Abstract
Salmonella enterica serovar Typhimurium peptidase E (PepE) is an N-terminal Asp-specific dipeptidase. PepE is not inhibited by any of the classical peptidase inhibitors, and its amino acid sequence does not place it in any of the known peptidase structural classes. A comparison of the amino acid sequence of PepE with a number of related sequences has allowed us to define the amino acid residues that are strongly conserved in this family. To ensure the validity of this comparison, we have expressed one of the most distantly related relatives (Xenopus) in Escherichia coli and have shown that it is indeed an Asp-specific dipeptidase with properties very similar to those of serovar Typhimurium PepE. The sequence comparison suggests that PepE is a serine hydrolase. We have used site-directed mutagenesis to change all of the conserved Ser, His, and Asp residues and have found that Ser120, His157, and Asp135 are all required for activity. Conversion of Ser120 to Cys leads to severely reduced (104-fold) but still detectable activity, and this activity but not that of the parent is inhibited by thiol reagents; these results confirm that this residue is likely to be the catalytic nucleophile. These results suggest that PepE is the prototype of a new family of serine peptidases. The phylogenetic distribution of the family is unusual, since representatives are found in eubacteria, an insect (Drosophila), and a vertebrate (Xenopus) but not in the Archaea or in any of the other eukaryotes for which genome sequences are available.
Peptide-hydrolyzing enzymes are required for the complete degradation of proteins to free amino acids and for the utilization of peptides as nutrient sources. Salmonella enterica serovar Typhimurium contains at least 14 peptidases with various substrate specificities, ensuring that these two processes are carried out fully (12). Four of these peptidases (peptidases N, A, B, and D) have a broad substrate specificity, and previous studies have shown that a mutant lacking all four enzymes is unable to completely degrade intracellular proteins to free amino acids (19). These broad-specificity peptidases do not, however, hydrolyze all peptides equally well. For example, none of the four can hydrolyze X-Pro peptides (where X can be any amino acid). Specialized enzymes (peptidases P and Q) that specifically hydrolyze X-Pro peptides have evolved to carry out this function (11). Only one of the broad-specificity enzymes, peptidase B, is able to hydrolyze Asp-X peptides (Z. Mathew, T. M. Knox, and C. G. Miller, submitted for publication), and cells contain an additional enzyme, peptidase E, that is specific for Asp-X dipeptides (5).
Peptidase E was first identified as an activity capable of hydrolyzing Asp-X peptides and was subsequently shown to have specificity for aspartyl dipeptides (5). It does not hydrolyze Glu-X, Asn-X, Gly-X, or any other nonaspartyl peptide that has been tested (5). Peptidase E requires a free N terminus and does not cleave N-blocked peptides. The only other Asp-X-specific peptidases are the caspases, which are endoproteases that hydrolyze an internal Asp-X peptide bond (8, 16). The aspartyl dipeptide specificity of peptidase E is therefore unique among characterized peptidases.
Little is known about the mechanism of action of peptidase E or about the structural basis for its specificity. Peptidase E is not sensitive to inhibitors of the major classes of peptidases (6), and its sequence does not clearly place it in any known class of enzymes (13). No structural motifs that could identify peptidase E as a member of a known family have been recognized. Peptidase E, unlike many Salmonella peptidases, lacks any apparent metal binding sites that would classify it as a metalloprotease. These observations suggest that peptidase E may define a new class of peptide hydrolases. As sequences from other organisms have become available, peptidase E homologs have been identified. Alignment of these sequences has revealed several highly conserved regions, one of which contains a Gly-X-Ser-X-Gly motif typical of serine hydrolases, suggesting that peptidase E may utilize a serine residue as a nucleophile. The results of site-directed mutagenesis experiments aimed at testing the hypothesis that peptidase E is a serine peptidase and at identifying the amino acid residues required for catalysis are presented.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All Salmonella serovar Typhimurium strains listed in Table 1 are derivatives of strain LT2. Escherichia coli DH5α was routinely used for DNA cloning experiments, with the exception of site-directed mutagenesis (described below), in which BMH71-18 mutS was used (Clontech). Strains were typically grown at 37°C in Lennox broth (L broth) (Gibco BRL). To supplement auxotrophy, leucine and proline were added at 0.3 and 2.0 mM, respectively. When peptides were used as leucine sources, strains were grown on E minimal medium (17) supplemented with 0.4% glucose and 0.3 mM peptide. For the growth of plasmid-containing strains, ampicillin was added to the medium at 50 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmida | Relevant genotype | Source or reference |
|---|---|---|
| Strains | ||
| S. enterica serovar Typhimurium | ||
| TN1246 | leuBCD485 pepN90 pepA16 pepB11 supQ302 Δ(proAB pepD) pepP1 pepQ1 | Laboratory collection |
| TN2540 | metE551 metA22 ilv-452 trpB2 hisC527 galE496 xyl-404 rpsL120 flaA66 hsdL6 hsdSA29 (r− m+) | Laboratory collection |
| TN2718 | leuBCD485 pepN90 pepA16 pepB11 supQ302 Δ(proAB pepD) pepP1 pepQ1 pepE8::MudJ | Laboratory collection |
| TN2719 | leuBCD485 pepE8::MudJ | Laboratory collection |
| TN5213 | leuBCD485 pepB11 pepN90 pepA16 supQ302 Δ(proAB pepD) pepP1 pepQ1 pepE8::MudJ/pCM389 | This study |
| TN5415 | leuBCD485 pepB11 pepN90 pepA16 supQ302 Δ(proAB pepD) pepP1 pepQ1 pepE8::MudJ/pCM441 | This study |
| TN5416 | leuBCD485 pepE8::MudJ/pCM441 | This study |
| E. coli | ||
| DH5α | Φ80lacZΔM15 Δ(lacZYA)U169 endA1 hsdR17 (rK− mK−) supE44 thi-1 recA1 gyrA96 relA1 | Gibco BRL |
| BMH71-18 mutS | thi supE Δ(lac-proAB) (mutS::Tn10) (F′ proAB lacIqZΔM15) | Clontech |
| B. subtilis DB104 | his | R. Switzer |
| Plasmids | ||
| pSE380 | Cloning vector with the ptrc inducible promoter; Ampr | 2 |
| pCM247 | 2.6-kb fragment containing pepE and flanking sequence in pBluescript II KS(+); Ampr | 6 |
| pCM389 | pepE from serovar Typhimurium cloned into the EcoRI and BamHI restriction sites of pSE380 | This study |
| pCM440 | pepE from B. subtilis cloned into the EcoRI and BamHI restriction sites of pSE380 | This study |
| pCM441 | pepE from X. laevis cloned into the EcoRI and BamHI restriction sites of pSE380 | This study |
Plasmids carrying mutated pepE are listed in Table 2, where the mutation is also indicated.
Analysis of sequence data.
Searches for sequences similar to the peptidase E sequence were carried out with the BLAST program (1) to search the GenBank database maintained by the National Center for Biotechnology Information. In addition to the nonredundant GenBank database, the dbest database (expressed sequence tag database) and the unfinished microbial genome database were also searched. Drosophila melanogaster peptidase E was found in dbest. Several sequences were found in the unfinished microbial genome database and were accessed from the contributing sites as follows: Deinococcus radiodurans and Shewanella putrefaciens from The Institute for Genomic Research; Pasteurella multocida from the University of Minnesota Computational Biology Center; and Actinobacillus actinomycetemcomitans from the University of Oklahoma Advanced Center for Genome Technology. Sequence alignments were constructed with the ClustalW algorithm.
Available GenBank accession numbers for peptidase E sequences are as follows: S. enterica serovar Typhimurium, P36936; E. coli, P32666; Haemophilus influenzae, P44766; Xenopus laevis, AAC59869; and D. melanogaster, AA536396 and AI260864.
DNA manipulations.
DNA manipulations were performed by standard techniques (10). DNA fragments were prepared for cloning by agarose gel extraction with a Qiaquick gel extraction kit (Qiagen). Restriction enzymes were either from Gibco BRL or from New England Biolabs. Taq polymerase and T4 DNA ligase were from Gibco BRL, and shrimp alkaline phosphatase was from Amersham Pharmacia Biotech. DNA sequencing was carried out on a Perkin-Elmer ABI 377A automated DNA sequencer at the W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois.
Plasmids were transferred between serovar Typhimurium strains with the generalized transducing phage P22 HT 12/4 int-3 (15).
Site-directed mutagenesis.
Oligonucleotide-directed mutagenesis was performed essentially as described for the Transformer Site-Directed Mutagenesis Kit (Clontech). The selection primer, PstI/XhoI, was used with each of the mutagenic primers, all of which are listed in Table 2. A pepE plasmid, pCM247 [pBluescript II KS(+) with a 2.6-kb serovar Typhimurium genomic DNA insert containing pepE and two adjacent genes], was used as the template (6). Mutated plasmids were screened for loss of a PstI site and gain of an XhoI site by corresponding restriction endonuclease digestion. Plasmids that were found to have acquired an XhoI site were then screened for pepE mutations by DNA sequencing. In the course of these experiments, we found that strains carrying pCM247 are not stable upon repeated subculturing. Each pepE mutant was therefore amplified by PCR with primers PepE5 and PepE6 (Table 2) and cloned into EcoRI/BamHI-digested pSE380 to produce plasmids in which pepE transcription is controlled by an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter. The presence of each mutation in the subclones was confirmed by DNA sequencing. Wild-type peptidase E and mutant peptidase E expressed from these pSE380 constructs were found to be completely stable, and strains containing these plasmids (Table 2) were analyzed for peptidase E activity. The instability of pCM247-derived plasmids presumably results from the overexpression of one of the genes adjacent to the peptidase E gene on pCM247 and not from the overexpression of peptidase E itself. Plasmids were electroporated into TN2540 and finally transduced into TN2719 for phenotypic analysis. The expression of peptidase E mutants was induced by growing each plasmid-containing strain overnight in L broth-ampicillin plus 1 mM IPTG. Soluble cell extracts of each were prepared by sonication and subsequent centrifugation. Activity was determined by measuring the rate of aspartyl p-nitroanilide (Asp-pNA) (Bachem) hydrolysis as described below.
TABLE 2.
Oligonucleotides used in this study
| Method | Primer | Sequencea | Plasmidb |
|---|---|---|---|
| PCR | PepE5 | GGAATTCTTTCCTGATGAGGTGGAGAA | |
| PepE6 | CGGGATCCAATATGGCCTGGGATTAA | ||
| BsPepE1 | GAGAATTCGGCGGGTGTGATGATGAAGCAGAT | ||
| BsPepE2 | CGGGATCCTTGATTTCGCACTCATCTTATCAT | ||
| Sequencing | PepE1 | CGTCTCTAAACGTGTTAAAT | |
| PepE3 | CAATATGGCCTGGGATTA | ||
| PepE4 | ATCCGTGTACTCGTCCCATGTT | ||
| pSE380 | AATTGTGAGCGGATAAC | ||
| Mutagenesis | PstI/XhoI | GATATCGAATTCCTCGAGCCCGGG | |
| PepE-S7A | GCTTTTATTAGCTAACTCGACGC | pCM442 | |
| PepE-S9A | GCTTTTATTAAGTAACGCGACGCTGCCGGG | pCM461 | |
| PepE-S120A | CGGCTGGGCCGCGGGCGCC | pCM437 | |
| PepE-S120C | CGGCTGGTGCGCGGGCGCC | pCM456 | |
| PepE-H19A | GCCTGGCTGGAAGCCGCTCTGCCGCTG | pCM447 | |
| PepE-H19Q | CTGGCTGGAACAAGCTCTGCCGC | pCM443 | |
| PepE-H68A | GTCACAGGGATTGCTCGTGTCGCCGATC | pCM448 | |
| PepE-H157A | GCAAATTAATCCTGCCTTCACTAACGCC | pCM439 | |
| PepE-H157Q | GCAAATTAATCCTCAATTCACTAACGCC | pCM457 | |
| PepE-H166A | CTTGCCGGAAGGTGCTAAGGGCGAGACC | pCM465 | |
| PepE-H166Q | GCCGGAAGGTCAAAAGGGCGAGAC | pCM466 | |
| PepE-D47A | CGCAAACATGGGCCGAGTACACGG | pCM450 | |
| PepE-D135A | GCACGACCAACGCTATGCCGATCG | pCM438 | |
| PepE-E172A | GAGACCCGCGCGCAGCGTATTC | pCM458 | |
| PepE-K95A | CCAGCTACTGGCAGAATCGCGTG | pCM462 | |
| PepE-K171E | GGGCGAGACCGAGGAGCAGCG | pCM453 | |
| PepE-R174A | CCGCGAGCAGGCTATTCGCGAAC | pCM454 | |
| PepE-R174E | CCGCGAGCAGGAGATTCGCGAAC | pCM455 |
The relevant mutagenized codon or restriction site is underlined.
Plasmids are derivatives of pCM389, which carries wild-type pepE.
Cloning of X. laevis pepE and Bacillus subtilis ygaJ.
A cDNA clone of gene D2, the X. laevis pepE gene, carried on pBluescript was kindly provided by D. Brown (18). With restriction enzymes EcoRI and XhoI, an 800-bp fragment including the pepE open reading frame was excised and cloned into the EcoRI/XhoI sites of vector pSE380. The resulting plasmid, pCM441, was then electroporated into serovar Typhimurium strain TN2540, from which it was transduced into TN2719 and TN2718 to make TN5416 and TN5415, respectively.
B. subtilis gene ygaJ was amplified by PCR from strain DB104 (kindly provided by R. Switzer) with primers BsPepE1 and BsPepE2 (Table 2). The 750-bp product was cloned into the EcoRI/BamHI sites of pSE380 to produce pCM440, which was then electroporated into TN2540 and transduced into TN2719 and TN2718. Each of the two resulting plasmids, pCM441 and pCM440, was sequenced with primer pSE380 (Table 2) and confirmed to carry the correct sequence.
Strain TN2718 carrying either pCM441 or pCM440 was used to determine whether or not expression from the plasmid would allow growth on peptides. Cells were spread on E minimal medium containing 0.4% glucose, 2 mM proline, and 1 mM IPTG. Crystals of either leucine or the following leucine-containing peptides were added to the surface of the medium to supplement the leucine auxotrophy: Asp-Leu, Glu-Leu, Lys-Leu, Tyr-Leu, Thr-Leu, and Asn-Leu.
Purification of serovar Typhimurium peptidase E.
Strain TN5213 was grown overnight in L broth supplemented with 50 μg of ampicillin per ml and 1 mM IPTG. Cells were harvested, washed with 0.1 M Tris-Cl (pH 7.5), and disrupted by sonication. The soluble fraction was subjected to anion-exchange chromatography on a Q Sepharose HiLoad 26/10 column at a flow rate of 4 ml/min and eluted with a linear gradient of 0 to 1.0 M NaCl in 20 mM Tris-Cl (pH 7.5) (in a total volume of 1,000 ml). Asp-Leu-hydrolyzing activity, which eluted at 0.2 M NaCl, was detected by the amino acid oxidase assay (5). Fractions containing this activity were pooled and concentrated with Centriprep 10 concentrator units (Amicon). After equilibration in 20 mM Tris-Cl (pH 7.5)–150 mM NaCl, the sample was subjected to gel filtration chromatography in the same buffer with a Superose 12 HR 10/30 column at a flow rate of 0.5 ml/min. Peptidase E eluted at a molecular mass of 25 kDa and was concentrated and desalted before chromatography on a DEAE-Sepharose Fast Flow 10/10 column at 2 ml/min. A linear gradient of 0 to 1.0 M NaCl in 20 mM Tris-Cl (pH 7.5) (total volume, 200 ml) was used for elution, and the active fractions were pooled, concentrated, and equilibrated in the gel filtration buffer. Material from the DEAE-Sepharose step was subjected to an additional gel filtration chromatography step on a Superose 12 HR 10/30 column at a flow rate of 0.5 ml/min and eluted with 20 mM Tris-Cl (pH 7.5)–150 mM NaCl.
All chromatography analyses were performed on a fast protein liquid chromatography system (Amersham Pharmacia Biotech). Protein concentrations were determined by the Coomassie dye binding assay with Coomassie Plus protein reagent (Pierce) and bovine serum albumin as the standard.
Purification of X. laevis peptidase E.
X. laevis peptidase E was purified from strain TN5416 essentially as described for serovar Typhimurium peptidase E but without the final gel filtration step. Asp-Leu-hydrolyzing activity eluted at approximately 0.1 M NaCl from the Q Sepharose column, at a predicted molecular mass of 40 kDa from the Superose 12 column, and at 0.08 M NaCl from the DEAE-Sepharose column.
SDS-PAGE.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was performed as described by Schaggar and von Jagow (14), typically with 10% separating and 4% stacking gels. Molecular weight standards were Mark 12 wide-range protein standards (Novex).
Assays of peptidase E activity.
Peptidase E activity was measured by determining the rate of Asp-pNA hydrolysis (6). The reaction mixture contained either purified enzyme or crude cell extract in 0.1 M Tris-Cl (pH 7.5) and 1 mM Asp-pNA (from a 50 mM stock solution in dimethylformamide). The rate of p-nitroaniline release was measured as an increase in the A410 on a Thermomax microplate reader (Molecular Devices) at room temperature for 30 min. When assays were done with protease inhibitors, enzyme and inhibitor were allowed to incubate for 15 min prior to the addition of substrate. The following inhibitors, which were obtained from Sigma, were used: 3,4-dichloroisocoumarin, l-trans-epoxysuccinyl-leucylamide-(4-guanido)-butane (E-64), and N-ethylmaleimide (NEM).
Hydrolysis of other peptides was carried out at 37°C with 0.1 mM Tris-Cl (pH 7.5), 1 mM substrate (or substrate concentrations of 0.02 to 1.0 mM and 0.06 to 1.0 mM for Km determinations of the X. laevis and S. typhimurium enzymes, respectively), and either 30 ng of purified serovar Typhimurium peptidase E or 400 ng of purified X. laevis peptidase E. Aliquots were removed from the reaction mixture and precipitated in 5% trichloroacetic acid (50-μl aliquot added to 5 μl of 50% trichloroacetic acid). Each supernatant was then derivatized as described by Carter (4) with 2,4,6-trinitrobenzenesulfonic acid (TNBS) (Pierce). Briefly, 20 μl of the reaction mixture was combined with 80 μl of 5% borax (1 g of sodium borate in 20 ml of 0.1 N KOH) and 0.25 mM isoleucine as the internal standard for chromatography. To derivatize the peptide and amino acids, 4 μl of TNBS (0.05% in H2O) was added, mixed well, and incubated for 5 min at room temperature. To stop the reaction by acidification, 5 μl of 6 N HCl was added to each tube and mixed well. The mixture was then diluted to a 1-ml final volume by adding 891 μl of a solution containing 95% of 0.1% trifluoroacidic acid (TFA) (Pierce) in H2O and 5% of 0.1% TFA in acetonitrile (Fisher). The resulting trinitrophenyl derivatives were analyzed by reversed-phase high-pressure liquid chromatography with a C18 Ultrasphere column (Beckman Instruments). The buffers used for chromatography were 0.1% TFA in H2O as the starting buffer and a 0 to 100% gradient of 0.1% TFA in acetonitrile for elution.
Western blot analysis.
Plasmid-containing strains were grown overnight in L broth containing 50 μg of ampicillin per ml and 1 mM IPTG. An aliquot of cells was then combined with 2× sample loading buffer (0.125 M Tris-Cl [pH 6.8], 4% SDS, 20% glycerol, 0.2 M dithiothreitol, 0.02% bromophenol blue) and heated to 100°C for 5 min before being subjected to Tris-Tricine-SDS-PAGE. Protein was transferred from the gel to polyvinylidene difluoride by semidry electroblotting (Hoefer), and blots were incubated sequentially in 3% bovine serum albumin, rabbit serum containing anti-peptidase E antibody at a 1/1,000 dilution, and anti-rabbit immunoglobulin G–alkaline phosphatase conjugate (Sigma) at a 1/1,000 dilution. Alkaline phosphatase activity was detected with Western Blue reagent (Promega). Rabbit anti-peptidase E antibody was raised against purified serovar Typhimurium peptidase E at the University of Illinois Immunological Resource Center.
RESULTS
Comparison of serovar Typhimurium peptidase E to homologs from other organisms.
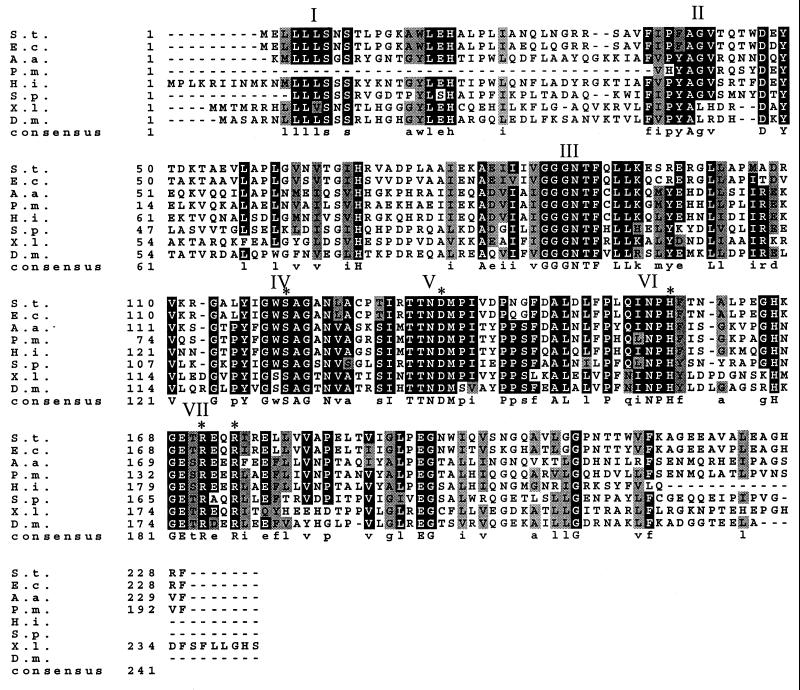
Figure 1 shows an alignment of sequences similar to serovar Typhimurium peptidase E. The organisms from which these sequences are derived and their percent identity and percent similarity (in parentheses) to serovar Typhimurium peptidase E are as follows: E. coli (89 and 92), H. influenzae (46 and 63), A. actinomycetemcomitans (42 and 59), S. putrefaciens (39 and 59), partial sequence from P. multocida (44 and 59), X. laevis (42 and 55), and D. melanogaster (43 and 60).
FIG. 1.
Multiple alignment of the amino acid sequences of the peptidase E family. The sequence alignment was determined with ClustalW, and Boxshade 3.1 was used to shade positions at which 70% of the sequences are identical (black) or similar (gray). The residues that are essential for enzyme activity (Ser120, Asp135, His157, Arg171, and Arg174) are indicated with asterisks. Blocks of conserved residues are labeled with roman numerals for clarity when mentioned in the text. Abbreviations are as follows: S.t., S. enterica serovar Typhimurium; E.c., E. coli; A.a., A. actinomycetemcomitans; P.m., partial sequence from P. multocida; H.i., H. influenzae; S.p., S. putrefaciens; X.l., X. laevis; and D.m., D. melanogaster.
A Blast search also found two more distantly related sequences, one from B. subtilis (37% identity and 48% similarity) and one from Deinococcus radiodurans (32% identity and 50% similarity). A plasmid expressing the B. subtilis pepE homolog (ygaJ), pCM440, was constructed, and its gene product was assayed for peptide-hydrolyzing activity. No in vitro hydrolysis of Asp-pNA was observed. In addition, this strain was no better than the parent strain in its ability to grow on X-Leu peptides (Asp-Leu, Glu-Leu, Lys-Leu, Tyr-Leu, Thr-Leu, and Asn-Leu) as sources of leucine. The B. subtilis gene product is not similar to the other peptidase E sequences in the C-terminal region, which contains two arginine residues that are essential for catalysis, as discussed below. Based on these observations, the B. subtilis ygaJ gene product sequence and the sequence from D. radiodurans, which is even less similar to serovar Typhimurium peptidase E, were not included in the alignment. Although the sequence from S. putrefaciens is 39% identical to that of serovar Typhimurium peptidase E, which is not significantly more similar than the B. subtilis homolog, it contains most of the strongly conserved residues, including the two essential arginines, and was therefore included in the alignment.
From the sequence alignment shown in Fig. 1, it is apparent that all the members of the peptidase E family are approximately the same size and that none of them has N- or C-terminal extensions or lengthy divergent regions. There are at least seven conserved blocks of sequence, the first being a leucine-rich block at the N terminus (region I). Inspection of the conserved blocks led to the identification of a potential serine protease motif, Gly-X-Ser-X-Gly, in region IV. Hypothesizing that peptidase E may therefore be a serine protease, we also identified conserved blocks containing an aspartate (region V) and a histidine (region VI) which, together with serine, could comprise a catalytic triad typical of serine proteases.
Analysis of purified X. laevis peptidase E.
In order to verify that the alignment in Fig. 1 includes sequences for enzymes with similar activities, we chose one of the proteins most distantly related to serovar Typhimurium peptidase E, that of X. laevis, for further characterization. The putative X. laevis pepE gene (called gene D in reference 3) was cloned into cloning vector pSE380 under the control of an inducible promoter (ptrc) and expressed in a multiply peptidase-deficient serovar Typhimurium strain (TN5415). The enzyme was purified from extracts of this strain (Fig. 2 and Table 3), and its activity and substrate specificity were compared to those of purified serovar Typhimurium peptidase E. As shown in Table 4, both serovar Typhimurium peptidase E and X. laevis peptidase E hydrolyze several aspartyl dipeptides, including Asp-Pro. Both enzymes also hydrolyze the chromogenic substrate Asp-pNA. In addition, both hydrolyze the tripeptide Asp-Gly-Gly at approximately 50% the rate for Asp-Leu. Asp-Gly-Gly is the smallest aspartyl tripeptide, which may explain why this peptide is a substrate for peptidase E, whereas other tripeptides are not. No significant activity was detected toward any larger tripeptides, such as Asp-Leu-Gly. In addition, no hydrolysis of any non-aspartyl peptides was observed for either serovar Typhimurium or X. laevis peptidase E. The apparent Michaelis constant for each enzyme toward Asp-Leu was determined to be approximately 0.3 mM. However, the kcat/Km of serovar Typhimurium peptidase E for Asp-Leu (2.08 × 103 M−1 s−1) is 10-fold higher than that of the X. laevis enzyme (0.24 × 103 M−1 s−1). These results indicate that X. laevis peptidase E is a strict Asp-X peptidase with a specificity very similar to that of serovar Typhimurium peptidase E.
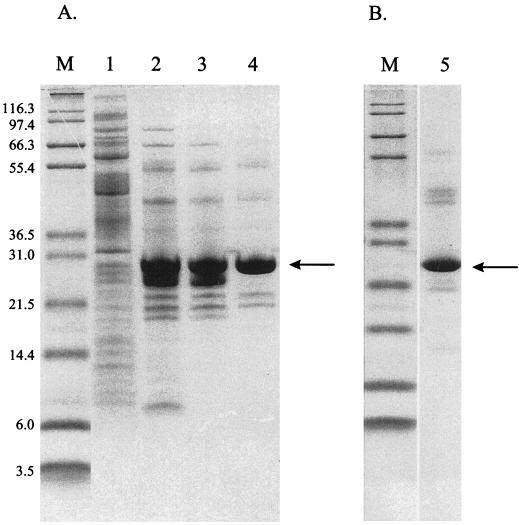
FIG. 2.
Peptidase E purification. (A) X. laevis peptidase E was purified from soluble cell extracts of strain TN5415 as described in Materials and Methods. Samples from each step in the purification were analyzed by SDS-PAGE and loaded as follows: crude cell extract (lane 1), pooled Q Sepharose fractions (lane 2), pooled Superose fractions (lane 3), and pooled DEAE fractions (lane 4). (B) Serovar Typhimurium peptidase E, which was purified as described in Materials and Methods, was subjected to SDS-PAGE and loaded in lane 5. Molecular mass markers were loaded in lanes labeled “M,” and their masses (in kilodaltons) are noted to the left of the gels. The band on each gel corresponding to peptidase E is indicated with an arrow.
TABLE 3.
X. laevis peptidase E purification
| Fraction | Total vol (ml) | Total protein (mg/ml) | Activity (nmol of pNA min−1) μg−1) | Total U (nmol of pNA min−1) (105) | Yield | Purification (fold) |
|---|---|---|---|---|---|---|
| Crude | 15.0 | 93.0 | 0.02 | 7.7 | 100 | |
| Q Sepharose | 0.5 | 0.9 | 1.00 | 3.2 | 42 | 43 |
| Superose | 1.0 | 0.5 | 1.13 | 1.8 | 23 | 49 |
| DEAE | 0.3 | 0.3 | 1.40 | 1.3 | 17 | 60 |
TABLE 4.
Comparison of the peptide-hydrolyzing activities of peptidase E from serovar Typhimurium and X. laevis
| Peptide | % Relative activity of peptidase E from:
|
|
|---|---|---|
| Serovar Typhimurium | X. laevis | |
| Asp-Leu | 100a | 100b |
| Asp-His | 100 | 129 |
| Asp-Pro | 4 | 11 |
| Asp-Gly-Gly | 56 | 40 |
| Asp-Leu-Gly | 0 | 0 |
| Asp-Leu-Lys | 0 | NDc |
| Asp-Leu-Phe | 0 | ND |
| Glu-Leu | 0 | 0 |
| Asn-Leu | 0 | 0 |
| Lys-Leu | 0 | 0 |
| Asp-pNAd | + | + |
One hundred percent is 186 nmol of Asp released min−1 mg−1.
One hundred percent is 115 nmol of Asp released min−1 mg−1.
ND, not determined.
Asp-pNA hydrolysis was determined by observing yellow color from released pNA (+).
Although the activities of peptidase E from these two different organisms are very similar, the surface-exposed residues must be quite different. Cross-reactivity of polyclonal antibody raised against serovar Typhimurium peptidase E was barely detectable toward X. laevis peptidase E, as determined by Western blotting (data not shown).
Site-directed mutagenesis of potential active-site of peptidase E.
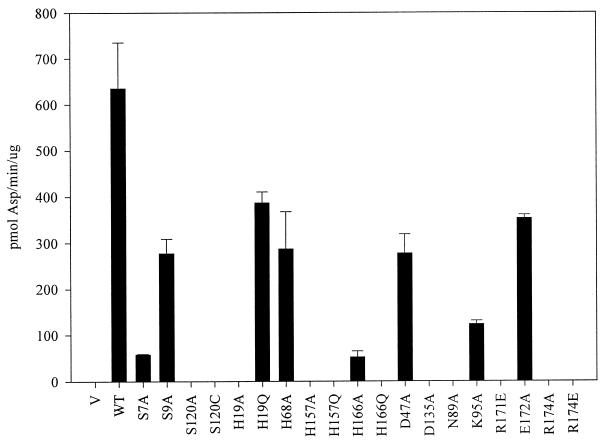
Site-directed mutagenesis was used to change each of the conserved serine (Ser7, Ser9, and Ser120), histidine (His19, His68, His157, and His166), and aspartate (Asp47 and Asp135) residues of serovar Typhimurium peptidase E individually to alanine, and the effects of these mutations on enzyme activity were assayed by measuring the rate of Asp-pNA hydrolysis. The results (Fig. 3) clearly show that although all of these residues are conserved, not all are required for enzyme activity. Of the three conserved serine residues, a mutation in only one, Ser120, abolished activity, indicating that neither Ser7 nor Ser9 can be the catalytic nucleophile.
FIG. 3.
Asp-pNA-hydrolyzing activity of peptidase E site-directed mutants compared to that of the wild type, Salmonella strain TN2719 (leuBCD485 pepE8::MudJ) carrying plasmid pSE380 (vector [V]), wild-type peptidase E plasmid (pCM389) (WT), or mutant peptidase E plasmids (listed in Table 2) was grown overnight in L broth containing 50 μg of ampicillin per ml and 1 mM IPTG. Soluble cell extracts of each strain were assayed for Asp-pNA-hydrolyzing activity as described in Materials and Methods. Error bars show standard deviations.
For some serine hydrolases it has been shown that replacement of the active-site serine by cysteine leads to an enzyme with greatly reduced but still detectable activity (7). To further investigate the role of Ser120 in peptidase E catalysis, a mutant with a Ser120Cys substitution was constructed, and its ability to hydrolyze Asp-pNA was tested. The Ser120Cys mutant hydrolyzed Asp-pNA at a 104-fold lower rate than the wild type, as measured from crude cell extracts of plasmid-containing strains (TN2719/pCM389 and TN2719/pCM456). The sensitivity of a purified preparation of the Ser120Cys mutant to cysteine protease inhibitors was compared to that of the wild-type enzyme (Table 5). The Ser120Cys mutant was expected to have the characteristics of a cysteine protease, including sensitivity to compounds which specifically inhibit cysteine proteases. Wild-type peptidase E was not affected by iodoacetate, NEM, or E-64, but the activity of the Ser120Cys mutant was significantly decreased by all three inhibitors. This result indicates that the cysteine residue in this mutant is important for catalysis and that it potentially acts as the nucleophile. The results of these experiments, together with the results for the Ser120Ala mutant, provide strong evidence that peptidase E is a serine peptidase and that Ser120 is the active-site nucleophile.
TABLE 5.
Effect of cysteine protease inhibitors on wild-type and Ser120Cys peptidase E
| Inhibitor | % Relative activity of peptidase E from:
|
|
|---|---|---|
| Wild type | Ser120Cys mutant | |
| None | 100a | 100b |
| Iodoacetate | 100 | 14 |
| NEM | 100 | 7 |
| E-64 | 100 | 30 |
One hundred percent is 1,083 pmol of Asp released min−1 mg−1.
One hundred percent is 0.11 pmol of Asp released min−1 mg−1.
Mutation in one of the two conserved aspartate residues led to the loss of enzymatic activity, indicating that only Asp135 is required for activity. Two of the conserved histidine residues (His68 and His166) could be changed to alanine without a loss of activity. Conversion of either His19 or His157 to alanine, however, led to a complete loss of enzymatic activity. To further investigate the role of the histidine residues, each of the four histidine residues was changed to glutamine, a more conservative change than a mutation to alanine. The His157Gln mutation resulted in an inactive enzyme, whereas His19Gln led to an active enzyme, indicating that His157 is the only one of the four conserved histidine residues required for activity.
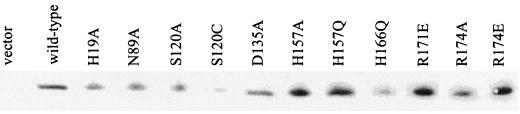
To determine whether or not the mutations affected activity directly or simply caused protein instability, the presence of peptidase E protein in cell extracts of each inactive mutant was detected by Western blotting with polyclonal antibody raised to wild-type serovar Typhimurium peptidase E. The results (Fig. 4) indicate that most of the mutations do not affect the stability of peptidase E. Both Ser120Cys and His166Gln mutants are present in significantly smaller amounts than the wild type. However, the Ser120Cys mutant can be purified, has measurable activity, and so is presumed to be stable, and another mutation at His166 (His166Ala) did not abolish activity, suggesting that this residue is not essential. The conclusion, therefore, is that the mutations that led to inactivity affected enzyme activity rather than enzyme stability.
FIG. 4.
Western blot analysis of peptidase E mutants. Extracts of strains overexpressing wild-type (from pCM389) or mutant (from plasmids listed in Table 2) peptidase E were analyzed by SDS-PAGE and Western blotting as described in Materials and Methods.
Analysis of potential substrate specificity mutations.
Because peptidase E recognizes negatively charged substrates (aspartyl peptides), it was hypothesized that positively charged residues may be required in the active site to interact with the P1 aspartate of the substrate. Site-directed mutagenesis was used to change each of the conserved positively charged amino acids to either alanine or glutamate. In the initial sequence alignment, Lys95 appeared to be conserved, so a Lys95Ala mutant was constructed and found to retain Asp-pNA hydrolytic activity. Two sequences that were recently added to the peptidase E family (from S. putrefaciens and D. melanogaster) do not have a conserved lysine at the position equivalent to Lys95, confirming that this residue is not essential. Two arginine residues located in region VII, Arg171 and Arg174, are completely conserved. Mutations were constructed in each residue, Arg171Glu, Arg174Ala, and Arg174Glu; all led to an inactive enzyme (Fig. 3). In addition, none of the arginine mutants appeared to acquire new activity toward non-aspartyl peptides. A mutation in a conserved glutamate, Glu172Ala, had little effect on peptidase E activity, even though it is in close proximity to the two conserved arginine residues. We conclude that Arg171 and Arg174 are essential for peptidase E activity, but we have not shown that either is responsible for Asp-X peptide specificity.
DISCUSSION
The results presented here define a new family of peptidases, the first member of which was peptidase E from serovar Typhimurium. An analysis of the catalytic residues of this family was based on a sequence comparison of all known peptidase E homologs which, at the start of this work, included only those from serovar Typhimurium, E. coli, H. influenzae, and X. laevis. Peptidase E from X. laevis was especially useful in the sequence alignment because, with this distantly related homolog, we were able to derive a meaningful consensus sequence for the family. The X. laevis pepE gene, called gene D, was one of 39 genes found to be up-regulated by thyroid hormone during the late stages of tail resorption in tadpoles (18). X. laevis peptidase E was purified, and its specificity was shown to be nearly identical to that of serovar Typhimurium peptidase E. The similarity in substrate specificity of these two enzymes in combination with the high degree of sequence conservation among all members of the family led to the conclusion that members of the peptidase E family not only are structurally related but also share an unusual strict specificity for small aspartyl peptides. The defining characteristics of the family, which now contains eight members from different organisms, are as follows: specificity for small aspartyl peptides, including dipeptides and Asp-Gly-Gly; a requirement for a free N-terminal aspartate; a molecular mass of approximately 25 kDa; and a catalytic serine.
There may be a broader peptidase E family including the product of the B. subtilis ygaJ gene, which has sequence similarity to peptidase E; however, ygaJ does not encode an aspartyl-specific peptidase. It is common for families of proteases to share structural features but to differ in substrate specificities; therefore, the gene products from both B. subtilis (37% identity) and D. radiodurans (32% identity) may be included in a family of proteins with the structural characteristics of peptidase E.
An alignment of the eight most similar sequences presumed to share substrate specificity was used to identify residues within conserved regions thought to be important for catalysis. Site-directed mutagenesis suggests that peptidase E is a serine protease that utilizes a catalytic triad of Ser120, Asp135, and His157. The recently solved crystal structure of serovar Typhimurium peptidase E confirms that Ser120 and His157 have the correct proximity and orientation for catalysis (K. Håkansson, A. Wang, and C. G. Miller, unpublished results). The distance of Asp135 from His157, however, suggests that Asp135 is not part of the catalytic triad. Because this residue is essential, it must still play an important role in peptidase E activity.
The proposal that Ser120 is the active-site nucleophile is supported by its location in a typical serine hydrolase motif (Gly-X-Ser-X-Gly) and by the importance of this residue for peptidase E activity. This proposal is also supported by the fact that substitution of Cys for Ser120 leads to a protein with low but detectable enzymatic activity and that this activity, but not that of the wild type, is inhibited by thiol reagents. Similar results have been found for other serine hydrolases, such as rat trypsin, in which a Ser195Cys mutant has different levels of activity depending upon the substrate tested, ranging from no detectable activity to activity 30-fold lower than that of the wild type (7). The serine hydrolase Tsp protease (tail-specific protease encoded by prc in E. coli) is also tolerant to a change from serine (Ser430) to cysteine, the Ser430Cys mutant having 5 to 10% wild-type activity (9). In addition, Ser-to-Cys mutants of both trypsin and Tsp protease are sensitive to Cys protease inhibitors, like the peptidase E Ser120Cys mutant.
The phylogenetic distribution of members of the peptidase E family is unusual. Peptidase E is present in the γ-proteobacteria but is also present in two eukaryotes. Although peptidase E from D. melanogaster has not been tested experimentally for aspartyl dipeptidase activity, the amino acid residues shown to be important for peptidase E activity are all conserved, implying that D. melanogaster peptidase E has the same activity as the others. Interestingly, neither of the two eukaryotes for which the complete genome sequences are available, Caenorhabditis elegans and Saccharomyces cerevisiae, has a peptidase E homolog. In addition, no homologs were found in any of the archaeal genomes that have been completely sequenced, including Methanococcus jannaschii, Methanobacterium thermoautotrophicum, Archaeoglobus fulgidus, Pyrococcus horikoshii, and Aeropyrum pernix. These observations indicate that peptidase E has a wide but sporadic phylogenetic distribution, being present in eubacteria, an insect, and an amphibian but not in other eukaryotes or in archaea.
Other peptidases, such as the broad-specificity aminopeptidases peptidase N, peptidase B, and peptidase A and the proline-specific peptidase peptidase P, are found in a wide variety of organisms representing all three domains of the phylogenetic tree. The fact that peptidase E is not so widespread leads to the speculation that it plays a more specialized role in organisms in which it is found. For example, in X. laevis, peptidase E is induced during a particular late stage of development, suggesting a function for this enzyme beyond that of generalized peptide hydrolysis. It will be interesting to learn the role of peptidase E in different organisms and to further analyze the distribution of new members of the peptidase E family as genome sequences become available.
ACKNOWLEDGMENTS
This work was supported by a grant (AI10333) from the National Institute of Allergy and Infectious Diseases. R.A.L.L. was supported in part by a training grant (5 T32 GMO7283) from the National Institute of General Medical Sciences.
We thank Kjell Håkansson for providing purified Ser120Cys peptidase E and for sharing unpublished results about the peptidase E crystal structure. We acknowledge the contributions of Lhing-Yew Li to this project. We thank Donald Brown for provided a cDNA clone encoding the X. laevis enzyme and Robert Switzer for providing B. subtilis.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Brosius J. Superpolylinkers in cloning and expression vectors. DNA. 1989;8:759–777. doi: 10.1089/dna.1989.8.759. [DOI] [PubMed] [Google Scholar]
- 3.Brown D D, Wang Z, Furlow J D, Kanamori A, Schwartzman R A, Remo B F, Pinder A. The thyroid hormone-induced tail resorption program during Xenopus laevis metamorphosis. Proc Natl Acad Sci USA. 1996;93:1924–1929. doi: 10.1073/pnas.93.5.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter T H. Ph.D. thesis. Cleveland, Ohio: Case Western Reserve University; 1982. [Google Scholar]
- 5.Carter T H, Miller C G. Aspartate-specific peptidases in Salmonella typhimurium: mutants deficient in peptidase E. J Bacteriol. 1984;159:453–459. doi: 10.1128/jb.159.2.453-459.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conlin C A, Håkansson K, Liljas A, Miller C G. Cloning and nucleotide sequence of the cyclic AMP receptor protein-regulated Salmonella typhimurium pepE gene and crystallization of its product, an α-aspartyl dipeptidase. J Bacteriol. 1994;176:166–172. doi: 10.1128/jb.176.1.166-172.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higaki J N, Evnin L B, Craik C S. Introduction of a cysteine protease active site into trypsin. Biochemistry. 1989;28:9256–9263. doi: 10.1021/bi00450a004. [DOI] [PubMed] [Google Scholar]
- 8.Howard A D, Kostura M J, Thornberry N, Ding G J, Limjuco G, Weidner J, Salley J P, Hogquist K A, Chaplin D D, Mumford R A, Schmidt J A, Tocci M J. IL-1-converting enzyme requires aspartic acid residues for processing of the IL-1 beta precursor at two distinct sites and does not cleave 31-kDa IL-1 alpha. J Immunol. 1991;147:2964–2969. [PubMed] [Google Scholar]
- 9.Keiler K C, Sauer R T. Identification of active site residues of the Tsp protease. J Biol Chem. 1995;270:28864–28868. doi: 10.1074/jbc.270.48.28864. [DOI] [PubMed] [Google Scholar]
- 10.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 11.McHugh G L, Miller C G. Isolation and characterization of proline peptidase mutants of Salmonella typhimurium. J Bacteriol. 1974;120:364–371. doi: 10.1128/jb.120.1.364-371.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller C G. Protein degradation and proteolytic modification. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 938–954. [Google Scholar]
- 13.Rawlings N D, Barrett A J. Evolutionary families of peptidases. Biochem J. 1993;290:205–218. doi: 10.1042/bj2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 15.Schmieger H. Phage P22 mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 16.Sleath P R, Hendrickson R C, Kronheim S R, March C J, Black R A. Substrate specificity of the protease that processes human interleukin-1 beta. J Biol Chem. 1990;265:14526–14528. [PubMed] [Google Scholar]
- 17.Vogel H J, Bonner D M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 18.Wang Z, Brown D D. Thyroid hormone-induced gene expression program for amphibian tail resorption. J Biol Chem. 1993;268:16270–16278. [PubMed] [Google Scholar]
- 19.Yen C, Green L, Miller C G. Peptide accumulation during growth of peptidase deficient mutants. J Mol Biol. 1980;143:35–48. doi: 10.1016/0022-2836(80)90123-0. [DOI] [PubMed] [Google Scholar]