Abstract
Enteropathogenic Escherichia coli expresses a type IV fimbria known as the bundle-forming pilus (BFP) that is required for autoaggregation and localized adherence (LA) to host cells. A cluster of 14 genes is sufficient to reconstitute BFP biogenesis in a laboratory strain of E. coli. We have undertaken a systematic mutagenesis of the individual genes to determine the effect of each mutation on BFP biogenesis and LA. Here we report the construction and analysis of nonpolar mutations in six genes of the bfp cluster, bfpG, bfpB, bfpC, bfpD, bfpP, and bfpH, as well as the further analysis of a previously described bfpA mutant strain that is unable to express bundlin, the pilin protein. We found that mutations in bfpB, which encodes an outer membrane protein; bfpD, which encodes a putative nucleotide-binding protein; and bfpG and bfpC, which do not have sequence homologues in other type IV pilus systems, do not affect prebundlin expression or processing but block both BFP biogenesis and LA. The mutation in bfpP, the prepilin peptidase gene, does not affect prebundlin expression but blocks signal sequence cleavage of prebundlin, BFP biogenesis, and LA. The mutation in bfpH, which is predicted to encode a lytic transglycosylase, has no effect on prebundlin expression, prebundlin processing, BFP biogenesis, or LA. For each mutant for which altered phenotypes were detected, complementation with a plasmid containing the corresponding wild-type allele restored the wild-type phenotypes. We also found that association of prebundlin or bundlin with sucrose density flotation gradient fractions containing both inner and outer membrane proteins does not require any accessory proteins. These studies indicate that many bfp gene products are required for biogenesis of functional type IV pili but that mutations in the individual genes do not lead to the identification of new phases of pilus assembly.
Enteropathogenic Escherichia coli (EPEC) is a leading cause of infantile diarrhea in developing countries (12). EPEC is recognized by two phenotypes that are apparent in vitro and in vivo, attaching and effacing and localized adherence (LA). The attaching and effacing phenotype is characterized by the destruction of microvilli and the formation of cup-like pedestals upon which the bacteria rest and is mediated by the products of genes present on a chromosomal pathogenicity island (15, 26). LA is the distinctive pattern of EPEC adherence to epithelial cells, in which the bacteria form tightly packed clusters (11, 37). LA is dependent upon the synthesis of a type IV fimbria, the bundle-forming pilus (BFP), the expression of which is dependent on genes found on a large plasmid (13, 19).
Type IV pili are expressed by a wide variety of pathogenic organisms. These fimbriae mediate adherence to epithelial cells and autoaggregation of bacteria, act as receptors for bacteriophages (8, 44), and mediate a type of surface translocation known as twitching motility (9). Although many organisms express type IV pili on their surfaces, very little is known about the biogenesis of these organelles. Type IV pili are usually composed of a single repeating pilin protein. With the exceptions of Neisseria gonorrhoeae, which has an adhesin protein located at the tip of the pilus (35), and E. coli strains that express the R64 thin pilus, which has a minor pilus component (48), type IV pili appear to be comprised solely of a single pilin protein. Pilin has a short leader sequence that is cleaved by a specific prepilin peptidase (43). Type IV pilus biogenesis systems also contain an outer membrane protein belonging to a family of proteins called secretins. These proteins assemble into rings composed of 12 to 14 monomers and are believed to form channels in the outer membrane through which pilin subunits pass (7). The functions of the other proteins involved in type IV pilus biogenesis are not known. The most extensively studied type IV pilus system is that of Pseudomonas aeruginosa. This system requires the protein products of more than 30 genes for formation of functional pili (4). Many of the genes that have been implicated in type IV pilus biogenesis in one system have homologues in other type IV systems, as well as in systems involved in protein secretion, DNA uptake, and filamentous phage assembly (21, 42). Some of these genes encode putative nucleotide-binding proteins and inner or outer membrane proteins. The precise functions of these genes in pilus biogenesis are not known.
LA by EPEC is dependent upon the presence of a 95-kb plasmid, termed the EPEC adherence factor (EAF) plasmid. A cluster of 14 genes from the EAF plasmid, when transformed along with an additional fragment of the EAF plasmid containing regulatory genes, is sufficient to confer both BFP biogenesis and LA on a laboratory strain of E. coli (39, 40). Thus, the BFP system is one of the few in which all the genes required for the biogenesis of a type IV pilus have been identified. The first gene in the bfp cluster is bfpA, which encodes prebundlin, the prepilin protein (13, 38). The bfpP gene encodes the prepilin peptidase which is believed to cleave prebundlin to its mature form, is homologous to prepilin peptidase genes in other type IV pilus systems, and is capable of complementing a P. aeruginosa prepilin peptidase mutant (51). The bfpB gene encodes an outer membrane lipoprotein (34) that belongs to the secretin family of outer membrane proteins including PilQ of the P. aeruginosa type IV pilus system. The bfpF gene encodes a putative nucleotide-binding protein which is homologous to the PilT protein of P. aeruginosa and is not required for BFP biogenesis or LA (5, 6). Although bfpF mutant strains autoaggregate, the aggregates are morphologically distinct from those of wild-type EPEC, and while the wild-type aggregates disperse over time, the bfpF mutant aggregates do not (6). Knutton et al. have shown that, when wild-type EPEC infects tissue culture cells in vitro, BFP undergoes a dramatic change in morphology, with the individual pilus filaments forming much longer and thicker bundles (23). BFP in a bfpF mutant strain does not undergo this transformation (23). In addition, volunteer studies have shown that the virulence of a bfpF mutant strain of EPEC is greatly attenuated compared to that of wild-type EPEC (6). Thus, while BfpF does not play a role in BFP biogenesis, it does play a role in BFP function. Additional genes of the bfp cluster are predicted to encode a putative nucleotide-binding protein, BfpD, which is homologous to PilB of P. aeruginosa (28); a transmembrane protein, BfpE, which is homologous to the PilC protein of P. aeruginosa (28); and a lytic transglycosylase, BfpH. The cluster also contains several genes that encode putative prepilin peptidase substrates (bfpI, bfpJ, and bfpK) (39, 40). The remaining genes of the cluster, bfpG, bfpC, bfpU, and bfpL, show no sequence homology to known genes (39, 40).
To date, mutations in genes involved in type IV pilus biogenesis in other organisms have resulted in nonpiliated bacteria with the accumulation of pilin protein within membrane fractions (1–3, 28), with the exception of a family of nucleotide-binding proteins related to and including the bfpF gene product, in which mutations result in hyperfimbriated bacteria (6, 47). In an effort to define the genes of the bfp cluster that are involved in BFP biogenesis or LA, we intend to create nonpolar mutations in each of the bfp genes. Here we report the mutation of six bfp genes, bfpG, bfpB, bfpC, bfpD, bfpP, and bfpH, and the effects of these mutations as well as that of a previously described bfpA mutation on BFP biogenesis, autoaggregation, and LA.
MATERIALS AND METHODS
Strains and plasmids.
Bacterial strains and plasmids used in these experiments are listed in Table 1. Strains were grown on Luria-Bertani (LB) agar or in LB broth at 37°C except where indicated and stored at −80°C in 50% LB broth–50% (vol/vol) glycerol stock. Antibiotics, when necessary, were added at the indicated concentrations: ampicillin, 200 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 20 μg/ml; and nalidixic acid, 50 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or features | Source or reference |
|---|---|---|
| Strains | ||
| E2348/69 | Prototype O127:H6 EPEC strain | 25 |
| UMD901 | E2348/69 bfpA C129S | 50 |
| UMD928 | E2348/69 bfpG::aphA3 | This study |
| UMD923 | E2348/69 bfpB::aphA3 | This study |
| UMD924 | E2348/69 bfpC::aphA3 | This study |
| UMD926 | E2348/69 bfpD::aphA3 | This study |
| UMD932 | E2348/69 bfpP::aphA3 | This study |
| UMD918 | E2348/69 bfpH::aphA3 | This study |
| DH5α | supE44 ΔlacU169(φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 36 |
| DH5αλpir | DH5α(λpir) | 27 |
| Plasmids | ||
| pBluescript | High-copy-number cloning vector | Stratagene |
| pMSD205 | bfpA gene cloned into PCR1000 under control of an isopropyl-β-d-thiogalactopyranoside-inducible promoter | 13 |
| pRPA100 | 1-kb BamHI-HindIII fragment of the bfp cluster containing bfpA cloned into pWKS30 | This study |
| pRPA101 | 3-kb BamHI-BalI fragment of the bfp cluster containing bfpA-bfpB cloned into pWKS30 | This study |
| pRPA103 | 4.1-kb BamHI-XbaI fragment of the bfp cluster containing bfpA-bfpC cloned into pWKS30 | This study |
| pRPA104 | 2.3-kb BamHI-HindIII fragment of the bfp cluster containing bfpA-bfpG cloned into pWKS30 | This study |
| pRPA106 | 6.5-kb BamHI-KpnI fragment of the bfp cluster containing bfpA-bfpD cloned into pWKS30 | This study |
| pRPA107 | 1.3-kb PstI-BamHI fragment of the bfp cluster containing bfpP cloned into pWKS30 | This study |
| pCVD442 | Positive-selection suicide vector | 14 |
| pRK2073 | pRK2 derivative suitable for mobilization of plasmids | 10 |
| pKDS302 | Entire bfp cluster cloned into pTRC99A under control of trc promoter | 40 |
| pMSD230 | bfpA cloned into pTRC99A under control of trc promoter | This study |
| pMSD233 | 5.3-kb BamHI fragment of the bfp cluster containing bfpA-bfpU cloned into pWKS30 | This study |
| pDN19PB | bfpP cloned into pDN19 | 51 |
| pDN19PBΔ | bfpP with a 93-bp deletion cloned into pDN19 | 51 |
| pUC18K | aphA3 gene cloned into pUC18, creating a nonpolar kanamycin cassette | 27 |
| pUC18K2 | pUC18K with 1 bp added after the ATG start codon | K. Jarvis and J. Kaper |
| pUC18K3 | pUC18K with 2 bp added after the ATG start codon | K. Jarvis and J. Kaper |
DNA cloning and mutant construction.
DNA restriction digestions, electrophoresis, and ligations were performed by standard procedures (36). Plasmids were introduced into strains by CaCl2 transformation or electroporation. Electroporation was carried out in 10% glycerol in a 0.1-cm cuvette, using an E. coli pulser (Bio-Rad, Hercules, Calif.) set at 1.8 kV.
Strain UMD901 contains a single missense mutation in bfpA that results in the substitution of serine for cysteine at position 129 and leads to an unstable product that is rapidly degraded (50). To complement the bfpA mutation in UMD901, a BamHI-PstI fragment of the bfp cluster containing bfpA with its native promoter (40) and ending within bfpG was subcloned into the low-copy-number vector pWKS30 (45) to make plasmid pRPA100. The construct was verified by restriction analysis and electroporated into UMD901, creating strain UMD901(pRPA100). As a negative control for complementation, pWKS30 was electroporated into UMD901, creating strain UMD901(pWKS30).
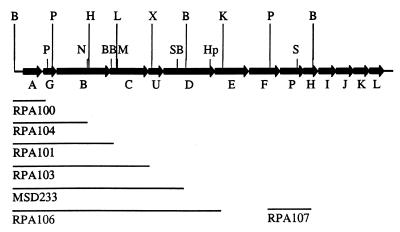
The remainder of the mutations described were constructed using a previously described nonpolar kanamycin cassette (27). The cassette is flanked by SmaI restriction sites which allow it to be inserted into any blunt-end restriction sites. The general mutation strategy was to subclone a portion of the bfp cluster containing the gene of interest into pBluescript II (Stratagene, La Jolla, Calif.) and to insert the appropriate kanamycin cassette into either a single restriction site or a deletion made by excising the fragment between two restriction sites. When required, either T4 polymerase or the Klenow fragment of DNA polymerase I was used to render overhanging ends blunt ended. Constructs modified this way were sequenced to ensure preservation of the reading frame. The mutated gene was then excised from pBluescript by cutting with SacI and SalI, which are on opposite ends of the pBluescript multiple cloning region, and cloned into the SacI and SalI sites of the positive-selection suicide vector pCVD442 (14). The resulting construct was then mobilized into wild-type EPEC by triparental conjugation, and allelic exchange was carried out as previously described (17). Potential mutant colonies were then screened by PCR. PCRs were performed on fresh bacterial colonies using Taq DNA polymerase (Gibco BRL, Gaithersburg, Md.) in 50-μl samples. The reactions were run for 30 cycles of denaturation (94°C for 1 min), annealing (45°C for 30 s), and extension (72°C for 1 min). Figure 1 shows the site in each gene where the kanamycin cassette was added and the fragments of the bfp cluster used to complement each mutation.
FIG. 1.
Location of bfp mutants and complementing fragments. Lines beneath the genes show the fragments of the cluster used to construct complementing and control plasmids. For each mutant except bfpA and bfpP mutants, a plasmid containing the fragment ending within the corresponding mutated gene served as the control plasmid and a plasmid containing the fragment ending within the gene immediately downstream of the mutated gene served as the complementing plasmid. For the bfpA and bfpP mutants, the vector served as the control plasmid and the vector containing the corresponding gene served as the complementing plasmid. The upper row of restriction sites shows the beginning and endpoint of each fragment. The lower row of restriction sites indicates where the kanamycin cassette was inserted into each gene. Restriction enzyme abbreviations: B, BamHI; BB, BstBI, H, HindIII; Hp, HpaI; K, KpnI; L, BalI; M, MfeI; N, NruI; P, PstI; S, SpeI; SB, SnaBI; X, XbaI.
To introduce a bfpG mutation into wild-type EPEC, a 2.3-kb BamHI-HindIII fragment of the bfp cluster was used. A 151-bp PstI deletion was made in bfpG, the PstI ends were blunt ended by the 3′→5′ exonuclease activity of T4 polymerase (New England Biolabs, Beverly, Mass.), and the kanamycin cassette was inserted. Primers Donne-2 (5′-CAA TGG GAA TAC CAC-3′) and Donne-60 (5′-GGG CGT ATT ATA TGG GAG GTA T-3′) were used to amplify the bfpG gene when screening potential mutant colonies. The bfpG mutant EPEC strain was named UMD928. To complement the bfpG mutation in UMD928, a BamHI-HindIII fragment of the bfp gene cluster beginning upstream of bfpA and ending within bfpB was cloned into pWKS30. This construct was called pRPA104 and was electroporated into UMD928, creating strain UMD928(pRPA104). As a negative control for complementation, pRPA100 was electroporated into UMD928, creating strain UMD928(pRPA100).
To introduce a bfpB mutation into wild-type EPEC, a 2.5-kb ScaI-XbaI fragment of the bfp cluster was used. The kanamycin cassette was inserted into a single NruI site in bfpB. Primers Donne-42 (5′-TAT TAA TAC ACT GAA TGA-3′) and Donne-61 (5′-CAT CAA CAC TTG TAT TGA CCT C-3′) were used to amplify the bfpB gene when screening potential mutant colonies. The resulting bfpB mutant strain was called UMD923. To complement the bfpB mutation in UMD923, a BamHI-BalI fragment of the bfp cluster beginning upstream of bfpA and ending within bfpC was cloned into the BamHI and EcoRV sites of pWKS30. The resulting construct was called pRPA101 and was electroporated into UMD923, creating strain UMD923(pRPA101). As a negative control for complementation, pRPA104 was electroporated into UMD923, creating strain UMD923(pRPA104).
To introduce a bfpC mutation into wild-type EPEC, the same ScaI-XbaI fragment was used as for bfpB. A 192-bp deletion was made in bfpC by cutting with BstBI and MfeI. The resulting ends were filled in by the 5′→3′ polymerase activity of the Klenow fragment of DNA polymerase I, and the kanamycin cassette was inserted. Primers Donne-42 and Donne-61 were used to amplify the bfpC gene when screening potential mutant colonies. The bfpC mutant EPEC strain was called UMD924. To complement the bfpC mutation in UMD924, a BamHI-XbaI fragment of the bfp cluster, beginning upstream of bfpA and ending within bfpU, was cloned into the BamHI and XbaI sites of pWKS30. The resulting construct, pRPA103, was electroporated into UMD924, creating strain UMD924(pRPA103). As a negative control for complementation, pRPA101 was transformed into UMD924, creating strain UMD924(pRPA101).
To introduce a bfpD mutation into wild-type EPEC, a 3.1-kb XbaI fragment was used. A 1,030-bp deletion was made in bfpD by cutting with SnaBI and HpaI, and the kanamycin cassette was inserted. Primers Donne-40 (5′-CTC GTT TTC GAC GTG AAT AGC AGT CTG-3′) and Donne-68 (5′-CAT CTG GTA CAG ATG TTA CGT G-3′) were used to amplify the bfpD gene when screening potential mutant colonies. The bfpD mutant strain was named UMD926. To complement the bfpD mutation in UMD926, a HindIII-KpnI fragment of the bfp cluster was cloned next to the BamHI-HindIII fragment in pRPA104 to make pRPA106, which begins upstream of bfpA and ends within bfpE. Plasmid pRPA106 was electroporated into UMD926, creating strain UMD926(pRPA106). As a negative control for complementation, a 5.3-kb BamHI fragment of the bfp cluster beginning with bfpA and ending in bfpD was cloned into pWKS30, creating pMSD233. This plasmid was electroporated into UMD926, creating strain UMD926(pMSD233).
To introduce a bfpP mutation into wild-type EPEC, a 3.3-kb XbaI-EcoRI fragment of the bfp cluster was used. The bfpP gene was cut with SpeI, the resulting ends were filled in with the Klenow fragment of DNA polymerase I, and the kanamycin cassette was inserted. Primers Donne-62 (5′-GCG AAG CTT TTA ATG ATA AAC TAA ACA TAT-3′) and Donne-63 (5′-CGC GGA TCC ATG CAA GAA AGT ATA TTT CTA-3′) were used to amplify bfpP to screen potential mutant colonies. The resulting bfpP mutant strain was called UMD932. To complement the bfpP mutation in UMD932, a PstI-BamHI fragment of the bfp cluster containing bfpP was cloned into pWKS30, creating pRPA107. This plasmid was electroporated into UMD932, creating strain UMD932(pRPA107). As a negative control for complementation, pWKS30 was electroporated into UMD932, creating strain UMD932(pWKS30).
To introduce a bfpH mutation into wild-type EPEC, the same XbaI-EcoRI fragment of the bfp cluster was used as for bfpP. The bfpH gene was cut with BamHI, the resulting ends were filled in with the Klenow fragment of DNA polymerase I, and the kanamycin cassette was inserted. Primers Donne-108 (5′-GGC CGG ATC CTA TGG GAC CAG CTC-3′) and Donne-109 (5′-GCC GAA GCT TCT TAG ATA TTC CTT TGA-3′) were used to amplify bfpH to screen potential mutant colonies. The resulting bfpH mutant strain was called UMD918.
Tissue culture and adherence assay.
Adherence assays were performed with HEp-2 cells (ATCC CCL 23) in eight-well chamber slides (Becton Dickinson, Franklin Lakes, N.J.) as previously described (16).
Autoaggregation assay.
Overnight bacterial cultures were diluted 1:100 in 20 ml of Dulbecco's modified Eagle's medium (DMEM)–F-12 with 15 mM HEPES and 2 mM glutamine and grown for at 37°C with shaking at 250 rpm until bacterial aggregates were visible in strains expressing BFP. A 1-ml aliquot of each culture was removed, and the A600 of each culture was measured. The samples were then vortexed for 30 s, and the A600 was measured again. The percent increase in A600 after vortexing was recorded as a quantitative aggregation index. Mean values were compared using Student's t test for paired samples (two tailed).
Immunoblotting.
Overnight bacterial cultures were diluted 1:100 in 20 ml of DMEM and grown for 5 h at 37°C with shaking at 250 rpm. Samples were centrifuged (3,400 × g, 4°C, 5 min) and resuspended in 350 μl of water. Fifty microliters of each sample was saved for determination of the protein concentration by the bicinchoninic acid protein assay (Pierce, Rockford, Ill.) in a multititer plate reader with bovine serum albumin as a standard. One hundred microliters of 4× sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) loading buffer (0.25 M Tris-HCl [pH 6.8], 8% SDS, 40% [vol/vol] glycerol, 0.008% bromphenol blue, 20% 2-mercaptoethanol) was added to the remaining 300 μl of each sample. Samples were denatured by boiling for 10 min in the SDS-PAGE buffer, and 5 μg of total protein per lane was loaded on 15% polyacrylamide gels and separated by electrophoresis. Samples were transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore, Bedford, Mass.). The blots were blocked with phosphate-buffered saline (PBS) containing 0.5% (vol/vol) Tween 20 and 5% milk, reacted with mouse antibundlin monoclonal antibody ICA4 (20) at a 1:5,000 dilution, incubated with a goat antimouse antiserum conjugated to horseradish peroxidase (Sigma, St. Louis, Mo.) at a 1:30,000 dilution, and developed by enhanced chemiluminescence (ECL) (Amersham Life Science, Arlington Heights, Ill.).
Transmission electron microscopy.
For viewing BFP, overnight bacterial cultures were diluted 1:100 in 20 ml of DMEM and grown at 37°C for 5 h. Samples were centrifuged (3,400 × g, 5 min) and resuspended in 1 ml of DMEM. A 10-μl aliquot of each sample was spotted on Formvar-carbon-coated copper grids (Electron Microscopy Sciences, Fort Washington, Pa.) for 5 min. Grids were blotted, washed with PBS, stained with phosphotungstic acid in PBS, and examined in a JEOL JEM-1200 EXII transmission electron microscope. Samples were coded and examined without knowledge of their identity.
Sucrose density flotation gradient fractionation.
EPEC strains were grown overnight in LB medium and diluted 1:100 into 10 50-ml conical tubes containing 20 ml of DMEM–F-12 medium with 15 mM HEPES and 2 mM glutamine. Recombinant HB101 strains were grown overnight in LB medium and diluted 1:100 into 1 liter of LB medium. Cultures were grown for 5 h, centrifuged (4,000 × g, 4°C, 15 min), and resuspended in 3 ml of 25 mM Tris-HCl, pH 7.4. Samples were then lysed in a French press at 20,000 lb/in2, centrifuged twice (3,000 × g, 4°C, 5 min), and stored at −20°C. Sucrose (3 g) was added to 2 ml of lysate to create a 60% (wt/wt) solution, and 1.3 ml of this solution was placed in the bottom of an ultracentrifuge tube. A sucrose density gradient was created by overlaying the sample with 1.3 ml of sucrose solutions of decreasing concentrations in 3% decrements, from 56 to 28% (wt/wt). Samples were centrifuged (245,000 × g, 10°C, 72 h), and 540-μl fractions were removed from the tubes and placed into microcentrifuge tubes. To determine the density, the tubes were weighed before and after removing 100 μl from fractions. These 100-μl aliquots were saved, and NADH oxidase activity was assayed as previously described (30). The rate of decline in A340 was used as an indicator of relative enzyme activity in each fraction. The remaining 440 μl was precipitated by adding an equal volume of cold 25% trichloroacetic acid, incubating on ice for 30 min, and centrifuging (6,000 × g, 4°C, 30 min). The samples were resuspended in 80 μl of 10% saturated Tris base in 4× SDS-PAGE loading buffer. Twenty-microliter aliquots of each fraction were loaded on SDS–15% PAGE gels and separated by electrophoresis. The gels were transferred as described above, and the blots were stained with Ponceau stain (Sigma) to identify fractions containing soluble and insoluble proteins and those containing the outer membrane protein OmpA (32). The blots were blocked and immunoblotted for bundlin as described above.
RESULTS
Construction of bfp mutants.
To examine the roles of the Bfp proteins in BFP biogenesis and function, the bfpG, bfpB, bfpC, bfpD, bfpP, and bfpH genes were mutated in this study. The genes were disrupted by the addition of a nonpolar kanamycin cassette (27). The cassette begins with stop codons in all three reading frames followed by the aphA3 gene. Immediately following this is a consensus ribosome binding site and an ATG start codon to reinitiate translation of the downstream portion of the interrupted gene. The cassette has been modified by the addition of either one or two nucleotides immediately following the start codon, so that it is available in all three reading frames. The disrupted gene was subcloned into pCVD442 (14), a positive-selection suicide vector, and mobilized into wild-type EPEC by triparental conjugation, and allelic exchange was carried out as previously described (17). PCR analysis of each mutant confirmed the addition of the 850 bp of the kanamycin cassette in the intended gene, with no changes in nearby loci (data not shown). Each mutation that resulted in a discernible phenotype was complemented by the addition of a wild-type copy of the mutant gene in trans on a low-copy-number vector to confirm that the mutations had no polar effects on prebundlin processing, BFP expression, or the ability to autoaggregate or perform LA. Control plasmids were also introduced into each mutant to verify that complementation was due to the presence of the wild-type allele of the gene (Fig. 1).
Effects of bfp mutations on expression and processing of prebundlin.
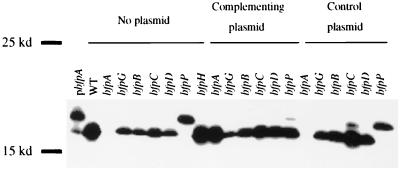
bfpA, the first gene in the bfp cluster, encodes bundlin, the major structural subunit of BFP. Bundlin is expressed as a preprotein with a short leader sequence. To determine if any of the mutations that we made affected prebundlin expression or processing, immunoblotting for bundlin was performed on all mutant strains and on wild-type EPEC (Fig. 2). In all mutant strains tested except the bfpP mutant strain UMD932, bundlin was expressed and processed to mature bundlin in a manner similar to that of wild-type EPEC. The addition of the bfpP gene on plasmid pRPA107 restored the ability of UMD932 to process prebundlin to bundlin, while a control plasmid had no effect. This indicates that only BfpP is required for the processing of prebundlin to mature bundlin. We found no consistent effect of any bfp mutation on the amount of bundlin expression in repeated experiments.
FIG. 2.
Expression and processing of bundlin in wild-type EPEC and mutant strains. Whole-cell lysates of cells grown in DMEM were prepared, separated by SDS-PAGE electrophoresis on a 15% polyacrylamide gel, and analyzed by immunoblotting with an antibundlin monoclonal antibody. Lanes 2 to 9 contain wild-type EPEC and mutant strains, lanes 10 to 15 contain complemented mutant strains, and lanes 16 to 21 contain mutants with control plasmids. Lane 1, pMSD205, unprocessed bundlin control; lane 2, wild-type (WT) EPEC strain E2348/69; lanes 3, 10, and 16, bfpA mutant strain UMD901; lanes 4, 11, and 17, bfpG mutant strain UMD928; lanes 5, 12, and 18, bfpB mutant strain UMD923; lanes 6, 13, and 19, bfpC mutant strain UMD924; lanes 7, 14, and 20, bfpD mutant strain UMD926; lanes 8, 15, and 21, bfpP mutant strain UMD932; lane 9, bfpH mutant strain UMD918.
Effects of bfp mutations on LA.
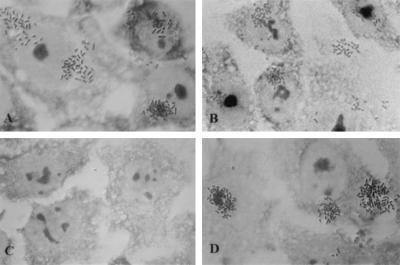
We tested the ability of each mutant strain to adhere to HEp-2 cells. LA is a qualitative adherence pattern recognized as compact microcolonies on the surface of epithelial cells. With the exception of the bfpH mutant strain, all mutants were unable to perform LA. In contrast, the bfpH mutant strain adhered in a manner indistinguishable from that of wild-type EPEC (Fig. 3). In all other mutant strains, addition of the wild-type allele to complement the mutation restored the ability to perform LA. Mutants containing negative control plasmids did not regain the ability to perform LA. These results indicate that, with the exception of bfpH, all the genes tested are required for EPEC to perform LA.
FIG. 3.
Localized adherence of wild-type EPEC and mutant strains to epithelial cells. HEp-2 cells were incubated with bacteria for 3 h, washed, fixed, and stained with Giemsa stain. Slides were examined by light microscopy with a 63× objective lens. (A) Wild-type EPEC strain E2348/69. (B) bfpH mutant strain UMD918. (C) bfpC mutant strain UMD924. (D) bfpC mutant strain UMD924 containing plasmid pRPA103. Mutant and complemented strains not shown were indistinguishable from strains UMD924 and UMD924(pRPA103).
Effects of bfp mutations on autoaggregation of EPEC.
EPEC strains grown under optimal conditions for BFP expression aggregate into large clusters, and this aggregation is dependent on BFP. When observed by microscopy, these clusters disaggregate in a matter of minutes as the culture cools (6). In contrast to LA, autoaggregation can be quantified, although the precise relationship between the aggregation index and the amount of pili produced is not known since we currently have no method of measuring the latter. Strains were grown in tissue culture medium and tested for BFP-mediated aggregation. To quantify the ability of mutant strains to aggregate in comparison to wild-type strains, the A600 of a 4-h culture was measured before and after vortexing and the percent increase after vortexing was calculated as an aggregation index. For strains that aggregate, disaggregation after vortexing was confirmed by microscopy. Table 2 shows the aggregation indices for all the strains tested. All mutant strains had aggregation indices significantly lower than that of wild-type EPEC, and in each case, these indices increased significantly upon addition of complementing plasmids. However, in no case did the aggregation indices of complemented mutants increase to near-wild-type levels, raising the possibility that this failure of full complementation was due to polar effects of the mutations. To determine whether complementing plasmids could interfere with BFP biogenesis, aggregation assays were performed on the wild-type EPEC strain, the wild-type EPEC strain containing pRPA103, the bfpC mutant strain UMD924, and the bfpC mutant strain complemented with pRPA103. Plasmid pRPA103, the complementing plasmid for the bfpC mutant strain UMD924, was chosen for these experiments because strain UMD924 containing this plasmid had the lowest aggregation index of the complemented mutants. In two experiments, each with quadruplicate samples, the wild-type EPEC strain had a mean aggregation index (± standard deviation) of 150.1% ± 70.7% while strain UMD924 had an aggregation index of 2.0% ± 0.3% (P = 0.005 versus wild type). Wild-type EPEC containing plasmid pRPA103 had an index of 13.8% ± 8.0% (P = 0.008 versus wild type), and UMD924 containing pRPA103 had an index of 8.7% ± 3.6% (P = 0.001 versus UMD924; P = 0.19 versus wild type containing pRPA103). Thus, the presence of the complementing plasmid pRPA103 reduced the aggregation index of the wild-type strain to a level similar to that of the bfpC mutant containing the same plasmid. We attribute the decreased aggregation indices seen in the complemented mutant strains not to polar effects of the mutations but rather to stoichiometry problems involving the extra copy of the genes on the complementing plasmids. In this case, pRPA103 introduces extra copies of bfpA, bfpG, bfpB, and bfpC. The resulting overexpression of one or more of these proteins may partially interfere with BFP biogenesis and result in the lower aggregation index. In contrast to the increased aggregation indices seen in all the mutants containing complementing plasmids, aggregation indices of mutants did not change upon addition of control plasmids (data not shown). The bfpH mutant was capable of both aggregation and spontaneous disaggregation (data not shown). These results indicate that all of the genes tested except bfpH are required for EPEC autoaggregation.
TABLE 2.
Autoaggregation of wild-type and bfp mutant EPEC strains
| Strain | Genotype | Aggregation index ± SDa | % of wild-type value | P value |
|---|---|---|---|---|
| E2348/69 | Wild type | 80.2 ± 19.3 | 100 | |
| UMD901 | bfpA | 2.0 ± 0.7 | 2.5 | 0.004b |
| UMD901(pRPA100) | bfpA (pbfpA+) | 43.8 ± 11.1 | 54.6 | 0.005c |
| UMD928 | bfpG | 0.8 ± 0.5 | 1.0 | 0.004b |
| UMD928(pRPA104) | bfpG (pbfpA+G+) | 17.0 ± 3.5 | 21.2 | 0.002c |
| UMD923 | bfpB | 0.8 ± 0.3 | 0.9 | 0.004b |
| UMD923(pRPA101) | bfpB (pbfpA+G+B+) | 42.3 ± 11.8 | 52.7 | 0.006c |
| UMD924 | bfpC | 2.0 ± 0.5 | 2.4 | 0.004b |
| UMD924(pRPA103) | bfpC (pbfpA+G+B+C+) | 6.4 ± 1.7 | 7.9 | 0.008c |
| UMD926 | bfpD | 1.0 ± 0.9 | 1.2 | 0.003b |
| UMD926(pRPA106) | bfpD ((pbfpA+G+B+C+D+) | 19.8 ± 11.1 | 24.7 | 0.051c |
| UMD932 | bfpP | 2.0 ± 0.9 | 2.5 | 0.004b |
| UMD932(pRPA107) | bfpP (pbfpP+) | 11.5 ± 1.4 | 14.3 | 0.001c |
| UMD918 | bfpH | 50.2 ± 5.8 | 62.6 | 0.03b |
Values shown are mean values from four separate experiments ± standard deviations (SDs).
Versus wild type.
Versus corresponding mutant.
Effects of bfp mutations on the biogenesis of BFP.
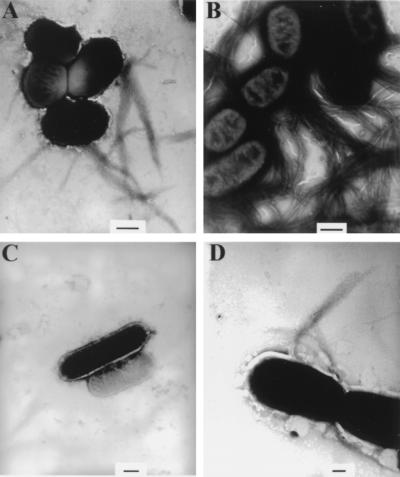
To examine the effects of the bfp mutations on BFP biogenesis, mutant strains and wild-type EPEC were examined by transmission electron microscopy. BFP was detected in samples of wild-type EPEC and bfpH mutant strain UMD918 (Fig. 4). In contrast, we could not detect BFP in the remaining mutant strains. In every case, the addition of the complementing plasmid containing the wild-type allele of the mutated gene to each strain restored the ability of each mutant strain to express BFP, while the addition of the control plasmid did not. These results indicate that bfpH is not required for BFP biogenesis while the remaining mutated genes are.
FIG. 4.
Expression of BFP by wild-type EPEC and mutant strains. Strains were grown in DMEM, spotted on Formvar-copper-coated grids, negatively stained with phosphotungstic acid, and examined by electron microscopy. (A) Wild-type EPEC strain E2348-69. (B) bfpH mutant strain UMD918. (C) bfpG mutant strain UMD928. (D) bfpG mutant strain UMD928 containing plasmid pRPA104. Mutant and complemented strains not pictured were indistinguishable from strains UMD928 and UMD928(pRPA104), respectively. Bars, 500 (A to C) and 200 (D) nm.
Prebundlin does not require processing or the presence of any other Bfp proteins to localize to both the inner and outer membranes.
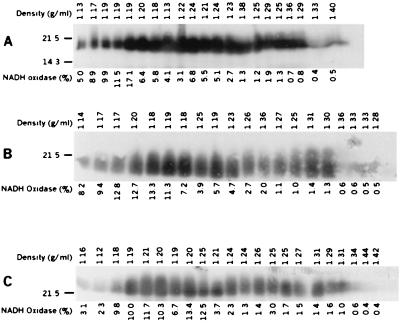
To determine the role of each Bfp protein in the localization of bundlin, sucrose density flotation gradients of wild-type EPEC and the various mutant strains were analyzed. In all strains tested, bundlin was found both in fractions that had high activity of NADH oxidase, an inner membrane protein, and in fractions that contained the outer membrane protein OmpA (data not shown). Thus, it appeared that the protein products of none of the bfp genes examined in this study were required for bundlin to localize to fractions containing inner and outer membrane proteins and in fact that prebundlin did not require processing to localize to these fractions. To determine whether localization to fractions containing both membranes is an intrinsic property of bundlin, three additional strains were tested. Sucrose density flotation gradient analyses were performed on recombinant E. coli strain DH5α containing the entire bfp gene cluster, containing only bfpA and bfpP, and containing bfpA alone. The results of these studies using recombinant E. coli are shown in Fig. 5. Peak NADH oxidase activity was found in fractions with densities of 1.17 to 1.20 g/ml. In contrast, Ponceau staining revealed that soluble and insoluble cellular proteins were found in fractions with densities of 1.33 to 1.44 g/ml and that the outer membrane protein OmpA was found in fractions with densities of greater than 1.26. Bundlin (or, in the case of the strain containing bfpA alone, prebundlin) was found in all fractions from the gradient, including those with high NADH oxidase activity, those containing outer membrane proteins, and those with intermediate densities (Fig. 5). Localization to fractions containing both inner and outer membrane proteins is thus independent of the presence of other bfp genes and appears to occur whether or not prebundlin is processed to mature bundlin.
FIG. 5.
Distribution of bundlin in membrane fractions from recombinant E. coli strain DH5α containing the entire bfp gene cluster on plasmid pKDS302 (A), containing bfpA on plasmid pMSD230 and bfpP on plasmid pDN19PB (B), and containing bfpA on plasmid pMSD230 and a bfpP deletion on plasmid pDN19PBΔ (C). Fractions collected from the sucrose flotation density gradients were analyzed by immunoblotting with antibodies against bundlin. NADH oxidase activity was measured and displayed as a percentage of the total NADH oxidase activity per fraction. Fractions were loaded on SDS–15% PAGE gels in order from the top to the bottom of the gradient (left to right, respectively). The density of each fraction is indicated above each blot. Variations from the linear trend of increasing density can be ascribed to pipetting error. Data are representative of two separate experiments with similar results. Numbers to the left of each panel indicate molecular mass(es) in kilodaltons.
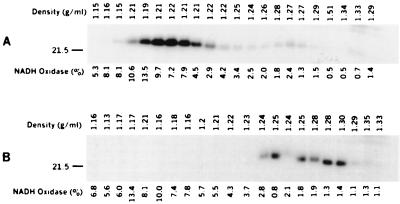
The presence of prebundlin in all fractions of the gradient raises the possibility that prebundlin may be able to form reversible complexes with itself or other proteins that may interact transiently with membrane vesicles. To determine whether the presence of prebundlin in fractions containing inner and outer membranes was an artifact of a reversible interaction, we separately pooled six low-density fractions and six high-density fractions from a sucrose flotation gradient fractionation of prebundlin from the recombinant strain containing bfpA alone. The pools were adjusted to a concentration of 60% sucrose and mixed with a lysate of strain DH5α containing no plasmid in 60% sucrose. These mixtures were then subjected to a second round of sucrose flotation gradient fractionation. Western blot analysis showed that the prebundlin from the low-density fractions migrated back to low-density fractions after the second fractionation and that the prebundlin from high-density fractions migrated back to high-density fractions (Fig. 6). This result suggests that the presence of prebundlin in fractions containing inner and outer membrane proteins is not due to reversible interactions between prebundlin and membrane vesicles. Electron microscopy of the pooled fractions showed that membrane vesicles were present in both the low-density and the high-density fractions. No fimbria-like structures were seen (data not shown).
FIG. 6.
Redistribution of previously fractionated prebundlin from low-density (A) and high-density (B) fractions after a second round of sucrose flotation density gradient fractionation. Fractions collected from the sucrose flotation gradient were analyzed by immunoblotting with antibodies against bundlin. NADH oxidase activity was measured and displayed as a percentage of the total NADH oxidase activity per fraction. Fractions were loaded on SDS–15% PAGE gels in order from the top to the bottom of the gradient (left to right, respectively). The density of each fraction is indicated above each blot. Variations from the linear trend of increasing density can be ascribed to pipetting error. Numbers to the left of the panels indicate molecular mass in kilodaltons.
DISCUSSION
Type IV pilus biogenesis is currently a poorly understood process. For many species, the essential genes for type IV pilus systems are scattered at several sites in the bacterial chromosome, making the study of all the genes required for pilus biogenesis very difficult. As all the genes required for the expression of BFP in a recombinant E. coli host are known, the BFP system is an attractive model system for the study of type IV pilus biogenesis. In this study, we examined the roles of seven bfp genes, bfpA, bfpG, bfpB, bfpC, bfpD, bfpP, and bfpH, in BFP biogenesis and LA. Previous studies have shown that mutations in bfpA block BFP biogenesis and LA (13). Ramer et al. reported that a bfpB mutation also blocked BFP biogenesis, autoaggregation, and LA (34), and Bieber et al. reported similar findings for a bfpD mutant (6). Our results confirm these findings and also show that bfpG, bfpC, and bfpP are required for BFP biogenesis and LA, while bfpH is not required for either BFP biogenesis or LA.
To examine the role of each Bfp protein in BFP biogenesis and related phenotypes, mutations were introduced into the wild-type EPEC background in the bfpG, bfpB, bfpC, bfpD, bfpP, and bfpH genes. The mutations were made by disrupting the genes with an aphA3 kanamycin cassette (27) which is designed to be nonpolar. Each mutation that resulted in a discernible phenotype was complemented by the addition of a wild-type copy of the mutant gene in trans on a low-copy-number vector. Fragments of the bfp cluster beginning 270 bp upstream of bfpA and ending within the gene directly downstream of the mutated gene were used to complement mutations in the bfpA, bfpG, bfpB, bfpC, and bfpD genes. This strategy was used to include the native bfp promoter in each complementation plasmid in an effort to restore wild-type levels of the gene products of the mutated genes, as earlier attempts to complement some bfp mutants with plasmids containing only a single gene were unsuccessful (data not shown). Complementation analysis of the mutant strains confirmed that all mutations constructed were nonpolar. All complemented mutants were able to express mature bundlin, and although we were unable to obtain a single Western blot showing equivalent amounts of bundlin in all strains, we have observed no consistent decreases in levels of bundlin in any of the mutants compared to the wild type. All complemented mutants were also able to perform LA and to express BFP. Assays for LA and BFP expression are qualitative, not quantitative. The autoaggregation index is an attempt to quantify a phenotype associated with BFP expression. Autoaggregation studies of wild-type EPEC containing plasmid pRPA103 suggest that stoichiometry problems posed by additional copies of bfp genes on the complementing plasmids are responsible for the lower aggregation indices of complemented mutant strains. These stoichiometry problems may result in decreased levels of BFP expression by altering the ratio of components of the Bfp biogenesis machinery and competing for required interactions. Negative control plasmids for complementation, containing fragments of the bfp cluster that end within the mutated gene, showed that the restoration of the wild-type phenotype was solely due to the addition of the wild-type copy of the mutated gene.
Western blotting showed that in the bfpG, bfpB, bfpC, bfpD, and bfpH mutant strains prebundlin is expressed and processed as in wild-type EPEC, while in the bfpA mutant strain no bundlin is seen and in the bfpP mutant strain prebundlin is expressed but not processed into mature bundlin. These results were expected, as the mutation in bfpA renders bundlin unstable (50) and bfpP encodes a prepilin peptidase (51). We previously showed that BfpP was capable of processing the P. aeruginosa prepilin. In the current report, we detected no processed bundlin in the bfpP mutant. Therefore, BfpP is the only protein in EPEC that processes prebundlin. The remaining mutations did not have a consistent effect on either bundlin expression or processing. These results indicate that the protein products of the bfpG, bfpB, bfpC, bfpD, and bfpH genes are not involved in the expression, stability, or processing of bundlin.
Studies of the abilities of the mutant strains to autoaggregate, perform LA, and express BFP all gave similar results. The bfpA, bfpG, bfpB, bfpC, bfpD, and bfpP mutant strains were unable to autoaggregate into clusters, perform LA, or express BFP. Since all mutations that disabled BFP biogenesis also disabled both autoaggregation and LA, it appears that both these phenotypes require mature pili. The addition of a wild-type copy of the mutated gene in trans restored the ability of the mutant strains to express these phenotypes. In contrast, the bfpH mutant strain was similar to wild-type EPEC in all three phenotypes. In addition, the autoaggregates formed by the bfpH mutant were able to disperse, thus distinguishing this mutant from bfpF mutants, which express pili and form aggregates that do not disperse (6).
Of the genes examined in this study, the functions of few are known. Prebundlin is encoded by the bfpA gene, and we show here that bfpP encodes the prepilin peptidase that cleaves prebundlin to its mature form. Ramer et al. reported that bfpB encodes a member of the secretin family of proteins (34). The functions of bfpG, bfpC, and bfpD are not known. BfpD contains functional motifs found in nucleotide-binding proteins, so it may be required to provide the energy required for BFP biogenesis. BfpG appears to be a small lipoprotein, and BfpC has a single putative transmembrane domain. Based on the phenotypes tested in this study, no function can be ascribed to BfpH. The modest reduction seen in the ability of the bfpH mutant to autoaggregate, while statistically significant, is of questionable biological significance. The bfpH gene is predicted to encode a lytic transglycosylase belonging to a large family. The presumed function of BfpH would be to hydrolyze a region of the peptidoglycan layer to allow for either bundlin or mature BFP to reach the outer membrane. However, several observations suggest that BfpH is not fully functional in EPEC strain E2348/69. First, results reported here show that a bfpH mutation does not disable BFP biogenesis, autoaggregation, or LA. Second, of all the BFP proteins, BfpP and BfpH are the only ones that we were unable to detect by T7 RNA polymerase expression (40). Furthermore, a comparison with other transglycosylases indicates that BfpH lacks several highly conserved residues (24). Thus, it is possible that bfpH has accumulated mutations because it is superfluous or poorly expressed. There are other transglycosylases encoded on the EPEC chromosome (18), and so it is possible that in the absence of BfpH, one of these proteins functionally replaces BfpH. An alternative explanation could be that BFP evolved from or still is a bacteriophage (22), which required or requires the action of the peptidoglycan hydrolase to enter the bacterial cell rather than for pilus biogenesis. The self-transmissible plasmid R64 encodes a type IV pilus and has a BfpH homologue, PilT. Mutation of pilT did not affect pilus biogenesis or sensitivity to pilus-specific bacteriophages (49), and thus, pilT appears to be similar to bfpH in that it is not required for pilus biogenesis. However, pilT mutants exhibit a reduced transfer frequency of the R64 plasmid. The possibility that bfpH is required for an as-yet-unidentified BFP function cannot be excluded.
The method that we chose to study membrane localization, sucrose flotation gradient centrifugation, is considered to be highly reliable, since membrane vesicles must float up to reach the fractions of equivalent density (31). Therefore, insoluble material does not contaminate membrane fractions as it can with sedimentation gradients, and proteins with unusual detergent solubility do not yield spurious results, as they can when differential detergent solubility is used. Previous studies indicated that prebundlin is at least transiently a cytoplasmic transmembrane protein accessible simultaneously to both BfpP and DsbA (50). However, its fate subsequent to the posttranslational modifications catalyzed by these enzymes and prior to incorporation into a pilus is unknown. The results of sucrose flotation gradients appear to show that bundlin localizes to both membrane fractions independent of any Bfp proteins including BfpP. Similar results were obtained in analysis of P. aeruginosa PAK pilin (41, 46) as well as the PulG and Xcp type IV pilus-like proteins from type II secretion systems (29, 33). The localization of prebundlin to fractions containing outer membrane proteins may be surprising since the protein would be predicted to be anchored in the inner membrane by two positively charged residues near the amino terminus followed by a stretch of hydrophobic amino acids. By subjecting fractions containing prebundlin to repeat flotation gradient centrifugation, we demonstrated that localization to inner and outer membrane fractions is not a transient, reversible phenomenon. However, it remains possible that the cell disruption techniques employed could result in cytoplasmic prebundlin or prebundlin complexes becoming irreversibly associated with membrane vesicles. As we are unable to prove that these results are not an artifact of cell disruption, we can conclude only that cell fractionation studies do not shed new light on the possible export of bundlin to the outer membrane during a phase of pilus biogenesis.
Based on the results of this study, we are able only to separate BFP biogenesis into three stages. The first stage is the synthesis of prebundlin. In the second stage, prebundlin simultaneously acquires a disulfide bond in the periplasmic space and is processed to bundlin through the loss of its leader sequence. The DsbA protein is required for the disulfide bond formation (50), and we have shown here conclusively that BfpP is required for the processing of prebundlin to bundlin. In the third stage, mature bundlin is assembled into BFP by a process that is not understood but which does not appear to involve sequential passage from inner to outer membrane. We have shown here that BfpG, BfpB, BfpC, and BfpD are involved in this process, as are BfpE and BfpU (T. E. Blank and M. S. Donnenberg, unpublished data). While our results do not elucidate the roles of any of these proteins in BFP biogenesis, they do show that these proteins do not have a role in the first or second stage of BFP biogenesis and probably act after processing by BfpP. Our results also indicate that BfpH does not play a critical role in BFP biogenesis or function for any of the phenotypes that we tested. While mutations in four bfp genes (bfpI, bfpJ, bfpK, and bfpL) which encode proteins with some resemblance to pilin subunits remain to be analyzed, it appears unlikely that analysis of the effects of single mutations using established approaches will yield new insights into type IV pilus biogenesis. Other approaches that examine interactions between individual components or the localization of Bfp components other than bundlin will be required to form a more coherent picture of the type IV pilus assembly machine.
ACKNOWLEDGMENTS
We thank Jim Kaper and Karen Jarvis for providing the kanamycin cassettes pUC18K2 and pUC18K3. We also thank Rick Blank and Barry McNamara for helpful suggestions over the course of these experiments and Rebecca Wade for technical assistance.
This work was supported by Public Health Service award AI27606 from the National Institutes of Health.
REFERENCES
- 1.Alm R A, Bodero A J, Free P D, Mattick J S. Identification of a novel gene, pilZ, essential for type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J Bacteriol. 1996;178:46–53. doi: 10.1128/jb.178.1.46-53.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm R A, Hallinan J P, Watson A A, Mattick J S. Fimbrial biogenesis genes of Pseudomonas aeruginosa: PilW and pilX increase the similarity of type 4 fimbriae to the GSP protein-secretion systems and pilY1 encodes a gonococcal PilC homologue. Mol Microbiol. 1996;22:161–173. doi: 10.1111/j.1365-2958.1996.tb02665.x. [DOI] [PubMed] [Google Scholar]
- 3.Alm R A, Mattick J S. Identification of a gene, pilV, required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa, whose product possesses a pre-pilin-like leader sequence. Mol Microbiol. 1995;16:485–496. doi: 10.1111/j.1365-2958.1995.tb02413.x. [DOI] [PubMed] [Google Scholar]
- 4.Alm R A, Mattick J S. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene. 1997;192:89–98. doi: 10.1016/s0378-1119(96)00805-0. [DOI] [PubMed] [Google Scholar]
- 5.Anantha R P, Stone K D, Donnenberg M S. The role of BfpF, a member of the PilT family of putative nucleotide-binding proteins, in type IV pilus biogenesis and in interactions between enteropathogenic Escherichia coli and host cells. Infect Immun. 1998;66:122–131. doi: 10.1128/iai.66.1.122-131.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bieber D, Ramer S W, Wu C Y, Murray W J, Tobe T, Fernandez R, Schoolnik G K. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 7.Bitter W, Koster M, Latijnhouwers M, De Cock H, Tommassen J. Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol Microbiol. 1998;27:209–219. doi: 10.1046/j.1365-2958.1998.00677.x. [DOI] [PubMed] [Google Scholar]
- 8.Bradley D E. The adsorption of Pseudomonas aeruginosa pilus-dependent bacteriophages to a host mutant with nonretractile pili. Virology. 1974;58:149–163. doi: 10.1016/0042-6822(74)90150-0. [DOI] [PubMed] [Google Scholar]
- 9.Bradley D E. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can J Microbiol. 1980;26:146–154. doi: 10.1139/m80-022. [DOI] [PubMed] [Google Scholar]
- 10.Chikami G K, Fierer J, Guiney D G. Plasmid-mediated virulence in Salmonella dublin demonstrated by use of a Tn5-oriT construct. Infect Immun. 1985;50:420–424. doi: 10.1128/iai.50.2.420-424.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cravioto A, Gross R J, Scotland S M, Rowe B. An adhesive factor found in strains of Escherichia coli belonging to the traditional infantile enteropathogenic serotypes. Curr Microbiol. 1979;3:95–99. [Google Scholar]
- 12.Donnenberg M S. Enteropathogenic Escherichia coli. In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press, Ltd.; 1995. pp. 709–726. [Google Scholar]
- 13.Donnenberg M S, Girón J A, Nataro J P, Kaper J B. A plasmid-encoded type IV fimbrial gene of enteropathogenic Escherichia coli associated with localized adherence. Mol Microbiol. 1992;6:3427–3437. doi: 10.1111/j.1365-2958.1992.tb02210.x. [DOI] [PubMed] [Google Scholar]
- 14.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donnenberg M S, Kaper J B. Enteropathogenic Escherichia coli. Infect Immun. 1992;60:3953–3961. doi: 10.1128/iai.60.10.3953-3961.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donnenberg M S, Nataro J P. Methods for studying adhesion of diarrheagenic Escherichia coli. Methods Enzymol. 1995;253:324–336. doi: 10.1016/s0076-6879(95)53028-2. [DOI] [PubMed] [Google Scholar]
- 17.Donnenberg M S, Yu J, Kaper J B. A second chromosomal gene necessary for intimate attachment of enteropathogenic Escherichia coli to epithelial cells. J Bacteriol. 1993;175:4670–4680. doi: 10.1128/jb.175.15.4670-4680.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elliott S J, Wainwright L A, McDaniel T K, Jarvis K G, Deng Y, Lai L-C, McNamara B P, Donnenberg M S, Kaper J B. The complete sequence of the locus of enterocyte effacement (LEE) of enteropathogenic E. coli E2348/69. Mol Microbiol. 1998;28:1–4. doi: 10.1046/j.1365-2958.1998.00783.x. [DOI] [PubMed] [Google Scholar]
- 19.Girón J A, Ho A S Y, Schoolnik G K. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science. 1991;254:710–713. doi: 10.1126/science.1683004. [DOI] [PubMed] [Google Scholar]
- 20.Girón J A, Qadri F, Azim T, Jarvis K J, Kaper J B, Albert M J. Monoclonal antibodies specific for the bundle-forming pilus of enteropathogenic Escherichia coli. Infect Immun. 1995;63:4949–4952. doi: 10.1128/iai.63.12.4949-4952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hobbs M, Mattick J S. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol Microbiol. 1993;10:233–243. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- 22.Karaolis D K, Somara S, Maneval D R, Jr, Johnson J A, Kaper J B. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature. 1999;399:375–379. doi: 10.1038/20715. [DOI] [PubMed] [Google Scholar]
- 23.Knutton S, Shaw R K, Anantha R P, Donnenberg M S, Zorgani A A. The type IV bundle-forming pilus of enteropathogenic Escherichia coli undergoes dramatic alterations in structure associated with bacterial adherence, aggregation and dispersal. Mol Microbiol. 1999;33:499–509. doi: 10.1046/j.1365-2958.1999.01495.x. [DOI] [PubMed] [Google Scholar]
- 24.Lehnherr H, Hansen A M, Ilyina T. Penetration of the bacterial cell wall: a family of lytic transglycosylases in bacteriophages and conjugative plasmids. Mol Microbiol. 1998;30:454–457. doi: 10.1046/j.1365-2958.1998.01069.x. [DOI] [PubMed] [Google Scholar]
- 25.Levine M M, Nataro J P, Karch H, Baldini M M, Kaper J B, Black R E, Clements M L, O'Brien A D. The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coli is dependent on a plasmid encoding an enteroadhesiveness factor. J Infect Dis. 1985;152:550–559. doi: 10.1093/infdis/152.3.550. [DOI] [PubMed] [Google Scholar]
- 26.McDaniel T K, Kaper J B. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on K-12 E. coli. Mol Microbiol. 1997;23:399–407. doi: 10.1046/j.1365-2958.1997.2311591.x. [DOI] [PubMed] [Google Scholar]
- 27.Ménard R, Sansonetti P J, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nunn D, Bergman S, Lory S. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J Bacteriol. 1990;172:2911–2919. doi: 10.1128/jb.172.6.2911-2919.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nunn D N, Lory S. Cleavage, methylation, and localization of the Pseudomonas aeruginosa export proteins XcpT, -U, -V, and -W. J Bacteriol. 1993;175:4375–4382. doi: 10.1128/jb.175.14.4375-4382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osborn M J, Gander J E, Parisi E, Carson J. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J Biol Chem. 1972;247:3962–3972. [PubMed] [Google Scholar]
- 31.Poquet I, Kornacker M G, Pugsley A P. The role of the lipoprotein sorting signal (aspartate +2) in pullulanase secretion. Mol Microbiol. 1993;9:1061–1069. doi: 10.1111/j.1365-2958.1993.tb01235.x. [DOI] [PubMed] [Google Scholar]
- 32.Possot O, Pugsley A P. Molecular characterization of PuIE, a protein required for pullulanase secretion. Mol Microbiol. 1994;12:287–299. doi: 10.1111/j.1365-2958.1994.tb01017.x. [DOI] [PubMed] [Google Scholar]
- 33.Pugsley A P, Possot O. The general secretory pathway of Klebsiella oxytoca: no evidence for relocalization or assembly of pilin-like PuIG protein into a multiprotein complex. Mol Microbiol. 1993;10:665–674. doi: 10.1111/j.1365-2958.1993.tb00938.x. [DOI] [PubMed] [Google Scholar]
- 34.Ramer S W, Bieber D, Schoolnik G K. BfpB, an outer membrane lipoprotein required for the biogenesis of bundle-forming pili in enteropathogenic Escherichia coli. J Bacteriol. 1996;178:6555–6563. doi: 10.1128/jb.178.22.6555-6563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudel T, Scheuerpflug I, Meyer T F. Neisseria PilC protein identified as type-4 pilus tip-located adhesin. Nature. 1995;373:357–359. doi: 10.1038/373357a0. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Scaletsky I C A, Silva M L M, Trabulsi L R. Distinctive patterns of adherence of enteropathogenic Escherichia coli to HeLa cells. Infect Immun. 1984;45:534–536. doi: 10.1128/iai.45.2.534-536.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sohel I, Puente J L, Murray W J, Vuopio-Varkila J, Schoolnik G K. Cloning and characterization of the bundle-forming pilin gene of enteropathogenic Escherichia coli and its distribution in Salmonella serotypes. Mol Microbiol. 1993;7:563–575. doi: 10.1111/j.1365-2958.1993.tb01147.x. [DOI] [PubMed] [Google Scholar]
- 39.Sohel I, Puente J L, Ramer S W, Bieber D, Wu C-Y, Schoolnik G K. Enteropathogenic Escherichia coli: identification of a gene cluster coding for bundle-forming pilus morphogenesis. J Bacteriol. 1996;178:2613–2628. doi: 10.1128/jb.178.9.2613-2628.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stone K D, Zhang H-Z, Carlson L K, Donnenberg M S. A cluster of fourteen genes from enteropathogenic Escherichia coli is sufficient for biogenesis of a type IV pilus. Mol Microbiol. 1996;20:325–337. doi: 10.1111/j.1365-2958.1996.tb02620.x. [DOI] [PubMed] [Google Scholar]
- 41.Strom M S, Lory S. Cloning and expression of the pilin gene of Pseudomonas aeruginosa PAK in Escherichia coli. J Bacteriol. 1986;165:367–372. doi: 10.1128/jb.165.2.367-372.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strom M S, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 43.Strom M S, Nunn D N, Lory S. A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc Natl Acad Sci USA. 1993;90:2404–2408. doi: 10.1073/pnas.90.6.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 45.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 46.Watts T H, Worobec E A, Paranchych W. Identification of pilin pools in the membranes of Pseudomonas aeruginosa. J Bacteriol. 1982;152:687–691. doi: 10.1128/jb.152.2.687-691.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolfgang M, Lauer P, Park H S, Brossay L, Hébert J, Koomey M. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol Microbiol. 1998;29:321–330. doi: 10.1046/j.1365-2958.1998.00935.x. [DOI] [PubMed] [Google Scholar]
- 48.Yoshida T, Furuya N, Ishikura M, Isobe T, Haino-Fukushima K, Ogawa T, Komano T. Purification and characterization of thin pili of IncI1 plasmids ColIb-P9 and R64: formation of PilV-specific cell aggregates by type IV pili. J Bacteriol. 1998;180:2842–2848. doi: 10.1128/jb.180.11.2842-2848.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshida T, Kim S R, Komano T. Twelve pil genes are required for biogenesis of the R64 thin pilus. J Bacteriol. 1999;181:2038–2043. doi: 10.1128/jb.181.7.2038-2043.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang H-Z, Donnenberg M S. DsbA is required for stability of the type IV pilin of enteropathogenic Escherichia coli. Mol Microbiol. 1996;21:787–797. doi: 10.1046/j.1365-2958.1996.431403.x. [DOI] [PubMed] [Google Scholar]
- 51.Zhang H-Z, Lory S, Donnenberg M S. A plasmid-encoded prepilin peptidase gene from enteropathogenic Escherichia coli. J Bacteriol. 1994;176:6885–6891. doi: 10.1128/jb.176.22.6885-6891.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]