SUMMARY
Purpose:
Advancing age is one of the strongest risk factors for osteoarthritis (OA). DNA methylation-based measures of epigenetic age acceleration may provide insights into mechanisms underlying OA.
Methods:
We analyzed data from the Multicenter Osteoarthritis Study in a subset of 671 participants ages 45–69 years with no or mild radiographic knee OA. DNA methylation was assessed with the Illumina Infinium MethylationEPIC 850K array. We calculated predicted epigenetic age according to Hannum, Horvath, PhenoAge, and GrimAge epigenetic clocks, then regressed epigenetic age on chronological age to obtain the residuals. Associations between the residuals and knee, hand, and multi-joint OA were assessed using logistic regression, adjusted for chronological age, sex, clinical site, smoking status, and race.
Results:
Twenty-three percent met criteria for radiographic hand OA, 25% met criteria for radiographic knee OA, and 8% met criteria for multi-joint OA. Mean chronological age (SD) was 58.4 (6.7) years. Mean predicted epigenetic age (SD) according to Horvath, Hannum, PhenoAge, and GrimAge epigenetic clocks was 64.9 (6.4), 68.6 (5.9), 50.5 (7.7), and 67.0 (6.2), respectively. Horvath epigenetic age acceleration was not associated with an increased odds of hand OA, odds ratio (95% confidence intervals) = 1.03 (0.99–1.08), with similar findings for knee and multi-joint OA. We found similar magnitudes of associations for Hannum epigenetic age, PhenoAge, and GrimAge acceleration compared to Horvath epigenetic age acceleration.
Conclusions:
Epigenetic age acceleration as measured by various well-validated epigenetic clocks based on DNA methylation was not associated with increased risk of knee, hand, or multi-joint OA independent of chronological age.
Keywords: Epigenetics, Aging, Osteoarthritis
Introduction
Osteoarthritis (OA) is the most common form of arthritis in the United States, affecting over 27 million adults and is a leading cause of physical disability in older individuals.1 Over half of the population has radiographic evidence of OA in one or more joints by the age of 60 and almost 100% have some evidence by the age of 80.2 Advancing age is one of the strongest predictors for OA development.
Manifestations of aging are related to epigenetic mechanisms3,4 that are needed to maintain tight control over transcription and translation to ensure tissue homeostasis and adaptation to the changing microenvironment.5 Of all epigenetic mechanisms, DNA methylation of cytosine-phosphate-guanine dinucleotides (CpGs) is the most well-characterized. Both age-related DNA hypomethylation and hypermethylation have been observed, where age-related hypermethylation occurs preferentially at CpG islands6 that results in gene silencing.7
Assessment of DNA methylation have been used to develop epigenetic clocks that measure biological aging. One of the most widely used is Horvath’s epigenetic clock, which is the combination of DNA methylation levels at 353 CpG sites. It has been shown to provide a robust measure of DNA methylation age across multiple tissue types.8 Horvath suggests that the epigenetic clock measures the cumulative effects of an epigenetic maintenance system.8 The more work that is needed to maintain epigenetic stability, the greater the DNA methylation age. Therefore, perturbations to the system and the corresponding protective response of the epigenetic maintenance system would accelerate DNA methylation age (i.e., an increase in the difference between predicted age based on DNA methylation and observed chronological age). This may be measured as intrinsic or extrinsic epigenetic age acceleration. Intrinsic measures are independent of blood cell composition, while extrinsic measures are dependent on blood cell composition, reflecting aspects of immune system aging.9
In addition to the Horvath clock, other epigenetic clocks have been developed that may capture other aspects of epigenetic aging. The epigenetic clock reported by Hannum consists of 71 CpG markers but only performs well in blood.10 Both Horvath and Hannum clocks were developed and trained on chronological age but do not necessarily correlate well with mortality risk. Therefore, other DNA methylation based epigenetic clocks, including PhenoAge and GrimAge, were developed to better capture correlates of morbidity and mortality, rather than chronological age alone. PhenoAge was trained on clinical characteristics including calendar age, albumin, creatinine, glucose, C-reactive protein, lymphocyte percentage, mean cell volume, red blood cell distribution weight, alkaline phosphatase, and white blood cell count to measure phenotypic age and includes 513 CpG sites that differentiate lifespan from healthspan.11 PhenoAge is associated with all-cause and cause-specific mortality, burden of comorbidities, and physical functioning. GrimAge is composed of 1030 CpG sites, which were identified based on seven aging-related plasma proteins, including plasminogen activation inhibitor 1 (PAI-1), growth differentiation factor 15 (GDF15), beta-2 microglobulin (B2M), adrenomedullin (ADM), Cystatin C, leptin, and tissue inhibitor metalloproteinase 1 (TIMP1), and smoking pack years; it is highly predictive of mortality and key hallmarks of aging including mitochondrial dysfunction and cellular senescence.12
Increased epigenetic age compared to chronologic age may provide insights into physiologic aging mechanisms underlying OA. We therefore conducted the first large-scale study of epigenetic aging in OA to determine whether epigenetic age acceleration was associated with knee, hand, and multi-joint OA.
Methods
Study cohort
We assessed data from the Multicenter Osteoarthritis Study (MOST), a longitudinal study of OA risk factors in older individuals who have or are at increased risk of developing knee OA based on weight, knee symptoms, or history of knee injuries or operations.13 Starting in 2003, men and women aged 50 to 79 years were recruited from a community-based sample at two clinical centers, University of Alabama at Birmingham and University of Iowa, and have been examined repeatedly since then.13 A newly recruited cohort of 1525 individuals ages 45 to 69 years was recruited in 2016. Recruitment was conducted in the same way as the original cohort, except individuals were not selected for OA risk factors. While many of the individuals originally recruited in 2003 are now much older and have advanced OA and/or knee replacements, this newly recruited cohort focused on identifying younger individuals with no or mild disease (i.e., Kellgren-Lawrence grade (KL) 0, 1, or 2 in the worse affected tibiofemoral or patellofemoral compartment) and no constant and severe pain in either knee. We acknowledge that findings may not be generalizable to the full spectrum of disease but allows us to study an earlier stage of disease unselected for OA risk factors and before the development of major comorbidities. We excluded individuals with advanced structural disease (KL ≥ 3) or knee replacement, rheumatoid or other inflammatory arthritis, or contraindications to magnetic resonance imaging (MRI) or who were unwilling to undergo x-rays or MRIs.
For an ancillary study of epigenetic aging and OA, we enrolled a subset of 689 new cohort participants to obtain blood draws and bilateral posteroanterior (PA) hand radiographs in addition to weight-bearing fixed flexion PA and lateral knee radiographs. These individuals were assessed in the latter half of the follow-up visit for the parent study. We measured DNA methylation in peripheral blood buffy coat with the Illumina Infinium MethylationEPIC 850K array in 671 participants who consented to genotyping. We removed poorly performing probes and samples (detection p-values > 0.01) and conducted within-array and between-sample normalization using stratified quantile normalization.14
Institutional review board approvals were obtained from University of California, San Francisco, Boston University, University of Alabama at Birmingham, The University of Iowa, and Hebrew SeniorLife. All participants provided written consent for study participation.
Radiographic definitions of OA
Bilateral PA knee and hand radiographs were read by two readers for KL grade and individual radiographic features including joint space narrowing grade (JSN) and osteophyte grade (OST) with an adjudication panel composed of three readers if there was disagreement. We defined knee OA as KL ≥ 2 in one or both knees. Based on previous work to identify a hand OA phenotype that would facilitate identification of multiple joint OA cases,15 we defined radiographic hand OA based on the presence of three required criteria: (1) three or more joints with KL grade ≥ 2 where at least two are part of the same distal interphalangeal (DIP), proximal interphalangeal (PIP), or carpometacarpal (CMC) joint groups across both hands, (2) KL grade ≥ 2 in at least one DIP, and (3) bilateral involvement in at least one of the DIP, PIP, or CMC joint groups.15 We defined multi-joint OA as the presence of both hand and knee OA with no history of joint injury.16
Statistical methods
We calculated DNA methylation-based predictors of epigenetic age according to the Horvath, Hannum, PhenoAge, and GrimAge epigenetic clocks using an online calculator with default settings (https://dnamage.genetics.ucla.edu/new). Age acceleration was calculated as the residual resulting from regressing predicted DNA methylation age on chronological age. Intrinsic and extrinsic epigenetic age acceleration were calculated as the residuals from regressing predicted DNA methylation age on chronological age with and without adjustment for estimated cell proportions,17 respectively. We calculated the Pearson correlations between epigenetic age and chronological age. We conducted logistic regression to determine the association between epigenetic age acceleration and presence/absence of OA, adjusting for chronological age, sex, clinical site, smoking status, and race. According to best practices, we included chronological age in the models to account for potential residual confounding.18 In secondary analyses, using the same online calculator mentioned above, we calculated DNA methylation-based predictors of the individual proteins that compose GrimAge to determine whether there may be specific proteins that contribute to OA. We used the Box-Tidwell test to check the linearity assumption between the continuous predictors and the logit. All relationships passed the linearity test (p-values > 0.05) except for hand OA vs. PhenoAge. Transformation of PhenoAge did not affect our findings, so only results from untransformed predictors are presented here. We considered p-values < 0.05 statistically significant. All analyses were conducted in R.
Results
We analyzed a total of 671 participants with mean age (SD) of 58.4 (6.7) years. About half (53%) were women; 16% and 3% were self-reported Black and other race, respectively (Table I). Mean (SD) body mass index (BMI) was 28.3 (5.5) kg/m2. Twenty-five percent met criteria for radiographic knee OA, 23% met criteria for radiographic hand OA, and 8% met criteria for multi-joint OA. Mean predicted epigenetic age (SD) in years according to Horvath, Hannum, PhenoAge, and GrimAge epigenetic clocks was 64.9 (6.4), 68.6 (5.9), 50.5 (7.7), and 67.0 (6.2), respectively, and was higher in individuals with OA compared to those who did not have OA. Correlation between epigenetic age and chronological age ranged from 0.66 to 0.80 (Fig. 1).
Table I.
Sample characteristics.
| Hand OA (n = 155) | No hand OA (n = 516) | Knee OA (n = 165) | No knee OA (n = 501) | Multi-site OA (n = 51) | No multi-site OA (n = 615) | Total (n = 671) | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| n (%) Women | 93 (60.0) | 265 (51.4) | 96 (58.2) | 259 (51.7) | 35 (68.6) | 320 (52.0) | 358 (53.4) |
| n (%) Black race | 13 (8.4) | 97 (18.8) | 28 (17.0) | 81 (16.2) | 1 (2.0) | 108 (17.6) | 110 (16.4) |
| n (%) Other non-White race | 8 (5.2) | 15 (2.9) | 5 (3.0) | 18 (3.6) | 3 (5.9) | 20 (3.3) | 23 (3.4) |
| Mean BMI (SD) in kg/m2 | 28.2 (5.1) | 28.3 (5.6) | 29.7 (5.9) | 27.9 (5.3) | 28.9 (5.3) | 28.3 (5.5) | 28.3 (5.5) |
| n (%) Current/former Smoker | 41 (26.4) | 127 (24.6) | 39 (23.6) | 127 (25.4) | 10 (19.6) | 156 (25.4) | 168 (25.0) |
| Mean age (SD) in years | 62.6 (5.6) | 57.1 (6.4) | 59.4 (6.4) | 58.1 (6.7) | 61.9 (5.8) | 58.1 (6.6) | 58.4 (6.7) |
| Mean Horvath age (SD) in years | 68.2 (5.6) | 63.9 (6.4) | 65.9 (5.8) | 64.5 (6.6) | 67.5 (5.4) | 64.6 (6.5) | 64.9 (6.4) |
| Mean Hannum age (SD) in years | 71.1 (5.8) | 67.8 (5.7) | 69.2 (5.4) | 68.4 (6.1) | 70.9 (5.0) | 68.4 (5.9) | 68.6 (5.9) |
| Mean PhenoAge (SD) in years | 53.6 (8.1) | 49.5 (7.3) | 51.6 (7.2) | 50.1 (7.8) | 53.6 (7.2) | 50.2 (7.7) | 50.5 (7.7) |
| Mean GrimAge (SD) in years | 69.5 (6.0) | 66.3 (6.1) | 67.5 (6.2) | 66.9 (6.2) | 69.0 (5.2) | 66.9 (6.3) | 67.0 (6.2) |
Fig. 1.
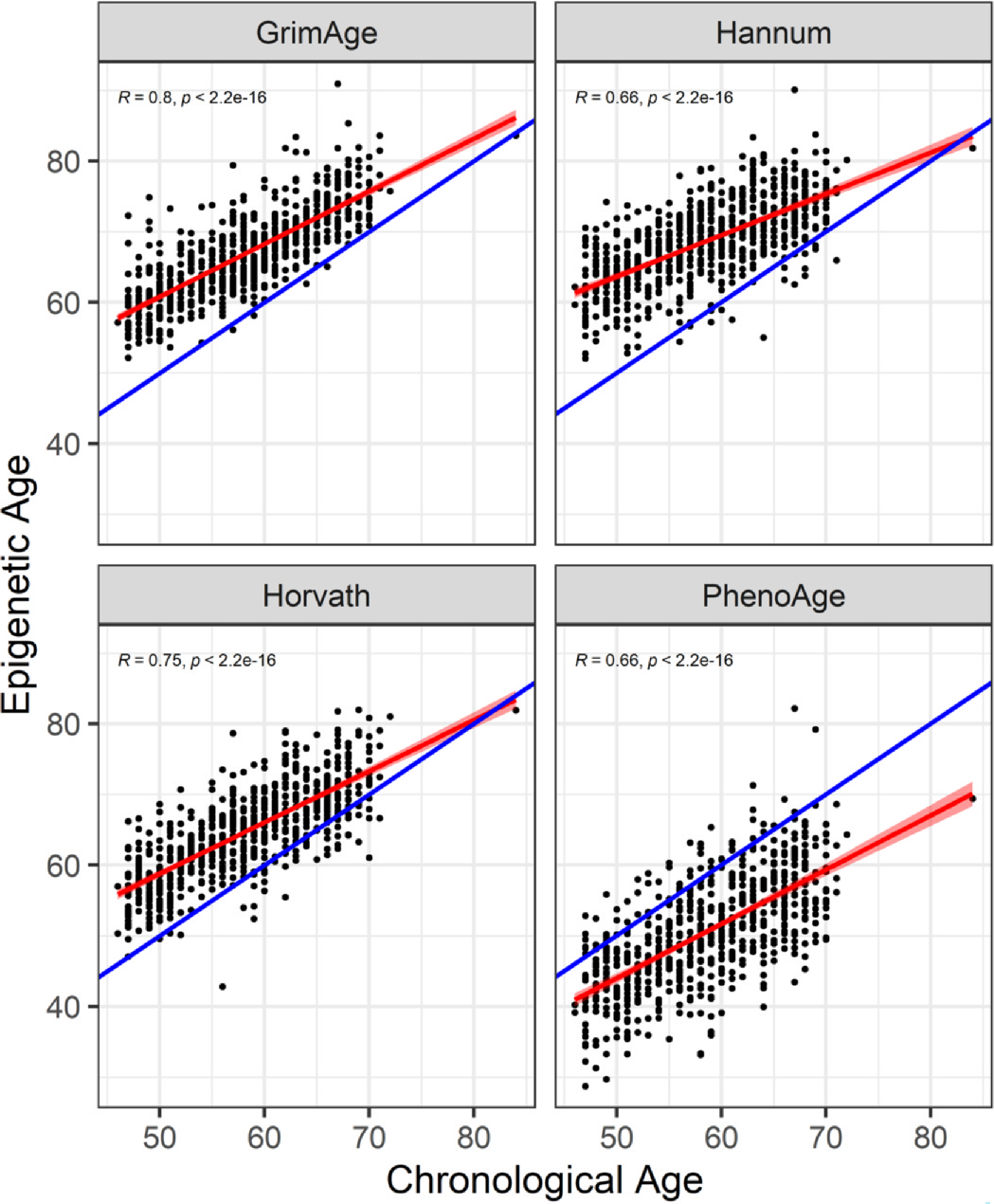
Pearson’s correlation between epigenetic age and chronological age. Scatterplots represent epigenetic age (GrimAge, Hannum, Horvath, PhenoAge) in years on the y axis and chronological age in years on the x axis along with their respective Pearson correlation coefficients and p-values. Red lines represent the best fit line and blue lines represent epigenetic age = chronological age.
After adjustment for chronological age, sex, clinical site, smoking status, and race, Horvath epigenetic age acceleration per year was associated with a 3% increased odds of hand OA, odds ratio (OR) (95% confidence intervals (CI) = 1.03 (0.99–1.08), with similar findings for knee and multi-joint OA (Fig. 2). Associations with Hannum epigenetic age acceleration were OR (95% CI) = 1.00 (0.96–1.05) for hand OA, OR (95% CI) = 1.01 (0.96–1.05) for knee OA, and OR (95% CI) = 1.02 (0.95–1.09) for multi-joint OA, with similar magnitudes of associations for PhenoAge epigenetic age acceleration. Associations with GrimAge epigenetic age acceleration were OR (95% CI) = 0.94 (0.88–1.01) for hand OA, OR (95% CI) = 0.99 (0.94–1.05) for knee OA, and OR (95% CI) = 1.02 (0.92–1.12) for multi-joint OA. Magnitudes of association between intrinsic epigenetic age acceleration and extrinsic epigenetic age acceleration were similarly small and none reached statistical significance.
Fig. 2.
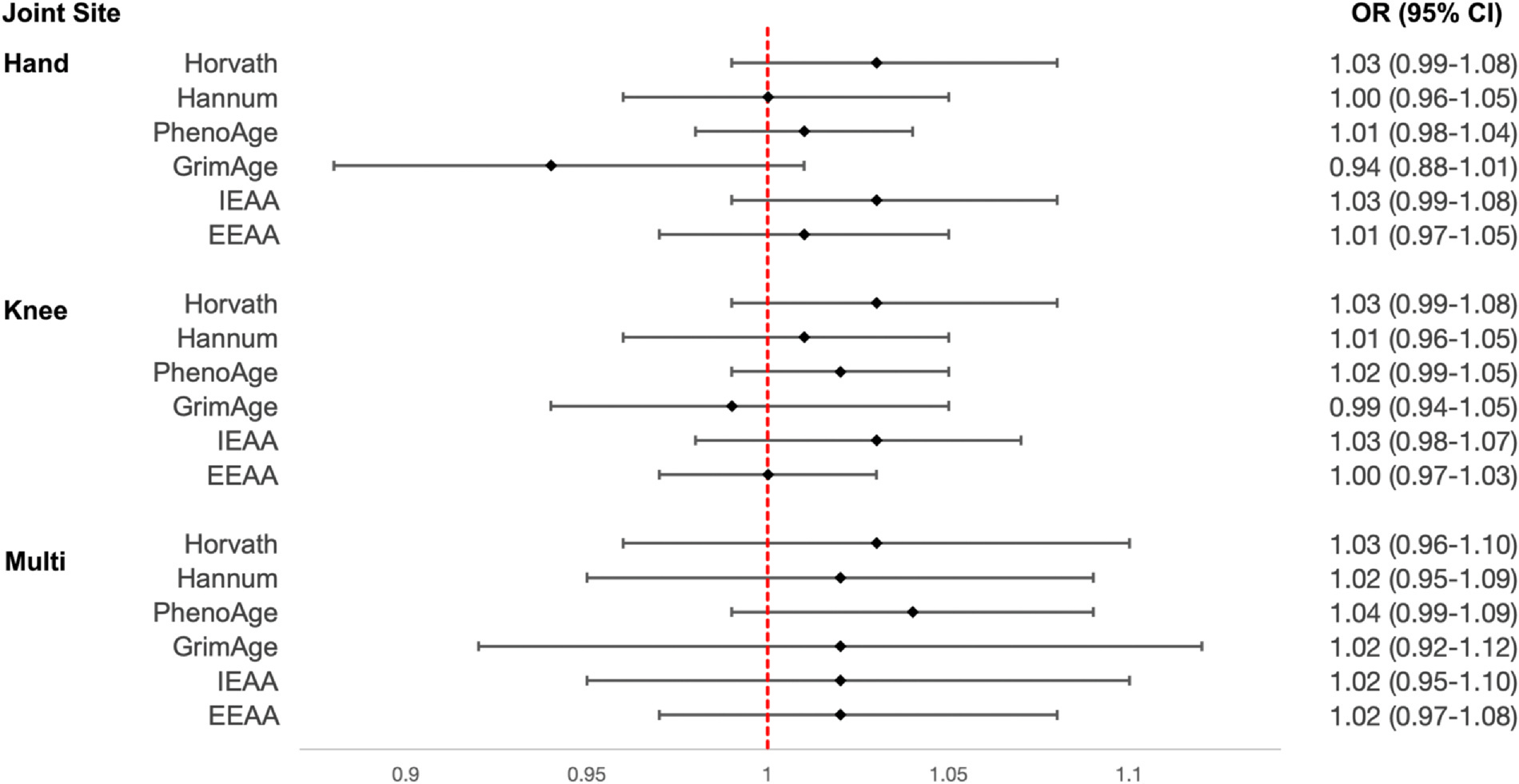
Associations between epigenetic age acceleration and hand, knee, and multi-joint OA. We calculated measures of epigenetic age acceleration based on Horvath, Hannum, PhenoAge, GrimAge epigenetic clocks, as well as intrinsic epigenetic age acceleration (IEAA) and extrinsic epigenetic age acceleration (EEAA). We then tested for associations between each of these measures (per year) and hand, knee, and multi-joint OA. All models were adjusted for chronological age, sex, clinical site, smoking status, and race. The forest plot reflects OR and corresponding 95% CI for each association.
In secondary analyses with individual predicted plasma protein biomarkers that compose GrimAge, we found a 27% increased odds of knee OA for each SD increase in PAI-1, OR (95% CI) = 1.27 (1.05–1.54) (Fig. 3). We also found a 44% increased odds of multi-joint OA for each SD increase in B2M, OR (95% CI) = 1.44 (0.95–2.14) and 28% increased odds of multi-joint OA for each SD increase in leptin, OR (95% CI) = 1.28 (0.84–1.91) (Fig. 3).
Fig. 3.
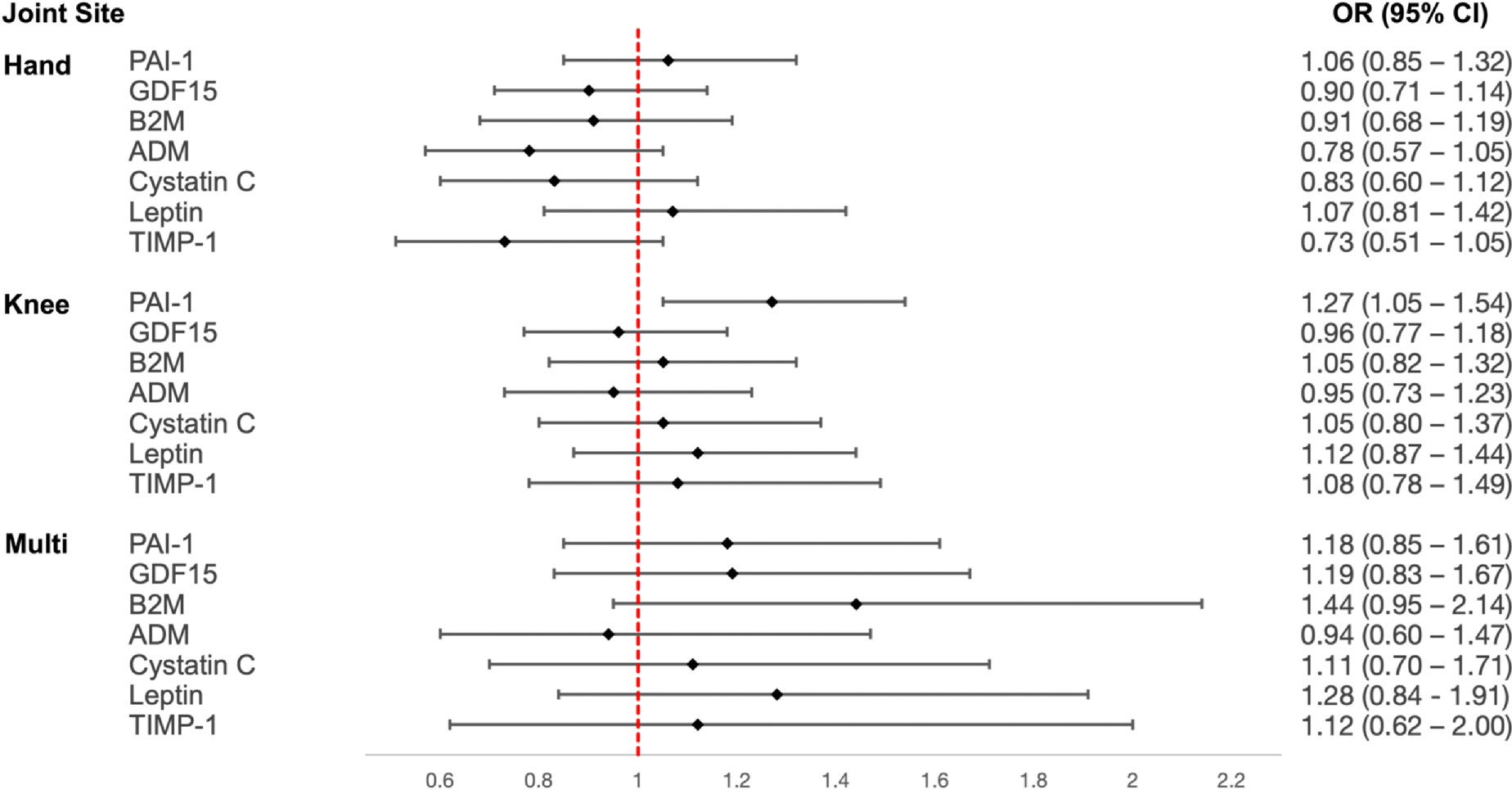
Associations between OA and GrimAge biomarkers. We calculated epigenetically predicted protein levels of PAI-1 = plasminogen activation inhibitor 1, GDF15 = growth differentiation factor 15, B2M = beta-2 microglobulin, ADM = adrenomedullin, Cystatin C, leptin, and TIMP1 = tissue inhibitor metalloproteinase 1. We then tested for associations between each of the predicted protein levels (per SD) and hand, knee, and multi-joint OA. All models were adjusted for chronological age, sex, clinical site, smoking status, and race. The forest plot reflects OR and corresponding 95% CI for each association.
Discussion
We assessed epigenetic age acceleration according to various epigenetic clocks, including Horvath, Hannum, PhenoAge, and GrimAge, and found little evidence for associations with knee, hand, or multi-joint OA independent of chronological age. We also did not find strong evidence for an association between either intrinsic or extrinsic epigenetic age acceleration and OA.
Previous studies of epigenetic age in OA bone, cartilage, and blood tissues have shown that compared to controls, OA patients have an accelerated aging of 3.7 years in cartilage. However, there is little evidence of accelerated aging in bone or blood, which were 0.04 years older and −0.6 years younger, respectively than chronological age.19 In our much larger study, we also did not find evidence for associations between epigenetic age acceleration and OA. We additionally examined multi-joint OA but all associations were null. We had hypothesized that multi-joint OA, a subtype of OA that is likely driven by systemic factors, would reveal accelerated epigenetic aging in blood. A previous study using blood from 64 OA patients showed that when compared to chronological age, the rate of epigenetic aging was decelerated by 4.9 years in rapidly progressive knee OA patients and by 0.07 years in non-progressive knee OA patients.20 The unexpected direction of effect may be attributed to the nature of the study population, which was enriched for individuals at risk for OA and could lead to bias and paradoxical effects.21,22
We tested the effects of both intrinsic and extrinsic epigenetic age acceleration because they capture different aspects of biological aging. Intrinsic epigenetic age acceleration captures cell-intrinsic properties of the aging process that are preserved across various cell types and organs. On the other hand, extrinsic epigenetic age acceleration captures age-related changes in blood cell composition and aspects of immunosenescence.9 Both measures predict all-cause mortality, with the strongest associations observed for measures of extrinsic epigenetic age acceleration. In our study, we found little evidence for associations between OA and either intrinsic or extrinsic epigenetic age acceleration. Confidence intervals were narrow and effect sizes were small, suggesting that even with a larger sample size, our results would likely be unchanged. Importantly, we assessed a subset of younger MOST participants who had no or mild knee OA and were not enriched for OA risk factors, avoiding bias that may have been present in previous studies.
GrimAge is a composite biomarker based on seven DNA methylation-based predictors of plasma proteins and smoking pack years.12 It may be superior to the other epigenetic clocks tested in its ability to predict time-to-death and time-to-comorbidities including coronary heart disease and cancer.12 While it is not clear whether OA is associated with increased mortality independent of cardiovascular disease,23,24 individual GrimAge proteins may provide insights into the relationship between OA and aging. We calculated the predicted levels of each individual GrimAge protein based on epigenetic markers and found that increased predicted levels of PAI-1 and leptin were associated with an increased odds of hand, knee, and multi-joint OA. ORs tended to be largest for multi-joint OA compared to hand and knee OA, possibly reflecting the systemic nature of the phenotype. However, except for PAI-1, no associations reached statistical significance, and this was specific to knee OA. PAI-1 inhibits plasmin and is part of the family of serpins, serine proteinase inhibitors, specifically SerpinE1.25 It is induced by pro-inflammatory cytokines,26 and responds to mechanical stress and sheer stress in OA cartilage.27,28 The biomechanical nature of knee OA may partially explain why we observed the strongest PAI-1 associations with knee OA. We also saw consistent associations between leptin and knee, hand, and multi-joint OA. Leptin plays a role in cartilage metabolism in OA,29 inducing the expression of proinflammatory factors30 and cartilage degradation through upregulation of matrix metalloproteinases.31 Leptin levels have been shown to increase the odds of hand and knee OA, partially mediating the association between adiposity and OA.32 Predicted levels of PAI-1 and leptin show that there may be specific aspects of aging, rather than aging itself, that are associated with OA.
There are a few limitations to our study. One limitation is that DNA methylation was only measured in blood. DNA methylation in blood may not necessarily reflect local effects on joint tissues but provides a global assessment of effects across all joints that may be local or systemic in nature. Cartilage and/or bone tissues have not been collected in this cohort and would typically not be possible until patients undergo end stage joint replacements. These end stage tissues may reflect different disease processes and may even be confounded by comorbidities, making validation in disease relevant tissues difficult. Functional analyses in disease-relevant tissues and replication in an independent cohort will be needed to confirm our findings and could be the focus of future studies. Additionally, our findings may not be generalizable to older individuals with severe OA but likely reflect processes at an earlier stage of disease, which is a unique feature of our cohort. Since the cohort excluded individuals with severe knee OA, the prevalence of hand OA may have been lower than expected. Despite this, 23% of individuals met criteria for radiographic hand OA, providing ample power to detect an effect. We also acknowledge that DNA methylation age is likely specific to mechanisms that maintain epigenetic stability8 and does not comprehensively reflect all aging mechanisms. For example, DNA methylation has not been shown to be associated with telomere attrition,33 though telomere shortening in leukocytes has been reported in OA.34 Finally, our study is cross-sectional, limiting causal inferences, and does not differentiate new onset from long-standing disease.
However, our study has many strengths including its large sample size in a well-characterized cohort that includes gold standard radiographic measures of knee and hand OA. Samples were collected using the same protocol, stored in the same facility, and run at the same time, which would reduce any technical artifacts that may affect results. The cohort assessed was also younger and unselected for OA risk factors, which may avoid collider bias. While we did not present race-stratified analyses, we did conduct sensitivity analyses and found that findings were similar across racial groups.
In conclusion, we found that epigenetic age acceleration as measured by various well-validated epigenetic clocks was not associated with increased risk of knee, hand, or multi-joint OA independent of chronological age. Rather, there may be specific aspects of aging processes as revealed by predicted levels of PAI-1 and leptin that increase the risk of OA.
Acknowledgments
We wish to acknowledge the contributions of the study participants, investigators, and research staff of the MOST Study. This study was supported by R01 AR075356 and P30 AR072571. MOST is comprised of four cooperative grants: U01 AG18820 David T. Felson (Boston University); U01 AG18832 James Torner (University of Iowa); U01 AG18947 Cora E. Lewis (University of Alabama at Birmingham); U01 AG19069 Michael C. Nevitt (University of California, San Francisco), funded by the National Institutes of Health, a branch of the Department of Health and Human Services, and conducted by MOST study investigators. This manuscript was prepared using data collected in MOST and does not claim, infer, or imply endorsement by MOST, by the MOST investigators and their respective institutions or by the University of California of the data recipients’ use of the data, of the entity or personnel conducting the research, or of any results of the research.
Footnotes
Competing interest statement
The authors have no relevant conflicts of interest.
CRediT authorship contribution statement
Conception and design: MSY. Data acquisition: MSY, IKH, JAL, MCN, CEL, JCT, DTF. Analysis and interpretation of the data: MSY, PCO. Drafting of the article: MSY. Critical revision of the article for important intellectual content: all authors. Final approval of the article: all authors.
References
- 1.Lawrence RC, Felson DT, Helmick CG, Arnold LM, Choi H, Deyo RA, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part II. Arthritis Rheum 2008;58:26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagge E, Bjelle A, Valkenburg HA, Svanborg A. Prevalence of radiographic osteoarthritis in two elderly European populations. Rheumatol Int 1992;12:33–8. [DOI] [PubMed] [Google Scholar]
- 3.Oberdoerffer P, Sinclair DA. The role of nuclear architecture in genomic instability and ageing. Nat Rev Mol Cell Biol 2007;8:692–702. [DOI] [PubMed] [Google Scholar]
- 4.Campisi J, Vijg J. Does damage to DNA and other macromolecules play a role in aging? If so, how? J Gerontol A Biol Sci Med Sci 2009;64:175–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loeser RF. Aging processes and the development of osteoarthritis. Curr Opin Rheumatol 2013;25:108–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, et al. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev 2009;130:234–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science 2007;315:1141–3. [DOI] [PubMed] [Google Scholar]
- 8.Horvath S DNA methylation age of human tissues and cell types. Genome Biol 2013;14:R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen BH, Marioni RE, Colicino E, Peters MJ, Ward-Caviness CK, Tsai PC, et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY) 2016;8:1844–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell 2013;49:359–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 2018;10:573–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu AT, Quach A, Wilson JG, Reiner AP, Aviv A, Raj K, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY) 2019;11:303–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Segal NA, Nevitt MC, Gross KD, Hietpas J, Glass NA, Lewis CE, et al. The Multicenter Osteoarthritis Study: opportunities for rehabilitation research. PM R 2013;5:647–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maksimovic J, Gordon L, Oshlack A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol 2012;13:R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kraus VB, Jordan JM, Doherty M, Wilson AG, Moskowitz R, Hochberg M, et al. The Genetics of Generalized Osteoarthritis (GOGO) study: study design and evaluation of osteoarthritis phenotypes. Osteoarthritis Cartilage 2007;15:120–7. [DOI] [PubMed] [Google Scholar]
- 16.Yau MS, Jonsson H, Lynch JA, Lewis CE, Torner JC, Nevitt MC, et al. Do associations with hand OA vary by knee osteoarthritis phenotype? Cross-sectional data from the Multicenter Osteoarthritis Study. Osteoarthr Cartil Open 2023;5, 100331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 2012;13, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krieger N, Chen JT, Testa C, Diez Roux A, Tilling K, Watkins S, et al. Use of correct and incorrect methods of accounting for age in studies of epigenetic accelerated aging: implications and recommendations for best practices. Am J Epidemiol 2023;192:800–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vidal-Bralo L, Lopez-Golan Y, Mera-Varela A, Rego-Perez I, Horvath S, Zhang Y, et al. Specific premature epigenetic aging of cartilage in osteoarthritis. Aging (Albany NY) 2016;8:2222–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rivas A, Andrews M, Jeffries MA. Genome-wide DNA methylation profiling of OA PBMCs reveals slowed epigenetic aging among rapid radiographic progressors: data from the Osteoarthritis Initiative (OAI). Arthritis Rheum 2017;69. [Google Scholar]
- 21.Niu J, Zhang YQ, Torner J, Nevitt M, Lewis CE, Aliabadi P, et al. Is obesity a risk factor for progressive radiographic knee osteoarthritis? Arthritis Rheum 2009;61:329–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi HK, Nguyen US, Niu J, Danaei G, Zhang Y. Selection bias in rheumatic disease research. Nat Rev Rheumatol 2014;10:403–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendy A, Park J, Vieira ER. Osteoarthritis and risk of mortality in the USA: a population-based cohort study. Int J Epidemiol 2018;47:1821–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Nguyen UDT, Lane NE, Lu N, Wei J, Lei G, et al. Knee osteoarthritis, potential mediators, and risk of all-cause mortality: data from the Osteoarthritis Initiative. Arthritis Care Res (Hoboken) 2021;73:566–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilkinson DJ. Serpins in cartilage and osteoarthritis: what do we know? Biochem Soc Trans 2021;49:1013–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Datta PK, Blake MC, Moses HL. Regulation of plasminogen activator inhibitor-1 expression by transforming growth factor-beta-induced physical and functional interactions between smads and Sp1. J Biol Chem 2000;275:40014–9. [DOI] [PubMed] [Google Scholar]
- 27.Houtman E, Tuerlings M, Riechelman J, Suchiman E, van der Wal RJP, Nelissen R, et al. Elucidating mechano-pathology of osteoarthritis: transcriptome-wide differences in mechanically stressed aged human cartilage explants. Arthritis Res Ther 2021;23:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai CH, Lee SS, Chang YC. Hypoxic regulation of plasminogen activator inhibitor-1 expression in human buccal mucosa fibroblasts stimulated with arecoline. J Oral Pathol Med 2015;44:669–73. [DOI] [PubMed] [Google Scholar]
- 29.Dumond H, Presle N, Terlain B, Mainard D, Loeuille D, Netter P, et al. Evidence for a key role of leptin in osteoarthritis. Arthritis Rheum 2003;48:3118–29. [DOI] [PubMed] [Google Scholar]
- 30.Gomez R, Scotece M, Conde J, Gomez-Reino JJ, Lago F, Gualillo O. Adiponectin and leptin increase IL-8 production in human chondrocytes. Ann Rheum Dis 2011;70:2052–4. [DOI] [PubMed] [Google Scholar]
- 31.Hui W, Litherland GJ, Elias MS, Kitson GI, Cawston TE, Rowan AD, et al. Leptin produced by joint white adipose tissue induces cartilage degradation via upregulation and activation of matrix metalloproteinases. Ann Rheum Dis 2012;71:455–62. [DOI] [PubMed] [Google Scholar]
- 32.Kroon FPB, Veenbrink AI, de Mutsert R, Visser AW, van Dijk KW, le Cessie S, et al. The role of leptin and adiponectin as mediators in the relationship between adiposity and hand and knee osteoarthritis. Osteoarthritis Cartilage 2019;27:1761–7. [DOI] [PubMed] [Google Scholar]
- 33.Marioni RE, Harris SE, Shah S, McRae AF, von Zglinicki T, Martin-Ruiz C, et al. The epigenetic clock and telomere length are independently associated with chronological age and mortality. Int J Epidemiol 2018;47:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhai G, Aviv A, Hunter DJ, Hart DJ, Gardner JP, Kimura M, et al. Reduction of leucocyte telomere length in radiographic hand osteoarthritis: a population-based study. Ann Rheum Dis 2006;65:1444–8. [DOI] [PMC free article] [PubMed] [Google Scholar]


