Abstract
Dormant Bacillus subtilis spores germinate in the presence of particular nutrients called germinants. The spores are thought to recognize germinants through receptor proteins encoded by the gerA family of operons, which includes gerA, gerB, and gerK. We sought to substantiate this putative function of the GerA family proteins by characterizing spore germination in a mutant strain that contained deletions at all known gerA-like loci. As expected, the mutant spores germinated very poorly in a variety of rich media. In contrast, they germinated like wild-type spores in a chemical germinant, a 1-1 chelate of Ca2+ and dipicolinic acid (DPA). These observations showed that proteins encoded by gerA family members are required for nutrient-induced germination but not for chemical-triggered germination, supporting the hypothesis that the GerA family encodes receptors for nutrient germinants. Further characterization of Ca2+–DPA-induced germination showed that the effect of Ca2+–DPA on spore germination was saturated at 60 mM and had a Km of 30 mM. We also found that decoating spores abolished their ability to germinate in Ca2+–DPA but not in nutrient germinants, indicating that Ca2+–DPA and nutrient germinants probably act through parallel arms of the germination pathway.
Bacillus subtilis cells form metabolically dormant spores when starved for one or more nutrients (7). In the presence of particular nutrients, called germinants, the spores break dormancy through the process of germination and, after going through outgrowth, eventually resume vegetative growth (15). It is currently believed that spores recognize nutrient germinants through receptor proteins encoded by three loci (gerA, gerB, and gerK). This hypothesis is based on genetic studies which showed that spores containing mutations at any one of these loci fail to germinate in response to specific germinants (14, 15, 19). Further support for the receptor hypothesis came from the finding that dominant mutations in gerB allow spores to germinate in novel germinants (17). The gerA, gerB, and gerK loci are each tricistronic operons encoding proteins which share significant homology across the three operons (4, 12, 25). Two of the three proteins encoded by each operon are predicted to be membrane proteins, which is again consistent with the idea that they encode germinant receptors (4, 25). The proteins, however, have been refractory to in vitro biochemical manipulation, and thus the receptor hypothesis remains untested.
Recent sequencing and genetic studies have identified members of the gerA family of operons in other endospore-forming bacteria including Bacillus cereus (3), Bacillus anthracis (9), Bacillus halodurans (23), Clostridium acetobutylicum (Genome Therapeutics Corporation), Clostridium difficile (Sanger Centre), and Clostridium pasteurianum (GenBank). However, thus far no gerA homologs have been identified in other bacterial groups. Furthermore, the B. cereus gerA operon homolog, gerI, has been implicated in inosine-induced spore germination in that organism (3). These findings are certainly consistent with the putative receptor function of GerA family proteins in spore germination.
To substantiate the receptor function of the gerA family of operons in spore germination, we have constructed a mutant B. subtilis strain that lacks all three gerA-like operons, gerA, gerB, and gerK, as well as two putative gerA homologs, yndDEF and yfkQRT, which were identified by the B. subtilis genome sequencing project (12). The quintuple-mutant spores germinated very poorly in a variety of rich media, but they germinated normally (like wild-type spores) in a nonnutrient chemical germinant, a 1-1 chelate of Ca2+ with pyridine-2,6-dicarboxylic acid (also known as dipicolinic acid, or DPA) (8). These observations support the idea that gerA-like operons encode receptors for nutrient germinants. We also characterized Ca2+–DPA-induced spore germination and show that at least one spore component appears to be uniquely required for this process. Thus, Ca2+–DPA and nutrient germinants seem to activate spore germination through independent pathways.
MATERIALS AND METHODS
Media, growth conditions, and transformation.
The Escherichia coli strains TG1 and DH5α were used for plasmid construction and grown at 37°C in LB medium (18) supplemented with 50 or 100 μg of ampicillin/ml as required. E. coli transformations were performed by the CaCl2 method (18).
The B. subtilis strains used in this study are shown in Table 1. B. subtilis strains were grown in one of four rich media, LB, 2×YT (18), nutrient broth (Difco), or 2×SG (16), or in the minimal medium TSS (5). Antibiotics (100 μg of spectinomycin/ml, 5 μg of chloramphenicol/ml, 5 μg of erythromycin/ml plus 25 μg of lincomycin/ml, 20 μg of tetracycline/ml, or 7 μg of neomycin/ml) were added as required. B. subtilis transformations were performed as described elsewhere (1).
TABLE 1.
B. subtilis strains
| Strain | Genotype (antibiotic resistance) | Sourcea |
|---|---|---|
| PS832 | Wild typeb | Laboratory stock |
| FB20 | ΔgerA::spc | pFE11→PS832 |
| FB60 | ΔgerB::cat | pFE107→PS832 |
| FB68 | ΔgerK::ermC | pFE143→PS832 |
| FB77 | ΔyndDEF::tet | pFE170→PS832 |
| FB83 | ΔyfkQRT::cat | pFE168→PS832 |
| FB84 | ΔyfkQRT::neo | pCm::Km→FB83 |
| FB61 | ΔgerA::spc ΔgerB::cat | FB60 DNA→FB20 |
| FB86 | ΔgerA::spc ΔgerK::ermC | FB68 DNA→FB20 |
| FB87 | ΔgerB::cat ΔgerK::ermC | FB68 DNA→FB60 |
| FB72 | ΔgerA::spc ΔgerB::cat ΔgerK::tet | FB68 DNA→FB61 |
| FB85 | ΔgerA::spc ΔgerB::cat ΔgerK::ermC ΔyndDEF::tet ΔyfkQRT::neo | FB77 and FB84 DNA→FB72 |
The construction and/or source of the individual plasmids is described in Materials and Methods.
The genetic background is 168, but this strain is trp+.
Preparation, cleaning, and decoating of spores and measurement of spore DPA content.
B. subtilis strains were sporulated by the resuspension method (22), as we found that different preparations of spores made by this method exhibited less variation in spore germination than did spores prepared in 2×SG medium, in particular in different batches of 2×SG medium. The spores were cleaned by daily cold-water washes over a period of 4 to 7 days and stored at 4°C in the dark (16). The cleaned spore preparations contained ≥95% phase-bright spores.
Spores were decoated as described elsewhere (24). Briefly, spore suspensions at an optical density at 600 nm (OD600) of 10 to 20 were incubated in 1 ml of 0.1 M NaOH–0.1 M NaCl–1% sodium dodecyl sulfate (SDS)–0.1 M dithiothreitol for 30 min at 70°C. The decoated spores were washed at least 10 times with distilled water to remove any traces of the decoating solution.
Spore DPA content was measured as described elsewhere (16). Ca2+–DPA treated spores were washed at least 10 times in 1 ml of distilled water prior to the assay.
Construction of multiple ger mutant strains.
All of the ger mutant constructions were designed to generate null mutations. Plasmid pFE11 was used to generate the ΔgerA::spc mutant strain (17). Plasmid pFE107 was used to construct the ΔgerB::cat strain and was derived from the ΔgerB::spc plasmid pFE106 (17) as follows. The cat marker was excised as a BamHI-SalI fragment from plasmid pDG364 (5) and cloned between the same sites in plasmid pUC18 (New England Biolabs) to generate plasmid pFE109. A BamHI-PstI fragment containing the cat marker from plasmid pFE109 was used to replace the BamHI-PstI fragment containing the spc marker in plasmid pFE106 to produce plasmid pFE107. The plasmids used to disrupt the gerK, yndDEF, and yfkQRT gene clusters were constructed as described below. The sequences of the primers used in these constructions will be provided on request.
Plasmid pFE143 was used to generate the ΔgerK::ermC strain. The macrolide-lincosamide-streptogramin B (MLS) resistance gene ermC was excised from plasmid pE194 (2) as an XbaI-ClaI fragment and cloned between the XbaI and ClaI sites in pBluescriptII SK(−) (Stratagene) to create plasmid pFE140. A DNA fragment containing the 3′ end of the gerK operon [+3796 to +4092 relative to the gerKA translation site (+1)] was PCR amplified from strain PS832 chromosomal DNA with primers ΔgerKC5 and ΔgerKC3, cleaved with EagI (site within ΔgerKC3) and XbaI (site within ΔgerKC5), and cloned between the same sites in plasmid pFE140 to generate plasmid pFE141. The 5′ end of the gerK operon (−91 to +170) was similarly amplified with primers ΔgerKN5 and ΔgerKN3, cut with XhoI (site within ΔgerKN5) and ClaI (site with ΔgerKN3), and cloned between the same sites in plasmid pFE141 to generate plasmid pFE143.
Plasmid pFE170 was used to disrupt the yndDEF gene cluster with the tetracycline resistance marker tet. The tet marker from plasmid pCm::Tc (21) was PCR amplified and cloned into the pCR2.1 vector (Invitrogen) to generate plasmid pFE149. The EagI and PstI sites flanking the insert in pCR2.1 were used to subclone the tet marker between the same sites in plasmid pBluescriptII SK(−) to create plasmid pFE152. The 5′ end of the yndDEF cluster (−147 to +92 relative to the start codon of the yndD gene) was PCR amplified from strain PS832 chromosomal DNA with primers ΔgerXN5 and ΔgerXN3, which contain HindIII and PstI sites, respectively. The PCR product was cut with HindIII and PstI and inserted between the same sites in plasmid pFE152 to generate plasmid pFE161. The 3′ end of the yndDEF cluster (+3826 to +4095) was PCR amplified with primers ΔgerXC5 and ΔgerXC3, cleaved with EagI (site within ΔgerXC5) and SstI (site within ΔgerXC3), and inserted between the same sites in plasmid pFE161 to generate plasmid pFE170.
The ΔyfkQRT::neo strain FB84 was constructed in two steps. First, a ΔyfkQRT::cat strain, FB83, was constructed using plasmid pFE168 (described below), and then the pCm::Nm plasmid (21) was used to convert the cat marker in strain FB83 to a neo marker. Plasmid pFE168 was derived from plasmid pBluescriptII SK(−) by sequentially subcloning the 5′ end of the yfkQRT gene cluster (−229 to +130 relative to the yfkQ start codon) between the HindIII and EcoRI sites, the cat gene from pFE109 (see above) between the BamHI and PstI sites, and the 3′ end of the yfkQRT cluster (+3814 to +4251) between the SpeI and NotI sites in the multiple cloning sequence. The 5′ end of the yfkQRT cluster was obtained by PCR amplification from strain PS832 chromosomal DNA with primers ΔgerYN5 and ΔgerYN3, which contain HindIII and EcoRI sites, respectively. The 3′ end of the yfkQRT cluster was PCR amplified from chromosomal DNA with primers ΔgerYC5 and ΔgerYC3, subcloned into the pCR2.1 vector, and recovered by cleavage with SpeI and NotI, whose sites flank the insert in the pCR2.1 plasmid.
Each single-mutant strain was constructed by transforming strain PS832 with a plasmid, and chromosomal integration of the plasmid by a double-crossover event was confirmed by Southern blot analysis. Multiple mutant strains were constructed as shown in Table 1.
Germination assays.
Unless otherwise noted, all spore preparations were heat activated at 70°C for 30 min prior to germination. Spore germination was assayed by measurements of the OD600 as described previously (17). Spore germination over extended times on agar plates was measured as follows. Aliquots containing 50 wild-type spores or 5,000 mutant spores were spread on 15-cm-diameter LB agar plates, which were wrapped in plastic bags to prevent drying, and incubated at 37°C. The plates were examined every 24 h for new colonies. The experiment was terminated after 8 days because the medium appeared to deteriorate by that time.
Spore germination in Ca2+–DPA was preceded by a heat shock as described above. Heat-shocked spores were diluted 10- to 50-fold in Ca2+–DPA (at 60 mM unless otherwise noted) at pH 8.0 and then incubated at room temperature. At various times aliquots were diluted 100-fold into distilled water and then heated for 1 h at 70°C, and survivors were measured as described below.
Germination over prolonged periods in broth cultures was measured as follows. Spore suspensions in water (1 OD600 unit/ml) were heat activated and diluted 10-fold into 5 ml of LB broth. Ampicillin (40 μg/ml) was added to the medium to prevent growth of germinated spores, and the samples were incubated with shaking at 37°C. At 24- to 48-h intervals, the samples were heated for 1 h at 70°C to kill any surviving germinated spores, and dilutions of a 0.1-ml aliquot were spread on LB agar plates, which were incubated for 24 hr prior to counting of colonies. Only dormant spores that germinate within the 24-h incubation on LB agar would be measured by this method. The remaining liquid culture was supplemented with ampicillin (20 μg/ml) and incubated with shaking at 37°C. The recovered colonies tested negative for ampicillin resistance, indicating that the regimen was not inadvertently selecting for antibiotic resistance.
Buoyant-density gradient separation of spores.
Mutant or wild-type spores were incubated in 60 mM Ca2+–DPA or water for 1 h at room temperature, washed with 1 ml of cold distilled water, and resuspended in a 33% metrizoic-acid solution. A step gradient was prepared in a 2.5-ml ultraclear TLS-55 centrifuge tube (Beckman, Fullerton, Calif.) by sequentially layering 0.1 ml of metrizoic acid solutions ranging in concentration from 66 to 52% in steps of 1% and from 50 to 44% in steps of 2%. The spore mixture was layered on top of the gradient and centrifuged in a TLS-55 rotor (TLA-100 ultracentrifuge) at 13,000 rpm for 45 min at 20°C with a deceleration rate setting of 8.
RESULTS
Germination of spores lacking all operons of the gerA family.
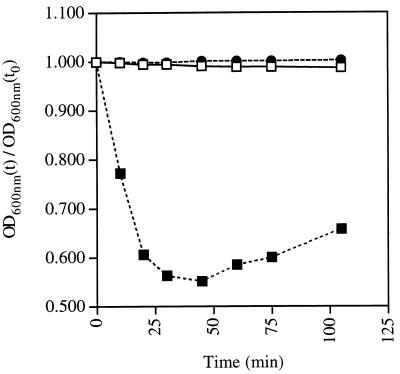
Genetic studies with B. subtilis have implicated three homologous operons, gerA, gerB, and gerK, in germinant recognition (13, 15). To better understand the role played by these operons in spore germination, we constructed a triple-mutant strain, FB72, that contained large deletions in gerA, gerB, and gerK. We also deleted two gene clusters, yndDEF and yfkQRT, from this strain because the predicted proteins encoded by these two clusters are similar to GerA family proteins (12). Although the latter two gene clusters are transcribed poorly if at all (A. Cabrera-Hernandez and P. Setlow, unpublished data, 1999), we believed that their deletion would preclude any recombinational repair of the gerA, gerB, and gerK deletions. The quintuple-mutant strain FB85 was viable and sporulated like the wild-type strain PS832. Spores prepared from the quintuple-mutant strain and the wild-type strain were assayed spectrophotometrically for germination in 2×YT medium (Fig. 1). As expected, the OD600 of the wild-type spore suspension dropped rapidly by 40%, indicating efficient and complete spore germination. In contrast, the OD600 of the quintuple-mutant spore suspensions remained unchanged even after 100 min (Fig. 1), suggesting that those spores did not germinate. This conclusion was supported by phase-contrast microscopy, which showed that the mutant spores (>99%) remained refractile even after 100 min in the germination medium. The germination defect was also observed in 2×SG medium, LB medium, and nutrient broth, suggesting that it was not specific to 2×YT medium. Thus, spores lacking all gerA family members were severely compromised for germination in a variety of rich media.
FIG. 1.
Germination of wild-type and quintuple-mutant spores in LB broth. Spores of the wild-type strain PS832 (squares) and the mutant strain FB85 (circles) were heat activated and incubated in water (open symbols) or 2×YT medium plus 5 mM l-alanine (solid symbols) at 37°C. The OD600 of the suspensions was monitored over time. There was no difference between the behavior of mutant and wild-type spores in water, and only one curve is shown.
Although OD600 measurement provides a rapid assay for spore germination, it is not a very sensitive method. Consequently, we used a colony formation assay to determine if a small percentage of the mutant spores germinated in rich medium. Wild-type and mutant spore suspensions were heat activated, adjusted to a concentration of 1 OD600 unit/ml (about 108 spores/ml when counted microscopically), and plated on LB agar. After overnight incubation at 37°C, the wild-type spore titers ranged between 0.7 × 108 and 1.3 × 108 CFU/ml/OD600 unit. Surprisingly, the quintuple-mutant spore titers ranged from 2 × 104 to 4 × 105 CFU/ml/OD600 unit, showing that 0.02 to 0.4% of the mutant spores germinated in 24 h. Treatment of the quintuple-mutant spores with detergent (1% SDS) or heat (70°C for 1 h) did not abolish the appearance of colonies (data not shown), suggesting that the colonies could not be attributed to vegetative cells or immature sporangia in the spore suspension. The percentage of germinating spores from strain FB85 was also too large to be accounted for by genetic reversion. Indeed, when the colonies that were obtained from quintuple-mutant spores were themselves sporulated, the resulting spores expressed the mutant germination phenotype (data not shown), demonstrating that the colonies were not the result of genetic reversion.
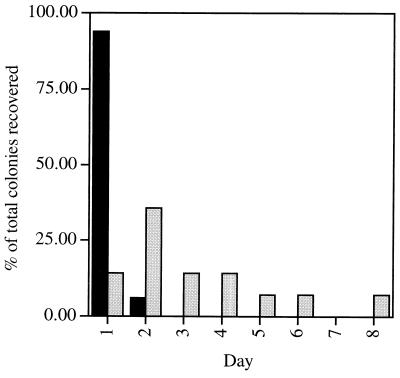
The small number of colonies that arose from the quintuple-mutant spores could represent either a small fraction of the population that had a tendency to germinate in rich medium or the summation over time of stochastic germination events in the spore population. In the latter case, the number of colonies recovered would be expected to increase upon longer incubation, whereas this would not be the situation in the former case. To distinguish between these two possibilities, dilutions of wild-type and mutant spore suspensions were spread on agar plates (15 cm), such that each plate would contain approximately 50 wild-type spores or 5,000 mutant spores. The plates were incubated at 37°C for a total of 8 days, and the number of new colonies that appeared in each 24-h period was determined. As expected, >97% of the wild-type colonies appeared within the first 2 days (Fig. 2). In contrast, new colonies on the mutant plates appeared at a relatively constant rate over the 8 days of the experiment (Fig. 2). These results indicate that stochastic germination events in the spore population were most likely responsible for the germination of the quintuple-mutant spores. Similar results were obtained when TSS minimal medium was used, indicating that the germination events did not require complex mixtures of nutrients.
FIG. 2.
Germination of spores over long periods on solid medium. Approximately 50 wild-type strain PS832 spores (solid bars) or 5,000 mutant strain FB85 spores (shaded bars) were spread on 15-cm LB agar plates and incubated at 37°C for 8 days. The number of new colonies that appeared every 24 h was recorded and expressed as a percentage of the total number of colonies recovered over the 8 days of the experiment (33 wild-type colonies and 14 mutant colonies). Similar data were obtained with two different batches of spores.
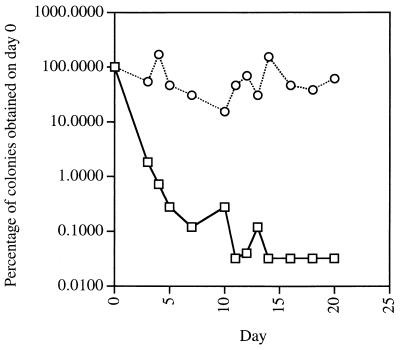
To confirm that the appearance of colonies from mutant spores was due to infrequent germination events and not delayed outgrowth, we performed a modification of the above experiment using liquid medium. Identical dilutions of wild-type and mutant spores were incubated in LB broth at 37°C, and the medium was supplemented with ampicillin (40 μg/ml) to kill any spores that germinated. At 24- to 48-h intervals, the sample was heated at 70°C for 1 h to kill surviving germinated spores, an aliquot was diluted and plated on LB agar, and the plates were incubated for 24 h at 37°C to determine the number of dormant spores that germinated during that particular 24-h period. In this experiment, the wild-type-spore titers dropped by 2 orders of magnitude within the first 3 days (Fig. 3), showing that less than 1% of those spores remained dormant after 3 days in LB medium. In contrast, the mutant-spore titers remained at a constant level over a period of 20 days (Fig. 3), demonstrating that the spores were breaking dormancy at a relatively constant rate over the entire duration of the experiment.
FIG. 3.
Germination of spores over long periods in liquid medium. Spores (0.1 OD600 unit/ml) from the wild-type strain PS832 (□) and the mutant strain FB85 (○) were inoculated into 5 ml of LB medium supplemented with ampicillin (40 μg/ml) and incubated at 37°C. An aliquot was withdrawn every 24 to 48 h for 20 days, heated at 70°C for 1 h, diluted, plated on LB agar, and incubated at 37°C for 24 h prior to colony counting. The titer (expressed as a percentage of the titer on day 0, which was 1.25 × 108 CFU/ml/OD600 unit for wild-type spores and 5 × 105 CFU/ml/OD600 unit for mutant spores) was plotted against the period of incubation in liquid medium.
Contribution of each gerA-like operon toward the quintuple-mutant spore phenotype.
As noted above, spores that lack all gerA-like operons germinate at a very low frequency in rich medium. To determine the contribution of each gerA family member toward this phenotype, we assessed the germination of spores from various single-, double-, and triple-mutant strains by analyzing colony formation on LB agar after a 12- to 16-h incubation at 37°C. In this assay, the quintuple-mutant spore titers were 4 × 104 CFU/ml/OD600 unit compared to wild-type spore titers of 1.3 × 108 CFU/ml/OD600 unit (Table 2). The gerA gerB gerK triple-mutant spores behaved identically to the quintuple-mutant spores (Table 2), suggesting that the products of these three operons were primarily responsible for spore germination in LB medium. The triple-mutant-spore phenotype was not seen in gerA gerB, gerA gerK, or gerB gerK spores and thus required the absence of all three operons (Table 2). Nevertheless, gerA gerK spores and gerA gerB spores gave titers that were 100- and 5-fold lower than those of wild-type spores (Table 2), respectively, suggesting that those double-mutant spores were compromised for germination in rich medium. These observations showed that the products of gerA, gerB, and gerK operons play major and probably overlapping roles in nutrient-induced spore germination in the media tested and that the predicted proteins encoded by yndDEF and yfkQRT play a minor role, if any, in this process.
TABLE 2.
Overnight colony formation by spores of various strainsa
| Mutant genotype (strain) | Titer (CFU/ml/OD600 unit)b |
|---|---|
| Wild type (PS832) | 1.3 × 108 |
| gerA (FB20) | 5.4 × 107 |
| gerB (FB60) | 1.6 × 108 |
| gerK (FB68) | 1.4 × 108 |
| gerA gerB (FB61) | 3.0 × 107 |
| gerA gerK (FB86) | 1.2 × 106 |
| gerB gerK (FB87) | 1.4 × 108 |
| gerA gerB gerK (FB72) | 4.0 × 104 |
| gerA gerB gerK yndDEF yfkQRT (FB85) | 4.0 × 104 |
Spores of various strains were heat activated in water and plated on LB agar, and colonies were counted after overnight incubation at 37°C.
Mean of two experiments. The titers from the two experiments were within 40% of each other, except in the case of spores from strains FB72 and FB85, where the titers varied 10-fold.
Effect of chemical germinants on quintuple-mutant spores.
Although our observations suggested that the low titers of the quintuple-mutant spores reflected infrequent germination, we wanted to confirm that the phenotype was not simply due to poor spore viability. For this purpose, we asked if the mutant spores could germinate in a previously described nonnutrient chemical germinant, a 1-1 chelate of Ca2+ and DPA, whose action might be independent of the germinant receptors (8). Quintuple-mutant-spore suspensions were heat activated, incubated at 1 OD600 unit/ml in water or freshly prepared 50 mM Ca2+–DPA (pH 8.0) for 45 min at room temperature, diluted, and plated on LB agar at 37°C. Whereas untreated spores had a titer of 8 × 104 CFU/ml/OD600 unit, Ca2+–DPA-treated quintuple-mutant-spore titers (3 × 107 to 5 × 107 CFU/ml/OD600 unit) were similar to those of wild-type spores (1 × 108 CFU/ml/OD600 unit), demonstrating that the viability of quintuple-mutant spores was comparable to that of wild-type spores. (The small difference between the mutant and wild-type spore titers vanishes at higher [Ca2+–DPA], as described below.) Thus, the low titers of the quintuple-mutant spores in rich medium are due to a defect in their germination rather than to lower spore viability.
Although Ca2+–DPA had been previously reported to trigger spore germination, some reports suggested that it might simply activate spores for germination (8). Because spore activation alone would not easily explain the ability of Ca2+–DPA to rescue the quintuple-mutant-spore phenotype, our observations were more consistent with Ca2+–DPA actually triggering spore germination. To confirm this possibility, we examined the effect of Ca2+–DPA treatment on spore refractility and buoyant density, both of which change during germination (15, 20). Heat-activated quintuple-mutant spores were treated with either 60 mM Ca2+–DPA or water for 1 h at room temperature, washed, layered on metrizoic-acid gradients (44 to 66%), and subjected to buoyant-density gradient centrifugation. Whereas the water-treated spores migrated as a single band in the 63 and 64% metrizoic-acid layers, the Ca2+–DPA-treated spores formed three distinct bands (labeled I, II, and III from the top of the gradient). Band I lay in the 44% metrizoic-acid layer and contained a majority of the spores (>60%), band II was less intense and spanned the 52 to 53% metrizoic-acid layers, and band III was located in the 63 to 64% metrizoic-acid layers and was very faint (about 10% of the spores). Phase-contrast microscopy showed that the spores in band I were phase dark, demonstrating that Ca2+–DPA was sufficient to trigger germination of quintuple-mutant spores. Interestingly, the spores in band II appeared phase bright (most of the spores were not as bright as dormant spores and could be more appropriately described as phase gray [15]) and might represent an intermediate stage in spore germination, while band III contained phase-bright spores which had probably not initiated germination.
Characterization of Ca2+–DPA-induced spore germination.
The ability of Ca2+–DPA to trigger the germination of quintuple-mutant spores suggested that this chelate could activate the germination pathway independently of the GerA family receptors. To determine if this mechanism was also present in wild-type spores, we repeated the buoyant-density gradient centrifugation experiment described above with wild-type spores. Wild-type spores treated with 60 mM Ca2+–DPA for 1 h formed three bands (I, II, and III) which were identical to those formed by Ca2+–DPA-treated mutant spores with respect to buoyant density, relative proportions, and spore refractility. Thus, Ca2+–DPA also induced germination in wild-type spores.
Nutrient-induced spore germination is accompanied by loss of spore heat resistance and excretion of DPA from the spore core (20). To determine if Ca2+–DPA treatment was sufficient to bring about a loss of spore heat resistance, wild-type and quintuple-mutant spores were treated with Ca2+–DPA at pH 8.0, diluted in water, incubated at either room temperature or 70°C for 1 h, diluted, and plated on LB agar. As shown in Table 3, only 7% of the Ca2+–DPA-treated wild-type spores survived heat treatment compared to 70% of the water-treated spores. The quintuple-mutant spores showed an even lower survival rate after heating than did wild-type spores, with less than 0.1% of the Ca2+–DPA-treated spores surviving heat treatment (Table 3). The reason for the much higher heat sensitivity of the Ca2+–DPA-treated quintuple-mutant spores is not known. One trivial possibility is that this simply reflects a higher level of germination of the mutant spores in this experiment compared to that of wild-type spores. However, both wild-type and quintuple-mutant spores germinate to similar extents in Ca2+–DPA as measured by other criteria, including density gradient centrifugation, phase-contrast microscopy, and DPA release (see above and below). Another possibility is that the much larger loss in heat resistance of the quintuple-mutant spores incubated in Ca2+–DPA reflects both spore germination and some deactivation of the Ca2+–DPA sensing system upon continued incubation in Ca2+–DPA, as this latter effect would lower the viability of quintuple-mutant spores but not that of wild-type spores. Clearly, further work is needed to clarify this issue.
TABLE 3.
Effect of Ca2+–DPA treatment on the heat resistance of wild-type and mutant sporesa
| Genotype (strain) and germination medium | Titer (CFU/ml/OD600 unit)b of spores
|
|
|---|---|---|
| Unheated | Heated | |
| Wild type (PS832) | ||
| Water | 6.5 × 107 | 4.5 × 107 |
| Ca2+–DPA | 6.5 × 107 | 4.5 × 106 |
| ger mutantc (FB85) | ||
| Water | 2.1 × 105 | 8 × 105 |
| Ca2+–DPA | 5.7 × 107 | 2.4 × 104 |
Spores were heat activated, incubated in water or 60 mM Ca2+–DPA for 60 min at room temperature, either heated (at 70°C for 1 h) or not heated, plated on LB agar, and incubated overnight at 37°C prior to colony counting.
The titers represent the means of two different experiments, and the values in these two experiments were within 40% of each other. Similar values were also obtained using two different preparations of spores.
The ΔgerA::spc ΔgerB::cat ΔgerK::ermC ΔyndDEF::tet ΔyfkQRT::neo mutant was used.
Dormant spores contain a large depot of DPA which is excreted during germination (20). To examine the effect of Ca2+–DPA on spore DPA content, wild-type and quintuple-mutant spores were treated at pH 8.0 with 60 mM Ca2+–DPA for 1 h at room temperature, harvested, washed, and assayed for DPA content. Whereas untreated wild-type and mutant spores contained 27 and 25 μg of DPA/OD600 unit, respectively, this content was reduced sevenfold (3.5 μg/OD600 unit) after Ca2+–DPA treatment, showing that Ca2+–DPA triggered DPA excretion. Together, these observations show that Ca2+–DPA is sufficient to induce a number of changes that are associated with nutrient-triggered spore germination, including loss of spore refractility, reduction in buoyant density, loss of heat resistance, and DPA excretion.
Requirements for Ca2+–DPA-triggered spore germination.
The effect of Ca2+–DPA on quintuple-mutant spores could be due to Ca2+ alone or DPA alone, or it might require both Ca2+ and DPA. To dissect the specifics of the requirements for Ca2+–DPA-induced germination, we assayed spore germination after incubation at pH 8.0 in either 50 mM CaCl2, 50 mM DPA, or 50 mM Ca2+–DPA (50 mM CaCl2 plus 50 mM DPA). Spore germination in 50 mM CaCl2 was comparable to that seen in water (Table 4), suggesting that calcium ions by themselves did not support the germination of the mutant spores. Spores incubated in 50 mM DPA showed titers 20-fold higher than those of the controls, but these titers were only 5 to 7% of those seen in spores incubated in 50 mM Ca2+–DPA (Table 4). Thus, DPA by itself triggered some spore germination, but the full effect was seen only in the presence of both Ca2+ and DPA. Because spores from some strains of Bacillus megaterium germinate in mixtures of DPA with ions other than Ca2+ (8), we tested if Mg2+ or K+ could substitute for Ca2+ in Ca2+–DPA. However, no significant (less than twofold) changes in spore germination were observed in Mg2+–DPA or K+–DPA compared to spore germination in DPA alone (Table 4).
TABLE 4.
Requirements for Ca2+–DPA-triggered germination of quintuple-mutant sporesa
| Germination medium | Titer (CFU/ml/OD600 unit)b |
|---|---|
| Water | 6 × 104 |
| 50 mM Ca2+–DPA | 2.2 × 107 |
| 50 mM CaCl2 | 5 × 104 |
| 50 mM DPA | 1.4 × 106 |
| 50 mM Mg2+–DPA | 8 × 105 |
| 50 mM K+–DPAc | 3 × 106 |
Spores of strain FB85 were heat activated, incubated in various germination media for 1 h at room temperature, plated on LB agar, and incubated overnight at 37°C prior to colony counting.
The titers represent the means of two different experiments, and the values in these two experiments were within 40% of each other. Similar values were also obtained using two different preparations of spores.
Identical results were obtained when [K+] was adjusted to 100 or 50 mM.
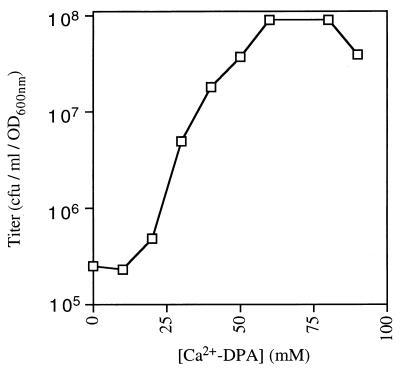
We also determined the relationship between Ca2+–DPA concentration and the efficacy of spore germination using the colony formation assay with quintuple-mutant spores. Spore suspensions were heat activated, incubated at 1 OD600 unit/ml in varying concentrations of Ca2+–DPA for 1 h at room temperature, and then spread on LB agar plates which were incubated overnight at 37°C prior to counting of the colonies. Strikingly, the triggering of spore germination exhibited a sigmoidal response to [Ca2+–DPA]; at concentrations below 20 mM, Ca2+–DPA had little effect on the spore titers, followed by a logarithmic increase in spore titers with [Ca2+–DPA], and a plateau at Ca2+–DPA concentrations beyond 60 mM (Fig. 4). (The lower titers observed at 90 mM Ca2+–DPA were probably caused by precipitation of the chelate.) The maximum titer of the mutant spores, 9 × 107 CFU/ml/OD600 unit, was comparable to that of wild-type spores (about 1 × 108 CFU/ml/OD600 unit). These observations showed that the effect of Ca2+–DPA was saturable at 60 mM, with an apparent Km, the [Ca2+–DPA] which produced a half-maximal spore titer (4.8 × 107 CFU/ml/OD600 unit), of 30 mM.
FIG. 4.
Concentration dependence of Ca2+–DPA-induced germination. Mutant spores from strain FB85 were heat activated, incubated in various concentrations of Ca2+–DPA for 1 h at room temperature, plated on LB agar, and incubated overnight at 37°C prior to colony counting. The titer of germinated spores was plotted against [Ca2+–DPA].
The sigmoidal nature of the dependence of spore germination on [Ca2+–DPA] suggested that Ca2+–DPA triggered germination by allosterically activating some spore component. Previously, such an allosteric effect of DPA has been observed on autoprocessing of the P46 zymogen of the germination protease to the active P41 form (10). Because the spore component that responds to Ca2+–DPA in stimulating spore germination would have to be accessible to externally applied Ca2+–DPA, we suspected that it might be located in a spore integument layer, such as the spore coat, cortex, or inner membrane. Thus, we investigated if spores stripped of their coats by hot-detergent treatment retained their ability to germinate in Ca2+–DPA. Strikingly, decoated quintuple-mutant spores gave similar low titers with or without 60 mM Ca2+–DPA treatment in the overnight germination assay (Table 5). In contrast, decoating did not affect the titer of water-treated quintuple-mutant spores, demonstrating that decoating did not simply reduce spore survival (Table 5). Similar results were obtained with gerA gerB gerK triple-mutant spores, showing that this effect was not due to the ΔyndDEF::tet or ΔyfkQRT::neo mutation. Thus, our findings suggest that the decoating treatment damages or removes at least one component required for Ca2+–DPA-induced spore germination.
TABLE 5.
Effect of decoating of quintuple-mutant spores on their Ca2+–DPA-triggered germinationa
| Spore treatment and germination medium |
Titer (CFU/ml/OD600 unit)b |
|---|---|
| No treatment | |
| Water | 2.1 × 105 |
| Ca2+–DPA | 5.7 × 107 |
| Decoating | |
| Water | 2.8 × 105 |
| Ca2+–DPA | 2.7 × 105 |
Decoated or untreated spores of strain FB85 were heat activated, incubated in water or 60 mM Ca2+–DPA for 1 h at room temperature, plated on LB agar, and incubated overnight at 37°C prior to colony counting. Eliminating the heat activation step did not alter the results.
The titers represent the means of two different experiments, and the values in these two experiments were within 40% of each other. Similar values were also obtained using two different preparations of spores.
To determine if decoating also abolished Ca2+–DPA-triggered spore germination in wild-type spores, we assessed the germination of decoated wild-type spores in Ca2+–DPA by phase-contrast microscopy. (The colony formation assay could not be used to measure germination because wild-type spores and decoated wild-type spores germinate well on rich medium.) In contrast to untreated wild-type spores, most of which (>60%) became phase dark after 1 h in 60 mM Ca2+–DPA, the decoated wild-type spores remained phase bright, suggesting that decoating interfered with Ca2+–DPA-induced germination in wild-type spores. This conclusion was tested by subjecting the spores to buoyant-density gradient centrifugation. No upward shift was seen in the Ca2+–DPA-treated decoated spores, which, like the water-treated controls, formed a single band at 64 to 65% metrizoic acid. In the same experiment, nondecoated spores migrated into the 44% metrizoic-acid layer after Ca2+–DPA treatment as described above. These observations show that the decoating treatment either extracts or irreversibly damages a spore component, presumably protein, that is essential for Ca2+–DPA-induced spore germination. Interestingly, the decoating treatment had no significant effect on the germination of wild-type spores in rich medium as measured by loss of OD600 and colony-forming ability (data not shown), indicating that the nutrient-induced spore germination pathway remained intact in these decoated spores. However, it is known that loss of individual spore coat proteins through mutation can have some, often subtle, effects on spore germination (6).
DISCUSSION
Bacterial spores, although metabolically inert, respond to nutrient-rich environments by breaking dormancy (20). Previous studies have suggested that spores detect nutrient germinants through specific receptors (11, 13), and a group of homologous operons, typified by the B. subtilis gerA operon, has been identified as likely encoding the putative receptors (15). In this study, we showed that B. subtilis spores lacking all five gerA-like operons had a severe defect in nutrient-induced germination. This defect was specific to nutrient germinants, as evidenced by the fact that the quintuple ger mutant spores germinated normally in the chemical germinant Ca2+–DPA. This phenotype is exactly what would be expected of spores that lack the proposed nutrient receptors, and therefore, our observations support the proposal that the gerA operon and its homologs encode germinant receptors.
Three gerA homologs, gerA, gerB, and gerK, have been genetically identified in B. subtilis, as mutations in each of these operons block germination in response to distinct nutrient germinants (14). Deletion of all three operons in the same strain eliminates the ability of spores from that strain to respond to nutrient germinants, suggesting that these three operons contain the genes that are primarily responsible for nutrient sensing. On the other hand, deletion of two gene clusters, yndDEF and yfkQRT, whose predicted products are homologous to the GerA family proteins (12) had no observable effect on the germination phenotype of the gerA gerB gerK triple-mutant spores, suggesting that the products of the former clusters do not contribute significantly to nutrient-triggered spore germination. This suggestion is consistent with the minimal (if any) expression of the yndDEF and ytkQRT clusters during sporulation (Cabrera-Hernandez and Setlow, unpublished data, 1999).
Besides nutrients, certain nonnutrient chemicals also trigger spore germination (8). However, the mechanism by which chemical germinants activate spore germination has not been investigated in great detail; indeed, there is some ambiguity as to whether chemical germinants simply activate spores for subsequent germination or actually trigger spore germination (8). Our studies show that the chemical Ca2+–DPA is sufficient to trigger spore germination, as assessed by changes in spore heat resistance, DPA content, refractility, and buoyant density. This effect of Ca2+–DPA was seen in the quintuple-mutant spores, showing that it does not require the GerA family of germinant receptors. Thus, Ca2+–DPA might either activate an effector that lies downstream of the germinant receptors or stimulate a parallel arm of the germination pathway. We favor the latter possibility because our studies suggest that at least one spore component, which is sensitive to decoating treatment, is uniquely required for Ca2+–DPA-induced spore germination. However, we still need to rule out the possibility that decoating simply makes the spore less permeable to Ca2+–DPA. Further studies aimed at identification of the spore component (or components) that is required for Ca2+–DPA-triggered germination should help clarify this issue.
An unexpected finding from this study was that mutant spores which lack all gerA-like operons can germinate in rich medium, albeit at a very low frequency. This finding can be explained in one of two ways. First, spores might have a very low efficiency receptor system that is distinct from the gerA family. Although the identity of this receptor system is not known, it must be distinct from the Ca2+–DPA-induced germination pathway because it is not affected by decoating. Alternatively, stochastic activation of effectors, for example, cortex-lytic enzymes or ion or water channels, that might act downstream from the germinant receptors (8, 11) could be responsible for the infrequent germination events. These two possibilities could be distinguished by examining the occurrence of the infrequent germination events in water, because stochastic activation events would be expected to take place even in distilled water. However, we have not been able to test this prediction because of the difficulty of detecting rare germinated spores in a population of dormant spores. Nevertheless, we favor the latter explanation because, as would be expected for chance events, a large variation was observed in titers of the same batch of mutant spores on rich medium (compare Tables 4 and 5). Such an interpretation also fits well with the finding that the germination of the quintuple-mutant spores continues to occur in a minimal medium.
In conclusion, our studies support the hypothesis that the GerA family proteins function as nutrient receptors. We also characterized Ca2+–DPA-induced germination in B. subtilis spores and show that Ca2+–DPA and nutrient germinants probably act through independent arms of the germination pathway. In addition to extending previous work regarding nutrient and nonnutrient triggering of spore germination, our findings suggest new strategies for genetic analysis of nutrient-triggered germination. For example, Ca2+–DPA might provide a means to recover mutant spores which could not be recovered thus far because they carried mutations that completely eliminated nutrient-triggered germination.
ACKNOWLEDGMENTS
We thank members of this laboratory for their suggestions and criticisms.
This work was supported by grant GM19698 from the National Institutes of Health.
REFERENCES
- 1.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:74–76. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bron S. Plasmids. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, England: John Wiley & Sons Ltd.; 1990. pp. 75–174. [Google Scholar]
- 3.Clements M O, Moir A. Role of the gerI operon of Bacillus cereus 569 in the response of spores to germinants. J Bacteriol. 1998;180:6729–6735. doi: 10.1128/jb.180.24.6729-6735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corfe B M, Sammons R L, Smith D A, Mauël C. The gerB region of the Bacillus subtilis 168 chromosome encodes a homologue of the gerA spore germination operon. Microbiology. 1994;140:471–478. doi: 10.1099/00221287-140-3-471. [DOI] [PubMed] [Google Scholar]
- 5.Cutting S M, Vander Horn P B. Genetic analysis. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, England: John Wiley & Sons Ltd.; 1990. pp. 27–74. [Google Scholar]
- 6.Driks A. The Bacillus subtilis spore coat. Microbiol Mol Biol Rev. 1999;63:1–20. doi: 10.1128/mmbr.63.1.1-20.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gould G W. Germination. In: Gould G W, Hurst A, editors. The bacterial spore. New York, N.Y: Academic Press; 1969. pp. 397–444. [Google Scholar]
- 9.Guidi-Rontani C, Pereira Y, Ruffie S, Sirard J C, Weber-Levy M, et al. Identification and characterization of a germination operon on the virulence plasmid pXO1 of Bacillus anthracis. Mol Microbiol. 1999;33:407–414. doi: 10.1046/j.1365-2958.1999.01485.x. [DOI] [PubMed] [Google Scholar]
- 10.Illades-Aguiar B, Setlow P. Autoprocessing of the protease that degrades small, acid-soluble proteins of spores of Bacillus species is triggered by low pH, dehydration, and dipicolinic acid. J Bacteriol. 1994;176:7032–7037. doi: 10.1128/jb.176.22.7032-7037.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnstone K. The trigger mechanism of spore germination: current concepts. J Appl Bacteriol Symp Suppl. 1994;76:17S–24S. doi: 10.1111/j.1365-2672.1994.tb04354.x. [DOI] [PubMed] [Google Scholar]
- 12.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 13.Moir A, Kemp E H, Robinson C, Corfe B M. The genetic analysis of bacterial spore germination. J Appl Bacteriol Symp Suppl. 1994;76:9S–16S. [PubMed] [Google Scholar]
- 14.Moir A, Lafferty E, Smith D A. Genetic analysis of spore germination mutants of Bacillus subtilis 168: the correlation of phenotype and map location. J Gen Microbiol. 1979;111:165–180. doi: 10.1099/00221287-111-1-165. [DOI] [PubMed] [Google Scholar]
- 15.Moir A, Smith D A. The genetics of bacterial spore germination. Annu Rev Microbiol. 1990;44:531–553. doi: 10.1146/annurev.mi.44.100190.002531. [DOI] [PubMed] [Google Scholar]
- 16.Nicholson W L, Setlow P. Sporulation, germination, and outgrowth. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, England: John Wiley & Sons Ltd.; 1990. pp. 391–450. [Google Scholar]
- 17.Paidhungat M, Setlow P. Isolation and characterization of mutations in Bacillus subtilis that allow spore germination in the novel germinant d-alanine. J Bacteriol. 1999;181:3341–3350. doi: 10.1128/jb.181.11.3341-3350.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 19.Sammons R L, Moir A, Smith D A. Isolation and properties of spore germination mutants of Bacillus subtilis 168 deficient in the initiation of germination. J Gen Microbiol. 1981;124:229–241. [Google Scholar]
- 20.Setlow P. Biochemistry of bacterial forespore development and spore germination. In: Levinson H S, Tipper D J, Sonenshein A L, editors. Sporulation and germination. Washington, D.C.: American Society for Microbiology; 1981. pp. 13–28. [Google Scholar]
- 21.Steinmetz M, Richter R. Plasmids designed to alter the antibiotic resistance expressed by insertion mutations in Bacillus subtilis, through in vivo recombination. Gene. 1994;142:79–83. doi: 10.1016/0378-1119(94)90358-1. [DOI] [PubMed] [Google Scholar]
- 22.Sterlini J M, Mandelstam J. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J. 1969;113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takami H, Nakasone K, Hirama C, Takaki Y, Masui N, et al. An improved physical and genetic map of alkaliphilic Bacillus sp. C-125. Extremophiles. 1999;3:21–28. doi: 10.1007/s007920050095. [DOI] [PubMed] [Google Scholar]
- 24.Vary J C. Germination of Bacillus megaterium spores after various extraction procedures. J Bacteriol. 1973;116:797–802. doi: 10.1128/jb.116.2.797-802.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuberi A R, Moir A, Feavers I M. The nucleotide sequence and gene organization of the gerA spore germination operon of Bacillus subtilis 168. Gene. 1987;51:1–11. doi: 10.1016/0378-1119(87)90468-9. [DOI] [PubMed] [Google Scholar]