Abstract
Background
Worldwide, coronary artery disease (CAD) is a leading cause of mortality and morbidity and remains to be a top health priority in many countries. A non-invasive imaging modality for diagnosis of CAD such as single photon emission computed tomography-myocardial perfusion imaging (SPECT-MPI) is usually requested by cardiologists as it displays radiotracer distribution in the heart reflecting myocardial perfusion. The interpretation of SPECT-MPI is done visually by a nuclear medicine physician and is largely dependent on his clinical experience and showing significant inter-observer variability.
Objective
The aim of the study is to apply a deep learning approach in the classification of SPECT-MPI for perfusion abnormalities using convolutional neural networks (CNN).
Methods
A publicly available anonymized SPECT-MPI from a machine learning repository (https://www.kaggle.com/selcankaplan/spect-mpi) was used in this study involving 192 patients who underwent stress-test-rest Tc99m MPI. An exploratory approach of CNN hyperparameter selection to search for optimum neural network model was utilized with particular focus on various dropouts (0.2, 0.5, 0.7), batch sizes (8, 16, 32, 64), and number of dense nodes (32, 64, 128, 256). The base CNN model was also compared with the commonly used pre-trained CNNs in medical images such as VGG16, InceptionV3, DenseNet121 and ResNet50. All simulations experiments were performed in Kaggle using TensorFlow 2.6.0., Keras 2.6.0, and Python language 3.7.10.
Results
The best performing base CNN model with parameters consisting of 0.7 dropout, batch size 8, and 32 dense nodes generated the highest normalized Matthews Correlation Coefficient at 0.909 and obtained 93.75% accuracy, 96.00% sensitivity, 96.00% precision, and 96.00% F1-score. It also obtained higher classification performance as compared to the pre-trained architectures.
Conclusions
The results suggest that deep learning approaches through the use of CNN models can be deployed by nuclear medicine physicians in their clinical practice to further augment their decision skills in the interpretation of SPECT-MPI tests. These CNN models can also be used as a dependable and valid second opinion that can aid physicians as a decision-support tool as well as serve as teaching or learning materials for the less-experienced physicians particularly those still in their training career. These highlights the clinical utility of deep learning approaches through CNN models in the practice of nuclear cardiology.
Keywords: SPECT-MPI, coronary artery disease, convolutional neural networks, deep learning
INTRODUCTION
Coronary artery disease (CAD) remains to be a leading cause of mortality and morbidity in many countries world-wide.1 In the recent years, the incidence of CAD has also increased even among young members of the population.2 Because of its huge public health impact particularly on health resource utilization and the associated costs, it remains to be one of the top health priorities in many regions around the world.3 CAD encompasses a lot of clinical problems from asymptomatic subclinical atherosclerosis to its serious complications such as angina pectoris, acute myocardial infarction, and sudden cardiac death.4 In CAD, there is a narrowing or blockage of coronary blood vessels caused primarily by atherosclerotic plaque formation within the intima of the vessel wall leading to an abnormality in blood flow, thus resulting to reduced delivery of oxygen to the myocardium.5-7 This obstruction in the coronary vessels leads to myocardial ischemia and subsequent impairment of the myocardial functions. In its severe form, the ischemia can further lead to unwanted cardiac events such as myocardial infarction, lengthy hospitalization, chronic heart failure, and sudden cardiac death.1,8 Cardiologists usually refer patients to undergo non-invasive imaging modalities for a diagnosis of CAD as well as determining options for revascularization procedures, assessment of prognosis, and evaluating acute coronary syndromes.9
Single Photon Emission Computed Tomography – Myocardial Perfusion Imaging (SPECT-MPI) is one of the most requested imaging tests to diagnose CAD and has been shown to be a cost-effective modality due to its ability to significantly reduce frequency of unwarranted angiographies and permits pertinent treatment options.10,11 SPECT-MPI shows details of the distribution of a radioactive tracer in the myocardium reflecting myocardial perfusion and as such, it is a cost-efficient imaging test for determining presence and extent of CAD.11 To determine areas of the myocardium with decreased blood flow, the heart is imaged twice: scanning at rest and another scanning during stress. Visual evaluation of the SPECT-MPI images in various dimensions (vertical long axis, horizontal long axis, short axis) is performed by a nuclear medicine physician. However, the interpretation is largely dependent on his clinical experience and shows significant inter-observer variations.12 It is in this area of classification of SPECT-MPI images for CAD where deep learning approaches using convolutional neural networks (CNN) can be utilized to assist physicians in the diagnostic assessment thus enabling patients to responsibly monitor their style of living, diminishing the risks of serious cardiac events.13-15 CNN is one of the various types of artificial neural networks and is specifically used for image recognition using processing of pixel data of medical images. It is differentiated with the traditional machine learning models as feature extraction and selection are done automatically through analysis of pixel data. Hence, CNNs can be applied on medical images such as the SPECT-MPI for assessment of any medical problem or conditions in patients, such as the presence of CAD. The use of CNN models enhances the SPECT-MPI diagnostic process leading to institution of the much-needed medical therapy to upgrade overall survival rates and/or quality of life of patients with CAD.13,15-18 The visual assessment of myocardial perfusion abnormalities in SPECT-MPI remains an important research challenge to both cardiologists and data scientists since many research studies in clinical medicine now involve applying a variety of machine learning algorithms, deep learning methods, and statistical techniques to improve CAD detection.19-23
In previous studies involving deep learning methods applied to nuclear medicine imaging, various types of CNN models were applied on bone scans of breast and prostate cancer patients to detect osseous metastasis.13,24–27 Results demonstrated superior performance of these CNN models with several advantages such as high reliability, valid, faster, simpler architecture, and short training even with a comparatively smaller image dataset.13,25,26 In line with these recent updates, this study is being expanded for the generalization capabilities of CNN models in nuclear cardiology.
The objective of the study is to develop a CNN in the classification of SPECT-MPI for perfusion abnormalities. An exploratory approach of hyperparameter selection was made to search for the optimum neural network model in terms of classification performance. The notable contribution of this research is clearly delineated by the establishment of valid and reliable CNN models for the identification of CAD with acceptable performance metrics which can be utilized as physician decision support tools in the clinical nuclear cardiology practice. This research study shall assess the diagnostic performance of these CNN models in the detection of SPECT-MPI scans for CAD.
Review of Related Literature
In recent years, various artificial intelligence (AI) techniques have flourished in healthcare as clinical data become more progressively complicated. These techniques have become very prominent in cardiovascular imaging by computerizing many processes or calculations, find new patterns automatically in the data and specify differential diagnoses.28 Papandrianos and Papageorgiou10 applied a deep learning method to classify SPECT-MPI images as normal or abnormal with an impressive 93.47±2.81 accuracy and 0.936 area under the receiver operating characteristics curve (AUC). They further reported their CNN model to be an efficient, and robust deep neural network in diagnosing blood flow abnormalities as seen in myocardial infarction and ischemia on SPECT-MPI images. In the study by Mostafapour et al., ResNet and UNet deep CNN models were utilized to generate a valid attenuation correction in SPECT-MPI images.29 In another study, authors employed transfer learning with pre-trained CNN to assess cardiac blood flow problems (myocardial infarction, myocardial ischemia) with 94% accuracy, 88% sensitivity, and 100% specificity and further concluded that the proposed CNN models could assist in physician decision making to make a diagnosis of infarction and ischemia in SPECT-MPI images.11 Chen et al. utilized CNN in the assessment of myocardial perfusion images for coronary heart diseases with 87.64% accuracy, 81.58% sensitivity, and 92.16% specificity, and concluded that use of the CNN model can significantly reduce the time needed for doctors to interpret the images, write reports and thus, can help physicians in the diagnosis of coronary heart diseases reliably in clinical practice.30
Several studies have also applied CNN models on SPECT-MPI polar maps for myocardial perfusion abnormalities. Teuho et al.31, utilized CNN in the classification of polar maps for myocardial ischemia with 82.61% accuracy, 0.8058 AUC, 76.47% F1-score, 65% sensitivity, 96.15% specificity, and 93.75% precision. CNN models using polar maps of SPECT-MPI was also analyzed in another study to forecast a patient’s 5-year survival rate after a cardiac event with a 0.77 accuracy. The CNN models used the frequency spectra as the input using fast Fourier transform on images instead of the usual raw images.32 In another study by Liu et al.33, authors applied CNN to assess for CAD from SPECT-MPI images with a 0.872 AUC, 82.7% accuracy, 74.4% sensitivity, and 84.9% specificity. A simple CNN for classification of SPECT-MPI images into binary categories (normal, ischemia) was used in another study.10 After the application of data augmentation technique, an exploratory approach utilizing different number of layers, batch sizes, pixel size, and dense nodes was performed with the CNN generating a 90.20% accuracy and a 0.9377 AUC. Authors further concluded that the use of CNN models offered additional benefits for diagnostic challenge, as these models generated more robust and dependable outcomes compared to the usual purely clinical methods.
In summary, most of the works focused on the diagnosis of CAD from SPECT-MPI using deep learning methods. Their results served as the backbone for our preliminary simulations to discover the most satisfactory network architecture and its corresponding network parameters.
MATERIALS AND METHODS
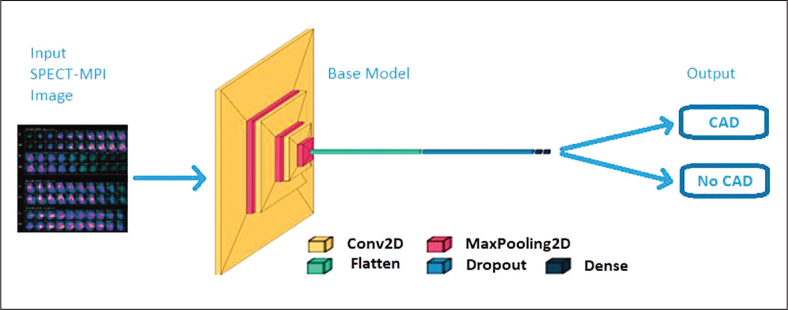
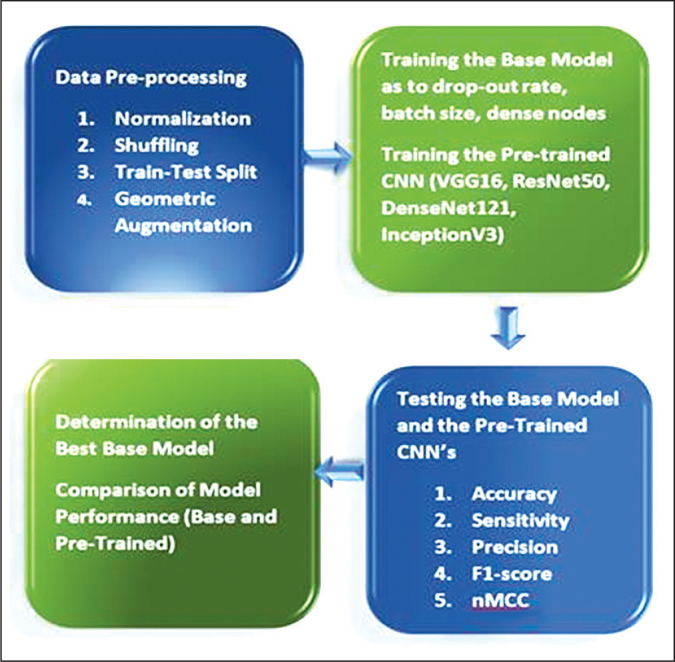
The SPECT-MPI data images were obtained from a publicly available machine learning image repository. The pre-processing steps consisting of image normalization, data shuffling, and geometric augmentation techniques were applied to the images. After a train-test split, the pre-processed data was then fed to the base CNN and other commonly used pre-trained CNNs on medical images (InceptionV3, DenseNet121, VGG16 and ResNet50). Classification performance of these CNN models was assessed using various metrics. All simulation experiments were performed in Kaggle because of the free use of NVIDIA TESLA P100 GPUs. TensorFlow 2.6.0., Keras 2.6.0, and Python language 3.7.10 were employed in this study. Figure 1 shows the deep learning pipeline for this research study.
Figure 1.
Deep learning pipeline in the classification of SPECT-MPI for CAD.
Dataset Description
A publicly available anonymized SPECT-MPI from a data repository (https://www.kaggle.com/selcankaplan/spect-mpi) was used in this study.34 The dataset contains 192 patients who underwent stress-test-rest Tc99m MPI. There was data imbalance with 150 patients (78%) having coronary artery disease while 42 patients (22%) do not have coronary artery disease.
Sample SPECT-MPI images with and without CAD are shown in Figures 2 and 3, respectively. The assessment of the SPECT-MPI images for perfusion defects is made by nuclear medicine physicians or nuclear cardiologists by comparing the stress images with rest images across all dimensions (vertical long axis, horizontal long axis, and short axis). A blood perfusion abnormality detected in stress images with no corresponding abnormality in rest images is classified as myocardial ischemia while an abnormality detected in both stress and rest images is labelled as myocardial infarction.11 For this study, SPECT-MPI images with ischemia and/or infarction is diagnosed as having CAD while those images without perfusion defects are classified as not having CAD. Figure 2 shows tracer inhomogeneity in the anterior and antero-lateral walls of the myocardium seen in stress images but not on the resting images. This finding is typical of stress-induced myocardial ischemia and hence, classified as having CAD. On the other hand, the image shown in Figure 3 demonstrates uniform radiotracer distribution in all of the myocardium in both stress and rest images which is indicative of the absence of CAD.
Figure 2.
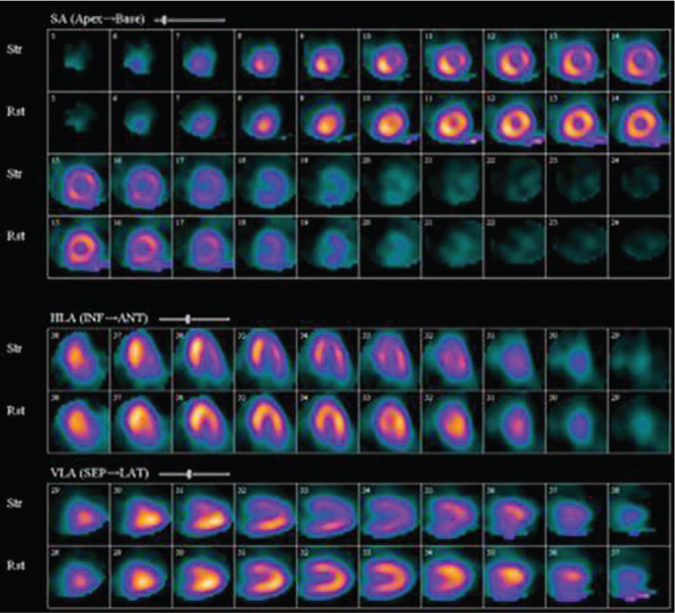
Sample SPECT-MPI image with CAD.
Figure 3.

Sample SPECT-MPI image without CAD.
Pre-processing Steps
The pre-processing steps applied to the dataset were Min-Max normalization of SPECT-MPI images, shuffling of images to generate random order of images, train – test split, and geometric augmentation.13 The dataset was divided in three parts: testing (20%) and the rest (80%) was further divided to 80% for training and 20% for validation. The following typical geometric augmentations were applied to the images in the training: zoom range = +0.15, rotation range = +15, shear range = +0.30, translation = +0.15, and horizontal flipping. To handle data imbalance, class weights were applied using inverse proportion of class frequencies.
Base CNN Model Architecture
An exploratory approach with bibliographic research was employed to create a CNN architecture for the assessment of CAD. The parameters needed for a good network architecture were examined as to batch size, drop-out rate, and the dense nodes. Similar to the previous studies in the literature9-11,13 and after a series of various preliminary model configurations for this study in finding the best model configuration as to the number of layers of a simple base CNN and its parameters such as the input size, the number of epochs, learning rate, activation function, etc., a final model configuration of a layered sequential neural network architecture consisting of three convolutional layers followed by max pooling was made. This was followed by flattening layer where it reshapes the multidimensional array of pixels to one-dimensional for extracting the output, a dropout layer, one dense layer followed by an output layer.10 The use of a simple neural network would be an impediment against overfitting which may possibly occur.13 Rectified Linear Unit (ReLU) activation function was employed in all convolutional and dense layers while sigmoid activation function was utilized in the final output layer for binary classification of with or without CAD. A filter size of 32, 64 and 128, sequentially and with kernel size of 3x3 was applied in the convolutional layers. Maximum values were utilized in the pooling layers which had 2x2 kernel size. The number of filters was increased in each layer to extract the more complicated trends and patterns of the SPECT-MPI image in the training network. The loss function used was binary cross entropy with Adam, an adaptive learning rate optimization method, as the optimizer.9 Figure 4 shows the architecture for this study.
Figure 4.
CNN architecture for SPECT-MPI classification for CAD.
Hyperparameters Search
The different combinations of batch sizes, dense nodes, and drop-out rates were analyzed in various simulation experiments to search for the optimum model configuration. An exploratory approach in the development of the base CNN model with optimum hyperparameters was utilized with examination of batch sizes (8, 16, 32, 64), drop-out rates (0.2, 0.5, 0.7), and dense nodes (32, 64, 128, 256). Based on the preliminary simulations, the input pixel size was fixed at 256 x 256 x 3 while the number of epochs was set at 50.
Comparison with Pre-Trained CNN Models
Comparison between the base CNN model and the pre-trained models typically used in medical images such as VGG16, InceptionV3, DenseNet121, and ResNet50 was also performed. This group of pre-trained models is a mixture of shallower and deeper layer CNN’s. VGG16 contains 16 weight layers and usually developed using 3 x 3 convolutional layers attached to each other architecture with 1.3 x 108 trainable free parameters and has specific network structure that is simple to change.25,26,35 ResNet50, involves a 50-weight layer version of ResNet (Residual neural Network) having 2.3 x 107 trainable parameters and has been introduced to address the vanishing degradation challenge of model training that causes performance deterioration in the models.25,26,36 DenseNet121 has 121 layers with more than 8 million parameters and is partitioned into DenseBlocks with feature maps having same dimensions within the block but with different number of filters. The transition layers between blocks apply batch normalization for down-sampling.37 InceptionV3 is a 48-layer deep network with fewer parameters (2.1 x 107 trainable free parameters).26
Performance Metrics
To evaluate the diagnostic performance of the CNN models, accuracy, recall (sensitivity), precision (positive predictive value), and F1-scores were computed. To determine the best performing model, normalized Matthews correlation coefficient (nMCC) was used because it provides more information and thus, more enlightening than accuracy and F1-scores since it considers all entries in the confusion matrix.38
RESULTS
There was a total of 192 patient images included in the CNN model building. The results of the various simulation experiments in the base CNN model with respect to the batch sizes, dense nodes, and drop-out rates to determine optimal parameters are shown in Tables 1 (drop-out = 0.2), 2 (drop-out = 0.5), and 3 (drop-out = 0.7). For dropout 0.2, the best configuration was obtained by batch size 16 and dense nodes 128 with a superior performance metrics of 93.75% accuracy, 100% sensitivity, 95.29% precision, 96.15% F1-score and a nMCC of 0.907. As shown in Table 1, generally, batch sizes 8 and 32 yielded lower performance metrics while batch size 64 fared poorly.
Table 1.
Performance Metrics of the Base CNN Model for Drop-Out = 0.2
| Batch Size | Dense Nodes | Accuracy | Recall | Precision | F1-score | nMCC |
|---|---|---|---|---|---|---|
| 8 | 32 | 81.25 | 92.00 | 85.19 | 88.46 | 0.698 |
| 64 | 78.12 | 100.00 | 78.13 | 87.72 | 0.500 | |
| 128 | 81.25 | 100.00 | 80.65 | 89.29 | 0.670 | |
| 256 | 78.12 | 100.00 | 78.13 | 87.72 | 0.500 | |
|
| ||||||
| 16 | 32 | 90.62 | 92.00 | 95.83 | 93.88 | 0.871 |
| 64 | 81.25 | 100.00 | 80.65 | 89.29 | 0.670 | |
| 128 | 93.75 | 100.00 | 95.29 | 96.15 | 0.907 | |
| 256 | 84.38 | 88.00 | 91.67 | 89.80 | 0.784 | |
|
| ||||||
| 32 | 32 | 75.00 | 92.00 | 79.30 | 85.19 | 0.545 |
| 64 | 84.38 | 100.00 | 83.33 | 90.91 | 0.744 | |
| 128 | 78.12 | 100.00 | 78.13 | 87.72 | 0.500 | |
| 256 | 78.12 | 100.00 | 78.13 | 87.72 | 0.500 | |
|
| ||||||
| 64 | 32 | 78.12 | 100.00 | 78.13 | 87.72 | 0.500 |
| 64 | 78.12 | 100.00 | 78.13 | 87.72 | 0.500 | |
| 128 | 78.12 | 100.00 | 78.13 | 87.72 | 0.500 | |
| 256 | 78.12 | 100.00 | 78.13 | 87.72 | 0.500 | |
Table 2 shows the performance metrics of the base CNN model for drop-out 0.5. Similarly, the best configuration was obtained by batch size 16 and dense node 256 as it generated the highest nMCC and accuracy at 0.857 and 90.62%, respectively. Batch sizes 8 and 32 obtained lower performance metrics while batch 64 classification performance was below par.
Table 2.
Performance Metrics of the Base CNN Model for Drop-Out = 0.5
| Batch Size | Dense Nodes | Accuracy | Recall | Precision | F1-score | nMCC |
|---|---|---|---|---|---|---|
| 8 | 32 | 84.38 | 100.00 | 83.33 | 90.91 | 0.744 |
| 64 | 87.50 | 92.00 | 92.00 | 92.00 | 0.817 | |
| 128 | 78.12 | 100.00 | 78.13 | 87.72 | 0.500 | |
| 256 | 81.25 | 100.00 | 80.65 | 89.29 | 0.670 | |
|
| ||||||
| 16 | 32 | 81.25 | 100.00 | 80.65 | 89.29 | 0.670 |
| 64 | 81.25 | 100.00 | 80.65 | 89.29 | 0.670 | |
| 128 | 78.12 | 100.00 | 78.13 | 87.72 | 0.500 | |
| 256 | 90.62 | 96.00 | 92.31 | 94.12 | 0.857 | |
|
| ||||||
| 32 | 32 | 81.25 | 100.00 | 80.65 | 89.29 | 0.670 |
| 64 | 78.12 | 100.00 | 78.13 | 87.72 | 0.500 | |
| 128 | 84.38 | 100.00 | 83.33 | 90.91 | 0.744 | |
| 256 | 81.25 | 100.00 | 80.65 | 89.29 | 0.670 | |
|
| ||||||
| 64 | 32 | 78.12 | 100.00 | 78.13 | 87.72 | 0.500 |
| 64 | 78.12 | 100.00 | 78.13 | 87.72 | 0.500 | |
| 128 | 78.12 | 100.00 | 78.13 | 87.72 | 0.500 | |
| 256 | 78.12 | 100.00 | 78.13 | 87.72 | 0.500 | |
The performance metrics of the base CNN model for dropout 0.7 is shown in Table 3. The optimum configuration was obtained by batch size 8 and dense node 32 with the highest nMCC and accuracy of 0.909 and 93.75%, respectively. Batch sizes 16 and 32 (except for batch size 16, dense node 256) generally have much lower performance as compared to batch size 8. Similarly, batch size 64 performed poorly for this dataset.
Table 3.
Performance Metrics of the Base CNN Model for Drop-Out = 0.7
| Batch Size | Dense Nodes | Accuracy | Recall | Precision | F1-score | nMCC |
|---|---|---|---|---|---|---|
| 8 | 32 | 93.75 | 96.00 | 96.00 | 96.00 | 0.909 |
| 64 | 84.38 | 92.00 | 88.46 | 90.20 | 0.760 | |
| 128 | 78.12 | 100.00 | 78.13 | 87.72 | 0.500 | |
| 256 | 84.38 | 100.00 | 83.33 | 90.91 | 0.744 | |
|
| ||||||
| 16 | 32 | 78.12 | 100.00 | 78.13 | 87.72 | 0.500 |
| 64 | 84.38 | 96.00 | 85.71 | 90.57 | 0.743 | |
| 128 | 87.50 | 92.00 | 92.00 | 92.00 | 0.817 | |
| 256 | 78.12 | 100.00 | 78.13 | 87.72 | 0.500 | |
|
| ||||||
| 32 | 32 | 84.38 | 100.00 | 83.33 | 90.91 | 0.744 |
| 64 | 81.25 | 100.00 | 80.65 | 89.29 | 0.670 | |
| 128 | 87.50 | 100.00 | 86.21 | 92.59 | 0.804 | |
| 256 | 81.25 | 96.00 | 82.76 | 88.89 | 0.674 | |
|
| ||||||
| 64 | 32 | 78.12 | 100.00 | 78.13 | 87.72 | 0.500 |
| 64 | 78.12 | 100.00 | 78.13 | 87.72 | 0.500 | |
| 128 | 78.12 | 100.00 | 78.13 | 87.72 | 0.500 | |
| 256 | 78.12 | 100.00 | 78.13 | 87.72 | 0.500 | |
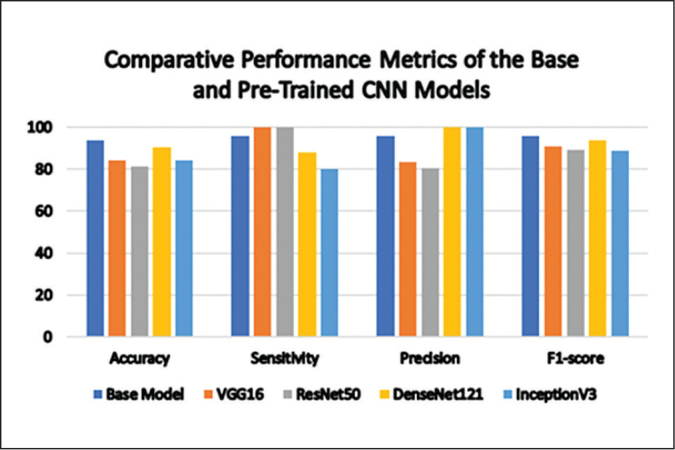
Overall, the top performing base CNN model configuration was obtained by batch size 8, dense nodes 32, drop-out 0.7. It generated the highest nMCC at 0.909 and obtained 93.75% accuracy, 96.00% sensitivity, 96.00% precision, and 96.00% F1-score. Table 4 and Figure 5 highlight the comparison of the performance metrics between the best base CNN model and the pre-trained architectures. The base model outperformed the other models as it yielded the highest nMCC, accuracy, and F1-scores. This was followed very closely by DenseNet121 with a 90.62% accuracy and 0.892 nMCC.
Table 4.
Comparative Performance of CNN Models
| CNN Models | Accuracy | Recall | Precision | F1-score | nMCC |
|---|---|---|---|---|---|
| Base | 93.75 | 96.00 | 96.00 | 96.00 | 0.909 |
| VGG16 | 84.38 | 100.00 | 83.33 | 90.91 | 0.744 |
| ResNet50 | 81.25 | 100.00 | 80.65 | 89.29 | 0.670 |
| DenseNet121 | 90.62 | 88.00 | 100.00 | 93.62 | 0.892 |
| InceptionV3 | 84.38 | 80.00 | 100.00 | 88.89 | 0.842 |
Figure 5.
Performance metrics of the CNN models for CAD classification.
DISCUSSION
In the past several years, there has been a significant increase in the number and variety of cardiovascular imaging seen in clinical practice to assist physicians in the detection of cardiac conditions as well as a guide for the choice of therapeutic options for the patients. Typically, a cardiologist would refer a patient initially for a less-invasive cardiac imaging modality such as SPECT-MPI to assess for coronary artery disease.9,39 The clinical and prognostic value of these cardiovascular imaging tests is constrained and challenged by the presence of significant intra and interobserver variability, suboptimal image quality, time-consuming process, operators’ fatigue, etc.12,40 Some of these issues can be addressed by applying AI in cardiovascular imaging which has also prospered in recent years. AI is being utilized in the processing of cardiac imaging data at different levels of increasing complexity.41 At the examination level, AI is being utilized to simplify image acquisition and processing with higher image resolution while at reading and reporting levels, AI focuses on automatic detection of lesions with description of features and corresponding assessment of extent and size in the images. Additionally, at the prediction and prescription levels, AI focuses on risk prediction and stratification, as opposed to merely detecting, measuring, and quantifying images such as an AI-based CT-fractional flow reserve modeling.
Deep Learning (DL) is a subset of machine learning based on the concept of artificial neural network algorithms.42 As such, DL has more extensive computations and higher number of parameters which in turn allow DL models to learn more from the data. Lately, many medical image processing procedures have adopted DL approaches ranging from fundus images, endoscopic images, ultrasound images, cardiovascular images, CT/MRI images, and pathological images. Currently, DL methods is mainly used in classification and segmentation in medical images.40,43,44 Further use of DL methods will largely depend on continuing accumulation of medical big data for training which is expected to happen soon. However, in cases where there is limited size of medical cohorts for training by the DL models and the cost of expert-annotated data sets is extremely prohibitive, these can be addressed by the use of transfer learning techniques.45 Transfer learning aims to generate high performance on target tasks by using the knowledge learned in advance from other source tasks. This allows the use of DL approaches even with a smaller medical dataset.13,25,26,45
In this study, a simple base CNN model for the classification of SPECT-MPI images for CAD was created with optimum parameters using an exploratory approach combined with bibliographic search. The base CNN model despite having a simpler and shallower architecture outperformed the deeper pre-trained architectures commonly applied on medical images. It is important to emphasize that simpler CNN architecture can be as effective as the deeper CNN architecture so long as the network parameters have been fine-tuned.
The performance metrics based on the accuracy rates and nMCC of the base CNN model were comparable with those reported in the literature.9-11 The outcomes suggest that this can be a valid and reliable tool for the evaluation of SPECT-MPI images for CAD even with a small dataset.10,11,13 Additionally, these models can serve as learning materials for the less experienced doctors particularly those still in their training career. These models can also be used as a valid and dependable second opinion that can aid physicians as a decision support tool.10,11,13,15 This highlights the clinical utility of these CNN models in nuclear cardiology. Nonetheless, it is important to underscore that while DL approaches through CNN models can be a helpful tool for the evaluation of images in cardiac imaging more particularly on the reporting and interpretation, these CNN models cannot replace human expert readers.40 These models are merely useful tools for enhanced implementation of a clinical workflow.
CONCLUSIONS AND RECOMMENDATIONS
The use of CNN has shown its capability to diagnose CAD in SPECT – MPI images. Several CNN models through an exploratory approach were investigated to determine the CNN architecture with the optimum network configuration. Simulation experiments showed the optimum model configuration had the following parameters: epochs = 50, input pixel size = 256 x 256 x 3, batch size = 8, dense node = 32, and drop-out = 0.7. It generated the highest nMCC at 0.909, 93.75% accuracy, 96.00% sensitivity, 96.00% precision, and 96.00% F1-score. Additionally, the base CNN model outperformed the commonly used pre-trained architectures (VGG16, InceptionV3, DenseNet121, and ResNet50) in medical images. The results suggest that CNN models can be valid tools to classify SPECT-MPI images, even with a small image dataset. This study showed encouraging outcomes which can be deployed in the daily workflow routine by nuclear medicine physicians in their clinical practice thereby augmenting their decision skills in the interpretation of SPECT-MPI tests. These models can also be utilized as valid and dependable second opinions that can aid physicians as a decision support tool as well as serve as teaching or learning materials for the less-experienced physicians particularly those still in their training career. These highlights the clinical utility of deep learning approaches through CNN models in the practice of nuclear cardiology. For future work, there is a need to further expand the performance of the models using large dataset and consider adding clinical data to perfusion data. It is also recommended to employ a dataset of SPECT-MPI images with coronary angiography as the gold diagnostic standard rather than an expert reader interpretation.
Statement of Authorship
Both authors certified fulfillment of ICMJE authorship criteria.
Author Disclosure
Both authors of this publication have no conflicts of interest to declare.
Funding Source
The authors received no financial support for the research, authorship, and/or publication of this article.
REFERENCES
- 1.Ties D, van Dorp P, Pundziute G, van der Aalst CM, Gratama JWC, Braam RL, et al. Early detection of obstructive coronary artery disease in the asymptomatic high-risk population: objectives and study design of the EARLY-SYNERGY trial. Am Heart J. 2022. Apr; 246: 16-77. doi: 10.1016/j.ahj.2022.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Juan-Salvadores P, Jiménez Díaz VA, Iglesia Carreño C, Guitián González A, Veiga C, Martínez Reglero C, et al. Coronary artery disease in very young patients: analysis of risk factors and long-term follow-up. J Cardiovasc Dev Dis. 2022. Mar;9(3):82. doi: 10.3390/jcdd9030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.King A, Rajpura J, Liang Y, Paprocki Y, Uzoigwe C. Impact of cardiovascular disease on health care economic burden and resource utilization: a retrospective cohort study in adults in the United States with type 2 diabetes with or without stroke, myocardial infarction, and peripheral arterial disease. Curr Med Res Opin. 2022. Nov;38(11): 1831-40. doi: 10.1080/03007995.2022.2125259. [DOI] [PubMed] [Google Scholar]
- 4.Butnariu LI, Florea L, Badescu MC, Țarcă E, Costache I-I, Gorduza EV. Etiologic puzzle of coronary artery disease: How important is genetic component? Life. 2022. Jun;:865. doi: 10.3390/life12060865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frąk W, Wojtasińska A, Lisińska W, Młynarska E, Franczyk B, Rysz J. Pathophysiology of cardiovascular diseases: new insights into molecular mechanisms of atherosclerosis, arterial hypertension, and coronary artery disease. Biomedicines. 2022. Aug;10(8):1938. doi: 10.3390/biomedicines10081938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shahjehan RD, Bhutta BS. Coronary Artery Disease. [Updated 2022 Nov 7]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2022. Jan [cited 2022 Aug]. Available from: https://www.ncbi.nlm.nih.gov/books/NBK564304/ [PubMed] [Google Scholar]
- 7.Zaric BL, Radovanovic JN, Gluvic Z, Stewart AJ, Essack M, Motwalli O, et al. Atherosclerosis linked to aberrant amino acid metabolism and immunosuppressive amino acid catabolizing enzymes. Front Immunol. 2020. Sep;11:551758. doi: 10.3389/fimmu.2020.551758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moran AE, Forouzanfar MH, Roth GA, Mensah GA, Ezzati M, Murray CJL, et al. Temporal trends in ischemic heart disease mortality in 21 world regions, 1980 to 2010: the Global Burden of Disease 2010 Study. Circulation. 2014. Apr;129(14):1483–92. doi: 10.1161/CIRCULATIONAHA.113.004042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papandrianos N, Papageorgiou E. Automatic diagnosis of coronary artery disease in SPECT myocardial perfusion imaging employing deep learning. Appl Sci. 2021;11(14):6362. doi: 10.3390/app11146362. [DOI] [Google Scholar]
- 10.Papandrianos N, Feleki A, Papageorgiou E. Exploring classification of SPECT MPI images applying convolutional neural networks. In 25th Pan-Hellenic Conference on Informatics (PCI 2021), November 2021, Volos, Greece. Association for Computing Machinery, New York, NY, USA. pp. 483-489. doi: 10.1145/3503823.3503911. [DOI] [Google Scholar]
- 11.Berkaya SK, Sivrikoz IA, Gunal S. Classification models for SPECT myocardial perfusion imaging. Comput Biol Med. 2020. Aug;123:103893. doi: 10.1016/j.compbiomed.2020.103893. [DOI] [PubMed] [Google Scholar]
- 12.Spier N, Nekolla S, Rupprecht C, Mustafa M, Navab N, Baust M. Classification of polar maps from cardiac perfusion imaging with graph-convolutional neural networks. Sci Rep. 2019. May;9(1): 7569. doi: 10.1038/s41598-019-43951-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magboo VPC, Abu PAR. Deep Neural Network for Diagnosis of Bone Metastasis. In: 2022 The 5th International Conference on Software Engineering and Information Management (ICSIM) (ICSIM 2022). Association for Computing Machinery, New York, USA. pp. 144-151. doi: 10.1145/3520084.3520107. [DOI] [Google Scholar]
- 14.Magboo VP, Magboo MS. Imputation Techniques and Recursive Feature Elimination in Machine Learning Applied to Type II Diabetes Classification. In: 2021 4th Artificial Intelligence and Cloud Computing Conference (AICCC '21). Association for Computing Machinery, New York, USA. pp. 201–207. doi: 10.1145/3508259.3508288. [DOI] [Google Scholar]
- 15.Magboo MSA, Magboo VPC. Detection of Brain Tumors from MRI Images using Convolutional Neural Networks. In: 2022 5th International Conference of Computer and Informatics Engineering (IC2IE), Jakarta, Indonesia. pp. 325 – 330. doi: 10.1109/IC2IE56416.2022.9970126. [DOI] [Google Scholar]
- 16.Magboo VPC, Magboo MSA. Classification Models for Autism Spectrum Disorder. In: Kumar A, Fister I Jr., Gupta PK, Debayle J, Zhang ZJ, Usman M. (eds) Artificial Intelligence and Data Science. ICAIDS 2021. Communications in Computer and Information Science. Springer, Cham.; 2022. p.1673. doi: 10.1007/978-3-031-21385-4_37. [DOI] [Google Scholar]
- 17.Magboo VPC, Magboo MSA. Prediction of late intrauterine growth restriction using machine learning models. Procedia Comput Sci. 2022;207:1427-36. doi: 10.1016/j.procs.2022.09.199. [DOI] [Google Scholar]
- 18.Magboo VPC, Magboo MSA. Important Features Associated with Depression Prediction and Explainable AI. In: Li H, Ghorbanian Zolbin M, Krimmer R, Kärkkäinen J, Li C, Suomi R, eds. Well-Being in the Information Society: When the Mind Breaks. WIS 2022. Communications in Computer and Information Science. Springer, Cham.; 2022. p.1626. 10.1007/978-3-031-14832-3_2. [DOI] [Google Scholar]
- 19.Magboo MS, Coronel AD. Data Mining Electronic Health Records to Support Evidence-Based Clinical Decisions. In: Chen YW., Zimmermann A., Howlett R., Jain L. (eds) Innovation in Medicine and Healthcare Systems, and Multimedia. Smart Innovation, Systems and Technologies. Springer, Singapore; 2019. p.145. doi: 10.1007/978-981-13-8566-7_22. [DOI] [Google Scholar]
- 20.Lopez KM, Magboo MS. A Clinical Decision Support Tool to Detect Invasive Ductal Carcinoma in Histopathological Images Using Support Vector Machines, Naïve-Bayes, and K-Nearest Neighbor Classifiers. Tallón-Ballesteros A and Chen CH, eds. Netherlands: IOS Press BV; 2020. pp. 46–53. [Google Scholar]
- 21.Magboo VPC, Magboo MSA. Machine learning classifiers on breast cancer recurrences. Procedia Comput Sci. 2021; 192:2742-52. doi: 10.1016/j.procs.2021.09.044. [DOI] [Google Scholar]
- 22.Magboo VPC, Magboo MSA. Prediction Models for COVID-19 in Children. In: Chen YW, Tanaka S, Howlett RJ, Jain LC, eds) Innovation in Medicine and Healthcare. Smart Innovation, Systems and Technologies. Springer, Singapore;2022. p. 308. doi: 10.1007/978-981-19-3440-7_2 [DOI] [Google Scholar]
- 23.Magboo MSA, Magboo VPC. Explainable AI for Autism Classification in Children. In: Jezic G, Chen-Burger YHJ, Kusek M, Šperka R, Howlett RJ, Jain LC, eds. Agents and Multi-Agent Systems: Technologies and Applications 2022. Smart Innovation, Systems and Technologies. Springer, Singapore; 2022. p. 306. doi: 10.1007/978-981-19-3359-2_17 [DOI] [Google Scholar]
- 24.Ntakolia C, Diamantis DE, Papandrianos N, Moustakidis S, Papageorgiou EI. A lightweight convolutional neural network architecture applied for bone metastasis classification in nuclear medicine: a case study on prostate cancer patients. Healthcare. 2020. Nov;8(4):493. doi: 10.3390/healthcare8040493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papandrianos NI, Papageorgiou E, Anagnostis A, Feleki A. A deep-learning approach for diagnosis of metastatic breast cancer in bones from whole-body scans. Appl Sci. 2020;10(3):997. doi: 10.3390/app10030997. [DOI] [Google Scholar]
- 26.Papandrianos N, Papageorgiou E, Anagnostis A, Papageorgiou K. Efficient bone metastasis diagnosis in bone scintigraphy using a fast convolutional neural network architecture. Diagnostics. 2020. Jul;10(8):532. doi: 10.3390/diagnostics10080532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng DC, Hsieh TC, Yen KY, Kao CH. Lesion-based bone metastasis detection in chest bone scintigraphy images of prostate cancer patients using pre-train, negative mining, and deep learning. Diagnostics. 2021. Mar;11(3):518. doi: 10.3390/diagnostics11030518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seetharam K, Shrestha S, Sengupta PP. Cardiovascular imaging and intervention through the lens of artificial intelligence. Interv Cardiol. 2021. Oct;16:e31. doi: 10.15420/icr.2020.04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mostafapour S, Gholamiankhah F, Maroufpour S, Momennezhad M, Asadinezhad M, Zakavi SR, et al. Deep learning-guided attenuation correction in the image domain for myocardial perfusion SPECT imaging. J Comput Des Eng. 2022. Apr;9(2):434 –47. doi: 10.1093/jcde/qwac008. [DOI] [Google Scholar]
- 30.Chen JJ, Su TY, Chen WS, Chang YH, Lu HHS. Convolutional neural network in the evaluation of myocardial ischemia from CZT SPECT myocardial perfusion imaging: comparison to automated quantification. Appl Sci. 2021;11(2):514. doi: 10.3390/app11020514. [DOI] [Google Scholar]
- 31.Teuho J, Schultz J, Klén R, Knuuti J, Saraste A, Ono N, et al. Classification of ischemia from myocardial polar maps in 15O–H2O cardiac perfusion imaging using a convolutional neural network. Sci Rep. 2022. Feb;12(1):2839. doi: 10.1038/s41598-022-06604-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng DC, Hsieh TC, Hsu YJ, Lai YC, Yen KY, Wang CCN, et al. Prediction of all-cause mortality based on Stress/Rest Myocardial Perfusion Imaging (MPI) using deep learning: a comparison between image and frequency spectra as input. J Pers Med. 2022. Jul;12(7): 1105. doi: 10.3390/jpm12071105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu H, Wu J, Miller EJ, Liu C, Liu Y, Liu YH. Diagnostic accuracy of stress-only myocardial perfusion SPECT improved by deep learning. Eur J Nucl Med Mol Imaging. 2021. Aug;48(9): 2793 – 800. doi: 10.1007/s00259-021-05202-9. [DOI] [PubMed] [Google Scholar]
- 34.SPECT-MPI dataset, Department of Nuclear Medicine , Eskisehir Osmangazi University from December 2018 to September 2019 for stress/rest Tc-99m MPI [Internet]. [cited 2022 Jun]. Available from: https://www.kaggle.com/selcankaplan/spect-mpi, last accessed 2022/2/28.
- 35.Yang D, Martinez C, Visuña L, Khandhar H, Bhatt C, Carretero J. Detection and analysis of COVID-19 in medical images using deep learning techniques. Sci Rep. 2021. Oct;11(1):19638. doi: 10.1038/s41598-021-99015-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hassan M, Ali S, Alquhayz H, Safdar K. Developing intelligent medical image modality classification system using deep transfer learning and LDA. Sci Rep. 2020. Jul;10(1):12868. doi: 10.1038/s41598-020-69813-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shazia A, Xuan TZ, Chuah JH, Usman J, Qian P, Lai KW. A comparative study of multiple neural network for detection of COVID-19 on chest X-ray. EURASIP J Adv Signal Process. 2021;2021(1):50. doi: 10.1186/s13634-021-00755-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chicco D, Jurman G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genomics. 2020. Jan;21(1):6. doi: 10.1186/s12864-019-6413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Canu M, Broisat A, Rious L, Vanzetto G, Fagret D, Ghezzi C, et al. Non-invasive multimodality imaging of coronary vulnerable patient. Front Cardiovasc Med. 2022. Feb;9:836473. doi: 10.3389/fcvm.2022.836473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muscogiuri G, Volpato V, Cau R, Chiesa M, Saba L, Guglielmo M, et al. Application of AI in cardiovascular multimodality imaging. Heliyon. 2022. Oct;8(10):e10872. doi: 10.1016/j.heliyon.2022.e10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma P, Suehling M, Flohr T, Comaniciu D. Artificial Intelligence in diagnostic imaging: status quo, challenges, and future opportunities. J Thorac Imaging. 2020. May; 35 Suppl 1:S11–S16. doi: 10.1097/rti.0000000000000499 [DOI] [PubMed] [Google Scholar]
- 42.Rouzrokh P, Khosravi B, Vahdati S, Moassefi M, Faghani S, Mahmoudi E, et al. Machine learning in cardiovascular imaging: a scoping review of published literature. Curr Radiol Rep. 2023;11(2):34 – 45. doi: 10.1007/s40134-022-00407-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai L, Gao J, Zhao D. A review of the application of deep learning in medical image classification and segmentation. Ann Transl Med. 2020. Jun;8(11):713. doi: 10.21037/atm.2020.02.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puttagunta M, Ravi S. Medical image analysis based on deep learning approach. Multimed Tools Appl. 2021;80(16):24365 –24398. doi: 10.1007/s11042-021-10707-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim HE, Cosa-Linan A, Santhanam N, Jannesari M, Maros ME, Ganslandt T. Transfer learning for medical image classification: a literature review. BMC Med Imaging. 2022. Apr;22(1):69. doi: 10.1186/s12880-022-00793-7. [DOI] [PMC free article] [PubMed] [Google Scholar]