
To overcome these challenges in data sharing and privacy, the cancer community has explored three primary options. The first is centralized learning161, where a group organizes multi-party legal agreements and creates a shared set of security protocols. Once all sites agree, data is transferred to a central, secure enclave where AI development and evaluation can commence. This is the standard, default approach used in most commercial and academic AI efforts to date. The second option is deidentification and public release of data, usually via a consortium model. For example, the CAMELYON45 and International Brain Tumor Segmentation (BraTS) (arXiv:1811.02629) challenges have both made large-scale de-identified data sets available to machine learning teams, and the recently created Nightingale Open Science Initiative165 aims to make additional de-identified data sets available. The third option is federated learning (FL), where data remains private at each institution, but machine learning models are jointly updated and shared in a distributed fashion166,167. While federated learning introduces a number of logistical and algorithmic complexities, progress has been made in narrowing the gap in model performance between federated learning and centralized learning160, where FL has now been successfully applied to multiple cancer applications. For example, Ogier du Terrail et al.83 used a FL approach to develop an AI model for predicting histological response to neoadjuvant chemotherapy in patients with triple-negative breast cancer. A study by Pati et al.161 also used federated learning across 71 sites to predict tumor boundaries within multi-parametric MRI scans of Glioblastoma patients.
In addition to data curation across clinical sites, there is a need for curation across data modalities to enable multimodal AI approaches. The majority of AI approaches described above have been unimodal, such as making predictions from a medical image alone. However, oncologists rely on the totality of information regarding a patient — including radiology images, pathology images, lab values, genomic data, family history, and prior clinical notes — to provide optimal care. These different data modalities are likely to contain distinct and complementary information; thus, multimodal AI is an active area of research in oncology168,169. For instance, Chen et al.75 developed an approach to fuse genomic data and histopathology images to predict survival in a pan-cancer cohort. This work leverages the TCGA database, which has become a critical multimodal data set to advance AI efforts. Boehm et al.170 used TCGA and an internal cohort to fuse genomic data, histopathology imaging, and radiology imaging to predict survival in high-grade serous ovarian cancer. Additionally, ArteraAI has developed a clinically-available laboratory developed test to predict prostate cancer prognosis by integrating digital pathology images with clinical metadata115. The increasing focus on multimodal approaches in oncology parallels similar efforts in natural images and text, such as OpenAI’s GPT-4 model (arXiv:2303.08774). Such “foundation models” (arXiv:2108.07258) that are trained on vast quantities of data have shown promise in facilitating downstream tasks with less amounts of labeled data, a common regime in oncology applications. Beyond using general purpose foundation models like GPT-4, there are increasing efforts building domain-specific foundation models for healthcare171, including pathology172 (arXiv:2309.07778, arXiv:2307.12914). These efforts further emphasize the need for robust data sets, as the same data set and even model architecture can be used to support multiple clinical tasks.
Bias and Fairness
An overarching challenge in AI in medicine is ensuring fairness in performance and use across populations173. Conversely, AI offers opportunities to mitigate healthcare disparities that are already known to exist174,175. Both of these considerations certainly apply in oncology, where potential sources of bias span the full patient care and AI lifecycle continuums. As an illustrative application, breast cancer screening has been heavily studied in the context of both general healthcare equity and AI. In clinical practice, there are known disparities in access to newer imaging technologies and patient outcomes for underserved populations, such as Black women176–178. From an AI perspective, there are examples of both bias perpetuation and potential mitigation. For instance, an ensemble of AI models trained on mammograms from a largely White population demonstrated lower performance when tested on a more diverse population179. Conversely, there are efforts to generate more diverse public data sets, such as EMBED180, and instances where AI performance has generalized well across different patient populations9,10,12. The diversity of public data sets is a challenge across other oncology applications, including popular genomics data sets, such as TCGA181 and GENIE182. Fairness and diversity have especially important considerations in such applications given that genetic ancestry may be causally related to some genetic traits and diseases178,183–186, as opposed to the social construct of race173. Altogether, there is growing awareness of the fairness of AI algorithms, including guidance from regulatory bodies, but there remains much room for improvement in reducing bias in AI and healthcare more broadly.
Regulation and Evaluation
Most of the described AI applications fall under the purview of regulatory agencies, depending on regional regulations and the intended use of the device. Neither the US nor the EU currently have AI-specific regulatory approval pathways for AI-based medical tools. Rather, both regulate AI healthcare applications as medical devices, and categorize such devices based on the level of risk posed to individual patients.
The US currently follows a centralized model of medical device regulation, overseen by the FDA. The FDA regulates AI applications under the umbrella of Software as a Medical Device (SaMD) with similar regulatory processes as other SaMD devices and non-AI algorithms187. The requirements for FDA clearance depend largely on the class and intended use of the device, with device classes ranging from I (lowest risk) to III (highest risk). The vast majority of AI devices thus far, including those in oncology, fall under Class II, for which the FDA 510(k) pathway applies and requires the device to be “substantially equivalent” to other tools already cleared by the FDA187. Importantly, given this classification, randomized controlled clinical trials are not typically required by the FDA, though this can depend on the device’s intended use. In contrast to US FDA’s single centralized model, the EU has two broad regulations that govern medical device safety and efficacy, including AI applications. These are the Medical Device Regulation (MDR), which governs devices implanted within the body, and the In Vitro Diagnostic Devices Regulation (IVDR), which governs devices that test specimens outside the human body. Within the EU, there is no one central agency that performs device assessments; rather, assessments are handled by accredited organizations, which are empowered to issue Conformité Européenne (CE) marks.
In the future, both the US and the EU are likely to evolve their regulatory frameworks to leverage the unique characteristics of AI and mitigate corresponding risks. In particular, the unique capacity of improving AI by continually learning from new data will likely require a shift from single product approvals to life-cycle change management and robust postmarket monitoring188. To this end, the US FDA is developing a pathway for a “Predetermined Change Control Plan” to enable AI updates in a device without requiring a resubmission189. Similarly, the EU AI Act will require device manufactures to document their strategy for modifying and subsequently testing and validating AI medical devices190. A particularly evolving and debated topic is the regulation of LLMs, and in which contexts and intended uses do they qualify as medical devices191,192.
One challenge in monitoring the progress of regulatory clearances is the lack of up-to-date, frequently-refreshing databases focused on AI devices and their specific characteristics. The FDA has released a list of AI-enabled devices193 on a yearly basis for the last few years, which is very valuable for the field, yet the rapid progress and value of aggregating more detailed characteristics creates a strong need for complementary solutions. To this end, multiple studies have also attempted to create snapshots of approved AI devices. For example, Luchini et al. 194 summarized 71 FDA-cleared devices in oncology (up to 2021), and found that radiology accounted for the majority of approved devices (54.9%), followed by pathology (19.7%). These algorithms are largely designed to assist clinicians in making diagnoses and/or quantify image-based properties (e.g., lesion size) but the clinician makes the final call. The same study also found that breast cancer was the most commonly cited cancer type (31%), most commonly associated with radiology, providing further evidence of the advancing maturity of AI in mammography. Muehlematter et al.195 surveyed all approved AI medical devices in both the US and the EU for the period 2015–2020, and also found that a majority of approved devices in both the US and the EU were radiology focused.
An aspect that is closely tied to regulation is the evaluation of devices to ensure safe and effective performance, where clinical trials remain the gold standard in assessing new clinical interventions. While clinical trials in AI oncology remain rare, we are now starting to see an uptick in such trials. A search of ClinicalTrials.gov of “cancer” and “artificial intelligence” at the time of writing finds 526 studies, 81 of which are classified as interventional and actively recruiting. These include trials involving AI-based chatbots, such as use in genetic counseling (NCT05562778, NCT04354675 RRID:SCR_002309). Multiple clinical trials have now also published results in the past two years, such as those described in the breast cancer and colorectal screening sections above.
A critical component of AI evaluation and development more generally is ensuring transparency and reproducibility, allowing independent research groups to review and build on prior scientific progress196. This is especially important for assessing potential biases in data sets and generalizability across different clinical settings. The EQUATOR (Enhancing the QUAlity and Transparency Of health Research) Network currently provides a growing set of reporting guidelines specific to AI in healthcare and oncology197. This includes MINIMAR (MINimum Information for Medical AI Reporting), which defines a minimal set of reporting elements covering study population and setting, patient demographics, model architecture, and model evaluation198. Additional guidelines include SPIRIT-AI (Standard Protocol Items: Recommendations for Interventional Trials - Artificial Intelligence), which provides an extended set of 15 items for AI focused protocol documents199, and CONSORT-AI (Consolidated Standards of Reporting of Trials - Artificial Intelligence), which provides an extended set of 14 items for reporting on AI focused clinical trials200. Finally, there is an increasing use of AI in analyzing real-world data (RWD), and the European Society for Medical Oncology Guidance for Reporting Oncology Real-World Evidence (ESMO-GROW) now provides publication guidelines regarding use of RWD in oncology, along with specific AI-focused reporting guidelines201.
Integrating into Clinical Workflows
Realizing the potential of AI in oncology requires effective integration into clinical workflows, which presents both logistical and scientific challenges202. AI applications require significant computational infrastructure (either on-premise or cloud-based) and highly skilled engineers – both of which are likely to extend already stretched healthcare information technology (IT) budgets. Beyond initial hardware considerations, data pipelines must also be developed to efficiently route data to AI systems and subsequently make the AI results available to clinicians. Such workflows require integration into clinical systems, which can be especially challenging for proprietary systems with limited customization. The precise form of the AI results requires careful consideration of the clinical end goal and the intended use of the device. To this end, explainability is an often cited challenge for “black box” AI models203,204. As such, explainable AI (XAI) is a highly active area of research focused on conveying how an AI model arrived at its prediction. Common approaches to XAI involve post-hoc explanations, such as generating saliency maps that highlight aspects of the input that most contribute to the model’s prediction204. There are ongoing efforts to provide more structured explainability, such as representing AI predictions in terms of human-interpretable concepts205–207 and language-based explanations208,209 (medRxiv 2023.06.07.23291119). However, the optimal form of explainability for clinical use is an important open question, with a recent report indicating that scope of explainability offered in FDA-cleared devices for medical imaging is often limited (medRxiv 2023.11.28.23299132).
Even with effective infrastructure and AI outputs, monetization and liability questions can hinder widespread adoption, where the vast majority of commercially-available AI devices are not currently reimbursed210. This lack of reimbursement also makes it challenging to assess the true extent of clinical integration, as traditional methods for measuring medical device use such as insurance claims data cannot be used except for a small number of applications211. Despite these challenges, notable efforts are underway to facilitate efficient clinical integration. In particular, there is a growing number of AI “orchestration” platforms that serve as an intermediary between clinical systems and AI algorithms. Rather than having each AI application interface separately with clinical systems, these platforms, such as those developed by Nvidia212 and CARPL213, serve as a single interface and orchestrate the execution of multiple AI algorithms based on rule-based logic.
Future Directions and Conclusions
The field of AI in oncology is developing at a rapid pace. This development encompasses both algorithmic advances and new clinical use cases. Over the next few years, we are likely to see a steady flow of new AI regulatory approvals and clinical trial results, including those going beyond the current prevalent use case of AI as an assistive detection/diagnostic aid. We are also likely to see multiple new LLM-based applications and those that utilize multimodal data.
Nonetheless, the future of AI oncology will not be driven by technology innovation alone. Rather, we see several fundamental requirements that will drive the future of AI in oncology (Figure 4). These requirements will need to be jointly assessed by multiple stakeholders, including research teams, clinicians, ethicists, hospital administrators, and IT teams as they consider the development and deployment of AI models in oncology. First, there is a strong need for objective assessment of AI impact on patient outcomes, defined in cancer-specific, clinically meaningful metrics. This will require a greater investment in real-world studies and ideally randomized clinical trials, with an emphasis on critically evaluating AI generalizability across patient populations and clinical sites. Second, the healthcare value of AI deployment needs to be rigorously assessed and combined with aligned payment structures. Healthcare value is defined as the health outcome achieved per dollar spent214, and is a critical metric for making informed healthcare investments. Critically, while AI will ideally improve healthcare outcomes, its biggest impact may ultimately be in improving resource allocation and access to quality care. Realizing these benefits may require structural changes and new clinical workflows. Relatedly, a third need is a clear, standardized process for AI clinical integration, which covers aspects ranging from technical standards to stakeholder involvement and training.
Figure 4: Important forward-looking considerations for AI in Oncology.
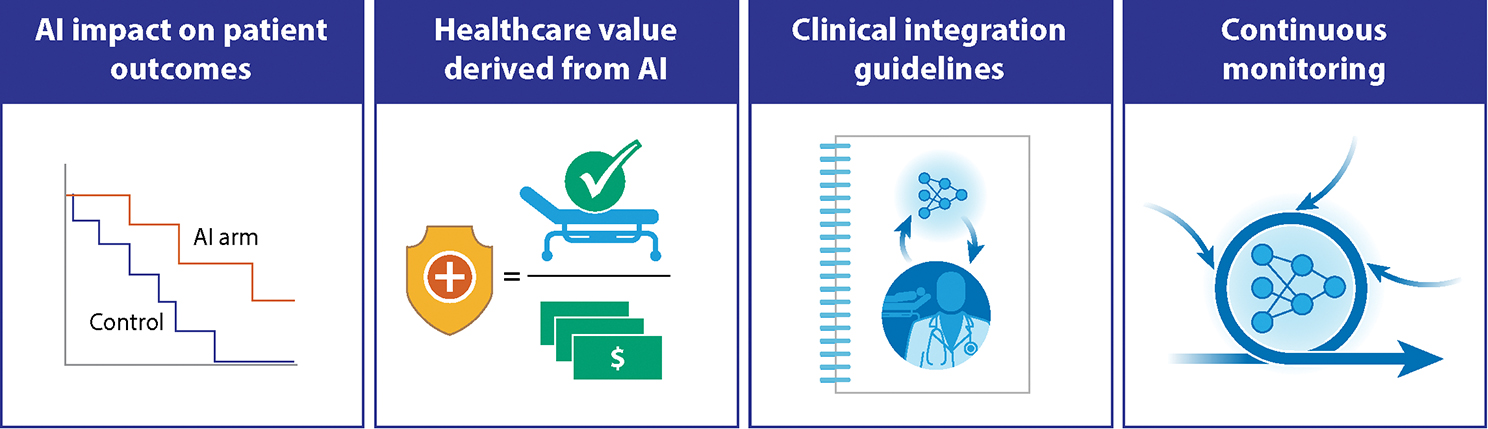
Effective clinical adoption of AI in oncology will require more than technology advances, and must include critical evaluation of AI impact on patient outcomes and healthcare value. Robust processes for integration across diverse clinical settings are also required, along with ongoing monitoring to ensure patient benefit and safety.
Finally, stakeholders must adopt objective processes for continuously monitoring AI impact and measuring patient safety. Arguably, the greatest strength of data-driven, deep learning approaches is the ability to continually improve over time. However, realizing these benefits requires well-defined regulatory processes and joint efforts to continuously monitor AI devices over the long-term. This necessity applies even for “frozen” AI devices that may experience data or population shifts over time. Altogether, these considerations will strongly shape the future of AI in oncology to help ensure effective, equitable, and sustainable use of AI for improving the care of cancer patients.
Statement of Significance.
Artificial Intelligence (AI) is increasingly being applied to all aspects of oncology, where several applications are maturing beyond research and development to direct clinical integration. This review summarizes the current state of the field through the lens of clinical translation along the clinical care continuum. Emerging areas are also highlighted, along with common challenges, evolving solutions, and potential future directions for the field.
Acknowledgements
The authors acknowledge financial support from the U.S. National Cancer Institute (P30CA008748 and P50CA272390), the Mark Foundation for Cancer Research, the Ellison Foundation, and the Nancy Lurie Marks Family Foundation.
Footnotes
Conflicts of Interest:
EVA reports the following potential conflicts of interest. Advisory/Consulting: Tango Therapeutics, Genome Medical, Genomic Life, Enara Bio, Manifold Bio, Monte Rosa, Novartis Institute for Biomedical Research, Riva Therapeutics, Serinus Bio; Research support: Novartis, BMS, Sanofi. Equity: Tango Therapeutics, Genome Medical, Genomic Life, Syapse, Enara Bio, Manifold Bio, Microsoft, Monte Rosa, Riva Therapeutics, Serinus Bio. Travel reimbursement: None. Patents: Institutional patents filed on chromatin mutations and immunotherapy response, and methods for clinical interpretation; intermittent legal consulting on patents for Foaley & Hoag. Editorial Boards: JCO Precision Oncology, Science Advances. All other authors declare no potential conflicts of interest.
References:
- 1.LeCun Y, Bengio Y & Hinton G Deep learning. Nature 521, 436–444 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Topol EJ High-performance medicine: the convergence of human and artificial intelligence. Nat. Med. 25, 44–56 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Niazi MKK, Parwani AV & Gurcan MN Digital pathology and artificial intelligence. Lancet Oncol. 20, e253–e261 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.AACR Project GENIE Consortium. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 7, 818–831 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garraway LA, Verweij J & Ballman KV Precision oncology: an overview. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 31, 1803–1805 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Bhinder B, Gilvary C, Madhukar NS & Elemento O Artificial Intelligence in Cancer Research and Precision Medicine. Cancer Discov. 11, 900–915 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Surveillance, Epidemiology, and End Results (SEER): Cancer Stat Facts: Common Cancer Sites. [cited 2024-02-21]. Available from: https://seer.cancer.gov/statfacts/html/common.html.
- 8.Schaffter T et al. Evaluation of Combined Artificial Intelligence and Radiologist Assessment to Interpret Screening Mammograms. JAMA Netw. Open 3, e200265 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKinney SM et al. International evaluation of an AI system for breast cancer screening. Nature 577, 89–94 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Lotter W et al. Robust breast cancer detection in mammography and digital breast tomosynthesis using an annotation-efficient deep learning approach. Nat. Med. 27, 244–249 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lång K et al. Artificial intelligence-supported screen reading versus standard double reading in the Mammography Screening with Artificial Intelligence trial (MASAI): a clinical safety analysis of a randomised, controlled, non-inferiority, single-blinded, screening accuracy study. Lancet Oncol. 24, 936–944 (2023). [DOI] [PubMed] [Google Scholar]
- 12.Yala A et al. Multi-Institutional Validation of a Mammography-Based Breast Cancer Risk Model. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 40, 1732–1740 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arasu VA et al. Comparison of Mammography AI Algorithms with a Clinical Risk Model for 5-year Breast Cancer Risk Prediction: An Observational Study. Radiology 307, e222733 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eriksson M, Czene K, Vachon C, Conant EF & Hall P Long-Term Performance of an Image-Based Short-Term Risk Model for Breast Cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 41, 2536–2545 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou D et al. Diagnostic evaluation of a deep learning model for optical diagnosis of colorectal cancer. Nat. Commun. 11, 2961 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang P et al. Effect of a deep-learning computer-aided detection system on adenoma detection during colonoscopy (CADe-DB trial): a double-blind randomised study. Lancet Gastroenterol. Hepatol. 5, 343–351 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Repici A et al. Efficacy of Real-Time Computer-Aided Detection of Colorectal Neoplasia in a Randomized Trial. Gastroenterology 159, 512–520.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Karsenti D et al. Effect of real-time computer-aided detection of colorectal adenoma in routine colonoscopy (COLO-GENIUS): a single-centre randomised controlled trial. Lancet Gastroenterol. Hepatol. S2468–1253(23)00104–8 (2023) doi: 10.1016/S2468-1253(23)00104-8. [DOI] [PubMed] [Google Scholar]
- 19.Ahmad A et al. Evaluation of a real-time computer-aided polyp detection system during screening colonoscopy: AI-DETECT study. Endoscopy 55, 313–319 (2023). [DOI] [PubMed] [Google Scholar]
- 20.Xu H et al. Artificial Intelligence-Assisted Colonoscopy for Colorectal Cancer Screening: A Multicenter Randomized Controlled Trial. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 21, 337–346.e3 (2023). [DOI] [PubMed] [Google Scholar]
- 21.Mangas-Sanjuan C et al. Role of Artificial Intelligence in Colonoscopy Detection of Advanced Neoplasias : A Randomized Trial. Ann. Intern. Med. 176, 1145–1152 (2023). [DOI] [PubMed] [Google Scholar]
- 22.Shaukat A et al. Computer-Aided Detection Improves Adenomas per Colonoscopy for Screening and Surveillance Colonoscopy: A Randomized Trial. Gastroenterology 163, 732–741 (2022). [DOI] [PubMed] [Google Scholar]
- 23.Ahmad OF Deep learning for colorectal polyp detection: time for clinical implementation? Lancet Gastroenterol. Hepatol. 5, 330–331 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Misawa M, Kudo S-E & Mori Y Computer-aided detection in real-world colonoscopy: enhancing detection or offering false hope? Lancet Gastroenterol. Hepatol S2468–1253(23)00166–8 (2023) doi: 10.1016/S2468-1253(23)00166-8. [DOI] [PubMed] [Google Scholar]
- 25.Mori Y, Kaminski MF, Hassan C & Bretthauer M Clinical trial designs for artificial intelligence in gastrointestinal endoscopy. Lancet Gastroenterol. Hepatol. 7, 785–786 (2022). [DOI] [PubMed] [Google Scholar]
- 26.US Preventive Services Task Force et al. Screening for Lung Cancer: US Preventive Services Task Force Recommendation Statement. JAMA 325, 962–970 (2021). [DOI] [PubMed] [Google Scholar]
- 27.National Lung Screening Trial Research Team et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 365, 395–409 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Field JK et al. Lung cancer mortality reduction by LDCT screening: UKLS randomised trial results and international meta-analysis. Lancet Reg. Health Eur. 10, 100179 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ardila D et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med. 25, 954–961 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Aidence and Google Health enter into collaboration to help improve lung cancer screening with AI. [cited 2024-02-21]. Available from: https://www.aidence.com/news/aidence-and-google-health/ (2022).
- 31.Nam JG et al. AI Improves Nodule Detection on Chest Radiographs in a Health Screening Population: A Randomized Controlled Trial. Radiology 307, e221894 (2023). [DOI] [PubMed] [Google Scholar]
- 32.Mikhael PG et al. Sybil: A Validated Deep Learning Model to Predict Future Lung Cancer Risk From a Single Low-Dose Chest Computed Tomography. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 41, 2191–2200 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.European Association of Urology: Guidelines on Prostate Cancer - Diagnostic Evaluation. [cited 2024-02-21]. Available from: https://uroweb.org/guidelines/prostate-cancer/chapter/diagnostic-evaluation.
- 34.American College of Radiology: Prostate MRI Model Policy. [cited 2024-02-21]. Available from: https://www.acr.org/-/media/ACR/Files/Advocacy/AIA/Prostate-MRI-Model-Policy72219.pdf.
- 35.Li H et al. Machine Learning in Prostate MRI for Prostate Cancer: Current Status and Future Opportunities. Diagn. Basel Switz. 12, 289 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sunoqrot MRS, Saha A, Hosseinzadeh M, Elschot M & Huisman H Artificial intelligence for prostate MRI: open datasets, available applications, and grand challenges. Eur. Radiol. Exp. 6, 35 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winkel DJ et al. A Novel Deep Learning Based Computer-Aided Diagnosis System Improves the Accuracy and Efficiency of Radiologists in Reading Biparametric Magnetic Resonance Images of the Prostate: Results of a Multireader, Multicase Study. Invest. Radiol. 56, 605 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Hamm CA et al. Interactive Explainable Deep Learning Model Informs Prostate Cancer Diagnosis at MRI. Radiology 307, e222276 (2023). [DOI] [PubMed] [Google Scholar]
- 39.Turkbey B et al. Prostate Imaging Reporting and Data System Version 2.1: 2019 Update of Prostate Imaging Reporting and Data System Version 2. Eur. Urol. 76, 340–351 (2019). [DOI] [PubMed] [Google Scholar]
- 40.You S et al. Real-time intraoperative diagnosis by deep neural network driven multiphoton virtual histology. NPJ Precis. Oncol. 3, 33 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hollon TC et al. Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat. Med. 26, 52–58 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nasrallah MP et al. Machine learning for cryosection pathology predicts the 2021 WHO classification of glioma. Med N. Y. N 4, 526–540.e4 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ozyoruk KB et al. A deep-learning model for transforming the style of tissue images from cryosectioned to formalin-fixed and paraffin-embedded. Nat. Biomed. Eng. 6, 1407–1419 (2022). [DOI] [PubMed] [Google Scholar]
- 44.Giuliano AE, Edge SB & Hortobagyi GN Eighth Edition of the AJCC Cancer Staging Manual: Breast Cancer. Ann. Surg. Oncol. 25, 1783–1785 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Ehteshami Bejnordi B et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer. JAMA 318, 2199–2210 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steiner DF et al. Impact of Deep Learning Assistance on the Histopathologic Review of Lymph Nodes for Metastatic Breast Cancer. Am. J. Surg. Pathol. 42, 1636–1646 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Challa B et al. Artificial Intelligence-Aided Diagnosis of Breast Cancer Lymph Node Metastasis on Histologic Slides in a Digital Workflow. Mod. Pathol. Off. J. U. S. Can. Acad. Pathol. Inc 36, 100216 (2023). [DOI] [PubMed] [Google Scholar]
- 48.U.S. FDA Grants Paige Breakthrough Device Designation for Cancer Detection in Breast Lymph Nodes. [cited 2024-02-21]. Available from: https://paige.ai/u-s-fda-grants-paige-breakthrough-device-designation-for-cancer-detection-in-breast-lymph-nodes/ (2023).
- 49.Sui D et al. A Pyramid Architecture-Based Deep Learning Framework for Breast Cancer Detection. BioMed Res. Int. 2021, 2567202 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sandbank J et al. Validation and real-world clinical application of an artificial intelligence algorithm for breast cancer detection in biopsies. NPJ Breast Cancer 8, 129 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Couture HD et al. Image analysis with deep learning to predict breast cancer grade, ER status, histologic subtype, and intrinsic subtype. NPJ Breast Cancer 4, 30 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Veta M, van Diest PJ, Jiwa M, Al-Janabi S & Pluim JPW Mitosis Counting in Breast Cancer: Object-Level Interobserver Agreement and Comparison to an Automatic Method. PloS One 11, e0161286 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Balkenhol MCA et al. Deep learning assisted mitotic counting for breast cancer. Lab. Investig. J. Tech. Methods Pathol. 99, 1596–1606 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Ibex Galen Breast [cited 2024-02-21]. Available from: https://ibex-ai.com/galen-breast/.
- 55.Aiforia Clinical Suite [cited 2024-02-21]. Available from: https://www.aiforia.com/aiforia-clinical-suite.
- 56.Mindpeak Solutions. [cited 2024-02-21]. Available from: https://mindpeak.ai//products/mindpeak-breast-er-pr-roi.
- 57.Panakeia Products. [cited 2024-02-21]. Available from: https://www.panakeia.ai/products.
- 58.Wang Y et al. Improved breast cancer histological grading using deep learning. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 33, 89–98 (2022). [DOI] [PubMed] [Google Scholar]
- 59.Stratipath Breast. [cited 2024-02-21]. Available from: https://www.stratipath.com/?page_id=4372.
- 60.Raciti P et al. Novel artificial intelligence system increases the detection of prostate cancer in whole slide images of core needle biopsies. Mod. Pathol. Off. J. U. S. Can. Acad. Pathol. Inc 33, 2058–2066 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.da Silva LM et al. Independent real-world application of a clinical-grade automated prostate cancer detection system. J. Pathol. 254, 147–158 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nagpal K et al. Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. NPJ Digit. Med. 2, 48 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bulten W et al. Automated deep-learning system for Gleason grading of prostate cancer using biopsies: a diagnostic study. Lancet Oncol. 21, 233–241 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Ström P et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: a population-based, diagnostic study. Lancet Oncol. 21, 222–232 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Ibex: Galen Prostate. [cited 2024-02-21]. Available from: https://ibex-ai.com/galen-prostate/.
- 66.Indica Labs: HALO Prostate AI [cited 2024-02-21]. Available from: https://indicalab.com/clinical-products/halo-prostate-ai/.
- 67.Kartasalo K et al. Detection of perineural invasion in prostate needle biopsies with deep neural networks. Virchows Arch. Int. J. Pathol. 481, 73–82 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coudray N et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 24, 1559–1567 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu MY et al. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. 5, 555–570 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shaban M et al. Context-Aware Convolutional Neural Network for Grading of Colorectal Cancer Histology Images. IEEE Trans. Med. Imaging 39, 2395–2405 (2020). [DOI] [PubMed] [Google Scholar]
- 71.Yu G et al. Accurate recognition of colorectal cancer with semi-supervised deep learning on pathological images. Nat. Commun. 12, 6311 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Korbar B et al. Deep Learning for Classification of Colorectal Polyps on Whole-slide Images. J. Pathol. Inform. 8, 30 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mobadersany P et al. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl. Acad. Sci. U. S. A. 115, E2970–E2979 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen RJ et al. Multimodal Co-Attention Transformer for Survival Prediction in Gigapixel Whole Slide Images. in 2021 IEEE/CVF International Conference on Computer Vision (ICCV) 3995–4005 (2021). doi:10.1109/ICCV48922.2021.00398. [Google Scholar]
- 75.Chen RJ et al. Pan-cancer integrative histology-genomic analysis via multimodal deep learning. Cancer Cell 40, 865–878.e6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jain MS & Massoud TF Predicting tumour mutational burden from histopathological images using multiscale deep learning. Nat. Mach. Intell. 2, 356–362 (2020). [Google Scholar]
- 77.Kather JN et al. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat. Cancer 1, 789–799 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tsai P-C et al. Histopathology images predict multi-omics aberrations and prognoses in colorectal cancer patients. Nat. Commun. 14, 2102 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fu Y et al. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nat. Cancer 1, 800–810 (2020). [DOI] [PubMed] [Google Scholar]
- 80.Devlin J, Chang M-W, Lee K & Toutanova K BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. in Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers) (eds. Burstein J, Doran C & Solorio T) 4171–4186 (Association for Computational Linguistics, Minneapolis, Minnesota, 2019). doi:10.18653/v1/N19–1423. [Google Scholar]
- 81.Brown T et al. Language Models are Few-Shot Learners. in Advances in Neural Information Processing Systems vol. 33 1877–1901 (Curran Associates, Inc., 2020). [Google Scholar]
- 82.Singhal K et al. Large language models encode clinical knowledge. Nature 620, 172–180 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ogier du Terrail J et al. Federated learning for predicting histological response to neoadjuvant chemotherapy in triple-negative breast cancer. Nat. Med. 29, 135–146 (2023). [DOI] [PubMed] [Google Scholar]
- 84.Amgad M et al. A population-level digital histologic biomarker for enhanced prognosis of invasive breast cancer. Nat. Med. 30, 85–97 (2024). [DOI] [PubMed] [Google Scholar]
- 85.Saltz J et al. Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images. Cell Rep. 23, 181–193.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Binder A et al. Morphological and molecular breast cancer profiling through explainable machine learning. Nat. Mach. Intell. 3, 355–366 (2021). [Google Scholar]
- 87.Shamai G et al. Deep learning-based image analysis predicts PD-L1 status from H&E-stained histopathology images in breast cancer. Nat. Commun. 13, 6753 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jiang X et al. An MRI Deep Learning Model Predicts Outcome in Rectal Cancer. Radiology 307, e222223 (2023). [DOI] [PubMed] [Google Scholar]
- 89.Wulczyn E et al. Interpretable survival prediction for colorectal cancer using deep learning. Npj Digit. Med. 4, 1–13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kather JN et al. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PLOS Med. 16, e1002730 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bychkov D et al. Deep learning based tissue analysis predicts outcome in colorectal cancer. Sci. Rep. 8, 3395 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu Z et al. Graph deep learning for the characterization of tumour microenvironments from spatial protein profiles in tissue specimens. Nat. Biomed. Eng. 6, 1435–1448 (2022). [DOI] [PubMed] [Google Scholar]
- 93.Foersch S et al. Multistain deep learning for prediction of prognosis and therapy response in colorectal cancer. Nat. Med. 29, 430–439 (2023). [DOI] [PubMed] [Google Scholar]
- 94.DoMore Diagnostics: Histotype Px® Colorectal [cited 2024-02-21]. Available from: https://www.domorediagnostics.com/products.
- 95.L’Imperio V et al. Pathologist Validation of a Machine Learning–Derived Feature for Colon Cancer Risk Stratification. JAMA Netw. Open 6, e2254891 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Skrede O-J et al. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet Lond. Engl. 395, 350–360 (2020). [DOI] [PubMed] [Google Scholar]
- 97.Echle A et al. Artificial intelligence for detection of microsatellite instability in colorectal cancer—a multicentric analysis of a pre-screening tool for clinical application. ESMO Open 7, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Echle A et al. Clinical-Grade Detection of Microsatellite Instability in Colorectal Tumors by Deep Learning. Gastroenterology 159, 1406–1416.e11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kather JN et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 25, 1054–1056 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li K, Luo H, Huang L, Luo H & Zhu X Microsatellite instability: a review of what the oncologist should know. Cancer Cell Int. 20, 16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Owkin: MSIntuit CRC | AI Cancer Testing. [cited 2024-02-21]. Available from: https://owkin.com/diagnostics/msintuitcrc/.
- 102.Yu K-H et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat. Commun. 7, 12474 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Saad MB et al. Predicting benefit from immune checkpoint inhibitors in patients with non-small-cell lung cancer by CT-based ensemble deep learning: a retrospective study. Lancet Digit. Health 5, e404–e420 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nam JG et al. Histopathologic Basis for a Chest CT Deep Learning Survival Prediction Model in Patients with Lung Adenocarcinoma. Radiology 305, 441–451 (2022). [DOI] [PubMed] [Google Scholar]
- 105.Sha L et al. Multi-Field-of-View Deep Learning Model Predicts Nonsmall Cell Lung Cancer Programmed Death-Ligand 1 Status from Whole-Slide Hematoxylin and Eosin Images. J. Pathol. Inform. 10, 24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Park S et al. Artificial Intelligence-Powered Spatial Analysis of Tumor-Infiltrating Lymphocytes as Complementary Biomarker for Immune Checkpoint Inhibition in Non-Small-Cell Lung Cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 40, 1916–1928 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rakaee M et al. Association of Machine Learning-Based Assessment of Tumor-Infiltrating Lymphocytes on Standard Histologic Images With Outcomes of Immunotherapy in Patients With NSCLC. JAMA Oncol. 9, 51–60 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vanguri RS et al. Multimodal integration of radiology, pathology and genomics for prediction of response to PD-(L)1 blockade in patients with non-small cell lung cancer. Nat. Cancer 3, 1151–1164 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Aiforia achieves CE-IVD mark for clinical PD-L1 AI model in lung cancer diagnostics. [cited 2024-02-21]. Available from: https://www.aiforia.com/blog/ce-ivd-ai-model-pd-l1.
- 110.Mindpeak Solutions: Plug-and-play AI solution to detect and quantify PD-L1 (SP263) within predefined ROIs in non-small-cell lung carcinoma (NSCLC). [cited 2024-02-21]. Available from: https://mindpeak.ai//products/mindpeak-lung-nsclc-pd-l1-sp263-roi.
- 111.Lunit Scope PD-L1 [cited 2024-02-21]. Available from: https://www.lunit.io/en/products/pdl1.
- 112.Elmarakeby HA et al. Biologically informed deep neural network for prostate cancer discovery. Nature 598, 348–352 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Esteva A et al. Prostate cancer therapy personalization via multi-modal deep learning on randomized phase III clinical trials. NPJ Digit. Med. 5, 71 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Spratt DE et al. Artificial Intelligence Predictive Model for Hormone Therapy Use in Prostate Cancer. NEJM Evid. 2, EVIDoa2300023 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.ArteraAI Prostate Test [cited 2024-02-21]. Available from: https://artera.ai/arteraai-prostate-cancer-test.
- 116.Li G, Wu X & Ma X Artificial intelligence in radiotherapy. Semin. Cancer Biol. 86, 160–171 (2022). [DOI] [PubMed] [Google Scholar]
- 117.Radici L et al. Implementation of a Commercial Deep Learning-Based Auto Segmentation Software in Radiotherapy: Evaluation of Effectiveness and Impact on Workflow. Life Basel Switz. 12, 2088 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.MD Anderson Cancer Center: Radiation Planning Assistant. [cited 2024-02-21]. Available from: https://rpa.mdanderson.org/.
- 119.Kawamura M et al. Revolutionizing radiation therapy: the role of AI in clinical practice. J. Radiat. Res. (Tokyo) 65, 1–9 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mascagni P et al. Computer vision in surgery: from potential to clinical value. NPJ Digit. Med. 5, 163 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Huynh E et al. Artificial intelligence in radiation oncology. Nat. Rev. Clin. Oncol. 17, 771–781 (2020). [DOI] [PubMed] [Google Scholar]
- 122.Khazeeva G et al. DeNovoCNN: a deep learning approach to de novo variant calling in next generation sequencing data. Nucleic Acids Res. 50, e97 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sahraeian SME et al. Achieving robust somatic mutation detection with deep learning models derived from reference data sets of a cancer sample. Genome Biol. 23, 12 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jongbloed EM et al. Machine learning-based somatic variant calling in cell-free DNA of metastatic breast cancer patients using large NGS panels. Sci. Rep. 13, 10424 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Luo P, Ding Y, Lei X & Wu F-X deepDriver: Predicting Cancer Driver Genes Based on Somatic Mutations Using Deep Convolutional Neural Networks. Front. Genet. 10, 13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.AlDubayan SH et al. Detection of Pathogenic Variants With Germline Genetic Testing Using Deep Learning vs Standard Methods in Patients With Prostate Cancer and Melanoma. JAMA 324, 1957–1969 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sherman MA et al. Genome-wide mapping of somatic mutation rates uncovers drivers of cancer. Nat. Biotechnol. 40, 1634–1643 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jumper J et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cheng J et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science 381, eadg7492 (2023). [DOI] [PubMed] [Google Scholar]
- 131.Li Q et al. CancerVar: An artificial intelligence-empowered platform for clinical interpretation of somatic mutations in cancer. Sci. Adv. 8, eabj1624 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Griffith M et al. CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat. Genet. 49, 170–174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chakravarty D et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis. Oncol. 2017, PO.17.00011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Penson A et al. Development of Genome-Derived Tumor Type Prediction to Inform Clinical Cancer Care. JAMA Oncol. 6, 84–91 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Moon I et al. Machine learning for genetics-based classification and treatment response prediction in cancer of unknown primary. Nat. Med. 29, 2057–2067 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Grewal JK et al. Application of a Neural Network Whole Transcriptome-Based Pan-Cancer Method for Diagnosis of Primary and Metastatic Cancers. JAMA Netw. Open 2, e192597 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Divate M et al. Deep Learning-Based Pan-Cancer Classification Model Reveals Tissue-of-Origin Specific Gene Expression Signatures. Cancers 14, 1185 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lu MY et al. AI-based pathology predicts origins for cancers of unknown primary. Nature 594, 106–110 (2021). [DOI] [PubMed] [Google Scholar]
- 139.Khattak A et al. Abstract CT001: A personalized cancer vaccine, mRNA-4157, combined with pembrolizumab versus pembrolizumab in patients with resected high-risk melanoma: Efficacy and safety results from the randomized, open-label Phase 2 mRNA-4157-P201/Keynote-942 trial. Cancer Res. 83, CT001 (2023). [Google Scholar]
- 140.Jurtz V et al. NetMHCpan-4.0: Improved Peptide-MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data. J. Immunol. Baltim. Md 1950 199, 3360–3368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sarkizova S et al. A large peptidome dataset improves HLA class I epitope prediction across most of the human population. Nat. Biotechnol. 38, 199–209 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sidhom J-W, Larman HB, Pardoll DM & Baras AS DeepTCR is a deep learning framework for revealing sequence concepts within T-cell repertoires. Nat. Commun. 12, 1605 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lu T et al. Deep learning-based prediction of the T cell receptor-antigen binding specificity. Nat. Mach. Intell. 3, 864–875 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Unger JM & Fleury M Nationally representative estimates of the participation of cancer patients in clinical research studies according to the commission on cancer. J. Clin. Oncol. 39, 74–74 (2021). [Google Scholar]
- 145.Murthy VH, Krumholz HM & Gross CP Participation in cancer clinical trials: race-, sex-, and age-based disparities. JAMA 291, 2720–2726 (2004). [DOI] [PubMed] [Google Scholar]
- 146.Kwiatkowski K, Coe K, Bailar JC & Swanson GM Inclusion of minorities and women in cancer clinical trials, a decade later: Have we improved? Cancer 119, 2956–2963 (2013). [DOI] [PubMed] [Google Scholar]
- 147.Stensland KD et al. Adult cancer clinical trials that fail to complete: an epidemic? J. Natl. Cancer Inst. 106, dju229 (2014). [DOI] [PubMed] [Google Scholar]
- 148.von Itzstein MS et al. Application of Information Technology to Clinical Trial Evaluation and Enrollment: A Review. JAMA Oncol. 7, 1559–1566 (2021). [DOI] [PubMed] [Google Scholar]
- 149.Chow R et al. Use of artificial intelligence for cancer clinical trial enrollment: a systematic review and meta-analysis. J. Natl. Cancer Inst. 115, 365–374 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Agrawal M, Hegselmann S, Lang H, Kim Y & Sontag D Large language models are few-shot clinical information extractors. in Proceedings of the 2022 Conference on Empirical Methods in Natural Language Processing (eds. Goldberg Y, Kozareva Z & Zhang Y) 1998–2022 (Association for Computational Linguistics, Abu Dhabi, United Arab Emirates, 2022). doi:10.18653/v1/2022.emnlp-main.130. [Google Scholar]
- 151.Benary M et al. Leveraging Large Language Models for Decision Support in Personalized Oncology. JAMA Netw. Open 6, e2343689 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Schulte B Capacity of ChatGPT to Identify Guideline-Based Treatments for Advanced Solid Tumors. Cureus 15, e37938 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Arndt BG et al. Tethered to the EHR: Primary Care Physician Workload Assessment Using EHR Event Log Data and Time-Motion Observations. Ann. Fam. Med. 15, 419–426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Robertson SL, Robinson MD & Reid A Electronic Health Record Effects on Work-Life Balance and Burnout Within the I3 Population Collaborative. J. Grad. Med. Educ. 9, 479–484 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.DeepScribe - AI-Powered Medical Scribe. [cited 2024-02-21]. Available from: https://www.deepscribe.ai/.
- 156.Nuance Communications: Ambient Clinical Intelligence | Automatically Document Care. [cited 2024–02-21]. Available from: https://www.nuance.com/healthcare/ambient-clinical-intelligence.html. [Google Scholar]
- 157.Chuang Y-N, Tang R, Jiang X & Hu X SPeC: A Soft Prompt-Based Calibration on Performance Variability of Large Language Model in Clinical Notes Summarization. J. Biomed. Inform. 151, 104606 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Weeks JC et al. Patients’ expectations about effects of chemotherapy for advanced cancer. N. Engl. J. Med. 367, 1616–1625 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Johnson SB et al. Using ChatGPT to evaluate cancer myths and misconceptions: artificial intelligence and cancer information. JNCI Cancer Spectr. 7, pkad015 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sheller MJ et al. Federated learning in medicine: facilitating multi-institutional collaborations without sharing patient data. Sci. Rep. 10, 12598 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pati S et al. Federated learning enables big data for rare cancer boundary detection. Nat. Commun. 13, 7346 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Karargyris A et al. Federated benchmarking of medical artificial intelligence with MedPerf. Nat. Mach. Intell. 5, 799–810 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sweeney SM et al. Challenges to Using Big Data in Cancer. Cancer Res. 83, 1175–1182 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lawlor RT The impact of GDPR on data sharing for European cancer research. Lancet Oncol. 24, 6–8 (2023). [DOI] [PubMed] [Google Scholar]
- 165.Mullainathan S & Obermeyer Z Solving medicine’s data bottleneck: Nightingale Open Science. Nat. Med. 28, 897–899 (2022). [DOI] [PubMed] [Google Scholar]
- 166.Rieke N et al. The future of digital health with federated learning. NPJ Digit. Med. 3, 119 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Saldanha OL et al. Swarm learning for decentralized artificial intelligence in cancer histopathology. Nat. Med. 28, 1232–1239 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Boehm KM, Khosravi P, Vanguri R, Gao J & Shah SP Harnessing multimodal data integration to advance precision oncology. Nat. Rev. Cancer 22, 114–126 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lipkova J et al. Artificial intelligence for multimodal data integration in oncology. Cancer Cell 40, 1095–1110 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Boehm KM et al. Multimodal data integration using machine learning improves risk stratification of high-grade serous ovarian cancer. Nat. Cancer 3, 723–733 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Moor M et al. Foundation models for generalist medical artificial intelligence. Nature 616, 259–265 (2023). [DOI] [PubMed] [Google Scholar]
- 172.Huang Z, Bianchi F, Yuksekgonul M, Montine TJ & Zou J A visual–language foundation model for pathology image analysis using medical Twitter. Nat. Med. 29, 2307–2316 (2023). [DOI] [PubMed] [Google Scholar]
- 173.Chen RJ et al. Algorithmic fairness in artificial intelligence for medicine and healthcare. Nat. Biomed. Eng. 7, 719–742 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Chen IY, Joshi S & Ghassemi M Treating health disparities with artificial intelligence. Nat. Med. 26, 16–17 (2020). [DOI] [PubMed] [Google Scholar]
- 175.Badal K, Lee CM & Esserman LJ Guiding principles for the responsible development of artificial intelligence tools for healthcare. Commun. Med. 3, 47 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Christensen EW et al. Relationship between Race and Access to Newer Mammographic Technology in Women with Medicare Insurance. Radiology 306, e221153 (2023). [DOI] [PubMed] [Google Scholar]
- 177.Rauscher GH, Allgood KL, Whitman S & Conant E Disparities in screening mammography services by race/ethnicity and health insurance. J. Womens Health 2002 21, 154–160 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Martini R, Newman L & Davis M Breast cancer disparities in outcomes; unmasking biological determinants associated with racial and genetic diversity. Clin. Exp. Metastasis 39, 7–14 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hsu W et al. External Validation of an Ensemble Model for Automated Mammography Interpretation by Artificial Intelligence. JAMA Netw. Open 5, e2242343 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Jeong JJ et al. The EMory BrEast imaging Dataset (EMBED): A Racially Diverse, Granular Dataset of 3.4 Million Screening and Diagnostic Mammographic Images. Radiol. Artif. Intell. 5, e220047 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Spratt DE et al. Racial/Ethnic Disparities in Genomic Sequencing. JAMA Oncol. 2, 1070–1074 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Cheung ATM et al. Racial and ethnic disparities in a real-world precision oncology data registry. NPJ Precis. Oncol. 7, 7 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zhang G et al. Characterization of frequently mutated cancer genes in Chinese breast tumors: a comparison of Chinese and TCGA cohorts. Ann. Transl. Med. 7, 179 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mahal BA et al. Racial Differences in Genomic Profiling of Prostate Cancer. N. Engl. J. Med. 383, 1083–1085 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gusev A et al. Atlas of prostate cancer heritability in European and African-American men pinpoints tissue-specific regulation. Nat. Commun. 7, 10979 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Nassar AH et al. Ancestry-driven recalibration of tumor mutational burden and disparate clinical outcomes in response to immune checkpoint inhibitors. Cancer Cell 40, 1161–1172.e5 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Harvey HB & Gowda V How the FDA Regulates AI. Acad. Radiol. 27, 58–61 (2020). [DOI] [PubMed] [Google Scholar]
- 188.Gerke S, Babic B, Evgeniou T & Cohen IG The need for a system view to regulate artificial intelligence/machine learning-based software as medical device. NPJ Digit. Med. 3, 53 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.FDA: Marketing Submission Recommendations for a Predetermined Change Control Plan for Artificial Intelligence/Machine Learning (AI/ML) - Enabled Device Software Functions: Draft Guidance for Industry and Food and Drug Administration Staff. [cited 2024-02-21]. Available from: https://www.fda.gov/media/166704/download.
- 190.Vokinger KN & Gasser U Regulating AI in medicine in the United States and Europe. Nat. Mach. Intell. 3, 738–739 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Meskó B & Topol EJ The imperative for regulatory oversight of large language models (or generative AI) in healthcare. NPJ Digit. Med. 6, 120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Gilbert S, Harvey H, Melvin T, Vollebregt E & Wicks P Large language model AI chatbots require approval as medical devices. Nat. Med. 29, 2396–2398 (2023). [DOI] [PubMed] [Google Scholar]
- 193.FDA: Artificial Intelligence and Machine Learning (AI/ML)-Enabled Medical Devices. [cited 2024-02-21]. Available from: https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-aiml-enabled-medical-devices. [DOI] [PMC free article] [PubMed]
- 194.Luchini C, Pea A & Scarpa A Artificial intelligence in oncology: current applications and future perspectives. Br. J. Cancer 126, 4–9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Muehlematter UJ, Daniore P & Vokinger KN Approval of artificial intelligence and machine learning-based medical devices in the USA and Europe (2015–20): a comparative analysis. Lancet Digit. Health 3, e195–e203 (2021). [DOI] [PubMed] [Google Scholar]
- 196.Haibe-Kains B et al. Transparency and reproducibility in artificial intelligence. Nature 586, E14–E16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Simera I et al. Transparent and accurate reporting increases reliability, utility, and impact of your research: reporting guidelines and the EQUATOR Network. BMC Med. 8, 24 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hernandez-Boussard T, Bozkurt S, Ioannidis JPA & Shah NH MINIMAR (MINimum Information for Medical AI Reporting): Developing reporting standards for artificial intelligence in health care. J. Am. Med. Inform. Assoc. JAMIA 27, 2011–2015 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Cruz Rivera S et al. Guidelines for clinical trial protocols for interventions involving artificial intelligence: the SPIRIT-AI extension. Lancet Digit. Health 2, e549–e560 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Liu X et al. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. Nat. Med. 26, 1364–1374 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Castelo-Branco L et al. ESMO Guidance for Reporting Oncology real-World evidence (GROW). Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. S0923–7534(23)04018–8 (2023) doi: 10.1016/j.annonc.2023.10.001. [DOI] [PubMed] [Google Scholar]
- 202.Chua IS et al. Artificial intelligence in oncology: Path to implementation. Cancer Med. 10, 4138–4149 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Rudin C Stop Explaining Black Box Machine Learning Models for High Stakes Decisions and Use Interpretable Models Instead. Nat. Mach. Intell. 1, 206–215 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.van der Velden BHM, Kuijf HJ, Gilhuijs KGA & Viergever MA Explainable artificial intelligence (XAI) in deep learning-based medical image analysis. Med. Image Anal. 79, 102470 (2022). [DOI] [PubMed] [Google Scholar]
- 205.Kim B et al. Interpretability Beyond Feature Attribution: Quantitative Testing with Concept Activation Vectors (TCAV). in Proceedings of the 35th International Conference on Machine Learning (eds. Dy J & Krause A) vol. 80 2668–2677 (PMLR, 2018). [Google Scholar]
- 206.Koh PW et al. Concept Bottleneck Models [cited 2024-02-27]. Proceedings of the 37th International Conference on Machine Learning Available from: https://openreview.net/forum?id=nA5AZ8CEyow (2020). [Google Scholar]
- 207.Yuksekgonul M, Wang M & Zou J Post-hoc Concept Bottleneck Models [cited 2024-02-27]. The Eleventh International Conference on Learning Representations Available from: https://openreview.net/forum?id=jlAjNL8z5cs (2023). [Google Scholar]
- 208.Agarwal S, Semenov YR & Lotter W Representing visual classification as a linear combination of words. in Proceedings of the 3rd Machine Learning for Health Symposium (eds. Hegselmann S et al.) vol. 225 27–38 (PMLR, 2023). [Google Scholar]
- 209.Menon S & Vondrick C Visual Classification via Description from Large Language Models. in The Eleventh International Conference on Learning Representations (2023). [Google Scholar]
- 210.Parikh RB & Helmchen LA Paying for artificial intelligence in medicine. NPJ Digit. Med. 5, 63 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wu K et al. Characterizing the Clinical Adoption of Medical AI Devices through U.S. Insurance Claims. NEJM AI 1, AIoa2300030 (2023). [Google Scholar]
- 212.NVIDIA Clara Supercharges Next Gen Medical Devices. [cited 2024-02-21]. Available from: https://www.nvidia.com/en-in/clara/.
- 213.CARPL Platform. [cited 2024-02-21]. Available from: https://carpl.ai/.
- 214.Porter ME What is value in health care? N. Engl. J. Med. 363, 2477–2481 (2010). [DOI] [PubMed] [Google Scholar]


