Abstract
wbdA is a mannosyltransferase gene that is involved in synthesis of the Escherichia coli O9a polysaccharide, a mannose homopolymer with a repeating unit of 2-αMan-1,2-αMan-1,3-αMan-1,3-αMan-1. The equivalent structural O polysaccharide in the E. coli O9 and Klebsiella O3 strains is 2-αMan-1,2-αMan-1,2-αMan-1,3-αMan-1,3-αMan-1, with an excess of one mannose in the 1,2 linkage. We have cloned wbdA genes from these O9 and O3 strains and shown by genetic and functional studies that wbdA is the only gene determining the O-polysaccharide structure of O9 or O9a. Based on functional analysis of chimeric genes and site-directed mutagenesis, we showed that a single amino acid substitution, C55R, in WbdA of E. coli O9 converts the O9 polysaccharide into O9a. DNA sequencing revealed the substitution to be conserved in other E. coli O9a strains. The reverse substitution, R55C, in WbdA of E. coli O9a resulted in lipopolysaccharide synthesis showing no ladder profile instead of the conversion of O9a to O9. This suggests that more than one amino acid substitution in WbdA is required for conversion from O9a to O9.
O polysaccharides of gram-negative bacteria are structurally polymorphic, and consequently, they are utilized as the O antigen for serological typing. The O polysaccharide is part of lipopolysaccharides (LPSs) and covalently binds to lipid A through the core-oligosaccharide portion. Each O polysaccharide consists of many repeating units of an oligosaccharide composed of several sugars with various linkages. We have been studying the synthetic mechanism of Escherichia coli O9a polysaccharides using the wb* gene cluster cloned from E. coli O9a strain F719. The wb* cluster is constituted of eight genes, two for GDP-mannose synthesis, two for the putative ABC transporter, three for mannosyltransferases, and one whose function is unknown (8). WbdA, one of the mannosyltransferases encoded in the cluster, is regarded as an enzyme that transfers two mannoses in the 1,2 linkage based on the results of a previous biochemical analysis (8) and the E. coli O9a polysaccharide structure. It consists of 815 amino acids and has two domains with almost the same molecular mass, designated WbdA(N) and WbdA(C) for the N- and C-terminal domains, respectively (7). An amino acid sequence comparison revealed the presence of a conserved amino acid sequence of SXXEGFGLPXXE in both domains (8). Similar sequences have been identified in several prokaryotic α-mannosyltransferases (2). Elimination of the C-terminal domain of WbdA by transposon insertion or gene deletion results in synthesis of an altered structural O polysaccharide consisting only of α-1,2-linked mannose. Both domains are necessary for the synthesis of the E. coli O9a polysaccharide. However, it is synthesized even when the C-terminal domain is present in trans (7). Moreover, genetic analysis of the wbdA gene suggests that it might have arisen by fusion of two independent genes (7).
The repeating unit of E. coli O9a is composed of four mannoses with the structure 2-αMan-1,2-αMan-1,3-αMan-1,3-αMan-1 (11). The strain has been recognized as an exceptional divergent E. coli O9 that has the repeating unit 2-αMan-1,2-αMan-1,2-αMan-1,3-αMan-1,3-αMan-1 with an excess of one mannose in the 1,2 linkage (10, 12). Klebsiella O3 strains have the same structural O polysaccharide (4). Recently we have produced a monoclonal antibody (MAb) recognizing the E. coli O9a polysaccharide but not E. coli O9 and named it O9a MAb. The minimum number of mannose residues needed to theoretically define the O9 and O9a polysaccharides is four, and the sequence 3-αMan-1,2-αMan-1,2-αMan-1,3-αMan-1 is the shortest candidate for the epitope bound to the MAb (6). Serological investigation of various conventional O9 strains using the O9a MAb revealed that the majority, 8 of 10, were the O9a type (6). This relatively large number is unexpected because only one strain is classified as O9a by conventional serotyping using polyclonal antibodies (10). This result prompted us to study the genetic relationship between the strains. In this report, we provide experimental proof that the O9 polysaccharide is converted into O9a by a single amino acid substitution in the WbdA mannosyltransferase. Based on genetic and functional analyses of wbdA genes, we suggest that a new O-serotype group, E. coli O9a, could have been generated from the E. coli O9 strain by this single amino acid substitution. We also discuss the effect of the amino acid substitution on the processive action of WbdA transferring two or three mannoses in 1,2 linkage.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids utilized in this study are listed in Table 1. To analyze the synthesized O polysaccharide, the E. coli K-12 strain HU1190 lacking the whole wb* locus was used as the host for plasmids. Bacterial strains were cultivated in LB broth or on LB agar plates supplemented with antibiotics. The gene nomenclature for wb* genes in this article follows the nomenclature in the review by Reeves et al. (14).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant property | Source or reference |
|---|---|---|
| Strains | ||
| E. coli K-12 strain HU1190 | Δ(sbcB-rfb) hsdR4 recA56 srl::Tn10 | 17 |
| Wild strains | ||
| E. coli Bi316-42 | O9:K9:H12 | 2 |
| E. coli Bi449-42 | O9a:K26:H− | 2 |
| E. coli K14a | O9a:K28:H− | 2 |
| E. coli F379 | O9a:K29−:H− | 2 |
| E. coli F719 | O9a:K31−:H−his | 2 |
| E. coli H509d | O9:K57:H32 | 2 |
| Klebsiella K49S | O3:K49− | 15 |
| Plasmids | ||
| pBluescript II SK(+) | Cloning vector; Apr | TOYOBO |
| pNKB26::Tn1000-52 | E. coli O9a wb* cluster but wbdA::Tn1000-52 | 7 |
| pBSC25-1 | pBluescript II SK(+) carrying wbdAF719 | 8 |
| pBSC25-1 (R55C) | pBSC25-1 carrying a point mutation giving the R55C substitution in WbdA | This work |
| pWBDABi316-42 | pBluescript II SK(+) carrying wbdABi316-42 | This work |
| pWBDABi316-42 (C55R) | pWBDABi316-42 carrying a point mutation giving the C55R substitution in WbdA | This work |
| pWBDAK49S | pBluescript II SK(+) carrying wbdAK49S | This work |
| pWBDAK49S (C55R) | pWBDAK49S carrying a point mutation giving the C55R substitution in WbdA | This work |
| pWBDA-BF | pBluescript II SK(+) carrying chimeric wbdABi316-42-wbdAF719 | This work |
| pWBDA-BF (C55R) | pWBDA-BF carrying a point mutation giving the C55R substitution in hybrid WbdA | This work |
| pWBDA-BK | pBluescript II SK(+) carrying chimeric wbdABi316-42-wbdAK49S | This work |
| pWBDA-FB | pBluescript II SK(+) carrying chimeric wbdAF719-wbdABi316-42 | This work |
| pWBDA-FB (R55C) | pWBDA-FB carrying a point mutation giving the R55C substitution in hybrid WbdA | This work |
| pWBDA-FK | pBluescript II SK(+) carrying chimeric wbdAF719-wbdAK49S | This work |
| pWBDA-KF | pBluescript II SK(+) carrying chimeric wbdAK49S-wbdAF719 | This work |
| pWBDA-KF (C55R) | pWBDA-KF carrying a point mutation giving the C55R substitution in WbdA | This work |
| pWBDA-KB | pBluescript II SK(+) carrying chimeric wbdAK49S-wbdABi316-42 | This work |
PCR amplification and cloning of the wbdA genes.
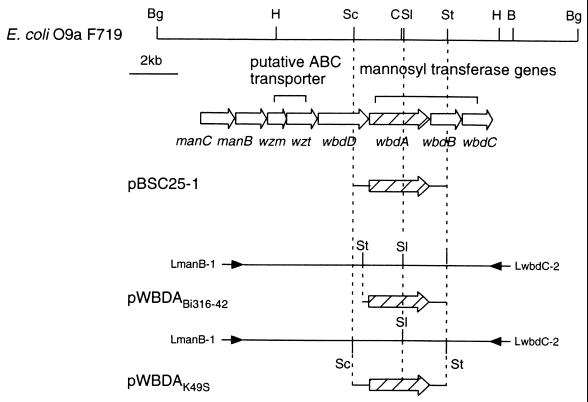
PCR amplification was performed using a mixture of 100 ng of chromosomal DNA and 10 pmol of each oligonucleotide primer for the manB-wbdC region. The manB-wbdC region was amplified with 30 cycles, with each cycle consisting of (i) a denaturation step of 20 s at 98°C and (ii) an annealing and a polymerization step of 15 min at 68°C. DNA polymerase TaKaRa ExTaq was purchased from Takara Shuzo Co., Ltd., Tokyo, Japan. Oligonucleotide primers for amplification were designed based on the DNA sequence of the E. coli F719 wb* (DDBJ accession no. D43637) as follows: LmanB-1 (5′-AGAGATTTACTTCGCCACTTTCCACCTC-3′) and LwbdC-2 (18). The amplified fragment from E. coli Bi316-42 was digested with StuI to give a fragment carrying only the wbdA gene and cloned into the EcoRV site of a cloning vector pBluescript II SK(+). The fragment from Klebsiella K49S was doubly digested with SacI and StuI and cloned into the SacI-EcoRV-digested pBluescript II SK(+). These plasmids with only wbdA were designated pWBDABi316-42 and pWBDAK49S, respectively. A schematic illustration of amplification and subcloning of wbdA genes from strains Bi316-42 and K49S is shown in Fig. 1. For the DNA sequencing of the wbdA genes from other E. coli O9a strains, PCR-amplified fragments were directly sequenced using fluorescein isothiocyanate (FITC)-labeled primers (5′-AATGGCCTATCGGGACCTGCTG-3′ or 5′-CGCCCATGCCAGACCAAGACGG-3′).
FIG. 1.
Schematic representation of PCR amplification and subcloning of wbdA genes from E. coli Bi316-42 and Klebsiella K49S. Restriction enzyme abbreviations: B, BamHI; Bg, BglII; C, ClaI; H, HindIII; Sc, SacI; Sl, SalI; St, StuI. Arrows indicate primers used for PCR.
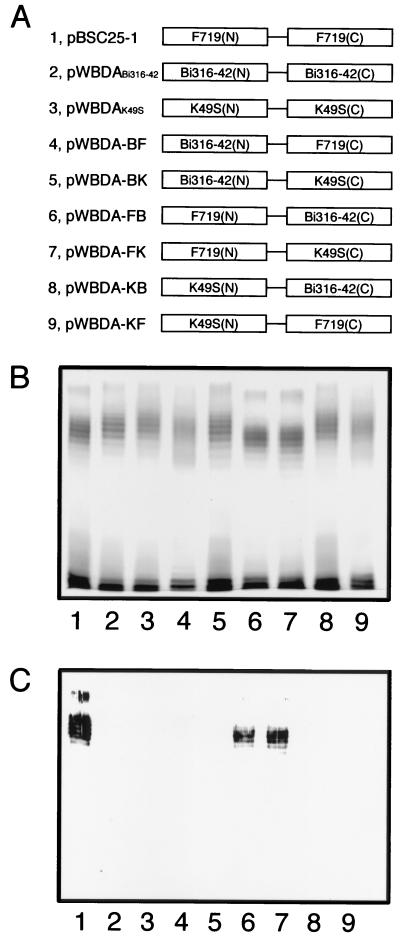
Chimeric plasmids carrying the 5′ region of the wbdA gene encoding WbdA(N) and the 3′ region encoding WbdA(C) of strains F719, Bi316-42, and K49S were constructed using the SalI site (Fig. 1; see Fig. 2A). Each plasmid was digested with SalI, and purified fragments from gels were ligated in various combinations. Plasmids carrying chimeric wbdA genes were selected and designated pWBDA-FB, pWBDA-FK, pWBDA-BF, pWBDA-BK, pWBDA-KF, and pWBDA-KB. Letters after the hyphen denote the origin of the wbdA genes and the order of the combination as follows: F, E. coli F719; B, E. coli Bi316-42; K, Klebsiella K49S. For instance, pWBDA-FB is the plasmid constructed from the 5′ region of wbdA of F719 and the 3′ region of wbdA of Bi316-42.
FIG. 2.
Schematic representation of the WbdA protein structure (A) and results of SDS-PAGE analysis of LPSs directed by the chimeric wbdA genes (silver-stained gel [B] and an immunoblot of the same gel using the O9a MAb [C]). LPSs examined were from E. coli HU1190(pNKB26::Tn1000-52) carrying the following plasmids: pBSC25-1 (lane 1), pWBDABi316-42 (lane 2), pWBDAK49S (lane 3), pWBDA-BF (lane 4), pWBDA-BK (lane 5), pWBDA-FB (lane 6), pWBDA-FK (lane 7), pWBDA-KB (lane 8), and pWBDA-KF (lane 9).
Site-directed mutagenesis.
Plasmids pBSC25-1, pWBDABi316-42, pWBDA-BF, pWBDA-KF, and pWBDA-FB were subjected to mutagenesis with a QuickChange site-directed mutagenesis kit obtained from Stratagene (La Jolla, Calif.). The conditions and buffers used were those recommended by the manufacturer. The primers employed are listed in Table 2. They were designated based on DNA sequences of wbdA genes and purified by high-pressure liquid chromatography before use. Mutations were confirmed by DNA sequencing of complete wbdA regions in the plasmids.
TABLE 2.
Primers used for site-directed mutagenesis in this study
| Amino acid substitution |
Primer sequencea |
|---|---|
| R55C | GCGCCAACAGCCTATTGTCATATTGAAAACC |
| C55R | GCCAACGGCCTACCGCAATATCGATAACCACGG |
| N56H | GCGCCAACGGCCTACTGCCATATCGATAACC |
| G92S | CTTCTTCGAAGGTCATAGCGATAGTTATACC |
| T206S | GATGAATTTATCCTATCGCTGGCGATGATTG |
| M225L | CATGCTTACAGCTTGCTGCCTGCTGAAT |
| H243Q | GGCGTATAAGGTGCAGCCGGAGGGTTTGGAG |
| I399L | GTGACTACCTCGCGCCTCCAGACGATGCAG |
| T401M | CCTCGCGCATCCAGATGATGCAGAAAATTG |
Only sense primers are shown. These were used with the complementary primers for mutagenesis. The nucleotides exchanged are underlined.
Analysis of LPS.
LPS was extracted by the phenol-water method and purified by ultracentrifugation (22). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed by the method of Tsai and Frasch (20), and silver staining of gels was performed by the method of Hitchcock and Brown (3). Immunoblot analysis was done as reported previously with rabbit polyclonal antiserum against E. coli O9 test strain Bi316-42 and an MAb against E. coli O9a strain F719 named O9a MAb (6). Peroxidase-conjugated goat immunoglobulin G fractions against mouse and rabbit immunoglobulin G were purchased from Cappel (Organon Teknika Corp., West Chester, Pa.).
Nuclear magnetic resonance (NMR) spectroscopy of the O polysaccharides.
The O-polysaccharide structures of LPS from E. coli HU1190(pNKB26::Tn1000-52) carrying the wbdA gene from strain Bi316-42 (wbdABi316-42) with the C55R substitution and the wbdAF719 gene with the R55C substitution were determined by two-dimensional homonuclear Hartman-Hahn spectroscopy (9). The O polysaccharides were prepared and purified as described previously (7). Spectra were recorded on a JEOL JNM-GSX 400 spectrometer (400 MHz), operating at a probe temperature of 45°C. Each sample was dissolved in D2O at 1% (wt/vol), and acetone was used as the internal standard (2.217 ppm).
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this article will appear in the DDBJ, EMBL, and GenBank nucleotide sequence databases with the accession numbers AB031867 and AB031868.
RESULTS
Cloning and functional analysis of wbdA genes from E. coli Bi316-42 and Klebsiella K49S.
Genes corresponding to wbdA, -B, and -D of E. coli F719 were cloned on pBluescript II SK(+) cloning vector from PCR-amplified fragments of E. coli O9 Bi316-42 and Klebsiella K49S. They are compatible with pACYC184-based clone pNKB26 in E. coli K-12 strain HU1190. They were introduced into HU1190 carrying pNKB26 derivatives which were inactivated in the corresponding genes by Tn1000 insertion (17). The O-polysaccharide structure was changed only when wbdA was involved. DNA sequencing of genes corresponding to wbdAF719, cloned from strains of Bi316-42 and K49S, revealed DNA identities of 91.0 (Bi316-42) and 96.5% (K49S). The percent similarities for the deduced amino acid sequences were 96.5 (F719 versus Bi316-42) and 97.5 (F719 versus K49S). Thus, genes from Bi316-42 and K49S were found to be highly homologous to wbdAF719 and were termed wbdABi316-42 and wbdAK49S, respectively.
Cloned wbdA genes were introduced into E. coli K-12 HU1190 carrying the wbdA-deficient plasmid, pNKB26::Tn1000-52, bearing all genes necessary for O9a polysaccharide synthesis but wbdA (7). LPS preparations from each strain were analyzed by SDS-PAGE and subsequently by immunoblot analysis (Fig. 2B and C, lanes 1 to 3). All LPSs from recombinant strains showed the typical ladder profile of smooth LPS, as found in the parental wild strains. Only ladders of LPSs from the recombinant strain carrying wbdAF719 stained with the O9a MAb (Fig. 2C, lane 1). Ladders of all preparations were stained with the polyclonal rabbit serum against the E. coli O9 test strain Bi316-42 (data not shown). Based on the O9a MAb specificity, we concluded that the strains carrying the wbdA genes from Bi316-42 and K49S synthesize the O9 polysaccharide while strains carrying wbdAF719 synthesize O9a. Because all the other genes for the O-polysaccharide synthesis in pNKB26::Tn1000-52 were cloned from the O9a-synthesizing strain F719, it was clear that only the wbdA gene determined the O9 polysaccharide structure.
Comparison of WbdA proteins.
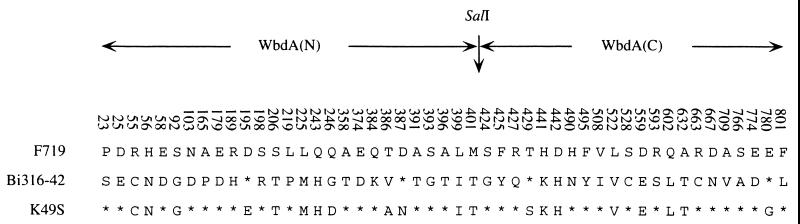
The newly cloned wbdA genes also encoded WbdA proteins with 815 amino acid residues and predicted secondary structures suggesting similarities to WbdAF719 with a few minor variations. Both had N- and C-terminal domains like those of WbdAF719. Amino acid sequences were compared with that of WbdAF719 in detail (Fig. 3). A total of 45 amino acid substitutions were identified in WbdABi316-42, and 20 substitutions were identified in WbdAK49S. Sixteen amino acid substitutions occurred at the same sites, with 14 amino acids identical in WbdABi316-42 and WbdAK49S. Eight of the fourteen were located in WbdA(N) and the remainder were located in WbdA(C). Because the amino acid substitutions are common to the O9-synthesizing strains of Bi316-42 and K49S, it is likely that one or more of these may be involved in the difference in O-polysaccharide structure.
FIG. 3.
Amino acid substitutions in the WbdA proteins. For the WbdA proteins of Bi316-42 and K49S, only the amino acids differing from those of the F719 protein are shown. The SalI site used for the construction of the chimeric wbdA genes is shown. The numbers above the residues indicate the positions of the amino acid residues. Amino acids identical to those of F719 are indicated by asterisks.
Function of chimeric wbdA genes.
To identify amino acid substitutions determining the O-polysaccharide structure, six chimeric wbdA genes were constructed as pairs of 5′ and 3′ halves of the genes from F719, Bi316-42, and K49S (Fig. 2A) and introduced into E. coli HU1190 carrying pNKB26::Tn1000-52. LPSs from the strains showed the ladder profile except for LPSs from two strains carrying either pWBDA-BF or pWBDA-KF (Fig. 2B, lanes 4 and 9). These two plasmids carried chimeric wbdA genes constructed with the 5′ half of the wbdA gene of the O9-synthesizing strain and the 3′ half of wbdAF719. The O polysaccharides synthesized by these chimeric genes stained very faintly with the O9a MAb. The O polysaccharides from strains carrying the chimeric genes of the 5′ half of wbdAF719 and the 3′ half of O9-synthesizing strains appeared to be slightly shorter and were positively stained with the O9a MAb (Fig. 2, lanes 6 and 7). Exchange of the 5′ and 3′ halves between wbdABi316-42 and wbdAK49S did not affect the LPS profile or the O antigenicity against the O9a MAb (Fig. 2, lanes 5 and 8). From these results, we conclude that the 5′ half of the wbdAF719 predominantly determines O9a polysaccharide synthesis.
Identification of the mutation that converts the O9 polysaccharide into O9a.
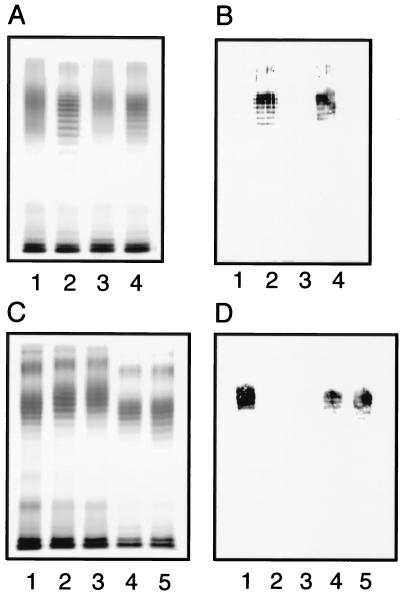
Site-directed mutagenesis was employed to identify mutations in the 5′ half of the wbdA gene resulting in amino acid substitution and alteration of the O-polysaccharide structure. Chimeric plasmids of pWBDA-BF and pWBDA-KF were subjected to mutagenesis to test for O-polysaccharide conversion. Plasmids carrying a series of point mutations in the chimeric wbdA genes were constructed using the DNA primers shown in Table 2 and introduced into E. coli HU1190(pNKB26::Tn1000-52), and LPSs were analyzed by SDS-PAGE. Only the mutation giving the C55R substitution altered the O-polysaccharide structure (Fig. 4). The original chimeric genes directed synthesis of LPS with no ladder profile (Fig. 4A, lanes 1 and 3), whereas plasmids with the C55R mutation directed the synthesis of LPS with a ladder profile (Fig. 4A, lanes 2 and 4). Staining with the O9a MAb (Fig. 4B) indicated that this mutation in the chimeric wbdA genes resulted in conversion into O9a. However, since the chimeric genes carried the 3′ half of the wbdAF719, it was not clear whether the single C55R mutation was sufficient for the O-polysaccharide conversion from O9 into O9a. To confirm that the single C55R substitution caused the conversion, the mutation was introduced into the intact wbdA genes of Bi316-42 and K49S. Synthesized LPS showed the ladder profile (Fig. 4C, lanes 4 and 5). These O polysaccharides appeared slightly shorter than those synthesized by the original wbdA genes and were positively stained with the O9a MAb while those of the parental strain were not (Fig. 4D).
FIG. 4.
SDS-PAGE analysis of LPSs. Silver-stained gels (A and C) and immunoblots of the same gels using the O9a MAb (B and D) are shown. LPSs directed by the chimeric wbdA genes in pWBDA-BF and pWBDA-KF and their C55R mutations are shown in panels A and B. LPSs extracted from E. coli HU1190(pNKB26::Tn1000-52) carrying the following plasmids are examined: pWBDA-BF (lane 1), pWBDA-BF (C55R) (lane 2), pWBDA-KF (lane 3), and pWBDA-KF (C55R) (lane 4). LPSs directed by wbdA from Bi316-42 and K49S strains and by those carrying the C55R mutation are shown in panels C and D. LPSs were from HU1190(pNKB26::Tn1000-52) carrying the following plasmids: pBSC25-1 (lane 1), pWBDABi316-42 (lane 2), pWBDAK49S (lane 3), pWBDABi316-42 (C55R) (lane 4), and pWBDAK49S (C55R) (lane 5).
Reverse mutation in the wbdAF719 gene.
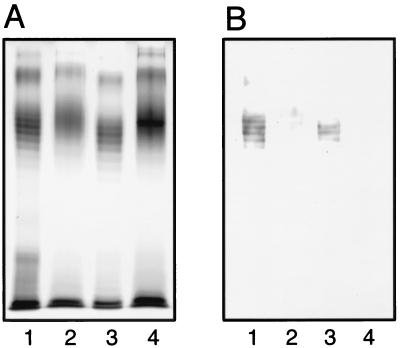
The point mutation giving the reverse R55C substitution in WbdAF719 was introduced into pBSC25-1 to determine whether the O9a polysaccharide was converted into O9. The LPS preparation from the strain carrying the mutated plasmid showed the no-ladder profile in the SDS-polyacrylamide gel (Fig. 5A, lane 2). Moreover, the R55C mutation in the chimeric plasmid pWBDA-FB also directed the synthesis of LPS with no-ladder profile, although a dense band in the region of LPS with O polysaccharides was seen (Fig. 5A, lane 4). These O polysaccharides were stained very faintly with the O9a MAb, while those from strains carrying the original plasmid were stained positively (Fig. 5B). Because the chimeric plasmid possessed the 3′ half of the wbdABi316-42 gene, the results suggested that the conversion of the O9a polysaccharide into the O9 requires more than one amino acid substitution in the WbdA(N) region of WbdAF719.
FIG. 5.
SDS-PAGE analysis of LPSs directed by wbdA with and without the R55C mutation. A silver-stained gel (A) and an immunoblot of the same gel using the O9a MAb (B) are shown. LPSs examined were from E. coli HU1190(pNKB26::Tn1000-52) carrying the following plasmids: pBSC25-1 (lane 1), pBSC25-1 (R55C) (lane 2), pWBDA-FB (lane 3), and pWBDA-FB (R55C) (lane 4).
NMR analysis of O-polysaccharide structures.
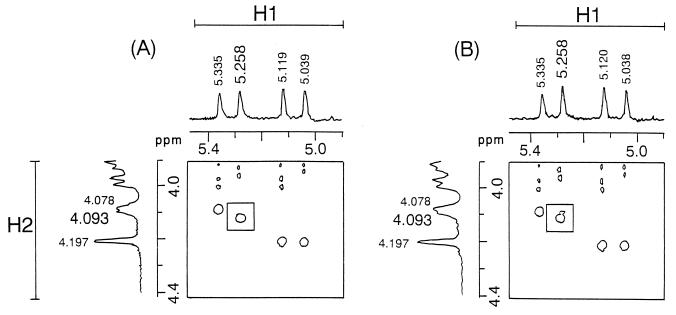
The conversion of the O9 polysaccharide into O9a was confirmed by NMR analysis of O polysaccharides purified from E. coli HU1190(pNKB26::Tn1000-52) carrying wbdABi316-42 with the C55R mutation (Fig. 6A). H1-H2 cross-peaks at 5.039 and 4.197 ppm, 5.119 and 4.197 ppm, 5.258 and 4.093 ppm, and 5.335 and 4.078 ppm were identified to be the four mannose units, -1,3αMan-1,2-, -1,3αMan-1,3-, -1,2αMan-1,2-, and -1,2αMan-1,3-, respectively, as reported previously (6). In addition, the intensities of the corresponding H1 signals, 25.7:27.0:24.8:22.5, indicated the molar ratio of the four mannose units to be approximately 1:1:1:1. Thus, the polysaccharides from the strain carrying the C55R mutation in wbdABi316-42 had the repeating unit of the O9a polysaccharide, 2-αMan-1,2-αMan-1,3-αMan-1,3-αMan-1.
FIG. 6.
Partial two-dimensional homonuclear Hartman-Hahn (H1-H2 region) spectra of O polysaccharides synthesized by WbdA with single amino acid substitutions. O polysaccharides purified from LPS of E. coli HU1190(pNKB26::Tn1000-52) carrying wbdABi316-42 with the C55R mutation (A) or carrying wbdAF719 with the R55C mutation (B) are shown.
Analyses of the O polysaccharides from E. coli HU1190(pNKB26::Tn1000-52) carrying wbdAF719 with the R55C mutation (Fig. 6B) revealed four distinct cross-peaks at 5.038 and 4.197 ppm, 5.120 and 4.197 ppm, 5.258 and 4.093 ppm, and 5.335 and 4.078 ppm in the H1-H2 field with intensities of 24.6:27.3:26.5:21.6 (approximately 1:1:1:1). In addition, a notable cross-peak at 5.258 and 4.078 ppm in the field near the cross-peak at 5.258 and 4.093 ppm was identified (compare the boxed regions of Fig. 6). This indicated the presence of a second -1,2αMan-1,2- unit (6) in the polysaccharides from the strain carrying the R55C mutation in wbdAF719, suggesting that two kinds of the repeating units of O9 and O9a were included in the fraction. The weak reactivity of the O polysaccharides with the O9a MAb suggests that two repeating units may exist in a single O-polysaccharide chain rather than both O9a and O9 polysaccharides being present.
Amino acid substitutions in other E. coli O9a and O9 strains.
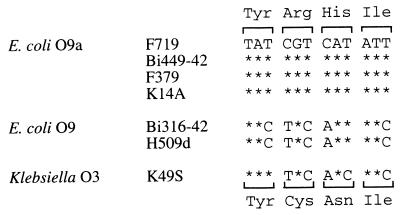
DNA sequences of the 120- to 419-bp segment of the wbdA genes from E. coli O9 and O9a strains were determined using PCR-amplified fragments. They were found to be completely conserved in the E. coli O9a strains investigated. Several nucleotides were different between E. coli O9 strains Bi316-42 and H509d. The latter is the other strain so far regarded serologically as E. coli O9 using the O9a MAb (6). Only the DNA sequence alignment of 12 nucleotides encoding tyrosine 54 to isoleucine 57 is shown in Fig. 7. The arginine residue at position 55 and its substitution by cysteine were completely conserved in O9a- and O9-synthesizing strains, respectively. Two nucleotides were found to differ in the codon for the amino acid residue 55 between O9 and O9a strains.
FIG. 7.
Nucleotide sequence alignment of the region encoding tyrosine at position 54 to isoleucine at position 57 of wbdA from E. coli O9a and O9 and Klebsiella O3 strains. Nucleotides identical to those in F719 are indicated by asterisks. Amino acids encoded are shown above (for E. coli O9a) and below (for E. coli O9 and Klebsiella O3) the DNA sequences.
DISCUSSION
The single amino acid substitution C55R in WbdABi316-42 was shown here to cause the conversion of O9 polysaccharides into O9a. The O-polysaccharide structure was ascertained serologically (Fig. 4) and by two-dimensional NMR analysis (Fig. 6). The critical role of the amino acid in the mannosyltransferase action is not clear. WbdA has the two conserved amino acid sequences deduced from the sequence comparison and the hydrophobic cluster analysis plot of several glycosyl transferases (2, 7, 8). The conserved sequences are located in segments of amino acids 292 to 300 and 725 to 733, so that the mutation point that converts the O9 polysaccharide into O9a is not included. Neutral cysteine exchange for positively charged arginine alters the electrophilic properties, and we can speculate that this affects the protein structure. No significant change in the predicted secondary structure and hydrophobicity of the WbdA protein was observed when the amino acid substitution was introduced into the molecule, and there was also no change in hydrophobic cluster analysis plots. This indicates the necessity of analysis by X-ray diffraction of crystallized proteins to reveal the effects of the amino acid substitution on the WbdA structure and function and for elucidation of the O-polysaccharide synthetic mechanism.
Reeves' group has suggested that the polymorphism of the O polysaccharide in Salmonella enterica was acquired by interspecific gene transfer between gene clusters responsible for O-polysaccharide synthesis (1, 13, 23). These involve genes for nucleotide-sugar synthesis, glycosyl transferases, and O-polysaccharide translocation. In addition to such relatively large-scale gene transfer, it has been demonstrated that point mutations can in some circumstances result in O-polysaccharide alteration (13, 21). The O polysaccharides of S. enterica groups A, B, and D differ only in a dideoxyhexose side chain sugar, paratose, abequose, and tyvelose, respectively. These side chain sugars are synthesized from the common precursor CDP-4-keto-3,6-dideoxy-d-glucose by prt, abe, and tyv genes. Abequose and paratose are synthesized from the precursor by the abe and prt genes directly. Paratose is subsequently converted to tyvelose by the tyv gene. Thus, the polymorphism is caused by diversity of the gene that synthesizes the dideoxyhexose on the common backbone of the O polysaccharide. The abe and prt genes probably diverged from a common ancestor. There is little information about the development of tyv, but the genetic mechanism giving the group A strains is clear. Southern hybridization analysis has revealed that the tyv gene is maintained in these strains, although the gene is inactivated and the Tyv protein is not expressed due to one base deletion in S. enterica group A strain IMVS1316. A frameshift mutation in the gene for CDP-tyvelose synthesis of S. enterica group D thus results in inactivation of the gene and possibly synthesis of the O polysaccharide of S. enterica group A. This point mutation in the tyv gene thus might cause generation of a new serogroup in S. enterica. In our case, the mutated gene was expressed, but its enzyme action might be modulated. In the case of E. coli O9 and related strains that possess a mannose homopolymer as the O polysaccharide, it has been suggested that gene transfer along the wb* region has also occurred (18, 19). Thus, the new serotype O9a was presumably generated from the O9 strain (or vice versa) by point mutations in genes, probably after the ancestral wb* gene cluster was constructed by interspecific gene transfer. One possible interpretation of the available data is that the primary bacterial strategy to acquire O-polysaccharide polymorphism is gene transfer with reconstruction of the synthetic gene cluster. Point mutations then accumulate to extend the O-polysaccharide variety. Only bacteria that survive natural selection and adapt to the surroundings will develop a new serotype group.
There are at least two types of glycosyltransferases: processive enzymes transferring multiple sugar residues to the acceptor and nonprocessive enzymes transferring a single sugar to the acceptor (16). Many of the glycosyltransferases involved in the O-polysaccharide synthesis are nonprocessive. Among the three mannosyltransferases involved in O9 and O9a polysaccharide synthesis, WbdC, which transfers a mannose residue to a lipid acceptor, is nonprocessive, and the others, WbdA and WbdB, are processive (8). This is the reason for the smaller number of transferase genes than mannose residues in the wb* cluster of the O9 and O9a strains. The conversion of O9 polysaccharide into O9a implies a change in the processive action of WbdA, the enzyme with cysteine at position 55 transferring three mannoses in 1,2 linkage, while the enzyme with the arginine substitution transfers only two. It is possible that the amino acid residue will be involved in the processive reaction of the mannosyltransferase, although the precise mechanism is unknown. However, the O9a-to-O9 conversion was not achieved by the single amino acid substitution of R55C in WbdAF719. NMR analysis of the O polysaccharides from the strain suggested that they are mixtures of O9 and O9a (Fig. 6). This indicates that the single amino acid substitution of R55C does not suffice for the O9a-to-O9 conversion, suggesting that more than one amino acid substitution in WbdA(N)F719 is required for the conversion.
ACKNOWLEDGMENT
This study was supported in part by a Grant-in-Aid for Scientific Research (10670253) from the Ministry of Education, Science, Sports and Culture, Japan.
REFERENCES
- 1.Curd H, Liu D, Reeves P R. Relationships among the O-antigen gene clusters of Salmonella enterica groups B, D1, D2, and D3. J Bacteriol. 1998;180:1002–1007. doi: 10.1128/jb.180.4.1002-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geremia R A, Petroni E A, Ielpi L, Henrissat B. Towards a classification of glycosyltransferases based on amino acid sequence similarities: prokaryotic α-mannosyltransferases. Biochem J. 1996;318:133–138. doi: 10.1042/bj3180133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hitchcock P J, Brown T M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gel electrophoresis. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jann K, Jann B. Structure and biosynthesis of O-antigens. In: Rietschel E T, editor. Handbook of endotoxin. 1. Chemistry of endotoxin. Amsterdam, The Netherlands: Elsevier Science Publishers; 1984. pp. 138–186. [Google Scholar]
- 5.Kido N, Ohta M, Iida K-I, Hasegawa T, Ito H, Arakawa Y, Komatsu T, Kato N. Partial deletion of the cloned rfb gene of Escherichia coli O9 results in synthesis of a new O-antigenic lipopolysaccharide. J Bacteriol. 1989;171:3629–3633. doi: 10.1128/jb.171.7.3629-3633.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kido N, Morooka N, Paeng N, Ohtani T, Kobayashi H, Shibata N, Okawa Y, Suzuki S, Sugiyama T, Yokochi T. Production of monoclonal antibody discriminating serological difference in Escherichia coli O9 and O9a polysaccharides. Microbiol Immunol. 1997;41:519–525. doi: 10.1111/j.1348-0421.1997.tb01887.x. [DOI] [PubMed] [Google Scholar]
- 7.Kido N, Sugiyama T, Yokochi T, Kobayashi H, Okawa Y. Synthesis of Escherichia coli O9a polysaccharide requires the participation of two domains of WbdA, a mannosyltransferase encoded within the wb* gene cluster. Mol Microbiol. 1998;27:1213–1221. doi: 10.1046/j.1365-2958.1998.00765.x. [DOI] [PubMed] [Google Scholar]
- 8.Kido N, Torgov V I, Sugiyama T, Uchiya K, Sugihara H, Komatsu T, Kato N, Jann K. Expression of the O9 polysaccharide of Escherichia coli: sequencing of the E. coli O9 rfb gene cluster, characterization of mannosyl transferases, and evidence for an ATP-binding cassette transport system. J Bacteriol. 1995;177:2178–2187. doi: 10.1128/jb.177.8.2178-2187.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi H, Suzuki J, Tanaka S, Kiuchi Y, Oyamada H, Iwadate N, Suzuki H, Shibata N, Suzuki S. Structure of a cell wall mannan from the pathogenic yeast, Candida catenulata: assignment of 1H nuclear magnetic resonance chemical shifts of the inner α-1,6-linked mannose residues substituted by a side chain. Arch Biochem Biophys. 1997;341:70–74. doi: 10.1006/abbi.1997.9939. [DOI] [PubMed] [Google Scholar]
- 10.Ørskov I, Ørskov F, Jann B, Jann K. Serology, chemistry, and genetics of O and K antigens of Escherichia coli. Bacteriol Rev. 1977;41:667–710. doi: 10.1128/br.41.3.667-710.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parolis L A S, Parolis H, Dutton G G S. Structural studies of the O-antigen polysaccharide of Escherichia coli O9a. Carbohydr Res. 1986;155:272–276. doi: 10.1016/s0008-6215(00)90158-7. [DOI] [PubMed] [Google Scholar]
- 12.Prehm P, Jann B, Jann K. The O9 antigen of Escherichia coli: structure of the polysaccharide chain. Eur J Biochem. 1976;67:41–55. doi: 10.1111/j.1432-1033.1976.tb10631.x. [DOI] [PubMed] [Google Scholar]
- 13.Reeves P. Evolution of Salmonella O antigen variation by interspecific gene transfer on a large scale. Trends Genet. 1993;9:17–22. doi: 10.1016/0168-9525(93)90067-R. [DOI] [PubMed] [Google Scholar]
- 14.Reeves P R, Hobbs M, Valvano M A, Skurnik M, Whitfield C, Coplin D, Kido N, Klena J, Maskell D, Raetz C, Rick P. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;4:495–503. doi: 10.1016/s0966-842x(97)82912-5. [DOI] [PubMed] [Google Scholar]
- 15.Saeki A, Kido N, Sugiyama T, Ohta M, Iwashita T, Uchiya K, Kato N. Isolation of rfb gene clusters directing the synthesis of O polysaccharides consisting of mannose homopolymers and serological analysis of lipopolysaccharides. Microbiol Immunol. 1993;37:601–606. doi: 10.1111/j.1348-0421.1993.tb01682.x. [DOI] [PubMed] [Google Scholar]
- 16.Saxena I M, Brown R M, Jr, Fevre M, Geremia R A, Henrissat B. Multidomain architecture of β-glycosyl transferases: implications for mechanism of action. J Bacteriol. 1995;177:1419–1424. doi: 10.1128/jb.177.6.1419-1424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugiyama T, Kido N, Komatsu T, Ohta M, Jann K, Jann B, Saeki A, Kato N. Genetic analysis of Escherichia coli O9 rfb: identification and DNA sequence of phosphomannomutase and GDP-mannose pyrophosphorylase genes. Microbiology. 1994;140:59–71. doi: 10.1099/13500872-140-1-59. [DOI] [PubMed] [Google Scholar]
- 18.Sugiyama T, Kido N, Kato Y, Koide N, Yoshida T, Yokochi T. Evolutionary relationship among rfb gene clusters synthesizing mannose homopolymer as O-specific polysaccharides in Escherichia coli and Klebsiella. Gene. 1997;198:111–113. doi: 10.1016/s0378-1119(97)00300-4. [DOI] [PubMed] [Google Scholar]
- 19.Sugiyama T, Kido N, Kato Y, Koide N, Yoshida T, Yokochi T. Generation of Escherichia coli O9a serotype, a subtype of E. coli O9, by transfer of the wb* gene cluster of Klebsiella O3 into E. coli via recombination. J Bacteriol. 1998;180:2775–2778. doi: 10.1128/jb.180.10.2775-2778.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai C M, Frasch C E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 21.Verma N, Reeves P. Identification and sequence of rfbS and rfbE, which determine antigenic specificity of group A and group D salmonellae. J Bacteriol. 1989;171:5694–5701. doi: 10.1128/jb.171.10.5694-5701.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weisgerber C, Jann B, Jann K. Biosynthesis of the O9 antigen of Escherichia coli: core structure of rfe mutant as indication of assembly mechanism. Eur J Biochem. 1984;140:553–556. doi: 10.1111/j.1432-1033.1984.tb08137.x. [DOI] [PubMed] [Google Scholar]
- 23.Xiang S-H, Hobbs M, Reeves P R. Molecular analysis of the rfb gene cluster of a group D2 Salmonella enterica strain: evidence for its origin from an insertion sequence-mediated recombination event between group E and D1 strains. J Bacteriol. 1994;176:4357–4365. doi: 10.1128/jb.176.14.4357-4365.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]