Abstract
Alanine aminotransferase (AlaAT) was purified from cell extracts of the hyperthermophilic archaeon Pyrococcus furiosus by multistep chromatography. The enzyme has an apparent molecular mass of 93.5 kDa, as estimated by gel filtration, and consists of two identical subunits of 46 kDa, as deduced by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and the gene sequence. The AlaAT displayed a broader substrate specificity than AlaATs from eukaryal sources and exhibited significant activity with alanine, glutamate, and aspartate with either 2-oxoglutarate or pyruvate as the amino acceptor. Optimal activity was found in the pH range of 6.5 to 7.8 and at a temperature of over 95°C. The N-terminal amino acid sequence of the purified AlaAT was determined and enabled the identification of the gene encoding AlaAT (aat) in the P. furiosus genome database. The gene was expressed in Escherichia coli, and the recombinant enzyme was purified. The pH and temperature dependence, molecular mass, and kinetic parameters of the recombinant were indistinguishable from those of the native enzyme from P. furiosus. The kcat/Km values for alanine and pyruvate formation were 41 and 33 s−1 mM−1, respectively, suggesting that the enzyme is not biased toward either the formation of pyruvate, or alanine. Northern analysis identified a single 1.2-kb transcript for the aat gene. In addition, both the aat and gdh (encoding the glutamate dehydrogenase) transcripts appear to be coregulated at the transcriptional level, because the expression of both genes was induced when the cells were grown on pyruvate. The coordinated control found for the aat and gdh genes is in good agreement with these enzymes acting in a concerted manner to form an electron sink in P. furiosus.
The hyperthermophilic archaea are a group of phylogenetically related microorganisms which by definition grow optimally at or above 80°C, with a maximal growth temperature of 90°C or higher (5). The majority of these are strictly anaerobic heterotrophs, most of which are obligately dependent upon the reduction of elemental sulfur (S0) to H2S. A limited number of facultative S0-reducing species are able to grow in the absence of S0 by means of an alternative fermentative-type metabolism. An example of this type of organism is Pyrococcus furiosus, which grows optimally at 100°C, with a temperature maximum of 105°C, by the fermentation of peptides and various carbohydrates, including starch, glycogen, β-glucans, cellobiose, and maltose (12). In addition, pyruvate can also be utilized as a carbon and energy source (9, 33). P. furiosus utilizes a modified Embden-Meyerhof pathway for the catabolism of sugars, which involves a pair of unprecedented ADP-dependent kinases (glucokinase and phosphofructokinase) and a unique glyceraldehyde-3-phosphate:ferredoxin oxidoreductase (18, 26, 35, 37). The main products produced during the fermentation of sugars include acetate, CO2, H2, and alanine (19).
During growth on either peptides or carbohydrates, reduced ferredoxin is generated (1, 20). Regeneration of oxidized ferredoxin is assumed to be accomplished by three mechanisms: either by S0 reduction to H2S, by proton reduction to H2, or by the formation of alanine. The third alternative, formation of alanine, is found when P. furiosus is grown in the absence of S0. In addition to acetate, a significant amount of alanine is excreted into the medium (19, 33). The amount of alanine produced varies, with an increase in the H2 partial pressure resulting in an increase in the amount of alanine produced. The transamination of pyruvate with glutamate by the action of an alanine aminotransferase (AlaAT) was detected in cell extracts of P. furiosus (19). Furthermore, this activity was shown to be affected by both the partial pressure of hydrogen as well as the available carbon source, suggesting some form of regulation. Glutamate must be replenished through the action of the NADP-dependent glutamate dehydrogenase (GDH). However, there is some controversy concerning the exact role of GDH in the metabolism, since it has been proposed to serve an anabolic role (7, 27), as well as a catabolic role (32). Interestingly, the activity of the GDH reacted similarly to that of AlaAT under the same growth conditions, further suggesting some coordinated regulation of these enzymes (19). The necessary NADPH can be generated by the transfer of reducing equivalents from reduced ferredoxin to NADP+ by the ferredoxin:NADP oxidoreductase activity of the sulfide dehydrogenase. These initial findings suggest that P. furiosus is able to shift its metabolism in response to its environment and in particular the redox potential of the available terminal electron acceptor.
In addition to P. furiosus, l-alanine production has been detected in the related archeaon Thermococcus profundus (21), as well as the hyperthermophilic bacteria belonging to the order of the Thermotogales (31). Pyrococcus and Thermococcus are considered to be one of the deepest branches in the domain of the Archaea, with Thermotogales being one of the deepest branches within the domain Bacteria. Based on this finding, it has been proposed that alanine production from sugar fermentation can be regarded as an ancestral metabolic characteristic (31). However, l-alanine production has also been reported during the fermentation of sugars under anaerobic conditions for the intestinal parasite Giardia lamblia (11) and a moderately thermophilic Clostridium species (28), suggesting that this pathway may be more common among the three domains than previously thought. Moreover, homoalanine fermentation was recently established by metabolic engineering in a lactic acid bacterium, indicating that this pathway may function as an electron sink in a wide range of organisms (16). Further analysis of this pathway may provide insight into not only the biology of these organisms, but also the evolution of fermentative metabolism.
MATERIALS AND METHODS
Growth of microorganisms.
For the purification of AlaAT, P. furiosus (DSM 3638) was grown at 95°C in a 200-liter fermentor with 40 mM pyruvate as a carbon source. A typical medium has the following composition (grams per liter): MgCl2, 2.7; MgSO4, 3.4; KCl, 0.33; NH4Cl, 0.25; KH2PO4, 0.14; CaCl2, 0.14; yeast extract, 1; NaCl, 25; Na2S, 0.25; cysteine-HCl, 0.5; Na2WO4, 0.0033. Vitamins and trace elements were added as described previously (35). All ingredients except Na2S were added to the fermentor and mixed before heating to 90°C. The fermentor was flushed with N2 gas, and the pH was kept constant at approximately pH 7.0. Just before inoculation with a 2-liter preculture of P. furiosus, the medium was reduced through the addition of Na2S. Growth was monitored by measuring the protein content of the culture according to the method of Bradford (6). As soon as growth ceased, cells were harvested to prevent lysis. After approximately 18 h of growth (0.28 mg of protein/ml), cells were harvested by continuous centrifugation and stored at −20°C. Approximately 1 g (wet weight) of cells per liter was obtained. For batch cultures, P. furiosus was grown in 250 ml of a sea salts medium which contained the following (per liter): 40 g of sea salts (Sigma), 3.1 g of PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], 1 g of yeast extract, and 1 g of tryptone. In addition, 1 ml of a trace elements stock, which contained the following (per 100 ml), was added: nitrilotriacetic acid, 1.50 g; FeCl2 × 6H2O, 0.50 g; Na2WO7 × 2H2O, 0.30 g; MnCl2 × 4 H2O, 0.40 g; NiCl2 × 6H2O, 0.2 g; ZnSO4 × 7H2O, 0.1 g; CoSO4 × 7H2O, 0.1 g; CuSO4 × 5H2O (10 mg/ml), 1.0 ml; and Na2MoO4 × 5H2O (10 mg/ml), 1.0 ml. Elemental sulfur, when used, was added at 1% (wt/vol). The pH of the medium was set to 6.8, flushed with N2, and reduced by the addition of Na2S. Cultures were routinely inoculated with a 1% inoculum from a freshly grown overnight preculture. Escherichia coli BL21(λDE3) and XL-1 were grown at 37°C in Luria-Bertani medium. When appropriate, the antibiotics kanamycin (50 μg/ml), ampicillin (50 μg/ml), and tetracycline (15 μg/ml) were included in the medium.
Enzyme assays.
AlaAT was routinely assayed at 80°C in a discontinuous assay (19). The amount of alanine converted to pyruvate was measured by the conversion of NADH in a separate lactate dehydrogenase (LDH) assay at 30°C. The AlaAT reaction mixture contained (in a volume of 1 ml) 50 mM MOPS (pH 7.2), 100 mM KCl, 50 μM pyridoxal 5′-phosphate (PLP), 20 mM α-ketoglutarate, and 50 mM l-alanine. The reaction mixture was preincubated at 80°C for 5 min, and the reaction was started by the addition of the enzyme or cell extract. The reaction was stopped by snap freezing in an ice-ethanol bath. Samples were taken (5 to 100 μl) and added to the LDH assay, which contained 100 mM potassium phosphate (pH 7.0), 0.2 mM NADH, and 3 μl of LDH from beef muscle. One unit of AlaAT or LDH is defined as the amount of enzyme that catalyzes the oxidation of 1 μmol of NADH/min. The discontinuous method was also used in the direction of alanine formation. In this case, the assay mixture contained 50 mM glutamate and 20 mM pyruvate instead of alanine and α-ketoglutarate. For the substrate specificity studies, the formation of glutamate was monitored in reactions that included 15 to 50 mM the amino acid and 20 mM α-ketoglutarate with the buffer described above. The reaction mixture was preincubated at 80°C for 5 min. The reaction was started by the addition of the enzyme and stopped by snap freezing in an ice-ethanol bath. Samples were taken (5 to 100 μl) and added to the GDH assay. The GDH assay contained 100 mM potassium phosphate (pH 7.0), 0.2 mM NADP+, and 5 U of P. furiosus GDH (23). The reaction was performed at 50°C, and the increase in A340 was monitored. To determine the GDH activity in cell extracts the assay described above was used, but now included 10 mM glutamate, and the reaction was initiated by the addition of the cell extract.
Purification of the P. furiosus AlaAT.
AlaAT was purified from P. furiosus as follows. Frozen cells (219 g [wet weight]) were thawed and resuspended in 520 ml of 10 mM Tris (pH 7.8). This resulted in approximately 700 ml of cells, of which 160 ml was used for the purification of the AlaAT. The cell suspension (160 ml) was passed through a French pressure cell (110 MPa) twice. The resulting extract was centrifuged at 13,000 × g for 1 h to remove any cellular debris. All steps were carried out at 23°C. In an effort to keep the enzyme in the PLP form, pyridoxal 5′ phosphate and α-ketoglutarate were added to all active fractions after each purification step at final concentrations of 0.1 and 2 mM, respectively. The supernatant was loaded onto a 300-ml Q-Sepharose (Pharmacia) column equilibrated with 20 mM Tris (pH 7.8). The column was eluted at a flow rate of 5 ml/min with a 1,000-ml linear gradient of 0 to 1.0 M NaCl in the same Tris buffer. The AlaAT eluted at an NaCl concentration of 0.42 to 0.52 M. The active fractions were combined (85 ml), and solid ammonium sulfate was added to a final concentration of 1 M. This solution was applied to a 20-ml phenyl Sepharose (Pharmacia) column equilibrated in 20 mM Tris (pH 7.8) containing 1 M ammonium sulfate. The column was eluted with a 750-ml gradient at a flow rate of 5 ml/min from 1.0 to 0 M ammonium sulfate. The AlaAT eluted at an ammonium sulfate concentration of 0.22 to 0.08 M. The active fractions were pooled (95 ml) and loaded onto a 200-ml hydroxyapatite (Pharmacia) column that had been equilibrated with 20 mM Tris (pH 7.8). The AlaAT was eluted from the column in a 1-liter linear gradient of 0 to 0.5 M potassium phosphate (pH 7.2) at a flow rate of 5 ml/min. The aminotransferase eluted from the column at a concentration of potassium phosphate of 0.2 to 0.25 M. The active fractions were pooled (80 ml) and concentrated, and the buffer was exchanged with 125 mM sodium citrate (pH 5.0). The protein was applied to a 25-ml S-Sepharose (Pharmacia) column that was preequilibrated with 125 mM sodium citrate (pH 5.0), and the aminotransferase was found in the flowthrough. The AlaAT-containing flowthrough was concentrated (5 ml), and the buffer exchanged with 20 mM Tris (pH 7.8). This was applied to a 1-ml Mono-Q (Pharmacia) column and eluted with a 20-ml linear gradient of 0 to 1.0 M with a flow rate of 0.5 ml/min. The active fractions from the Mono-Q were concentrated and applied to a column of Superdex 200 (Pharmacia) equilibrated with 20 mM Tris (pH 7.8) and 0.1 M NaCl at a flow rate of 0.5 ml/min. The active fractions were pooled and stored at 4°C (until required).
Cloning and expression of the gene encoding AlaAT.
The AlaAT gene was amplified from P. furiosus genomic DNA with the oligonucleotides BG432 (5′-CGCGCCATGGCCACTGTTATGATAAGGGCCTCA-3′), which contains an NcoI site, and BG433 (5′-CGCGGGATCCAGAAGTATCATTCTTTCAGTC-3′), which contains a BamHI site. PCR amplification was carried out with Pfu polymerase (Promega), and the resulting 1.2-kb PCR product was cloned into the T7 expression vector pET-24d (Novagen). The resulting plasmid, pLUW770, was transformed into E. coli BL21(λDE3). For expression of the recombinant AlaAT (rAlaAT), 1 liter of E. coli BL21(λDE3), harboring pLUW770, was grown at 37°C for 16 to 18 h, at which point, the cells were harvested for purification of the recombinant enzyme. The addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to the culture resulted in the rAlaAT being found in the insoluble fraction, probably as inclusion bodies, and for this reason it was omitted from the culture.
Purification of rAlaAT.
The rAlaAT was purified in a two-step purification. A 1-liter overnight culture of E. coli BL21(λDE3) harboring pLUW770 was harvested and resuspended in 10 ml of 20 mM Tris (pH 7.8), 0.1 mM pyridoxal 5′-phosphate, and 2 mM α-ketoglutarate. The cells were lysed by sonication, and the cellular debris was removed by centrifugation (13,000 × g for 30 min). The supernatant was then incubated at 80°C for 20 min, and the denatured E. coli proteins were removed by centrifugation (13,000 × g for 30 min). The supernatant (12 ml) was applied to a Mono-Q column equilibrated in Tris buffer. The aminotransferase was eluted with a 20-ml linear gradient of 0 to 1.0 M NaCl at a flow rate of 0.5 ml/min and eluted at a concentration of NaCl from 0.22 to 0.27 M. The recombinant protein was stored at 4°C until required.
RNA isolation and Northern analysis.
Total RNA of P. furiosus was isolated as described previously with the following modifications (10). A 250-ml culture of early- to mid-exponential-phase-grown cells were harvested and washed once in Tris-EDTA (TE). The cells were then resuspended in 0.5 ml of ice-cold TE to which 3.75 ml of guanidine thiocyanate solution was added. After a 5-min incubation at room temperature, 375 μl of 2 M sodium acetate (pH 4.5) and an equal volume of phenol-chloroform (5:1 [pH 4.5]) were added. The mixture was vortexed and then incubated on ice for 5 min, and phase separation was obtained by centrifugation (10,000 × g for 20 min at 4°C). The aqueous phase was extracted twice with an equal volume of phenol–chloroform–isoamyl alcohol (25:24:1 [pH 8]) and once with chloroform-isoamyl alcohol (24:1). The aqueous phase was removed, and RNA was precipitated by the addition of a 1/10 volume of 3 M sodium acetate (pH 5.5) and 2.5 volumes of 96% ethanol followed by a 2-h incubation at −80°C. The pelleted RNA was washed three times with 70% ethanol, dried, and resuspended in 10 mM Tris (pH 8.5). For Northern blot analysis, 15 μg of total RNA was separated on a 1.5% formaldehyde agarose gel and following electrophoresis was transferred to a Hybond N+ membrane. Probes were generated by PCR with the primers BG432 and BG433 for the AlaAT gene and BG34 (5′-ATTGTTATTAAGCAACTTGAAAGAG) and BG173 (5′-GTCTATGTTCTTCTCCTTTGCTATGTTGTAGACGTCG) for the GDH. The PCR product was purified by Qiaquick (Qiagen) and labelled by nick translation.
Other methods.
Molecular weights were estimated by gel filtration with a column (1 by 27 cm) of Superdex 200 (Pharmacia) with catalase (232,000), chymotrypsinogen (25,000), and RNase A (13,700) as standard proteins. The N-terminal sequence of the native AlaAT was determined with an Applied Biosystem 477 Sequencer (courtesy of Emile Schultz at the Institute of Organic Chemistry and Biochemistry, Freiburg, Germany).
Nucleotide sequence accession number.
The nucleotide accession number for the P. furiosus AlaAT is AF163769.
RESULTS
Purification of P. furiosus AlaAT.
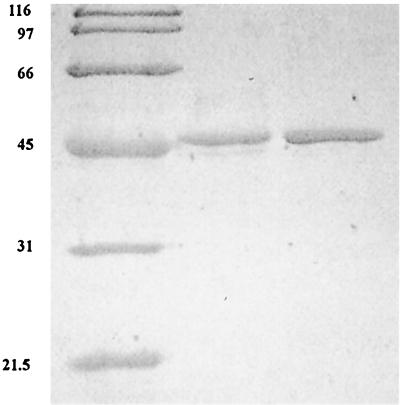
Extracts of P. furiosus cells grown in an 8-liter fermentor with pyruvate as the primary carbon and energy source contained high AlaAT activity (approximately 2.5 U/mg). In addition, the inclusion of 5-fold higher levels of NH4+ in the medium (20 mM final concentration) resulted in a further 1.5-fold increase in activity (4 U/mg). For this reason, P. furiosus was grown in a 200-liter fermentor in artificial seawater medium supplemented with tungsten, yeast extract, 20 mM NH4Cl, vitamins, and 40 mM pyruvate as the carbon source. The fermentor was sparged continuously with N2. The AlaAT was purified 51-fold with a yield of 7% and a specific activity of 158 U/mg (Table 1). In the absence of pyridoxal 5′-phosphate in the assay mixture, the activity was reduced twofold, suggesting that the coenzyme is only partly lost during the purification. The purified AlaAT migrated as a single band in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with an apparent molecular mass of 46 kDa (Fig. 1). The molecular mass of the native enzyme as determined by gel filtration on Superdex-200 was 93.5 kDa, suggesting that the active form of the enzyme exists as a dimer of identical subunits. This is similar to that observed with AlaATs from mesophilic sources (22, 34), as well as the aromatic aminotransferases (AroATs) from Thermococcus litoralis (3) and P. furiosus (2), which exist as homodimers. The N-terminal sequence was determined, and the sequence MIRASKRALSVEYAIR was obtained. This sequence was used to search the P. furiosus genomic database (http://combdna.umbi.umd.edu/bags.html). A single gene was identified that when translated contained an N terminus that matched exactly that determined from the purified enzyme. The structural gene encoding AlaAT (aat) was retrieved (http://www.genome.utah.edu/), which consisted of 1,203 bp and encoded a protein of 400 residues with a predicted molecular mass of 45.5 kDa.
TABLE 1.
Purification of the native AlaAT and rAlaAT from P. furiosus and E. coli
| Purification step | Activity (U) | Protein (mg) | Sp act (U/mg) | Recovery (%) | Relative purification |
|---|---|---|---|---|---|
| AlaAT | |||||
| Cell extract | 8,928 | 2,880 | 3.1 | 100 | 1 |
| Q-Sepharose | 2,940 | 773 | 3.8 | 33 | 1.2 |
| Phenyl-Sepharose | 2,945 | 427 | 6.8 | 33 | 2.2 |
| Hydroxyapatite | 2,992 | 60 | 50 | 33 | 16 |
| S-Sepharose | 1,984 | 44 | 45 | 22 | 15 |
| Mono-Q | 1,411 | 14 | 101 | 16 | 33 |
| Superdex 200 | 616 | 3 | 158 | 7 | 51 |
| rAlaAT | |||||
| Cell extract | 5,660 | 396 | 14.2 | 100 | 1 |
| Heat incubation | 4,400 | 25.8 | 170 | 78 | 12 |
| Mono-Q | 2,386 | 9.8 | 243 | 42 | 17 |
FIG. 1.
SDS-PAGE of purified AlaAT and rAlaAT. Lanes (from left to right): 1, molecular mass (kilodaltons) markers; 2, native AlaAT (2 μg); 3, rAlaAT (2 μg).
Comparison of the primary structure of P. furiosus AlaAT with other AlaATs.
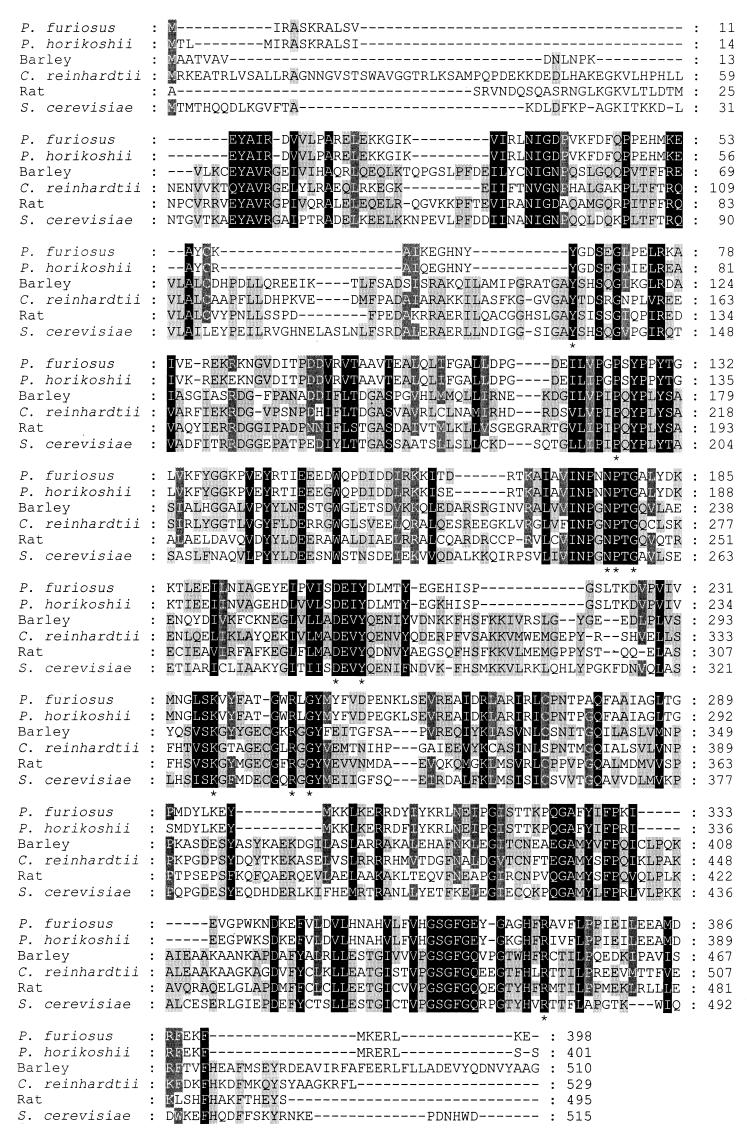
TFASTA and BLAST analyses of the translated sequence revealed moderate identity (30 to 40%) to the aminotransferases belonging to subgroup 1, which includes alanine, aspartate, and AroATs (25). A high level of identity, 91%, was found with a putative aspartate aminotransferase from Pyrococcus horikoshii, which is likely to be an AlaAT (17). A multiple sequence alignment among the known AlaATs clearly shows significant regions of homology (Fig. 2). The 11 invariant residues in this subgroup known to be involved in binding of either the substrate or the coenzyme pyridoxal 5′-phosphate are also conserved (25). An exception was found, however, with Tyr 127 in the P. furiosus enzyme, which is conserved in the AspAT and AroATs as a Trp (Trp 140). This substitution is in all of the AlaATs investigated. In aspartate aminotransferase, Arg 292 is involved in binding of the side chain carboxylate (8). It is also found in the dual substrate AroATs from E. coli and Paracoccus denitrificans (14, 29). This residue is absent from the Chlamydomonas reinhardtii AlaAT, which has no activity with aspartate (22). The P. furiosus enzyme does, however, have two arginines (Arg 270 and Arg 272) in this region, which could explain the low, but significant, activity with aspartate that is usually not observed with other AlaATs (22, 34).
FIG. 2.
Multiple sequence alignment of P. furiosus AlaAT to known AlaATs. The alignment was performed with Clustal W. The GenBank accession numbers for the other aminotransferases are as follows: P. horikoshii, BAA30428; rat, P25409; Saccharomyces cerevisiae, P52892; C. reinhardtii, AAB01685; and barley, P52894. Invariant residues found within the subclass 1 aminotransferases are designated with a star.
Production and purification of rAlaAT.
Production of rAlaAT was successfully achieved in E. coli BL21(λDE3), harboring pLUW770, by growing the cells for 18 h at 37°C in the absence of IPTG. The presence of IPTG resulted in rAlaAT being in an inactive and insoluble form, most likely as inclusion bodies. rAlaAT was purified 17-fold with a yield of 42% and a specific activity of 243 U/mg (Table 1). The specific activity of rAlaAT was 1.5-fold higher than that of the native enzyme. Similar results were observed when comparing the native to recombinant forms of the prolidase from P. furiosus (13) and aspartate aminotransferase from Sulfolobus solfataricus (4). The reason for this is not known. The difference is not due to the heat incubation of the recombinant enzyme, since similar treatment of the native enzyme did not result in any increase in activity. Analysis by SDS-PAGE and gel filtration gave predicted molecular masses of 46 kDa for the subunit and 93.4 kDa for the recombinant enzyme, which are identical to those observed for the native enzyme.
Physical and catalytic properties of the native AlaAT and rAlaAT.
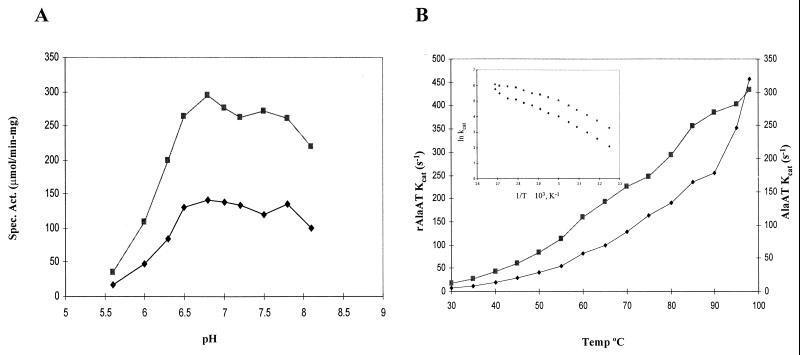
The specific activities of AlaAT and rAlaAT exhibited similar behaviors in response to changes in either temperature or pH (Fig. 3). The activities of both enzymes increased with increasing temperature from 30°C to 95°C, with the temperature optimum appearing to be above 95°C. An Arrhenius plot displays a break in the slope at 60°C for both enzymes. The calculated activation energies for AlaAT and rAlaAT are 58 and 27 kJ/mol and 64 and 44 kJ/mol, respectively. This shift in activation energies is often attributed to a change from one rate-limiting step to another, possibly due to a conformational change. The reason for the difference in activation energies between the native and recombinant enzymes is not clear. Both enzymes were stable over a broad pH range and retained complete activity from pH 6.5 to 7.8 (Fig. 3).
FIG. 3.
The effects of pH (A) and temperature (B) on the activities of AlaAT and rAlaAT. The activities of AlaAT (●) and rAlaAT (■) were determined in the temperature range of 30 to 95°C and the pH range of 6.0 to 8.5 in the presence of saturating concentrations of alanine and α-ketoglutarate. For both AlaAT and rAlaAT, the amount of enzyme in the assay was 3.0 μg. For the determination of the effects of pH, 100 mM potassium phosphate was used, and the assays were performed at 80°C.
The kinetic parameters of AlaAT and rAlaAT were determined at 80°C by varying the substrate concentrations of alanine, α-ketoglutarate, glutamate, and pyruvate (Table 2). As expected, double-reciprocal plots of the initial velocity against the concentration of alanine and α-ketoglutarate in the presence of various fixed concentrations of α-ketoglutarate and alanine gave sets of parallel lines indicating that the reaction proceeds via the “ping-pong bi-bi” mechanism (38). The Km values for the α-keto acids α-ketoglutarate and pyruvate were higher than those observed for other AlaATs. The kinetic parameters of rAlaAT were comparable to those of the native enzyme, with the exception of kcat, which was approximately 50% higher, consistent with the results obtained during the purification. The kcat/Km values for alanine and pyruvate formation were 41 and 33 s−1 mM−1, respectively, suggesting that the enzyme is not biased towards the formation of either pyruvate (forward reaction) or alanine (reverse reaction).
TABLE 2.
Kinetic parameters of P. furiosus AlaAT and rAlaATa
| Substrate pair | Apparent Km (mM) | Apparent Vmax (μmol/min/mg) | kcat (s−1) | kcat/Km (s−1 mM−1) |
|---|---|---|---|---|
| AlaAT | ||||
| Alanine | 2.8 | 151 | 114 | 41 |
| α-Ketoglutarate | 0.9 | |||
| Glutamate | 4.3 | 187 | 142 | 33 |
| Pyruvate | 5.4 | |||
| rAlaAT | ||||
| Alanine | 3.2 | 245 | 186 | 58 |
| α-Ketoglutarate | 1.1 | |||
| Glutamate | 4.7 | 285 | 216 | 46 |
| Pyruvate | 5.6 |
Activities were determined at 80°C as described in the Materials and Methods. For alanine and glutamate, 5 mM α-ketoglutarate and pyruvate were used as the amino acceptors. For α-ketoglutarate and pyruvate, 20 mM alanine and glutamate were used as the amino donors.
The ability of the AlaAT to catalyze the transamination between various amino acids and α-ketoglutarate or pyruvate as the amino acceptor was examined in the presence of saturating amounts (20 to 50 mM) of the substrates. A high specificity was found for the transamination of alanine with α-ketoglutarate and glutamate with pyruvate (Table 3). This has also been reported for other AlaATs (22, 34). However, the enzyme did exhibit significant activity toward aspartic acid and, to a much lesser extent, the branched-chain amino acids with α-ketoglutarate. With pyruvate as the amino acceptor, only aspartic acid could be used in addition to glutamate. No activity could be detected with the aromatic amino acids regardless of the amino acceptor.
TABLE 3.
Substrate specificity of rAlaAT toward various amino acids with α-ketoglutarate or pyruvate as the amino acceptor
| Amino acid | Relative activity (%)a
|
|
|---|---|---|
| α-Ketoglutarate | Pyruvate | |
| Alanine | 100 | |
| Glutamate | 100 | |
| Aspartate | 14 | 10 |
| Valine | 4 | <0.1 |
| Isoleucine | 4 | <0.1 |
| Leucine | 4 | <0.1 |
| Tyrosine | <0.1 | <0.1 |
| Phenylalanine | <0.1 | <0.1 |
For α-ketoglutarate, 100% activity is 150 U/mg, and for pyruvate, 100% activity is 180 U/mg.
Regulation of AlaAT expression in P. furiosus.
The possible effect of the carbon source on the activity of the AlaAT and GDH in crude extracts of P. furiosus was investigated, and the results are shown in Table 4. P. furiosus was grown with either 10 mM cellobiose, 40 mM pyruvate, or 5 g of tryptone per liter as the primary carbon and energy source in either the presence or absence of S0. For both AlaAT and GDH, a significant increase in activity was observed when comparing cellobiose-grown cells to pyruvate-grown cells with a 3.5-fold increase in activity for AlaAT and a 4.5-fold increase for GDH. To determine if this difference in activities was controlled at the level of transcription, Northern analyses were carried out. The levels of expression of both aat and gdh were found to be dependent on the carbon source (Fig. 4). A single 1.2-kb transcript was observed for aat. There was a low level of expression when grown on cellobiose and an approximately threefold increase in the level of transcript in pyruvate-grown cells (Fig. 4A). A similar induction, approximately sixfold, on pyruvate-grown cells was also observed with the gdh gene (Fig. 4C). These patterns of expression are in excellent agreement with the enzyme activities of the AlaAT and GDH measured in cell extracts (Table 4). The addition of S0 (1 g/liter) in the medium, a potential electron acceptor in P. furiosus, had little effect on the expression of either the aat or gdh gene.
TABLE 4.
Activities of AlaAT and GDH in crude extracts of P. furiosus grown on various carbon sources
| Carbon source | Sp act (U/mg)
|
|
|---|---|---|
| AlaAT | GDH | |
| Cellobiose (10 mM) | 0.49 | 0.88 |
| Pyruvate (40 mM) | 1.81 | 3.63 |
| Cellobiose (10 mM) + S0 | 0.43 | 0.85 |
| Pyruvate (40 mM) + S0 | 1.30 | 3.60 |
| Tryptone (5 g/liter) + S0 | 0.88 | 1.44 |
FIG. 4.
Northern analysis of the P. furiosus aat (A and B) and gdh (C) transcripts. RNA was isolated from cells grown in the presence of 10 mM cellobiose (C), 40 mM pyruvate (P), 10 mM cellobiose with S0(CS), 40 mM pyruvate with S0(PS), and tryptone (5 g/liter) with S0(TS).
DISCUSSION
The coordinated activities of AlaAT and GDH have been proposed to play an important role in the maintenance of the redox balance during fermentative growth of P. furiosus (19). These activities result in a change in the relative flux of pyruvate to acetate formation toward alanine formation. Pyruvate is therefore used as a catabolic electron sink. Due to the important role AlaAT plays in this pathway, this enzyme was purified from P. furiosus and represents the first AlaAT purified from either an archaeon or a hyperthermophile.
Similar to the AlaAT from mesophilic sources, the active form of the enzyme was found to be a homodimer with a subunit molecular mass of 43.5 kDa (22, 34, 36). It has been reported that the AlaATs have a high substrate specificity and are only able to transaminate alanine or glutamate (22, 34, 36). The P. furiosus enzyme, however, was capable of utilizing aspartate and, to a much lesser extent, the branched-chain amino acids with α-ketoglutarate as the amino acceptor, clearly distinguishing it from the other AlaATs. This activity on the branched-chain amino acids is most likely not significant from a metabolic standpoint. It has been shown that P. furiosus has a strict requirement for the presence of the amino acids valine, isoleucine, and, to a lesser extent, leucine in the medium (15). The absence of a branched-chain aminotransferase in P. furiosus is probably one of the reasons for this strict requirement for valine and isoleucine in the medium. Apparently the activity of the AlaAT on the branched-chain amino acids is not able to compensate for the absence of a branched-chain aminotransferase. While the kinetic parameters for the various AlaATs do vary, a common feature is that the Km for the amino acceptor is lower than that of the amino donor (34). While this trend is also present in the pyrococcal enzyme for the alanine–α-ketoglutarate pair, it is not true for the glutamate-pyruvate pair.
An important question to be addressed relates to the metabolic role of AlaAT in P. furiosus and the factors involved in its control. The enzyme in plants plays pivotal roles in the biosynthesis of alanine, degradation of alanine, and the intercellular carbon shuttle associated with C4 photosynthesis (34). The AlaAT from P. furiosus may have multiple roles as well. During proteolytic fermentation, it is feasible that the P. furiosus AlaAT may function in the catabolism of alanine, thereby generating pyruvate. The pyruvate would then be converted to acetate and ATP by the combined actions of the pyruvate:ferredoxin oxidoreductases and the acetyl coenzyme A synthase I (24). This is supported by both the presence of AlaAT activity in crude extracts and the detection of the aat transcript of P. furiosus grown with tryptone as the primary carbon and energy source. Alternatively, it also plays a role in the maintenance of the redox balance via the formation of alanine. If the P. furiosus AlaAT has a dual role, then the enzyme should not exhibit a preference for either the degradation or production of alanine. While the Km for alanine is almost twofold lower than that for glutamate, the overall levels of efficiency (kcat/Km) of the two reactions are quite similar. This would suggest that the enzyme is fully capable of performing the dual roles proposed here.
Alanine formation in P. furiosus has been proposed to be due to the coordinated actions of the AlaAT and GDH (19). This coordinated activity was shown to be controlled at the level of transcription, with the highest levels of the aat and gdh transcripts found in pyruvate-grown cells, which is in perfect agreement with the observed enzyme activities measured in crude extracts. It is possible that pyruvate is the inducer of expression of the aat and gdh. Because insights into the control of archaeal gene expression are only starting to emerge, the mechanism by which pyruvate may act as an inducer is not known. In E. coli, pyruvate has been shown to be the inducer of expression of the pyruvate dehydrogenase complex in E. coli and is mediated through the regulatory protein PdhR (30). While pyruvate is produced during the catabolism of cellobiose, the amount in the cell is most likely significantly lower then when grown on pyruvate, resulting in the lower level of expression. What was also of interest was the observation that the transcript was present when the cells were grown in the presence of S0. This suggests that the regulation of expression is not mediated by fluctuations of the intracellular redox potential. Nevertheless, P. furiosus is capable of shifting its metabolism in response to the availability of the terminal electron acceptor. This brings about the question of the identity of the actual signal that controls the shift in metabolism from a mixed acetate-alanine fermentation, when grown in the absence of S0, to an almost strict acetate fermentation when grown in the presence of S0. This control must be occurring at the level of the enzymes, since the aat and gdh are still expressed in the presence of S0. The concentration of pyruvate inside the cell may fluctuate substantially, reflecting the relative redox potential of the available electron acceptor. In the absence of S0, there may be a decrease in the flux of pyruvate to acetate through the pyruvate: ferredoxin oxidoreductases due to limited availability of oxidized ferredoxin resulting in a buildup of pyruvate. This buildup of pyruvate may cause the switch to alanine formation. It is clear that more detailed analyses in a chemostat under steady-state conditions are necessary to test this hypothesis, which will lead to a better understanding of the role this pathway plays in the fermentative metabolism. These studies are currently under way.
ACKNOWLEDGMENTS
This research was supported by the EC BIOTECH Programme “Extremophiles as cell Factories” (BIO2-CT93-0274) and the Brite EuRam III Programme “Promofilm” (BRPR-CT97-0484).
We thank Emile Schiltz of the Institute of Organic Chemistry and Biochemistry, Freiburg, Germany, for the determination of the N terminus, as well as Huub Haaker and E. J. Crane for critical reading of the manuscript.
REFERENCES
- 1.Adams M W W, Kletzin A. Oxidoreductase-type enzymes and redox proteins involved in fermentative metabolisms of hyperthermophilic Archaea. Adv Protein Chem. 1996;48:101–180. doi: 10.1016/s0065-3233(08)60362-9. [DOI] [PubMed] [Google Scholar]
- 2.Andreotti G, Cubellis M V, Nitti G, Sannia G, Mai X, Adams M W W, Marino G. An extremely thermostable aromatic aminotransferase from the hyperthermophilic archaeon Pyrococcus furiosus. Biochim Biophys Acta. 1995;1247:90–96. doi: 10.1016/0167-4838(94)00211-x. [DOI] [PubMed] [Google Scholar]
- 3.Andreotti G, Cubellis M V, Nitti G, Sannia G, Mai X, Marino G, Adams M W W. Characterization of aromatic aminotransferases from the hyperthermophilic archaeon Thermococcus litoralis. Eur J Biochem. 1994;220:543–549. doi: 10.1111/j.1432-1033.1994.tb18654.x. [DOI] [PubMed] [Google Scholar]
- 4.Arnone M I, Birolo L, Cubellis M V, Nitti G, Marino G, Sannia G. Expression of a hyperthermophilic aspartate aminotransferase in Escherichia coli. Biochim Biophys Acta. 1992;1160:206–212. doi: 10.1016/0167-4838(92)90009-3. [DOI] [PubMed] [Google Scholar]
- 5.Baross J A, Holden J F. Overview of hyperthermophiles and their heat-shock proteins. Adv Protein Chem. 1996;48:1–34. doi: 10.1016/s0065-3233(08)60360-5. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 7.Consalvi V, Chiaraluce R, Politi L, Vaccaro R, De Rosa M, Scandurra R. Extremely thermostable glutamate dehydrogenase from the hyperthermophilic archaebacterium Pyrococcus furiosus. Eur J Biochem. 1991;202:1189–1196. doi: 10.1111/j.1432-1033.1991.tb16489.x. [DOI] [PubMed] [Google Scholar]
- 8.Cronin C N, Kirsch J F. Role of arginine-292 in the substrate specificity of aspartate aminotransferase as examined by site-directed mutagenesis. Biochemistry. 1988;27:4572–4579. doi: 10.1021/bi00412a052. [DOI] [PubMed] [Google Scholar]
- 9.de Vos W M, Kengen S W M, Voorhorst W G B, van der Oost J. Sugar utilization and its control in hyperthermophiles. Extremophiles. 1998;2:201–205. doi: 10.1007/s007920050061. [DOI] [PubMed] [Google Scholar]
- 10.DiRuggiero J, Achenbach L A, Brown S H, Kelly R M, Robb F T. Regulation of ribosomal RNA transcription by growth rate of the hyperthermophilic Archaeon, Pyrococcus furiosus. FEMS Microbiol Lett. 1993;111:159–164. doi: 10.1111/j.1574-6968.1993.tb06379.x. [DOI] [PubMed] [Google Scholar]
- 11.Edwards M R, Gilroy F V, Jimenez B M, O'Sullivan W J. Alanine is a major end product of metabolism by Giardia lamblia: a proton nuclear magnetic resonance study. Mol Biochem Parasitol. 1989;37:19–26. doi: 10.1016/0166-6851(89)90098-4. [DOI] [PubMed] [Google Scholar]
- 12.Fiala G, Stetter K O. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch Microbiol. 1986;161:168–175. [Google Scholar]
- 13.Ghosh M, Grunden A M, Dunn D M, Weiss R, Adams M W W. Characterization of native and recombinant forms of an unusual cobalt-dependent proline dipeptidase (prolidase) from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 1998;180:4781–4789. doi: 10.1128/jb.180.18.4781-4789.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi H, Inoue K, Nagata T, Kuramitsu S, Kagamiyama H. Escherichia coli aromatic amino acid aminotransferase: characterization and comparison with aspartate aminotransferase. Biochemistry. 1993;32:12229–12239. doi: 10.1021/bi00096a036. [DOI] [PubMed] [Google Scholar]
- 15.Hoaki T, Wirsen C O, Hanzawa S, Maruyama T, Jannasch H W. Amino acid requirements of two hyperthermophilic archaeal isolates from deep-sea vents, Desulfurococcus strain SY and Pyrococcus strain GB-D. Appl Environ Microbiol. 1993;59:610–613. doi: 10.1128/aem.59.2.610-613.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hols P, Kleerebezem M, Schanck A N, Ferain T, Hugenholtz J, Delcour J, de Vos W M. Conversion of Lactococcus lactis from homolactic to homoalanine fermentation through metabolic engineering. Nat Biotechnol. 1999;17:588–592. doi: 10.1038/9902. [DOI] [PubMed] [Google Scholar]
- 17.Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, Sakai M, Ogura K, Otsuka R, Nakazawa H, Takamiya M, Ohfuku Y, Funahashi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Kikuchi H. Complete sequence and gene organization of the genome of a hyperthermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 18.Kengen S W M, de Bok F A, van Loo N D, Dijkema C, Stams A J M, de Vos W M. Evidence for the operation of a novel Embden-Meyerhof pathway that involves ADP-dependent kinases during sugar fermentation by Pyrococcus furiosus. J Biol Chem. 1994;269:17537–17541. [PubMed] [Google Scholar]
- 19.Kengen S W M, Stams A J M. Formation of L-alanine as a reduced end product in carbohydrate fermentation by the hyperthermophilic archaeon Pyrococcus furiosus. Arch Microbiol. 1994;161:168–175. [Google Scholar]
- 20.Kengen S W M, Stams A J M, de Vos W M. Sugar metabolism of hyperthermophiles. FEMS Microbiol Rev. 1996;18:119–137. [Google Scholar]
- 21.Kobayashi T, Higuchi S, Kimura K, Kudo T, Horikoshi K. Properties of glutamate dehydrogenase and its involvement in alanine production in a hyperthermophilic archaeon, Thermococcus profundus. J Biochem. 1995;118:587–592. doi: 10.1093/oxfordjournals.jbchem.a124950. [DOI] [PubMed] [Google Scholar]
- 22.Lain Guelbenzu B, Cardenas J, Munoz Blanco J. Purification and properties of L-alanine aminotransferase from Chlamydomonas reinhardtii. Eur J Biochem. 1991;202:881–887. doi: 10.1111/j.1432-1033.1991.tb16447.x. [DOI] [PubMed] [Google Scholar]
- 23.Lebbink J H, Eggen R I, Geerling A C, Consalvi V, Chiaraluce R, Scandurra R, de Vos W M. Exchange of domains of glutamate dehydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus and the mesophilic bacterium Clostridium difficile: effects on catalysis, thermoactivity and stability. Protein Eng. 1995;8:1287–1294. doi: 10.1093/protein/8.12.1287. [DOI] [PubMed] [Google Scholar]
- 24.Mai X, Adams M W W. Purification and characterization of two reversible and ADP-dependent acetyl coenzyme A synthetases from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 1996;178:5897–5903. doi: 10.1128/jb.178.20.5897-5903.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehta P K, Hale T I, Christen P. Aminotransferases: demonstration of homology and division into evolutionary subgroups. Eur J Biochem. 1993;214:549–561. doi: 10.1111/j.1432-1033.1993.tb17953.x. [DOI] [PubMed] [Google Scholar]
- 26.Mukund S, Adams M W W. Glyceraldehyde-3-phosphate ferredoxin oxidoreductase, a novel tungsten-containing enzyme with a potential glycolytic role in the hyperthermophilic archaeon Pyrococcus furiosus. J Biol Chem. 1995;270:8389–8392. doi: 10.1074/jbc.270.15.8389. [DOI] [PubMed] [Google Scholar]
- 27.Ohshima T, Nishida N. Purification and properties of extremely thermostable glutamate dehydrogenases from two hyperthermophilic archaebacteria, Pyrococcus woesei and Pyrococcus furiosus. Biosci Biotechnol Biochem. 1993;57:945–951. doi: 10.1271/bbb.57.945. [DOI] [PubMed] [Google Scholar]
- 28.Orlygsson J, Anderson R, Svensson B H. Alanine as an end product during fermentation of monosaccharides by Clostridium strain P2. Antonie Leeuwenhoek. 1995;68:273–280. doi: 10.1007/BF00874136. [DOI] [PubMed] [Google Scholar]
- 29.Oue S, Okamoto A, Nakai Y, Nakahira M, Shibatani T, Hayashi H, Kagamiyama H. Paracoccus denitrificans aromatic amino acid aminotransferase: a model enzyme for the study of dual substrate recognition mechanism. J Biochem Tokyo. 1997;121:161–171. doi: 10.1093/oxfordjournals.jbchem.a021561. [DOI] [PubMed] [Google Scholar]
- 30.Quail M A, Haydon D J, Guest J R. The pdhR-aceEF-lpd operon of Escherichia coli expresses the pyruvate dehydrogenase complex. Mol Microbiol. 1994;12:95–104. doi: 10.1111/j.1365-2958.1994.tb00998.x. [DOI] [PubMed] [Google Scholar]
- 31.Ravot G, Ollivier B, Fardeau M-L, Patel B K C, Andrews K T, Magot M, Garcia J-L. l-Alanine production from glucose fermentation by hyperthermophilic members of the domains Bacteria and Archaea: a remnant of an ancestral metabolism? Appl Environ Microbiol. 1996;62:2657–2659. doi: 10.1128/aem.62.7.2657-2659.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robb F T, Park J B, Adams M W W. Characterization of an extremely thermostable glutamate dehydrogenase: a key enzyme in the primary metabolism of the hyperthermophilic archaebacterium, Pyrococcus furiosus. Biochim Biophys Acta. 1992;1120:267–272. doi: 10.1016/0167-4838(92)90247-b. [DOI] [PubMed] [Google Scholar]
- 33.Schaefer T, Xavier K B, Santos H, Schoenheit P. Glucose fermentation to acetate and alanine in resting cell suspensions of Pyrococcus furiosus: proposal of a novel glycolytic pathway based on 13C labelling data and enzyme activities. FEMS Microbiol Lett. 1994;121:107–114. [Google Scholar]
- 34.Son D, Jo J, Sugiyama T. Purification and characterization of alanine aminotransferase from Panicum miliaceum leaves. Arch Biochem Biophys. 1991;289:262–266. doi: 10.1016/0003-9861(91)90470-4. [DOI] [PubMed] [Google Scholar]
- 35.Tuininga J E, Verhees C H, van der Oost J, Kengen S W M, Stams A J M, de Vos W M. Molecular and biochemical characterization of the ADP-dependent phosphofructokinase from the hyperthermophilic archaeon Pyrococcus furiosus. J Biol Chem. 1999;274:21023–21028. doi: 10.1074/jbc.274.30.21023. [DOI] [PubMed] [Google Scholar]
- 36.Umemura I, Yanagiya K, Komatsubara S, Sato T, Tosa T. Purification and some properties of alanine aminotransferase from Candida maltosa. Biosci Biotechnol Biochem. 1994;58:283–287. doi: 10.1271/bbb.58.283. [DOI] [PubMed] [Google Scholar]
- 37.van der Oost J, Schut G, Kengen S W, Hagen W R, Thomm M, de Vos W M. The ferredoxin-dependent conversion of glyceraldehyde-3-phosphate in the hyperthermophilic archaeon Pyrococcus furiosus represents a novel site of glycolytic regulation. J Biol Chem. 1998;273:28149–28154. doi: 10.1074/jbc.273.43.28149. [DOI] [PubMed] [Google Scholar]
- 38.Velick S F, Vavra J. A kinetic and equilibrium analysis of the glutamate oxaloacetate transaminase mechanism. J Biol Chem. 1962;237:2109–2122. [PubMed] [Google Scholar]